Computer vision allows the identification of genes controlling uncharacterized lesion traits in Arabidopsis.
Abstract
Plant resistance to generalist pathogens with broad host ranges, such as Botrytis cinerea (Botrytis), is typically quantitative and highly polygenic. Recent studies have begun to elucidate the molecular genetic basis of plant-pathogen interactions using commonly measured traits, including lesion size and/or pathogen biomass. However, with the advent of digital imaging and high-throughput phenomics, there are a large number of additional traits available to study quantitative resistance. In this study, we used high-throughput digital imaging analysis to investigate previously poorly characterized visual traits of plant-pathogen interactions related to disease resistance using the Arabidopsis (Arabidopsis thaliana)/Botrytis pathosystem. From a large collection of visual lesion trait measurements, we focused on color, shape, and size to test how these aspects of the Arabidopsis/Botrytis interaction are genetically related. Through genome-wide association mapping in Arabidopsis, we show that lesion color and shape are genetically separable traits associated with plant disease resistance. Moreover, by employing defined mutants in 23 candidate genes identified from the genome-wide association mapping, we demonstrate links between loci and each of the different plant-pathogen interaction traits. These results expand our understanding of the functional mechanisms driving plant disease resistance.
The ability to resist biotic attackers, including plant pathogens, is central to a plant’s survival and fitness. However, evolving resistance is complicated by the immense variety of pathogenic species that can attack an individual plant. Pathogens differ in their lifestyle and host range, with each having different optimal virulence mechanism(s). Specialist biotrophic pathogens often coevolve with host plants and develop virulence strategies that target specific resistance mechanisms within the host. These pathogens must strike a delicate balance between keeping the host plant cells alive while absorbing nutrients, avoiding detection, and suppressing plant defenses. Plant defenses against coevolving specialists are tailored to the specific pathogen, often involving gene products that directly or indirectly recognize effectors or molecular patterns specific to that pathogen. This coevolutionary link between host and specialist pathogen frequently leads to large-effect, gene-for-gene resistance involving specific pathogen recognition genes (Jones and Dangl, 2006). In contrast, necrotrophic pathogens often are generalists, deploying a diverse array of virulence mechanisms designed to disrupt and kill host plant cells (e.g. diverse phytotoxins, enzymes, and microRNAs; Laluk and Mengiste, 2010; Veloso and van Kan, 2018). Coevolution is more complicated in host-generalist pathogen interactions because the pathogen must generalize to infect numerous hosts. In turn, host resistance to generalist necrotrophs is highly quantitative and polygenic, involving a complex network of traits, including physical barriers, such as cuticle formation and cell wall modification; chemical barriers, such as secondary metabolites or reactive oxygen species; and inducible defenses (Rowe and Kliebenstein, 2008; Laluk and Mengiste, 2010; Windram et al., 2012; Corwin et al., 2016a).
Plant-pathogen interactions are highly complex systems that can affect a wide array of traits, from visual to chemical. However, most molecular plant pathology research relies either on lesion size or pathogen biomass within the infected plant tissue to quantify the severity of infection. Whereas these traits are easily measured and reflect plant susceptibility to a pathogen, they do not fully describe all aspects of disease resistance within the plant host. For instance, comparing the molecular and quantitative genetic basis of resistance using lesion size versus the underlying biochemical responses found that different genes could contribute to these traits. This suggests that lesion size measurements do not provide the full picture of the resistance response (Rowe and Kliebenstein, 2008; Bock et al., 2010; Li et al., 2015; Corwin et al., 2016a; Schwanck and Del Ponte, 2016; Barbedo, 2017; Matsunaga et al., 2017). There has been recent interest in extending our understanding of plant-pathogen resistance through more extensive phenotyping of disease symptoms, including hyperspectral imaging of lesions (Montes et al., 2007; Kuska et al., 2015; Mutka and Bart, 2015; Leucker et al., 2016). This analysis has differentiated between diseases on the same plant using light spectra from 400 to 1,000 nm to detect biochemical responses (Mahlein et al., 2012). These spectra provide a blend of symptoms characterized not only by the extent of necrosis but also by water content, pigment modification, tissue function, and the appearance of fungal structures (Mahlein et al., 2012). However, the molecular mechanisms controlling these visual and biochemical aspects of the plant-pathogen interaction and their relation to pathogen biomass/lesion size mechanisms are largely unstudied. Measuring plant-pathogen interactions from this multidimensional view could reveal new insights into the physiology of pathogen strategy and host response.
One developing model system to directly study the complexity of molecular plant resistance to generalist pathogens is the Arabidopsis (Arabidopsis thaliana)/Botrytis cinerea (Botrytis) pathosystem (Corwin and Kliebenstein, 2017). Botrytis is a generalist necrotrophic pathogen with a broad range of hosts from bryophytes through gymnosperms and angiosperms (Jarvis, 1977; Staats et al., 2005; Choquer et al., 2007; Elad et al., 2016). Intraspecific genetic variation facilitates this broad host range and quantitatively influences the interaction with genetically varied hosts (Giraud et al., 1997; Alfonso et al., 2000; Váczy et al., 2008; Kretschmer et al., 2009; Stefanato et al., 2009; Rowe and Kliebenstein, 2010; Valiuskaite et al., 2010; Amselem et al., 2011; Aguileta et al., 2012; Atwell et al., 2015; Corwin et al., 2016b). Classical forward genetic screens have shown that Arabidopsis resistance to Botrytis involves key signaling pathways, such as jasmonate signaling and the BIK/BAK response system (Thomma et al., 1999; Veronese et al., 2006). Our recent use of high-throughput digital imaging analysis for lesion size in combination with host genome-wide association (GWA) mapping showed that the Arabidopsis/Botrytis interaction relied on natural genetic variation in thousands of host genes; however, most of these host genes were associated with altering the interaction of only a subset of Botrytis isolates (Corwin et al., 2016b). Critically, GWA does not impart a priori assumptions on the genes or mechanisms that may play a role in controlling the trait in question, thus reducing experimental bias. This is exemplified by the observation that many of the genes identified by GWA mapping in Arabidopsis have not been linked previously to this pathosystem (Jarvis, 1977; Staats et al., 2005; Choquer et al., 2007; Rowe and Kliebenstein, 2008; Mengiste et al., 2010; Valiuskaite et al., 2010; Amselem et al., 2011; Corwin et al., 2016a; Corwin and Kliebenstein, 2017). For example, LATE ELONGATED HYPOCOTYL (LHY; At1g01060), a key developmental and circadian-regulated gene not linked previously to disease resistance, was identified as having a significant effect on Botrytis resistance (Corwin et al., 2016a). In addition to known genes, our GWA approach identified disease resistance effects in previously uncharacterized genes, such as At4g01860, which encodes a poorly characterized Cul4-RING E3 component. This suggests that extending GWA analysis to other traits may allow the identification of additional new disease resistance loci. Furthermore, a reanalysis of the digital images collected from our previous study can directly facilitate the exploration of other traits related to disease resistance.
In this study, we reanalyzed the images collected from our previous GWA experiments to measure visual aspects of lesion development that are not commonly measured, specifically lesion shape, color, and texture, and relate these to disease resistance (Corwin et al., 2016a). A GWA mapping analysis of select traits in Arabidopsis revealed a large compendium of genes associated with lesion size, yellowness, greenness, and shape. Gene Ontology (GO) enrichment analysis showed that there were biological processes specific to subsets of traits rather than processes affecting all lesion-associated traits. To validate the results of the GWA mapping, we selected 23 genes associated with one or more of the lesion traits and compared their T-DNA knockout mutant phenotypes. The validation rate was highest for lesion size, at 60%, and lower for lesion color and shape traits, specifically 33% for lesion yellowness, 38% for lesion greenness, and 20% for lesion eccentricity. This further demonstrated that the genes affected subsets of lesion traits rather than being common to all traits. This confirms that expanding our phenotypic analysis of plant-pathogen interactions to novel visual traits like lesion shape and color, rather than focusing solely on lesion size, can expand our knowledge of plant-pathogen interactions and identify different mechanisms.
RESULTS
Identification of Alternative Disease-Related Traits
Using a custom digital image-analysis pipeline, we previously measured lesion size (area) to identify Arabidopsis genes controlling lesion size within the Arabidopsis/Botrytis pathosystem using four diverse pathogen genotypes (Corwin et al., 2016a). That previous study used 96 diverse Arabidopsis accessions chosen based on similar flowering time to minimize ontogenic variation, which also decreased population structure and rare alleles within the collection. The host-pathogen interaction was recorded digitally within a randomized complete block design over two experiments with four independent biological replicates per genotype per experiment. Using the same images from the previous experiment, we adapted our imaging pipeline to quantify 75 different lesion parameters for each infected leaf. For example, these parameters included the number of pixels for specific hues within the lesion, lesion perimeter, proportion of the leaf the lesion occupied, length of the major and minor axes of the lesion, eccentricity of the perimeter, and Haralick texture features within the lesion (Fig. 1; Corwin et al., 2016b). As reported previously, there is a wide range of lesion sizes resulting from different plant-pathogen interactions (Fig. 1, A versus C). In addition to lesion size, there is variation in color within both the lesion and the residual leaf. For example, some plant-pathogen interactions result in large chlorotic sectors surrounding the lesions (Fig. 1, A, C, and F), whereas others cause less chlorosis surrounding the developing lesion (Fig. 1, B and E). In our previous work, we have shown that the pathogen is limited solely to the developing lesion and that these chlorotic regions are systemic plant responses to the infection (Rowe and Kliebenstein, 2008; Corwin et al., 2016b). Even within the lesion there are color variants, with some exhibiting more yellow coloring (Fig. 1A), whereas others have a greener hue (Fig. 1D). In addition to lesion color, there is variation in lesion shape, with some lesions exhibiting high symmetry and circularity (Fig. 1, D and F), whereas others are elongated along the major axis of the leaf, possibly due to extended pathogen growth along the midvein (Fig. 1, A and B). This preferential midvein growth was seen previously as genetically variable among Botrytis isolates that infect Arabidopsis (Corwin et al., 2016b). We proceeded to study if these additional measurements of the Arabidopsis/Botrytis interaction provided unique genetic insight into the virulence outcome that was not obtained readily from lesion size alone
Figure 1.
Illustration of different lesion traits. Representative images show Arabidopsis leaves infected with Botrytis illustrating the varying combinations of lesion traits observed. Lesions were observed to be more circular or more eccentric, with pointed ends growing along the midvein of the leaf. Greenness and yellowness of both areas within the lesion and in the surrounding tissue also were highly variable. The water-soaked grayish sectors represent the actual infected lesion. A, Large eccentric lesion with extensive leaf senescence. B, Moderate eccentric lesion with minimal leaf senescence. C, Small lesion with extensive leaf senescence. D, Moderate circular lesion with moderate leaf senescence. E, Small lesion with moderate leaf senescence. F, Moderate circular lesion with extensive leaf senescence.
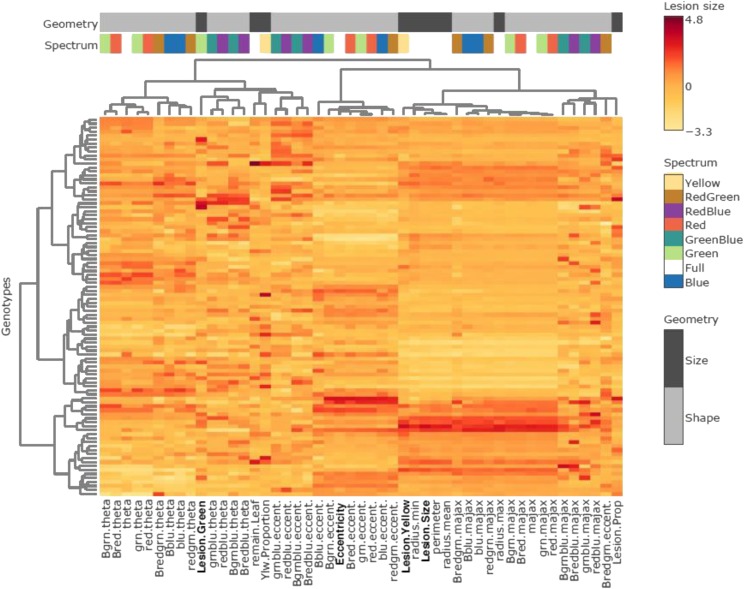
Hierarchical Clustering Analysis for Trait Selection
To better understand the relationship among the measured visual traits and select a subset of traits for further study, we conducted a hierarchical clustering analysis (HCA) of the lesion traits across the 96 Arabidopsis accessions infected with the different Botrytis isolates (Fig. 2; Supplemental Table S1; Supplemental Fig. S1). This analysis allowed us to assess the interrelatedness of these infection traits both with each other and with lesion size to determine whether the traits provide distinct or similar genetic signatures. The HCA showed extensive variation in all the traits dependent on Arabidopsis accessions and Botrytis isolates, leading to a wide range of trait measurements (Supplemental Fig. S1). Interestingly, in these clusters, lesion color and shape traits distinctly clustered apart from full-spectrum lesion size and varied genetically across plant accessions. Using the analysis among all isolates, we chose to proceed with three distinct traits that are wholly distinct from lesion size (mm2). Specifically, these traits are lesion yellowness (the proportion of yellow pixels within the lesion to total pixels within the lesion), lesion greenness (total mm2 of green pixels within the lesion), and lesion eccentricity (maximum lesion diameter – minimum diameter/maximum lesion diameter).
Figure 2.
HCA of lesion traits. The heat map shows the collection of lesion measurements using the Apple517 Botrytis isolate on the collection of 96 Arabidopsis accessions (rows). The bars at the top show if the trait is linked to either size or shape of the lesion and in which spectrum it was measured, according to the legends on the right. The four traits used in this study are shown on the bottom of the plot in boldface. HCA was conducted using the Ward algorithm with z-scaling within each trait.
The HCA shows that lesion yellowness is partially associated with lesion size within each isolate (Fig. 2), but it also shows a discrete pattern distinct from lesion size when all isolates are compared, suggesting that lesion yellowness may describe a distinct genetic and molecular architecture of the plant-pathogen interaction (Supplemental Fig. S1). Lesion greenness and lesion eccentricity were selected to represent the two other distinct branches of the HCA and traits uncorrelated with lesion size (Fig. 2; Supplemental Fig. S1). Lesion greenness may be related to the presence of green islands that were noted in other plant-pathogen interactions but have not been commonly studied (Walters et al., 2008). Lesion eccentricity is a lesion shape variation trait that previous work in our lab tentatively linked to genetic variation in the pathogen’s ability to identify and grow directionally along the midvein of the host leaf (Corwin et al., 2016b). This trait may provide unique genetic insight into the pathogen’s growth orientation preference along the primary leaf vasculature within the host. These four traits represent a broad sampling of the identified variation in the lesion traits, revealing natural genetic variation within both the plant and the pathogen, and provide a diverse array of measurable aspects of the Arabidopsis/Botrytis interaction.
Genetic Variation in New Lesion Traits
We utilized the phenotypic measurements for all four traits across all genotypic interactions of Arabidopsis and Botrytis to quantify the role of host and pathogen genetic variation in controlling these traits. We used linear models to analyze the variance across the 96 Arabidopsis accessions and the four Botrytis isolates. The genetic variation in both the pathogen and the host significantly influenced all four resistance-associated traits (Table 1). The proportion of variance attributable to the Arabidopsis accession was significant for these four traits, although relatively small. The heritability across the entire data set that was attributable solely to Arabidopsis was relatively low, although significant for each trait tested (Table 1). The proportion of heritability attributable to pathogen genetic variation was higher for the color and size traits, ranging from 10% to 40% (Table 1). In contrast, eccentricity was controlled by the interaction of genetic variation in the host and pathogen (Table 1). Thus, there is significant genetic variation for all traits that is dependent upon both host and pathogen genomic diversity.
Table 1. Heritability of lesion traits.
For each of the four traits, the table displays the sum of squares (SS) for each term in an ANOVA, the P value for each term, and the calculated heritability (proportion of total variance) attributed to the specific model terms. The analysis used the model Lesion Trait ∼ Experiment + Accession: Isolate.
| Trait | SS | P | Heritability |
|---|---|---|---|
| Size | |||
| Experiment | 1,224 | 0.002 | |
| Plant | 179,962 | <0.001 | |
| Accession | 25,273 | <0.001 | 3.0% |
| Isolate | 333,466 | <0.001 | 39.5% |
| Accession:isolate | 57,552 | <0.001 | 6.8% |
| Yellow | |||
| Experiment | 2,387,113 | 0.036 | |
| Plant | 964,232,385 | <0.001 | |
| Accession | 70,751,623 | 0.014 | 2.5% |
| Isolate | 557,388,090 | <0.001 | 20.0% |
| Accession:isolate | 176,639,269 | 0.083 | 6.3% |
| Green | |||
| Experiment | 3 | 0.281 | |
| Plant | 3,958 | <0.001 | |
| Accession | 756 | <0.001 | 6.1% |
| Isolate | 1,232 | <0.001 | 10.0% |
| Accession:isolate | 1,084 | <0.001 | 8.8% |
| Eccentricity | |||
| Experiment | 0.00070 | 0.842 | |
| Plant | 13.04996 | 0.003 | |
| Accession | 2.34077 | 0.008 | 4.3% |
| Isolate | 0.00387 | 0.974 | <0.1% |
| Accession:isolate | 6.00843 | 0.022 | 11.1% |
In our further analysis, we obtained the model-corrected least squared means from these linear models to control for the environmental stochasticity. This was designed to focus the analysis on the genetic influence of the plant host in the downstream genetic mapping. Given the significance of the interaction between plant accession and pathogen isolate, we analyzed each lesion trait individually for each pathogen genotype across the diverse Arabidopsis accessions as a separate trait to maximize our ability to identify causal loci.
Analysis of Covariance of Lesion Traits
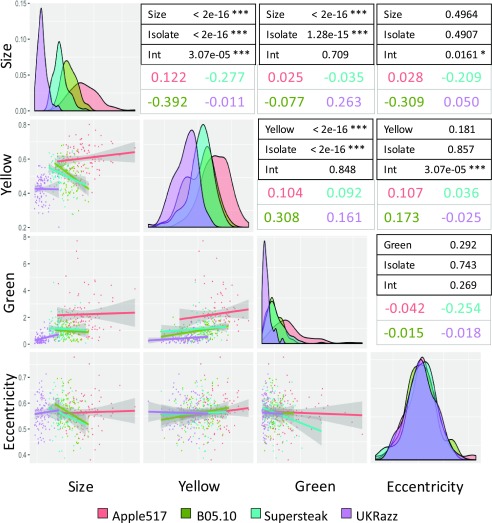
To test how the lesion size, color, and shape traits may correlate across the different plant-pathogen combinations, we conducted an analysis of covariance (ANCOVA) comparing each pair of traits across the 96 diverse Arabidopsis accessions for each of the four Botrytis isolates (Fig. 3). Significant correlations (P < 0.05) between the color and size traits indicate that these three traits are generally related. Lesion yellowness and greenness show a positive correlation, both overall and within each Botrytis isolate group, and this correlation is statistically consistent between groups (Fig. 3). In contrast, lesion color (both greenness and yellowness) and size display different relationships within each Botrytis isolate, in that B05.10 and Supersteak isolates show a negative correlation between lesion yellowness and size whereas Apple517 and UKRazz show a neutral or slightly positive correlation (Fig. 3). Additionally, eccentricity does not display uniform correlation with other lesion traits across all the isolates. Instead, the correlation is highly dependent on the plant-pathogen interaction (Fig. 3). For example, for eccentricity and lesion size, there was a strong negative correlation for the Botrytis isolates B05.10 and Supersteak, whereas there was a very slight positive correlation for Apple517 and UKRazz.
Figure 3.
Isolate dependency of trait correlations across the isolates. Trait correlations between each pair of traits across the 96 Arabidopsis accessions were tested via ANCOVA using each of the four different Botrytis isolates. On the left of the plots are scatterplots showing each trait correlation in each isolate, with the gray areas around the lines showing the 95% confidence interval for the prospective correlation. The dot and line color show the specific isolate tested. The diagonal contains histograms for the distributions of each specific trait across the four Botrytis isolates. The tables on the right show the P values from the ANCOVA for the trait-trait correlations, the Botrytis isolate dependency of these correlations, and the interaction of Botrytis isolate on the trait-trait correlation (Int). The estimated slopes for each trait-trait interaction for each Botrytis isolate are shown by the colored text.
This suggests that genetic variation at the origin of the Arabidopsis response to these diverse Botrytis isolates can be both shared and trait specific. Whereas we found some correlation between the four lesion traits, the host and pathogen genotypes alter these relationships, indicating that there is not a simple mechanistic basis for these relationships. Furthermore, the four lesion traits represent measures of distinct and nuanced interactions between the plant and the pathogen. The genes associated with these traits likely influence different and complementary mechanisms in a multifaceted defense response.
GWA Mapping of Lesion Traits
To identify Arabidopsis genes that are associated with these new lesion traits, we conducted a GWA study. We used the model-corrected trait least squares means of each trait for every Arabidopsis accession infected independently with the four different Botrytis isolates across the traits of lesion size, eccentricity (shape), and yellow and green color. We employed a publicly available database of genomic polymorphisms across the 96 Arabidopsis accessions consisting of 115,301 single-nucleotide polymorphisms (SNPs) with minor allele frequency > 0.2 (Atwell et al., 2010; Corwin et al., 2016a). The model for GWA mapping used a previously published ridge regression approach with a permuted-effects threshold, which has been used to successfully identify causal loci for both plant-pathogen interactions and plant metabolic variation (Shen et al., 2013; Corwin et al., 2016a; Francisco et al., 2016; Kooke et al., 2016). We conducted GWA mapping for each combination separately for each isolate across the Arabidopsis accessions, resulting in four separate GWA maps, one for each trait. Because the variance and mean of each lesion phenotype differed according to pathogen genotype, the resulting effect sizes for Arabidopsis SNPs also varied in magnitude. To create comparative Manhattan plots of the results for a single trait across all four Botrytis isolates, we z-scaled the SNP effect sizes within each pathogen and overlaid the results (Figs. 4 and 5). We calculated the significance threshold for each individual trait by permuting the data 1,000 times and running the ridge regression on the permuted data and locating the 95th percentile (Supplemental Tables S3–S6).
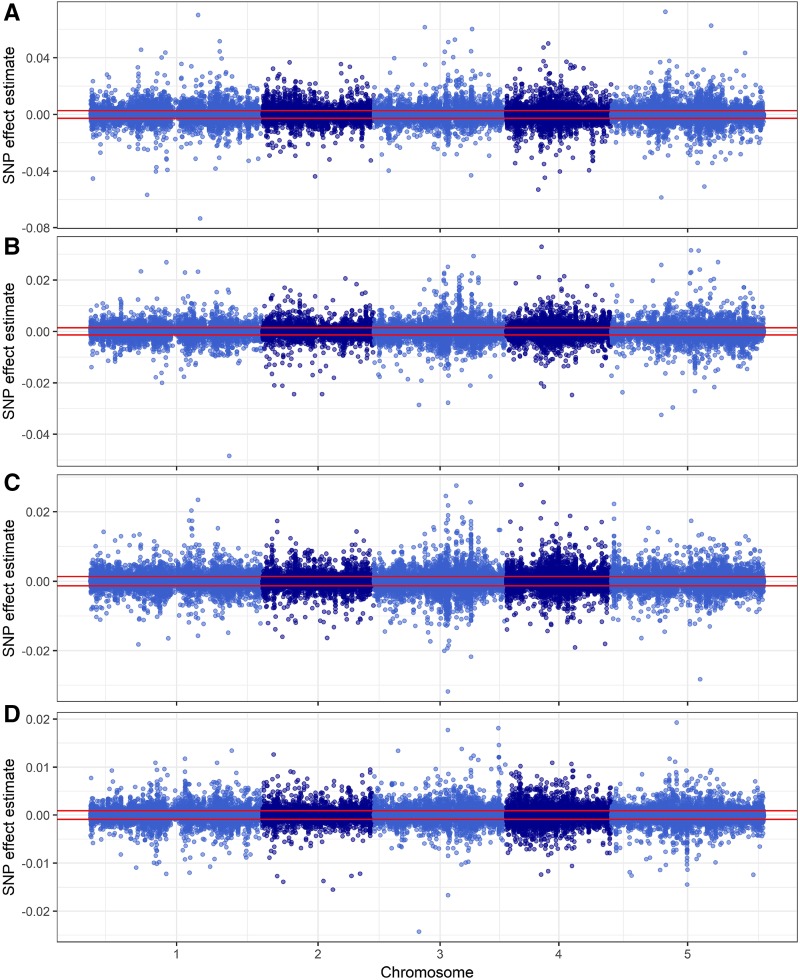
Figure 4.
Lesion size GWA for four Botrytis isolates. Manhattan plots show lesion size trait GWA results as measured on four Botrytis isolates. A, Apple517. B, B05.10. C, Supersteak. D, UKRazz. The horizontal red lines show the significance threshold as estimated by permutation. The effect sizes are measured as cm2 of lesion area.
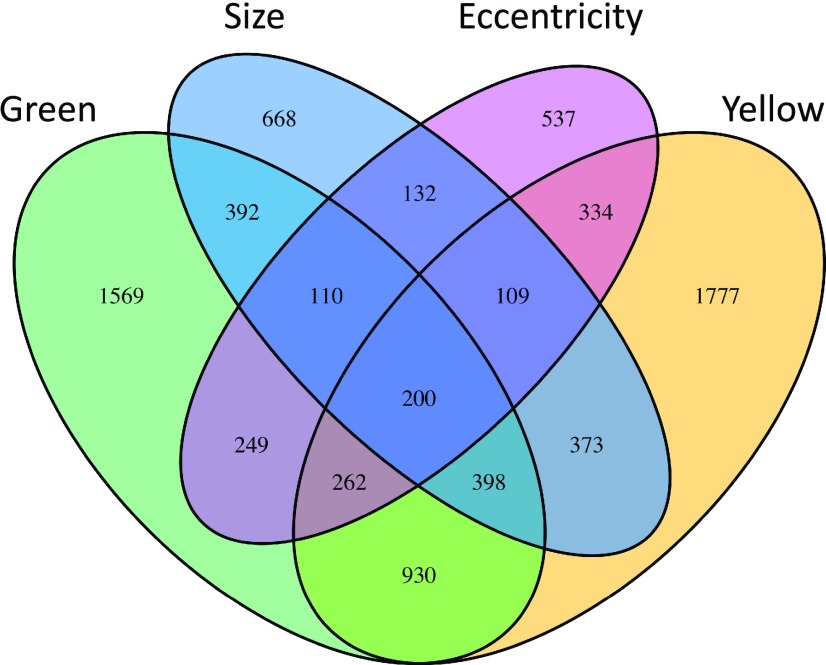
Figure 5.
Venn diagram of trait-associated genes found in GWA mapping. The Venn diagram shows the candidate Arabidopsis genes found to associate with each lesion trait via GWA. The number of genes listed for each lesion trait reflects the number of genes with two or more SNPs above the 95th percentile threshold for that particular trait.
The comparative Manhattan plots (Fig. 4; Supplemental Figs. S2 and S3) showed that all four lesion-associated traits lacked large, distinct SNP peaks. Instead, there are numerous significant SNPs with small to medium effect sizes distributed across the genome, suggesting a highly polygenic nature underlying the genetic variation in these traits. Furthermore, the significant SNPs differed between the Botrytis isolates, suggesting that most SNPs are dependent upon the interaction between plant and pathogen genotypes (Figs. 4 and 5). For lesion yellowness and eccentricity, only Apple517 had significant SNPs, possibly because this isolate had the highest level of phenotypic diversity in these traits (Fig. 3). We next proceeded to identify putative causal loci in the GWA results by calling a gene as significantly associated with a trait if two or more SNPs within the coding region and 1 kb of the 5′ upstream and 3′ downstream regions had effect sizes above the 95th percentile threshold (Chan et al., 2011; Corwin et al., 2016a). This approach has been shown previously to reduce false positives within the data set (Corwin et al., 2016a). Combining the analyses of each trait across the Botrytis isolates for which there were significant SNPs identified 2,382 genes associated with lesion size, 4,383 with lesion yellowness, 4,110 with lesion greenness, and 1,933 with lesion eccentricity. In agreement with the hypothesis that these traits can identify new cellular mechanisms, the majority of the genes identified are associated with only one or a few traits (Fig. 5). Few genes (2.5% of 7,940 total genes identified) were associated with all four lesion-related traits (Fig. 5). Of the genes associated with lesion size, 8% were not associated with any of the other three lesion traits. Likewise, 22% of genes associated with yellowness were associated exclusively with that phenotype, 20% for greenness, and 7% for eccentricity. Overall, 57% of the genes found in this study associated with only one of the lesion traits and 31% associated with exactly two of the lesion traits. These results reinforce the idea that, although the traits are partially correlated, they are quantitative traits that represent different measures and identify different genetic components of the plant-pathogen interaction.
GO Category Enrichment Analysis
We performed a GO category enrichment analysis on biological processes of genes associated with each of the four traits to identify Arabidopsis mechanisms that may influence variation in lesion traits. We performed GO enrichment analysis for each set of candidate genes (Table 2). Both lesion color traits linked to genes involved in defense against pathogens and insects (Table 2). They also identified unique enrichment categories for each trait. Specifically, lesion greenness showed enrichment in maintenance of shoot apical meristem identity and lesion yellowness showed enrichment in lignin biosynthetic processes. In contrast, eccentricity-associated candidate genes were enriched in a variety of unexpected processes, such as negative regulation of flower development and negative regulation of reproductive process, in addition to known pathogen-associated processes, such as cell wall thickening and callose deposition in cell walls. Genes controlling cell wall modification play a role in plant defense strategies to contain the infected area but have unknown effects on lesion shape (Cantu et al., 2008; Mengiste et al., 2010; Bethke et al., 2016). Thus, these novel visual traits identify known defense mechanisms while also extend the analysis out to new pathways.
Table 2. GO category enrichment.
The table displays the top 10 enrichment categories (biological processes) associated with each of the four lesion traits.
| GO Identifier | Term | P | Trait |
|---|---|---|---|
| GO:0006952 | Defense response | 1.40E-05 | Size |
| GO:1901264 | Carbohydrate derivative transport | 0.0003 | Size |
| GO:0009617 | Response to bacterium | 0.0007 | Size |
| GO:0006749 | Glutathione metabolic process | 0.0027 | Size |
| GO:0009556 | Microsporogenesis | 0.0028 | Size |
| GO:0006897 | Endocytosis | 0.0043 | Size |
| GO:0030029 | Actin filament-based process | 0.0048 | Size |
| GO:0048236 | Plant-type spore development | 0.0050 | Size |
| GO:0072523 | Purine-containing compound catabolic process | 0.0061 | Size |
| GO:1903409 | Reactive oxygen species biosynthetic process | 0.0081 | Size |
| GO:0009910 | Negative regulation of flower development | 0.0011 | Eccentricity |
| GO:2000242 | Negative regulation of reproductive process | 0.0015 | Eccentricity |
| GO:0006349 | Regulation of gene expression by genetic imprinting | 0.0021 | Eccentricity |
| GO:0071514 | Genetic imprinting | 0.0021 | Eccentricity |
| GO:0052386 | Cell wall thickening | 0.0055 | Eccentricity |
| GO:0052543 | Callose deposition in cell wall | 0.0055 | Eccentricity |
| GO:0009617 | Response to bacterium | 0.0093 | Eccentricity |
| GO:0010150 | Leaf senescence | 0.0093 | Eccentricity |
| GO:0006821 | Chloride transport | 0.0107 | Eccentricity |
| GO:0015700 | Arsenite transport | 0.0107 | Eccentricity |
| GO:0006952 | Defense response | 0.0002 | Green |
| GO:0010492 | Maintenance of shoot apical meristem identity | 0.0046 | Green |
| GO:0002218 | Activation of innate immune response | 0.0051 | Green |
| GO:0002253 | Activation of immune response | 0.0051 | Green |
| GO:0042742 | Defense response to bacterium | 0.00520 | Green |
| GO:0009617 | Response to bacterium | 0.0068 | Green |
| GO:0006928 | Movement of cell or subcellular component | 0.0092 | Green |
| GO:0030048 | Actin filament-based movement | 0.0092 | Green |
| GO:0034614 | Cellular response to reactive oxygen species | 0.0093 | Green |
| GO:0048236 | Plant-type spore development | 0.0095 | Green |
| GO:0006952 | Defense response | 0.0002 | Yellow |
| GO:0016568 | Chromatin modification | 0.0011 | Yellow |
| GO:0044699 | Single-organism process | 0.0012 | Yellow |
| GO:0009625 | Response to insect | 0.0021 | Yellow |
| GO:0044710 | Single-organism metabolic process | 0.0022 | Yellow |
| GO:0006325 | Chromatin organization | 0.0029 | Yellow |
| GO:0016569 | Covalent chromatin modification | 0.0033 | Yellow |
| GO:0009809 | Lignin biosynthetic process | 0.0039 | Yellow |
| GO:0042886 | Amide transport | 0.0039 | Yellow |
| GO:0006857 | Oligopeptide transport | 0.0045 | Yellow |
In contrast to the individual trait candidate gene lists, the top enrichment categories for the 200 Arabidopsis genes that associate with all four lesion traits were defense response, response to stress, immune response, plant-type hypersensitive response, programmed cell death, chromatin remodeling, and response to salicylic acid (Supplemental Table S2). Thus, the core genes associated with all four lesion-related traits linked in part to known defense genes. Overall, the enrichment categories of genes associated with only a single trait uncovered several distinct and unique biological processes that are tangentially associated with defense, whereas genes shared across four traits were more conservatively associated with defense. These uniquely represented enrichment categories according to trait and reflect varying, nonoverlapping plant mechanisms involved in the multifaceted disease resistance against generalists. This analysis revealed four portraits of plant defense that showed some overlap but also revealed the extent of biochemical and molecular mechanisms involved in lesion traits.
Validation of Genes Associated with Lesion Phenotypes
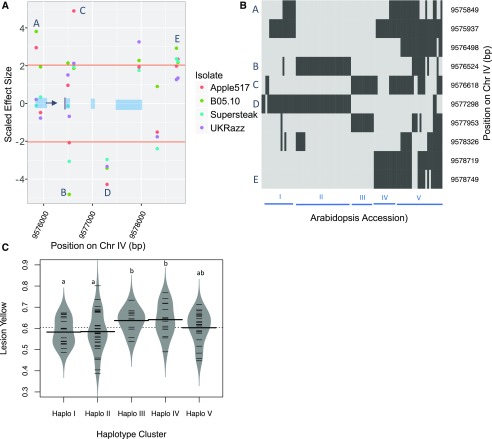
To test whether the trait-associated genes we found in the GWA mapping were causal, we measured the infection-related phenotypes on knockout mutants of genes associated with each trait of interest. We chose 23 genes (Table 3) based on GWA effect-size ranking, specificity, or overlap of traits affected and the availability of viable homozygous lines of T-DNA knockout mutants. The mutant lines were grown, infected, and phenotyped using the same image-analysis software as for the initial Arabidopsis accessions. All mutants were tested for resistance against each isolate using 20 independent biological replicates per genotype spread across two experiments. All of the traits were measured for each lesion, and each trait was tested independently for a difference between that in the wild type and that in the mutant using ANOVA (Table 3). The validation rate was 60% for lesion size, 33% for lesion yellowness, 38% for lesion greenness, and 20% for lesion eccentricity. Interestingly, this correlates with the overall fraction of each trait controlled by genetic variation (Table 1). This suggests that the lower validation rates for these alternative visual traits may be partially a power-to-detect effect issue and, thus, require increased replication. Another complication is that, in our GWA mapping, we observed that effect sizes for individual SNPs within a single gene could be both positive and negative in comparison with the reference Columbia-0 (Col-0) accession (Fig. 6; Table 3). Thus, the natural loci likely have multiple haplotypes with differing functionalities, whereas the validation mutants are unidirectional loss-of-function mutants within the reference Col-0 accession. We illustrate this with AT4G17010, encoding a component of the transcription factor III B complex, where significantly associated SNPs have both positive and negative effects in comparison with that in the reference Arabidopsis accession Col-0 (Fig. 6; Table 3). Three SNPs have effect sizes that fall above the 95th percentile threshold in the positive direction for lesion yellowness, and two have a similarly large magnitude in the negative direction, indicating that the different haplotypes of this gene uniquely affect lesion yellowness (Fig. 7). Clustering the accessions according to SNPs within this particular gene separates the haplotypes into five major groups, with groups I and II containing the SNPs with significant negative effect size, groups III and IV containing the SNPs of greatest positive effect size, and group V containing the remaining diverse haplotypes (Fig. 7). Even in the face of this complication, the T-DNA knockout mutant for AT4G17010 did exhibit altered lesion yellowness but not altered lesion size (Table 3). Our validation rates are likely influenced by the environment, as one mutant genotype known to affect Botrytis resistance, tga3-2, had no significant effect in our experiment (Windram et al., 2012). We also should note that these are preliminary validations, as multiple mutant alleles or complementation analysis would be required to finalize all of these validations. Thus, we can use GWA from high-throughput digital imaging to identify and validate new genes affecting visual lesion traits beyond lesion size.
Table 3. Candidate gene assessment.
The table shows the validation tests on mutants in 23 potential candidate genes for each of the four measures of lesion formation: size, yellowness, greenness, and eccentricity. The number of SNPs classified as significant above the 95th percentile threshold for that trait for that gene is shown as well as the significance of the difference between the insertion mutant and the wild type across all Botrytis isolates (P – Plant) or an interaction with Botrytis isolates (P – Plant:Isolate). Significance was tested using the models mentioned in “Materials and Methods.” Nominal P values between 0.1 and 0.05 are provided if at least one other trait showed significance below 0.05. NS, Not significant.
| Arabidopsis Genome Initiative Code | Gene | Mutant | No. of SNPs in the Coding Region | Size | Yellow | Green | Eccentricity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Unique SNPs above 95 | P – Plant | P – Plant:Isolate | No. of Unique SNPs above 95 | P – Plant | P – Plant:Isolate | No. of Unique SNPs above 95 | P – Plant | P – Plant:Isolate | No. of Unique SNPs above 95 | P – Plant | P – Plant:Isolate | ||||
| AT2G25540 | CESA10 | cesA10-1 | 11 | 2 | 0.005 | NS | 4 | 0.053 | NS | 2 | NS | NS | 3 | NS | NS |
| AT2G39940 | COI1 | coi1-1 | 4 | 0 | <0.001 | 0.096 | 4 | <0.001 | NS | 0 | <0.001 | NS | 0 | NS | NS |
| AT1G01060 | LHY | lhy-20 | 2 | 2 | 0.006 | 0.093 | 2 | NS | NS | 2 | 0.028 | 0.014 | 1 | NS | NS |
| AT3G26830 | PAD3 | pad3-1 | 1 | 1 | <0.001 | 0.014 | 0 | 0.004 | NS | 0 | 0.008 | 0.069 | 0 | NS | NS |
| AT3G52430 | PAD4 | pad4-1 | 6 | 0 | 0.002 | 0.062 | 0 | NS | NS | 1 | 0.010 | 0.019 | 4 | NS | NS |
| AT4G39350 | CESA2 | CS818491 | 8 | 3 | 0.020 | NS | 2 | NS | NS | 2 | 0.067 | NS | 0 | 0.090 | NS |
| AT1G22070 | TGA3 | tga3-2 | 1 | 0 | NS | NS | 0 | NS | NS | 0 | NS | NS | 1 | NS | NS |
| AT4G17010 | AT4G17010 | CS861804 | 7 | 3 | NS | NS | 4 | 0.048 | NS | 2 | NS | NS | 1 | 0.020 | NS |
| AT1G14690 | MAP65 | CS872894 | 10 | 0 | NS | NS | 0 | NS | 0.076 | 0 | NS | NS | 0 | NS | NS |
| AT4G01883 | – | CS67496 | 6 | 0 | 0.083 | NS | 0 | NS | NS | 2 | 0.056 | NS | 0 | 0.043 | NS |
| AT4G14420 | AT4G14420 | N508498 | 6 | 2 | 0.046 | NS | 1 | NS | NS | 2 | 0.003 | NS | 2 | NS | NS |
| AT4G14400 | ACD6 | N559132 | 20 | 3 | NS | NS | 5 | NS | NS | 15 | 0.047 | NS | 0 | NS | NS |
| AT4G14368 | – | N574598 | 16 | 3 | NS | NS | 0 | NS | NS | 6 | NS | NS | 0 | NS | NS |
| AT1G31260 | ZIP10 | N604586 | 9 | 5 | NS | NS | 3 | NS | NS | 0 | NS | NS | 6 | NS | NS |
| AT4G01860 | – | N606172 | 13 | 1 | NS | NS | 0 | NS | NS | 6 | NS | NS | 0 | NS | NS |
| AT2G32150 | – | N640067 | 6 | 0 | NS | NS | 0 | NS | NS | 1 | NS | NS | 0 | NS | NS |
| AT4G01880 | – | CS857701 | 4 | 0 | 0.012 | NS | 1 | 0.037 | NS | 1 | 0.079 | NS | 0 | NS | NS |
| N608769 | |||||||||||||||
| AT3G46640 | LUX | lux | 5 | 0 | NS | NS | 0 | NS | NS | 4 | NS | NS | 0 | 0.046 | NS |
| AT3G45780 | PHOT1 | phot1 | 6 | 3 | NS | 0.052 | 1 | NS | NS | 5 | NS | NS | 0 | NS | NS |
| AT4G18130 | PHYE | phyE-2 | 3 | 3 | 0.002 | NS | 0 | 0.024 | NS | 0 | NS | NS | 0 | NS | NS |
| AT4G39030 | SID1 | sid1 | 9 | 0 | NS | 0.014 | 0 | NS | NS | 8 | NS | NS | 0 | NS | NS |
| AT5G67370 | CGLD27 | N529971 | 4 | 0 | NS | NS | 3 | NS | NS | 1 | NS | NS | 0 | NS | NS |
| AT5G45950 | – | N582692 | 5 | 1 | NS | NS | 4 | NS | NS | 0 | NS | NS | 0 | NS | NS |
| AT1G20910 | – | N641443 | 5 | 0 | NS | 0.023 | 0 | NS | NS | 5 | NS | NS | 1 | NS | NS |
| AT5G13550 | SULTR4;1 | N662135 | 5 | 0 | NS | NS | 0 | NS | NS | 0 | NS | NS | 5 | 0.094 | NS |
Figure 6.
Haplotype diversity effects on trait-to-genotype linkages using AT4G17010. A, Plot of z-scaled SNP effect size across all four Botrytis isolates on lesion yellowness within 1,000 bp of the AT4G17010 coding region (represented in blue blocks). The arrow indicates the transcriptional start site. The horizontal orange lines indicate the positive and negative permutation thresholds for the Botrytis isolate Apple517. The letters show the SNPs that are associated significantly with lesion yellowness in Botrytis Apple517. B, Hierarchical clustering of 95 Arabidopsis accessions based on SNPs within AT4G17010. Haplotypes are assigned into five major groups, denoted by Roman numerals. Light gray indicates that the SNP is the Col-0 allele, whereas dark gray indicates that it is the opposite allele. The SNPs are presented in their genomic order rather than the haplotype grouped structure. C, Distributions of lesion yellowness across the Arabidopsis accessions infected with Botrytis Apple517. The Arabidopsis accessions are grouped by approximate SNP haplotypes in B. The model-corrected mean value of lesion yellowness for Arabidopsis reference accession Col-0 was 0.545. Significant differences (P < 0.05) between the groups are indicated by different lowercase letters.
Figure 7.
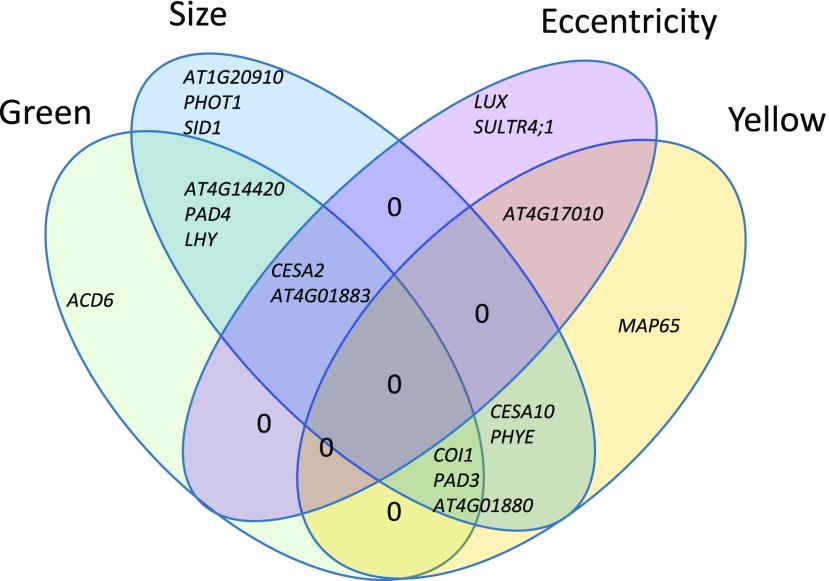
Lesion traits affected by T-DNA insertional mutants. The Venn diagram shows the lesion traits affected significantly by each of the T-DNA knockout mutants in comparison with that in the wild-type background. The gene identifiers are located within the appropriate section of the diagram showing all the phenotypes altered by mutations in that gene.
Pleiotropy among Validation Mutants
The validation tests showed that the genes were highly specific in their effects on the lesion traits by affecting only one or two traits. Only two genes, CESA2 and AT4G01883, were linked to three traits (Fig. 7). Critically, no mutant affected all of the measured traits (Fig. 7). This analysis identified new genes not linked previously to resistance to Botrytis. A knockout mutant disrupted in PHYTOCHROME E (phyE), previously linked to shade avoidance, germination, seedling deetiolation, and flowering time (Halliday and Whitelam, 2003), showed a statistically significant increase in lesion size and lesion yellowness compared with the wild type. Previous work showed that phyE affects stomatal conductance and photorespiration, which play an important role in signaling programmed cell death and systemic acquired resistance defense response (Muhlenbock et al., 2008; Szechyńska-Hebda et al., 2010). Other phytochromes, such as phyB, have been linked with defense responses in Arabidopsis via this pathway (Griebel and Zeier, 2008; Zhao et al., 2014), but there is no prior evidence in the literature of phyE affecting resistance. sultr4;1, a knockout mutant of SULFATE TRANSPORTER4.1, showed greater lesion eccentricity. SULTR4;1 transports sulfur from storage in the vacuole, facilitating the synthesis of sulfur-containing amino acids and glutathione, a compound involved in balancing cellular redox (Zuber et al., 2010). SULTR4;1 has not been linked previously with pathogen resistance, yet somehow it plays a role in defining the shape of a Botrytis lesion. This gene was neither associated with nor found to affect any of the other lesion traits, highlighting the importance of expanding the visual information used in the investigation of lesions. This highlights the utility of measuring different lesion-related traits beyond biomass or lesion size to identify new resistance mechanisms.
DISCUSSION
In the analysis of plant-pathogen interactions, there is a dominant focus on measuring pathogen success either as lesion area or pathogen biomass to quantify the level of resistance/virulence in the interaction. However, this is merely one measure of the interaction, and active infections can affect many different traits within the plant host (Agrios, 2005). New, nondestructive imaging methods are beginning to illuminate other aspects of the host-pathogen interaction that have been largely unexplored in mechanistic studies. Here, we utilized a simple digital imaging platform to show that it is possible to identify the genetic basis of these other defense-related traits, such as the color and shape of the developing lesion, in addition to the traditional lesion size trait. This shows that the generation of these visual traits is genetically determined by variation in the host and in the pathogen and the interaction of genetic variation in the two organisms (similar to lesion size), which can identify genes not known to influence lesion size.
Conducting GWA within the Arabidopsis accessions and subsequent mutant validation studies showed that the four lesion traits we quantified had a blend of distinct and overlapping genetic mechanisms controlling their generation. Classical Botrytis resistance genes, like COI1 and PAD3, influenced both the color and size of the developing lesion but not the shape of the lesion, showing the role of known resistance mechanisms in these new lesion traits (Fig. 7; Thomma et al., 1998; Ferrari et al., 2003; Kliebenstein et al., 2005; Rowe et al., 2010). In contrast, some known resistance genes like ACD6 affected only the color but not the size or shape of the lesion (Fig. 7; Rate et al., 1999; Song et al., 2004; Lu et al., 2009). More importantly, using new lesion-related traits for GWA identified new genes involved in resistance to Botrytis. This included genes involved in light signal transduction, LUX, PHOT1, and PHYE, as well as those that play a role in sulfur transport (Clack et al., 1994; Devlin et al., 1998; Briggs and Christie, 2002; Halliday and Whitelam, 2003; Kataoka et al., 2004; Hazen et al., 2005; Zuber et al., 2010; Nusinow et al., 2011; Sánchez-Lamas et al., 2016). The three light-related genes were associated with different lesion traits, with PHOT1 related specifically to lesion size and PHYE playing a role in lesion size and yellowing, whereas LUX, alongside the sulfur transporter, functioned specifically in lesion size (Fig. 7). Thus, even these three light-related genes that might have been expected to have overlapping roles exhibit distinct effects. This shows that there are specific mechanisms for each set of lesion traits and that more than simply measuring pathogen growth is required to fully understand the plant-pathogen interaction.
Lesion Eccentricity as an Indicator of Pathogen Success
One difficulty of developing new traits from high-throughput digital imaging is comprehending their possible biological role. For example, lesion eccentricity (i.e. deviation from circularity) is a trait that is controlled by variation in both pathogen and host genes. The heritability of lesion eccentricity was 4.3% for plant accession and less than 0.1% for pathogen isolate but jumped to 11.1% for the plant-pathogen interaction, indicating that variation in this trait is highly dependent on the interaction of plant and pathogen genotypes. In two of the pathogen genotypes, namely B05.10 and Supersteak, there was a negative correlation between lesion size and eccentricity, suggesting that eccentric lesions tend to be smaller (Fig. 3). In previous work, we showed that eccentric lesions are associated with preferential growth along the primary vasculature, which implies that there is a shift from general radial outward growth to directed growth along the leaf midrib (Corwin et al., 2016b). Purely radial growth can maximize local growth in microbial organisms, and this would argue that directed growth might forsake potential local growth. A possible reason for forsaking local growth is that this could enable some pathogen isolates to move systemically through the vasculature of the plant. The fact that there are plant genes specific to affecting this trait indicates that Arabidopsis has specific mechanisms geared toward altering the pattern of pathogen growth. As such, the success of the pathogen in the host-pathogen interaction would depend on a combination of the ability to infect local tissue and to move rapidly to distal tissue. Equally, if the pathogen has different growth strategies to optimize fitness, the plant would have to defend against local growth and growth geared toward spread of the pathogen.
Lesion Color Genetic Mechanisms and Agriculture
It is harder to assess the potential ecological or evolutionary role of specific genetic mechanisms influencing the color of a lesion. However, lesion color is a potentially key aspect of the plant-pathogen interaction for vegetable and fruit crops. For example, lesions that give rise to a highly noticeable color (Fig. 1C) on a fruit or vegetable would greatly decrease the consumer acceptance of that product and lead to postharvest crop loss if the lesion was large. As such, understanding the specific genetic mechanisms influencing the different color traits can have significant agricultural importance, and they are not studied at present. Our study showed that, as described previously, COI1 plays a key role in controlling the development of lesion yellowness in the plant-pathogen interaction (Rowe et al., 2010). This agrees with the known connection of jasmonate signaling to photosynthetic/plastid functioning (Kazan and Manners, 2011; Attaran et al., 2014; Campos et al., 2016). Interestingly, this also showed that additional genes not known previously to affect Botrytis resistance also function in determining the level of lesion yellowness, like PHYE (Fig. 7).
Genes associated with lesion greenness were enriched in activation of the immune system, activation of the innate immune system, and movement of cellular or subcellular components. These biological processes have obvious roles in the plant response to a fungal infection, but their links with lesion greenness are less obvious. Previous studies had identified green islands, which are dead green tissue within pathogen lesions, in some plant-pathogen interactions as associated with increased levels of cytokinins and polyamines that delay senescence as well as a variety of fungal toxins (Walters et al., 2008). We previously attempted to extract green pigment from lesions using both methanol and hexane but were unsuccessful, indicating that the source of this greenness is not free chlorophyll (Corwin et al., 2016b). Furthermore, greener lesions had no living plant cells, from what we could assess with Trypan Blue staining (Corwin et al., 2016b). However, the genes associated with greenness included known Botrytis resistance genes like COI1, PAD3, and PAD4 (Fig. 7). Interestingly, the only gene associated solely with lesion greenness was ACD6, a gene that is key to regulating plant cell death in response to pathogen attack. This suggests that there may be a role for how the plant cell dies in controlling lesion color development. Whereas our results do not provide an immediate direct mechanism for lesion color traits, they do show that there are potential mechanisms specific to lesion color traits in plant-pathogen interactions that may be useful for understanding the infection process.
Infection Methodology and Trait Analysis
The mode of infection, a major influencing factor, affects these results as well as all published results and any field virulence mechanisms. The mode of infection refers to whether the inoculum is a localized droplet or a diffuse spray, the location of the inoculation on leaves, and the spore density of the inoculation. In this analysis, we chose to utilize a single droplet with low spore concentration placed near the midrib for several reasons. A single droplet was utilized because previous work with sprayed inoculations revealed interactions between the lesions such that outgrowth was limited (Mengiste et al., 2003; Veronese et al., 2004, 2006; Zheng et al., 2006). Thus, a single droplet allows for quantification of the effects of both lesion establishment and outgrowth. A low spore inoculum was utilized to ensure that the pathogen did not merely overwhelm the plant’s defenses and that we could better quantify the interaction (Denby et al., 2004; Kliebenstein et al., 2005). This specific spore concentration allows for a high efficiency of lesion establishment with modest outgrowth (Rowe and Kliebenstein, 2007, 2008). Finally, the location of the droplet by the midrib facilitated the measurement of preferential growth along the primary but not the secondary vasculature (Rowe et al., 2010; Corwin et al., 2016b). Placing the droplet near the primary vasculature ensured that this would create an elliptical growth pattern allowing easy measurement of eccentricity. Moving the droplet away from the midrib would still allow for measuring preferential growth but would require a different method of quantification. The highly polygenic nature of the Arabidopsis/Botrytis pathosystem means that altering the inoculation parameters could shift how the pathosystem is measured and which causal genes are identified (Corwin et al., 2016a; Corwin and Kliebenstein, 2017; Zhang et al., 2017). Equally, given the diverse field infection strategies of this pathogen, there is no single optimal system to find the important genes. Understanding the complete network of genes that are critical for this interaction requires a diverse range of strategies (Elad et al., 2016).
Expanding the visualization platform to utilize different spectra, as enabled by high-resolution multispectral or hyperspectral platforms, would open up an even wider range of phenotypes (Bock et al., 2010; Mahlein et al., 2012; Matsuda et al., 2012). The inclusion of additional wavelengths would allow for direct links to specific biochemical processes like photosynthesis and phenolic or lipid metabolism. Furthermore, the use of higher resolution imaging would allow for treatment of the lesion as a two-dimensional fine-scale map and create the potential for distance measurements and other measures that the lower resolution imaging did not enable. Whereas the analytical challenges to these assays are significant, the technical challenge is potentially greater. Although these new platforms would provide more traits, the sample throughput would have to be as high, if not higher, than in this experiment to provide the necessary statistical power (∼750 measured lesions per isolate for GWA). Given the cost and throughput of multispectral platforms, measuring more than 3,000 lesions in a single experiment is a significant impediment.
Polygenic Trait Architecture, Model Choice, and True Effects
The genetic architecture of all the traits that we measured in the Arabidopsis/Botrytis interaction was highly polygenic (Corwin et al., 2016a). This situation is a difficult setting for standard GWA models like SNP-by-SNP mixed models, which work better for situations with at least some moderate effect loci (Atwell et al., 2010; Zhang et al., 2010). Instead, GWA on lesion traits requires polygenic models, like the ridge regression approach applied here, that facilitate the identification of hundreds to thousands of SNPs with similar effect sizes (Shen et al., 2013; Corwin et al., 2016a; Francisco et al., 2016; Kooke et al., 2016). However, this approach is limited, as it does not readily identify SNPs with truly no effect on the trait, suggesting that there is room to optimize models for polygenic trait-SNP identification. It should be noted that, whereas the individual SNPs appeared to have small effects, the individual gene validation efforts showed, in agreement with other studies (Chan et al., 2011; Corwin et al., 2016a), that most of these individual genes can have 25% or larger effects on the trait (Supplemental Table S7). This apparent discrepancy likely is due to the difficulty in generating accurate effect size estimates in the GWA data with thousands of causal SNPs, more than 100,000 SNPs that are not causal, and a paucity of genotypes from which to derive these estimates. Thus, it remains to be determined whether the GWA effect estimate or the single gene trait effect is the better estimate of the true potential for that specific causal locus to affect trait evolution in a polymorphic population.
CONCLUSION
This study has utilized high-throughput digital imaging of plant-pathogen interactions to uncover the genetics for previously undercharacterized lesion traits. Many of the candidate genes that were tested using insertional mutants affected more than one aspect of the plant-pathogen interaction, as measured by lesion color, size, or shape. However, no gene affected all of these lesion traits. As most studies of quantitative resistance focus on pathogen biomass or lesion size measurements and omit lesion shape and color, our findings suggest that there are potentially unrecognized mechanisms that may be important for plant-pathogen interactions. Further validation studies could reveal more plant genes that have a greater effect on quantitative resistance, filling gaps in our knowledge of the cellular mechanisms involved in lesion traits and pathogen virulence and revealing new gene targets that deserve attention in plant breeding programs. Equally, analysis of these alternative lesion traits highlights the need for broader life history studies of plant-pathogen interactions in the field to identify the potential ecological or evolutionary drivers of these traits. GWA mapping coupled with high-throughput digital measurement of virulence-associated traits will be a useful tool in understanding broader plant-pathogen interactions.
MATERIALS AND METHODS
Population Selection for GWA
We used a previously published collection of 96 Arabidopsis accessions that was chosen for pathogen resistance GWA based on similarity in flowering time (63.1 ± 0.95 [se] d to flowering). This population minimizes the indirect effects of ontogenic variation caused by a wide range of flowering times while also reducing the effect of rare, medium-effect alleles in order to inflate genetically related residuals (Corwin et al., 2016a). Selection based on flowering time also removed genetic outliers and minimized population structure. The selected population extends broadly across the known phylogeny of Arabidopsis (Arabidopsis thaliana) accessions, increasing the resolution of the association study (Corwin et al., 2016a).
Growth Conditions and Pathogen Infection
Seeds were cold stratified in 1% (w/v) Phytagar at 4°C for 7 d prior to sowing. Seeds then were sown in a randomized complete block design with three seeds per cell into four 104-cell flats containing standard potting soil (Sunshine Mix #1; Sun Gro Horticulture). Plants were covered with a transparent humidity dome, placed in short-day (10 h of full-spectrum light) conditions in a controlled environment at 22°C, and watered biweekly as needed. After 1 week, seedlings were thinned to one seedling per pot. Sowing was repeated 2 weeks later to create another balanced randomized experiment so that eight biological replicates per accession were present across the two independent experiments in a randomized complete block design.
After 5 weeks of growth, mature leaves were excised and placed on agar flats (Denby et al., 2004; Rowe and Kliebenstein, 2008). Individual leaves were inoculated with one of four phenotypically and genotypically diverse isolates of Botrytis (Botrytis cinerea; Apple517, B05.10, Supersteak, and UKRazz) or a mock-inoculated control (Atwell et al., 2015; Corwin et al., 2016a, 2016b). Spores of 5-d-old sporulation cultures grown on organic peach (Prunus persica) slices were collected in sterilized one-half-strength organic grape juice, counted with a hemocytometer, and standardized to a solution of 10 spores μL−1. Individual leaves were inoculated with a single 4-μL droplet of one of the Botrytis isolates or mock inoculated with a control of one-half-strength organic grape juice for approximately 40 spores per leaf. The inoculated leaves were kept under transparent humidity domes at room temperature to allow lesions to grow for 3 d. At 72 h post inoculation, photographs were taken of infected leaves along with a 1-cm2 scale for size reference using an 18-megapixel high-resolution T3i Canon camera outfitted with an EF-S 10-22mm f/3.5-4.5 USM ultra-wide angle lens, achieving a resolution of approximately 10 pixels mm−1 (Corwin et al., 2016a).
Image Analysis
A semiautomated image-analysis script using the open-source R statistical environment (R Development Core Team, 2016) and the Bioconductor packages EBImage and CRImage (Failmezger, 2010; Pau et al., 2010) were used to measure lesion traits such as lesion size, shape, and color (Corwin et al., 2016a). Briefly, the script identified leaves as objects that have a green hue and are highly saturated at their perimeter, whereas lesions were brown objects of low saturation within the leaf perimeter. Lesion and leaf mask images were generated and refined manually or corrected where faulty. The area in pixels of these leaves and lesions, as well as the numbers and proportions of pixels of certain colors and dimensions such as major and minor axes, perimeter, and eccentricity, were recorded. These values were converted to mm2 using a 1-cm reference standard contained within each image.
GWA Mapping
For each trait, we modeled heritability and obtained the model-corrected least squared means using the linear model:
where E is the experimental replicate, P is the individual plant, A is the genotype of the Arabidopsis accession, and B is the genotype of the Botrytis isolate. Experimental replicate was modeled as a random effect, whereas individual plant, Arabidopsis accession, and Botrytis isolate were categorical. Publicly available data on genomic polymorphisms of the selected Arabidopsis accessions were collected from the Arabidopsis 1,001 Genomes project (Ossowski et al., 2008; Cao et al., 2011; Gan et al., 2011; Schneeberger et al., 2011; Long et al., 2013; Alonso-Blanco et al., 2016). SNPs with minor allele frequency < 0.2 were filtered out, resulting in a set of 115,310 SNPs for the 95 accessions. Association mapping was performed using the bigRR package in the R statistical environment (Shen et al., 2013). This previously published and validated method uses ridge regression, an appropriate approach given the small proportion of phenotypic variance attributable to genotype (Corwin et al., 2016a; Francisco et al., 2016). The ridge regression models the effects of all polymorphisms in a single model, treating each SNP as a random effect and introducing a bias to the regression estimates to reduce se. Thus, each polymorphism is assigned a heteroscedastic effect size rather than a P value, which is difficult to determine for random variables. Instead, a significant effect threshold value is delineated by permuting the phenotype data as they correspond to the polymorphism data 1,000 times and taking the 95th and 99th percentiles. A gene is considered associated with a trait when two or more SNPs within the coding region have an effect size greater than the 95th percentile threshold. This is a functional genome-wide prediction method that has been shown to provide similar results to Efficient Mixed-Model Association-based GWAS (Kooke et al., 2016). Furthermore, this GWAS pipeline has yielded a high rate of gene validation for diverse traits within Arabidopsis (Corwin et al., 2016a; Francisco et al., 2016).
GO Enrichment Analysis
GO enrichment analysis for biological processes was performed on all genes significantly associated with each of the four lesion traits using the Bioconductor packages org.At.tair.db, topGO, and goProfiles in the R statistical environment. Genes within Arabidopsis that contain at least two significant SNPs that were associated with the trait of interest in the data set were used as the genomic background sample for the analysis.
Validation Tests
Based on the availability of viable homozygous validated T-DNA knockout lines, 23 Arabidopsis genes were selected that were found to be highly associated with lesion size, greenness, yellowness, and eccentricity. Seventeen of the lines had been identified previously and tested for lesion size, and an additional six genes were chosen based on the magnitude of predicted effect size. The genotypes were sourced from a number of origins, based on the availability of validated homozygous genotypes, and then regrown in conjunction with the control wild types prior to analysis (Supplemental Table S7). Plants were grown as described above, and no genotypes showed any evidence of flowering within this time frame in these conditions. At 5 weeks of age, the six first fully mature leaves were excised and inoculated with the four isolates of Botrytis mentioned above at the same concentrations and imaged at 72 h post inoculation. These leaves showed no visual evidence of senescence at the start of the experiment, and the control grape juice mock-inoculated leaves showed no developing lesion during the image analysis. The experiment was carried out in a randomized complete block design, with two experimental replicates each containing 10 biological replicates, amounting to 20 replicates for each Arabidopsis mutant/Botrytis isolate combination. Statistical differences between the wild-type Col-0 background and each mutant genotype were assessed using the linear model:
where E is the experimental replicate, P is the plant genotype, and I is the fungal isolate genotype. Experimental replicate was modeled as a random effect, whereas plant and fungal isolate genotypes were categorical.
Accession Numbers
The TAIR numbers for the genes discussed in this work are as follows: CESA10, AT2G25540; COI1, AT2G39940; LHY, AT1G01060; PAD3, AT3G26830; PAD4, AT3G52430; CESA2, AT4G39350; TGA3, AT1G22070; AT4G17010, AT4G17010; MAP65, AT1G14690; AT4G14420, AT4G14420; ACD6, AT4G14400; ZIP10, AT1G31260; LUX, AT3G46640; PHOT1, AT3G45780; PHYE, AT4G18130; SID1, AT4G39030; CGLD27, AT5G67370; and SULTR4;1, AT5G13550.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HCA of lesion traits.
Supplemental Figure S2. Lesion greenness Manhattan plots.
Supplemental Figure S3. Lesion yellowness and eccentricity shape Manhattan plots for Apple517.
Supplemental Table S1. Least squared means for all measured lesion traits.
Supplemental Table S2. GO enrichment test using the 200 candidate genes associated with all four lesion traits.
Supplemental Table S3. Lesion area GWA.
Supplemental Table S4. Lesion greenness GWA.
Supplemental Table S5. Lesion yellowness GWA.
Supplemental Table S6. Lesion eccentricity GWA.
Supplemental Table S7. Validation genotype information and trait means.
Footnotes
Financial support for this work was provided by the National Research Foundation DNRF grant 99, US NSF grants IOS 1339125, MCB 1330337, and IOS1021861, and the USDA National Institute of Food and Agriculture, Hatch project number CA-D-PLS-7033-H.
Articles can be viewed without a subscription.
References
- Agrios G. (2005) Plant Pathology. Elsevier Academic Press, San Diego, CA, USA [Google Scholar]
- Aguileta G, Lengelle J, Chiapello H, Giraud T, Viaud M, Fournier E, Rodolphe F, Marthey S, Ducasse A, Gendrault A, et al. (2012) Genes under positive selection in a model plant pathogenic fungus, Botrytis. Infect Genet Evol 12: 987–996 [DOI] [PubMed] [Google Scholar]
- Alfonso C, Raposo R, Melgarejo P (2000) Genetic diversity in Botrytis cinerea populations on vegetable crops in greenhouses in south-eastern Spain. Plant Pathol 49: 243–251 [Google Scholar]
- Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, Cao J, Chae E, Dezwaan TM, Ding W, et al. (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran E, Major IT, Cruz JA, Rosa BA, Koo AJK, Chen J, Kramer DM, He SY, Howe GA (2014) Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol 165: 1302–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Corwin JA, Soltis NE, Subedy A, Denby KJ, Kliebenstein DJ (2015) Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front Microbiol 6: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbedo JGA. (2017) A new automatic method for disease symptom segmentation in digital photographs of plant leaves. Eur J Plant Pathol 147: 349–364 [Google Scholar]
- Bethke G, Thao A, Xiong G, Li B, Soltis NE, Hatsugai N, Hillmer RA, Katagiri F, Kliebenstein DJ, Pauly M, et al. (2016) Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 28: 537–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock CH, Poole GH, Parker PE, Gottwald TR (2010) Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit Rev Plant Sci 29: 59–107 [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell ALT (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci USA 105: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer M, Fournier E, Kunz C, Levis C, Pradier JM, Simon A, Viaud M (2007) Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol Lett 277: 1–10 [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Corwin JA, Kliebenstein DJ (2017) Quantitative resistance: more than just perception of a pathogen. Plant Cell 29: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin JA, Copeland D, Feusier J, Subedy A, Eshbaugh R, Palmer C, Maloof J, Kliebenstein DJ (2016a) The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet 12: e1005789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin JA, Subedy A, Eshbaugh R, Kliebenstein DJ (2016b) Expansive phenotypic landscape of Botrytis cinerea shows differential contribution of genetic diversity and plasticity. Mol Plant Microbe Interact 29: 287–298 [DOI] [PubMed] [Google Scholar]
- Denby KJ, Kumar P, Kliebenstein DJ (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38: 473–486 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elad Y, Pertot I, Cotes Prado AM, Stewart A (2016) Plant hosts of Botrytis spp. In Fillinger S, Elad Y, eds, Botrytis: The Fungus, the Pathogen and Its Management in Agricultural Systems. Springer International Publishing, Cham, Switzerland, pp 413–486 [Google Scholar]
- Failmezger H, Yuan Y, Rueda O, Markowetz F (2010) CRImage: CRImage a Package to Classify Cells and Calculate Tumour Cellularity.
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco M, Joseph B, Caligagan H, Li B, Corwin JA, Lin C, Kerwin RE, Burow M, Kliebenstein DJ (2016) Genome wide association mapping in Arabidopsis thaliana identifies novel genes involved in linking allyl glucosinolate to altered biomass and defense. Front Plant Sci 7: 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Fortini D, Levis C, Leroux P, Brygoo Y (1997) RFLP markers show genetic recombination in Botryotinia fuckeliana (Botrytis cinerea) and transposable elements reveal two sympatric species. Mol Biol Evol 14: 1177–1185 [DOI] [PubMed] [Google Scholar]
- Griebel T, Zeier J (2008) Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol 131: 1913–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis WR. (1977) Botryotinia and Botrytis Species: Taxonomy, Physiology, and Pathogenicity: A Guide to the Literature. Canada Department of Agriculture, Ottawa [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2011) The interplay between light and jasmonate signalling during defence and development. J Exp Bot 62: 4087–4100 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Rowe HC, Denby KJ (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J 44: 25–36 [DOI] [PubMed] [Google Scholar]
- Kooke R, Kruijer W, Bours R, Becker F, Kuhn A, van de Geest H, Buntjer J, Doeswijk T, Guerra J, Bouwmeester H, et al. (2016) Genome-wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in Arabidopsis. Plant Physiol 170: 2187–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer M, Leroch M, Mosbach A, Walker AS, Fillinger S, Mernke D, Schoonbeek HJ, Pradier JM, Leroux P, De Waard MA, et al. (2009) Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog 5: e1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuska M, Wahabzada M, Leucker M, Dehne HW, Kersting K, Oerke EC, Steiner U, Mahlein AK (2015) Hyperspectral phenotyping on the microscopic scale: towards automated characterization of plant-pathogen interactions. Plant Methods 11: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Mengiste T (2010) Necrotroph attacks on plants: wanton destruction or covert extortion? The Arabidopsis Book 8: e0136, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucker M, Mahlein AK, Steiner U, Oerke EC (2016) Improvement of lesion phenotyping in Cercospora beticola-sugar beet interaction by hyperspectral imaging. Phytopathology 106: 177–184 [DOI] [PubMed] [Google Scholar]
- Li B, Hulin MT, Brain P, Mansfield JW, Jackson RW, Harrison RJ (2015) Rapid, automated detection of stem canker symptoms in woody perennials using artificial neural network analysis. Plant Methods 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, Nizhynska V, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Salimian S, Gamelin E, Wang G, Fedorowski J, LaCourse W, Greenberg JT (2009) Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J 58: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlein AK, Steiner U, Hillnhütter C, Dehne HW, Oerke EC (2012) Hyperspectral imaging for small-scale analysis of symptoms caused by different sugar beet diseases. Plant Methods 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda O, Tanaka A, Fujita T, Iba K (2012) Hyperspectral imaging techniques for rapid identification of Arabidopsis mutants with altered leaf pigment status. Plant Cell Physiol 53: 1154–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga TM, Ogawa D, Taguchi-Shiobara F, Ishimoto M, Matsunaga S, Habu Y (2017) Direct quantitative evaluation of disease symptoms on living plant leaves growing under natural light. Breed Sci 67: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Laluk K, AbuQamar S (2010) Mechanisms of induced resistance against B. cinerea. In Prusky D, Gullino M eds, Postharvest Pathology: Plant Pathology in the 21st Century. Springer, Dordrecht, The Netherlands, pp 13–30 [Google Scholar]
- Montes JM, Melchinger AE, Reif JC (2007) Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci 12: 433–436 [DOI] [PubMed] [Google Scholar]
- Muhlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356; erratum Muhlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S (2008)Plant Cell 20: 3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka AM, Bart RS (2015) Image-based phenotyping of plant disease symptoms. Front Plant Sci 5: 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau G, Fuchs F, Sklyar O, Boutros M, Huber W (2010) EBImage: an R package for image processing with applications to cellular phenotypes. Bioinformatics 26: 979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT (1999) The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2016) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rowe HC, Kliebenstein DJ (2007) Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol Plant Microbe Interact 20: 1126–1137 [DOI] [PubMed] [Google Scholar]
- Rowe HC, Kliebenstein DJ (2008) Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180: 2237–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Kliebenstein DJ (2010) All mold is not alike: the importance of intraspecific diversity in necrotrophic plant pathogens. PLoS Pathog 6: e1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Walley JW, Corwin J, Chan EKF, Dehesh K, Kliebenstein DJ (2010) Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog 6: e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Lamas M, Lorenzo CD, Cerdán PD (2016) Bottom-up assembly of the phytochrome network. PLoS Genet 12: e1006413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Ott F, Klein JD, Wang X, Lanz C, Smith LM, Cao J, Fitz J, Warthmann N, et al. (2011) Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA 108: 10249–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanck AA, Del Ponte EM (2016) Measuring lesion attributes and analysing their spatial patterns at the leaf scale using digital image analysis. Plant Pathol 65: 1498–1508 [Google Scholar]
- Shen X, Alam M, Fikse F, Rönnegård L (2013) A novel generalized ridge regression method for quantitative genetics. Genetics 193: 1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Lu H, McDowell JM, Greenberg JT (2004) A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J 40: 200–212 [DOI] [PubMed] [Google Scholar]
- Staats M, van Baarlen P, van Kan JAL (2005) Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol Biol Evol 22: 333–346 [DOI] [PubMed] [Google Scholar]
- Stefanato FL, Abou-Mansour E, Buchala A, Kretschmer M, Mosbach A, Hahn M, Bochet CG, Métraux JP, Schoonbeek HJ (2009) The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J 58: 499–510 [DOI] [PubMed] [Google Scholar]
- Szechyńska-Hebda M, Kruk J, Górecka M, Karpińska B, Karpiński S (2010) Evidence for light wavelength-specific photoelectrophysiological signaling and memory of excess light episodes in Arabidopsis. Plant Cell 22: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KFMJ, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váczy KZ, Sándor E, Karaffa L, Fekete E, Fekete E, Arnyasi M, Czeglédi L, Kövics GJ, Druzhinina IS, Kubicek CP (2008) Sexual recombination in the Botrytis cinerea populations in Hungarian vineyards. Phytopathology 98: 1312–1319 [DOI] [PubMed] [Google Scholar]
- Valiuskaite A, Surviliene E, Baniulis D (2010) Genetic diversity and pathogenicity traits of Botrytis spp. isolated from horticultural hosts. Zemdirbyste 97: 85–90 [Google Scholar]
- Veloso J, van Kan JAL (2018) Many shades of grey in Botrytis-host plant interactions. Trends Plant Sci 23: 613–622 [DOI] [PubMed] [Google Scholar]
- Veronese P, Chen X, Bluhm B, Salmeron J, Dietrich R, Mengiste T (2004) The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J 40: 558–574 [DOI] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DR, McRoberts N, Fitt BD (2008) Are green islands red herrings? Significance of green islands in plant interactions with pathogens and pests. Biol Rev Camb Philos Soc 83: 79–102 [DOI] [PubMed] [Google Scholar]
- Windram O, Madhou P, McHattie S, Hill C, Hickman R, Cooke E, Jenkins DJ, Penfold CA, Baxter L, Breeze E, et al. (2012) Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24: 3530–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Corwin JA, Copeland D, Feusier J, Eshbaugh R, Chen F, Atwell S, Kliebenstein DJ (2017) Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 29: 2727–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhou J, Xing D (2014) Phytochrome B-mediated activation of lipoxygenase modulates an excess red light-induced defence response in Arabidopsis. J Exp Bot 65: 4907–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zuber H, Davidian JC, Wirtz M, Hell R, Belghazi M, Thompson R, Gallardo K (2010) Sultr4;1 mutant seeds of Arabidopsis have an enhanced sulphate content and modified proteome suggesting metabolic adaptations to altered sulphate compartmentalization. BMC Plant Biol 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]