Physiological, biochemical, and proteomic analyses of Arabidopsis mutants reveal the relationship between Deg1 and Deg5-Deg8 protease complexes and demonstrate a more prominent role in growth for Deg1.
Abstract
Deg proteases are involved in protein quality control in prokaryotes. Of the three Arabidopsis (Arabidopsis thaliana) homologs, Deg1, Deg5, and Deg8, located in the thylakoid lumen, Deg1 forms a homohexamer, whereas Deg5 and Deg8 form a heterocomplex. Both Deg1 and Deg5-Deg8 were shown separately to degrade photosynthetic proteins during photoinhibition. To investigate whether Deg1 and Deg5-Deg8 are redundant, a full set of Arabidopsis Deg knockout mutants were generated and their phenotypes were compared. Under all conditions tested, deg1 mutants were affected more than the wild type and deg5 and deg8 mutants. Moreover, overexpression of Deg5-Deg8 could only partially compensate for the loss of Deg1. Comparative proteomics of deg1 mutants revealed moderate up-regulation of thylakoid proteins involved in photoprotection, assembly, repair, and housekeeping and down-regulation of those that form photosynthetic complexes. Quantification of protein levels in the wild type revealed that Deg1 was 2-fold more abundant than Deg5-Deg8. Moreover, recombinant Deg1 displayed higher in vitro proteolytic activity. Affinity enrichment assays revealed that Deg1 was precipitated with very few interacting proteins, whereas Deg5-Deg8 was associated with a number of thylakoid proteins, including D1, OECs, LHCBs, Cyt b6f, and NDH subunits, thus implying that Deg5-Deg8 is capable of binding substrates but is unable to degrade them efficiently. This work suggests that differences in protein abundance and proteolytic activity underlie the differential importance of Deg1 and Deg5-Deg8 protease complexes observed in vivo.
The chloroplast proteolytic machinery is composed of ATP-dependent and -independent proteases that are distributed in all major subcompartments of the organelle: the chloroplast envelope, stroma, thylakoid membrane, and thylakoid lumen. It is involved in either limited cleavage of targeting sequences following protein import and sorting or processive degradation of entire proteins, thus supporting the proper biogenesis of chloroplasts and the maintenance of their function in the face of ever-changing physiological and environmental conditions (for review, see Sakamoto, 2006; Adam, 2007; Adam and Sakamoto, 2014; van Wijk, 2015; Nishimura et al., 2017). Given the prokaryotic evolutionary origin of chloroplasts, it is not surprising that chloroplast proteases are orthologs of known bacterial proteases. However, whereas the different bacterial proteases are mostly products of single genes, their plant orthologs have been multiplied into families (Adam et al., 2001; Sokolenko et al., 2002), raising the question of whether these gene multiplications have a functional significance. The best-characterized chloroplast proteases to date are the Clp protease system operating in the stroma (Nishimura and van Wijk, 2015) and the thylakoid-membrane FtsH protease (Kato and Sakamoto, 2018), both of which are classified as self-compartmentalizing proteases (Baumeister et al., 1998). As such, their active sites are located within barrel-like structures, secluded from their surrounding environment, allowing highly selective proteolysis. Degradation of their substrates requires their recognition, ATP-dependent unfolding, and feeding into the proteolytic chamber (for review, see Olivares et al., 2016).
The ATP-independent Deg proteases, also known as HtrA proteases, constitute a family of Ser proteases found in bacteria and those organelles of prokaryotic origin, where they are involved in protein quality control and responses to different stress conditions (for review, see Clausen et al., 2011). These proteins are characterized by an N-terminal protease domain with a Ser-His-Asp catalytic triad, a C-terminal PDZ domain(s) involved in protein-protein interaction, and an oligomeric structure associated with their active state. The Arabidopsis (Arabidopsis thaliana) genome contains 16 Deg genes, some of which are suspected to be pseudogenes (Schuhmann and Adamska, 2012; Tanz et al., 2014). The remaining gene products are distributed among chloroplasts, mitochondria, peroxisomes, and the nucleus. Within the chloroplast, Deg2 and Deg7 have been identified in the stroma (Haussühl et al., 2001; Sun et al., 2010a), whereas Deg1, Deg5, and Deg8 are located in the thylakoid lumen (Itzhaki et al., 1998; Peltier et al., 2002; Schubert et al., 2002). Deg1 has been shown to form an active homohexamer at acidic pH, whereas at basic or neutral pH, the protein assumes a monomeric form that is inactive (Chassin et al., 2002; Kley et al., 2011). Hexamerization of Deg1 at acidic pH is dependent on the protonation of a specific His residue, His-244, in the proteolytic domain. Upon protonation, the N-terminal α-helix of the protein is stabilized in an orientation that allows its interaction with a neighboring monomer, thus forming a trimer that can readily dimerize to a hexamer. In the hexameric structure, the residues of the catalytic triad are stabilized in a proper orientation for catalysis (Kley et al., 2011). Deg5, unlike all other Deg proteases, lacks a PDZ domain and interacts in the lumen with Deg8 to form an active heterohexamer (Sun et al., 2007). Although Deg5 and Deg8 have been crystalized individually and their hexameric structures determined (Sun et al., 2013), the structural details of the heterohexamer have not been demonstrated yet.
The chloroplast Deg proteases on both sides of the thylakoid membrane have been implicated in the degradation of photosynthetic proteins, especially the D1 protein of the PSII reaction center following its oxidative damage, in relation to the PSII repair cycle operating during photoinhibition (Itzhaki et al., 1998; Haussühl et al., 2001; Kapri-Pardes et al., 2007; Sun et al., 2007, 2010b; Huesgen et al., 2009; Kley et al., 2011; Luciński et al., 2011a, 2011b; Schuhmann and Adamska, 2012; Zienkiewicz et al., 2013). Cleavage of the soluble domains of highly hydrophobic photosynthetic proteins such as the D1 protein apparently facilitates their further degradation by the ATP-dependent, stroma-oriented thylakoid FtsH protease complex (Kapri-Pardes et al., 2007; Kato and Sakamoto, 2009; Kato et al., 2015). In addition, Deg1 was proposed to participate in PSII assembly by virtue of a chaperone activity (Sun et al., 2010b), an activity that also was attributed to the bacterial DegP (Clausen et al., 2011).
The location of both Deg1 and Deg5-Deg8 in the thylakoid lumen, together with the reports on their involvement in D1 protein degradation in the context of response to photoinhibition (Kapri-Pardes et al., 2007; Sun et al., 2007; Kato et al., 2012), raise the question of whether they fulfill the same physiological functions or not. To answer this, we generated a full set of single, double, and triple Deg knockout (KO) mutants affected in the three thylakoid lumen Deg proteins and compared their mutant phenotypes under different environmental conditions. In combination with biochemical and proteomic analyses, we reveal that differences in protein abundance and proteolytic activity underlie the differential importance of Deg1 and Deg5-Deg8 in vivo.
RESULTS
Deg5 and Deg8 Are Dispensable under Optimal Growth Conditions
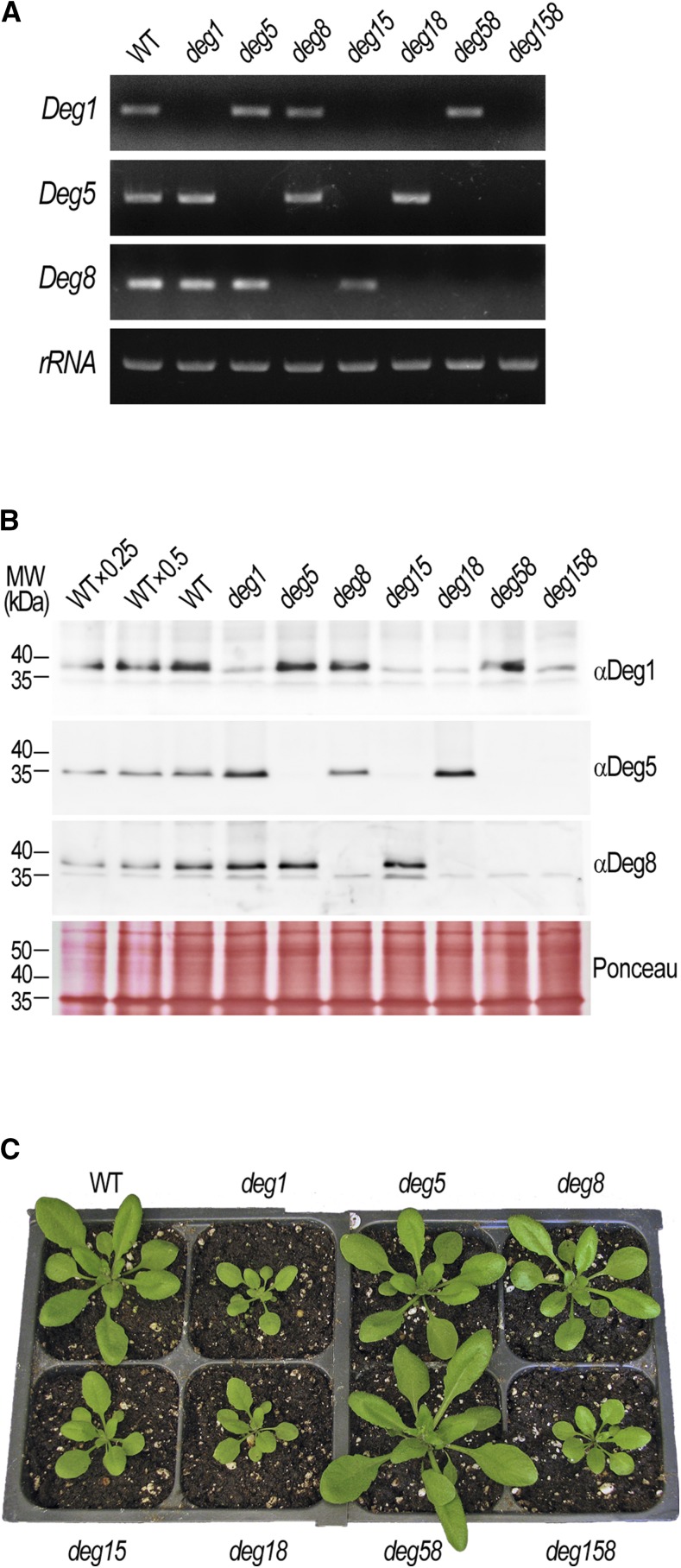
Previous studies, primarily in transgenic plants accumulating lower levels of Deg1 (Kapri-Pardes et al., 2007) or complete Deg5-Deg8 KO mutants (Sun et al., 2007), have suggested that the Deg1 and Deg5-Deg8 thylakoid lumen-located protease complexes fulfill the same function, specifically degradation of the D1 protein of the PSII reaction center in the context of its repair during photoinhibition. To test whether the two complexes play equal functional roles in this process, we sought to compare them side by side in a comprehensive manner. To that end, we generated a full set of single, double, and triple Deg KO mutants affected in the three lumenal Deg proteases. The only relevant T-DNA insertion line of Deg1 available at the time (Gabi-Kat_414D07) and the two published lines of Deg5 (SALK_099162) and Deg8 (SALK_004770; Sun et al., 2007; see schematic maps in Supplemental Fig. S1A) were crossed, thus generating seven different genotypes: the single mutants deg1, deg5, and deg8, the double mutants deg15, deg18, and deg58, and the triple mutant deg158 (for genotyping, see Supplemental Fig. S1B). As expected, the different genotypes were devoid of the corresponding one-, two-, or three-gene transcripts (Fig. 1A). Immunoblot analysis with specific antibodies revealed the absence of the protein products of the mutated genes (Fig. 1B). Occasionally, we observed some nonspecific cross reactivity of the Deg1 and Deg8 antibodies with proteins of slightly smaller size. However, Deg1 or Deg8 could not be identified in the corresponding mutant lines in the mass spectrometry (MS) analyses described below.
Figure 1.
Characterization of thylakoid Deg protease KO mutants. A, Transcript levels in leaves of the wild type (WT) and homozygous deg single, double, and triple mutants (genotypes are indicated above the gel images), as determined by semiquantitative reverse transcription-PCR using gene-specific primer pairs (target genes are indicated on the left; see Supplemental Materials and Methods S1). rRNA was used as a loading control. B, Immunoblot analysis of Deg protein levels in the wild type and homozygous deg mutants (genotypes are indicated above the immunoblots). Thylakoid membranes were isolated from leaves of the different genotypes, separated by SDS-PAGE, and detected with specific antibodies (target proteins are indicated on the right). The migration of molecular mass (MW) markers is indicated on the left. Five micrograms of chlorophyll was loaded in each lane. Ponceau staining of membranes was used as a loading control. C, Representative images of the wild type and homozygous deg mutants (genotypes are indicated adjacent to the images) grown in soil for 24 d under 18 h of light/6 h of dark, 22°C/18°C, at ∼160 µmol photons m−2 s−1.
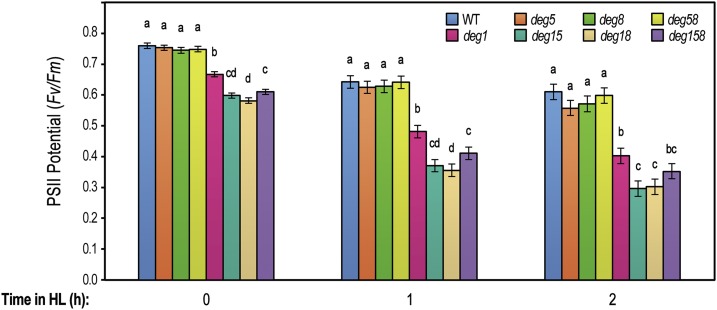
When grown under optimal conditions, the deg1 allele was associated with reduced plant size and height compared with the wild type (Fig. 1C). On the other hand, the absence of Deg5, Deg8, or both proteins had no effect on the visual appearance of the respective mutant plants. Similar effects were evident following the quantitation of other growth parameters, such as fresh weight, number of leaves, and rosette area, as well as the level of chlorophyll (Supplemental Table S1). The effect of Deg1 absence on the overall plant size correlated with growth light intensity, ranging from causing intermediate to substantial size reduction (Supplemental Fig. S2). Interestingly, statistical analysis of the growth parameters revealed that, for some of them, deg15, deg18, and deg158 mutants were grouped separately from deg1, displaying a slight augmentation of the mutant phenotype (Supplemental Table S1). Since the lumenal Deg proteases were linked individually to the repair of PSII under photoinihibitory conditions (Kapri-Pardes et al., 2007; Sun et al., 2007), PSII activity and its sensitivity to high-light conditions were measured in the seven genotypes by pulse amplitude modulation. As shown in Figure 2, all deg1 mutants had lower PSII activity compared with that in the wild type or deg5 and deg8 mutants even prior to experimental treatment. Upon exposure to higher light intensity for either 1 or 2 h, all genotypes, including the wild type, demonstrated a typical decrease in the activity of PSII, but this decrease was more pronounced in genotypes lacking Deg1. Moreover, deg15, deg18, and deg158 appeared to be slightly more sensitive to high-light treatment than deg1. Subjecting maximum efficiency of PSII photochemistry data to statistical analysis revealed that, in the three time points, the wild type, deg5, deg8, and deg58 grouped together, deg1 was significantly different, and deg15, deg18, and deg158 formed a third group (Fig. 2).
Figure 2.
deg1 mutants are more sensitive to high light than deg5 and deg8 mutants. Maximum efficiency of PSII photochemistry (Fv/Fm) was measured in the wild type (WT) and homozygous deg single, double, and triple mutants prior to experimental treatment (time 0) and after a 1- or 2-h exposure to 3-fold higher light intensity (200 versus 600 µmol photons m−2 s−1). Values are means ± se of four replicates. Data were analyzed using one-way ANOVA and contrast tests for each time point. Multiple comparisons were adjusted using the false discovery rate (α = 0.05). Shared letters indicate no statistically significant difference. Note that letters are applicable only within individual time points. Similar trends were observed in three repetitions of the experiment.
Together, these results suggest that the contribution of Deg1 to PSII repair and overall plant growth is higher than that of Deg5 and Deg8, providing a possible explanation for the differential visual phenotypes observed in Figure 1C. However, in the absence of Deg1, the contribution of Deg5 and Deg8 also was evident.
Deg5 and Deg8 Contribute to Plant Growth under Harsh Conditions
To further assess the possible physiological role of Deg5 and Deg8, we subjected the entire set of mutants to several suboptimal growth conditions. Generally, the addition of a stress factor, such as growing the plants in high-light or high-temperature conditions, augmented the mutant phenotype in all mutant plants that lacked Deg1 (Supplemental Fig. S2). Interestingly, when different stress treatments were combined, the phenotypical difference between deg1 and the double and triple mutants became more pronounced. When seedlings were grown at a very high density and at 30°C, deg15, deg18, and deg158 barely survived (Fig. 3A). In contrast, deg1, albeit with delayed plant development compared with the wild type and deg5 and deg8 mutants, grew under these severe conditions. Similarly, when individual plants were grown at 28°C and under high light intensity, deg15, deg18, and deg158 were considerably smaller than deg1 (Fig. 3B). These results suggest that Deg5 and Deg8 play a more substantial role under stress conditions than in optimal conditions. However, their contribution to overall plant performance under unfavorable growth conditions was smaller than that of Deg1, as deg5, deg8, and deg58 behaved more like the wild type than the different deg1 mutants (Fig. 3). It is interesting that, when the different genotypes were subjected to even harsher growth conditions, such as 34°C plus high light intensity, deg1 did not survive (Supplemental Fig. S3). This effect is consistent with the well-known role of bacterial Deg proteases in the response to heat (Clausen et al., 2011).
Figure 3.
Deg5 and Deg8 play a role in stress tolerance under severe conditions. A, Representative images of the wild type (WT) and homozygous deg mutants (genotypes are indicated adjacent to the images), sown at high density, that were grown in soil for 15 d under optimal conditions (growth light [GrL]: ∼160 µmol photons m−2 s−1, 18 h of light/6 h of dark, 22°C/18°C), then transferred to 30°C for an additional 13 d. B, Representative images of the wild type and homozygous deg mutants that were grown in soil for 21 d under optimal conditions, then transferred for 7 d into a greenhouse at 28°C with natural diurnal fluctuating light intensity (daylight [DL]: reaching up to ∼1,600 µmol photons m−2 s−1 at midday).
Proteomic Consequences of Knocking Out Deg Proteases
To unravel the molecular consequences of knocking out lumenal Deg proteases, which may underlie the aforementioned phenotypic differences, a comparative proteomic analysis was performed on the single deg1 and the triple deg158 mutants, lacking the Deg1 complex or both the Deg1 and Deg5-Deg8 complexes, respectively. Total leaf extracts were prepared from 25-d-old mutant and wild-type plants grown under optimal conditions (plants similar to those shown in Fig. 1C). These extracts were subjected to trypsin digestion followed by MS analysis and quantification of the identified proteins. Altogether, 2,569 different proteins were identified and quantified, with 1,698 identified by more than one unique peptide (Supplemental Table S2; Supplemental Data Set S1). Whereas quantification based on a single peptide tends to be less precise, identification is typically accurate (Shalit et al., 2015), which is the reasoning for the inclusion of single-peptide identifications in our data set. Of the entire list of proteins, 954 were annotated as chloroplast proteins, including 160 thylakoid proteins (cellular locations were according to the Plant Proteome Database [http://ppdb.tc.cornell.edu]; Sun et al., 2009). The levels of 1,105 proteins, 392 of which were chloroplast proteins, were altered significantly (P < 0.05) in mutants lacking the Deg1 complex or both the Deg1 and Deg5-Deg8 complexes when compared with the wild type (Supplemental Table S2; Supplemental Data Set S1B).
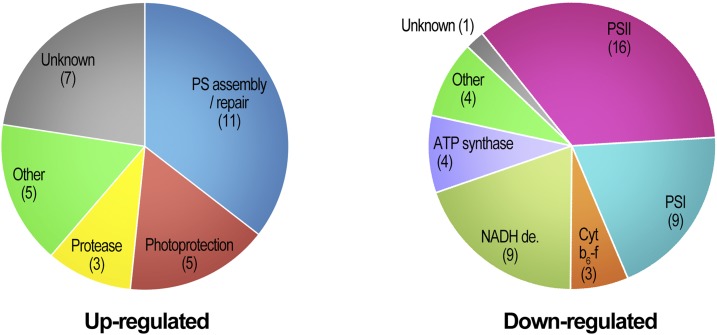
Of the chloroplast proteins whose levels were changed in the mutants, 75 were soluble (and peripheral) lumen proteins and integral thylakoid membrane proteins that may be exposed to the lumenal Deg proteases, which were of primary interest to us. Thirty-one of these proteins were found to be significantly up-regulated in deg mutants compared with wild-type plants (Fig. 4; Table 1; Supplemental Data Set S1C). Seventeen were up-regulated in both mutant genotypes. Ten of the proteins were lumenal proteins, and the rest were integral membrane proteins. The extent of overaccumulation was moderate, with most proteins showing a 15% to 50% increase in their level in the mutants compared with the wild type. Ten proteins demonstrated a greater than 1.5-fold increase in their steady-state level in at least one of the mutant genotypes. A number of up-regulated proteins were related to thylakoid biogenesis, PSI and PSII assembly, and repair (Table 1). Among these were TATB, involved in the translocation of precursor proteins across the thylakoid membrane (Jakob et al., 2009); ALB3, which is required for the insertion of LHCBs into the thylakoid membrane (Sundberg et al., 1997); the PSI assembly factor Y3IP1 (Albus et al., 2010); and LPA2, which is involved in the assembly of PSII (Ma et al., 2007). Proteins involved in photoprotection, such as CHL, which prevents thylakoid membrane lipid peroxidation (Levesque-Tremblay et al., 2009), components of the thylakoid FtsH protease complex, and a number of unknown proteins likewise were up-regulated. Of note, among the latter was STR4A, a rhodanese-like domain-containing protein, which was undetectable in the wild type and could be identified only in deg mutants (Table 1). All these up-regulated proteins might be Deg substrates, or alternatively, their up-regulation may result from a secondary effect of Deg depletion.
Figure 4.
Overview of up- and down-regulated thylakoid proteins in deg1 and deg158 mutants. The distribution among functional categories of proteins exhibiting significant changes in abundance in deg mutant backgrounds (Tables 1 and 2) is shown. Numbers of proteins in each category are indicated in parentheses.
Table 1. Thylakoid lumen and integral membrane proteins significantly up-regulated in deg1 and deg158 mutants compared with the wild type.
Values in boldface indicate greater than 50% increase in the steady-state level in the mutants.
| Gene Identifiera | Nameb | Functionb | deg1/Wild Type (P Value)c | deg158/Wild Type (P Value)c | Locationd |
|---|---|---|---|---|---|
| AT4G09010 | TL29 | PSII associated | ns | 1.21 (0.02) | L |
| AT1G06680 | PsbP1 | Oxygen evolution | 1.15 (0.05) | ns | L |
| AT5G44650 | Y3IP1 | PSI assembly | 2.50 (0.01) | 3.22 (0.00) | M |
| AT5G23120 | HCF136 | PSII assembly | 1.25 (0.00) | 1.27 (0.03) | L |
| AT1G02910 | LPA1 | PSII assembly | 1.30 (0.00) | 1.42 (0.01) | M |
| AT5G51545e | LPA2 | PSII assembly | 1.85 (0.01) | 1.65 (0.02) | M |
| AT4G28660 | Psb28 | PSII assembly | 1.35 (0.00) | 1.36 (0.02) | L |
| AT1G54500 | RubA | PSII assembly | 1.36 (0.02) | 1.48 (0.04) | M |
| AT1G03600 | Psb27-H1 | PSII assembly | ns | 1.13 (0.05) | L |
| AT1G54780 | TLP18.3 | PSII repair | 1.15 (0.03) | ns | L |
| AT3G55330 | PPL1 | PSII repair | 1.16 (0.02) | ns | L |
| AT2G28800 | ALB3 | Protein translocation/insertion | 1.53 (0.00) | 1.98 (0.01) | M |
| AT5G52440 | TATB | Protein translocation/insertion | 1.50 (0.00) | 1.39 (0.02) | M |
| AT4G01150 | CURT1A | Thylakoid architecture | ns | 1.20 (0.05) | M |
| AT3G47860 | CHL | Photoprotection | 1.22 (0.04) | 1.61 (0.00) | L |
| AT5G02120e | OHP1 | Photoprotection | 1.33 (0.03) | ns | M |
| AT1G34000 | OHP2 | Photoprotection | 1.12 (0.01) | ns | M |
| AT1G08550 | NPQ1 | Photoprotection | 1.34 (0.03) | 1.45 (0.04) | L |
| AT1G44575 | NPQ4 | Photoprotection | ns | 1.20 (0.02) | M |
| AT1G50250 | FtsH1 | Protease | 1.46 (0.00) | 1.59 (0.04) | M |
| AT2G30950 | FtsH2 | Protease | 1.20 (0.01) | 1.18 (0.03) | M |
| AT5G42270 | FtsH5 | Protease | 1.24 (0.00) | 1.41 (0.00) | M |
| AT4G09350e | NDHT | NADH dehydrogenase | 1.39 (0.02) | ns | M |
| AT4G17600 | LIL3.1 | Chlorophyll synthesis | 1.15 (0.05) | 1.31 (0.01) | M |
| AT5G17170 | ENH1 | Unknown | ns | 1.37 (0.03) | M |
| AT3G25480e | STR4A | Unknown | >106 (0.01) | >106 (0.00) | M |
| AT1G12250 | TL20.3 | Unknown | 1.37 (0.00) | 1.32 (0.00) | L |
| AT1G14345e | AT1G14345 | Unknown | 1.25 (0.04) | ns | M |
| AT4G02725e | AT4G02725 | Unknown | 1.57 (0.00) | ns | M |
| AT5G02160 | AT5G02160 | Unknown | 2.63 (0.02) | 3.11 (0.05) | M |
| AT5G51010e | AT5G51010 | Unknown | ns | 1.62 (0.02) | M |
According to TAIR (https://www.arabidopsis.org).
According to TAIR (https://www.arabidopsis.org), PPDB (http://ppdb.tc.cornell.edu), AtChloro (http://at-chloro.prabi.fr/at_chloro/), and/or UniProt (http://www.uniprot.org).
Student’s t test. ns, Nonsignificant change with P > 0.05.
Suborganellar location. L, Soluble in the lumen and/or peripheral to the membrane; M, thylakoid integral membrane protein.
Identified by a single peptide.
The 49 significantly down-regulated thylakoid proteins were dominated by components of the major integral membrane protein complexes responsible for photosynthetic electron transport, PSII, Cyt b6f, and PSI, and the NADH dehydrogenase and ATP synthase complexes (Fig. 4; Table 2; Supplemental Data Set S1C). Interestingly, multiple components of each complex demonstrated a similar trend of reduced accumulation. Here again, the extent of decrease was relatively moderate, approximately 15% to 30% less compared with wild-type levels of individual proteins. Nevertheless, this down-regulation leads most likely to a decrease in the photosynthetic capacity that is manifested in the smaller size of the deg1 and deg158 mutant plants. Other down-regulated proteins were RIQ1 and RIQ2, involved in grana stacking and LHCII dynamics (Yokoyama et al., 2016), two peptidyl-prolyl cis-trans-isomerases (Schubert et al., 2002), and one unknown protein.
Table 2. Thylakoid lumen and integral membrane proteins significantly down-regulated in deg1 and deg158 mutants compared with the wild type.
Values in boldface indicate greater than 30% decrease in the steady-state level in the mutants.
| Gene Identifiera | Nameb | Functionb | deg1/Wild Type (P Value)c | deg158/Wild Type (P Value)c | Locationd |
|---|---|---|---|---|---|
| AT3G27925 | Deg1 | Protease | <10−6(0.00) | <10−6(0.00) | L |
| AT4G18370 | Deg5 | Protease | 0.79 (0.02) | <10−6(0.00) | L |
| AT5G39830 | Deg8 | Protease | 0.63 (0.00) | 0.01 (0.00) | L |
| ATCG00020 | PsbA | PSII reaction center | ns | 0.75 (0.03) | M |
| ATCG00680 | PsbB | PSII reaction center | 0.79 (0.01) | 0.73 (0.02) | M |
| ATCG00280 | PsbC | PSII reaction center | 0.82 (0.01) | 0.75 (0.01) | M |
| ATCG00270 | PsbD | PSII reaction center | 0.82 (0.01) | 0.79 (0.01) | M |
| ATCG00580 | PsbE | PSII reaction center | 0.72 (0.01) | 0.65 (0.00) | M |
| ATCG00710 | PsbH | PSII reaction center | ns | 0.67 (0.03) | M |
| ATCG00560 | PsbL | PSII reaction center | 0.78 (0.05) | 0.57 (0.01) | M |
| AT1G79040 | PsbR | PSII reaction center | 0.78 (0.01) | 0.74 (0.04) | M |
| AT4G05180 | PsbQ2 | Oxygen evolution | 0.84 (0.04) | 0.77 (0.04) | L |
| AT3G21055 | PsbTN | Oxygen evolution | ns | 0.56 (0.03) | L |
| AT5G11450 | PPD5 | Oxygen evolution | 0.80 (0.01) | 0.69 (0.00) | L |
| AT3G61470 | LHCA2 | PSII antenna | 0.77 (0.01) | ns | M |
| AT5G54270 | LHCB3 | PSII antenna | 0.65 (0.00) | 0.56 (0.01) | M |
| AT5G01530 | LHCB4.1 | PSII antenna | 0.91 (0.04) | ns | M |
| AT2G40100 | LHCB4.3 | PSII antenna | 0.60 (0.01) | 0.46 (0.00) | M |
| AT4G10340 | LHCB5 | PSII antenna | 0.90 (0.03) | 0.84 (0.02) | M |
| ATCG00350 | PsaA | PSI reaction center | 0.67 (0.00) | 0.57 (0.01) | M |
| ATCG00340 | PsaB | PSI reaction center | 0.83 (0.02) | 0.67 (0.00) | M |
| AT4G02770 | PsaD1 | PSI reaction center | ns | 0.60 (0.03) | PS |
| AT1G31330 | PsaF | PSI reaction center | 0.73 (0.00) | 0.68 (0.00) | M |
| AT1G55670 | PsaG | PSI reaction center | 0.66 (0.03) | 0.40 (0.01) | M |
| AT1G08380e | PsaO | PSI reaction center | 0.02 (0.02) | ns | M |
| AT3G54890 | LHCA1.1 | PSI antenna | 0.75 (0.00) | 0.75 (0.01) | M |
| AT1G19150e | LHCA1.2 | PSI antenna | 0.57 (0.00 | 0.75 (0.02) | M |
| AT1G45474e | LHCA5 | PSI antenna | 0.44 (0.02) | ns | M |
| ATCG00540 | PETA | Cyt b6f complex | 0.76 (0.00) | 0.81 (0.01) | M |
| ATCG00720 | PETB | Cyt b6f complex | 0.74 (0.01) | 0.72 (0.01) | M |
| AT4G03280 | PETC | Cyt b6f complex | 0.76 (0.01) | 0.70 (0.02) | M |
| ATCG01100e | NDHA | NADH dehydrogenase | 0.60 (0.01) | 0.48 (0.01) | M |
| ATCG01010 | NDHF | NADH dehydrogenase | 0.85 (0.02) | 0.78 (0.03) | M |
| ATCG00430 | NDHK | NADH dehydrogenase | 0.82 (0.03) | ns | PS |
| AT5G58260 | NDHN | NADH dehydrogenase | 0.82 (0.02) | 0.65 (0.01) | PS |
| AT4G23890 | NDHS | NADH dehydrogenase | 0.68 (0.00) | 0.65 (0.00) | PS |
| AT5G21430 | NDHU | NADH dehydrogenase | 0.79 (0.04) | 0.71 (0.04) | M |
| AT2G04039 | NDHV | NADH dehydrogenase | 0.61 (0.02) | 0.51 (0.043) | PS |
| AT1G15980 | PnsB1 | NADH dehydrogenase | 0.72 (0.01) | 0.73 (0.01) | L |
| AT5G43750 | PnsB5 | NADH dehydrogenase | 0.73 (0.00) | 0.73 (0.01) | L |
| ATCG00120 | ATPA | ATP synthase complex | ns | 0.90 (0.03) | PS |
| AT4G09650 | ATPD | ATP synthase complex | 0.76 (0.01) | ns | PS |
| AT4G32260 | ATPG | ATP synthase complex | 0.82 (0.03) | ns | M |
| ATCG00150e | ATPI | ATP synthase complex | 0.57 (0.01) | ns | M |
| AT5G08050 | RIQ1 | Thylakoid architecture | 0.40 (0.00) | 0.33 (0.00) | M |
| AT1G74730e | RIQ2 | Thylakoid architecture | 0.63 (0.00) | 0.58 (0.01) | M |
| AT1G18170 | FKBP17-2 | Protein folding | ns | 0.81 (0.04) | L |
| AT5G13410 | FKBP19 | Protein folding | ns | 0.66 (0.04) | L |
| AT5G42765 | AT5G42765 | Unknown | 0.59 (0.00) | 0.56 (0.00) | M |
According to TAIR (https://www.arabidopsis.org).
According to TAIR (https://www.arabidopsis.org), PPDB (http://ppdb.tc.cornell.edu), AtChloro (http://at-chloro.prabi.fr/at_chloro/), and/or UniProt (http://www.uniprot.org).
Student’s t test. ns, Nonsignificant change with P > 0.05.
Suborganellar location. L, Soluble in the lumen and/or peripheral to the membrane; M, thylakoid integral membrane protein; PS, peripheral on the stromal side of the thylakoid membrane.
Identified by a single peptide.
The effect of Deg depletion in the lumen was transduced also to other chloroplast subcompartments, most likely as an indirect effect. More than 60 chloroplast proteins, either peripheral to the thylakoid membrane on its stromal side, stromal soluble proteins, or envelope proteins, were up-regulated in the mutant plants by more than 50% (Supplemental Data Set 1C). These included nucleoid proteins required for plastid gene expression, like PTAC2, PTAC5, PTAC7, and PTAC12; proteins involved in translation, like RPL15, RPL21, and RPL36; or ATAB2. Other overaccumulating proteins were those involved in lipid metabolism (PES1 and MGD1), proteases (PREP1, PREP2, and FtsH11), and chaperones and assembly factors (CR88, RAF1, and SRP43), as well as many other less characterized proteins. Only 10 proteins were down-regulated by more than 50%, the best characterized of which were proteins involved in starch debranching (ISA1), amino acid biosynthesis (IPMI2 and TSB), and ferritin.
Deg5 and Deg8 Can Only Partially Substitute for the Lack of Deg1
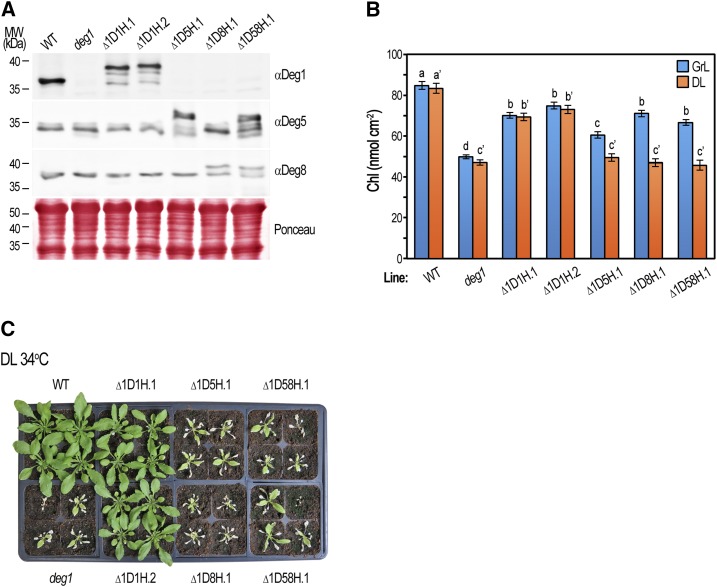
The aforementioned observation that Deg5 and Deg8 contribute to growth under suboptimal conditions raised the question of whether they are functionally equivalent to Deg1. To answer this, we attempted to complement the phenotype of the deg1 mutant by overexpressing Deg5 and Deg8. The deg1 line was transformed with genomic DNA constructs containing Deg1, Deg5, or Deg8, all with C-terminal His tags, under the control of their respective native promoters (Supplemental Tables S3 and S4). As shown in Figure 5A, the different transgenic lines accumulated the corresponding gene products. As expected, the mobility of the introduced proteins was slower compared with that of the endogenous variants due to the addition of the C-terminal tags. In the ∆1D5H, ∆1D8H, and ∆1D58H lines, the presence of both the transgene and the corresponding endogenous gene resulted in the overexpression of Deg5, Deg8, or both, respectively.
Figure 5.
Deg5 and Deg8 only partially complement the deg1 mutant. The wild type (WT), deg1 mutant, and deg1 mutant complemented with Deg1-His (∆1D1H.1, ∆1D1H.2), Deg5-His (∆1D5H.1), Deg8-His (∆1D8H.1), or Deg5-His/Deg8-His (∆1D58H.1) were germinated on MS agar with or without antibiotic selection (Supplemental Table S4) for 8 d, then seedlings were transferred to soil and grown under optimal conditions for 13 d (growth light [GrL]: ∼160 µmol photons m−2 s−1, 18 h of light/6 h of dark, 22°C/18°C). Experimental plants were either transferred to a greenhouse at 22°C or 34°C with natural diurnal fluctuating light intensity (daylight [DL]: maximum of ∼1,600 μmol photons m−2 s−1 at midday) or kept under growth light conditions for an additional 7 d. A, Immunoblot analysis of protein content in the various genotypes (genotypes are indicated above the immunoblots) following 20 d of growth under optimal conditions (Supplemental Fig. S4A). Total protein was extracted from rosette leaves, separated by SDS-PAGE, and detected with the specific antibodies indicated on the right. The migration of molecular mass (MW) markers is indicated on the left. Five micrograms of chlorophyll was loaded in each lane. Ponceau staining of membranes was used as a loading control. B, Chlorophyll (Chl) content in plants grown at 22°C under growth light (blue bars) and daylight (orange bars) conditions (representative images of plants are shown in Supplemental Fig. S4, A and B, respectively). Values are means ± se of 16 replicates (four plants per genotype, four rosette leaves per plant). Data were analyzed using one-way ANOVA for each lighting condition. Shared letters indicate no statistically significant difference. Note that letters are applicable only within individual lighting treatments. C, Representative images of the wild type, deg1 mutant, and deg1 complemented plants (genotypes are indicated adjacent to the images) after 7 d of growth at 34°C in the greenhouse under daylight conditions.
Two independent ∆1D1H lines, expressing Deg1-His, demonstrated phenotypic complementation of the deg1 phenotype in all examined conditions, attributed to the presence of the transgene (Fig. 5C; Supplemental Fig. S4). The lines overexpressing Deg5, Deg8, or both were comparable to the deg1 line complemented with Deg1-His when grown under low light intensity (Supplemental Fig. S4A), suggesting that Deg5 and Deg8 can functionally substitute for the lack of Deg1. However, under stress conditions, such as growth under high light at 34°C, Deg1 alone, and not Deg5 and Deg8, was able to rescue the deg1 lethal phenotype (Fig. 5C). Using chlorophyll content as a quantifiable parameter, it was evident that Deg5 and Deg8 overexpression could complement the effect of the deg1 allele under low-light conditions, although not to the level of the Deg1 transgene, but they could not substitute for the lack of Deg1 under high light, even at the optimal temperature of 22°C (Fig. 5B; Supplemental Fig. S4B). Incomplete deg1 complementation by the Deg1-His transgene may have resulted from germination of the transgenic lines on antibiotic selection, which caused a delay in plant growth (data not shown).
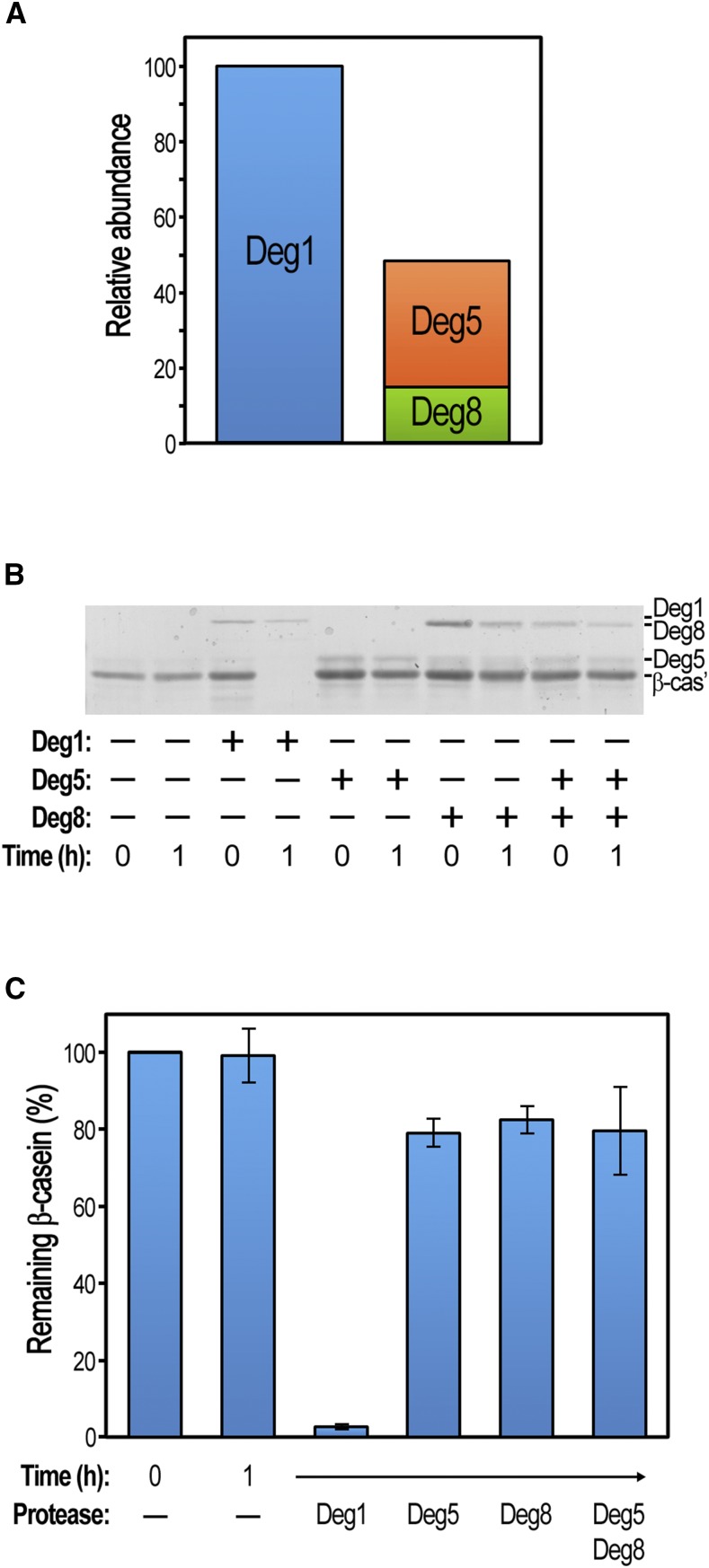
Deg1 Is 2-Fold More Abundant Than Deg5 and Deg8
One possible reason for the weaker effect of knocking out Deg5 and Deg8 compared with Deg1 could be their differential accumulation in the wild type. To test this possibility, the levels of the three proteins in wild-type plants, grown under optimal conditions, were determined by MS. Using known quantities of diagnostic synthetic peptides as a reference, we calculated the levels of Deg1, Deg5, and Deg8 to be 8.4, 2.81, and 1.24 mol × 10−14 per 100 µg of total protein, respectively (Supplemental Table S3; these numbers correspond to 3, 0.8, and 0.5 ng per 100 µg of total protein, respectively). Assuming that Deg5 and Deg8 form a heterocomplex (Sun et al., 2007), this complex is expected to be 2-fold less abundant than the Deg1 homocomplex (Fig. 6A). Thus, this difference in protein abundance could contribute to the differential effect of knocking out either Deg1 or Deg5-Deg8.
Figure 6.
Deg1 displays greater abundance and activity compared with Deg5-Deg8. A, Absolute amounts of the Deg proteases in rosette leaves of 25-d-old wild-type plants as determined by liquid chromatography-MS. Values are means of two independent replicates (Supplemental Table S3). The relative abundances of Deg1 and Deg5-Deg8 complexes are presented; 100% represents 8.41 × 10−14 mol per 100 μg of total protein. B, In vitro proteolytic degradation assay. Fifty picomoles of β-casein was mixed with 5 pmol of recombinant Deg proteases. After 1 h of incubation at 37°C, proteins in the reaction mixtures were separated by SDS-PAGE and the gels were stained with Coomassie Blue. Band identity is indicated on the right. A representative gel is shown. C, Comparison of remaining β-casein following proteolytic degradation assays, determined by quantifying protein bands as depicted in B. Values are means ± se of three independent experiments.
Recombinant Deg1 Protease Is More Active Than Deg5 and Deg8
Another possible source for the differential contribution of Deg1, Deg5, and Deg8 to plant growth and survival could be the differences in their proteolytic activity. Whereas the proteolytic activity of recombinant Deg1 has been demonstrated in a number of studies (Chassin et al., 2002; Kapri-Pardes et al., 2007; Kley et al., 2011), recombinant Deg8 was reported to be less active than Deg1 (Sun et al., 2013), and Deg5 was apparently inactive (Sun et al., 2007). These fragmentary reports prompted us to compare the activity of all three proteases in the same experiment. To this end, the three recombinant proteins were expressed and purified using the same protocol. A typical in vitro degradation assay, using β-casein as a model substrate, is presented in Figure 6B. When a 10-fold molar excess of β-casein over enzyme was present in the reaction mixture, almost no β-casein remained in the presence of Deg1, similar to previous reports (Chassin et al., 2002; Kley et al., 2011; Zienkiewicz et al., 2012). On the other hand, very little β-casein degradation could be observed upon incubation of the substrate with Deg5, Deg8, or both. Quantification of a typical experiment showed that only 20% to 30% of β-casein could be degraded by Deg5, Deg8, or both, whereas, under the same conditions, Deg1 degraded more than 95% of β-casein (Fig. 6C). Similar trends were observed in degradation assays using α-lactalbumin as a substrate (Supplemental Fig. S5). Thus, Deg1 indeed displays greater proteolytic activity than Deg5 and Deg8, which are considerably less efficient and do not display higher activity levels when combined compared with individual proteins.
Potential Substrates Are Trapped in Vivo by Deg5 and Deg8 But Not by Deg1
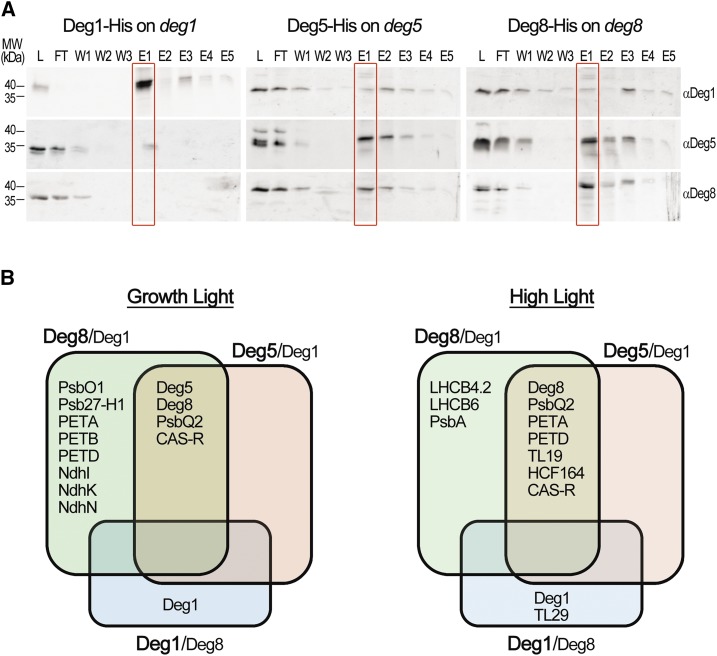
A previous in vitro study demonstrated that purified active Deg1 protein could coprecipitate multiple thylakoid proteins from a plant protein extract (Zienkiewicz et al., 2012). Together with the aforementioned impaired proteolytic activity of Deg5 and Deg8, this prompted us to attempt trapping substrates in vivo using the affinity enrichment mass spectrometry (AE-MS) approach (Keilhauer et al., 2015). To this end, deg1, deg5, and deg8 mutants, each complemented with the corresponding His-tagged Deg protein, were generated and analyzed (Supplemental Table S3). First, total protein extracts were prepared from the three different lines, detergent solubilized, and affinity precipitated (see “Materials and Methods”). Samples from the affinity precipitation procedure were subjected to immunoblot analysis with antibodies against the three Deg proteases. Figure 7A demonstrates that the precipitated material from each complemented line contained the respective His-tagged Deg protein. Whereas considerable amounts of Deg1 could be detected only in the material precipitated and eluted from the Deg1-tagged line, Deg5 and Deg8 were coprecipitated from Deg8- and Deg5-tagged lines, respectively (Fig. 7A), consistent with the notion that Deg5 and Deg8 form a heterocomplex.
Figure 7.
Potential substrates are trapped by Deg5 and Deg8 in epitope-tagged lines. Transgenic plants expressing His-tagged Deg proteins in their corresponding mutant backgrounds were subjected to an affinity enrichment procedure using Ni-IDA beads (see “Materials and Methods”). A, Immunoblot analysis of the enriched proteins from 25-d-old plants grown under 160 µmol photons m−2 s−1 (growth light). Protein samples were taken from each step of the affinity enrichment procedure, then resolved by SDS-PAGE and detected using specific antibodies as indicated on the right. The migration of molecular mass (MW) markers is indicated on the left. L, Proteins loaded on the beads; FT, flow through (unbound proteins); W1 to W3, washed proteins; E1 to E5, eluted proteins. B, Summary of the AE-MS results presented in Supplemental Table S4. Plants were grown under either growth light or 80 µmol photons m−2 s−1 and then transferred to 300 µmol photons m−2 s−1 for 7 h (high light). Plants were subjected to the same affinity enrichment procedure described above, and the precipitated proteins were subjected to MS analysis. Levels of proteins associated with Deg5 and Deg8 were compared with those associated with Deg1, and those associated with Deg1 were compared with those associated with Deg8.
After confirming that the tagged proteases could indeed be precipitated as expected, AE-MS experiments were carried out on total protein extracts from the three complemented lines. The samples were collected either under optimal lighting conditions or after exposing the plants to 7 h of elevated light intensity (∼4×). All proteins identified and quantified in the precipitated material from each line are presented in Supplemental Data Set S2. To look for coprecipitated proteins enriched in each sample, we compared their level in the bait sample (Deg1-His, Deg5-His, or Deg8-His) with that found in their negative controls (i.e. the wild type or another Deg-His line). We considered as significant interactors only thylakoid proteins whose fold change was greater than 2 and whose P value in Student’s t test was less than 0.05. A summary table depicting all thylakoid proteins that passed this statistical threshold (but also those with the less stringent cutoff of greater than 1.5) can be found in Supplemental Table S4. Thylakoid proteins coprecipitated with Deg5 and Deg8 proteases versus those coprecipitated with Deg1 are presented in Figure 7B. This analysis highlights a number of points. (1) The association between Deg5 and Deg8, and the lack of significant association between Deg1 and either Deg5 or Deg8, are reconfirmed. (2) All proteins precipitated from the tagged Deg5 line also were precipitated from the Deg8-His line. (3) In addition to shared proteins, a number of other proteins were coprecipitated with Deg8. (4) Only one additional protein was coprecipitated with Deg1, and only from plants grown under high light. It should be noted that, when the wild type rather than His-tagged Deg proteins was used as a control, several proteins were not enriched significantly (Supplemental Table S4). We believe that this stems from differential affinities to the matrix among nontagged proteins and that the more relevant control is that of His-tagged proteins rather than that of wild-type proteins.
Proteins interacting with Deg5 and Deg8 proteases included both membrane and soluble proteins. Three subunits of the Cyt b6f complex, PETA, PETB, and PETD, all integral membrane proteins, were recovered together with the Deg5-Deg8 complex from plants grown under optimal conditions or exposed to high-light conditions (Fig. 7B). Similarly, PsbQ2, the 16-kD subunit of the peripheral oxygen-evolving complex of PSII, was associated with the precipitated complex. Another protein found under both conditions was CAS-R, a thylakoid calcium-sensing receptor (Vainonen et al., 2008; Weinl et al., 2008). The coprecipitation of these proteins with both Deg5 and Deg8 under both growth conditions suggests that they are likely to be trapped substrates. Additional proteins coprecipitating with Deg8 from growth light plants include PsbO1, the 33-kD subunit of the oxygen-evolving complex; Psb27-H1, a protein involved in the repair of PSII from oxidative damage (Chen et al., 2006); and three different subunits of the NADH dehydrogenase complex, NdhI, NdhK, and NdhN. Exposure to high light resulted, as expected, in coprecipitation of the D1 protein of the PSII reaction center (PsbA) and also two antenna proteins of PSII: CP24 and CP29 (LHCB6 and LHCB4.2, respectively). This suggests that Deg complexes are involved not only in the repair of PSII but also in the adjustment of antenna size to elevated light intensities. Two other proteins that were coprecipitated under these conditions were TL19, a thylakoid lumen protein (Schubert et al., 2002) of unknown function, and HCF164, involved in the biogenesis of the Cyt b6f complex (Lennartz et al., 2001). The only protein associated with Deg1 under high light, but not with Deg5-Deg8, was TL29, a thylakoid lumen soluble protein whose function is unknown (Granlund et al., 2009). It is interesting that two of the coprecipitated proteins identified in these assays, TL29 and Psb27-H1, were found to be slightly up-regulated in the triple deg158 mutant (Table 1), suggesting that these proteins might be in vivo substrates of Deg proteases.
DISCUSSION
The presence of homologous proteins in the same cellular compartment raises the question of whether their function is redundant or they fulfill distinct functions. Previous studies identified two separate Deg protease complexes in the thylakoid lumen, Deg1 and Deg5-Deg8, and demonstrated their function in the repair cycle of PSII in response to photoinhibitory growth conditions (Kapri-Pardes et al., 2007; Sun et al., 2007). To explore the relative contribution of these complexes to plant growth, single, double, and triple Deg KO Arabidopsis mutants were generated and their phenotypes compared under different growth conditions. In all of the examined growth conditions, lines lacking Deg1 displayed a more affected mutant phenotype compared with deg5, deg8, and deg58 mutants; however, Deg5-Deg8 also was shown to contribute to plant growth, particularly under different suboptimal conditions (Figs. 1–3).
The differing physiological importance of the two homologous Deg complexes may be for several reasons. First, Deg1 has much stronger proteolytic activity compared with Deg5, Deg8, or the Deg5-Deg8 heterocomplex (Fig. 6, B and C). The inefficient proteolytic activity of Deg5 and Deg8 was reported previously (Sun et al., 2007, 2013) and can be attributed to both the distorted conformation of the catalytic triad in both enzymes and the twisted orientation of the PDZ domain of Deg8, as revealed by x-ray crystallography (Sun et al., 2013). In the latter work, it was suggested that substrate binding might confer an allosteric conformational change at the Deg8 active site, thus activating it. Second, Deg1, at least under optimal growth conditions, is 2-fold more abundant than Deg5-Deg8 (Fig. 6A); therefore, Deg1 loss may have a more detrimental consequence than the loss of Deg5-Deg8. This suggestion is in line with the results of the complementation assays, where overexpression of Deg5 and Deg8 could partially compensate for the loss of Deg1, but only under optimal growth conditions (Fig. 5). Thus, the observed partial complementation indicates that the two complexes perform analogous functions, probably by degrading similar substrates, but the levels of their individual contributions are different. In this context, it should be noted that at least part of the effects observed in the Deg1 KO mutant might result from a loss of chaperone activity rather than proteolytic activity. This issue requires further investigation in future studies.
The lack of a visual phenotype in the deg5, deg8, and deg58 mutants grown under optimal conditions (Fig. 1C) is reminiscent of the original report on the same mutant lines (Sun et al., 2007). However, Sun et al. (2007) have shown that, when grown in a greenhouse, these mutants were much smaller than the wild type, whereas no such differences were observed in any of our growth experiments under adverse conditions (Fig. 3; Supplemental Fig. S3). We cannot offer any plausible explanation for this discrepancy, except that light regimes were different in the two studies. Yet, the comparison of all possible mutant combinations of thylakoid Deg proteases in the same study does allow us to assess the relative contribution of each Deg complex to growth under different environmental conditions.
Despite fundamental differences between the lumenal Deg proteases and the stroma-oriented thylakoid FtsH protease complex (Ser versus metalloprotease, ATP independent versus ATP dependent, peripheral versus integral membrane proteins), the cooperation between the two proteases in the degradation of integral thylakoid membrane proteins is now well accepted (Kato et al., 2012). Nevertheless, their individual contributions to the overall functioning of chloroplasts are different, as can be inferred from an analysis of mutants. We demonstrate in this study that plants can grow and complete their life cycle without lumenal Deg proteases unless they are challenged with severe stress conditions. This is not the case with the thylakoid FtsH protease. Mutants lacking this thylakoid complex are lethal and can grow only on Suc as albino plants. These observations place the thylakoid Deg proteases in a second tier of functional importance for the proper development and operation of chloroplasts. This hierarchy also is consistent with the proposed roles of Deg and FtsH proteases in the degradation of multispanning integral membrane proteins; Deg proteases cleave connecting loops and termini exposed to the lumen, thus facilitating the extraction of transmembrane helices by the oppositely oriented FtsH complex.
Subunits of the major complexes of the thylakoid membrane, specifically PSII, Cyt b6f, PSI, PSII and PSI antenna, ATP synthase, and NADH dehydrogenase, were down-regulated in both deg1 and deg158 mutants (Fig. 4; Table 2). It should be noted that the stroma-exposed peripheral subunits of these complexes were down-regulated as well, indicating that the regulation is on the whole-complex level. This down-regulation, of 20% to 30% on average, probably is the reason for the observed reduced growth of the mutants even when grown under optimal conditions. The loss of Deg1 in the single and triple mutants was accompanied by an increase in the level of FtsH complex components (Fig. 4; Table 1). This most likely represents a compensating response to the decrease in proteolytic capacity in thylakoids rather than the stabilization of substrates in the absence of a protease responsible for their degradation. Other up-regulated thylakoid proteins included proteins involved in photoprotection, proteins facilitating the insertion of other proteins into the thylakoid membrane, and proteins that mediate the assembly and repair of photosystems. Here again, the increase in the level of these may reduce oxidative damage and increase the capacity of photosystem repair. It is thus tempting to hypothesize that reduced proteolytic capacity in the lumen leads to the down-regulation of photosynthetic complexes, which, in turn, results in the up-regulation of folding, assembly, and repair factors. How the reduction in proteolytic capacity is sensed and transduced to result in specific changes in protein accumulation is currently unknown. These questions will be explored in future studies.
The up-regulated proteins also may be Deg substrates, as they are exposed physically to Deg proteases in the wild type. Indeed, one of these proteins, NPQ4 (PsbS), was identified previously as a Deg1 substrate based on in vitro experiments (Zienkiewicz et al., 2012). Furthermore, several up-regulated thylakoid proteins were found to interact with the Deg proteins in the pull-down assays (Fig. 7B; Supplemental Table S4; Supplemental Data Set S2E), suggesting that they might be substrates of Deg proteases. These included Psb27-H1, PsbP1, TLP18.3, HCF136, and TL29. The lumenal protein TL18.3, whose function is unknown (Granlund et al., 2009), interacted with Deg5 or Deg8 both under growth-light conditions and following a photoinhibitory high-light treatment. On the other hand, Psb27-H1, which is involved in PSII assembly and repair (Chen et al., 2006), and the oxygen-evolving complex subunit PsbP1 were pulled down with Deg5 or Deg8 only under normal conditions. The identification of these proteins by two independent approaches suggests that they are in vivo substrates of lumenal Deg proteases.
It was not surprising to identify the D1 protein of the PSII reaction center (PsbA) associated with Deg8 in plants exposed to high light (Fig. 7B). D1 was identified previously as a substrate for both Deg8 (Sun et al., 2007) and Deg1 (Kapri-Pardes et al., 2007; Zienkiewicz et al., 2012), the latter demonstrating degradation only following high-light treatment. Interestingly, PSII antenna proteins also were coprecipitated under these conditions, suggesting that Deg proteases participate in the proteolytic adjustment of antenna size to increasing light intensities.
PsbQ2, a component of the OEC of PSII, subunits of the Cyt b6f complex, and the calcium-sensing receptor CAS-R (Vainonen et al., 2008; Weinl et al., 2008) interacted with the Deg5 and Deg8 proteases both in plants grown under growth light and those exposed to an additional increased light intensity (Fig. 7B). In contrast, three different subunits of the thylakoid NADH dehydrogenase complex were associated with Deg8 in plants grown under optimal conditions but not after exposure to high light. The reason for this differential behavior is not clear to us at this time; however, it should be noted that these proteins were down-regulated or unchanged in the mutant plants.
It is interesting that several of the proteins identified as Deg5-Deg8 interactors in our AE-MS experiment, specifically D1, D2 (PsbD), PsbS (NPQ4), LHCB6 (CP24), LHCB4 (CP29), Cyt b6 (PetB), PsbO, TLP18.3, and Ptac-16, were shown previously to interact with Deg1 as well (Zienkiewicz et al., 2012). This may explain the ability of Deg5-Deg8 overexpression to compensate partially for Deg1 loss (Fig. 5B). The fact that none of these proteins coprecipitated with Deg1-His in our study is likely due to different experimental settings: in vitro interaction between recombinant Deg1-His and solubilized thylakoid proteins in the study of Zienkiewicz et al. (2012) versus in vivo interaction in this report. The effective proteolytic activity of Deg1 may leave it devoid of substrates in the in vivo experiment, whereas the less potent Deg5-Deg8 complex behaves like a substrate trap. Further studies are needed to determine which of the proteins identified here are indeed direct substrates of lumenal Deg proteases and how they are distinguished from nonsubstrate proteins.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type and mutant Arabidopsis (Arabidopsis thaliana) ecotype Columbia plants were grown under long-day conditions (16 h of light/8 h of dark, 22°/18°C) with a photon flux density of ∼160 µmol m−2 s−1. For testing suboptimal growth conditions, plants were transferred to a temperature-controlled greenhouse, where they were exposed to natural diurnal light intensities, reaching a maximum of ∼1,600 µmol photons m−2 s−1 at midday and constant temperatures of 22°C, 28°C, or 34°C.
To select for antibiotic-resistant plants, surface-sterilized seeds were sown on sterile 0.5× Murashige and Skoog medium with 1% (w/v) Suc and B5 vitamins, stratified for 2 d at 4°C in the dark, and then grown under long-day conditions for 14 to 16 d (six to eight rosette leaves). The resistant seedlings were transferred to pots filled with moistened Kekkila peat. Alternatively, seeds were sown directly on peat and treated as above.
Generation of Mutant and Transgenic Lines, Genotyping, and Expression Analysis
The T-DNA insertion line Gabi-Kat_414D07 (Deg1; At3g27925) was obtained from the Nottingham Arabidopsis Stock Centre. The identity of the line was confirmed by PCR using gene- and T-DNA-specific primers (Supplemental Materials and Methods S1). Single mutants were backcrossed to the wild-type plants three times to remove additional mutations, and deg1 homozygous plants for the T-DNA insertions (identified by PCR analyses) were isolated.
The T-DNA insertion lines SALK_099162 (Deg5; At4g18370), SALK_004770 (Deg8; At5g39830), and a deg58 double mutant line, described previously by Sun et al. (2007), were a kind gift of Lixin Zhang (Chinese Academy of Sciences). The deg15 and deg18 double mutants and the deg158 triple mutant were obtained by crossing the single mutant deg1 with the double mutant deg58 and screening the F2 population by PCR using the gene- and T-DNA-specific primers described in Supplemental Materials and Methods S1.
Transgenic lines expressing His-tagged Deg proteins were generated using directional cloning of PCR products with Gateway technology. The Deg1, Deg5, and Deg8 promoter and coding sequences were amplified from wild-type genomic DNA with attB-modified custom primers, as specified in Supplemental Materials and Methods S1. The PCR products were recombined with the pDONR221 vector using Gateway BP-Clonase II Enzyme Mix (Invitrogen), yielding entry clones. Plasmids expressing the Deg genomic DNA with C-terminal His tags were constructed by performing LR-trans reactions between the pDONR221-Deg entry vectors and the pGWB407/pGWB607 destination vectors using the Gateway LR-Clonase II Enzyme Mix (Invitrogen). All inserted sequences and orientations in the plasmids were confirmed by DNA sequencing. All the generated vectors were propagated in DH5α Escherichia coli cells (Stratagene). The sequence-verified constructs were introduced into Agrobacterium tumefaciens strain GV3101 pMP90 (Koncz and Schell, 1986). Transformation of the Arabidopsis deg1, deg5, and deg8 mutants was performed by the floral dip method (Clough and Bent, 1998). Independent BASTA- or kanamycin-resistant T1 plants were obtained and confirmed to contain the His-tagged Deg inserts by PCR. The genotypes of the transgenic lines prepared in this work are summarized in Supplemental Table S4.
For expression analysis, total RNA was extracted using the Total RNA Isolation System (Promega), residual genomic DNA was removed by TURBO DNase (Ambion), and cDNA was generated as described previously (Katz et al., 2004) using SuperScript II Reverse Transcriptase (Invitrogen), all according to the manufacturer’s instructions.
Protein Extraction and Immunoblot Analysis
To extract thylakoid proteins, fresh leaf tissues (200 mg) were ground to fine powder that was suspended in 1 mL of ice-cold 10 mm HEPES, pH 8, supplemented with 2 mm EDTA. The homogenate was filtered through four to six gauze layers in a 1-mL syringe and centrifuged at 5,000g and 4°C for 5 min. The pellet was resuspended in 0.3 mL of ice-cold 10 mm HEPES, pH 8, layered onto 0.5 mL of 40% (v/v) Percoll in 10 mm HEPES, pH 8, and centrifuged at 2,500g and 4°C for 10 min. The thylakoid-containing band (at the interface between Percoll and HEPES) was collected, resuspended in 7 mL of ice-cold 10 mm HEPES, pH 8, and centrifuged at 5,000g and 4°C for 15 min. The pellet was resuspended in 50 μL of 4× SDS sample buffer (0.2 m Tris-HCl, pH 6.8, 5 m urea, 8% [w/v] SDS, 10% [v/v] glycerol, and 20% [v/v] β-mercaptoethanol) supplemented with 0.5 mm PMSF and 0.5 mm DTT and incubated for 5 min at room temperature. Samples were centrifuged at 5,000g and 4°C for 2 min, and the supernatant was subjected to SDS-PAGE.
Total protein extracts were obtained by grinding 180 mg of leaf tissue in 300 µL of 4× SDS sample buffer supplemented with 0.5 mm PMSF and 0.5 mm DTT. Samples were incubated at room temperature for 15 min and then centrifuged at 13,000g for 10 min, and the supernatant was subjected to SDS-PAGE. Blots were reacted with an antibody generated against the mature Deg1 (Chassin et al., 2002), Deg5, and Deg8 proteins (Agrisera), diluted 1:1,000, and an anti-rabbit horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich), diluted 1:10,000. All blots were developed using a homemade ECL solution (1.25 mm luminol, 0.198 mm paracumaric acid, 0.1 m Tris-Cl, pH 8.5, and 0.01% [v/v] H2O2).
Chlorophyll Content and Chlorophyll Fluorescence Measurements
Chlorophyll content was measured spectroscopically as described previously (Adam et al., 2011), using the equations of Porra (2002). Measurements of chlorophyll fluorescence and the determination of PSII maximum quantum yield were done essentially as described by Zaltsman et al. (2005), with the following changes: dark adaptation was for 30 min; the pulse amplitude-modulated fluorometer used was PAM-2000 (Heinz Walz); and F0 was determined at 0.8 µmol photons m−2 s−1 and Fm during a 1-s pulse of 3,000 µmol photons m−2 s−1.
Affinity Enrichment of Thylakoid Proteins Interacting with His-Tagged Deg Proteases
Proteins were extracted from transgenic lines by grinding 250 mg of rosette leaf tissue in 750 µL of lysis buffer (0.3 m sorbitol, 150 mm NaCl, and 50 mm HEPES-KOH, pH 8) supplemented with 1% (w/v) n-dodecyl-β-d-maltoside (Tivan-Biotech) and Protease Inhibitor Cocktail for plant (Sigma-Aldrich) and incubating on a tube rotator for 30 min at 60 rpm and 4°C. To remove insoluble material, extracts were centrifuged for 5 min at 14,000g and 4°C before the supernatants were centrifuged again for 15 min at 29,000g and 4°C. The cleared extracts were incubated on a tube rotator for 1 h at 60 rpm and 4°C with 100 µL of Ni-IDA beads slurry (Adar Biotech), which was prewashed with binding buffer (0.02 m sodium phosphate and 0.5 m NaCl, pH 7.4). The beads were precipitated by centrifugation at 500g and 4°C for 3 min and washed three times with 1 mL of binding buffer supplemented with protease inhibitors.
For immunoblot analysis, the proteins were eluted from the beads by incubation in 100 µL of elution buffer (0.02 m sodium phosphate, 0.5 m NaCl, and 0.5 m imidazole, pH 7.4) for 5 min. Equal volumes of bound and unbound proteins were used for immunoblotting. The membranes were then incubated with anti-Deg antibodies.
MS-Based Proteomics
For comparative proteomics analysis of wild-type and deg mutant plants, freeze-dried leaf tissue samples were prepared using the filter-aided sample preparation method (Shalit et al., 2015). Briefly, samples were dissolved in 500 µL of SDT buffer (4% [w/v] SDS, 100 mm Tris-HCl, pH 7.6, and 0.1 m DTT) and lysed for 3 min at 95°C. The lysate was then centrifuged at 16,000g for 10 min. A total of 100 µg of extracted proteins was mixed with 200 µL of UA buffer (8 m urea in 100 mm Tris-HCl, pH 8, and 50 mm ammonium bicarbonate) and loaded onto 30-kD cutoff filters and spun down. A total of 200 µL of UA buffer was added to the filter unit and centrifuged at 14,000g for 40 min. Alkylation of Cys was performed using iodoacetamide with two subsequent washes with ammonium bicarbonate, followed by incubation with trypsin (Promega) at a 1:50 trypsin:protein ratio for 16 h at 37°C.
For the analysis of proteins coprecipitated with Deg-His, the samples obtained by affinity enrichment were subjected to on-bead tryptic digestion. The beads were washed three times with 1 mL of PBS buffer. Urea at 8 m in 0.1 m Tris, pH 7.9, was added onto PBS-washed beads and incubated for 15 min at room temperature. Proteins were reduced by incubation with DTT (5 mm; Sigma) for 60 min at room temperature and alkylated with 10 mm iodoacetamide (Sigma) in the dark for 30 min at room temperature. Urea was diluted to 2 m with 50 mm ammonium bicarbonate. A total of 250 ng of trypsin (Promega) was added and incubated overnight at 37°C followed by the addition of 100 ng of trypsin for 4 h at 37°C. Digestions were stopped by the addition of trifluoroacetic acid (1% [v/v] final concentration). Following digestion, peptides were desalted using Oasis HLB μElution format (Waters), vacuum dried, and stored at −80°C until further analysis.
Samples were analyzed by liquid chromatography/mass spectrometric analysis using MS1 intensity-based label-free quantification (Shalit et al., 2015). They were subjected to splitless nano-ultra performance liquid chromatography (10 kpsinanoAcquity; Waters). The mobile phase was as follows: water + 0.1% (v/v)] formic acid + 2% (v/v)] DMSO (A) and acetonitrile + 0.1% ([v/v)] formic acid + 2% (v/v)] DMSO (B); no DMSO was used for the affinity-enriched analysis. Desalting of the samples was performed online using a reverse-phase C18 trapping column (180-µm i.d., 20-mm length, 5-µm particle size; Waters). The peptides were then separated using a T3 HSS nano-column (75-µm i.d., 150-mm length, 1.8-µm particle size; Waters) at 0.35 µL min−1. Peptides were eluted from the column into the mass spectrometer using the following gradients: (1) for the comparative proteomics analysis, 2% to 25% B in 140 min, 25% to 90% B in 25 min, maintained at 90% for 5 min, and then back to initial conditions; (2) for the affinity enrichment analysis, 4% to 20% B in 50 min, 20% to 90% B in 5 min, maintained at 90% for 5 min, and then back to initial conditions. The nano-liquid chromatograph was coupled online through a nano-electrospray ionization emitter (7-cm length, 10-mm tip; New Objective) to an orbitrap (Q Exactive plus or HF; Thermo Scientific) using a FlexIon nanospray apparatus (Proxeon). The comparative proteomics samples were run on a Q Exactive plus, while the affinity-enriched samples were analyzed on a Q Exactive HF. Data were acquired in data-dependent acquisition mode, using a Top20 method. MS1 resolution was set to 70,000 or 120,000 (at 400 m/zfor the Q Exactive plus or HF, respectively), mass range of 300 to 1,650 m/z, automatic gain control (AGC) of 3e6, and maximum injection time was set to 20 ms. MS2 resolution was set to 17,500 or 30,000 (for the Q Exactive plus or HF, respectively), quadrupole isolation 1.7 m/z, AGC of 1e6, dynamic exclusion of 30 or 60 s (for the affinity-enriched or comparative proteome analysis, respectively), and maximum injection time of 60 ms.
Raw data were imported into Expressionist software (Genedata) and processed as described previously (Shalit et al., 2015). The software was used for retention time alignment and peak detection of precursor peptides. A master peak list was generated from all MS/MS events and sent for database searching using Mascot version 2.5.1 (Matrix Sciences). Data were searched against a customized TAIR10 database (ftp://ftp.ensemblgenomes.org/pub/plants/release-34/fasta/arabidopsisthaliana/pep/) appended with 125 common laboratory contaminant proteins. The TAIR10 database was formatted in the following manner. All isoforms of the same gene were concatenated under one entry according to their gene identifier after removal of the first Met of each sequence in the order they appeared in the database, except for the first sequence that retained its Met. A fixed modification was set to carbamidomethylation of Cys, and variable modifications were set to oxidation of Met and deamidation of Asn or Gln. Search results were then filtered using the PeptideProphet algorithm (Keller et al., 2002) to achieve a maximum false discovery rate of 1% at the protein level. Peptide identifications were imported back to Expressionist to annotate identified peaks. Quantification of proteins from the peptide data was performed using an in-house script (Shalit et al., 2015). Data were normalized based on the total ion current. Protein abundance was obtained by summing the three most intense, unique peptides per protein. A pairwise Student’s t test, after logarithmic transformation, was used to identify significant differences in the levels of specific proteins between a given mutant and the wild type across the biological replicate. Fold changes were calculated based on the ratio of arithmetic means of the case versus control samples. The goal of the experiment was to discover as many potential hits as possible. Therefore, we chose not to use a correction for multiple hypothesis testing to minimize the false negative rate. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the data set identifier PXD008920 and 10.6019/PXD008920.
For absolute quantification of Deg proteins in wild-type leaf samples, custom-made stable isotope-labeled peptides of Deg1 (VGDEVTVEVLR and LVLGDIITSVNGTK), Deg5 (IVGLDPDNDLAVLK), and Deg8 (IVPQLIQFSK, ILDEYSVGDK, and VDAPETLLKPIK) were synthesized (JPT). The peptides were spiked into the protein samples that were prepared and processed as above, and data were acquired in parallel reaction monitoring using Xcalibur (Thermo Scientific), where MS resolution was set to 35,000, AGC target of 2e5, and maximum injection time was set to 300 ms. Data were then imported into Skyline software (MacLean et al., 2010; Stergachis et al., 2011) for final processing and evaluation. Quantification was based on the area under the curve of extracted ion chromatograms from the most intense transition per peptide. At least seven overlapping parallel reaction monitoring transitions were acquired per peptide.
Expression, Purification, and Activity Assay of Recombinant Deg Proteases
The expression and purification of N-terminal mature His-tagged Deg1 were described previously (Chassin et al., 2002). The constructs for expressing Deg5 and Deg8 were generated as follows. Sequences corresponding to mature Deg5 (amino acids 74–C terminus) and Deg8 (amino acids 91–C terminus) were amplified from wild-type cDNA using custom primers specified in Supplemental Materials and Methods S1. The Deg5 PCR product was ligated into the pDrive cloning vector (PCR Cloning Kit; Qiagen) and then subcloned in frame to the 3′ end of a His×6 sequence into the NdeI and XhoI restriction sites in the pET15b plasmid (Novagen). The Deg8 PCR product, amplified in frame with an N-terminal His×6 sequence, was ligated into the pCRII-TOPO cloning vector (TOPO TA Cloning kit; Invitrogen) and then subcloned into the NcoI and NotI restriction sites in the pETnH plasmid (modified pET15b vector; see Supplemental Materials and Methods S1). All inserted sequences and orientations in the plasmids were confirmed by DNA sequencing.
To express the His-tagged Deg proteins, the expression vectors containing the Deg constructs were transformed into E. coli BL21 (DE3) competent cells. The transformed bacteria were grown in 350 mL of Luria-Bertani medium at 37°C to an OD600 of 0.35, and protein expression was induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside. Cells were harvested 3 h after induction by centrifugation at 7,000g for 5 min. Induced cells were subjected to SDS-PAGE, and the identity of the expressed Deg proteins was confirmed by MS analysis. For extraction of the expressed proteins, 2 g of bacteria pellets was resuspended in 22 mL of ice-cold buffer A (50 mm NaH2PO4 and 300 mm NaCl, pH 8) supplemented with 10 mm imidazole. Cells were sonicated on ice three times for 2 min. The lysate was cleared by filtering through a 0.45-µm Millipore filter, and the His-tagged proteins were purified using an FPLC system (AKTA Explorer; Amersham) on an 8-mL column packed with Ni-NTA agarose resin (Qiagen). After loading the column, it was washed with 30 mL of buffer A containing 10 mm imidazole and then with 40 mL of buffer A with 20 mm imidazole. The His-tagged proteins then were eluted in 5-mL fractions with 30 mL of buffer A containing 250 mm imidazole. Protein-containing fractions were pooled, concentrated by ultrafiltration, and loaded onto a Sephacryl S-200 column. The column was then eluted with buffer B (10 mm Tris-HCl, 150 mm NaCl, and 1 mm EDTA, pH 8), and the absorbance of the eluted material was monitored at 280 nm.
The proteolytic activity of the recombinant Deg proteases was assayed as described previously (Chassin et al., 2002; Kley et al., 2011). Briefly, 5 pmol of recombinant protease was mixed with 50 pmol of β-casein or 40 pmol dithiothreitol-denatured α-lactalbumin as a substrate in 50 mm MES, pH 6, and incubated for 1 h at 37°C. The reaction was terminated by adding an equal volume 4× SDS sample buffer and resolved by SDS-PAGE. Gels were stained with Coomassie Blue R-250, and degradation of β-casein or α-lactalbumin was quantified using the ImageJ analysis software. It should be noted that similar results to those presented here were obtained when the reaction was carried out at 26°C.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Deg1, At3g27925; Deg5, At4g18370; and Deg8, At5g39830.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Generation of single, double, and triple Deg KO mutants.
Supplemental Figure S2. Phenotypes of Deg KO mutants grown at different light intensities.
Supplemental Figure S3. Phenotypes of Deg KO mutants grown at high light intensity and high temperature.
Supplemental Figure S4. Phenotypes of complemented Deg1 KO mutant lines.
Supplemental Figure S5. In vitro proteolytic degradation assay on α-lactalbumin.
Supplemental Table S1. Growth parameters of deg mutants.
Supplemental Table S2. Summary of comparative MS analysis results.
Supplemental Table S3. Absolute quantification of Deg proteins.
Supplemental Table S4. Summary of affinity enrichment experiments.
Supplemental Data Set S1. Comparative MS analysis of wild-type, deg1, and deg158 plants grown under optimal conditions.
Supplemental Data Set S2. MS analysis of affinity-enriched proteins from epitope-tagged lines.
Acknowledgments
We thank Dr. Shlomi Dagan for help and advice on statistical analyses.
Footnotes
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (no. AD 92/12-1 to Iwona Adamska and Z.A.), by grants from the Israel Science Foundation (no. 2585/16 to Z.A. and no. 1082/17 to Z.R.), and by a grant from the Hohenheim University-Hebrew University Collaborative Research Program to Z.A.
References
- Adam Z. (2007) Protein stability and degradation in plastids. Trends Curr Genet 19: 315–338 [Google Scholar]
- Adam Z, Sakamoto W (2014) Plastid proteases. In Theg SM, Wollman FA, eds, Plastid Biology, Vol 5 Springer, New York, pp 359–389 [Google Scholar]
- Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K, et al. (2001) Chloroplast and mitochondrial proteases in Arabidopsis: a proposed nomenclature. Plant Physiol 125: 1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z, Frottin F, Espagne C, Meinnel T, Giglione C (2011) Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell 23: 3745–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albus CA, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22: 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zühl F, Seemüller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 [DOI] [PubMed] [Google Scholar]
- Chassin Y, Kapri-Pardes E, Sinvany G, Arad T, Adam Z (2002) Expression and characterization of the thylakoid lumen protease DegP1 from Arabidopsis. Plant Physiol 130: 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang D, Guo J, Wu H, Jin M, Lu Q, Lu C, Zhang L (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61: 567–575 [DOI] [PubMed] [Google Scholar]
- Clausen T, Kaiser M, Huber R, Ehrmann M (2011) HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 12: 152–162 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Granlund I, Storm P, Schubert M, García-Cerdán JG, Funk C, Schröder WP (2009) The TL29 protein is lumen located, associated with PSII and not an ascorbate peroxidase. Plant Cell Physiol 50: 1898–1910 [DOI] [PubMed] [Google Scholar]
- Haussühl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J 20: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Schuhmann H, Adamska I (2009) Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Res Microbiol 160: 726–732 [DOI] [PubMed] [Google Scholar]
- Itzhaki H, Naveh L, Lindahl M, Cook M, Adam Z (1998) Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J Biol Chem 273: 7094–7098 [DOI] [PubMed] [Google Scholar]
- Jakob M, Kaiser S, Gutensohn M, Hanner P, Klösgen RB (2009) Tat subunit stoichiometry in Arabidopsis thaliana challenges the proposed function of TatA as the translocation pore. Biochim Biophys Acta 1793: 388–394 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E, Naveh L, Adam Z (2007) The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2009) Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J Biochem 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2018) FtsH protease in the thylakoid membrane: physiological functions and the regulation of protease activity. Front Plant Sci 9: 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sun X, Zhang L, Sakamoto W (2012) Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol 159: 1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Ozawa S, Takahashi Y, Sakamoto W (2015) D1 fragmentation in photosystem II repair caused by photo-damage of a two-step model. Photosynth Res 126: 409–416 [DOI] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Keilhauer EC, Hein MY, Mann M (2015) Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS). Mol Cell Proteomics 14: 120–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kley J, Schmidt B, Boyanov B, Stolt-Bergner PC, Kirk R, Ehrmann M, Knopf RR, Naveh L, Adam Z, Clausen T (2011) Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat Struct Mol Biol 18: 728–731 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lennartz K, Plücken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque-Tremblay G, Havaux M, Ouellet F (2009) The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J 60: 691–702 [DOI] [PubMed] [Google Scholar]
- Luciński R, Misztal L, Samardakiewicz S, Jackowski G (2011a) Involvement of Deg5 protease in wounding-related disposal of PsbF apoprotein. Plant Physiol Biochem 49: 311–320 [DOI] [PubMed] [Google Scholar]
- Luciński R, Misztal L, Samardakiewicz S, Jackowski G (2011b) The thylakoid protease Deg2 is involved in stress-related degradation of the photosystem II light-harvesting protein Lhcb6 in Arabidopsis thaliana. New Phytol 192: 74–86 [DOI] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, van Wijk KJ (2015) Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta 1847: 915–930 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W (2017) Essentials of proteolytic machineries in chloroplasts. Mol Plant 10: 4–19 [DOI] [PubMed] [Google Scholar]
- Olivares AO, Baker TA, Sauer RT (2016) Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol 14: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Söderberg L, Roepstorff P, von Heijne G, et al. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Sakamoto W. (2006) Protein degradation machineries in plastids. Annu Rev Plant Biol 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schröder WP, Kieselbach T (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Adamska I (2012) Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol Plant 145: 224–234 [DOI] [PubMed] [Google Scholar]
- Shalit T, Elinger D, Savidor A, Gabashvili A, Levin Y (2015) MS1-based label-free proteomics using a quadrupole orbitrap mass spectrometer. J Proteome Res 14: 1979–1986 [DOI] [PubMed] [Google Scholar]
- Sokolenko A, Pojidaeva E, Zinchenko V, Panichkin V, Glaser VM, Herrmann RG, Shestakov SV (2002) The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr Genet 41: 291–310 [DOI] [PubMed] [Google Scholar]
- Stergachis AB, MacLean B, Lee K, Stamatoyannopoulos JA, MacCoss MJ (2011) Rapid empirical discovery of optimal peptides for targeted proteomics. Nat Methods 8: 1041–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Gao F, Fan H, Shan X, Sun R, Liu L, Gong W (2013) The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases. Acta Crystallogr D Biol Crystallogr 69: 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Peng L, Guo J, Chi W, Ma J, Lu C, Zhang L (2007) Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Fu T, Chen N, Guo J, Ma J, Zou M, Lu C, Zhang L (2010a) The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ouyang M, Guo J, Ma J, Lu C, Adam Z, Zhang L (2010b) The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. Plant J 62: 240–249 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Small I, Millar AH (2014) Using the SUBcellular database for Arabidopsis proteins to localize the Deg protease family. Front Plant Sci 5: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen JP, Sakuragi Y, Stael S, Tikkanen M, Allahverdiyeva Y, Paakkarinen V, Aro E, Suorsa M, Scheller HV, Vener AV, et al. (2008) Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ. (2015) Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol 66: 75–111 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S, Held K, Schlücking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J (2008) A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Yamamoto H, Kondo M, Takeda S, Ifuku K, Fukao Y, Kamei Y, Nishimura M, Shikanai T (2016) Grana-localized proteins, RIQ1 and RIQ2, affect the organization of light-harvesting complex II and grana stacking in Arabidopsis. Plant Cell 28: 2261–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Feder A, Adam Z (2005) Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts: implications for thylakoid formation and photosystem II maintenance. Plant J 42: 609–617 [DOI] [PubMed] [Google Scholar]
- Zienkiewicz M, Ferenc A, Wasilewska W, Romanowska E (2012) High light stimulates Deg1-dependent cleavage of the minor LHCII antenna proteins CP26 and CP29 and the PsbS protein in Arabidopsis thaliana. Planta 235: 279–288 [DOI] [PubMed] [Google Scholar]
- Zienkiewicz M, Kokoszka N, Bacławska I, Drożak A, Romanowska E (2013) Light intensity and quality stimulated Deg1-dependent cleavage of PSII components in the chloroplasts of maize. Plant Physiol Biochem 67: 126–136 [DOI] [PubMed] [Google Scholar]