A leucine-rich repeat protein related to animal cell signaling components is important for gametogenesis, with gametophytespecific expression achieved by alternative mRNA splicing.
Abstract
Plant intracellular Ras-group leucine-rich repeat (LRR) proteins (PIRLs) are related to Ras-interacting animal LRR proteins that participate in developmental cell signaling. Systematic knockout analysis has implicated some members of the Arabidopsis (Arabidopsis thaliana) PIRL family in pollen development. However, for PIRL6, no bona fide knockout alleles have been recovered, suggesting that it may have an essential function in both male and female gametophytes. To test this hypothesis, we investigated PIRL6 expression and induced knockdown by RNA interference. Knockdown triggered defects in gametogenesis, resulting in abnormal pollen and early developmental arrest in the embryo sac. Consistent with this, PIRL6 was expressed in gametophytes: functional transcripts were detected in wild-type flowers but not in sporocyteless (spl) mutant flowers, which do not produce gametophytes. A genomic PIRL6-GFP fusion construct confirmed expression in both pollen and the embryo sac. Interestingly, PIRL6 is part of a convergent overlapping gene pair, a scenario associated with an increased likelihood of alternative splicing. We detected multiple alternative PIRL6 mRNAs in vegetative organs and spl mutant flowers, tissues that lacked the functionally spliced transcript. cDNA sequencing revealed that all contained intron sequences and premature termination codons. These alternative mRNAs accumulated in the nonsense-mediated decay mutant upf3, indicating that they are normally subjected to degradation. Together, these results demonstrate that PIRL6 is required in both male and female gametogenesis and suggest that sporophytic expression is negatively regulated by unproductive alternative splicing. This posttranscriptional mechanism may function to minimize PIRL6 protein expression in sporophyte tissues while allowing the overlapping adjacent gene to remain widely transcribed.
Ras-group Leu-rich repeat proteins take part in developmental and physiological signaling in animals and yeast (Claudianos and Campbell, 1995; Buchanan and Gay, 1996). As members of the large and functionally diverse leucine-rich repeat (LRR) superfamily, they feature a domain consisting of tandem repeats of 22- to 24-amino acid Leu-rich motifs, which mediate specific protein-protein interactions and serve as a basis for LRR protein classification (Kajava, 1998; Kobe and Kajava, 2001). Most Ras-group LRR proteins function in intracellular signaling pathways. One, SUR-8/SHOC2, mediates interactions between components in the Ras/Raf pathway, which regulates growth and is activated in a high percentage of human cancers (Sieburth et al., 1998; Li et al., 2000; McKay and Morrison, 2007; Young et al., 2013). Mammalian RSU1/RSP1 is a candidate tumor-suppressor protein that acts downstream of Ras to regulate kinase activities and cell migration (Cutler et al., 1992; Dougherty et al., 2008; Gonzalez-Nieves et al., 2013). Another example, FLIGHTLESS, is multifunctional, taking part in chromatin regulation in the nucleus and Ras-mediated cytoskeletal regulation (Davy et al., 2001; Lee et al., 2004; Jeong et al., 2009). Thus, despite diverse biochemical functions, developmental signaling emerges as a common theme for this class of LRR proteins.
Plants harbor a small class of proteins structurally related to Ras-group LRRs (Forsthoefel et al., 2005; You et al., 2010). Plant intracellular Ras-group LRRs (PIRLs) feature an internal Leu-rich domain that is most closely related to those of animal Ras-group proteins, suggesting that they may interact with similar targets in related pathways. However, outside of their Leu-rich domain, PIRLs feature divergent hydrophilic N- and C-terminal domains that set them apart as a novel, plant-specific class of proteins. At least one, PIRL9, features a conserved phosphorylation site in its N-terminal domain that is phosphorylated in maize (Zea mays) pollen (Chao et al., 2016).
To define the contexts in which PIRLs function, we have undertaken a systematic reverse genetic investigation using Arabidopsis (Arabidopsis thaliana) knockout mutants. The PIRL family in Arabidopsis comprises nine genes that group into three subfamilies. Knockout alleles have been identified for eight of these genes (Forsthoefel et al., 2005, 2013), and phenotypes have revealed that mutations in PIRL1, PIRL3, and PIRL9 affect pollen development (Forsthoefel et al., 2010, 2013; Forsthoefel and Vernon, 2011). However, for one, PIRL6 (At2g19330), true knockout alleles have not been recovered. Two prospective knockout alleles were identified in available insertion mutant populations, but PIRL6 transcripts were detected routinely in mutant homozygotes, indicating that neither allele was a genuine knockout (Forsthoefel et al., 2013). One allele, pirl6-1, harbored an insert in exon I, suggesting that expression persisted due to a transformation-related duplication event or similar aberration (Tax and Vernon, 2001; Clark and Krysan, 2010). Therefore, we deemed these putative insertion lines unsuitable for further study. In contrast to what was found for PIRL6, knockout alleles for the two most closely related Arabidopsis genes, PIRL7 and PIRL8, have been identified; neither results in an obvious phenotype (Forsthoefel et al., 2005; Chen et al., 2010b).
The lack of knockout alleles raises the possibility that PIRL6 has an essential gametophyte function. Fully penetrant null mutations cannot be recovered for genes required in both male and female gametophytes (referred to below as dual gametophyte-essential genes), because such alleles cannot be transmitted through the haploid phase by either parent and so cannot persist in mutagenized populations (Berg et al., 2005; Meinke et al., 2008; Drews and Koltunow, 2011). For this reason, the precise number of such genes in plant genomes is difficult to determine, but gametophyte genes have long been thought to be underrepresented in Arabidopsis insertion mutant populations (Bonhomme et al., 1998). Despite the production of hundreds of thousands of sequence-tagged insertion mutations in Arabidopsis, approximately 12% of annotated loci are still not yet tagged (O’Malley and Ecker, 2010); at least some of these are likely to be dual gametophyte-essential genes. Thus, essential gametophyte genes pose a challenge for the identification of the minimal plant gene set and the full functional characterization of plant genomes, two goals of postgenomic plant biology (O’Malley and Ecker, 2010; Muralla et al., 2011; Lloyd and Meinke, 2012).
Taking into account the established roles for some PIRLs in pollen development, and the lack of pirl6-knockout alleles, we hypothesized that PIRL6 may function in both male and female gametophytes. Here, we present molecular and reverse-genetic evidence that this is indeed the case. PIRL6 is expressed during development in both gametophytes, and RNA interference (RNAi)-mediated gene knockdown results in frequent developmental arrest and the failure of gametogenesis in both pollen and the embryo sac. The approach employed here, identification of a gene lacking knockout alleles followed by straightforward RNAi knockdown, could represent a promising strategy for functional annotation of other gametophyte-essential loci that have so far escaped identification by insertion mutagenesis or traditional forward genetics. Interestingly, the PIRL6 transcription unit overlaps with a widely expressed adjacent gene, and PIRL6 mRNA undergoes unproductive alternative splicing that prevents its expression in sporophyte tissues. We propose that this posttranscriptional regulation serves to tightly restrict PIRL6 expression while allowing widespread transcription of the adjacent gene at this locus. Other overlapping transcription units in both plant and animal genomes may be divergently regulated in a similar manner.
RESULTS
The PIRL6 Locus and Polymorphisms
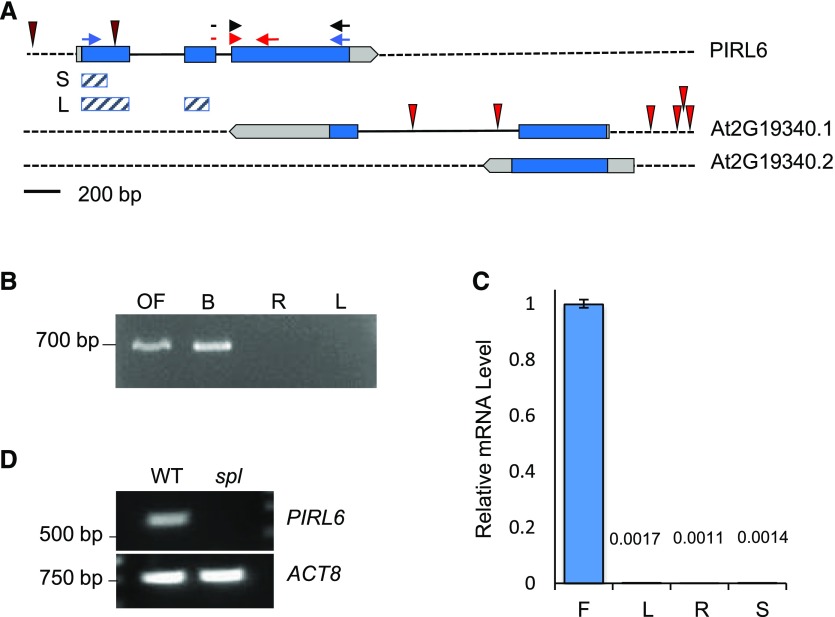
PIRL6 (At2g19330; GenBank accession no. AY849576) has the three-exon structure shared by most Arabidopsis PIRLs (Forsthoefel et al., 2005). The transcription unit is part of a convergent overlapping gene pair, with its 3′ end overlapping that of the neighboring At2g19340 gene, which encodes a putative oligosaccharyltransferase (Fig. 1). The overlap extends into the 3′ end of the PIRL6-coding region. There are no pirl6-knockout mutants in available Arabidopsis collections, including the original nonindexed Wisconsin mutant pools (Forsthoefel et al., 2005, 2013). However, there are six insertions documented for the adjacent At2g19340 gene, indicating that this chromosomal region is readily accessible to insertion mutagenesis (TAIR; http://www.arabidopsis.org). None of the At2g19340 insertions are in the region of overlap with PIRL6. We investigated other polymorphisms cataloged at TAIR and found that none had a major effect on the protein sequence. Of 13 nucleotide polymorphisms within the PIRL6-coding region, 12 are silent third-position transitions, and the one coding polymorphism results in a conservative Asn-to-Ser change (http://www.arabidopsis.org).
Figure 1.
PIRL6 gene structure and mRNA expression. A, PIRL6 (At2g19330) overlaps with the neighboring gene At2g19340, which generates two transcripts (0.1 and 0.2) with different polyadenylation sites (www.arabidopsis.org). Blue, Translated regions; light gray, untranslated regions; solid black lines, introns. T-DNA insertion sites in both loci are indicated by red spikes; neither PIRL6 insertion is a bona fide knockout (Forsthoefel et al., 2013). Small arrows indicate primer positions for PCR experiments shown in subsequent figures. Red arrows represent the primer pair used for qPCR; the forward primer straddles the exon II-III splice junction. Blue arrows represent primers used in full-length RT-PCR to detect the alternatively spliced cDNA species shown in Figure 4. Black arrows represent the primer pair used for RT-PCR experiments shown in Figure 2, A and C. Cross-hatched bars indicate exon regions included in short (S) and long (L) inverted repeat constructs for RNAi knockdown. B, PIRL6 RT-PCR carried out using a forward primer specific to the exon II-III splice junction, with RNA from open flowers (OF), developing inflorescences (buds; B), roots (R), and rosette leaves (L). C, PIRL6 RT-qPCR carried out on RNA from inflorescence (F), leaf (L), root (R), and germinated seedling (S) using the splice junction-specific primer used in B. Values are means from three replicate reactions; means for leaf, root, and seedling samples are provided above the bars. se is shown. D, RT-PCR of PIRL6 in flowers from wild-type (WT) and sterile spl homozygotes, which do not produce gametophytes. ACTIN8 (ACT8) was included as a positive control.
PIRL6 Is Expressed Primarily in Gametophytes
The lack of deleterious mutations in PIRL6 is consistent with an essential role in both male and female gametophytes. To first determine if PIRL6 is expressed in reproductive contexts, we surveyed its mRNA expression in wild-type inflorescences, leaves, and roots by reverse transcription-PCR (RT-PCR). The forward primer for these experiments spanned the exon II-exon III splice junction and, thus, was specific for PIRL6 mRNA from which intron II had been properly spliced. PIRL6 transcripts were detected by these primers in open and developing flowers but not in leaves or roots (Fig. 1B). Similarly, quantitative RT-PCR (RT-qPCR) with the same splice junction-specific forward primer did not detect significant transcript levels in leaves, roots, or whole seedlings (Fig. 1C). In contrast to PIRL6, transcripts derived from the adjacent At2g19340 gene were detected routinely by RT-PCR in leaves, roots, and inflorescences (Supplemental Fig. S1; documented at http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi).
We compared PIRL6 expression in wild-type flowers and sterile flowers from plants homozygous for the sporocyteless (spl) mutation, which lack both male and female gametophytes due to the arrest of reproductive differentiation prior to meiosis (Yang et al., 1999). Transcripts were again detected in wild-type flowers but not in flowers from spl homozygotes (Fig. 1D). This showed that spliced PIRL6 mRNA was detectable only in flowers that contained gametophytes.
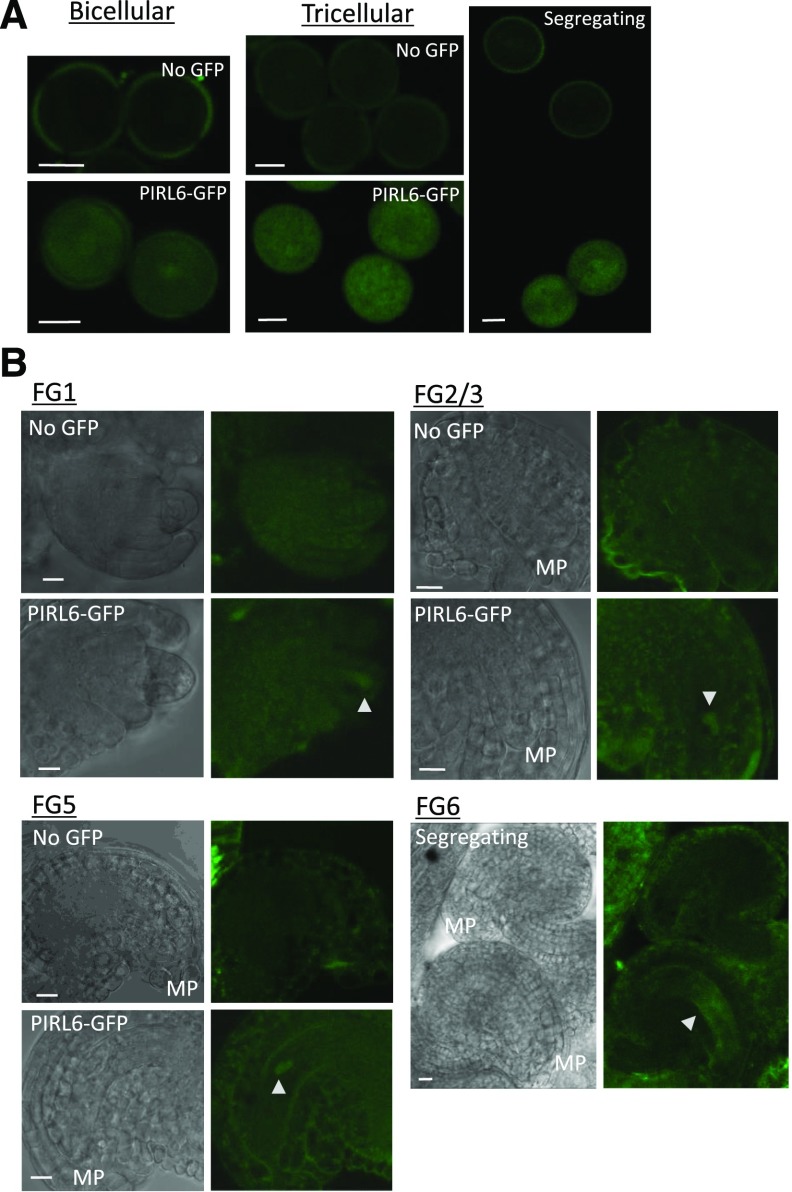
To determine if PIRL6 protein is expressed in both male and female gametophytes, we transformed wild-type Arabidopsis with a GFP fusion construct consisting of a PIRL6 genomic clone containing 1.1 kb of 5′ flanking sequence and the full transcription unit (including introns), ligated in frame to GFP. Male and female gametophytes were viewed at various developmental stages. Expression was detected in both developing pollen and the embryo sac. In pollen, expression was first visible at the bicellular stage, and it persisted through the tricellular stage (Fig. 2). In ovules, expression was evident in the embryo sac as early as stage FG1, and it persisted through megagametogenesis, ultimately localizing centrally within the embryo sac by stage FG5/6 (Fig. 2; stages defined as in Christensen et al., 1998). These results were consistent with published transcriptome results (Honys and Twell, 2004; Wuest et al., 2010; Loraine et al., 2013), including RNA sequencing (RNA-seq) that detected expression in the central cell of the female gametophyte (Schmid et al., 2012).
Figure 2.
Expression of PIRL6-GFP in both male and female gametophytes. Gametophytes segregating for a full-length PIRL6-GFP fusion construct were viewed at the indicated developmental stages by confocal and differential interference contrast (DIC) microscopy. Plants were hemizygous for the reporter construct, producing pollen and embryo sacs that segregated 1:1 for the PIRL6-GFP construct; for each developmental stage shown above, pollen and ovules were from the same anthers and ovaries, respectively. A, Expression in bicellular and tricellular stage pollen; the right-most image is a single image illustrating the meiotic segregation of the reporter construct in pollen from a hemizygous anther. B, Expression during female gametophyte development. Triangles mark locations of PIRL-GFP within the embryo sac. Female gametophyte (FG) developmental stages were defined by Christensen et al. (1998). To provide a reference point, the micropylar end of each ovule is labeled on the DIC panels (MP). The FG6 stage image is a single image illustrating the segregation of the reporter construct in adjacent ovules. Bars = 10 μm.
PIRL6 Transcripts Undergo Unproductive Alternative Splicing
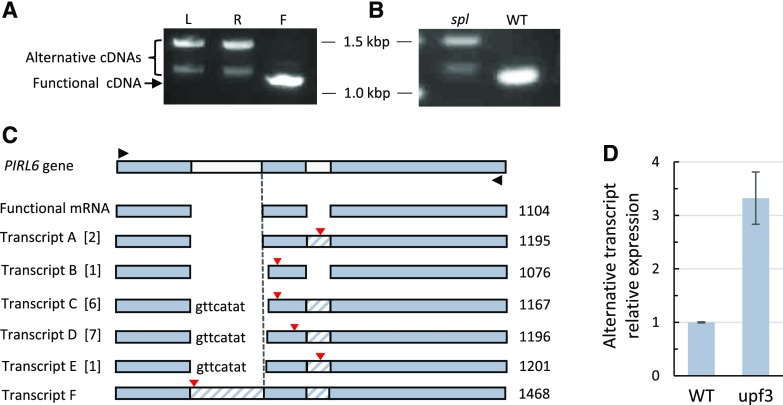
Overlapping gene pairs in plants have been reported to exhibit a higher-than-average frequency of alternative splicing (Jen et al., 2005). Alternative splicing can impact a gene’s regulation and lead to the misinterpretation of transcriptome data. Therefore, we investigated the possibility of alternative splicing for PIRL6. We first examined mRNA expression in flowers, leaves, and roots by RT-PCR, using primers that amplified the full PIRL6-coding region, which includes both intron sites (Fig. 1). cDNA was produced with an oligo(dT) primer, so RT-PCR products were derived from polyadenylated RNA. The results are shown in Figure 3. Flower RNA yielded primarily a 1.1-kb cDNA, and gel purification and sequencing confirmed that it was produced from the spliced transcript containing the predicted functional PIRL6 open reading frame. With these reactions targeting the full-length transcript, leaves and roots yielded other cDNA products, including an approximately 1.5-kb product corresponding to the predicted unspliced RNA, plus intermediate-length transcripts approximately 100 bp longer than the fully spliced band obtained from flowers. Sequencing confirmed that they were transcribed from PIRL6 (see below). Therefore, vegetative organs produce polyadenylated, alternatively spliced PIRL6 RNAs longer than those detected in flowers.
Figure 3.
PIRL6 undergoes unproductive alternative splicing outside of gametophytes. PIRL6 transcripts were amplified from polyadenylated RNA from wild-type and spl mutant homozygotes using primers designed to amplify the full-length coding region. A, RT-PCR products from wild-type leaves (L), roots (R), and flowers (F). Sequencing confirmed that the predominant flower cDNA was derived from the translatable, spliced PIRL6 mRNA. Larger alternative products are evident in leaves and roots; the largest band corresponds in size to the unspliced transcript (1,468 bp). B, RT-PCR of PIRL6 transcripts from wild-type (WT) and sterile spl mutant flowers, showing the predominance of alternative transcripts in flowers that lack gametophytes. To detect transcripts present in spl, the spl lane was loaded with 10× the volume of the reaction product than the wild-type lane. C, Structures of alternative polyadenylated PIRL6 transcripts sequenced from roots and leaves. Gel-resolved leaf and root RT-PCR products between approximately 1.1 and 1.4 kb were cloned in Escherichia coli, and 17 independent cDNAs were sequenced. Black triangles indicate primer positions used for PCR following cDNA synthesis primed with oligo(dT). Functional mRNA represents the flower RT-PCR product, corresponding to the annotated spliced PIRL6 mRNA. Transcripts A to F are alternative mRNA species. Numbers in brackets indicate the number of independent cDNA clones obtained for each transcript species. Lowercase letters indicate residual unspliced nucleotides from intron I; cross-hatched regions indicate unspliced introns. Red triangles mark the positions of premature termination codons. The dashed line marks the position of the normal 5′ end of exon II. The sizes of RT-PCR products corresponding to each mRNA species are indicated in bp at far right. D, qPCR showing increased accumulation of PIRL6 alternative transcripts in the NMD-deficient mutant upf3. qPCR was carried out on leaf RNA using an intron II-specific forward primer, allowing the detection of alternative transcripts A, C, D, E, and F. Values are means of two biological replicates, with three reaction replicates for each; se is shown.
We also carried out full-length RT-PCR on inflorescences from spl mutant homozygotes (Fig. 3B). spl1 flowers lacked detectable levels of the properly spliced mRNA detected in wild-type flowers but expressed high-molecular-weight transcripts similar to those detected in leaf and root. These transcripts were somewhat harder to detect than the 1.1-kb spliced transcripts detected in wild-type flowers (the spl1 lane shown in Fig. 3B is loaded with a 10× higher volume of reaction products than the wild-type lane). These spl1 products were derived from diploid flower tissues, because spl1 reproductive development arrests before meiosis. The predominance of these abnormal transcripts in spl1 flowers provided additional evidence that the PIRL6 mRNA detected in wild-type flowers was present primarily, possibly exclusively, in gametophytes.
To further investigate the transcripts seen in leaves and roots, we cloned gel-resolved PIRL6 RT-PCR products and sequenced 17 cDNAs. The results are compiled in the maps shown in Figure 3C. The 17 clones harbored five different aberrantly spliced PIRL6 cDNA species (labeled A–E), all of which contained intron fragments and/or entire introns. Transcripts C and D were represented by six and seven independent clones, respectively, indicating that they were the most abundant PIRL6 mRNA species in vegetative tissues. Both contained intron II in its entirety. A sixth alternative transcript (transcript F) corresponding in size to unspliced PIRL6 transcript was detected by RT-PCR. All these alternative transcripts contained premature termination codons (PTCs) that would disrupt translation and truncate the protein early in the conserved LRR domain (Fig. 3C). Furthermore, these transcripts are subjected to nonsense-mediated decay (NMD): in leaves of NMD-impaired upf3 mutants (Arciga-Reyes et al., 2006), levels of alternative transcripts were approximately 300% higher than in those of the wild type (Fig. 3D). The combination of truncated open reading frames and NMD degradation, and the absence of detectable functional transcripts in vegetative organs, whole seedlings, and spl, suggest that alternative splicing serves a negative regulatory function to minimize PIRL6 protein expression outside of gametophytes.
PIRL6 Knockdown Disrupts Gametophyte Development
We predicted that RNAi against PIRL6 would result in gametophyte developmental defects. An RNAi-inducing construct, PIRL6-KD(S), containing inverted repeats of the 5′-most 139 nucleotides of the PIRL6-coding region, was generated in the pFGC5941 RNAi vector. The 5′ end of the gene was chosen to minimize the chances of off-target effects, for the following reasons: (1) it is distal from the region of 3′ overlap with the adjacent At2g19340 gene; (2) it is from a unique sequence region, sharing only 66% nucleotide sequence identity with PIRL7; and (3) the longest continuous stretch of alignment with any other predicted transcripts is only 9 bp, well below the similarity threshold required to trigger off-target RNAi. Furthermore, the RNAi pathway has considerably less tolerance for target sequence mismatches in 5′ coding sequences than in 3′ untranslated regions (Wei et al., 2012). A second construct, PIRL6-KD(L), with inverted repeats of the 5′-most 458 nucleotides of the coding region, also was produced. We sought to drive construct expression broadly in reproductive tissues, including both male and female gametophytes as well as premeiotic and adjacent sporophytic cells. Therefore, constructs were expressed under the control of the 35S promoter, which is widely expressed and has proven effective for RNAi knockdown in Arabidopsis, including the knockdown of gametophyte gene expression (McGinnis et al., 2005; León et al., 2007).
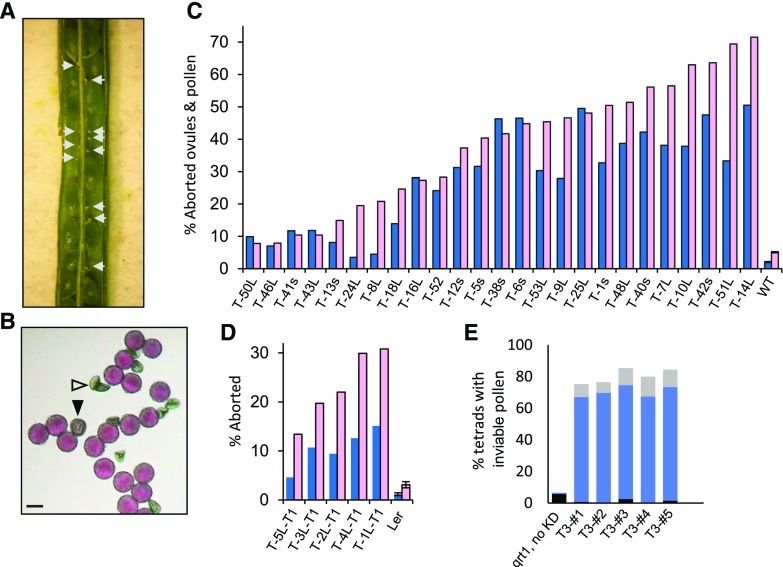
PIRL6-KD constructs were introduced into wild-type Arabidopsis (ecotype Wassilewskija [WS]) by Agrobacterium tumefaciens-mediated transformation, and 44 T1 seedlings were identified by glufosinate resistance. The presence of the PIRL6-KD transgene was confirmed in seedlings by genomic PCR on excised leaf tissue. Because of the potential for gametophyte lethality and, thus, decreased heritability, ovules and pollen produced by T1 plants were scored directly for segregating gametophyte defects. For initial phenotype observation and quantification, ovules were scored by dissection microscopy of developing siliques, and mature pollen was observed by light microscopy after staining with Alexander’s reagent, which rapidly identifies aborted pollen (Johnson-Brousseau and McCormick, 2004) as well as small abnormal pollen that still stains for the presence of cytoplasm (Forsthoefel and Vernon, 2011). We observed a high frequency of both ovule and pollen defects segregating in T1 plants (Fig. 4). Severely shrunken ovules were distributed randomly throughout developing siliques. In contrast, the few unfertilized ovules scored in wild-type controls were located at the ends of siliques. Pollen defects included early lethality as well as abnormally small grains that still stained as viable.
Figure 4.
Aborted ovules and pollen resulting from PIRL6 knockdown. PIRL6 inverted repeat constructs were introduced into wild-type WS Arabidopsis by A. tumefaciens-mediated transformation; transgenic T1 plants harboring the RNAi construct were identified by glufosinate resistance and confirmed by genomic PCR. T1 plants were hemizygous and were scored directly for segregating ovule and pollen defects. A, A silique from a T1 plant (T-48L-T1) containing seeds from successfully fertilized ovules segregating with aborted ovules (white arrows). B, Alexander-stained pollen produced by T1 plant T-48L-T1, showing the segregation of aborted (white triangle) and stunted (black triangle) grains. Bar = 20 µm. C, Percentages of abnormal or aborted pollen (blue) and aborted ovules (pink) in 25 T1 plants independently transformed with PIRL6 RNAi constructs and in wild-type controls (WT). The ranges of n values are as follows: pollen, 61 to 1,468; ovules, 105 to 191. L or s in plant line labels indicates the PIRL6-RNAi construct introduced in that line (see text). D, Replication of the RNAi-induced phenotype in the Ler ecotype. Percentages are shown for abnormal or aborted pollen (blue) and aborted ovules (pink) in five Ler T1 plants independently transformed with the PIRL6-RNAi(L) construct. The n value ranges are as follows: pollen, 352 to 1,124; ovules, 142 to 178. In C and D, means and se are shown only for untransformed controls; se was not determined for T1 samples because values were obtained, by definition, from individual transformed plants. E, Gametophytic basis of PIRL6-KD pollen defects, shown by meiotic segregation in pollen tetrads produced by five individual PIRL6-KD hemizygotes in a qrt1 background. Black bars, percentage of tetrads with one dead pollen grain; solid blue bars, percentage of tetrads with two dead pollen grains; light gray bars, percentage of tetrads with more than two dead pollen grains (the range of n values is as follows: 89–198 tetrads per plant).
The observed ovule and pollen defects were prevalent in the T1 population: 38 of the 44 T1 transformants exhibited aborted ovules and/or abnormal pollen at more than double the frequency scored in wild-type controls, most at substantially higher frequencies. The frequencies of pollen defects and ovule lethality from 25 T1 plants are compiled in Figure 4C. As expected in any population of transgenic plants generated by independent transformation events, phenotype severity varied. Pollen and ovule lethality ranged from low levels to approximately 50%. Two plants exhibited approximately 70% ovule lethality, and subsequent genetic analysis (described below) showed that these plants harbored more than one copy of the transgene, accounting for the high segregation frequency of their gametophyte defects. No consistent differences in severity were observed between plants harboring the PIRL6-KD(S) and PIRL6-KD(L) inverted repeat constructs. Notably, the frequency of effects on ovules and pollen was generally correlated: transformants with the highest rates of ovule abortion also tended to have high frequencies of pollen defects. The results of PIRL6 knockdown were replicated in a second Arabidopsis ecotype, Landsberg erecta (Ler). Ler plants harboring the PIRL6-KD constructs displayed both pollen and ovule defects resembling those produced by WS transformants (Fig. 4D).
Genetic experiments confirmed the gametophytic basis of the observed pollen and ovule phenotypes. For pollen, we introduced the PIRL6-KD(L) construct into a qrt1 mutant background, which is in the Ler ecotype (Johnson-Brousseau and McCormick, 2004). qrt1 tetrads produced by F2 plants hemizygous for the knockdown construct segregated for pollen defects, with a large majority of tetrads exhibiting two abnormal pollen grains (Fig. 4E). Disruption of megagametophyte function was demonstrated in two crosses in which PIRL6-KD hemizygotes were fertilized with wild-type pollen. With full transmission through the megagametophyte, F1 progeny would be predicted to segregate 1:1 for the glufosinate resistance encoded by the knockdown construct. However, in both such crosses, we observed significantly reduced frequencies of glufosinate resistance in F1 progeny, with ratios of 0.31:1 and 0.06:1 resistant:sensitive F1 seedlings (P < 0.02 and P < 0.001, respectively, as determined by χ2 tests). Such segregation distortion is a characteristic of disrupted gametophyte gene function, as a reproductive defect based in sporophytic cells would not specifically hinder transmission by gametophytes with the PIRL6-KD genotype (Feldmann et al., 1997; Drews and Koltunow, 2011).
Similarly, segregation distortion from reduced gametophytic transmission was evident in the progeny of self-fertilized PIRL6-KD transgenics. In F1 progeny of self-fertilized hemizygotes, glufosinate resistance should segregate at 3:1 (or 15:1 if a parent plant harbors two unlinked copies of the transgene) if there is normal Mendelian inheritance. We determined the ratio of glufosinate-resistant to -sensitive plants in 11 T2 progeny populations produced by selfed T1 hemizygotes. The results are shown in Table 1. Segregation distortion was observed in each case, with resistant:sensitive ratios substantially below 3:1 in nine of the 11 lines, indicating reduced transmission by gametophytes harboring the knockdown construct. In the two remaining lines, the resistant:sensitive ratio was considerably above 3:1, indicating that these lines contained more than one copy of the knockdown construct. Consistent with this, the T1 founders of both these lines also had exhibited substantially greater than 50% ovule lethality (Fig. 4). Reduced transmission occurred in these lines as well, with resistant:sensitive ratios substantially below the 15:1 predicted for plants containing two unlinked copies of the PIRL6-KD transgene.
Table 1. Reduced transmission of the PIRL6-KD constructs.
| Plant Line | Germinated | Glufosinate R:Sa (Ratio) | Expected R:Sb (Ratio) | χ2 (P)c | Interpretation |
|---|---|---|---|---|---|
| T-40s-T2 | 409 | 239:170 (1.4:1) | 307:102 (3:1) | 59.85 (<0.0001) | One insert, reduced transmission |
| T-48L-T2 | 250 | 163:87 (1.9:1) | 188:62 (3:1) | 12.8 (0.0003) | One insert, reduced transmission |
| T-53L-T2 | 235 | 141:94 (1.5:1) | 176:59 (3:1) | 28.2 (<0.0001) | One insert, reduced transmission |
| T-5s-T2 | 170 | 109:61 (1.8:1) | 128:42 (3:1) | 10.74 (0.0011) | One insert, reduced transmission |
| T-1s-T2 | 69 | 43:26 (1.7:1) | 52:17 (3:1) | 5.92 (0.015) | One insert, reduced transmission |
| T-9L-T2 | 63 | 40:23 (1.7:1) | 47:16 (3:1) | 4.45 (0.035) | One insert, reduced transmission |
| T-38s-T2 | 183 | 121:62 (1.95:1) | 137:46 (3:1) | 7.7 (0.005) | One insert, reduced transmission |
| T-12s-T2 | 92 | 58:34 (1.7:1) | 69:23 (3:1) | 7.01 (0.008) | One insert, reduced transmission |
| T-16s-T2 | 79 | 0:79 (0) | 59:20 (3:1) | 237 (<0.0001) | No transmission |
| T-14L-T2 | 102 | 83:19 (4.4:1) | 96:6 (15:1) | 26.67 (<0.0001) | Two inserts, reduced transmissiond |
| T-51L-T2 | 57 | 46:11 (4.2:1) | 53:4 (15:1) | 16.56 (<0.0001) | Two inserts, reduced transmissiond |
T2 seedling populations produced by self-fertilized PIRL6-KD T1 transformants were scored for glufosinate resistance (R) encoded by the selectable marker gene cointegrated with the RNAi construct. S, Glufosinate sensitivity.
Glufosinate resistance was predicted to segregate at a Mendelian ratio of 3:1 resistant (R) to sensitive (S) in each line if fully transmitted to T2 progeny and encoded by a single transgene locus and at 15:1 if the parent T1 harbored two fully transmitted unlinked inserts. Gametophyte lethality would result in significant distortion of these ratios (Feldmann et al., 1997).
χ2 calculations were determined using e-values based on a 3:1 Mendelian ratio for the first nine lines and a 15:1 ratio for T-14L-T2 and T-51L-T2, based on the apparent presence of more than one transgene. P < 0.05 indicates significant segregation distortion from the Mendelian ratio predicted for full transmission.
These lines each produced more than 50% aborted ovules (Fig. 5), consistent with the meiotic segregation of more than one transgene insert.
Backcrosses to the wild type further confirmed the heritability of the pollen and ovule phenotypes induced by PIRL6 knockdown. T1 transformant T-53L was backcrossed to the wild type, and pollen and siliques produced by F1 progeny were scored for defects. High frequencies of both pollen and ovule lethality were observed segregating in glufosinate-resistant F1s (Supplemental Fig. S2). Two of the F1 progeny from this cross were then used for a second (serial) backcross, and the resulting F1 progeny were again scored for gametophyte defects. Again, glufosinate-resistant backcross progeny consistently produced high percentages of both inviable pollen and aborted ovules (Supplemental Fig. S2B). A backcross with a T2 individual from another transformed line, T-5s, yielded similar results: 10 glufosinate-resistant F1 individuals from that cross were scored, and pollen and ovules from all of them segregated for high frequencies of defects (Supplemental Fig. S2C). This and the serial backcrosses shown in Supplemental Figure S2B both demonstrated the persistence of gametophyte defects through three generations in lines harboring the PIRL6-KD constructs.
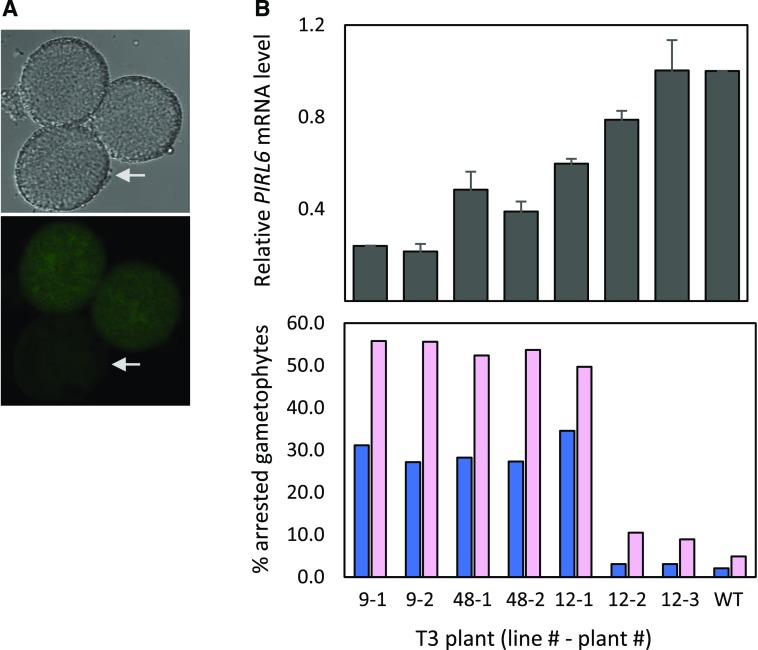
Effective knockdown of PIRL6 expression in PIRL6-KD transgenic plants was validated by two different methods. First, we introduced the PIRL6-KD constructs into plants harboring the PIRL6-GFP reporter and observed pollen produced by plants homozygous for PIRL6-GFP and hemizygous for the PIRL6-KD construct. Those pollen, which segregated for the knockdown construct, also showed the segregation of reduced PIRL6-GFP expression (Fig. 5), confirming PIRL6 knockdown within the male gametophyte. Second, RT-qPCR demonstrated reduced PIRL6 mRNA levels in PIRL6-KD lines, and knockdown efficacy in individual plants showed a general correlation with phenotype severity, as gauged by the frequency of gametophyte defects (Fig. 5B).
Figure 5.
Reduction of PIRL6 mRNA levels by PIRL6-KD. A, Reduced PIRL6-GFP expression in pollen produced by a plant homozygous for the genomic PIRL6-GFP reporter construct and hemizygous for the PIRL6-KD(L) construct, such that pollen all contained the GFP reporter but were segregating for the knockdown construct. Mature pollen were viewed by DIC (top) and confocal fluorescence (bottom) microscopy. Arrows indicate pollen with visibly reduced PIRL6-GFP signal. B, Relationship between PIRL6 mRNA expression and phenotype severity in wild-type controls (WT) and individual PIRL6-KD plants from three independent transgenic lines. Top graph, PIRL6 RT-qPCR carried out on developing flowers from individual T3 plants from three independent transgenic lines. Plants were homozygous for the PIRL6-KD construct; means and se from three replicate RT-qPCRs are shown. Bottom graph, Phenotype severity in the same individual T3 plants, gauged by the percentage of inviable pollen (dark blue) or aborted ovules (pink). The range of n values is as follows: 124 to 1,773 (pollen); 87 to 192 (ovules)
PIRL6 Is Required for Male and Female Gametogenesis
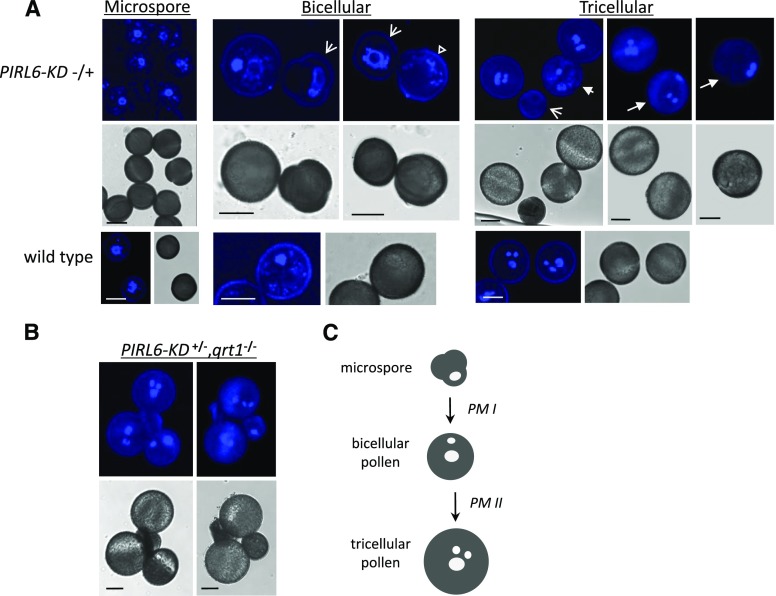
To better define the timing and impact of PIRL6 disruption in gametophytes, we further investigated the development of PIRL6-KD gametophytes. Gametophytes produced by PIRL6-KD hemizygotes, which segregate at approximately 50% for PIRL6 knockdown, were observed so that wild-type and knockdown gametophytes of identical chronological age could be compared directly. To observe pollen development, anthers at various developmental stages were stained with 4′,6-diamino-phenylindole (DAPI) and viewed by confocal microscopy (Fig. 6). Obvious developmental differences were not observed segregating in microspores but were evident as anther development progressed beyond pollen mitosis I (PM1). A variety of defects were seen in bicellular stage anthers, including arrested microspores that had failed to undergo PM1 and larger grains similar in size to bicellular pollen that nevertheless lacked a visible compact generative nucleus. Variation persisted in anthers that had progressed through both PM1 and PM2, including degenerating microspores, bicellular pollen that had arrested prior to PM2, and abnormal tricellular pollen in which sperm and vegetative nuclei failed to group into the centrally localized male germ unit characteristic of Arabidopsis pollen. A similar range of pollen abnormalities also was observed segregating in the qrt1 background (Fig. 6B). Therefore, PIRL6 functions in pollen development as early as PM1 and is required for progression through pollen mitotic stages and the subsequent formation of a spatially organized male germ unit.
Figure 6.
PIRL6 knockdown disrupts male gametogenesis. Microspores and pollen produced by plants hemizygous for the PIRL6-KD construct, which segregate at 50% for PIRL6 knockdown, were stained with DAPI and viewed by confocal fluorescence or DIC microscopy. A, Microspores and pollen from anthers at the indicated developmental stages, viewed by confocal fluorescence (top row) or DIC (middle row). Arrows indicate pollen segregating for developmental defects of varying severity, including arrested microspores (broad arrows), enlargement with absence of germinal nucleus (hollow triangle), arrest after PM1 (short arrow), or abnormal male germ unit configuration (long arrows). The bottom row shows wild-type control samples at corresponding developmental stages. Bars = 10 µm. B, Tetrads from tricellular stage anthers, showing meiotic segregation of developmentally arrested PIRL6-KD pollen in the qrt1 background. Bars = 10 µm. C, Simplified diagram of the major stages of wild-type Arabidopsis pollen development, provided for reference. PM, Pollen mitoses. White ovals represent idealized nuclear configurations observable with DAPI staining: large ovals, vegetative cell nuclei; small ovals, generative nucleus (bicellular stage) and sperm nuclei (tricellular stage). The tricellular stage nuclei illustrate the triangular configuration of the male germ unit characteristic of mature Arabidopsis pollen.
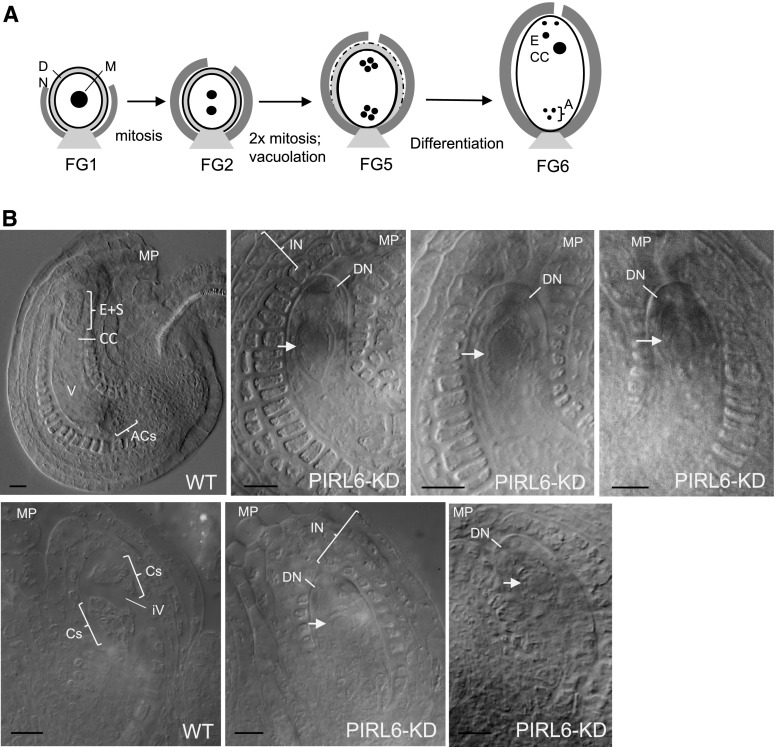
PIRL6 also was required for proper progression through mitotic stages in the female gametophyte. DIC microscopy of developing ovules was carried out with PIRL6-KD hemizygotes, which produce gametophytes segregating at approximately 50% for the knockdown construct. At the megaspore stage (FG1), ovules harboring the PIRL6-KD construct were not readily distinguishable from the wild type (n = 54), but aberrant development became evident as wild-type gametophytes within the same ovary progressed. Through the mitotic stages of embryo sac development, presumptive knockdown embryo sacs appeared arrested, with only one large, centrally located cell. Examples from segregating FG5 and FG6 stage ovaries are shown in Figure 7. A second notable abnormality reflecting early gametophyte arrest was the persistence of a discernible and well-defined distal nucellus, regardless of the chronological age of the ovule. This cell layer, which surrounds the megaspore and initial gametophytic mitotic progeny very early in embryo sac development, normally serves as a source of nutrients for the gametophyte, and it consequently degenerates as embryo sac mitoses progress. Typically, the distal nucellus is absent at FG6 (Ingram, 2017; illustrated in Fig. 7A). With the arrest of embryo sac development with PIRL6 knockdown, this did not occur, and the cell layer consistently remained intact and robust through FG6 (Fig. 7; n = 46). With mitotic arrest, PIRL6-KD gametophytes also did not grow and elongate, but surrounding sporophyte-derived integument cell layers continued to develop such that the overall ovule size ultimately was similar to that of wild-type ovules. Thus, in ovaries in which wild-type gametophytes had completed mitosis (stages FG5 and FG6), PIRL6-KD embryo sacs were routinely identifiable segregating as structures resembling stalled FG1-like gametophytes fully enveloped within integuments characteristic of older ovules. Normally, single-celled (FG1 stage) embryo sacs are flanked by short, undeveloped integuments (Fig. 7A; for an example of integuments typically associated with wild-type gametophytes at the single-cell stage, see Fig. 2B). In summary, as in pollen development, PIRL6 knockdown resulted in defects in gametogenesis, hindering progress through mitosis and differentiation. Notably, the onset of visible defects corresponded well with the timing of detectible PIRL6-GFP expression.
Figure 7.
PIRL6 knockdown disrupts female gametogenesis. A, Simplified diagram summarizing selected aspects of Arabidopsis female gametophyte development at the indicated developmental stages (Christensen et al., 1998), pictured with the micropyle at top and the chalazal end at bottom. The embryo sac is represented as a white oval and constituent nuclei as black circles. Adjacent sporophytic tissues are included: light gray, nucellus; dark gray, inner integument. A, Antipodal cell nuclei in their characteristic triangular configuration; CC, central cell nucleus; DN, distal nucellus region; M, megaspore nucleus. B, Ovules from ovaries at postmitotic developmental stages, cleared as whole mounts in Hoyer’s solution and viewed by DIC microscopy. The micropylar region (MP) of each ovule is labeled to indicate the orientation of the image. Parent plants were hemizygous for the PIRL6-KD construct, and embryo sacs segregated approximately 50% for PIRL6 knockdown. The top row shows a wild-type ovule (WT) and three examples of segregating PIRL6-KD ovules from FG6 stage ovaries. Antipodal cells (ACs), a large vacuole (V), the central cell (CC), and the cluster of egg cell and synergids (E+S) were discernible in segregating wild-type ovules (nuclei are not visible in the selected focal plane). PIRL6-KD embryo sacs each feature single, apparently mitotically arrested cells (white arrows) with large nuclei, surrounded by an abnormally persistent, sharply defined distal nucellus layer (DN). Integument (IN) layers fully envelope the PIRL6-KD gametophytes, despite their developmental arrest as single cells (in contrast to the integuments flanking the wild-type single-cell FG1 ovule shown in Fig. 2B). The bottom row shows wild-type and PIRL6-KD ovules from FG5 stage ovaries. Yet-undifferentiated cells produced by mitotic divisions (Cs) and an initiating central vacuole (iV) are discernible in the FG5 wild-type ovule. PIRL6-KD embryo sacs resembled those observed in PIRL6-KD FG6 ovaries, with single, arrested cells (white arrows) enveloped by a persistent distal nucleus (DN) and expanded integuments (IN) that resembled those of wild-type ovules. Bars = 10 µm.
DISCUSSION
Based on the established gametophyte functions for PIRL1 and PIRL9 and the lack of bona fide pirl6 knockout alleles in available Arabidopsis populations, we hypothesized that PIRL6 may be essential in both pollen and embryo sac development. The haploid nature of plant gametophytes can make it difficult to obtain unequivocal genetic evidence for essential genes in this class, since null or strong alleles may not be transmitted and, thus, may not persist in mutagenized populations. However, the PIRL6 expression and knockdown phenotypes reported here strongly support the hypothesis that this novel Ras-group LRR protein has a role early in the formation of both gametophytes.
Despite the theoretical difficulties posed by gametophyte developmental genetics, forward screens have successfully identified hundreds of genes important for gametophyte function (Feldmann et al., 1997; Christensen et al., 1998; Howden et al., 1998; Johnson et al., 2004; Pagnussat et al., 2005; Boavida et al., 2009; for review, see McCormick, 2004; Yadegari and Drews, 2004; Borg et al., 2009; Berger and Twell, 2011; Drews et al., 2011). Many of these genes were identifiable because phenotypes were specific to either pollen or the embryo sac, so disrupted alleles could be transmitted through the unaffected gametophyte. Yet, dual-gametophyte genes also were identified (Christensen et al., 1998; Howden et al., 1998; Johnson et al., 2004; Pagnussat et al., 2005; Oh et al., 2016; D’Ippólito et al., 2017). This was possible because mutant alleles did not entirely eliminate gene functions or the disrupted functions were not fully essential for gametophyte viability, resulting in some transmission of mutant alleles. Despite these many successes, from a functional genomics standpoint, gametophyte genes still pose a challenge (Berg et al., 2005; Meinke et al., 2008; Muralla et al., 2011). Such genes may be underrepresented in mutant populations and missed in forward mutant screens (Bonhomme et al., 1998; Christensen et al., 1998; Pagnussat et al., 2005); accordingly, an unexpectedly low percentage of developmentally characterized Arabidopsis loss-of-function mutations reside in gametophytic genes (Lloyd and Meinke, 2012). Approximately 12% of annotated Arabidopsis loci still are not represented among the hundreds of thousands of sequence-tagged Arabidopsis insertion mutant lines generated by large-scale functional genomics efforts (O’Malley and Ecker, 2010). For at least some of these loci, this is likely due to the transmission failure of mutant alleles caused by dual gametophytic lethality. Thus, PIRL6 is a member of a class of genes that has proven difficult to comprehensively define through classic forward genetics or gene-tagging efforts. The strategy we employed here, identification of a gene for which knockout alleles are not available, followed by straightforward knockdown (or, alternatively, targeted nonlethal disruption by CRISPR), could perhaps be applied more broadly for the systematic identification of additional gametophyte-essential genes in Arabidopsis.
PIRL6 Expression
The expression analyses reported here support a role for PIRL6 in gametophytes. RT-PCR using an exon II-III splice junction-specific primer detected transcripts in flowers, and this product was not detected in spl mutant flowers, which contain sporophytic flower tissues but lack both male and female gametophytes (Yang et al., 1999). Gametophyte expression of a translated product was confirmed with the PIRL6-GFP construct. Because this construct was under the control of the PIRL6 promoter and contained introns, it required both transcription and proper splicing for its expression. Therefore, it confirmed the gametophyte location and translatability of the flower mRNA detected by RT-PCR. Furthermore, reporter expression in pollen and embryo sacs produced by PIRL-GFP hemizygotes exhibited clear segregation (Fig. 2), indicating gametophytic expression. Available transcriptome data also support PIRL6 expression in both male and female gametophytes. Microarrays (Honys and Twell, 2004; Wuest et al., 2010) and RNA-seq (Schmid et al., 2012; Loraine et al., 2013) both have detected PIRL6 expression within the central cell of the embryo sac and in pollen. While those transcriptomic studies did not distinguish between the differentially expressed splice variants we identified, these transcriptome findings are all consistent with our expression data.
A gene need not be expressed solely in gametophytes to be important in gametophyte development. However, our results with the sterile spl1 mutant suggest that, at least in flowers, the expression of translatable PIRL6 mRNA may be gametophyte specific. In addition, RT-PCR experiments, including those with whole seedlings, did not detect fully spliced PIRL6 transcripts in vegetative organs (Fig. 1). Nevertheless, we cannot completely rule out PIRL6 expression in some limited sporophytic contexts.
Gene Overlap, Alternative Splicing, and Implications for Regulation
Overlapping genes such as those at the PIRL6 locus are found at an unexpectedly high frequency in eukaryotic genomes (Boi et al., 2004). Perhaps surprisingly, despite the potential for the production of antisense transcripts from opposite-strand transcription units, these gene pairs are not strongly associated with the production of small regulatory RNAs (Jen et al., 2005; Henz et al., 2007). Many overlapping genes are thought to be coexpressed due to their shared presence in euchromatic domains (Zhan and Lukens, 2013), but PIRL6 and At2g19340 do not have correlated expression patterns: At2g19340 transcripts are expressed widely in Arabidopsis development, as determined by both available transcriptome data (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi/) and RT-PCR in our laboratory (Supplemental Fig. S1).
Overlapping transcription units in Arabidopsis have been associated with a higher likelihood of alternative splicing (Jen et al., 2005; Henz et al., 2007), and we found that this association applied to PIRL6. We identified seven PIRL6 transcripts, including the functional transcript as well as six with residual intron sequence(s) and consequent PTCs. These alternative cDNAs were not simply preprocessed artifacts, for the following reasons: (1) the cDNAs were generated from polyadenylated transcripts; (2) they were present in plants at routinely detectable levels; and (3) they shared consistent sequence features, such as the residual 5′-GTTCATAT-3′ octanucleotide from intron I, found in four of the six alternate mRNAs. These cDNA structures also were consistent with RNA-seq products deposited in Plant MPSS transcriptome databases (Nakano et al., 2006). PTCs in alternative PIRL6 transcripts all were located within the highly conserved LRR domain, suggesting that the alternative transcripts go untranslated and/or encode nonfunctional truncated products. Unproductive splicing is a common phenomenon in both plants and animals (Lewis et al., 2003; Filichkin et al., 2010; Reddy et al., 2012). PTC-containing alternative transcripts reduce protein expression both by preventing full translation and by concomitantly reducing mRNA abundance via NMD (Marchant and Bennett, 1998; Palusa and Reddy, 2010; Reddy et al., 2012). This appears to be the case for PIRL6, based on our results with the NMD mutant upf3 (Fig. 3). This posttranscriptional negative regulation of PIRL6 may reflect a need for strict control in tissues where it is not required.
It is not known why overlapping gene pairs are associated with alternative splicing (Jen et al., 2005; Henz et al., 2007). We propose that alternative splicing in some cases allows for differential regulation of the genes at such loci. Overlapping genes may be difficult to selectively regulate via transcription, due to their close association within a single chromatin domain. Posttranscriptional regulation would allow for the divergent expression of protein products of those cotranscribed genes. This appears to be the case for PIRL6: its alternative transcripts are produced primarily in contexts in which translatable PIRL6 transcripts were not detected, including vegetative organs and the full suite of diploid tissues present in spl1 flowers. This unproductive splicing allows for selective down-regulation of PIRL6 protein expression, perhaps to ensure that it is minimized in nongametophyte tissues while the overlapping At2g19340 transcription unit remains transcriptionally active. Although studies of closely associated gene pairs have suggested that overlapping pairs often are coexpressed (Zhan et al., 2006), that observation, based on large-scale transcriptomics, may not fully take into account all forms of posttranscriptional regulation, including subtly different splice isoforms of the type we describe here. It would be interesting to investigate whether unproductive splicing of transcripts from overlapping loci is a widespread means of differential gene regulation in eukaryotes.
PIRL6 Knockdown
RNAi knockdown provided strong reverse genetic evidence of PIRL6’s importance in gametophytes. Both male and female gametophytes were affected following the introduction of the inverted repeat constructs. Multiple lines of evidence support a causal relationship between the presence of the PIRL6-KD constructs and the observed gametophyte defects. 35S-driven inverted repeat constructs similar to those used in this study have proven effective in both male and female gametophytes in Arabidopsis (León et al., 2007). RT-qPCR confirmed the reduced PIRL6 transcript levels in plants homozygous for the inverted repeat construct (Fig. 5). Those transcript reduction levels were consistent with the RNAi knockdown efficiencies observed in previous studies for many Arabidopsis genes (Kerschen et al., 2004), and they generally correlated to PIRL6-KD phenotype severity in the plants we tested. Genetic experiments showed that both male and female defects were heritable and persisted through backcrossing (Supplemental Fig. S2). We did not observe any separation of the pollen and ovule defects after backcrossing, and there was a consistent correlation between the severity of male and female defects within each line (i.e. lines with high frequencies of ovule lethality also tended to exhibit high pollen lethality). Finally, the knockdown construct resulted in reduced transmission of glufosinate resistance, demonstrating the association between PIRL6-KD and gametophyte failure.
It is unlikely that the observed consistent gametophyte dysfunction resulted from nonspecific or transformation-related effects. The inverted repeat constructs used in these experiments were designed to maximize specificity in two ways: by incorporating a region of PIRL6 that lacks extended sequence homology with other PIRLs and by targeting the 5′ end of the mRNA, where the RNAi pathway is less tolerant of mismatches between small interfering RNAs and target sequence (Wei et al., 2012). More importantly, knockout mutations in PIRL7 and PIRL8, the two genes most closely related to PIRL6, have been identified (Forsthoefel et al., 2005; Chen et al., 2010b), and they do not result in reproductive defects resembling those reported here for PIRL6. It is also extremely unlikely that the PIRL6-KD phenotype resulted indirectly from T-DNA transformation. While one study reported increased rates of nonspecific pollen lethality following transformation with inverted repeat constructs (Xing and Zachgo, 2007), that phenomenon did not resemble what we describe here, for the following reasons: (1) we observed pollen lethality in an overwhelming majority (38 of 44) of T1 plants; (2) the PIRL6-KD pollen defects were accompanied consistently by arrested megagametophyte development, not just pollen lethality; and (3) the phenotype we report was heritable through multiple generations and after backcrosses. T-DNA transformation also has been associated with chromosomal translocations, which can cause gametophyte lethality (Tax and Vernon, 2001; Clark and Krysan, 2010). However, translocation-induced lethality differs in two major respects from the PIRL6-KD phenotype reported in this study: it does not occur in a large majority of transgenic lines, and it tends to be fully penetrant in pollen (Clark and Krysan, 2010).
Gametophyte Function of PIRL6
Gametophyte development is dependent on the sporophyte and can be affected by sporophytic gene activity in premeiotic cells or in tissues neighboring the gametophytes (Olmedo-Monfil et al., 2010; Bencivenga et al., 2011; Forsthoefel and Vernon, 2011; Carter et al., 2016). However, both the expression and the segregation characteristics of PIRL6 fit the profile for a gametophyte gene. The PIRL6-KD phenotype exhibited meiotic segregation in pollen (Fig. 4E) and resulted in reduced female gametophyte transmission of the knockdown construct. Furthermore, as discussed above, translatable PIRL6 transcripts were not detected in the spl1 inflorescences, which contain the full suite of sporophytic flower tissues (Fig. 1D). Consistent with these results, PIRL6 expression has not been documented in published transcriptomes from male or female meiocytes (Chen et al., 2010a; Libeau et al., 2011; Yang et al., 2011; Zhao et al., 2014).
Gametophyte development involves male- and female-specific events as well as processes common to both (McCormick, 2004; Yadegari and Drews, 2004; Palanivelu and Johnson, 2010; Berger and Twell, 2011; Drews et al., 2011). Many male- or female-specific gametophyte functions are fertilization related and involve distinct late-gametophyte phases of gene expression (Wang et al., 2008; Qin et al., 2009; Palanivelu and Johnson, 2010). In contrast, PIRL6 functions in a process common to both gametophytes, and it acts early. In pollen, knockdown resulted in arrested gametogenesis, in some cases as early as the microspore stage, or it led to irregular configuration of the male germ unit (Fig. 6). In the embryo sacs, knockdown appeared to stall development prior to mitosis (Fig. 7). Our developmental analysis was carried out using knockdown lines with a highly penetrant phenotype. We cannot rule out that some PIRL6-KD embryo sacs can progress further, given the observed variable severity of phenotypes and variation in knockdown efficacy in these transgenic lines. Nevertheless, the frequent premitotic arrest observed in both male and female gametophytes indicates that PIRL6 is important very early in the haploid phase.
Taking these aspects of the knockdown phenotype into account along with the PIRL6 expression results, we propose that PIRL6 contributes to a pathway or process critical for progression through mitotic stages and differentiation in both male and female gametophytes. Its mode of posttranscriptional negative regulation suggests that its expression may need to be tightly restricted, a feature that is consistent with a regulatory function. PIRL6’s relationship to animal Ras-group LRRs, and the presence of a conserved phosphorylation site in PIRL9 (Chao et al., 2016), suggest that PIRLs also may have a role in cell signaling. However, the distinct N- and C-terminal regions outside of the Ras-group LRR domain may reflect a distinct role for this protein family in plants. Plant gametogenesis, the context in which at least four PIRLs operate, has no parallel in animals and involves novel cellular events. Processes critical for both male and female gametogenesis include mitosis, its initiation or regulation, cell positioning, differentiation, and gene regulation required for the proper timing and spatial coordination of such processes. The regulation of fundamental cell processes such as endomembrane and microtubules also is required early in both gametophytes (Oh et al., 2016; D’Ippólito et al., 2017).
Obtaining a full integrative functional understanding of truly novel genes is an incremental process. It is also important, given the enormous number of yet-uncharacterized genes in annotated plant genomes. Here, we have defined PIRL6 as an essential gene whose expression is restricted chiefly to gametophytes by alternative splicing. More detailed examination of developmental phenotypes at the cellular level, and the identification of PIRL protein interaction partners, will be essential for the next level of functional analysis of PIRL6 and other members of this intriguing gene family.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) plants were grown on soil from seed under 16-h-light/8-h-dark conditions as described by Tax and Vernon (2001) or were plated on media (Forsthoefel et al., 2010) as described below for specific experiments. Seeds for the spl-1, upf3-1, upf3-2, and qrt1 mutant lines (Yang et al., 1999; Johnson-Brousseau and McCormick, 2004; Arciga-Reyes et al., 2006) were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (stocks CS6586, CS9900, and CS8050, respectively). For spl and upf3, homozygotes were identified by their characteristic abnormal seedling phenotypes and used for RT-PCR analyses as described below.
Constructs and Transgenic Plants
To make the PIRL6-GFP fusion construct, a genomic PCR fragment containing the entire PIRL6 transcription unit and approximately 1.1 kb of adjacent upstream sequence was amplified with the following two primers: PIRL6-GFP-forward (5′-CGGGATCCTGGATGTTCTTTTCCAGTTCAG-3′, containing a BamHI site) and PIRL6-GFP-reverse (5′-AACTGCAGAGGTTCTTGGAGAAAAGAGACG-3′, containing a PstI site) and cloned into plasmid pORE R3 (Coutu et al., 2007), obtained from the ABRC.
Two knockdown constructs were generated using the pFGC5941 RNAi vector (GenBank accession no. AY310901) obtained from the ABRC (stock no. CD3-447). One, the PIRL6-KD(S) construct, contained inverted repeats of the 5′-most 139 bp of the PIRL6-coding region. This region shares only 66% sequence identity with the closest Arabidopsis gene (PIRL7), with the longest stretch of identical sequence only nine nucleotides long. This was amplified by PCR using the following primers: PIRL6-forward (5′-GCTCTAGAGGCGCGCCATGATATGCGAGGAGGCATAT-3′, containing restriction sites for XbaI and AscI) and PIRL6-reverse (5′-CGGGATCCATTTAAATATGGAGACGATGGTGATGATG-3′, containing restriction sites for BamHI and SwaI). The second, the PIRL6-KD(L) construct, contained inverted repeats of the 5′-most 458 bp of the PIRL6-coding region amplified by PCR using the same forward primer as above with the PIRL6-reverse-long primer (5′-CGGGATCCATTTAAATCTGCAATGTTGGATAGATTTGG-3′, containing restriction sites BamHI and SwaI). Inverted repeats of each of these fragments were generated by ligation into the pFGC5941 RNAi vector, first in the sense orientation after digesting with AscI and SwaI and then in the antisense orientation after digestion with BamHI and XbaI. The resulting plasmids were referred to as pFGC5941-PIRL6S and pFGC5941-PIRL6L. To ensure broad expression, constructs were under the control of the 35S promoter provided in pFGC5941, which has been used successfully to drive a similar inverted repeat construct in developing male and female gametophytes (León et al., 2007).
Transgenic lines for RNAi and GFP analysis were produced using Agrobacterium tumefaciens LBA4404-mediated germline transformation of Arabidopsis accession WS using the floral dip method (Clough and Bent, 1998). PIRL6-KD constructs also were introduced into Ler, including plants that also contained the qrt1-1 mutation (Johnson-Brousseau and McCormick, 2004). Seeds were collected from A. tumefaciens-treated plants and screened for vector-encoded marker phenotypes. Transgenic plants transformed with pFGC5941 PIRL6-KD constructs were identified by germination in soil and successive sprayings with glufosinate (Finale; Farnam). Plants with PIRL6-GFP in pORE R3 were identified by germination on medium containing 50 µg mL−1 kanamycin (Forsthoefel et al., 2010).
Transgene presence in all lines in this study was verified by genomic PCR with excised leaf tissue, using construct-specific primer combinations as described previously (Tax and Vernon, 2001). PIRL6-KD transgenes were confirmed in glufosinate-resistant seedlings using primers from the 35S promoter (5′-TTCGCAAGACCCTTCCTCTA-3′) and the pFGC5941 inverted repeat spacer region (5′-CTAGCTCGCTGGGAAACATC-3′). The presence of the PIRL6-GFP construct in kanamycin-resistant plants was confirmed with a PIRL6-specific forward primer (5′-TAGTGAGAAACAATCCTTTTGATTTGGTCA-3′) and a GFP-specific reverse primer (5′-TGCCCATTAACATCACCATC-3′).
To assess transgene copy numbers and construct segregation, progeny from selfed T1 plants were subjected to selection by glufosinate (for RNAi lines) or kanamycin (for PIRL6-GFP transformants) to determine the ratios of resistant to sensitive T2 progeny. Marker segregation was used to identify homozygotes and hemizygotes used for genetic, RT-qPCR, or microscopy experiments as described in “Results.”
Microscopy and Phenotype Analysis
For the analysis of RNAi-induced phenotypes, pollen collection, viability staining, and DAPI staining were carried out as described by Forsthoefel et al. (2010) and Forsthoefel and Vernon (2011). Viability staining and light microscopy were used to score aborted or abnormal pollen in WS transformants; for scoring, pollen were observed using bright-field microscopy with an Olympus BX60 microscope (20× and 40× objectives) and imaged using an Olympus E-510 camera.
Ler transformants were scored by DAPI staining and confocal microscopy as described below. Ovules were scored in excised siliques from self-fertilized plants using a Wild-Heerbrugg M8 dissection microscope, and images were captured on a Nikon E4500 digital camera. Developing embryo sacs were observed by dissecting developing ovaries, clearing ovules without fixation at 4°C for 0.5 to 22 h in Hoyer’s solution (Vernon and Meinke, 1994), and viewing as whole mounts using 20×, 40×, or 100× (oil immersion) objectives on an Olympus BX60 microscope equipped with DIC optics. Ovules were imaged using Olympus E-510 or Canon T5i/EOS700D digital cameras.
For GFP analysis, pollen or ovule samples were viewed in 0.1 m sodium phosphate buffer (pH 7) with 1 mm EDTA and 0.1% Triton X-100. Confocal imaging was performed with an SP5 II (Leica Microsystems) confocal laser scanning microscope, with 40× objective lens (HCX PL APO 0.85) for ovules or 63× oil-immersion lens (HCX PL APO CS 1.4) for ovules and pollen, using bright-field, DIC, or fluorescence microscopy as indicated in “Results.” GFP was viewed using a 488-nm line of an argon ion laser with emission spectra collected between 505 and 530 for pollen samples and between 505 and 599 for ovule samples to maximize signal. DAPI fluorescence was collected between 433 and 463 nm using a 405-nm laser. Images were collected and analyzed using Metamorph software (version 7.7.3.0; Molecular Devices).
RNA Expression
RNA was extracted from 50 to 100 mg of selected Arabidopsis tissues ground in liquid nitrogen using a Norgen Plant/Fungi Total RNA Purification Kit with RNase-free DNaseI. Immediately following RNA extraction, isolates were used for polyadenylated RNA purification (Takara-Clontech NucleoTrap mRNA Kit; Takara-Clontech) or cDNA synthesis (AffinityScript cDNA Synthesis Kit; Agilent Technologies), as indicated for experiments described below. Specificity for all RT-PCR and RT-qPCR primers was confirmed by BLASTN analysis against the Arabidopsis genome and, in some cases, by sequencing of gel-purified PCR products, as indicated below.
For full-length PIRL6 RT-PCR and analysis of alternative transcripts, polyadenylated RNA was isolated from wild-type leaf, root, flower, and siliques, transcribed from oligo(dT) primer, and subjected to PCR as described previously by Cushing et al. (2005), using primers designed to amplify the full PIRL6 translated region, which encompasses both intron splice sites: FP (5′-ATGCGAGGAGGCATATCATC-3′) and RP (5′-CGACGTGGAGAGAACATTCC-3′). To confirm RT-PCR product identity and purity, the RT-PCR product from flower tissue was gel purified and sequenced using FP and RP primers.
For RT-PCR and RT-qPCR specific to translatable spliced PIRL6 transcripts or for transcripts from the neighboring At3g19340 gene, total RNA was isolated from open flowers, developing flowers, leaves, and roots, reverse transcribed from oligo(dT) primers, and subjected to PCR as described by Cushing et al. (2005). For PIRL6 mRNA, a forward primer specific to the exon II-exon III splice junction was used to ensure that products were derived from spliced RNA: QFP (5′-CAACATTGCAGGTCATTGGA-3′) along with reverse primer QRP (5′-GTCAAGGACACGGAGAGATGT-3′). In the RT-qPCR performed to compare functional spliced PIRL6 levels in selected wild-type organs, TUB6 (At5g12250) and APT1 (At1g27450) served as normalizers: TUB6.QFP (5′-TGCAGATGACGAAGGCGAGTATGA-3′), TUB6.QRP (5′-CCGACCTCACTATTACACACAGACCA-3′), APT1.QFP (5′-GTTGCAGGTGTTGAAGCTAGAGGT-3′), and APT1.QRP (5′-TGGCACCAATAGCCAACGCAATAG-3′). Reactions were carried out on total RNA isolated from roots, leaves, or inflorescences pooled from three to six separate plants; the results were replicated in two experiments with different RNA preparations from two sets of plants. To examine At3g19340 expression, each primer pair included a reverse primer specific for the 3′ untranslated region of the target transcript: Alt FP (5′-AACCGGCTCCAGTGTATCAG-3′), Alt RP (5′-ATCTCTCCGTGTCCTTGACG-3′), and Alt2 RP (5′-GCTTCAAATTGGTGACTACGG-3′). For comparison of alternative PIRL6 transcript levels in upf and wild-type plants, we used the QRP primer (see above) and an intron II-specific forward primer (5′-TCACCCCATAACTTCCACAAA-3′) capable of amplifying six of the seven alternative PIRL6 transcripts, including the overwhelmingly predominant species, transcripts C and D (Fig. 4). A minimum of three reaction replicates were performed on each of two independent total leaf RNA isolates from upf3-1 and upf3-2 mutants and wild-type controls.
For RT-PCR comparison of transcripts in spl and wild-type plants, total RNA was collected from open and developing flowers from each genotype. The QFP/RP primer pair was used to specifically detect functional spliced transcript, and the full-length FP/RP primer pair was used to amplify all PIRL6 transcripts. ACTIN8 primers used for positive control reactions were: 5′-CCTTGCTGGTCGTGACCTTA-3′ (forward) and 5′-GTAAGAGCGAGAGCGGGTTT-3′ (reverse).
For the determination of knockdown efficacy in transgenic plants, total RNA was isolated from developing flowers from homozygous T3 plants from each of three independent transgenic lines (Fig. 5). To allow comparison of PIRL6 transcript levels with phenotype severity, RNA was isolated individually from two to three plants from each line, and the same individual plants were scored for the frequency of aborted ovules and pollen, as described above. A minimum of three replicate RT-qPCRs were carried out for each individual plant, using the primers specific for the functional spliced transcript described above.
All RT-qPCRs were run with Brilliant III SYBR Green Real-Time PCR Master Mix in an MX3000 QPCR System (Agilent Technologies) according to the manufacturer’s protocol. Primers were pretested for specificity and efficiency determined by reactions on 4× serial dilutions of target cDNAs. The 2−∆∆CT method was used to calculate the relative expression level of each gene using the MX3000 QPCR System software; values were later transferred to Microsoft Excel to create figures.
Cloning and Sequencing of PIRL6 Transcripts
To characterize the alternative PIRL6 RT-PCR products, cDNA was generated from leaf and root poly(A+) RNA. Synthesis was primed with oligo(dT) primer, and products were amplified with the full-length FP and RP PIRL6 primers described above for RT-PCR or with FP and an alternative reverse primer specific to intron 2, specific for alternative transcripts (5′-GGGAGTGTTGTCTTGTGAGAGC-3′). For the analysis of individual RT-PCR products, the resulting cDNAs from the 1.1- to 1.4-kb size range were resolved, gel purified on a 1.5% agarose gel, ligated into pPCR-Script AmpSK+ Vector (Stratagene), and cloned in Escherichia coli. Seventeen independent cDNA clones from this size range were sequenced using the FP and RP primers plus an additional primer specific to the exon 3 minus strand (5′-TCAAAACCAATGGAGTCTGG-3′), used to ensure sequence accuracy in the alternatively spliced regions. All sequencing was performed by the University of Arizona Genetics Core Facility in Tucson. Translation predictions and PTC identification were performed using the MacVector software (Accelrys).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY849576, MH618667, MH618668, MH618669, MH618670, MH618671, and MH618672.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transcripts from adjacent overlapping gene At2g19340 are widely expressed in plants.
Supplemental Figure S2. Heritability of PIRL6-KD developmental defects.
Acknowledgments
We thank Amelia Lampron-York for work with upf3, Trayvon Foy for assistance with ovule microscopy, and Dr. Arielle Cooley (Whitman College) and the Whitman Biology 342 class for helpful comments on the article.
Footnotes
This work was supported by the National Science Foundation (IOB-0616166 and MRI-1039958 to D.M.V.) and by Whitman College Louis B. Perry and S.A. Abshire awards.
Articles can be viewed without a subscription.
References
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B (2006) UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J 47: 480–489 [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S (2011) Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod 24: 113–121 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Berger F, Twell D (2011) Germline specification and function in plants. Annu Rev Plant Biol 62: 461–484 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Shuai B, Yu HJ, Pagnussat GC, Sundaresan V, McCormick S (2009) A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 181: 1369–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boi S, Solda G, Tenchini ML (2004) Shedding light on the dark side of the genome: overlapping genes in higher eukaryotes. Curr Genomics 5: 509–524 [Google Scholar]
- Bonhomme S, Horlow C, Vezon D, de Laissardière S, Guyon A, Férault M, Marchand M, Bechtold N, Pelletier G (1998) T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol Gen Genet 260: 444–452 [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Buchanan SG, Gay NJ (1996) Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol 65: 1–44 [DOI] [PubMed] [Google Scholar]
- Carter B, Henderson JT, Svedin E, Fiers M, McCarthy K, Smith A, Guo C, Bishop B, Zhang H, Riksen T, et al. (2016) Cross-talk between sporophyte and gametophyte generations is promoted by CHD3 chromatin remodelers in Arabidopsis thaliana. Genetics 203: 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Gao ZF, Wang YF, Li Z, Huang XH, Wang YC, Mei YC, Zhao BG, Li L, Jiang YB, et al. (2016) The proteome and phosphoproteome of maize pollen uncovers fertility candidate proteins. Plant Mol Biol 91: 287–304 [DOI] [PubMed] [Google Scholar]
- Chen C, Farmer AD, Langley RJ, Mudge J, Crow JA, May GD, Huntley J, Smith AG, Retzel EF (2010a) Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol 10: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Nayak N, Majee SM, Lowenson J, Schäfermeyer KR, Eliopoulos AC, Lloyd TD, Dinkins R, Perry SE, Forsthoefel NR, et al. (2010b) Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase 1 identified using phage display and biopanning. J Biol Chem 285: 37281–37292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, Subramanian S, Drews GN (1998) Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev Biol 202: 136–151 [DOI] [PubMed] [Google Scholar]
- Clark KA, Krysan PJ (2010) Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J 64: 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudianos C, Campbell HD (1995) The novel flightless-I gene brings together two gene families, actin-binding proteins related to gelsolin and leucine-rich-repeat proteins involved in Ras signal transduction. Mol Biol Evol 12: 405–414 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781 [DOI] [PubMed] [Google Scholar]
- Cushing DA, Forsthoefel NR, Gestaut DR, Vernon DM (2005) Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221: 424–436 [DOI] [PubMed] [Google Scholar]
- Cutler ML, Bassin RH, Zanoni L, Talbot N (1992) Isolation of rsp-1, a novel cDNA capable of suppressing v-Ras transformation. Mol Cell Biol 12: 3750–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ippólito S, Arias LA, Casalongué CA, Pagnussat GC, Fiol DF (2017) The DC1-domain protein VACUOLELESS GAMETOPHYTES is essential for development of female and male gametophytes in Arabidopsis. Plant J 90: 261–275 [DOI] [PubMed] [Google Scholar]
- Davy DA, Campbell HD, Fountain S, de Jong D, Crouch MF (2001) The flightless I protein colocalizes with actin- and microtubule-based structures in motile Swiss 3T3 fibroblasts: evidence for the involvement of PI 3-kinase and Ras-related small GTPases. J Cell Sci 114: 549–562 [DOI] [PubMed] [Google Scholar]
- Dougherty GW, Jose C, Gimona M, Cutler ML (2008) The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur J Cell Biol 87: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Koltunow AM (2011) The female gametophyte. The Arabidopsis Book 9: e0155, doi/10.1199/tab.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Wang D, Steffen JG, Schumaker KS, Yadegari R (2011) Identification of genes expressed in the angiosperm female gametophyte. J Exp Bot 62: 1593–1599 [DOI] [PubMed] [Google Scholar]
- Feldmann KA, Coury DA, Christianson ML (1997) Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147: 1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel NR, Vernon DM (2011) Effect of sporophytic PIRL9 genotype on post-meiotic expression of the Arabidopsis pirl1;pirl9 mutant pollen phenotype. Planta 233: 423–431 [DOI] [PubMed] [Google Scholar]
- Forsthoefel NR, Cutler K, Port MD, Yamamoto T, Vernon DM (2005) PIRLs: a novel class of plant intracellular leucine-rich repeat proteins. Plant Cell Physiol 46: 913–922 [DOI] [PubMed] [Google Scholar]
- Forsthoefel NR, Dao TP, Vernon DM (2010) PIRL1 and PIRL9, encoding members of a novel plant-specific family of leucine-rich repeat proteins, are essential for differentiation of microspores into pollen. Planta 232: 1101–1114 [DOI] [PubMed] [Google Scholar]
- Forsthoefel NR, Klag KA, Simeles BP, Reiter R, Brougham L, Vernon DM (2013) The Arabidopsis Plant Intracellular Ras-group LRR (PIRL) family and the value of reverse genetic analysis for identifying genes that function in gametophyte development. Plants (Basel) 2: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Nieves R, Desantis AI, Cutler ML (2013) Rsu1 contributes to regulation of cell adhesion and spreading by PINCH1-dependent and -independent mechanisms. J Cell Commun Signal 7: 279–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henz SR, Cumbie JS, Kasschau KD, Lohmann JU, Carrington JC, Weigel D, Schmid M (2007) Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol 144: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, Twell D (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram GC. (2017) Dying to live: cell elimination as a developmental strategy in angiosperm seeds. J Exp Bot 68: 785–796 [DOI] [PubMed] [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KW, Lee YH, Stallcup MR (2009) Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J Biol Chem 284: 29298–29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Kajava AV. (1998) Structural diversity of leucine-rich repeat proteins. J Mol Biol 277: 519–527 [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Müller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566: 223–228 [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11: 725–732 [DOI] [PubMed] [Google Scholar]
- Lee YH, Campbell HD, Stallcup MR (2004) Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol 24: 2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA 100: 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Han M, Guan KL (2000) The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev 14: 895–900 [PMC free article] [PubMed] [Google Scholar]
- Libeau P, Durandet M, Granier F, Marquis C, Berthomé R, Renou JP, Taconnat-Soubirou L, Horlow C (2011) Gene expression profiling of Arabidopsis meiocytes. Plant Biol (Stuttg) 13: 784–793 [DOI] [PubMed] [Google Scholar]
- Lloyd J, Meinke D (2012) A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol 158: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine AE, McCormick S, Estrada A, Patel K, Qin P (2013) RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol 162: 1092–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bennett MJ (1998) The Arabidopsis AUX1 gene: a model system to study mRNA processing in plants. Plant Mol Biol 36: 463–471 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell (Suppl) 16: S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K, Chandler V, Cone K, Kaeppler H, Kaeppler S, Kerschen A, Pikaard C, Richards E, Sidorenko L, Smith T, et al. (2005) Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol 392: 1–24 [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK (2007) Integrating signals from RTKs to ERK/MAPK. Oncogene 26: 3113–3121 [DOI] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC (2006) Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res 34: D731–D735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Ecker JR (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Oh SA, Jeon J, Park HJ, Grini PE, Twell D, Park SK (2016) Analysis of gemini pollen 3 mutant suggests a broad function of AUGMIN in microtubule organization during sexual reproduction in Arabidopsis. Plant J 87: 188–201 [DOI] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Johnson MA (2010) Functional genomics of pollen tube-pistil interactions in Arabidopsis. Biochem Soc Trans 38: 593–597 [DOI] [PubMed] [Google Scholar]
- Palusa SG, Reddy AS (2010) Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol 185: 83–89 [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS, Göhring J, Barta A (2012) Localization and dynamics of nuclear speckles in plants. Plant Physiol 158: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MW, Schmidt A, Klostermeier UC, Barann M, Rosenstiel P, Grossniklaus U (2012) A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS ONE 7: e29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth DS, Sun Q, Han M (1998) SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell 94: 119–130 [DOI] [PubMed] [Google Scholar]
- Tax FE, Vernon DM (2001) T-DNA-associated duplication/translocations in Arabidopsis: implications for mutant analysis and functional genomics. Plant Physiol 126: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW (1994) Embryogenic transformation of the suspensor in twin, a polyembryonic mutant of Arabidopsis. Dev Biol 165: 566–573 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Zhang L, Huang H, Chen Y, Zheng J, Zhou X, Yi F, Du Q, Liang ZC (2012) siRNA has greatly elevated mismatch tolerance at 3 '-UTR sites. PLoS ONE 7: e49309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S (2007) Pollen lethality: a phenomenon in Arabidopsis RNA interference plants. Plant Physiol 145: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell (Suppl) 16: S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Lu P, Wang Y, Ma H (2011) The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: the complexity and evolution of the meiotic process. Plant J 65: 503–516 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Dai X, Li X, Wang L, Chen G, Xiao J, Wu C (2010) Molecular characterization, expression pattern, and functional analysis of the OsIRL gene family encoding intracellular Ras-group-related LRR proteins in rice. Plant Mol Biol 74: 617–629 [DOI] [PubMed] [Google Scholar]
- Young LC, Hartig N, Muñoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, Vijayakumar V, Vietri Rudan M, Gewinner C, Henderson S, et al. (2013) An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol Cell 52: 679–692 [DOI] [PubMed] [Google Scholar]
- Zhan S, Lukens L (2013) Protein-coding cis-natural antisense transcripts have high and broad expression in Arabidopsis. Plant Physiol 161: 2171–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Horrocks J, Lukens LN (2006) Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J 45: 347–357 [DOI] [PubMed] [Google Scholar]
- Zhao L, He J, Cai H, Lin H, Li Y, Liu R, Yang Z, Qin Y (2014) Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J 80: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]