The SNARE protein VAMP711 is involved in regulating abscisic acid-mediated inhibition of plasma membrane H+-ATPase activity in response to drought stress.
Abstract
Drought stress is a limiting environmental factor that affects plant growth and development. The plant hormone abscisic acid (ABA) plays an important role in plant drought responses. Previous studies have indicated that ABA inhibits plasma membrane H+-ATPase (PM H+-ATPase) activity, and the decrease in PM H+-ATPase activity promotes stomatal closure under drought stress, thereby reducing water loss. However, the underlying molecular mechanisms are not well understood. In this study, we found that in Arabidopsis (Arabidopsis thaliana), ABA induces an N-ethylmaleimide-sensitive factor attachment protein receptor protein, namely, VESICLE-ASSOCIATED MEMBRANE PROTEIN 711 (VAMP711), to interact with the Arabidopsis PM H+-ATPases AHA1 and AHA2. The interaction occurs at their C-termini and inhibits PM H+-ATPase activity. Deletion of VAMP711 in Arabidopsis results in a higher PM H+-ATPase activity and slower stomatal closure in response to ABA and drought treatments. In addition, overexpression of VAMP711 partially rescues the drought-sensitive phenotype of ost2-2D, a mutation in AHA1 resulting in a constitutive activated PM H+-ATPase. Our results demonstrate that VAMP711 is involved in regulating ABA-mediated inhibition of PM H+-ATPase activity and stomatal closure in response to drought stress.
Plasma membrane (PM) H+-ATPase regulates many cellular activities by producing electrochemical gradients across the plasma membrane. Therefore, the activity of PM H+-ATPase must be tightly controlled. In plants, PM H+-ATPase plays regulatory roles in many cellular and development processes, such as cell expansion, intracellular pH homeostasis, and response to saline and drought stresses (Palmgren, 2001; Rober-Kleber et al., 2003; Fuglsang et al., 2007; Merlot et al., 2007; Yang et al., 2010; Yamauchi et al., 2016). The Arabidopsis (Arabidopsis thaliana) genome contains 11 PM H+-ATPase genes (AHA1 to AHA11; Palmgren, 2001; Falhof et al., 2016; Haruta et al., 2018).
The C terminus (∼100 residues) of PM H+-ATPase contains the autoinhibitory domain of the enzyme activity (Palmgren et al., 1991). The phosphorylation of the C terminus is critical for the regulation of PM H+-ATPase activity. The phosphorylation of the penultimate residue (Thr-947) of Arabidopsis H+-ATPase 2 (AHA2) generates a 14-3-3 biding site to activate PM H+-ATPase activity (Svennelid et al., 1999; Camoni et al., 2000; Gévaudant et al., 2007); the phosphorylation of the C terminus of AHA2 (Ser-931) by a Ser/Thr protein kinase (PKS5) inhibits the interaction between AHA2 and 14-3-3 to decrease PM H+-ATPase activity (Kinoshita and Shimazaki, 2002; Fuglsang et al., 2007; Yang et al., 2010).
Abscisic acid (ABA) negatively regulates PM H+-ATPase activity (Merlot et al., 2007). It promotes the dephosphorylation of the Thr-947 site in AHA2 (Yin et al., 2013; Falhof et al., 2016; Cai et al., 2017) and Snf1-related protein kinase 2.6 (SnRK2.6)/open stomata1 (OST1) protein is involved in this regulation (Merlot et al., 2007). SnRK2.2, but not SnRK2.6, directly phosphorylates the C terminus of AHA2 (Planes et al., 2015). The dominant mutants of ost2-1D and ost2-2D (AHA1) show constitutive higher PM H+-ATPase activity and abolish guard cell response to ABA (Merlot et al., 2007). However, the underlying molecular mechanisms are not well understood.
In addition to the regulation of PM H+-ATPase activity by direct phosphorylation, the regulation of the amount of PM H+-ATPase protein is an important mechanism that affects its activity (Hashimoto-Sugimoto et al., 2013). H+-ATPase translocation control1 (PATROL1) is a Munc13-like protein related to synaptic vesicle priming (Basu et al., 2005), which regulates AHA1 translocation to the plasma membrane and mediates stomatal movement in response to environmental signals such as light and CO2 (Hashimoto-Sugimoto et al., 2013).
SNARE proteins play an essential role in vesicle trafficking by facilitating the fusion of vesicles and the target membrane in eukaryotes (Uemura et al., 2004). SNARE proteins are classified into R- and Q-types according to the similarity and specific amino acid sequence (Uemura et al., 2005). One R-SNARE protein and three Q-SNARE proteins form a complex to promote membrane fusion (Pratelli et al., 2004; Jahn and Scheller, 2006). Vesicle-associated membrane protein7 (VAMP7)-like proteins are one group of R-SNAREs, which comprises two subgroups: VAMP71 and VAMP72 (Sanderfoot, 2007). The structure of SNARE proteins is conserved: a longin domain in the N terminus and a SNARE domain and a transmembrane domain (which anchors SNARE proteins to the membrane) in the C terminus in plants (Uemura et al., 2004; Hong, 2005).
In Arabidopsis, VAMP7s are involved in endosome trafficking and are highly conserved and localized to the vacuolar membrane and plasma membrane (Uemura et al., 2004; Hong, 2005). Increasing evidence shows that SNARE-mediated pathways are involved in response to biotic and abiotic stresses in plants (Collins et al., 2003; Leshem et al., 2006, 2010; Sugano et al., 2016). In rice (Oryza sativa), the intracellular SNARE protein, OsVAMP714, plays a role in resistance to rice blast disease (Sugano et al., 2016). In Arabidopsis, the decreased expression of VAMP711 inhibits the fusion of vesicles to the tonoplast, resulting in plant tolerance to salt and drought stresses (Leshem et al., 2006, 2010). In general, VAMP7-mediated cellular activities are linked to the function of vesicle fusion. However, the SNARE protein VAMP721 interacts with and inhibits the activity of inward-rectifying K+ channels KAT1 and KC1 to regulate the vegetative growth of Arabidopsis (Zhang et al., 2015).
In this study, we identified a SNARE family protein, VAMP711, as a negative regulator of PM H+-ATPase. VAMP711 directly interacted with and inhibited AHA1 and AHA2 activity, and this interaction was induced by ABA treatment. The drought-hypersensitive phenotype and ABA-mediated stomatal closure defect of the ost2-2D mutant were partially reduced by overexpression of VAMP711. Our results suggest that ABA-mediated PM H+-ATPase activity inhibition occurs, at least partially, through the interaction between VAMP711 and AHA1/AHA2, and provide evidence for an ABA-mediated regulatory mechanism for PM H+-ATPase activity.
RESULTS
VAMP711 Interacts with PM H+-ATPase AHA1
ABA inhibits PM H+-ATPase activity (Merlot et al., 2007; Planes et al., 2015). To identify components involved in this inhibition, we first confirmed this result. Four-week-old Arabidopsis Col-0 plants were treated with 200 mm NaHCO3 for 2 d to induce PM H+-ATPase activity and then treated with or without 10 µm of ABA for 1 d. The plasma membrane-enriched vesicles were isolated and used for measuring PM H+-ATPase activity. The ABA treatment significantly reduced H+-transport and ATP hydrolysis activities of PM H+-ATPase (Supplemental Figure S1, A–D). To identify possible regulators, we generated more than 30 Pro35S:GFP-AHA1 transgenic plants in Col-0 and selected stable transgenic plants in which the GFP-AHA1 signal was localized to the plasma membrane for further study. The 10-d-old transgenic plants were treated with or without 50 μm of ABA for 6 h, the plasma membrane-enriched fraction was collected, the GFP-AHA1 was immunoprecipitated with anti-GFP antibody-conjugated agarose, and putative AHA1-interacting proteins were detected by mass spectrometry. A SNARE family protein, VAMP711, was identified as a putative AHA1-interacting protein.
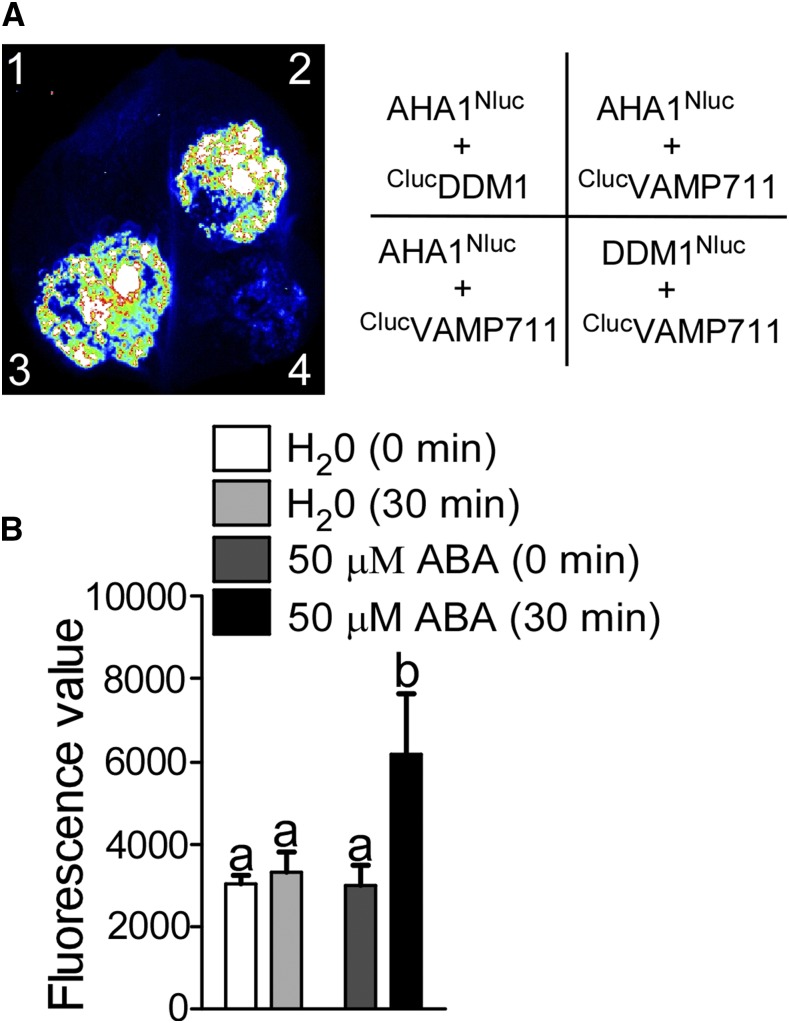
To verify this interaction, we performed a luciferase complementation (LUC) assay by constructing AHA1Nluc and ClucVAMP711 vectors, and transformed them into Nicotiana benthamiana leaves to collect luminescence signals after a 3-d incubation. The luminescence signal was detected in leaves expressing AHA1Nluc and ClucVAMP711; as a control, the expression of AHA1Nluc and ClucDDM1 (DDM1, DECREASED DNA METHYLATION1), and DDM1Nluc and ClucVAMP711 showed a very low signal (Fig. 1A); however, the indicated genes were detected in N. benthamiana leaves at similar levels (Supplemental Fig. S2A). We also cotransformed ClucAHA1 and VAMP711Nluc into N. benthamiana leaves. The luminescence signal was not detected after the 3-d incubation (Supplemental Fig. S2B), possibly because Nluc and Cluc are not close enough due to the specific fusion position.
Figure 1.
VAMP711 interacts with PM H+-ATPase AHA1. A, Luciferase complementation assay. The indicated constructs were expressed in Nicotiana benthamiana leaves. 1, AHA1Nluc and ClucDDM1; 2 and 3, AHA1Nluc and ClucVAMP711; 4, DDM1Nluc and ClucVAMP711. The luciferase signal was detected using a cooled CCD camera (iKon-L936; Andor Tech). B, ABA induced the interaction between AHA1 and VAMP711. AHA1Nluc and ClucVAMP711 were transformed into N. benthamiana leaves. After a 3-d incubation, the leaves were treated with or without ABA. The luciferase value was collected using a spectrophotometer.Error bars represent sd (small leaves acquired by a leaf punch, number, 10); Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters.
To determine whether the interaction between VAMP711 and AHA1 is induced by ABA, the N. benthamiana leaves transformed with AHA1Nluc and ClucVAMP711 were treated with or without 50 μm ABA for 6 h, and the fluorescence value was collected. The results showed that ABA could significantly promote the interaction between VAMP711 and AHA1 (Fig. 1B). Together, these results suggest that the SNARE protein VAMP711 interacts with AHA1, and the interaction is induced by ABA treatment.
To determine the possible subcellular location where these proteins interact, we constructed AHA1YCE and YNEVAMP711, and AHA1YNE and YCEVAMP711 for bimolecular fluorescence complementation (BIFC) assay. The combinations of AHA1YCE and YNEVAMP711, AHA1YCE and YNEDDM1, DDM1YCE and YNEVAMP711, AHA1YNE and YCEVAMP711, YCESYP22 and YNEVAMP711, and YCESYP22 and YNEDDM1 were transiently expressed in N. benthamiana leaves, respectively. The YFP (yellow fluorescent protein) fluorescence signal was detected by confocal microscopy in leaves of expressed AHA1YCE and YNEVAMP711,and AHA1YNE and YCEVAMP711 (Supplemental Fig. S2, C and D). The YFP signal partially colocalized with the signal of FM4-64 dye, which was used to label the plasma membrane and endosomes (Supplemental Fig. S2, C and D). VAMP711 has been reported to be a vacuolar membrane-localized protein and AHA1 is a plasma membrane-located protein (Leshem et al., 2006; Hashimoto-Sugimoto et al., 2013). Consistent with these notions, our results showed that VAMP711 predominately localized to the vacuolar membrane and prevacuolar compartments (PVCs), and AHA1 was localized to the plasma membrane (Supplemental Fig. S2, E and F). The fluorescence signal of GFP-VAMP711 was not colocalized with FM4-64 dye-labeled plasma membrane in the ProUBQ10:GFP-VAMP711 transgenic seedlings without ABA treatment. However, when the ProUBQ10:GFP-VAMP711 transgenic seedlings were treated with ABA, some weak fluorescence signal of GFP-VAMP711 was detected on the plasma membrane, and the Pearson correlation coefficient was much higher in ProUBQ10:GFP-VAMP711 transgenic seedlings treated with ABA (Supplemental Fig. S2G). In addition, the result of plasma membrane isolation and immunoblot analysis showed that the amount of VAMP711 proteins on the plasma membrane was greater in ProUBQ10:GFP-VAMP711 transgenic seedlings subjected to ABA treatment than in seedlings not subjected to ABA treatment (Supplemental Fig. S2H). These results indicated that ABA treatment might affect the localization of VAMP711. On the basis of these results, we speculated that the interaction between VAMP711 and AHA1 might partially occur on the plasma membrane.
VAMP711 Interacts with the C Terminus of AHA1 and AHA2
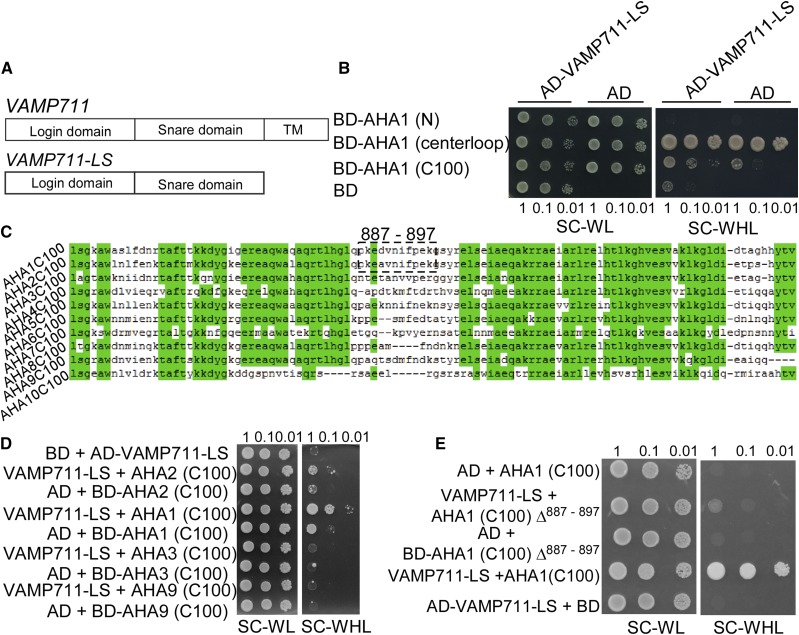
The SNARE protein VAMP711 has three conserved domains: the longin domain, SNARE domain, and transmembrane domain (Fig. 2A; Uemura et al., 2005; Fujiwara et al., 2014), and AHA1 contains three intracellular parts: the N terminus, centerloop, and C terminus (C100; Falhof et al., 2016). We used the VAMP711-LS (longin and SNARE domains of VAMP711) as a bait to detect the VAMP711-interacting portion of AHA1 in the yeast two-hybrid system. The indicated combinations were transformed into the yeast strain AH109. The yeast cells containing pGBKT7-AHA1 (centerloop) and pGADT7 vector could grow on synthetic complete (SC) medium lacking Trp, Leu, and His (WHL). We then tested the interaction between pGBKT7-AHA2 (centerloop) and pGADT7-VAMP711-LS, and no interaction was detected (Supplemental Fig. S3B). The yeast cells containing the combinations of pGBKT7-AHA1 (C100) and pGADT7-VAMP711-LS showed better growth than those containing the combinations pGBKT7-AHA1 (C100) and pGADT7 on SC medium lacking WHL. These results suggest that VAMP711 interacts with the C terminus of PM H+-ATPase (Fig. 2, A and B), which is a conserved domain (Fig. 2C).
Figure 2.
VAMP711 interacts with the C terminus of AHA1 and AHA2. A, Schematic structure of VAMP711 protein structure: longin domain, SNARE domain, and transmembrane domain (TMD). The longin and SNARE domains of VAMP711 (VAMP711-LS) were used in a yeast two-hybrid assay. B, Yeast two-hybrid assay to detect the interaction among the AHA1 N terminus, centerloop, C terminus, and VAMP711. C, The sequence alignment analysis of the C terminus of PM H+-ATPase (AHA) in Arabidopsis. D, Yeast two-hybrid assay to detect the interaction of VAMP711 with the C terminus of AHA family members. E, Yeast two-hybrid assay to detect the region of interaction between AHA1 C terminus, AHA1 C terminus minus amino acid residues 887 to 897 (AHA1 (C100)Δ887–897), and VAMP711. Yeast strains expressing the indicated plasmids were grown on synthetic complete medium without Trp and Leu (SC-WL, left) and on synthetic complete medium without Trp, Leu, and His (SC-WHL, right). Photographs were taken after 3 to 5 d of growth on the indicated medium. Panels show yeast serial decimal dilutions. Experimental details are provided in the “Materials and Methods.”
To determine the specificity of interaction, we cotransformed the C terminus of AHA1, AHA2, AHA3, and AHA9 with VAMP711-LS into a yeast strain. Only AHA1 and AHA2 interacted with VAMP711 (Fig. 2D). AHA1 and AHA2 are two major members of the AHA family (Palmgren, 2001). We found that 11 amino acid residues (887–897) in AHA1 and AHA2 are distinct from those in other AHA members (Fig. 2C). We speculated that these 11 amino acid residues may be required for VAMP711 to interact with AHA1 and AHA2 (Fig. 2C). When the 11 residues in AHA1 and AHA2 were deleted, the interaction between AHA1 and VAMP711 was abolished (Fig. 2E; Supplemental Fig. S3A). Together, our results suggest that the interaction between the VAMP711 and the C terminus of AHA1 and AHA2 requires these 11 amino acid residues.
VAMP711 Negatively Regulates PM H+-ATPase Activity
To further investigate whether VAMP711 could regulate PM H+-ATPase activity by direct interaction, we generated two CRISPR/Cas9 vamp711 mutants, vamp711-6 and vamp711-7. A two-base deletion (GA, 98–99 bp) in the second exon was detected in the vamp711-6 mutant, and a one-base insertion (C between 16 and 17 bp) in the first exon was detected in the vamp711-7 mutant. Both these mutations led to frameshift and early termination of VAMP711 (Supplemental Fig. S4A).
We also constructed complementation lines of vamp711-6 (com-1, com-2) by transferring a plasmid containing the VAMP711 genomic sequence that included the promoter region (1,500 bp from the translational start site) and 3′ untranslated region (500 bp downstream of the stop codon), more than 30 transgenic plants were obtained and two independent T4 transgenic lines (com-1, com-2) with VAMP711 expression levels similar to those in the wild type were used for further study (Supplemental Fig. S4B).
We also generated VAMP711-overexpressing lines (ProUBQ10:GFP-VAMP711) in the wild-type background and named these lines OE-VAMP711-15/27. In the OE-VAMP711 lines, the GFP-VAMP711 signal was localized at vacuolar membrane and PVCs. Reverse-transcription PCR (RT-PCR) detected increased expression of VAMP711 in both overexpressing lines (Supplemental Fig. S4C).
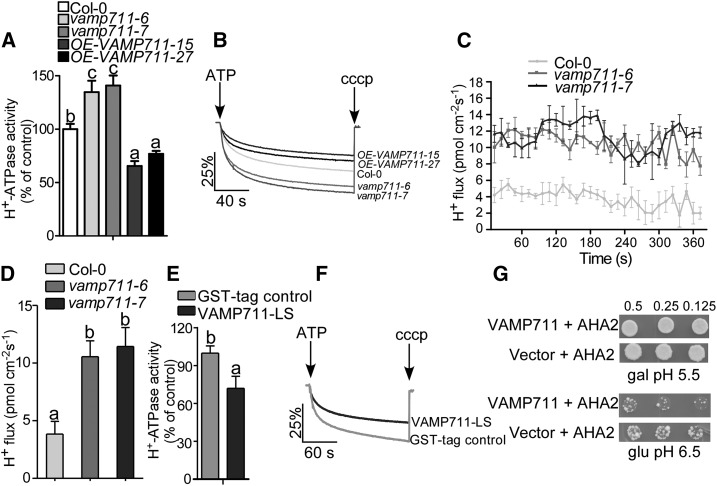
To investigate the PM H+-ATPase activity in Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27, we isolated plasma membrane vesicles from 4-week-old plants treated with 200 mm of NaHCO3 for 3 d. The H+ transport activity in vamp711-6/7 was 30% higher than that in Col-0; while the activity in VAMP711-overexpressing lines were 10%–15% lower than that in Col-0 (Fig. 3, A and B). To detect the PM H+-ATPase protein level on the plasma membrane in Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27 plants under NaHCO3 treatment, plasma membrane vesicles were isolated and detected by immunoblotting with anti-PM H+-ATPase antibody. No obvious difference was detected in Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27 plants (Supplemental Fig. S4D).
Figure 3.
VAMP711 negatively regulates PM H+-ATPase activity. A, PM H+-ATPase activity was measured in Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27. Plasma membrane vesicles of Col-0, vamp711-6, vamp711-7, OE-VAMP711-15/27 were isolated from 4-week-old soil-grown plants treated with 200 mm of NaHCO3 for 3 d. Error bars represent sd; Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters. The experiment was repeated three times independently. B, Comparison of PM H+-ATPase activity from A. C, Net H+ fluxes in different plant materials. Five-day-old seedlings of Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27 were treated with a buffer (0.5 mm KCl, 0.1 mm CaCl2, 75 mm NaCl, and 0.03 mm HEPES, pH 7.8) for 30 min, and then the H+ fluxes in the root tips was detected by the noninvasive microtest technique. Error bars represent sd (seedling number ≥10); Student’s t test was used to assess statistical significance (P ≤ 0.05). Experiment was repeated three times independently. D, Calculated net H+ fluxes from C. Error bars represent SD; Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters. E, VAMP711 inhibits PM H+-ATPase activity in vitro. Fifty nanograms of plasma membrane vesicles from Col-0 was incubated with 500 ng of VAMP711-LS protein for 15 min at room temperature. Error bars represent SD; Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters. The experiment is described in the “Materials and Methods” and repeated three times independently. F, Comparison of PM H+-ATPase activity from E. G, VAMP711 inhibits the PM H+-ATPase activity of AHA2 in the RS72 yeast strain. VAMP711 and an empty vector were transferred into the RS72 yeast strain containing AHA2. The endogenous H+-ATPase gene in RS72 was induced by Gal (gal). When AHA2 and VAMP711 were transferred into the RS72 yeast strain, the growth of the yeast depended on AHA2 activity on Glc (glu) medium. Photographs were taken after 3 to 5 d of growth on the indicated medium. Panels show yeast serial half dilutions. Experimental details are provided in the “Materials and Methods.” The experiment was repeated at least three times independently.
To further confirm the effect of VAMP711 on H+ fluxes, the noninvasive microtest technique was used to monitor H+ flux in the root. Five-day-old seedlings of Col-0, vamp711-6/7, and complementation lines com-1/2 were incubated in buffer at pH 7.8 for 30 min, then their root apexes were selected to detect the H+ flux for about 360 s. The H+ fluxes of vamp711-6/7 were significantly higher than those of Col-0 (Fig. 3, C and D). There was no obvious difference in the H+ fluxes between com-1/2 lines and the wild type (Supplemental Fig. S4, E and F). These results indicate that VAMP711 negatively regulates PM H+-ATPase activity in plants.
Arabidopsis mutants with a higher PM H+-ATPase activity display resistance to high pH stress (Fuglsang et al., 2007; Yang et al., 2010). To investigate whether mutation in VAMP711 has a similar effect, we transferred 6-d-old seedlings of Col-0 and vamp711-6/7 from Murashige and Skoog (MS) medium at pH 5.8 to MS medium at pH 5.8 or 8.0. There was no significant difference in the fresh weight and primary root length of Col-0 and vamp711-6/7 for 1 week after the transfer. However, because of their PM H+-ATPase activity levels, the mutants of vamp711-6/7 were much more resistant to pH 8.0 than the wild type, displaying higher fresh weight and longer primary root length for 10 d after the transfer (Supplemental Fig. S5, A and C). These results further suggest that VAMP711 negatively regulates PM H+-ATPase activity.
To investigate whether VAMP711 directly inhibits PM H+-ATPase activity, we purified glutathione S-transferase-tagged VAMP711-LS recombinant protein expressed in Escherichia coli BL21. The PM H+-ATPase activity of the plasma membrane vesicles was measured after incubation with 500 ng of VAMP711-LS recombinant proteins for 15 min at room temperature. When VAMP711-LS protein was added in the plasma membrane vesicles, H+-ATPase activity decreased by about 20% compared to the activity observed when a negative control or a glutathione S-transferase protein was added (Fig. 3, E and F).
We then used the yeast strain RS72, the growth of which depends on expressed heterologous AHA2 activity in Glc medium (Fuglsang et al., 2007). We transferred VAMP711 into the RS72 yeast strain expressing heterologous AHA2 to investigate the effect of VAMP711 on RS72 yeast growth. The VAMP711 inhibited RS72 yeast growth on Glc medium at pH 6.5 (Fig. 3G), which indicates that VAMP711 inhibits AHA2 activity. The immunoblot analysis showed that the expression of VAMP711 did not affect the PM H+-ATPase protein levels in the RS72 yeast strain (Supplemental Fig. S4G). Our results indicate that VAMP711 inhibits PM H+-ATPase activity by direct interaction.
VAMP711 Is Required for ABA-Mediated Inhibition of PM H+-ATPase Activity
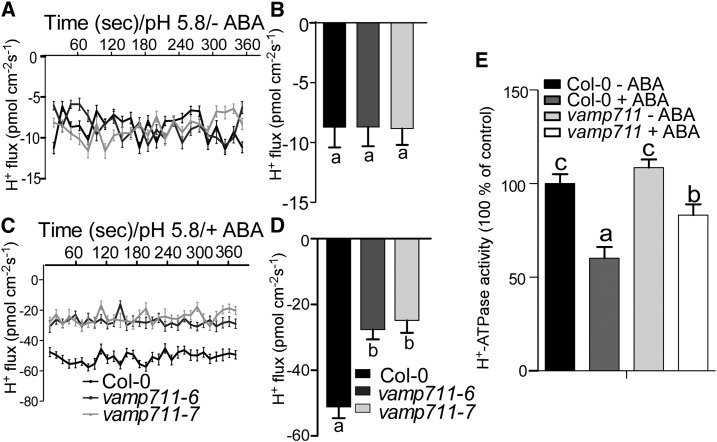
To study whether VAMP711 is required for ABA-mediated PM H+-ATPase activity inhibition, we detected the H+ fluxes and PM H+-ATPase activity in Col-0 and vamp711 mutants under ABA treatment. The H+ influxes were not obviously different between vamp711-6/7 mutants and wild type in the absence of ABA treatment (Fig. 4, A and B). The ABA treatment significantly increased the H+ influxes in both Col-0 and vamp711-6/7 mutants; however, the H+ influxes in the seedlings of vamp711-6/7 mutants were less sensitive to ABA treatment than those in the wild-type seedlings (Fig. 4, C and D). We then isolated the plasma membrane vesicles from 4-week-old plants treated with or without 10 µm of ABA and measured the PM H+-ATPase activity. The PM H+-ATPase activity was similar between Col-0 and vamp711 mutant without ABA treatment (Fig. 4E). After ABA treatment, the PM H+-ATPase activity decreased in both the wild type and the vamp711 mutant; however, greater reduction was detected in the wild type than in the vamp711 mutant (Fig. 4E). The immunoblot analysis showed that the PM H+-ATPase protein levels were no different between Col-0 and vamp711 mutants with or without ABA treatment (Supplemental Fig. S6A). These results together suggested that ABA inhibits PM H+-ATPase activity at least partially through VAMP711 interaction with the enzyme directly.
Figure 4.
VAMP711 is involved in ABA-mediated inhibition of PM H+-ATPase activity. A, Net H+ fluxes in root tips of Col-0, vamp711-6, and vamp711-7. Five-day-old seedlings were treated with a buffer (0.5 mm KCl, 0.1 mm CaCl2, and 0.03 mm MES, pH 5.8) for 30 min, and then the H+ flux was detected in root tips. Error bars represent sd (seedling number ≥10); Student’s t test was used to assess statistical significance (P ≤ 0.05).The experiment was repeated three times independently. B, Net H+ fluxes calculated from A. Significant differences were represented by different lowercase letters. C, Net H+ fluxes in the root tips in the presence of ABA. Five-day-old seedlings of Col-0, vamp711-6, and vamp711-7 were treated with a buffer (0.5 mm KCl, 0.1 mm CaCl2, and 0.03 mm MES, 2 μm ABA, pH 5.8) for 30 min, and the H+ fluxes were detected in the root tips. Error bars represent sd (seedling number ≥10); Student’s t test was used to assess statistical significance (P ≤ 0.05). The experiment was repeated three times independently. D, Net H+ fluxes calculated from C. Significant differences were represented by different lowercase letters. E, PM H+-ATPase activity was measured in Col-0 and vamp711-6 with or without ABA treatment. Plasma membrane vesicles of Col-0, vamp711-6 were isolated from 4-week-old plants treated with 10 μm of ABA for 1 d. Error bars represent sd; Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters. The experiment was repeated three times independently.
VAMP711 Inhibits AHA1-Mediated Stomatal Closure and Drought Response
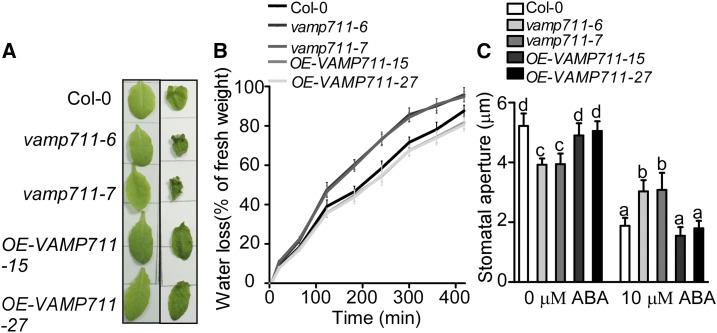
Previous studies have shown that vamp711 mutants are sensitive to drought stress and stomatal movement is impaired in the vamp711 mutant (Leshem et al., 2010). Consistent with these findings, the water loss in vamp711-6/7 mutants was more rapid than that in the wild type (Fig. 5, A and B), and both KCl-light-induced stomatal opening and ABA-induced stomatal closure were also impaired in vamp711-6/7 mutants (Fig. 5C).
Figure 5.
VAMP711 is involved in drought stress response. A. Detached rosette leaves from Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27. Photographs were taken at 0 (left) and 3 h (right). B, Water-loss measurement of detached leaves. Col-0, vamp711-6, vamp711-7, and OE-VAMP711-15/27 were grown in short-day conditions for 5 weeks. Fresh weight of detached leaves was monitored at the indicated times. The experiment was repeated three times independently. C, Stomatal aperture assay. Detached rosette leaves from Col-0, vamp711-6, and vamp711-7 were incubated in KCl buffer for about 2 h and then transferred in a buffer containing 10 μm of ABA for 2 h. Error bars represent sd (stomata number, 70); Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters. The experiment was repeated three times independently.
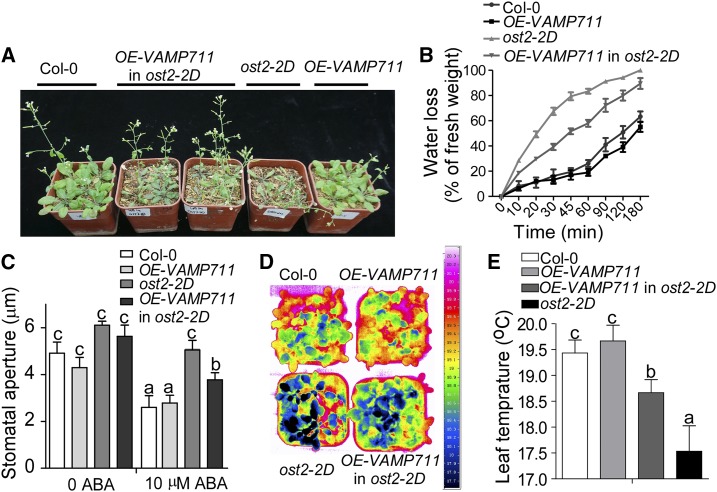
The ost2-2D mutant contains two point mutations in the AHA1 gene, resulting in AHA1 becoming constitutively hyperactive and the mutant becoming hypersensitive to drought stress (Merlot et al., 2007). To investigate whether the VAMP711-mediated plant ABA/drought response occurs through regulation of PM H+-ATPase activity, we crossed OE-VAMP711-15 into the ost2-2D genetic background and obtained ost2-2D-OE-VAMP711 plants. The Col-0, ost2-2D, ost2-2D-OE-VAMP711, and Col-0:OE-VAMP711 plants were grown in a greenhouse with 12 h light/12 h dark, and watering was stopped for 5-week-old plants for 2 weeks. The wilting rate of the leaves in the ost2-2D mutant was much higher than that in other plants, and overexpression of VAMP711 in ost2-2D partially rescued its drought-sensitive phenotype (Fig. 6A), and the decreased chlorophyll and relative water contents in ost2-2D mutant were also partially rescued in Col-0:OE-VAMP711 plants (Supplemental Fig. S7, A and B).
Figure 6.
VAMP711 regulates AHA1-mediated stomatal closure. A, Drought phenotype of the Col-0, ost2-2D, OE-VAMP711, in ost2-2D and OE-VAMP711 grown in soil. Watering was stopped for 5-week-old plants grown in soil under 12 h light/12 h dark for 2 weeks, and then photographs were taken. B, Measurement of water loss from detached leaves. Col-0, ost2-2D, OE-VAMP711, in ost2-2D and OE-VAMP711 were grown under short-day conditions for 5 weeks, and the fresh weight of detached seedlings was monitored at the indicated times. The experiment was repeated three times independently. C, ABA-induced stomatal closure assay. Detached rosette leaves from Col-0, ost2-2D, OE-VAMP711 in ost2-2D, and OE-VAMP711 were incubated in KCl buffer for about 2 h and then transferred in a buffer containing 10 μm of ABA for 2 h. Error bars represent sd (stomata number, 70); Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters. The experiment was repeated three times independently. D, Pseudocolor infrared images of Col-0, ost2-2D, OE-VAMP711 in ost2-2D, and OE-VAMP711.The images of 5-week-old plants in soil were taken using an infrared camera. E, Leaf temperature measurement from D using infrared camera software. Error bars represent sd (20 leaves were used); Student’s t test was used to assess statistical significance (P ≤ 0.05). Significant differences were represented by different lowercase letters.
Consistent with this observation, the water loss of detached leaves in ost2-2D-OE-VAMP711 was slower than that in ost2-2D and faster than that in Col-0 and Col-0:OE-VAMP711 (Fig. 6B). In order to determine whether the overexpression of VAMP711 represses ABA insensitivity of stomatal closure in ost2-2D, we performed the ABA-induced stomatal closure assay. Indeed, stomatal closure was insensitive to ABA in the ost2-2D mutant, and the overexpression of VAMP711 partially rescued the ost2-2D stomatal closure phenotype in response to ABA (Fig. 6C).
Since water loss by transpiration leads to temperature change in the leaf surface, we measured the leaf temperature of Col-0, ost2-2D, ost2-2D-OE-VAMP711, and OE-VAMP711 plants. The leaf temperature of ost2-2D-OE-VAMP711 was higher than that of ost2-2D mutant (Fig. 6, D and E). We also transformed the ProUBQ10:GFP-VAMP711 plasmid into the ost2-2D background and obtained two T4 homologous lines, ost2-2D-OE-VAMP711-1#/2#, in which the level of VAMP711 expression was higher than that in ost2-2D (Supplemental Fig. S7C). The ost2-2D-OE-VAMP711-1#/2# displayed a phenotype similar to that of the ost2-2D-OE-VAMP711 plants (Supplemental Fig. S7, E and F). Our results suggest that VAMP711 interacts with and represses AHA1 activity.
DISCUSSION
Drought is an adverse environmental factor that limits plant growth and crop productivity. Stomatal pores act as the gates for gas exchange (CO2, O2, and H2O) and play an important role in the control of water loss in response to drought stress (Yamaguchi-Shinozaki and Shinozaki, 2006; Kim et al., 2010). Regulation of stomatal closure is the first step in response to water loss (Yamaguchi-Shinozaki and Shinozaki, 2006). Although a complex signal network regulates the process of stomatal closure, the only well-studied pathway is the ABA-mediated signaling pathway in drought stress response (Munemasa et al., 2015). The regulation of stomatal closure by ABA causes a series of physiological changes, such as cytoplasm alkalization, ion rebalance (Ca2+, K+), P-type H+-ATPase suppression (Pei et al., 1998; Fujii et al., 2009; Joshi-Saha et al., 2011). However, it is not well understood how ABA-mediated PM H+-ATPase activity inhibition is regulated. In this study, we provide evidence that VAMP711 represses PM H+-ATPase activity to regulate ABA-dependent stomatal closure. Under drought stress, accumulation of ABA leads to VAMP711 physically interacting with AHA1 and AHA2 and inhibits their activities to promote stomatal closure (Figs. 1B, 3A, and 6C). VAMP711 negatively regulates vacuolar H+-ATPase activity in yeast strains (Leshem et al., 2006), which suggests that VAMP711 may also play a role in regulating cytoplasm alkalization.
VAMP711 is highly expressed in guard cells in Arabidopsis, and down-regulation of VAMP711 in Arabidopsis impairs stomatal closure during drought stress or ABA treatment (Leshem et al., 2010). However, the underlying mechanism is not clear. Our results demonstrate that VAMP711 inhibits PM H+-ATPase activity through direct interaction to promote stomatal closure, indicating that VAMP711 and AHA1/AHA2 work together to regulate stomatal closure in response to drought stress. VAMP711 protein is involved in endosome trafficking and vesicle fusion and is reported to localize on the vacuolar membrane and PVC (Leshem et al., 2006); however, AHA1 and AHA2 are plasma membrane localized proteins (Harper et al., 1989). It is thus interesting to know the interacting location of these two proteins. The BiFC assay showed that the interaction between AHA1 and VAMP711 was on both the plasma membrane and punctate structures, indicating that VAMP711 could interact with AHA1 at least partially at the plasma membrane, although we could not exclude VAMP711 interaction with AHA1 in other locations. Intriguingly, studies with other proteins also showed similar phenomena. For example, catalase proteins localize in peroxisomes (Apel and Hirt, 2004); however, under salt and drought stresses, the plasma membrane proteins salt tolerance receptor-like cytoplasmic kinase1 (Zhou et al., 2018) and Calcium Protein Kinase8 (Zou et al., 2015) interact with, phosphorylate, and activate Catalase C (in rice) and Catalase 3 (in Arabidopsis) on the plasma membrane, respectively. Therefore, it is possible that the VAMP711 protein could be translocated to the plasma membrane through certain unclear mechanisms or under certain physiological conditions. Indeed, our results indicated that ABA treatment promoted the interaction of VAMP711 with AHA1 and might partially affect the localization of VAMP711 to the plasma membrane. However, the observation and mechanism need to be further clarified. A recent study reported that VAMP711 is mislocalized to the plasma membrane in the ap-3 mutant, providing a possibility for future study (Feng et al., 2017). In addition, other possibilities for the physical interaction between VAMP711 and AHA1 cannot be excluded. For example, it has been observed that prevacuolar compartments/multivesicular bodies (PVC/MVB) could fuse with the plasma membrane during pathogen attack (An et al., 2006a, 2006b). Thus, it is possible that PVCs with a small portion of VAMP711 could move close to or fuse with the plasma membrane to promote the interaction and inhibit PM H+-ATPase activity under drought stress (Vida and Emr, 1995). This is supported by the fact that ABA treatment did not dramatically induce the plasma membrane localization of VAMP711, as determined by an immunoblot assay. Another possibility is the potential direct membrane contact site or stress-induced membrane contact site between the tonoplast and plasma membrane, which is yet to be observed (Pérez-Sancho et al., 2016). However, we also cannot exclude the possibility that VAMP711 represses PM H+-ATPase activity by regulating AHA1/AHA2 trafficking under drought stress.
In Arabidopsis, AHA1 and AHA2 are two highly expressed proteins and share higher similarity with other members of the AHA family. AHA1 and AHA2 are essential for plant growth and development, and the aha1 aha2 double mutant shows embryonic lethality (Harper et al., 1990). The AHA1 open stomata2-2D (ost2-2D) mutant plants with constitutive higher PM H+-ATPase activity abolishes the stomatal closure capacity under ABA treatment and are hypersensitive to drought stress (Merlot et al., 2007), indicating that the repression of PM H+-ATPase activity is essential for plant drought responses. SnRK2.2 but not OST1/SnRK2.6 directly phosphorylates AHA2 in vitro, and the ABA-mediated PM H+-ATPase activity inhibition is largely reduced in overexpression lines of the protein phosphatase gene Hypersensitive to ABA1 (HAB1), snrk2.2 snrk2.3 double mutant, and ABA sensors pyrabactin resistance/pyrabactin resistance-like (pyr/pyl) sextuple mutant plants (Planes et al., 2015). OST1/SnRK2.6 is required for the dephosphorylation of the Thr-947 site of AHA2 to inhibit PM H+-ATPase activity (Merlot et al., 2007; Planes et al., 2015). These results suggest that ABA signal is involved in inhibition of PM H+-ATPase activity in response to drought stress. However, the detailed mechanism is not understood.
In our study, we demonstrated that VAMP711 is involved in the ABA-mediated inhibition of PM H+-ATPase activity. However, ABA treatment also induced a decrease in PM H+-ATPase activity in the vamp711 mutant, although the decrease was not as great as in the wild type, suggesting that VAMP711 functions redundantly with VAMP711 homologous proteins. In Arabidopsis, the VAMP7C family contains four genes—VAMP711–714. It is reported that they are all involved in regulating drought stress (Leshem et al., 2010). It is possible that all VAMP7s are involved in the regulation of ABA-mediated PM H+-ATPase activity inhibition.
Our results and previous reports show that overexpression of VAMP711 does not significantly improve plant drought resistance, suggesting that the expression level of wild-type VAMP711 is sufficient to regulate stomatal switch in Arabidopsis (Leshem et al., 2010). In the AHA1 ost2-2D mutant, it required more VAMP711 molecules to bind more OST2-2D proteins to sufficiently inhibit AHA1 activity. On the other hand, other regulators, such as OST1/SnRK2.6, SnRK2.2, and SnRK2.3, are required to regulate PM H+-ATPase activity in response to drought stress.
MATERIALS AND METHODS
Plant Materials and Growth
The wild-type ecotype used in this study was Arabidopsis (Arabidopsis thaliana) Col-0. The mutant ost2-2D was described previously (Merlot et al., 2007). The AHA1-overexpresing transgenic plants (Pro35S:GFP-AHA1), VAMP711-overexpressing transgenic plants (ProUBQ10:GFP-VAMP711), and the complementary materials (ProVAMP711:VAMP711; com-1, 2) were generated through Agrobacterium tumefacience GV3101-mediated transformation. The VAMP711-overexpressing transgenic plants in the ost2-2D mutant were generated by crossing.
Seeds were sterilized in a solution containing 20% (v/v) sodium hypochlorite and 0.1% (v/v) Triton X-100 for 10 min, washed five times with sterilized distilled water, sown on MS medium containing 2.5% (w/v) Suc and 0.3% (for horizontal growth) or 0.5% (for vertical growth) Phytagel agar (Sigma-Aldrich), and grown in a growth chamber at 23°C after keeping them at 4°C for 3 d in darkness.
Arabidopsis and Nicotiana benthamiana plants were grown in soil under 8 h light/16 h dark at 23°C and 60% relative humidity.
Generation of vamp711 CRISPR/Cas9 Mutants
A pair of selected small guide RNA primers (C1, CCTCGTGGCTCGTGGCACGG; and C2, CGCCAAACAGATCCTCGAGA) in the VAMP711 gene were cloned into the PHEC401 vector as described previously (Wang et al., 2015). The construct was transformed into wild-type Arabidopsis. The homozygous vamp711 mutants (vamp711-6, vamp711-7) were identified by sequencing.
Split-Luciferase Complementation Assays
For the split-luciferase assays, the coding sequence of VAMP711 was amplified and cloned into pCM1300-Cluc and pCM1300-Nluc vectors between the KpnI and SalI sites, respectively, and the coding sequence of AHA1 was also amplified and cloned into pCM1300-Cluc and pCM1300-Nluc vectors between the KpnI and SalI sites, respectively. The constructs were transformed into the Agrobacterium tumefacience GV3101 and then infiltrated into N. benthamiana leaves. LUC signal was collected after 3 d by using a cooled CCD camera (iKon-L936; Andor Tech) after spraying 1 mm of d-luciferin on the leaves (Promega; Chen et al., 2008). Related LUC activity was calculated using the Winvie32 software. Primer sequences are listed in Supplemental Table S1.
BIFC Assay
BIFC assay was used to detect the location of interaction between AHA1 and VAMP711. The coding sequences of AHA1 and VAMP711 were cloned into pSYCE (MR) and pSYNE (R) 173 (for split YFP C-terminus/N-terminus fragment expression) vectors at the BamHI/SalI and SalI/KpnI sites, respectively. The coding sequence of AHA1 was cloned into pSYCE (MR) and pSYNE (R) 173 vectors by using BamHI/SalI, respectively; the coding sequence of VAMP711 was also cloned into pSYNE (R) 173 and pSYCE (MR) vectors at the SalI/KpnI sites, respectively; and the coding sequence of SYP22 was cloned into pSYCE (MR) vector by using BamHI/XhoI (Walter et al., 2004; Waadt et al., 2008). Primer sequences are listed in Supplemental Table S1. The constructs were transformed into GV3101 and then infiltrated into N. benthamiana leaves. The YFP fluorescence signal was detected using a Zeiss LSM 710 META confocal microscope.
Yeast Two-Hybrid Assays
The intracellular coding sequences of AHA1; AHA1 (N), AHA1 (centerloop), AHA1 (C100), AHA1 (C100)Δ(887–897), and AHA2 (C100)Δ(887–897) were cloned into the pGBKT7 vector between the EcoRI and BamHI sites. The C terminus coding sequences of AHA3, AHA6, and AHA9 were cloned into the pGBKT7 vector between the EcoRI and BamHI sites. The coding sequence of VAMP711, which contains a longin domain and a SNARE domain, was cloned into the pGADT7 vector between the EcoRI and BamHI sites. Primer sequences are listed in Supplemental Table S1. The indicated constructs were transformed into the yeast strain AH109 and growth assays were based on Yeast Protocols Handbook (Clontech).
RT-PCR
Total RNA of 10-d-old seedlings grown on MS medium was extracted with Trizol reagent (Invitrogen). RNase-free DNase I (Takara) was used to remove genomic DNA in total RNA. Total RNA was reverse-transcribed with Moloney Murine Leukemia Virus reverse transcriptase (Promega). The cDNA was used for RT-PCR analysis.
Confocal Microscopy Images
Confocal images were captured using a Zeiss LSM 710 META confocal microscope with a 40/1.4 oil objective. Five-day-old seedlings were stained with 5 μm of FM4-64 (Invitrogen) for 1 or 5 min. GFP and FM4-64 fluorescent signals were captured using multitrack function (488 nm for GFP, 561 nm for mCherry and FM4-64). Zeiss LSM image-processing software was used to extract confocal images.
The PSC colocalization plug-in in the ImageJ program (http://rsb.info.nih.gov/ij/) was used to analyze the colocalization of two fluorescent signals. The values of the linear Pearson correlation coefficient and nonlinear Spearman correlation coefficient were representative for the extent of colocalization.
Plasma Membrane Vesicle Isolation
Four-week-old plant materials were prepared for isolating plasma membrane vesicles using the aqueous two-phase separation method (Qiu et al., 2002; Yang et al., 2010). Plants were homogenized with an isolation buffer (0.33 m Suc, 0.2% [w/v] bovine serum albumin, 10% [w/v] glycerol, 5 mm dithiothreitol (DTT), 5 mm EDTA, 5 mm ascorbate, 0.6% [w/v] polyvinylpyrrolidone, 0.2% [w/v] casein, 1 mm phenylmethylsulfonyl fluoride, and 50 mm HEPES-KOH, pH 7.5). The homogenate was centrifuged at 10,000g for 10 min, and then the supernatant was filtered through two layers of Miracloth. The microsomal pellet was obtained by centrifugation for 1 h at 100,000g from the supernatant. The pellet was then resuspended in buffer I (0.33 m Suc, 3 mm KCl, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 0.1 mm EDTA, 5 mm potassium phosphate, and 1× protease inhibitor, pH 7.8). The prepared two-phase mixture (6.2% [w/w] Dextran T-500, 0.33 m Suc, 3 mm KCl, and 6.2% [w/w] polyethylene glycol 3350 in 5 mm potassium phosphate, pH 7.8) was used to separate the plasma membrane (up phase) from other membranes (down phase). The final phases were collected and diluted with buffer II (0.33 m Suc, 10% [w/v] glycerol, 0.1% [w/v] bovine serum albumin, 2 mm DTT, 0.1 mm EDTA, 20 mm HEPES-KOH, and 1× protease inhibitor, pH 7.5), and then centrifuged for 1 h at 100,000g. The pellet was resuspended with buffer I containing 1 mm EDTA. Finally, the plasma membrane vesicles were used for PM H+-ATPase activity and western blot analysis.
PM H+-ATPase Activity
The H+-transport activity of PM H+-ATPase was measured as described previously (Qiu et al., 2002). Transport of H+ by PM H+-ATPase forms an inside-acid pH gradient (ΔpH) in the vesicles, and it was measured as a decrease (quench) in the fluorescence of quinacrine (a pH-sensitive fluorescence probe). The measurement buffer (2 mL) containing 5 mm quinacrine, 3 mm MgSO4, 100 mm KCl, and 25 mm 1, 3-Bis [Tris(hydroxylmethyl)methylamino] propane-HEPES, pH 6.5, 250 mm mannitol, and 50 μg/mL of plasma membrane protein was used to detect the H+-ATPase activity. The mixtures were incubated at 25°C in a cuvette and were placed in a fluorescence spectrophotometer (Hitachi F-7000). Fluorescence reading was begun when the mixtures were being stirred in dark for 5 min. H+-ATPase activity was initiated by adding 3 mm of ATP, and the ΔpH was measured at 430 nm excitation and 500 nm emission wavelengths. To dissipate the remaining pH gradient, 20 mm m-chlorophenylhydrazone (cccp), a protonophore, was added at the end of each reaction. The PM H+-ATPase hydrolysis activity was measured as described previously (Qiu et al., 2002).
Mensuration of H+ Flux
The H+ fluxes were measured in the meristematic zone (approximately 100 μm from the root tip) using the noninvasive microtest technique (Younger; Xuyue [Beijing] Sci &Tech; Yang et al., 2010). The H+ selective micropipettes were filled with 10-μm columns of H+-selective liquid exchange cocktails (Fluka 95293). An Ag/AgCl wire electrode was inserted into the micropipettes from the back such that it made contact with the electrolyte buffer. The H+ concentration was evaluated by moving the H+ microelectrode between two positions at 20 to 40 μm from the root. Five-day-old Arabidopsis seedlings were treated with a pH-7.8 buffer (0.1 mm CaCl2, 0.1 mm KCl, 0.03 mm HEPES, and 75 mm NaCl) and a pH-5.8 buffer (0.1 mm CaCl2, 0.1 mm KCl, 0.03 mm 2-N-morpholinoe thanesulfonate (MES). To detect the effect of ABA on H+ flux, 2 μm of ABA was added into above-mentioned buffer.
Water Loss and Stomatal Aperture Assays
The rosette leaves from 5-week-old plants in soil were detached to examine water loss, and the photographs of leaves were taken 0 and 3 h after detachment. To measure the rate of water loss, the 5-week-old plants in soil were used to perform water loss measurement. The weight of the detached leaves was measured every 0.5 h.
The rosette leaves of 5-week-old plants in soil were used to detect the stomatal closure under the ABA treatment. The leaves were incubated in opening buffer (50 mm KCl, 10 mm CaCl2, and 10 mm MES, pH 6.15) in a growth chamber for 2 h to open stomatal pores completely. Stomatal apertures were measured after adding 10 of mM ABA for 2 h. More than 70 stomatal pores were observed to measure the aperture in three independent experiments.
Chlorophyll Content Analysis
The rosette leaves from 5-week-old plants in soil with 12 h light/12 h dark were used to determine chlorophyll content. The rosette leaves were weighed and added in 1 mL of 80% (v/v) acetone, and then the samples were incubated in darkness for 12 h at room temperature. The absorption of each sample was measured at 663 and 645 nm with a spectrophotometer. The chlorophyll content was calculated as follows: total chlorophyll = Chla + Chlb.
Relative Water Content
The 5-week-old plants in soil with 12 h light/12 h dark were used to detect relative water content. The fresh weight (Wfresh) was determined, and then the samples were dried at 65°C for 2 d and weighed to determine the dry weight (Wdry). The relative water content was calculated as follows: relative water content (%) = (Wfresh − Wdry)/Wfresh * 100%.
Infrared Thermograph Imaging
Thermal imaging was used to monitor the leaf temperature as described previously (Xie et al., 2006). The plants were well watered, and watering was stopped for 1 week to decrease plant humidity. Five-week-old plants in soil were used to measure leaf temperature by thermal imaging camera.
Accession Numbers
Sequence data in this study can be found in the Arabidopsis Genome Initiative database under the following accession numbers: VAMP711, At4g32150; AHA1, At2g18960; AHA2, At4g30190.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of ABA on PM H+-ATPase activity in Arabidopsis.
Supplemental Figure S2. Analysis of the localization of and the interaction between AHA1 and VAMP711.
Supplemental Figure S3. VAMP711 interacts with the C terminus of AHA2.
Supplemental Figure S4. The vamp711 mutants are generated by CRISPR/Cas9 technology.
Supplemental Figure S5. The vamp711 mutants are tolerant to high pH.
Supplemental Figure S6. The analysis of PM H+-ATPase protein level in the plasma membrane in Col-0 and vamp711 mutant seedlings,
Supplemental Figure S7. The drought-stress phenotype and expression level of VAMP711 in the indicated plants.
Supplemental Table S1. Primers used in this study.
Footnotes
This work was supported by the National Genetically Modified Organisms Breeding Major Projects (2016ZX08009002) and the National Natural Science Foundation of China (31430012, U1706201, 31670260).
Articles can be viewed without a subscription.
References
- An Q, Ehlers K, Kogel KH, van Bel AJ, Hückelhoven R (2006a) Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol 172: 563–576 [DOI] [PubMed] [Google Scholar]
- An Q, Hückelhoven R, Kogel KH, van Bel AJ (2006b) Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol 8: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Basu J, Shen N, Dulubova I, Lu J, Guan R, Guryev O, Grishin NV, Rosenmund C, Rizo J (2005) A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol 12: 1017–1018 [DOI] [PubMed] [Google Scholar]
- Cai S, Chen G, Wang Y, Huang Y, Marchant DB, Wang Y, Yang Q, Dai F, Hills A, Franks PJ, et al. (2017) Evolutionary conservation of ABA signaling for stomatal closure. Plant Physiol 174: 732–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L, Iori V, Marra M, Aducci P (2000) Phosphorylation-dependent interaction between plant plasma membrane H(+)-ATPase and 14-3-3 proteins. J Biol Chem 275: 9919–9923 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Falhof J, Pedersen JT, Fuglsang AT, Palmgren M (2016) Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol Plant 9: 323–337 [DOI] [PubMed] [Google Scholar]
- Feng QN, Song SJ, Yu SX, Wang JG, Li S, Zhang Y (2017) Adaptor Protein-3-Dependent vacuolar trafficking involves a subpopulation of COPII and HOPS tethering proteins. Plant Physiol 174: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Uemura T, Ebine K, Nishimori Y, Ueda T, Nakano A, Sato MH, Fukao Y (2014) Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol 55: 781–789 [DOI] [PubMed] [Google Scholar]
- Gévaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M (2007) Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol 144: 1763–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Surowy TK, Sussman MR (1989) Molecular cloning and sequence of cDNA encoding the plasma membrane proton pump (H+-ATPase) of Arabidopsis thaliana. Proc Natl Acad Sci USA 86: 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Manney L, DeWitt ND, Yoo MH, Sussman MR (1990) The Arabidopsis thaliana plasma membrane H(+)-ATPase multigene family. Genomic sequence and expression of a third isoform. J Biol Chem 265: 13601–13608 [PubMed] [Google Scholar]
- Haruta M, Tan LX, Bushey DB, Swanson SJ, Sussman MR (2018) Environmental and genetic factors regulating localization of the plant plasma membrane H+-ATPase. Plant Physiol 176: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Sugimoto M, Higaki T, Yaeno T, Nagami A, Irie M, Fujimi M, Miyamoto M, Akita K, Negi J, Shirasu K, et al. (2013) A Munc13-like protein in Arabidopsis mediates H+-ATPase translocation that is essential for stomatal responses. Nat Commun 4: 2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. (2005) SNAREs and traffic. Biochim Biophys Acta 1744: 493–517 [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C, Leung J (2011) Abscisic acid signal off the STARting block. Mol Plant 4: 562–580 [DOI] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol 43: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Leshem Y, Melamed-Book N, Cagnac O, Ronen G, Nishri Y, Solomon M, Cohen G, Levine A (2006) Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc Natl Acad Sci USA 103: 18008–18013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y, Golani Y, Kaye Y, Levine A (2010) Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J Exp Bot 61: 2615–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, et al. (2007) Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H(+)-ATPase. J Biol Chem 266: 20470–20475 [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287–290 [DOI] [PubMed] [Google Scholar]
- Pérez-Sancho J, Tilsner J, Samuels AL, Botella MA, Bayer EM, Rosado A (2016) Stitching organelles: organization and function of specialized membrane contact sites in plants. Trends Cell Biol 26: 705–717 [DOI] [PubMed] [Google Scholar]
- Planes MD, Niñoles R, Rubio L, Bissoli G, Bueso E, García-Sánchez MJ, Alejandro S, Gonzalez-Guzmán M, Hedrich R, Rodriguez PL, et al. (2015) A mechanism of growth inhibition by abscisic acid in germinating seeds of Arabidopsis thaliana based on inhibition of plasma membrane H+-ATPase and decreased cytosolic pH, K+, and anions. J Exp Bot 66: 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Sutter JU, Blatt MR (2004) A new catch in the SNARE. Trends Plant Sci 9: 187–195 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rober-Kleber N, Albrechtová JTP, Fleig S, Huck N, Michalke W, Wagner E, Speth V, Neuhaus G, Fischer-Iglesias C (2003) Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiol 131: 1302–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. (2007) Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol 144: 6–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Hayashi N, Kawagoe Y, Mochizuki S, Inoue H, Mori M, Nishizawa Y, Jiang CJ, Matsui M, Takatsuji H (2016) Rice OsVAMP714, a membrane-trafficking protein localized to the chloroplast and vacuolar membrane, is involved in resistance to rice blast disease. Plant Mol Biol 91: 81–95 [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H(+)-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Uemura T, Sato MH, Takeyasu K (2005) The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett 579: 2842–2846 [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Wang Y, Williamson L, Holroyd GH, Tagliavia C, Murchie E, Theobald J, Knight MR, Davies WJ, Leyser HMO, et al. (2006) The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol 16: 882–887 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yamauchi S, Takemiya A, Sakamoto T, Kurata T, Tsutsumi T, Kinoshita T, Shimazaki K (2016) The plasma membrane H+-ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiol 171: 2731–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS, et al. (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Adachi Y, Ye W, Hayashi M, Nakamura Y, Kinoshita T, Mori IC, Murata Y (2013) Difference in abscisic acid perception mechanisms between closure induction and opening inhibition of stomata. Plant Physiol 163: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Karnik R, Wang Y, Wallmeroth N, Blatt MR, Grefen C (2015) The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27: 1697–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YB, Liu C, Tang DY, Yan L, Wang D, Yang YZ, Gui JS, Zhao XY, Li LG, Tang XD, Yu F, Li JL, et al. (2018) The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30: 1100–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JJ, Li XD, Ratnasekera D, Wang C, Liu WX, Song LF, Zhang WZ, Wu WH (2015) Arabidopsis Calcium-dependent protein kinase 8 and Catalase 3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27: 1445–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]