Improvements to amiRNA technology, including amiRNA design, transgenic screening for maximal gene-silencing mutants, and multiplex performance, facilitate its applications in plants.
Abstract
Artificial microRNA (amiRNA) technology offers reversible and flexible gene inactivation and complements genome-editing technologies. However, obtaining transgenic plants with maximal gene silencing remains a major technical challenge in current amiRNA applications. Here, we incorporated an empirically determined feature of effective amiRNAs to the amiRNA design and in silico generated a database containing 533,429 gene-specific amiRNAs for silencing 27,136 genes in Arabidopsis (Arabidopsis thaliana), with a genome coverage of 98.87%. In both single-gene and multiple-gene silencing, we observed an overall improvement in performance by amiRNAs designed using our strategy in Arabidopsis protoplasts and transgenic plants. In addition, the endogenous tRNA-processing system was used to generate multiple amiRNAs from tRNA-pre-amiRNA tandem repeats for multiplex gene silencing. An intronic amiRNA-producing fluorescent reporter was explored as a visual screening strategy for transgenic Arabidopsis and rice (Oryza sativa) plants with maximal whole-plant or cell type-specific gene silencing. These improvements enable the amiRNA technology to be a functional gene knockout tool for basic and applied plant research.
Genome sequences across the plant kingdom are accumulating at a staggering rate due to the advent of whole-genome sequencing technologies, creating the need for flexible and versatile genetic tools to generate loss-of-function mutants to study gene functions. In plants, traditional genetic tools such as T-DNA insertion and ion/chemical-induced mutagenesis, which often cause random gene inactivation, are being replaced by the CRISPR-Cas technology, which can confer sequence-specific gene knockout (Zhang et al., 2016a). However, the CRISPR-Cas technology also has limitations. First, it is not able to generate homozygous mutants for essential genes whose knockout gives rise to plant lethality. Second, it cannot create homozygous mutants with cell-, tissue-, or developmental stage-specific gene knockouts. Third, numerous plant genes undergo alternative splicing (AS; e.g. 61% of multiexonic genes in Arabidopsis [Arabidopsis thaliana]; Marquez et al., 2012). However, CRISPR-Cas technology cannot be used to inactivate one AS isoform specifically without affecting others. Finally, the CRISPR-Cas system may be inefficient in editing genes located within the heterochromatin (Jensen et al., 2017) or chromosomal regions with high nucleosome occupancy (Horlbeck et al., 2016).
Gene-silencing technologies represent an invaluable complement to genome-editing tools, providing flexibility and reversibility in gene inactivation by acting on the target mRNA instead of genomic DNA (Zhang, 2014; Teotia et al., 2016). Gene silencing can be controlled tightly by a chemically inducible promoter to examine gene functions associated with lethal mutants or by a cell type- or developmental stage-specific promoter to study gene functions with spatial or temporal resolution. Currently, the hairpin RNA (hpRNA)-induced RNA interference (RNAi) is the predominant approach for gene silencing. However, because the generation and action of a multitude of hpRNA-derived small interfering RNAs (siRNAs) are not fully predictable, off-target effects remain a serious concern in this approach (Xu et al., 2006). By contrast, artificial microRNA (amiRNA)-mediated RNAi produces a single 21-nucleotide amiRNA (analogous to a single siRNA) that only recognizes a target sequence with less than five mismatches (Schwab et al., 2006; Ossowski et al., 2008). This feature not only ensures a higher silencing specificity for amiRNAs than hpRNAs but also offers unique advantages. An amiRNA can be designed to silence multiple genes sharing a short conserved sequence simultaneously, to silence individual AS isoforms, or to silence an endogenous small noncoding RNA (e.g. miRNA). amiRNA technology has been applied successfully to engineer a wide range of crop species to obtain desirable agronomic traits (Butardo et al., 2011; Toppino et al., 2011; Chi et al., 2014) or enhanced resistance to viruses (Niu et al., 2006; Duan et al., 2008; Kis et al., 2016), fungi (Wang et al., 2016; Zhang et al., 2016b), nematodes (Tian et al., 2016), and insects (Guo et al., 2014). Notably, the amiRNA-mediated viral resistance remains effective even at low temperatures (e.g. 15°C) that inhibit siRNA-mediated gene silencing (Niu et al., 2006; Kis et al., 2016). Therefore, amiRNA technology appears to be an attractive option for gene silencing in terms of specificity and versatility.
Different amiRNA candidates targeting the same gene often exhibit variable silencing efficiencies (Schwab et al., 2006; Duan et al., 2008; Li et al., 2013a). We previously developed a protoplast-based amiRNA screen, named the ETPamir assay, to identify the most efficient amiRNA for silencing a single gene or multiple homologous genes before conducting the time- and labor-consuming transgenic work (Li et al., 2013a). However, when gene silencing generates no visible growth phenotype, screening transgenic plants for optimal gene silencing could be complicated by unpredictable amiRNA action mechanisms (i.e. mRNA degradation or translational inhibition) and transgenic variations in amiRNA expression levels. Moreover, in the case of cell type (e.g. guard cell)-specific gene silencing, it will be challenging to identify transgenic plants with optimal silencing because screening techniques (e.g. reverse transcription [RT]-PCR) are applied to the whole plant. In this study, we explored a solution to tackle this issue by embedding a potent amiRNA into a portable intron within a fluorescent reporter (e.g. GFP). As the fluorescent reporter and amiRNA are coproduced from the same transcript, the fluorescent reporter can serve as a visible surrogate for amiRNA performance in planta. Indeed, transgenic plants selected by fluorescence consistently showed maximal silencing of target genes. In addition, we evaluated an alternative strategy to produce multiple amiRNAs for multiplex gene silencing by leveraging the endogenous tRNA-processing system.
RESULTS
An Alternative Strategy for amiRNA Design by Incorporating an Empirically Determined Feature of Potent amiRNAs
Different computer-designed amiRNAs can silence the same gene with variable efficiencies, and our previous evaluation of 63 amiRNAs designed by WMD3-AmiRNA Designer (wmd3.weigelworld.org; Schwab et al., 2006) for silencing 16 Arabidopsis genes revealed that perfect complementarity is a key common feature of active amiRNAs (Li et al., 2013a). Based on this observation, we proposed a new way for amiRNA design by selecting 21-nucleotide reverse complements from the target gene coding sequence (CDS) as amiRNA candidates and subsequently filtering out those nonspecific ones by the Target Search function at the WMD3 Web site.
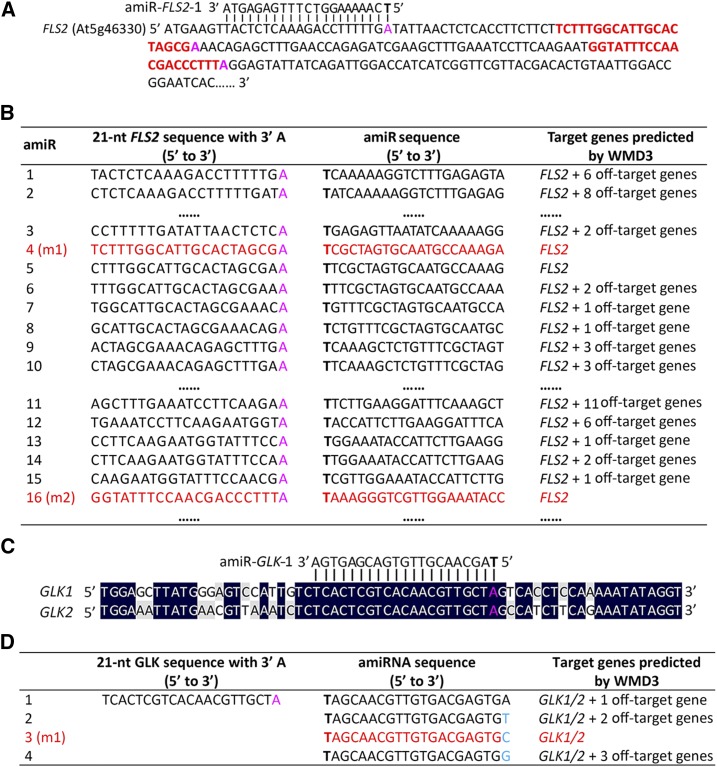
For single-gene silencing, we present an example of amiRNA design for silencing Arabidopsis FLS2 (Fig. 1). Because amiRNAs all possess a 5′ T at position 1 to satisfy the requirement of thermodynamic instability, we selected 21-nucleotide CDSs ending with A as target sequences (Fig. 1A), so that the thermoinstability requirement of amiRNAs and a perfect complementarity between the resulting amiRNAs and the target sequences can be satisfied simultaneously. We utilized the WMD3-Target Search to evaluate the specificity of these amiRNAs, because our previous work has experimentally validated its robustness among seven computational algorithms for predicting the most complete list of endogenous target genes for a given miRNA (Li et al., 2013a). The mismatch tolerance of Target Search was set to five nucleotides to capture all possible target genes for the query amiRNA, because neither endogenous miRNAs nor amiRNAs can silence a gene with more than five mismatches (Llave et al., 2002; Schwab et al., 2005, 2006). Only when the WMD3-Target Search reported the intended gene as a specific hit without other off targets was the query amiRNA considered a valid candidate. The vast majority of amiRNAs designed in this way, except amiR-FLS2 candidates 4, 5, and 16, were filtered out due to off-target possibilities (Fig. 1B). We prefer to design several amiRNA candidates with distinct target sites within the target gene to avoid the ineffectiveness caused by target site inaccessibility (Li et al., 2013a). Therefore, if two target-specific amiRNA candidates (e.g. amiR-FLS2 candidates 4 and 5; Fig. 1B) share overlapping target sequences, we select only one of them for further experimental screening. Notably, since a single mismatch at position 21 of an amiRNA has little negative effect on gene silencing (Li et al., 2013a), one may substitute the last nucleotide of a nonspecific amiRNA with any nucleotide, which likely changes its target profile and creates an amiRNA derivative with the intended specificity. To examine the genome-wide applicability of this amiRNA design strategy, we in silico scanned the CDSs of 27,445 Arabidopsis nuclear genes annotated by Araport11 (www.araport.org) to identify those gene-specific amiRNA candidates. These efforts led to a total of 533,429 amiRNA candidates for silencing 27,136 Arabidopsis genes (Supplemental Data Set S1), with a genome coverage of 98.87%. These results confirmed that our design approach is highly applicable to the Arabidopsis genome.
Figure 1.
amiRNA design for silencing Arabidopsis FLS2 or GLK1/2. A, Design of amiRNA candidates for FLS2. Reverse complements of 21-nucleotide (nt) target sequences with 3′ A (magenta) from the CDS of FLS2 were designed sequentially as amiRNA candidates. The first candidate, amiR-1, is shown as an example. Two FLS2-specific target sequences (targeted by amiR-4 and amiR-16 in B, respectively) are highlighted in red. B, Specificity inspection by WMD3-Target Search identifies FLS2-specific amiRNA candidates. amiR-4 and amiR-16 (red) were identified by WMD3-Target Search as FLS2-specific amiRNAs and were renamed amiR-FLS2-m1 and amiR-FLS2-m2 for the ETPamir assay. Note that amiR-5, despite being an FLS2-specific amiRNA candidate, was abandoned because it shares overlapping target sequences with amiR-4. C, Design of a single amiRNA to target the most conserved sequences between GLK1 and GLK2. D, Specificity inspection by WMD3-Target Search identifies GLK1/2-specific amiRNA candidates. amiR-3 (red) was identified by WMD3 as a GLK1/2-specific amiRNA and was renamed amiR-GLK-m1 for the ETPamir assay. All amiRNAs are initiated by T (boldface) to satisfy the thermoinstability requirement.
Gene function studies often require the silencing of more than one gene to obtain visible phenotypes. For multigene silencing, a sequence alignment between the intended target genes could identify the most conserved region, where the amiRNA candidates can be designed to target. For example, we designed amiRNAs to silence two homologous Arabidopsis genes, namely GLK1/2 (Fig. 1, C and D). The reverse complement of a 21-nucleotide target sequence ending with A within the conserved region was selected as a potentially effective amiRNA to silence both target genes (Fig. 1, C and D; Supplemental Fig. S1). However, very frequently, the WMD3-assisted specificity inspection would predict such a designed amiRNA to be nonspecific. If this was the case, one also could change the last nucleotide of the amiRNA candidate, and it is likely that one of the derived amiRNAs may become specific. The amiR-GLK1/2 candidates 1 to 4 (Fig. 1D) belong to this category. Only candidate 3 was predicted to be specific to GLK1/2, whereas the other candidates all have off-target genes. This design strategy can ensure that the resulting amiRNA candidate contains either no mismatch or only a single, tolerable mismatch to the target genes.
amiRNAs Designed by the New Strategy Exhibit Better Performance Than Those Designed by WMD3-AmiRNA Designer
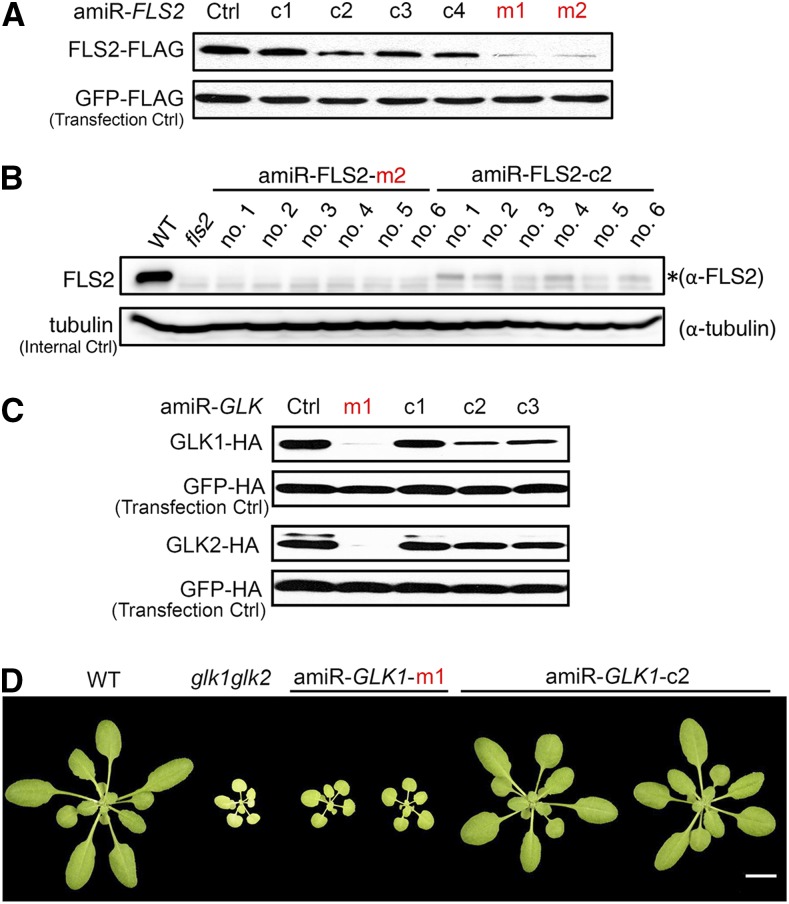
Next, we experimentally compared the efficiencies of amiRNA candidates designed by the new strategy or by the WMD3-AmiRNA Designer for silencing Arabidopsis FLS2 or GLK1/2. For silencing FLS2, we selected four top-ranking amiRNA candidates designed by WMD3-AmiRNA Designer (i.e. amiR-FLS2-c1 to amiR-FLS2-c4) in addition to the two amiRNA candidates (i.e. amiR-FLS2-m1 and amiR-FLS2-m2; Fig. 1B) that we designed earlier. Interestingly, amiR-FLS2-m1 and amiR-FLS2-m2 were not found in the candidate list generated by the AmiRNA Designer function of WMD3 (Schwab et al., 2006) or P-SAMS (Fahlgren et al., 2016). We first compared the activities of these amiRNAs using the ETPamir assay. In this assay, a target gene encoding an epitope-tagged target protein is coexpressed with individual amiRNAs in plant protoplasts, and the activity of each amiRNA is inversely reflected by the accumulation of target proteins, which can be monitored by immunoblotting using anti-tag antibodies (Li et al., 2014). In ETPamir assays, amiR-FLS2-m1 and amiR-FLS2-m2 both were more active than the WMD3-designed candidates (Fig. 2A). We further compared the silencing efficiency of amiR-FLS2-m2 with that of amiR-FLS2-c2, the most effective amiRNA designed by WMD3, in transgenic plants. Using anti-FLS2 antibodies, we could barely detect any FLS2 protein in six transgenic silencing lines overexpressing amiR-FLS2-m2 (Fig. 2B), suggesting that amiR-FLS2-m2 is able to knock out FLS2 expression. By contrast, six silencing lines overexpressing amiR-FLS2-c2 showed only reduced FLS2 abundance (Fig. 2B), suggesting that this amiRNA could only knock down FLS2 expression.
Figure 2.
amiRNAs designed by the new strategy perform better than those designed by WMD3-AmiRNA Designer. A, amiRNAs designed by the new strategy (m1 and m2) are more effective in silencing Arabidopsis FLS2 than WMD3-designed top-ranking amiRNAs (c1–c4) in ETPamir assay. B, Comparison of the performance of amiR-FLS2-m2 and amiR-FLS2-c2 in transgenic plants. Note that amiR-FLS2-m2 could knock out FLS2 expression in multiple transgenic silencing lines, whereas amiR-FLS2-c2 only led to gene knockdown, as revealed by immunoblotting using anti-FLS2 antibodies. FLS2 is indicated by the asterisk. C, amiR-GLK-m1 is more effective in silencing Arabidopsis GLK1/2 than WMD3-designed amiRNAs (c1–c3) in ETPamir assay. D, Comparison of the performance of amiR-GLK-m1 and amiR-GLK-c2 in transgenic plants. Note that amiR-GLK-m1 could almost knock out GLK1/2 expression in transgenic silencing lines, whereas amiR-GLK-c2 only led to gene knockdown, as reflected by silencing phenotypes. Bar = 1 cm. In A and C, target genes encoding FLAG- or HA-tagged target proteins and the same tagged GFP constructs (transfection control) were coexpressed with the indicated amiRNAs in Arabidopsis protoplasts for 36 h. Three independent repeats were conducted with similar results. Optimal amiRNAs are highlighted in red. Ctrl, Control without amiRNA coexpression; fls2, FLS2 null mutant; glk1 glk2, GLK1/2 double mutant; WT, wild type.
For silencing GLK1/2, we tested all three amiRNA candidates designed by WMD3-AmiRNA Designer (i.e. amiR-GLK-c1 to amiR-GLK-c3) along with amiR-GLK-m1 that we designed using the new strategy (Fig. 1D). Similarly, amiR-GLK-m1 was not on the candidate list generated by WMD3 or P-SAMS algorithms. The ETPamir assay indicated that only amiR-GLK-m1 was able to silence both GLK1 and GLK2, whereas amiR-GLK-c2, the most effective amiRNA designed by WMD3, only down-regulated GLK1/2 expression (Fig. 2C). Consistently, transgenic Arabidopsis plants overexpressing amiR-GLK-m1 exhibited pale green and dwarf phenotypes, resembling the glk1 glk2 T-DNA null mutant (Fig. 2D). By contrast, plants overexpressing amiR-GLK-c2 phenotypically resembled the wild type, albeit slightly smaller (Fig. 2D).
To further validate that the amiRNAs designed by the new strategy tend to work more efficiently than those designed with existing strategies, we conducted the ETPamir assay to compare the efficiencies of amiRNAs designed by the two strategies for silencing other paralogous genes, such as APK2A/2B and SERK1/2/3/4/5. For silencing APK2A/2B, we designed one amiRNA candidate (i.e. amiR-APK2-m1) and selected four WMD3-designed top-ranking candidates (i.e. amiR-APK2-c1 to amiR-APK2-c4). Although amiR-APK2-m1 could maximally silence both APK2A and APK2B, amiR-APK2-c1 to amiR-APK2-c4 could only effectively silence either APK2A or APK2B (Supplemental Fig. S2A).
For silencing SERK1/2/3/4/5, we designed one amiRNA candidate (i.e. amiR-SERK-m1) and compared it with the only available amiRNA candidate designed by WMD3 (i.e. amiR-SERK-c1). Although amiR-SERK-m1 and amiR-SERK-c1 demonstrated comparable efficiencies in silencing SERK3 and SERK5, the former was more active in silencing SERK1, SERK2, and SERK4 than the latter (Supplemental Fig. S2B). These results suggested that the amiRNA candidates designed by the new strategy tend to be more effective than the WMD3-designed candidates, particularly for multiple-gene silencing.
amiRNAs Designed by the New Strategy Can Silence a Specific Alternatively Spliced Isoform
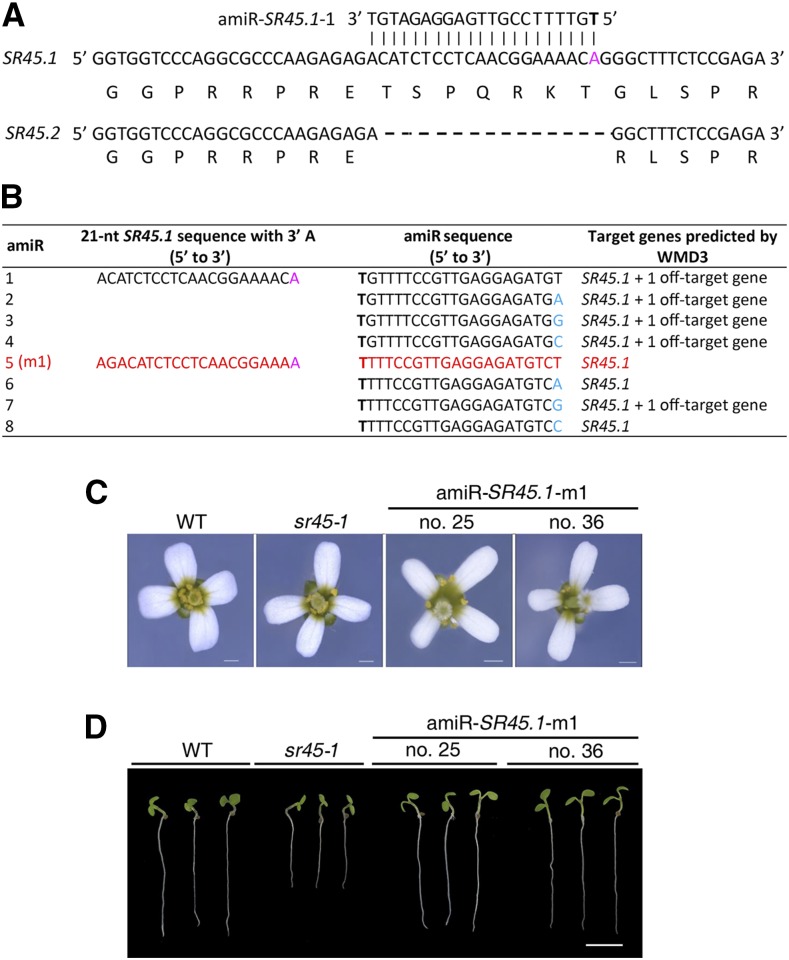
Since different AS isoforms are transcribed from the same genomic DNA, it is impossible to use the CRISPR-Cas technology to knock out a specific isoform without affecting others. However, amiRNA can be used to achieve this goal, since it functions posttranscriptionally. As a proof of principle, we designed an amiRNA to specifically target the AS isoform 1 of Arabidopsis SERINE-ARGININE-RICH45 (SR45.1). SR45.1 is distinguishable from a second isoform, namely SR45.2, by a 21-nucleotide AS fragment that confers seven additional amino acids to SR45.1 (Fig. 3A). These two isoforms have comparable expression levels in many tissues such as root and inflorescence. However, SR45.1 plays a crucial role in the normal development of flower petals but not roots, whereas SR45.2 plays an opposite role (Zhang and Mount, 2009). Using our design strategy, we obtained an amiRNA candidate, namely amiR-SR45.1-m1, to target the SR45.1-specific sequence (Fig. 3B). Transgenic Arabidopsis plants overexpressing amiR-SR45.1-m1 exhibited narrower flower petals compared with the wild type, resembling the flower phenotype of the sr45-1 T-DNA null mutant (Fig. 3C). However, seedlings overexpressing amiR-SR45.1-m1 exhibited root growth similar to the wild type, unlike the sr45-1 mutant seedlings, which showed delayed root growth (Fig. 3D), implying that this amiRNA could knock out SR45.1 specifically without affecting SR45.2.
Figure 3.
amiRNA designed by the new strategy for silencing a specific alternatively spliced transcript. A, Designing amiRNA for silencing Arabidopsis SR45.1 but not SR45.2 by targeting SR45.1-specific sequence. B, Specificity inspection by WMD3-Target Search identifies SR45.1-specific amiRNA candidates. amiR-5 (red) was identified by WMD3-Target Search as an SR45.1-specific amiRNA and was renamed amiR-SR45.1-m1 for transgenic expression. Note that amiR-6 and amiR-8, despite being designated as SR45.1-specific amiRNA candidates, were abandoned because they share overlapping target sequences with amiR-5. C, Transgenic SR45.1-silencing plants exhibit a similar flower petal phenotype to the sr45-1 null mutant. Bars = 0.5 mm. D, Transgenic SR45.1-silencing plants are different from the sr45-1 null mutant regarding the root growth phenotype. Bar = 5 mm. sr45-1,null mutant of both SR45.1 and SR45.2; WT, wild type.
Simultaneous Production of Multiple amiRNAs for Multiplex Gene Silencing Using the Endogenous tRNA-Processing System
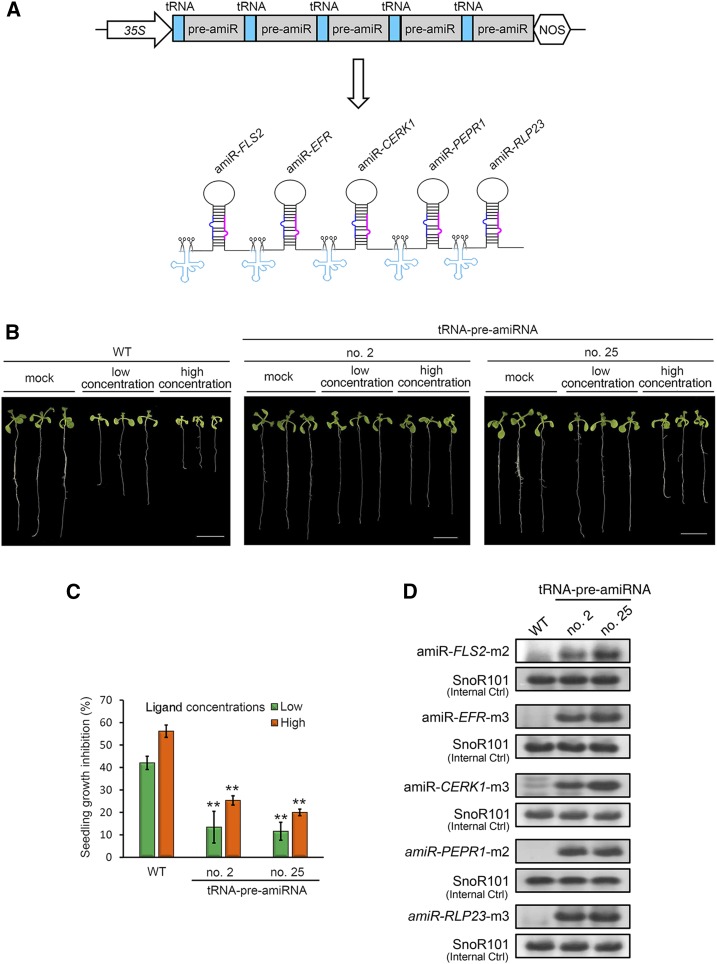
Several amiRNAs need to be coproduced to silence multiple, unrelated genes or homologous genes with insufficient sequence identity. Inspired by the efficient generation of multiple Cas9-associated guide RNAs through the endogenous tRNA-processing system in rice (Oryza sativa; Xie et al., 2015), we evaluated whether a similar strategy could be used to produce five amiRNAs for cosilencing five Arabidopsis immune receptor genes, namely FLS2, EFR, CERK1, PEPR1, and RLP23.
We first designed and screened for a potent amiRNA to silence each receptor gene using the ETPamir assay (Supplemental Fig. S3). Together with amiR-FLS2-m2 for silencing FLS2 (Fig. 2A), these five pre-amiRNAs were assembled into tRNA-pre-amiRNA tandem repeats under the control of the 35S promoter (Fig. 4A; Supplemental Fig. S4). Five immune ligands, namely flg22, elf18, chitin, Pep3, and nlp20, are perceived through these five immune receptors, respectively (Yu et al., 2017), all triggering plant growth inhibition. Therefore, only simultaneous silencing of all five receptor genes would desensitize plants and relieve their growth arrest in response to a cocktail of the five elicitors. Indeed, transgenic lines overexpressing these tRNA-pre-amiRNA tandem repeats demonstrated significantly suppressed seedling growth inhibition in the presence of the cocktail, particularly at a low concentration (Fig. 4, B and C). By contrast, the growth inhibition of fls2 null mutant seedlings by the elicitor cocktail was similar to that of the wild type (Supplemental Fig. S5). These results suggested that the five receptor genes have been cosilenced efficiently using the polycistronic tRNA-pre-amiRNA strategy, leading to the disruption of the growth inhibition triggered by all five elicitors. Consistently, using the stem-loop RT-PCR technique specialized for the sensitive detection of mature miRNA (Varkonyi-Gasic et al., 2007), we could detect the five mature amiRNAs in these silencing lines (Fig. 4D), which further evidenced the proper expression and processing of the five amiRNAs through the endogenous tRNA-processing system.
Figure 4.
Multiplex gene silencing using the polycistronic tRNA-pre-amiRNA strategy. A, Schematic diagram of tRNA-pre-amiRNA tandem repeats and their in vivo processing by the endogenous tRNA-processing system. B, Cosilencing of five immune receptor genes by tRNA-pre-amiRNA tandem repeats alleviates the growth inhibition of transgenic seedlings in response to a mixture of five immune ligands. The five immune receptor genes are FLS2, EFR, CERK1, PEPR1, and RLP23. Their cognate ligands are flg22 (10 or 100 nm), elf18 (10 or 100 nm), chitin (20 or 200 μg mL−1), pep3 (10 or 100 nm), and nlp20 (10 or 100 nm) and were tested as a mixture of low concentration or high concentration. Seedlings were germinated without immune ligands for 5 d and then treated with immune ligands or mock for 7 d. Bars = 1 cm. C, Quantification of seedling growth inhibition. Growth inhibition was determined by comparing the total fresh weight of four elicitor-treated seedlings with that of four mock-treated seedlings in one experiment, and data are presented as means ± sd of three experimental repeats. **, P < 0.01 (Student’s t test). D, Detection of five mature amiRNAs in transgenic silencing plants overexpressing tRNA-pre-amiRNA tandem repeats. Mature amiRNAs were detected using stem-loop RT-PCR, and individual PCR amplicons were subjected to 12% PAGE analysis. Arabidopsis SnoR101 served as an internal control. WT,wild type.
Notably, we could still detect considerable amounts of transcripts of these five immune receptor genes in the silencing lines that were blind to the cocktail of five immune ligands (Supplemental Fig. S6A). By contrast, using anti-FLS2 antibodies, we confirmed that the accumulation of FLS2 proteins in these lines was largely abolished (Supplemental Fig. S6B), suggesting a remarkable contribution of amiRNA-mediated translational inhibition to the silencing of FLS2.
An Intronic amiRNA-Producing Fluorescent Reporter for Screening Transgenic Arabidopsis for Optimal Gene Silencing
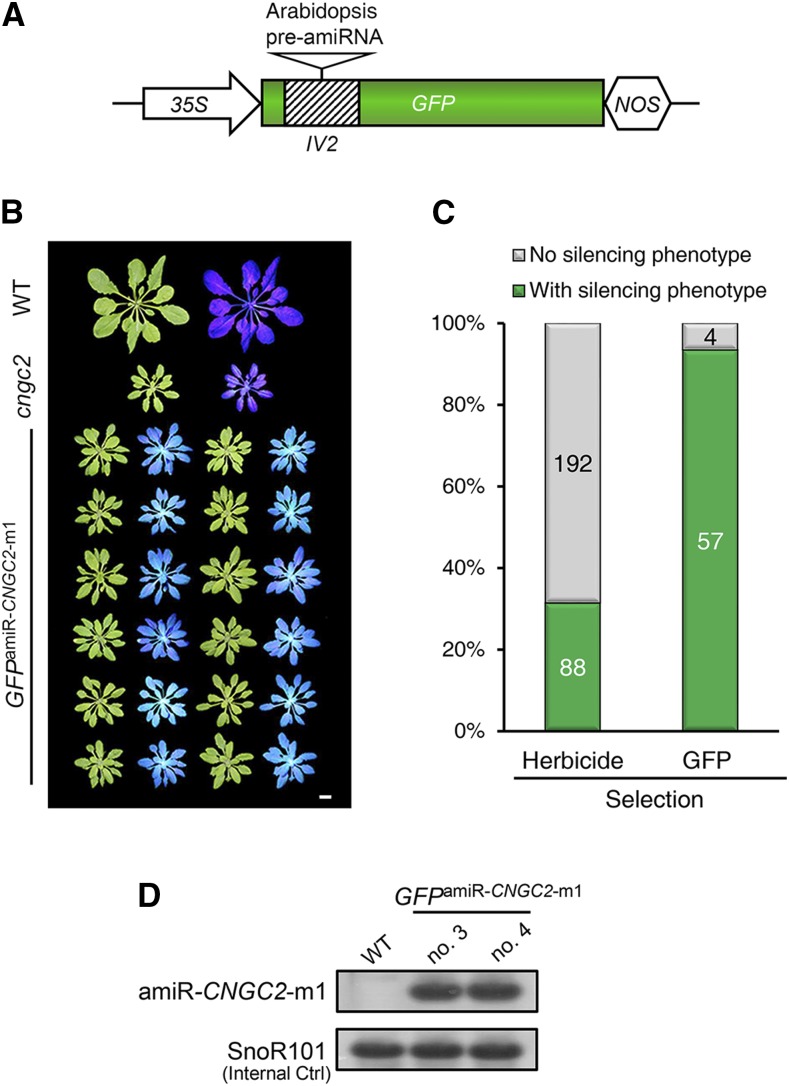
If target gene silencing generates no detectable phenotypic change, screening for transgenic plants with optimal silencing can be demanding. As demonstrated in the case of FLS2 silencing (Supplemental Fig. S6), without specific antibodies to assess the target protein level, the quantification of target transcript level alone may underestimate the silencing state in transgenic plants. We noticed that a few plant miRNAs in nature are produced from introns (Zhu et al., 2008; Meng and Shao, 2012; Yang et al., 2012). This prompted us to test whether an active amiRNA could be produced from an intron within a fluorescent reporter (e.g. GFP) and whether this reporter could serve as an easy readout for amiRNA activity. We selected Arabidopsis CNGC2 as a target gene since its loss-of-function mutant exhibits a readily detectable dwarf phenotype, thus allowing us to evaluate whether the reporter’s fluorescence could be used as a visible marker for selecting transgenic silencing lines. A potent amiRNA, namely amiR-CNGC2-m1, was prescreened for silencing CNGC2 using the ETPamir assay (Supplemental Fig. S7A). Its pre-amiRNA was then placed into a potato (Solanum tuberosum) IV2 intron (Pang et al., 1996), which, in turn, was inserted into GFP under the control of the 35S promoter (Fig. 5A; Supplemental Fig. S7B). We verified that both the normal amiR-CNGC2-m1 and the intronic amiR-CNGC2-m1 (referred to as GFPamiR-CNGC2-m1) could efficiently silence CNGC2 in protoplasts (Supplemental Fig. S7C). When screening transgenic Arabidopsis plants overexpressing GFPamiR-CNGC2-m1, we noticed that GFP per se could serve as an easy and economical selectable marker to distinguish transgenic from nontransgenic plants under blue light (Supplemental Fig. S8A). Importantly, over 93% (57 out of 61) of transgenic T1 plants selected by GFP fluorescence exhibited strong cngc2 null phenotypes (Fig. 5, B and C). By contrast, only 31% (88 out of 280) of transgenic T1 plants selected by the routine herbicide screen showed similar silencing phenotypes (Fig. 5C). In addition, we could detect mature amiR-CNGC2-m1 by stem-loop RT-PCR in representative transgenic silencing lines with GFP fluorescence (Fig. 5D), which further validated the proper expression and processing of intronic amiRNAs. Furthermore, a clear dwarf phenotype could be observed for the GFP-positive plants at the T2 generation (Supplemental Fig. S8B). These data suggested that the fluorescent reporter expressing intronic amiRNA could be used as a visible surrogate to ease the otherwise complicated screening of transgenic plants for optimal gene silencing.
Figure 5.
Facilitating transgenic screen of Arabidopsis plants with optimal silencing by an intronic amiRNA-producing fluorescent reporter. A, Schematic diagram of an intronic amiRNA-producing GFP reporter. B, An intronic amiR-CNGC2-producing GFP reporter (GFPamiRCNGC2-m1) facilitates the identification of transgenic plants with optimal CNGC2 silencing. Note that transgenic lines with GFP fluorescence show a cyan color when irradiated by a blue light flashlight in the dark, whereas the wild type (WT) and the cngc2 null mutant show a dark purple color. Bar = 1 cm. C, Comparison of the success rates to obtain optimal silencing plants by different transgenic selection strategies. D, Detection of mature amiR-CNGC2-m1 in representative transgenic plants with GFP fluorescence. Mature amiRNAs were detected using stem-loop RT-PCR, and the PCR amplicons were subjected to 12% PAGE analysis. Arabidopsis SnoR101 served as an internal control.
In a second case, we targeted the Arabidopsis gene At1g55000 as a representative of genes with unknown function. An effective amiRNA, namely amiR-At1g55000-m5, was prescreened for silencing At1g55000 using the ETPamir assay (Supplemental Fig. S9A), and its precursor was inserted into the IV2 intron of a GFP reporter. Because the silencing of At1g55000 resulted in no detectable phenotype and its protein antibody was not available, we quantified the transcripts of At1g55000 as gene-silencing readouts in transgenic T1 lines overexpressing GFPamiR-At1g55000-m5. Five randomly selected GFP-positive transgenic lines showed a consistent reduction in At1g55000 transcripts by approximately 80% (Supplemental Fig. S9B). By contrast, six randomly selected herbicide-resistant transgenic lines exhibited a variable reduction in At1g55000 transcripts by 40% to 80% (Supplemental Fig. S9B). These results again suggested that the fluorescent reporter expressing intronic amiRNA could help identify transgenic plants with maximal gene silencing.
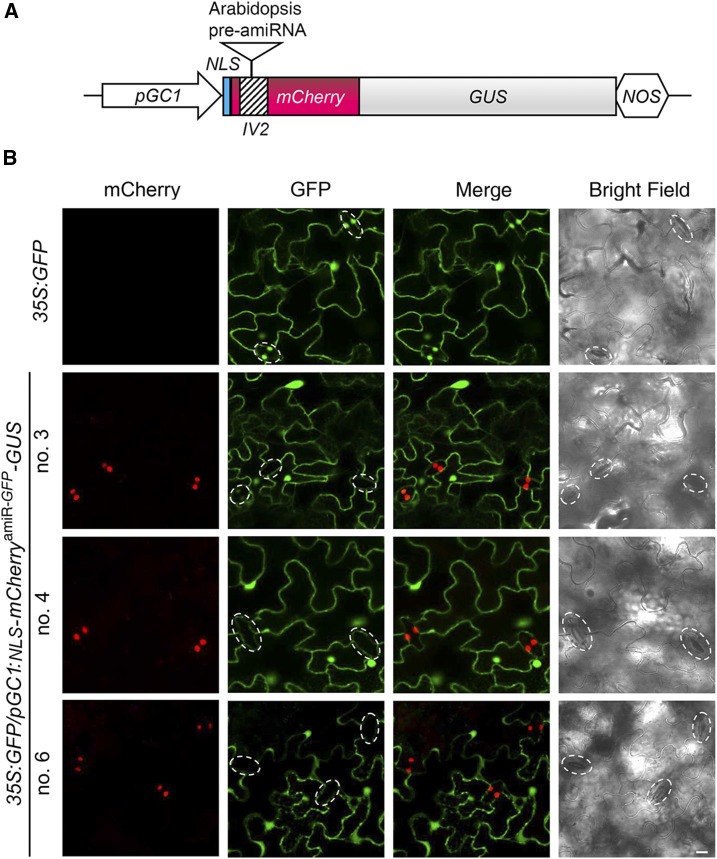
The amiRNA technology is a powerful tool to address cell type-specific gene function in plants. However, gene silencing in a specific cell type may not cause an obvious phenotypic change or target transcript reduction at the whole-plant level, making the screening of transgenic plants with optimal silencing a technical challenge. In a third case, we tested whether the intronic amiRNA-producing fluorescent reporter could be used to simplify cell type-specific gene silencing. As a proof of concept, we attempted to silence GFP in guard cells of transgenic Arabidopsis plants overexpressing GFP. The precursor of amiR-GFP (Li et al., 2013a) was inserted into the IV2 intron within an NLS-mCherry-GUS reporter under the control of the Arabidopsis guard cell-specific GC1 promoter (Yang et al., 2008; Fig. 6A; Supplemental Fig. S10A). We verified that both the normal amiR-GFP and the intronic amiR-GFP (referred to as NLS-mCherryamiR-GFP-GUS) could silence GFP equally well in protoplasts (Supplemental Fig. S10B). Indeed, we observed in several transgenic lines that the nucleus-localized mCherry fluorescence coincided with the diminished GFP fluorescence in guard cells, while the GFP fluorescence in surrounding pavement cells was not affected (Fig. 6B). These results suggested that the intronic amiRNA-producing fluorescent reporter can be used as a robust and visual marker for screening plants with optimal gene silencing not only at the whole-plant level but also in a specific cell type.
Figure 6.
Facilitating the transgenic screen of Arabidopsis plants with optimal silencing in a specific cell niche by an intronic amiRNA-producing fluorescent reporter. A, Schematic diagram of an intronic amiRNA-producing mCherry reporter under the control of a guard cell-specific promoter (pGC1). The fusion of mCherry with a nuclear localization signal (NLS) and GUS facilitates the easy detection of mCherry fluorescence in guard cells. B, Transgenic 35S:GFP/pGC1:NLS-mCherryamiR-GFP-GUS lines with nuclear mCherry fluorescence exhibit silenced GFP expression exclusively in guard cells (marked by dashed borders). Note that all epidermal cells, including guard cells, in 35S:GFP control plants have nuclei labeled by GFP, while only the pavement cells, but not the guard cells, in transgenic silencing plants have nuclei labeled by GFP. Bar = 10 μm.
An Intronic amiRNA-Producing Fluorescent Reporter for Screening Transgenic Rice for Optimal Gene Silencing
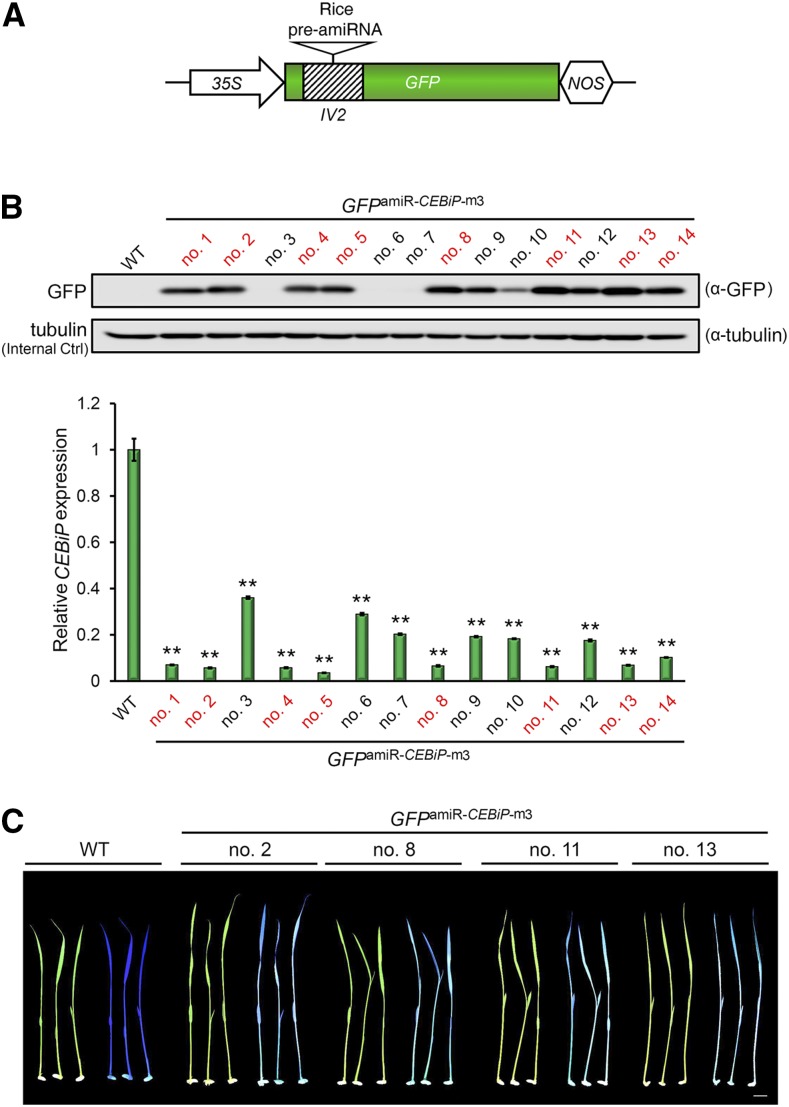
We showed previously that the Arabidopsis pre-miR319a backbone is not efficient in monocot species (Li et al., 2013a). To develop a suitable intronic amiRNA-producing fluorescent reporter for rice, we evaluated whether the frequently used rice amiRNA precursor, namely rice pre-miR528 (Warthmann et al., 2008), is compatible with the fluorescent reporter containing the IV2 intron. We designed five amiRNA candidates to target the rice immune receptor gene CEBiP and identified amiR-CEBiP-m3 as the most efficient candidate for silencing CEBiP in a rice protoplast-based ETPamir assay (Supplemental Fig. S11A). The precursor of amiR-CEBiP-m3 was embedded subsequently into the IV2 intron of a GFP reporter under the control of the 35S promoter (Fig. 7A; Supplemental Fig. S11B), and transgenic rice plants overexpressing this intronic amiRNA were generated. Although silencing of CEBiP led to no growth phenotype in rice, we could observe that GFP abundance (Fig. 7B) and fluorescence (Fig. 7C) coincided with the reduction of CEBiP transcripts in transgenic rice plants. Eight out of 10 transgenic rice lines with high GFP accumulation showed consistent declines of CEBiP transcripts by more than 90% (Fig. 7B). By contrast, four rice lines with undetectable or low GFP abundances had variable reductions of CEBiP transcripts by 60% to 80% (Fig. 7B). These results suggest that the intronic amiRNA-producing fluorescent reporter also can facilitate the identification of transgenic rice plants with maximal gene silencing. Notably, the rice pre-miR528 backbone also is able to cause gene silencing in wheat (Triticum aestivum) cells (Supplemental Fig. S12A). Similarly, the rice intronic amiRNA-producing GFP reporter also could generate functional GFP and amiRNA in wheat cells (Supplemental Fig. S12, B and C), suggesting that the rice amiRNA reporter system also may work for other monocot species such as wheat.
Figure 7.
Facilitating the transgenic screen of rice plants with optimal silencing by an intronic amiRNA-producing fluorescent reporter. A, Schematic diagram of an intronic amiRNA-producing GFP reporter for rice. B, An intronic amiR-CEBiP-producing GFP reporter (GFPamiR-CEBiP-m3) facilitates the identification of transgenic rice plants with optimal CEBiP silencing. Eight GFP-positive lines with optimal CEBiP silencing are highlighted in red. GFP abundances and CEBiP transcript levels were determined by immunoblotting and RT-quantitative PCR (qPCR), respectively. The CEBiP transcript level in wild-type (WT) rice plants was arbitrarily set as 1. RT-qPCR data are shown as means ± sd (n = 3). **, P < 0.01 (Student’s t test). C, Representative transgenic rice lines with optimal CEBiP silencing show a cyan color when irradiated by a blue light flashlight in the dark, whereas wild-type rice plants show a dark blue color. Bar = 1 cm.
DISCUSSION
The amiRNA design is a crucial step, where a rational approach can ensure that the designed amiRNA is specific to a target gene and has a better chance to be effective. By evaluating 63 amiRNAs, we noticed previously that most of the active amiRNAs tend to have perfect or nearly perfect complementarity to their targets (Li et al., 2013a). In this study, we incorporated this experimentally determined feature of potent amiRNAs into the amiRNA design and were convinced further that amiRNAs with such a feature are more effective in general (Fig. 2). Park et al. (2009) also described similar findings when using an amiRNA to silence the Arabidopsis AP1 gene. The most plausible reason behind these observations is that amiRNAs with perfect complementarity to their targets may require significantly less hybridization energy to anneal with the targets. Nevertheless, we would like to emphasize the necessity that any amiRNA candidate needs to be evaluated experimentally (e.g. using the ETPamir assay) to confirm its activity in plant cells or to identify the most potent one, because the target accessibility of a given amiRNA in a cellular context is the prerequisite for efficient gene silencing but is not predictable.
To date, several strategies have been explored to produce multiple amiRNAs simultaneously. First, multiple pre-amiRNAs in tandem repeats can be expressed as a single transcript with one promoter to generate up to three amiRNAs (Niu et al., 2006; Ji et al., 2011; Liang et al., 2012; Li et al., 2013a; Kis et al., 2016), but the production of five amiRNAs in this way has not been validated experimentally. Second, a naturally occurring polycistronic pre-miRNA, namely rice pre-miR395, has been harnessed to generate five amiRNAs in wheat (Fahim et al., 2012), but it remains unknown whether the pre-miR395 backbone can be used to produce multiple amiRNAs in other monocot or dicot species. Third, the Arabidopsis TAS gene has been engineered to produce five artificial trans-acting siRNAs in the presence of miR173 for multiplex gene silencing in Arabidopsis (de la Luz Gutiérrez-Nava et al., 2008; Felippes et al., 2012). However, the application of such an approach in other plant species may require the coexpression of miR173 (Felippes et al., 2012). In this study, we demonstrated an alternative strategy to produce five amiRNAs as tRNA-pre-amiRNA tandem repeats (Fig. 4A), which takes advantage of the efficient and conserved tRNA-processing machineries across monocot and dicot species. The native tRNA tandem repeats are transcribed by the RNA polymerase III (Pol III) complex (Xie et al., 2015). However, we found that tRNA-pre-amiRNA tandem repeats also can be expressed efficiently by the Pol II complex (Fig. 4D). This observation opens up the possibility that multiplex gene silencing can be regulated by Pol II promoters, such as a chemically inducible promoter or a cell type/tissue-specific promoter using the polycistronic tRNA-pre-amiRNA strategy. The maximum number of amiRNAs that can be expressed and processed through this strategy remains to be determined.
It is of note that several amiRNAs (e.g. amiR-FLS2-m2) mediated potent gene silencing mainly through translational repression rather than mRNA decay (Supplemental Fig. S6), which was reminiscent of our earlier observations (Li et al., 2013a) and similar reports on plant miRNAs (Brodersen et al., 2008; Li et al., 2013b). These results suggest that the prevalent practice of quantifying target mRNA abundances as an indicator of amiRNA-mediated gene silencing may underestimate the real levels of gene silencing. In addition, targeted gene silencing in a specific cell type cannot be screened based on whole-plant phenotypic changes, RT-PCR, or immunoblot assays. In the absence of visible silencing phenotypes, or in the case of cell type-specific gene silencing, it is a great challenge to identify transgenic plants with optimal gene silencing. Antibiotic- or herbicide-based selection of transgenic plants also cannot guarantee the optimal expression of the amiRNA (Fig. 5C), so the silencing of the target gene varies between individual transgenic lines. Here, we demonstrate that an intronic amiRNA-producing fluorescent reporter can be used to predict amiRNA performance, serving as a visual indicator of amiRNA activity in both dicot and monocot species, either in whole plants or in a specific cell type.
In summary, the amiRNA technology remains a useful genetic tool even in the CRISPR era, due to its excellent specificity, reversibility, and flexibility (Schwab et al., 2006; Hauser et al., 2013; Jover-Gil et al., 2014). It offers particular advantages for manipulating essential genes whose loss-of-function mutants are lethal or need to be created in a specific tissue, cell type, or developmental stage. Notably, multiplex gene silencing by amiRNAs is a convenient solution for the technical dilemma in gene functional studies when multiple-gene knockout results in plant lethality but single-gene knockout is barely informative due to gene redundancy. However, the unpredictable silencing efficacy of a designed amiRNA and the variation of its expression levels in transgenic plants (Schwab et al., 2006) render the screening for transgenic silencing lines challenging, limiting the applications of the amiRNA technology in plant research. In this study, we showcased that efficient amiRNA-mediated gene silencing in plants can be streamlined by (1) using an improved strategy to design odds-on amiRNA candidates; then (2) using the ETPamir assay (Li et al., 2013a, 2014) to identify the most potent amiRNA; and finally (3) using a fluorescent reporter system to screen transgenic plants with maximal target silencing. We envision that the strategies presented here can help amiRNA users in their gene-silencing endeavor in basic and applied plant research.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) fls2 (SALK_141277) and cngc2 (SALK_019922) mutant seeds were obtained from the Arabidopsis Biological Resource Center. The glk1 glk2 double mutant (NASC N9807) seeds were obtained from the European Arabidopsis Stock Center. The sr45-1 (SALK_004132) mutant seeds were described previously (Zhang and Mount, 2009). Wild-type Columbia-0, mutant, or transgenic Arabidopsis or wheat (Triticum aestivum) plants were grown on Jiffy soil (Jiffy Group) in a plant growth room with 65% humidity and 75 μmol m−2 s−1 light intensity under a photoperiod of 12 h of light at 23°C and 12 h of dark at 20°C. Wild-type or transgenic Zhonghua 11 rice (Oryza sativa) plants were grown on Jiffy soil in a plant growth chamber with 70% humidity and 200 μmol m−2 s−1 light intensity under a photoperiod of 12 h of light at 30°C and 12 h of dark at 27°C.
Plasmid Construction
Routine molecular cloning procedures were followed for plasmid construction. HBT plasmids containing the 35S promoter, a pre-miRNA (Arabidopsis pre-miR319a or rice pre-miR528), and the NOS terminator were used as starting vectors for amiRNA cloning and transient expression in protoplasts. To facilitate one-step cloning of amiRNAs, the original Arabidopsis pre-miR319a or rice pre-miR528 vector was subjected to PCR-based mutagenesis to generate a derived vector containing an engineered precursor. The engineered pre-miR319a and pre-miR528 contain EcoRI/XbaI and StuI/EcoRI restriction sites, respectively. The amiRNAs tested in this study (Supplemental Table S1) were cloned into the engineered pre-miR319a or pre-miR528 as follows. Briefly, mega-primers containing customized amiRNA/amiRNA* sequences were used for PCR amplification of a stem-loop fragment containing a new amiRNA/amiRNA* duplex using the original pre-miR319a or pre-miR528 as PCR template. PCR amplicons were digested by EcoRI/XbaI or StuI/EcoRI and inserted into the same digested HBT vector harboring the engineered pre-miR319a or pre-miR528. If necessary, the pre-amiRNAs were cut out from HBT plasmids by BamHI/PstI and inserted into the same digested pCB302 binary vector for transgenic expression.
To express a target gene encoding double HA- or FLAG-tagged target proteins in protoplasts, the full-length CDS of the target gene was amplified by RT-PCR, digested by BamHI/StuI, and inserted into the same digested HBT-2HA or HBT-2FLAG vector, where the target gene expression is driven by the 35S promoter.
For plasmid containing five tRNA-pre-amiRNA tandem repeats (Supplemental Sequences S1), the first tRNA-pre-amiRNA fragment was synthesized as a gBlock dsDNA (Synbio Technologies) flanked with BamHI/PstI sites and was digested and inserted into the BamHI/PstI window of the HBT vector between the 35S promoter and the NOS terminator. The second to fifth tRNA-pre-amiRNA fragments were assembled individually by overlapping PCR and flanked with NsiI/PstI sites. Taking advantage of the compatibility between the DNA overhangs after NsiI and PstI digestion and the disruption of both restriction sites after ligation, the second to fifth tRNA-pre-amiRNA fragment was digested by NsiI/PstI and inserted sequentially into the PstI site, which was always maintained at the junction between the last tRNA-pre-amiRNA repeat and the NOS terminator. The tRNA-pre-amiRNA tandem repeats finally were cut out by BamHI/PstI and inserted into the same digested pCB302 binary vector for transgenic expression.
To construct the intronic amiRNA-producing GFP reporter (Supplemental Sequences S1), an IV2 intron-containing GFP was synthesized as a gBlock dsDNA (Synbio Technologies) flanked with BamHI/PstI sites and was digested and inserted into the BamHI/PstI window of the HBT vector. The PCR amplicons of pre-amiRNA were digested by NotI and inserted into the NotI site within the IV2 intron. The GFPpre-amiRNA sequence was cut out by BamHI/PstI and inserted into the same digested pCB302 binary vector for transgenic expression in Arabidopsis or into the pCAMBIA binary vector for transgenic expression in rice. To construct the intronic amiRNA-producing NLS-mCherry-GUS reporter (Supplemental Sequences S1) driven by the guard cell-specific GC1 promoter (pGC1), an IV2 intron containing mCherry with an NLS was assembled by overlapping PCR and inserted into the BamHI/PstI sites of pUC119. The pre-amiR-GFP then was inserted into the NotI site of the IV2 intron. The GUS gene and the Arabidopsis pGC1 sequence were PCR amplified, digested, and inserted into the SmaI site and the EcoRI/BamHI sites, respectively. Finally, the pGC1:NLS-mCherryamiR-GFP-GUS:NOS expression cassette was cut out with AscI from pUC119 and inserted into the AscI site of the pBIN binary vector.
Protoplast Isolation
Four-week-old Arabidopsis and 10-d-old rice or wheat seedlings were used for protoplast isolation according to the protocol described previously (Zhang et al., 2011; Li et al., 2014). Briefly, leaves were cut into 0.5-mm strips with a sterile razor blade and digested in 10 mL of enzyme solution (1.5% [w/v] cellulase R10, 0.2% [w/v] macerozyme R10, 0.4 m mannitol, 20 mm KCl, 20 mm MES, pH 5.7, 10 mm CaCl2, and 0.1% BSA) for 3.5 h. After mixing with 10 mL of W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES, pH 5.7), the digestion mixture was filtered through a 75-μm FALCON cell strainer. Protoplasts were collected by centrifugation for 2 min at 100g for Arabidopsis and wheat or for 5 min at 200g for rice. Cells were resuspended with 10 mL of W5 solution and rested on ice for 30 min. Before transfection, protoplasts were pelleted by centrifugation and resuspended with MMg solution (0.4 m mannitol, 15 mm MgCl2, and 4 mm MES, pH 5.7) to a final concentration of 2 × 105 cells mL−1.
Protoplast Transfection and ETPamir Assay
DNA transfection was performed in a 2-mL round-bottom microcentrifuge tube, where 200 μL of protoplasts was mixed with 21 μL of DNA cocktail (2 μg μL−1) and 220 μL of polyethylene glycol solution (40% [v/v] PEG4000, 0.2 m mannitol, and 0.1 m CaCl2). After incubation at room temperature for 5 min for Arabidopsis and wheat or for 15 min for rice, transfection was quenched by adding 800 μL of W5 solution. Transfected protoplasts were collected by centrifugation for 2 min at 100g for Arabidopsis and wheat or for 5 min at 200g for rice and were resuspended with 100 μL of W5 solution. Cells then were transferred into 1 mL of WI solution (0.5 m mannitol, 4 mm MES, pH 5.7, and 20 mm KCl) on a six-well plate and were incubated in the dark. The ETPamir assay was conducted as described previously (Li et al., 2014). Briefly, 200 μL of protoplasts was transfected with a DNA cocktail (2 μg μL−1) containing 16 μL of amiRNA expression construct, 4 μL of target gene-HA/FLAG expression construct, and 1 μL of transfection control plasmid expressing GFP-HA/FLAG or LYK5-HA. In parallel, a negative control was set up by replacing the amiRNA expression construct with an equal amount of empty vector. After cotransfection, protoplasts were incubated for 18 to 36 h and the amiRNA performance was determined by immunoblot analysis of target protein accumulation.
RNA Extraction and RT-qPCR Analysis
A total of 400 μL of Arabidopsis protoplasts (8 × 104 cells) or 20 mg of Arabidopsis or rice seedlings was used for RNA extraction. Total RNA was extracted using the RNAiso Plus reagent (TaKaRa) according to the manufacturer’s instructions. For the quantification of target transcripts, total RNA of 1 μg was converted into the first-strand cDNA using the RT Primer Mix containing Oligo dT Primer and random 6-mers from the PrimeScript RT Reagent Kit with genomic DNA Eraser (TaKaRa) according to the manufacturer’s instructions. RT-qPCR was performed in a LightCycler 96 Instrument (Roche) using SYBR Premix Ex Taq (TaKaRa). Transcript levels of target genes were normalized to those of endogenous ACT1 (for Arabidopsis) or Actin-1 (for rice). Primers used for RT-qPCR are listed in Supplemental Table S2.
Mature amiRNA Detection
The protocol described earlier (Varkonyi-Gasic et al., 2007) was followed with minor modifications to detect mature amiRNAs in plant cells. Briefly, total RNA of 1 μg was converted into the first-strand cDNA using stem-loop RT primers (Supplemental Table S2). Mature amiRNAs produced in transgenic Arabidopsis plants expressing tRNA-pre-amiRNA tandem repeats or intronic amiRNA-producing GFP reporter were detected by resolving the stem-loop RT-PCR products on a 12% (w/v) polyacrylamide gel using endogenous SnoR101 as an internal control. Primers used for stem-loop RT-PCR are listed in Supplemental Table S2.
Seedling Growth Inhibition
Wild-type Columbia-0 or transgenic Arabidopsis seedlings overexpressing tRNA-pre-amiRNA tandem repeats were germinated and grown vertically on one-half-strength Murashige and Skoog (1/2 MS) medium containing 0.8% agar and 0.5% Suc for 5 d, then transferred to 1/2 MS liquid medium on a six-well culture plate containing 0.5% Suc and a cocktail of immune ligands (flg22, efl18, chitin, pep3, and nlp20) or mock. After growing for another 7 d, seedlings were photographed and weighed.
Immunoblot
Protoplasts were lysed with lysis buffer (10 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, and 10% glycerol). Leaves or seedlings were frozen by liquid nitrogen, ground into powder, and then lysed with lysis buffer. The lysates were mixed with 6× SDS protein loading buffer and heated at 95°C for 5 min. Total proteins were subjected to SDS-PAGE and immunoblotting with anti-HA (Roche), anti-FLAG (Sigma-Aldrich), anti-FLS2 (Genscript), anti-tubulin (Hopebio), or anti-GFP (Roche) antibodies.
Generation of Transgenic Plants
The recombinant pCB302 or pBIN binary plasmids were introduced individually into Agrobacterium tumefaciens GV3101 cells by electroporation, which, in turn, were used for floral dip-mediated Arabidopsis transformation. Transgenic Arabidopsis plants were selected on 1/2 MS medium containing 12.5 mg L−1 glufosinate ammonium or 50 mg L−1 kanamycin. Transgenic Arabidopsis plants expressing GFPamiR-CNGC2-m1 also were selected by using a specialized blue-light flashlight (LUYOR) in a darkroom. The binary plasmid pCAMBIA-GFPamiR-OsCEBiP-m3 was introduced into A. tumefaciens strain EHA105 cells by electroporation, which, in turn, were used for rice callus transformation. Transgenic rice plants were selected on 1/2 MS medium containing 50 mg L−1 hygromycin (Roche).
Fluorescence Microscopy
Confocal images were acquired on a Zeiss LSM 800 microscope using a Plan-Apochromat 20×/0.8 M27 objective. To excite GFP and mCherry, 488- and 561-nm lasers were used, respectively. Emitted fluorescence was detected by GaAsP-Detector, set to detect 490 to 561 nm for GFP and 575 to 665 nm for mCherry. The laser power was set between 9% and 29.9% to image GFP or mCherry. A scan average of two was used for the majority of experiments. Images of protoplasts were acquired on a Leica DMi8 fluorescence microscope using a 20×/0.4 objective. GFP and mCherry fluorescence were detected using the GFP-T filter set (excitation filter, 475/40 nm; dichromatic mirror, 500 nm; suppression filter, 530/50 nm) and the DSRED filter set (excitation filter, 546/11 nm; dichromatic mirror, 560 nm; suppression filter, 605/75 nm), respectively.
Generation of the Arabidopsis amiRNA Database
All 21-nucleotide target sequences ending with A were in silico extracted from the CDSs of Arabidopsis nuclear genes included in the Araport 11 database (www.arabidopsis.org). If alternatively spliced isoforms of a gene exist, only the number 1 isoform was used for target sequence extraction. Reverse complements of these target sequences then were generated as amiRNA candidates. Based on the observation that mismatch at position 21 of an amiRNA would not dramatically compromise its silencing efficiency (Li et al., 2013a), amiRNA derivatives with the last nucleotide substituted by one of the other three nucleotides were generated to quadruple the size of the amiRNA candidate library. Each amiRNA candidate in this library then was submitted to the WMD3-Target Search (wmd3.weigelworld.org) to identify target genes in Arabidopsis with a setting of 5 mismatch tolerance. All amiRNA candidates with more than one target gene were eliminated from the candidate library, resulting in a final list of gene-specific amiRNAs for 27,136 Arabidopsis nuclear genes (Supplemental Data Set S1).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: FLS2, At5g46330; GLK1, At2g20570; GLK2, At5g44190; APK2A, At1g14370; APK2B, At2g02800; SERK1, At1g71830; SERK2, At1g34210; SERK3, At4g33430; SERK4, At2g13790; SERK5, At2g13800; SR45, At1g16610; EFR, At5g20480; CERK1, At3g21630; PEPR1, At1g73080; RLP23, At2g32680; CNGC2, At5g15410; and CEBiP, Os03g04110.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence alignment between GLK1 and GLK2 identifies the most conserved sequences for amiRNA design.
Supplemental Figure S2. Comparison of the activities of amiRNA candidates designed by two different strategies for silencing the Arabidopsis APK2 or SERK family using the ETPamir assay.
Supplemental Figure S3. ETPamir screen identifies the most effective amiRNA for silencing Arabidopsis EFR, CERK1, PEPR1, or RLP23.
Supplemental Figure S4. Sequence of the tRNA-preamiRNA tandem repeats for cosilencing five Arabidopsis immune receptor genes.
Supplemental Figure S5. Growth inhibition of the fls2 mutant is comparable to that of the wild type in the presence of a cocktail of five immune ligands.
Supplemental Figure S6. amiRNA-mediated silencing of five immune receptor genes is largely due to translational inhibition instead of transcript degradation.
Supplemental Figure S7. Intronic amiR-CNGC2-m1 is active for silencing Arabidopsis CNGC2 in the ETPamir assay.
Supplemental Figure S8. Intronic amiRNA-producing GFP could serve as a selectable marker for transgenic plants with amiRNA expression.
Supplemental Figure S9. Intronic amiRNA-producing GFP reporter facilitates the screen of transgenic Arabidopsis plants with optimal silencing of At1g55000.
Supplemental Figure S10. Intronic amiR-GFP and normal amiR-GFP are equally active for silencing GFP in the ETPamir assay.
Supplemental Figure S11. Intronic amiR-CEBiP-m3 for silencing rice CEBiP.
Supplemental Figure S12. Rice amiRNA precursor and intronic amiRNA-producing GFP reporter are both functional in wheat cells.
Supplemental Table S1. Summary of the amiRNAs tested in this study.
Supplemental Table S2. Primers for RT-PCR or stem-loop RT-qPCR used in this study.
Supplemental Data Set S1. Gene-specific amiRNA candidates for Arabidopsis nuclear genes.
Supplemental Sequences S1. Sequences of polycistronic tRNA-pre-amiRNAs and intronic amiRNAs used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yang Yu for technical assistance on the stem-loop RT-PCR technique.
Footnotes
This work was supported by the National Natural Science Foundation of China grant 31522006 and start-up funds from China’s Thousand Young Talents Program to J.-F.L.
References
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Butardo VM, Fitzgerald MA, Bird AR, Gidley MJ, Flanagan BM, Larroque O, Resurreccion AP, Laidlaw HK, Jobling SA, Morell MK, et al. (2011) Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J Exp Bot 62: 4927–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi M, Bhagwat B, Lane WD, Tang G, Su Y, Sun R, Oomah BD, Wiersma PA, Xiang Y (2014) Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol 14: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Luz Gutiérrez-Nava M, Aukerman MJ, Sakai H, Tingey SV, Williams RW (2008) Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol 147: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CG, Wang CH, Fang RX, Guo HS (2008) Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J Virol 82: 11084–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahim M, Millar AA, Wood CC, Larkin PJ (2012) Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol J 10: 150–163 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Hill ST, Carrington JC, Carbonell A (2016) P-SAMS: a web site for plant artificial microRNA and synthetic trans-acting small interfering RNA design. Bioinformatics 32: 157–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Wang JW, Weigel D (2012) MIGS: miRNA-induced gene silencing. Plant J 70: 541–547 [DOI] [PubMed] [Google Scholar]
- Guo H, Song X, Wang G, Yang K, Wang Y, Niu L, Chen X, Fang R (2014) Plant-generated artificial small RNAs mediated aphid resistance. PLoS ONE 9: e97410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Chen W, Deinlein U, Chang K, Ossowski S, Fitz J, Hannon GJ, Schroeder JI (2013) A genomic-scale artificial microRNA library as a tool to investigate the functionally redundant gene space in Arabidopsis. Plant Cell 25: 2848–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Witkowsky LB, Guglielmi B, Replogle JM, Gilbert LA, Villalta JE, Torigoe SE, Tjian R, Weissman JS (2016) Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife 5: e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KT, Fløe L, Petersen TS, Huang J, Xu F, Bolund L, Luo Y, Lin L (2017) Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett 591: 1892–1901 [DOI] [PubMed] [Google Scholar]
- Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B, et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover-Gil S, Paz-Ares J, Micol JL, Ponce MR (2014) Multi-gene silencing in Arabidopsis: a collection of artificial microRNAs targeting groups of paralogs encoding transcription factors. Plant J 80: 149–160 [DOI] [PubMed] [Google Scholar]
- Kis A, Tholt G, Ivanics M, Várallyay É, Jenes B, Havelda Z (2016) Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol Plant Pathol 17: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J (2013a) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Zhang D, Sheen J (2014) Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat Protoc 9: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013b) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Yu D (2012) A new strategy for construction of artificial miRNA vectors in Arabidopsis. Planta 235: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22: 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Shao C (2012) Large-scale identification of mirtrons in Arabidopsis and rice. PLoS ONE 7: e31163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pang SZ, DeBoer DL, Wan Y, Ye G, Layton JG, Neher MK, Armstrong CL, Fry JE, Hinchee MAW, Fromm ME (1996) An improved green fluorescent protein gene as a vital marker in plants. Plant Physiol 112: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Zhai J, Lee JY (2009) Highly efficient gene silencing using perfect complementary artificial miRNA targeting AP1 or heteromeric artificial miRNA targeting AP1 and CAL genes. Plant Cell Rep 28: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotia S, Singh D, Tang X, Tang G (2016) Essential RNA-based technologies and their applications in plant functional genomics. Trends Biotechnol 34: 106–123 [DOI] [PubMed] [Google Scholar]
- Tian B, Li J, Oakley TR, Todd TC, Trick HN (2016) Host-derived artificial microRNA as an alternative method to improve soybean resistance to soybean cyst nematode. Genes (Basel) 7: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppino L, Kooiker M, Lindner M, Dreni L, Rotino GL, Kater MM (2011) Reversible male sterility in eggplant (Solanum melongena L.) by artificial microRNA-mediated silencing of general transcription factor genes. Plant Biotechnol J 9: 684–692 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Weiberg A, Lin FM, Thomma BP, Huang HD, Jin H (2016) Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants 2: 16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS (2006) Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GD, Yan K, Wu BJ, Wang YH, Gao YX, Zheng CC (2012) Genomewide analysis of intronic microRNAs in rice and Arabidopsis. J Genet 91: 313–324 [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55: 109–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ. (2014) Artificial trans-acting small interfering RNA: a tool for plant biology study and crop improvements. Planta 239: 1139–1146 [DOI] [PubMed] [Google Scholar]
- Zhang XN, Mount SM (2009) Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol 150: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li Z, Li JF (2016a) Targeted gene manipulation in plants using the CRISPR/Cas technology. J Genet Genomics 43: 251–262 [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua CL, Ding SW, Guo HS (2016b) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2: 16153. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C (2008) A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18: 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]