Dynamic changes in the transcriptome of developing seedless fruits reveals key regulators of fruit growth and maturation.
Abstract
Fruits protect the developing seeds of angiosperms and actively contribute to seed dispersion. Furthermore, fruit and seed development are highly synchronized and require exchange of information between the mother plant and the developing generations. To explore the mechanisms controlling fruit formation and maturation, we performed a transcriptomic analysis on the valve tissue of the Arabidopsis (Arabidopsis thaliana) silique using RNA sequencing. In doing so, we have generated a data set of differentially regulated genes that will help to elucidate the molecular mechanisms that underpin the initial phase of fruit growth and, subsequently, trigger fruit maturation. The robustness of our data set has been tested by functional genomic studies. Using a reverse genetics approach, we selected 10 differentially expressed genes and explored the consequences of their disruption for both silique growth and senescence. We found that genes contained in our data set play essential roles in different stages of silique development and maturation, indicating that our transcriptome-based gene list is a powerful tool for the elucidation of the molecular mechanisms controlling fruit formation in Arabidopsis.
In angiosperms fruit formation is triggered by fertilization, which occurs in the ovule. As a consequence of fertilization, the ovule develops into a seed while the ovary differentiates into a fruit (Coombe, 1975). Fruits evolved to protect and provide nutrients for the embryos, which grow and differentiate within the seeds (Müntz et al., 1978). Once embryos have completed their development, fruits contribute to their dispersal. To perform these functions, fruits and seeds must communicate throughout their development. Coordination between the mother plant and the developing generation is a highly complex process that relies on a variety of chemical mediators, such as hormones, proteins, peptides, mRNAs, small RNAs, and noncoding RNAs (Lindsey, 2001; Jones-Rhoades et al., 2006; Corbesier et al., 2007; Tamaki et al., 2007; Kalantidis et al., 2008; McAtee et al., 2013). Moreover, signals derived from pollen (O’Neill, 1997; O’Neill and Nadeau, 2010), ovules (Gillaspy et al., 1993), or other vegetative tissues (Nitsch, 1952) can stimulate fruit development. Indeed, the occurrence of seedless fruits, whether parthenocarpic (in which case they develop in the absence of fertilization) or stenospermocarpic (in which case fertilization occurs but seeds abort precociously), indicates that fruit and seed communication can be outflanked (Mazzucato et al., 1998; Varoquaux et al., 2000; Pandolfini, 2009; Rojas-Gracia et al., 2017).
The fruit of the model plant species Arabidopsis (Arabidopsis thaliana), known as the silique, is a dry dehiscent fruit that mechanically opens at maturity, releasing the seeds. Siliques develop from a gynoecium composed of two fused carpels linked by a central tissue named the septum (Rollins, 1993). Fertilization induces a rapid transformation of the gynoecium, leading to the development of the silique; this process can be divided into two distinct phases, encompassing growth and maturation. The first phase begins immediately after fertilization and determines the final size of the silique, which is reached at 6 to 7 DPA (Vivian-Smith and Koltunow, 1999). Growth of the silique in length and width involves cell division and cell expansion (Vivian-Smith and Koltunow, 1999). Once the silique has reached its final size, it quickly enters the maturation phase, which leads to the ripening and the subsequent senescence of the fruit. The effect of maturation is clearly visible by around 12 DPA, when the green color of the silique begins to turn yellowish. However, this visible phenotype is anticipated by several molecular events, such as the decline in chlorophyll content that begins before the yellowing becomes apparent.
The transcriptomic data on Arabidopsis fruit development currently available derive from a few surveys based on microarray analysis (Wagstaff et al., 2009; Carbonell-Bejerano et al., 2010; Jaradat et al., 2014). Furthermore, these studies analyzed mutant lines (eceriferum) or explored relatively late developmental stages (10 and 20 DPA) or siliques including the seeds, making it almost impossible to distinguish between fruit and seed development at the molecular level.
In our investigation, we have used next-generation sequencing to extend our knowledge of the molecular mechanisms that control the early stages of silique formation and the sequence of maturation events. We provide a comprehensive transcriptomic analysis of Arabidopsis fruits, devoid of seeds, at 3, 6, 9, and 12 DPA, which allows us to identify genes with potential roles in silique formation and maturation. Through a reverse genetics approach, we demonstrate that transcription factors, cytoskeletal proteins, and enzymes that modulate hormone homeostasis are intimately involved in these processes.
RESULTS
Expression Analysis Identifies Three Classes of Differentially Expressed Genes
In order to determine which genes are involved in Arabidopsis silique development, siliques at 3, 6, 9, and 12 DPA from Columbia-0 (Col-0) plants (Fig. 1A) were collected. The first two time points (3 and 6 DPA) are representative of the growth stage of the siliques, while the other two (9 and 12 DPA) correspond to the silique’s maturation phase. Seeds were removed manually, such that the poly(A) RNA extracted and subjected to Illumina sequencing was derived solely from maternal fruit tissues. The number of reads for each replicate is reported in Supplemental Figure S1A.
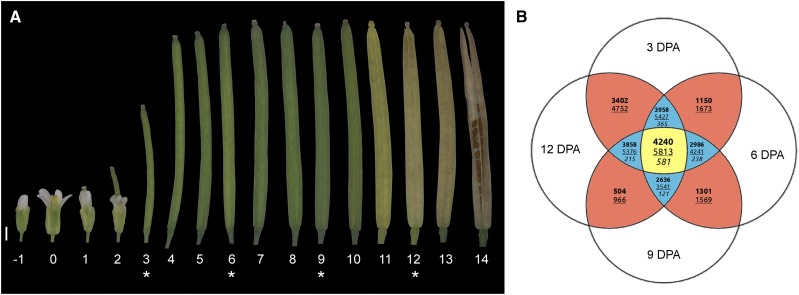
Figure 1.
RNA sequencing (RNAseq) strategy used to explore the mechanisms controlling fruit formation and maturation. A, Developmental series of Arabidopsis siliques from the flower bud (−1 DPA) to the mature fruit (14 DPA). Numbers below the siliques indicate the DPA. The siliques at 3, 6, 9, and 12 DPA, marked with asterisks, were opened and the seeds were removed prior to use for RNAseq analysis. The image shown is a composite image. Bar = 1 mm. B, Venn diagram shows the number of up-regulated (boldface), down-regulated (underlined), and alternative behavior (italics) genes across all comparisons performed using the selected cutoff (fold change of 2.5 and P = 0.001; see “Materials and Methods”). The four pairwise comparisons (6 versus 3 DPA, 9 versus 6 DPA, 12 versus 9 DPA, and 12 versus 3 DPA) show only two values, since, by definition, genes can only be either up-regulated or down-regulated. The center of the Venn diagram (i.e. the yellow portion) shows the number of differentially expressed genes considered in this work: 4,240 up-regulated genes (Supplemental Table S1), 5,813 down-regulated genes (Supplemental Table S2), and 581 genes that showed an alternative behavior (Supplemental Table S3–S6).
Principal component analysis of the sequences sampled at the four time points shows that they are well separated along the first component, which explains 56% of the variability (Supplemental Fig. S1B). Biological replicates were grouped together, with the second and third time points (6 and 9 DPA, respectively) showing higher variability than the first and last time points (3 and 12 DPA).
Two computational packages were used to identify differentially expressed genes, edgeR and Limma, which have been shown previously to have a Spearman correlation coefficient of 0.9 through the analysis of mouse data sets (Seyednasrollah et al., 2015). In our case, the two packages also gave very similar results, although Limma was more stringent (Supplemental Fig. S2). Therefore, we decided to consider the Limma outputs for further analyses, even though all the genes analyzed by reverse genetics in this study were differentially expressed according to both computational tools.
Based on the adopted cutoff (fold change of 2.5 and P = 0.001; see “Materials and Methods”), genes were grouped into three classes (Fig. 1B). Up-regulated genes consistently and significantly increased their transcription rate between the two end points in the time series; conversely, the expression of down-regulated genes diminished steadily from the first time point to the last. All genes displaying both up- and down-regulated profiles, depending on the time points considered, were defined as exhibiting alternative behavior (Fig. 1B). Among the alternative behavior genes, we performed a visual inspection of the hierarchical clustering results to identify meaningful subgroups; thus, four different clusters were defined (Supplemental Fig. S3; see “Materials and Methods”). With this approach, we defined 4,240 up-regulated genes (Supplemental Table S1), 5,813 down-regulated genes (Supplemental Table S2), and 581 genes with alternative behaviors (Supplemental Table S3–S6). In all, 11,274 genes showed no significant change in their expression level over the four stages analyzed.
Gene Ontology Analysis Reveals the Functions of the Differentially Expressed Genes
Changes in gene expression underlie differences in biological functions; using the agriGO software (Du et al., 2010), we explored the gene ontologies (GO) that were enriched among the previously defined groups (up- and down-regulated genes and genes showing alternative behaviors; see Supplemental Figs. S4, S5, and S6, respectively). Within the group of up-regulated genes, we identified GO categories for biological processes (GO:0050789/GO:0065007), cell communication (GO:0007154), regulation of cellular processes (GO:0050794), response to stimuli (GO:0050896), together with some of their child terms, and participation in secondary metabolism (GO:0019748). Functions in secondary metabolism (GO:0019748) also were associated with genes assigned to the alternative behavior sets. This reflects the fact that siliques are sink organs (Robinson and Hill, 1999), in which secondary metabolites, like trehalose and ascorbic acid, are produced soon after fertilization, whereas others, such as maltose, sorbitol, and galactinol, accumulate during silique maturation (Watanabe et al., 2013).
Among the down-regulated genes, GO terms related to cellular component organization (GO:0016043), cell cycle (GO:0007049), photosynthesis (GO:0015979), and carbohydrate metabolic processes (GO:0005975) were enriched. The down-regulation of these gene sets reflects alterations in cell division and cell expansion rates, which are high during the early phases of fruit growth and diminish in later stages of fruit development and maturation (Vivian-Smith and Koltunow, 1999). Furthermore, chlorophylls are degraded, without being replaced, during silique maturation (Wagstaff et al., 2009; Jaradat et al., 2014); consequently, photosynthesis-related genes are found consistently in this group.
Expression Analysis of Genes with Known Functions Proves the Robustness of the Transcriptome Data Set
The expression profiles of genes already known to be involved in silique formation and maturation were used to verify the quality of our data set (Table 1). For instance, the MADS box gene AGAMOUS (AG), which encodes a transcription factor that is responsible for determining the fate of the fourth whorl of the flower and later orchestrates silique shattering (Ferrándiz, 2002; Dinneny et al., 2005), is highly expressed at all of the four time points analyzed. Furthermore, among genes known to promote cell expansion, LONGIFOLIA1 (LNG1), LNG2, ROTUNDIFOLIA3 (ROT3), ANGUSTIFOLIA (AN), and ATHB13 (Homeobox-Leu zipper protein ATHB-13; Tsuge et al., 1996; Kim et al., 1998, 1999, 2002, 2005; Hanson et al., 2001; Lee et al., 2006; Prasad et al., 2010) are present in our data set. More specifically, ROT3 expression increases between 3 and 6 DPA and then decreases until 12 DPA; therefore, ROT3 is found among the alternative behavior genes. In contrast, LNG1, LNG2, AN, and ATHB13 are among the down-regulated group, since they are strongly expressed at 3 DPA and their expression decreases with the onset of maturation, in agreement with their role in promoting cell expansion, which is concluded at 6 DPA.
Table 1. Expression of genes known from the literature to be involved in fruit formation or maturation.
| AtG Code | Name | Acronym | 3 DAP | 6 DAP | 9 DAP | 12 DAP | Group |
|---|---|---|---|---|---|---|---|
| AT5G15580 | LONGIFOLIA1 | LNG1 | 6.385 | 3.924 | 1.427 | −0.115 | Down |
| AT3G02170 | LONGIFOLIA2 | LNG2 | 6.329 | 3.373 | 2.947 | 3.794 | Down |
| AT4G36380 | ROTUNDIFOLIA3 | ROT3 | 4.098 | 5.461 | 4.864 | 3.525 | Alternative |
| AT1G01510 | ANGUSTIFOLIA | AN | 5.622 | 5.192 | 4.414 | 4.099 | Down |
| AT1G69780 | ATHB13 | ATHB13 | 5.629 | 4.127 | 3.247 | 1.261 | Down |
| AT4G18960 | AGAMOUS | AG | 5.511 | 5.126 | 5.213 | 5.274 | |
| AT5G67110 | ALCATRAZ | ALC | 6.802 | 6.989 | 7.099 | 5.845 | |
| AT5G60910 | FRUITFULL | FUL | 7.216 | 6.413 | 7.079 | 7.079 | |
| AT4G00120 | INDEHISCENT | IND | 3.698 | 3.950 | 4.413 | 3.982 | |
| AT2G45190 | FILAMENTOUS FLOWER | FIL | 3.439 | 3.864 | 3.822 | 2.664 | |
| AT1G68480 | JAGGED | JAG | −1.461 | −0.659 | −4.440 | −4.592 | Down |
| AT5G02030 | REPLUMLESS | RPL | 5.176 | 5.458 | 5.825 | 6.368 | |
| AT4G09960 | SEEDSTICK | STK | 6.365 | 5.582 | 6.691 | 6.001 | |
| AT2G42830 | SHATTERPROOF2 | SHP2 | 4.358 | 5.042 | 4.400 | 3.565 | |
| AT3G58780 | SHATTERPROOF1 | SHP1 | 6.315 | 5.088 | 5.933 | 7.010 | Up |
| AT4G00180 | YABBY3 | YAB3 | 1.451 | 2.668 | 2.796 | 1.663 | |
| AT1G69490 | NAC-LIKE, ACTIVATED BY AP3/PI | AtNAP | −0.021 | 2.244 | 3.231 | 4.413 | Up |
| AT1G04580 | ALDEHYDE OXIDASES4 | AAO4 | 3.422 | 8.012 | 9.495 | 9.552 | Up |
Fruit development and maturation are strictly regulated by hormones (Kumar et al., 2014). In Supplemental Table S7, a list of genes involved in hormone metabolism, and differentially expressed in our samples, is provided. Among the five classical hormones, ethylene is known to play a key role in silique maturation (Kou et al., 2012). Ethylene is synthesized from Met, which is converted into S-adenosylmethionine by ACS (1-aminocyclopropane-1-carboxylate synthase) enzymes. Of the nine Arabidopsis ACS genes, only a few were found to be up-regulated in our data sets (Supplemental Table S7), such as ACS2 (Kou et al., 2012) and ACS7 (Tsuchisaka et al., 2009). ACS6 is found in the down-regulated gene set, although it is robustly transcribed during all the stages of fruit development taken into consideration. The ACC oxidase (ACO) enzymes, which are involved in the final step of the ethylene biosynthetic pathway, convert 1-aminocyclopropane-1-carboxylic acid (ACC) into ethylene. Among the five Arabidopsis ACO enzymes, ACO2 and ACO4 are assigned to the group of up-regulated genes, whereas ACO3 transcription increases quickly and reaches its maximum at 9 DAP before decreasing slightly by 12 DAP. Interestingly, AtNAP (NAC-LIKE, ACTIVATED BY AP3/PI, NAC029), a NAC transcription factor whose expression increases over the course of silique development and whose product orchestrates ethylene biosynthesis (Kou et al., 2012), also is represented in the list of up-regulated genes. Besides AtNAP,41 more NAC-encoding genes are assigned to the up-regulated group, 4 NAC genes are listed as alternative behavior, and 15 are part of the down-regulated gene set (Supplemental Table S8). NAC DNA-binding proteins are plant-specific transcription factors; in Arabidopsis, the NAC family so far includes 138 genes whose products play pivotal roles in plant development, hormone signal transduction, and stress responses (Shao et al., 2015). In agreement with the up-regulation of AtNAP, the transcription factor EIN3 (ETHYLENE INSENSITIVE3), a positive regulator of ethylene signal (Kou et al., 2012), also is found among the up-regulated genes. AtNAP expression is triggered by EIN3 (Kim et al., 2014), which modulates the expression of several ethylene-responsive factors that act in response to ethylene, abscisic acid, and jasmonic acid as well as to biotic and abiotic stresses. Indeed, several ethylene-responsive factor genes are differentially expressed in our data sets (Supplemental Table S8), including ABI4 (ABSCISIC ACID INSENSITIVE4; Dong et al., 2016) among the up-regulated genes and CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF4 among the down-regulated genes. ABI4 is an intriguing transcription factor, since it antagonizes ethylene production (Dong et al., 2016) and is involved in plastid-to-nucleus and mitochondrion-to-nucleus retrograde signaling (Giraud et al., 2009; León et al., 2013).
The latter phase of silique development also is characterized by events relating to dehiscence, which together result in seed release upon pod shattering. Silique shattering includes the gradual breakdown of silique tissues, and this process is tightly regulated (for review, see Matilla, 2007). However, since the last time point considered here is 12 DPA, before any signs of shattering become manifest, we do not expect the genes involved in this process to be differentially expressed yet. Indeed, all the genes known to be involved in pod shattering are present in our data set, but only SHATTERPROOF1 (SHP1) shows a statistically significant increase in its expression during silique development (Liljegren et al., 2000). SHP1 is a MIKC MADS box transcription factor (Masiero et al., 2011), and nine other MIKC MADS box genes are included in the list of up-regulated genes; among them are AGAMOUS-LIKE15 (AGL15), AGL18, and SHORT VEGETATIVE PHASE, which might cooperate to slow down fruit maturation (Fernandez et al., 2014).
To further assess the reliability of the RNAseq data set, the expression profiles of 202 nuclear genes known to be involved in chloroplast-related functions, such as photosynthesis (42 genes), tetrapyrrole biosynthesis (26 genes), plastid protein translation (31 genes), RNA metabolism (13 genes), chlorophyll degradation (nine genes), plastid protein chaperone and degradation (35 genes), plastid protein import (20 genes), plastid membrane metabolism (five genes), nucleus-to-plastid anterograde signaling (seven genes), and plastid-to-nucleus retrograde signaling (14 genes), were investigated (Fig. 2A; Supplemental Fig. S7; Supplemental Table S8). As expected, the expression profiles of genes coding for components of the photosynthetic machinery, including PSI and PSII, the light-harvesting complexes a (Lhca) and b (Lhcb), cytochrome b6f, the small subunit of Rubisco (RbcS), and subunits of ATP synthase, showed no significant change between 3 and 6 DPA before exhibiting a marked decrease at 9 and 12 DPA. The tetrapyrrole biosynthesis pathway, including its chlorophyll and heme branches, followed a similar general trend, with a slight decrease between 3 and 6 DPA and a marked reduction at 9 and 12 DPA. A similar expression pattern also was exhibited by genes encoding proteins involved in plastid gene expression, such as plastid ribosomes and plastid factors with a role in RNA metabolism. In particular, the expression of nuclear genes coding for plastid ribosomal proteins, for plastid RNA helicases of the DEAD/DEAH box family, and for plastid pentatricopeptide repeat proteins displayed a general decrease over the sampling time frame from 3 to 12 DPA. Conversely, the expression profiles of genes involved in chlorophyll degradation were characterized by a continuous increase from 3 to 12 DPA, in accordance with the marked structural alterations that chloroplasts undergo in the course of their metamorphosis into gerontoplasts during fruit maturation. Chloroplasts as well as the other plastid types, such as gerontoplasts and chromoplasts, need to communicate with the nucleus to modulate and harmonize the expression of the nuclear and plastid genomes. Our data show that the expression of nuclear genes involved in nucleus-to-chloroplast communication (i.e. anterograde signaling), including the nucleus-encoded RNA polymerase and the RNA polymerase Sigma factors (SigA–SigF), was down-regulated once chloroplasts were committed to turn into gerontoplasts, the only exception being SigE, whose expression doubled from 3 to 12 DPA (Supplemental Fig. S7; Supplemental Table S8). In contrast, the expression of genes involved in chloroplast-to-nucleus communication (i.e. retrograde signaling pathway) remained rather stable during fruit aging, and only small differences in the expression of STN7 (up-regulated by 33% from 3 to 12 DPA) and PRIN2 (down-regulated by 31% from 3 to 12 DPA) could be detected. The transition from chloroplast to gerontoplast also is accomplished through modifications of plastid membranes and the plastid proteome. As expected, nuclear genes encoding the CURT proteins, involved in the maintenance of thylakoid membrane ultrastructure, were down-regulated, whereas the expression of the VIPP1 gene, encoding VESICLE-INDUCING PROTEIN IN PLASTIDS1, remained constant. In general, genes for the plastid import apparatus (i.e. TIC and TOC proteins) were slightly down-regulated from 3 to 12 DPA. Only in a few cases were larger changes in gene expression observed, such as for TIC20-IV (up-regulated by about 4-fold from 3 to 12 DPA) and TIC62 (down-regulated by 4-fold). In contrast, no major changes in expression were observed among genes encoding plastid chaperones or proteases, indicating that the plastid proteome also is controlled at the posttranscriptional level.
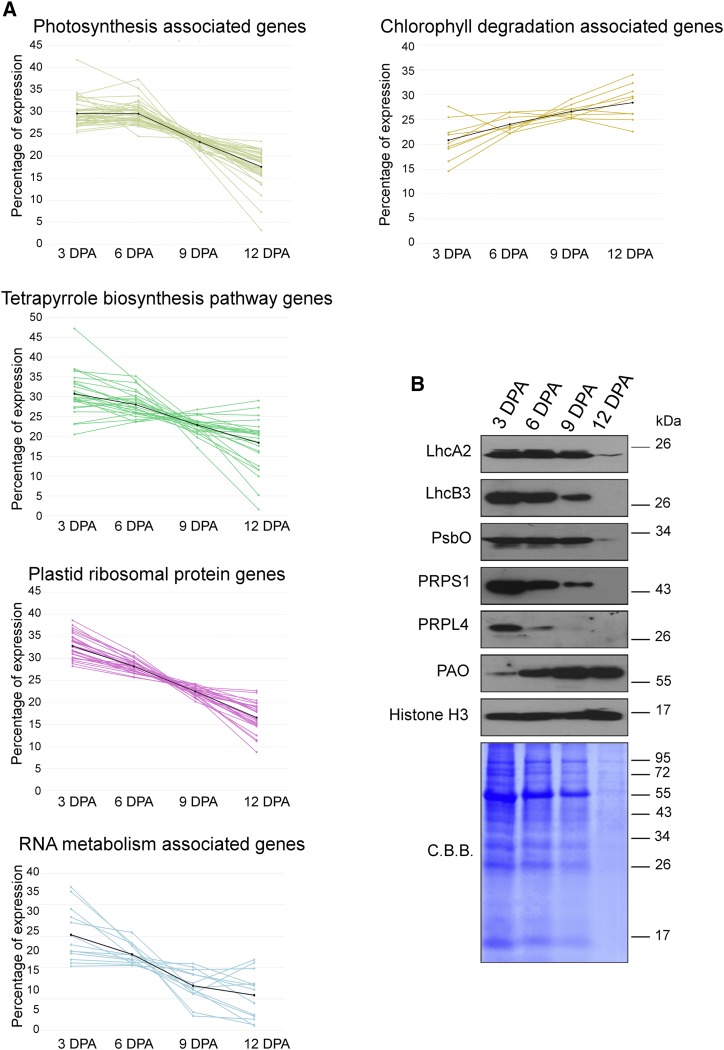
Figure 2.
Expression patterns of genes involved in chloroplast-related functions. A, Expression of genes involved in photosynthesis (i.e. encoding subunits of the thylakoid electron transport chain, tetrapyrrole biosynthesis, plastid ribosomal protein, RNA metabolism, and chlorophyll degradation). Each line describes the expression pattern of a single gene; the core values for each cluster are plotted in black. The percentage of expression was defined as the ratio between expression values at every single time point and the sum of the values of all the time points, given as 100%. The expression values of all the genes are reported in Supplemental Table S9. B, Immunoblot analyses of total protein extracts from 3-, 6-, 9-, and 12-DPA developing Col-0 siliques, whose seeds have been removed manually. Total protein extracts obtained from identical silique fresh weight were fractionated by SDS-PAGE (12% polyacrylamide) and transferred on polyvinylidene difluoride filters. Filters were then immunodecorated using antibodies specific for LhcA2, LhcB3, PsbO, PRPS1, PRPL4, PAO, and Histone H3 proteins. A replicate SDS-PAGE gel stained with Coomassie Brilliant Blue (C.B.B.) is shown as a loading control. One out of three immunoblots for each antibody is shown.
To verify the collinearity between transcript levels and photosynthesis-related protein accumulation, as shown previously for the Calvin cycle and amino acid biosynthesis pathways in the chloroplast (Piechulla et al., 1985; Kleffmann et al., 2004), immunoblot analyses were performed (Fig. 2B). Using Lhca2- and Lhcb3-specific antibodies, we measured the accumulation of the proteins that harvest the light energy and transfer it to the photosystems. In agreement with the transcriptomic data, Lhc accumulation decreased in old siliques at 9 and 12 DPA. The same behavior was observed for PsbO, a subunit of the oxygen-evolving complex and part of the PSII (Suorsa et al., 2016), and for the plastid ribosomal proteins of both the small (30S) and large (50S) subunits (PRPS1 [PLASTID RIBOSOMAL PROTEIN SMALL1] and PRPL4 [PLASTID RIBOSOMAL PROTEIN LARGE4]; Romani et al., 2012). Furthermore, Coomassie Brilliant Blue staining of total protein extracts demonstrated that, at 12 DAP, both the large and small subunits of Rubisco accumulate poorly (Fig. 2B), in agreement with a previous observation in leaves, where the degradation of Rubisco was proposed at the basis of senescence (Breeze et al., 2011). In contrast, proteins with a key role in senescence-associated changes accumulate in large amounts in fruits at 12 DAP, such as the enzyme PHEOPHORBIDE A OXYGENASE (PAO), which catalyzes the key reaction of chlorophyll catabolism (Ghandchi et al., 2016). A small amount of PAO mRNA is present at 3 DPA but a lot is accumulated at 12 DPA, showing a perfect collinearity with the enzyme amount. Filters also were immunodecorated with an anti-H3 antibody, as a control for loading (Chen et al., 2016).
RNAseq Data Validation through Promoter Expression Analysis
To further validate the RNAseq data, the silique transcriptome data sets were analyzed using promoter-driven GUS assays. In particular, we analyzed the promoter activity of the FLOWERING LOCUS T (pFT), LIPOXYGENASE3 (pLOX3), and LOX4 (pLOX4) genes from the up-regulated group, ABA INSENSITIVE5 (pABI5) from the alternative behavior group, and RESPONSE REGULATOR15 (pARR15) and LOX2 (pLOX2) genes as representative of the down-regulated gene set (Fig. 3; Supplemental Table S9). The putative promoter regions of these genes were cloned upstream of the GUS reporter gene, and the siliques of the corresponding transgenic lines were analyzed at 3, 6, 9, and 12 DPA, the same time points used for the RNAseq analysis (Fig. 3). These genes were selected because they display high levels of expression or pronounced variations among at least three of the four different time points, facilitating the observation and quantification of the differences.
Figure 3.
Promoter-driven GUS expression analysis to validate RNAseq data. The expression of the promoters of FT, LOX3, LOX4, ABI5, LOX2, and ARR15 fused to the GUS reporter gene was evaluated on the basis of UidA enzymatic activity. Silique images are representative of analyses performed on 10 siliques for each stage and transgenic line. At right, the level of expression of each gene (according to the RNAseq data reported in Supplemental Table S10) is shown. The images shown are composite images. Bars = 1 mm.
In fruits at 3 and 6 DPA, the enzymatic activity of the GUS protein expressed under the control of pFT is detected in the replum, in the septum, and in the pedicel, while at 9 and 12 DPA, the blue staining also is visible in the valves, thus confirming the transcriptome data, which show a consistent increase of FT transcripts during the development of the silique.
For pLOX3 and pLOX4, the signal at 3, 6, and 9 DPA is restricted to the basal part of the siliques, in the pedicel for pLOX4 and in the abscission zone for pLOX3. At 12 DPA, pLOX3 also drives GUS activity in the valves, in the replum, in the septum, and in the pedicel, while the signal of pLOX4 is visible only in some areas of the valves. The expression of pLOX3 and pLOX4 in the two early stages and at 12 DPA confirm the transcriptome data. Nevertheless, we could not detect any GUS activity at 9 DPA, although the transcriptome data set indicates that both genes are expressed strongly at this stage, suggesting the lack of important regulatory elements in the cloned DNA fragments.
By contrast, in pABI5::GUS fruits at 3 DPA, the enzymatic activity of the GUS protein is detected in the valves, in the style, and in the pedicel. After this stage, the signal dropped off drastically, remaining only in the pedicel, in the apical part of the valve tissue, in the style, and in some funiculi. These data confirm the trend of transcriptome levels detected by RNAseq between 3 and 6 DPA, while at 9 and 12 DPA, the GUS assay failed to reveal the ABI5 transcript increase.
In the case of pLOX2, GUS activity is detected in the whole silique and in the pedicel from 3 to 12 DPA, in agreement with the high level of transcript accumulation observed by RNAseq, although the enzyme assay failed to reveal the decrease in LOX2 transcript observed in the transcriptome data set, possibly due to the lower sensitivity of the GUS staining.
On the other hand, ARR15 promoter activity is observed in the entire silique and in the pedicel at 3 DPA, but its expression is restricted to the apical part of the silique and the pedicel at 6 DPA and persists only in the pedicel at 9 and 12 DPA, which is consonant with the transcriptome data.
Novel Genes Involved in Fruit Development and Maturation
Our data set indicates that more than 10,000 genes are differentially expressed among the four time points we chose, and the extent of the differences confirms the notion that our samples represent four different developmental stages. Furthermore, the robustness of our RNAseq-based silique transcriptome could be confirmed by several independent approaches, as detailed above, prompting us to query the data set for novel genes with key roles in fruit formation. To this end, we selected a few genes whose expression levels change substantially among the selected time points. These genes (Table 2) were chosen from all three classes of differentially expressed genes and are associated with different GO classes, including transcription factors (AGL15, SPL15, NAC058, and AT3G60670), hormone signaling (AT2G30810), metabolic processes (UGT73C1), histone modification (HDA3), housekeeping roles (TUB6), and genes of unknown function (AT5G10625 and AT3G01323).
Table 2. Expression of selected genes analyzed in this study.
| AtG Code | Name | Acronym | 3 DAP | 6 DAP | 9 DAP | 12 DAP | References |
|---|---|---|---|---|---|---|---|
| Cluster: up-regulated | |||||||
| AT5G13790 | AGAMOUS-LIKE15 | AGL15 | 1,607 | 3,617 | 4,514 | 3,233 | Fang and Fernandez (2002) |
| AT2G36750 | UDP-GLUCOSYL TRANSFERASE73C1 | UGT73C1 | 1,752 | 1,005 | 2,022 | 3,629 | Hou et al. (2004) |
| AT3G60670 | PLATZ transcription factor family gene | −3,388 | 1,290 | 2,847 | 4,129 | ||
| AT5G10625 | Unknown function | −2,349 | −1,859 | 2,712 | 7,879 | ||
| Cluster: down-regulated | |||||||
| AT3G44750 | HISTONE DEACETYLASE2A/HISTONE DEACETYLASE3 | HDA2A/HDA3 | 4,790 | 4,728 | 2,935 | 3,370 | Wu et al. (2000) |
| AT5G12250 | BETA-6 TUBULIN | TUB6 | 8,886 | 8,821 | 6,727 | 4,167 | Czechowski et al. (2005); Gutierrez et al. (2008) |
| AT3G57920 | SQUAMOSA PROMOTER BINDING PROTEIN-LIKE15 | SPL15 | 2,635 | 0,085 | −0,857 | −1,661 | Schwarz et al. (2008) |
| AT2G30810 | GA-regulated family gene | 7,416 | 2,513 | −2,858 | −3,818 | ||
| AT3G01323 | Unknown function | 4,008 | −0,564 | −1,851 | −4,592 | ||
| Cluster: alternative behavior | |||||||
| AT3G18400 | NAC DOMAIN CONTAINING PROTEIN58 | NAC058 | −4,691 | −3,143 | 4,759 | 1,802 | Coego et al. (2014) |
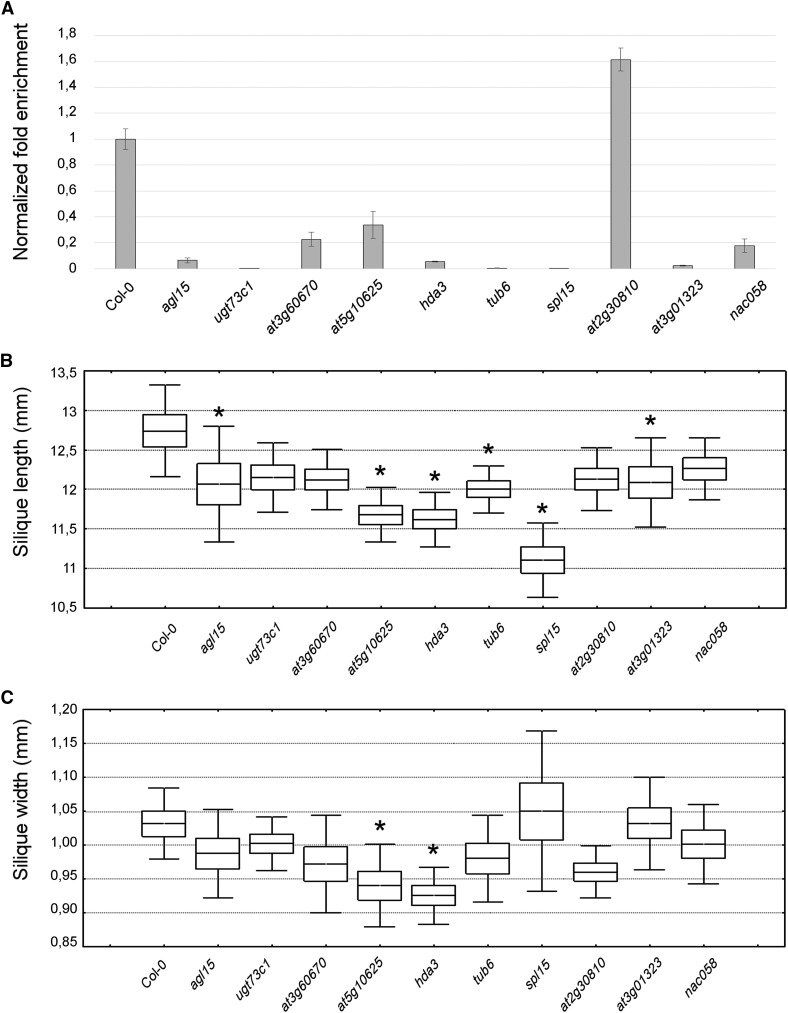
The expression profile of each of the selected genes was first validated by reverse transcription quantitative PCR (RT-qPCR) analysis (Fig. 4). However, the most common reference genes used for normalization in Arabidopsis, such as ACTIN2/ACT2, ACT7, ACT8, ELONGATION FACTOR 1α/EF1α, EUKARYOTIC TRANSLATION INITIATION FACTOR4A-1/eIF4A, TUBULIN2/TUB2, TUB6, TUB9, UBIQUITIN4/UBQ4, UBQ5, UBQ10, UBQ11, and GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE/GAPDH (Czechowski et al., 2005; Gutierrez et al., 2008), showed quite variable and stage-dependent expression in our data set (Supplemental Table S11). For instance, ACT2, TUB2, and TUB6 were in the group of down-regulated genes. Therefore, stably expressed genes that could be used as internal reference genes first had to be identified. In particular, we selected UBC9 and PP2A as internal reference genes (already suggested by Czechowski et al. [2005] as two of the best housekeeping genes for developmental series), which are highly and stably expressed at all four time points considered in our transcriptome analysis (Supplemental Table S10). The RT-qPCR results (Fig. 4) support our transcriptome data, with the sole exception of UGT73C1 at the 12-DPA stage. This gene belongs to a multigene family encoding UDP-glycosyltransferases, whose members share a high degree of similarity (Li et al., 2001), which complicates the expression analyses. Insertion mutant lines for the selected genes also were isolated, and the transcript levels in mutant and Col-0 siliques were compared by RT-qPCR. The sampling time points for RT-qPCR analysis were chosen based on the gene expression patterns (Fig. 4) in order to detect the gene expression peak; thus, the down-regulated genes were analyzed at 3 DPA while all the up-regulated genes and NAC058 were examined at 9 DPA. As shown in Figure 5A, nine T-DNA insertion lines exhibited marked reductions in transcript abundance relative to Col-0, while the T-DNA insertion into the AT2G30810 gene caused an overaccumulation of the corresponding transcripts.
Figure 4.
Expression patterns of selected genes by RT-qPCR analysis and RNAseq. The x axis indicates the four selected time points, while the y axis shows the mRNA levels: the scale at left indicates gene expression level based on RT-qPCR, and the scale at right is based on RNAseq. The blue lines correspond to RT-qPCR and the orange ones to RNAseq. RT-qPCR was performed on cDNA extracted from Col-0 siliques devoid of seeds and collected at the same time points chosen for RNAseq. RT-qPCR was conducted in triplicate using PP2A and UBC9 as internal reference genes (Supplemental Table S11). The graphs depict the results obtained using PP2A as the internal reference gene; identical results were obtained using UBC9 as the internal reference gene. Error bars represent the propagated sd value using three biological replicates.
Figure 5.
Molecular and morphological characterization of the phenotypic effects of the selected mutations on silique formation. A, RT-qPCR analysis to verify transcript accumulation in the mutant lines examined (Supplemental Table S12). RT-qPCR was conducted in triplicate using PP2A and UBC9 as internal reference genes (Supplemental Table S11). The graphs depict the results obtained using PP2A as the internal reference gene; identical results were obtained using UBC9 as the internal reference gene. Error bars represent the propagated sd value using three replicates. B and C, Measurements of the length (B) and width (C) of the siliques produced by the selected lines. The length and width of the mature siliques were evaluated by measuring 40 siliques for each genotype in two different biological replicates. Boxes indicate means plus se, while error bars indicate sd. One-way ANOVA combined with Dunnett’s comparison test indicated that the mutants marked with asterisks were significantly different from the Col-0 control (P < 0.05; n = 40).
The length and width of Col-0 and mutant siliques were then measured to deduce possible roles for the corresponding gene products in fruit growth (Fig. 5, B and C). Forty siliques for each genotype were measured in two different biological replicates, and interestingly, the siliques of agl15, hda3, tub6, spl15, at5g10625, and at3g01323 were reduced significantly in length with respect to Col-0, whereas reductions in silique width were observed in hda3 and at5g10625 mutant lines. Thus, HDA3 and AT5G10625 appear to be involved in the silique growth program in terms of both length and width, while AGL15, TUB6, SPL15, and AT3G01323 seem to be involved in functions that affect only one of these dimensions.
We also considered silique maturation, and this trait was evaluated by three different approaches that are used routinely to detect leaf senescence: (1) Trypan Blue assay (van Wees, 2008); (2) measurements of the quantum efficiency of PSII electron transport (Wingler et al., 2004); and (3) visual assessment of yellowing, combined with chlorophyll a and b measurements, during silique senescence (Ougham et al., 2008).
The viability of the silique cells was explored using the Trypan Blue dye-exclusion method (Fig. 6A; Supplemental Figs. S8 and S9), which allows one to visualize dead and dying cells (van Wees, 2008). In Col-0 siliques, we detected cell death in the stigma and in the gynophore, starting from 3 DPA and continuing to 12 DPA (Fig. 6A). Moreover, at later stages (9 and 12 DPA), portions of the funiculi and septum were found to be positive for Trypan Blue staining. A similar staining pattern was observed in eight of the 10 mutants analyzed, the exceptions being hda3 and at2g30810. More specifically, in hda3 fruits at 12 DPA, most of the funiculi and the septum displayed a markedly intense coloration, indicating widespread cell death in these regions. In contrast, in at2g30810, portions of the valves were already blue at 6 DPA, suggesting premature senescence of the silique; moreover, the blue cells are grouped in spots with a typical pattern (Fig. 6A; Supplemental Fig. S8). The precise quantification of the blue area (Supplemental Fig. S9) revealed that hda3 differs from Col-0 siliques only at 12 DPA, while at2g30810 siliques already show differences at 3 DPA and in all the other considered time points.
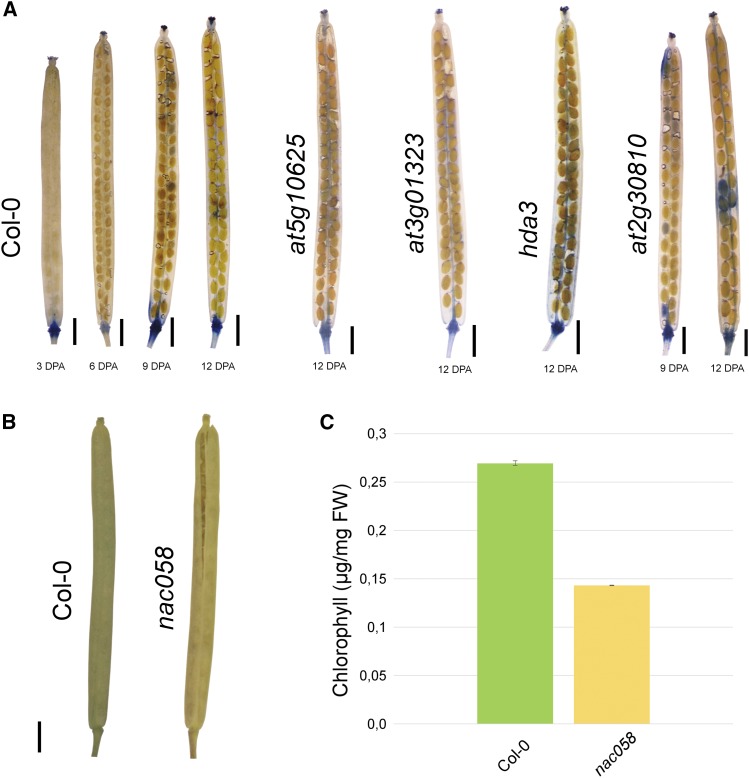
Figure 6.
Phenotypic effects of the selected mutations on silique maturation. A, The Trypan Blue dye-exclusion method allows the detection of cell death. A complete analysis is reported in Supplemental Figure S8. Trypan Blue also was quantified, and the quantification is reported in Supplemental Figure S9. The image shown is a composite image. B, Visual evaluation of the yellowing of Col-0 and nac058 siliques at 12 DPA. C, Measurements of chlorophyll content in Col-0 and nac058. Error bars represent sd of three independent measurements. FW, Fresh weight. Bars in A and B = 1 mm.
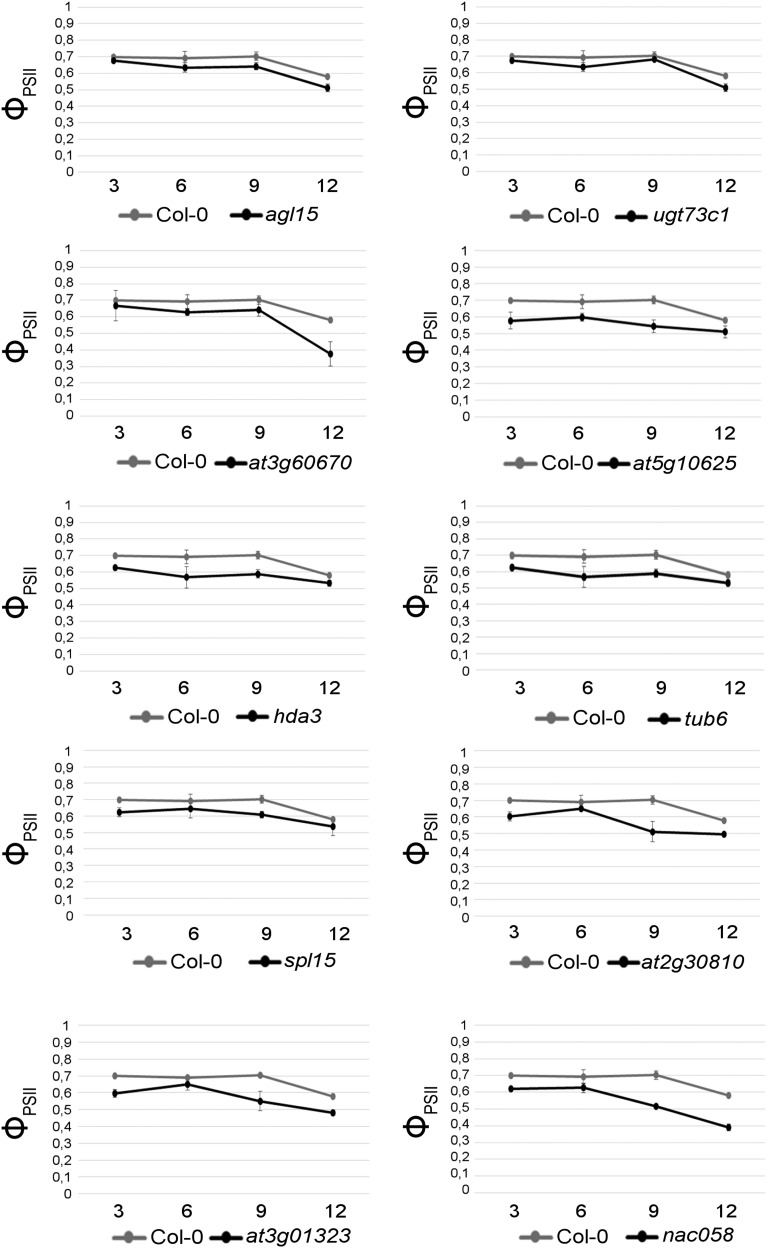
In order to monitor fruit senescence, we also measured the photosynthetic performance of PSII in the siliques with a Dual-PAM chlorophyll fluorometer (Wingler et al., 2004; Brooks and Niyogi, 2011; Scarpeci et al., 2017). Before analyzing the selected mutants with respect to the quantum efficiency of PSII, we conducted a preliminary experiment to exclude any contribution to PSII photosynthetic performance from embryo chloroplasts, as proplastids differentiate into photosynthetically active chloroplasts during embryo development in Arabidopsis (Mansfield and Briarty, 1991). To this end, we chose arf8-4 plants, which can develop parthenocarpic fruits if the anthers are removed manually (Vivian-Smith et al., 2001; Goetz et al., 2006). Measurements of the quantum efficiency of PSII at 3 and 6 DPA were performed on the Landsberg erecta (Ler) ecotype and compared with the data for self-pollinated and emasculated arf8-4 flowers, respectively. Moreover, to exclude the possibility that the removal of all of the floral organs surrounding the carpel in the emasculated arf8-4 siliques might affect the photosynthetic efficiency of the mutant, we also removed all the floral organs prior to manual pollination of Ler and arf8-4 pistils (we called these samples Ler naked and arf8-4 naked). Interestingly, measurements of the quantum efficiency of PSII did not reveal any differences among the mutants and the control, supporting the notion that the measured PSII efficiency refers to silique chloroplasts and does not include any contribution from the chlorophyll in embryos (Supplemental Fig. S10A). To further validate this observation, we used PRPL28/prpl28 heterozygotes (Romani et al., 2012), in which one-fourth of the embryos that develop inside the siliques are albinos. Once again, we failed to detect any differences in photosynthetic efficiency between the mutant and Col-0 siliques (Supplemental Fig. S10B). The analysis of the photosynthetic ability of the mutant siliques revealed several degrees of phenotype (Fig. 7): in agl15, ugt73c1, tub6, and spl15, photosynthetic efficiency was identical to that in the Col-0 siliques, while hda3, at5g10625, at2g30810, and at3g01323 displayed a slight reduction in photosynthetic ability with respect to Col-0. at3g60670 and nac058 mutants instead presented a marked reduction in photosynthetic efficiency at 12 DPA, which also was observed at 9 DPA in the case of nac058 (Fig. 7) and in agreement with the precocious senescence phenotype shown in Figure 6B, while visual assessment of yellowing during silique senescence failed to uncover any differences between Col-0 and at3g60670. To further validate the precocious yellowing observed in nac058, we extracted and quantified the total chlorophyll content in the valves of Col-0 and nac058 (Fig. 6C). This analysis confirmed that nac058 contains half of the chlorophyll present in Col-0 valves at 12 DPA, suggesting the premature breakdown of chloroplasts in this mutant.
Figure 7.
Photosynthetic efficiency (ΦPSII) of wild-type and mutant siliques. Three measurements for each stage were performed (using five siliques for each developmental stage for a single measurement), and three biological replicates were used. Mean values ± sd are reported.
DISCUSSION
Time-Course Transcriptome Profiling of Developing Seedless Fruits
Fruits are essential for humans, as an integrative part of our diet, and they also are an essential element in the plant life cycle, serving first to protect seeds and then facilitating their dispersal (Van der Pijl, 1982; Seymour et al., 2013). Beyond their role in the protection of the offspring, fruits also contain active chloroplasts that contribute to plant fitness (Blanke and Lenz, 1989; Raghavan, 2003; Gnan et al., 2017).
To analyze the molecular networks responsible for mediating and coordinating fruit growth and maturation in Arabidopsis, we have carried out a transcriptomic analysis of Col-0 valve tissues. We decided to manually remove the seeds to exclude any embryo/seed contribution to our data set, in order to increase the probability of pinpointing genes involved specifically in fruit development and maturation. Fruit growth is controlled by signals, most probably produced by the female gametophyte, aimed to communicate that fertilization has occurred successfully and to promote ovule conversion into seeds and pistils into fruits (Dorcey et al., 2009). As a matter of fact, in the absence of fertilization, pistils undergo senescence after a few days (Carbonell-Bejerano et al., 2010). Furthermore, fruit growth relies on seed development, and some evidence pinpoints that hormones are at the basis of seed-fruit communication. Because of that, the siliques used for our analyses were the result of natural, and not of manual, fertilization, to avoid flower organ removal and, as a consequence, hormone homeostasis perturbation.
We have identified thousands of genes differentially expressed among the four chosen time points. Based on previously published microarray data (de Folter et al., 2004; Wagstaff et al., 2009; Carbonell-Bejerano et al., 2010; Jaradat et al., 2014), RNAseq appears to be a more powerful strategy, since almost 7,000 genes annotated in the TAIR10 genome are not represented on the Affymetrix ATH1 microarray platform (Giorgi et al., 2013). Thus, RNAseq can probe portions of transcripts never analyzed before. For instance, AT3G01323, AT3G60670, and AT5G10625 are not present in the ATH1 platform, while our analyses pinpoint that they are differentially expressed and that their disruption affects fruit photosynthetic efficiency.
In our experimental design, we set the zero time point to coincide with flower anthesis, and we collected the material at 3, 6, 9, and 12 DPA in order to cover both the growing and the maturation phases. These time points also allow us to explore events that occur in the phase immediately following fertilization, which is characterized by rapid changes in the silique structure, since fruit size increases 5-fold within a few days (Fig. 1A; Kleindt et al., 2010). Indeed, at least three out of the four selected time points are included in the classical stage 17 of silique formation (Ferrándiz et al., 1999). However, our four data sets are clearly different from each other, suggesting that fruits normally annotated as stage 17 are undergoing highly dynamic development or that development is quite asynchronous. Our results support the idea that stage 17 should be disassembled into several different classes, in agreement with the data reported in some previous publications (Kleindt et al., 2010; He et al., 2018). In total, we have been able to detect 21,908 protein-coding genes, while 25,706 differentially expressed genes were reported by Klepikova et al. (2016), where RNAseq data of 79 Arabidopsis samples, including organs at different developmental stages, were analyzed, thus supporting the robustness of our approach. According to our analyses, 4,240 genes are consistently up-regulated and 5,813 genes are consistently down-regulated in Arabidopsis siliques from the first time point to the last, while 581 exhibit alternative behaviors (i.e. transient changes in expression within the period examined; Supplemental Tables S1–S6). Interestingly, 11,274 genes do not change their expression significantly, although we cannot exclude the possibility that some of these genes code for regulators of fruit development and maturation, since posttranscriptional regulation also plays an important role in generating rapid responses to environmental and intracellular signals.
New Genes Involved in Silique Growth and Maturation
Given the large numbers of genes differentially expressed in developing siliques, the main question is whether one can readily uncover information about new regulatory networks and genes involved in silique formation and maturation. Therefore, we selected 10 genes based on the following criteria: (1) their expression changes substantially among the selected time points; (2) they cover each of the three classes of differentially expressed genes; and (3) they are associated with different GO terms. The selected genes encode transcription factors (AGL15, SPL15, NAC058, and AT3G60670), proteins with a role in hormone signaling (AT2G30810), in metabolic processes (UGT73C1), in histone modification (HDA3), and in housekeeping functions (TUB6), and proteins of unknown function (AT5G10625 and AT3G01323). The corresponding T-DNA insertional mutants were isolated, and their involvement in determining fruit size and maturation was investigated. In particular, the disruption of two of the four genes encoding transcription factors affected fruit size or maturation, indicating that our transcriptome data sets allow the identification of master regulators of important developmental cascades. For instance, the agl15 knockout mutant develops shorter siliques but fruit maturation seems unaffected. Previous studies indicate that AGL15 overexpression (Fernandez et al., 2000; Fang and Fernandez, 2002) delays sepal and petal falls and that AGL15 is responsible, together with AGL18, for organ senescence (Adamczyk et al., 2007). Indeed, AGL18 also is included in the up-regulated gene data set and probably compensates for the absence of AGL15, thus explaining why agl15 siliques are not positive to the Trypan Blue staining and show wild-type-like photosynthetic performance (Fig. 7; Supplemental Figs. S8 and S9). Also in the case of the spl15 mutant, siliques were shorter, implying that silique length is a rather complex trait controlled by multiple regulatory pathways. SPL15 is a transcription factor that belongs to the SQUAMOSA Promoter Binding Protein (SBP) family. Four more SBP genes are found to be down-regulated in our data sets, while only two genes are included in the group of up-regulated genes, and no SBP gene is listed in the alternative behavior group. SBP genes are involved in organ formation (Wang et al., 2008; Usami et al., 2009), nutritional changes (Jung et al., 2011), copper metabolism (Yamasaki et al., 2009), and GA response (Zhang et al., 2007). On the other hand, the nac58 mutant displayed a reduced photosynthetic performance in fruits at 9 DPA, but no alteration in silique growth was observed. Many more NAC genes, 60 out of the 138, are differentially expressed in our data sets. Previous works (Breeze et al., 2011; Kim et al., 2014, 2016) pinpointed a pivotal role for the NAC genes in leaf aging, and several NAC-encoding genes have been found to be differentially expressed in aging leaves at different time points (Balazadeh et al., 2008; Breeze et al., 2011; Christiansen and Gregersen, 2014).
Overall, seven out of the 10 selected mutants showed either altered morphology in silique length and silique width or precocious senescence of mature siliques. These findings, together with the fact that several other transcription factors, such as the Auxin-Responsive Factor and the Ethylene-Responsive Factor families (Supplemental Table S8), and many genes already known to have a role in silique growth, like CKX5 (cytokinin oxidase/dehydrogenase), are present in our list of differentially regulated genes (Bartrina et al., 2011) or are among the genes expressed at high levels in all the analyzed stages, such as FATA2 (Wang et al., 2013), strongly indicate that our data set can be considered a reservoir from which genes with a role in fruit formation can be drawn.
Coordination of Chloroplast and Nuclear Gene Expression Is Fundamental during Fruit Development and Maturation
The unifying theory, based on the observation of a Jurassic plant with free central placentation, suggests that carpels are composed of an ovule-bearing shoot (the placenta) and its enclosing foliar structure (Wang, 2010). Cytological and molecular observations also confirm the ontogenic relationship between leaves and fruits (Gillaspy et al., 1993). However, unlike leaves, silique maturation is strictly regulated by seed formation and strictly controlled by the progression of embryogenesis.
Chlorophyll breakdown is the clearest marker of both leaf senescence and fruit ripening, but the pathways that control these processes differ between the two organs. For instance, the disruption of PHEOPHYTINASE causes a stay-green phenotype in leaves, although greening of seeds and fruits is not affected (Guyer et al., 2014; Zhang et al., 2014; Chen et al., 2017). By contrast, AtNAP plays a pivotal role during leaf natural and dark-induced senescence, since it modulates the transcription of several genes, including PAO (Yang et al., 2014), whose accumulation at later stages of silique development and maturation was confirmed by our RNAseq data sets and by immunoblot analysis. Indeed, the AtNAP transcription factor couples plastid physiological status to hormone metabolism, since it modulates ethylene metabolism in both senescing leaves and maturating siliques (Kou et al., 2012) and abscisic acid in senescing leaves (Yang et al., 2014). In particular, ethylene promotes or inhibits growth and senescence processes depending on its concentration and the timing of its production or application. Therefore, leaf senescence and silique senescence might vary in relation to the growth conditions. For instance, differences might be expected between plants grown in greenhouses as opposed to growth chambers. Under the latter conditions, the small volumes of the growth chambers might facilitate ethylene accumulation, which, in turn, might accelerate senescence mechanisms. Therefore, chlorophyll accumulation in Arabidopsis might reach its peak on different DPAs, depending on the precise growth conditions. In different studies, chlorophyll accumulation in the siliques has been reported to reach its maximum level between 6 and 8 DPA (Wagstaff et al., 2009; Kou et al., 2012). In this respect, our data set is extremely useful, since it contains specific genes, such as NAC058 and AT2G30810, that can be used as reliable molecular markers for the accurate staging of fruit maturation in place of chlorophyll amount. For instance, the tomato (Solanum lycopersicum) mutants lutescent1 and lutescent2 display a precocious loss of chlorophyll in leaves and fruits (Barry et al., 2012), but the onset of fruit ripening is delayed by approximately 1 week, indicating a major role for the chloroplast in mediating the onset of fruit ripening.
Furthermore, it was demonstrated recently that the chloroplast transcriptome, but not the mitochondrial transcriptome, undergoes major changes during leaf aging, which are accompanied by a largely unchanged expression pattern of nuclear genes coding for plastid-targeted proteins (Woo et al., 2016). Thus, unlike animal aging, leaf senescence proceeds with tight temporal and distinct interorganellar coordination of various transcriptomes, which presumably are critical for the highly regulated degeneration process and the recycling of nutrients, which contributes to plant fitness and productivity. This is in agreement with the observation that the cellular degeneration process during leaf aging begins with the decay of chloroplasts, while nuclei and mitochondria remain intact until the end of the leaf’s lifespan (Lim et al., 2007). The expression pattern of nuclear genes encoding plastid proteins involved in photosynthesis, protein translation, tetrapyrrole biosynthesis, RNA metabolism, and chlorophyll degradation (Fig. 2) observed in our data set resembles the behavior of these genes during the leaf senescence process, indicating that fruit maturation and leaf senescence have several features in common and pointing to a major role of the chloroplast in both processes. Furthermore, the strong linear correlation between the physiological state of chloroplasts and plastid-associated nuclear gene expression implies, also in the case of fruits, the need for chloroplast-nucleus retrograde and anterograde signaling to coordinate the activities of the two genomes (Pesaresi et al., 2014). Nevertheless, the importance of photosynthesis in fruit metabolism is still under debate and deserves further investigation (Piechulla et al., 1987; Wanner and Gruissem, 1991; Schaffer and Petreikov, 1997; Carrari et al., 2006; Steinhauser et al., 2010). For instance, several studies used tomato cultivars carrying the uniform ripening (u) light green fruit phenotype (Powell et al., 2012). The U gene encodes a transcription factor named GOLDEN2-LIKE, which belongs to the AP2 family. The u mutants produce fewer chloroplasts, which are abnormal and whose thylakoid grana are reduced. Furthermore, u fruits are paler than normal and they have less sugars and lycopene, but they have normal size and ripen normally, suggesting that photosynthesis is not essential for fruit formation. On the other hand, experiments on Brassica napus plants shaded or defoliated soon after fertilization suggest a close relationship among leaf area, pod number, and seed per pod number (Pechan and Morgan, 1985). 14C-labeling experiments indicated that the fruit photosynthates are transported to the developing seeds; thus, the photosynthetic ability of the siliques strongly contributes to B. napus seed oil content (Hua et al., 2012).
Overall, the analysis of our RNAseq data clearly justifies our decision to monitor transcript accumulation in siliques devoid of seeds at four different developmental stages. This is shown by the fact that several genes known to have a role in fruit development and maturation are represented in our data set. Moreover, by exploring the consequences of the disruption of a few of the genes contained in our data set, we have detected significant alterations in fruit development and maturation, proving that our transcriptomic data sets are highly enriched in genes with specific roles in fruit formation and, therefore, should be very useful in the elucidation of the molecular mechanisms controlling fruit growth and maturation in Arabidopsis.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) wild-type (Col-0 and Ler) and mutant plants were grown under controlled growth chamber conditions at 22°C with 16 h of light and 8 h of darkness, as described previously (Mizzotti et al., 2012). For RNAseq, RT-qPCR, and all the characterization analyses, plants were inspected daily, and flowers at anthesis (time zero) were marked. Fruits were then collected 4 h after the lights were turned on at 3, 6, 9, and 12 DPA. For RNAseq and RT-qPCR, seeds were removed manually: siliques were opened along the dehiscence zone using a needle attached to a syringe that also was used to remove the seeds.
All mutants described in this article are in the Col-0 ecotype, except for arf8-4 (Goetz et al., 2006), which is in the Ler background. Mutant alleles were identified by searching the T-DNA Express database (http://signal.salk.edu/cgi-bin/tdnaexpress), and mutant lines were obtained from the SALK (Alonso et al., 2003) and GABI-KAT collections (Rosso et al., 2003; Supplemental Table S12). T-DNA insertions were confirmed by sequencing PCR products obtained using gene- and T-DNA-specific primers (Supplemental Table S13).
Nucleic Acid Isolation, cDNA Library Preparation, RNAseq, and RT-qPCR Analysis
For RNAseq analysis, total RNA was extracted from Col-0 siliques at 3, 6, 9, and 12 DPA from three biological replicates using the NucleoSpin RNA kit (Macherey Nagel), according to the supplier’s instructions. RNA concentrations and integrity were determined using the 2100 Bioanalyzer (Agilent Technologies). For each biological replicate, we grew together five different Col-0 plants, and we collected the material from at least 10 siliques deprived of seeds.
Sequencing libraries were prepared using the TruSeq RNA Sample Prep kit (Illumina), according to the manufacturer’s instructions, and sequenced on an Illumina HiSeq2000 (50-bp single read). The processing of fluorescent images into sequences, base calling, and quality value calculations were performed using the Illumina data-processing pipeline (version 1.8).
For RT-qPCR analyses, 0.5-μg samples of total RNA were used for first-strand cDNA synthesis with the ImProm-II Reverse Transcription System, as already described by Mizzotti et al. (2017). RT-qPCR profiling was carried out using the iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad iCycler iQ Optical System (software version 3.0a), with the primers listed in Supplemental Table S13. UBC9 and PP2A transcripts were used as internal standards (Czechowski et al., 2005). Data from two biological and three technical replicates were analyzed as reported previously by Mizzotti et al. (2014). For each biological replicate, we grew at the same time five different plants (Col-0 or mutant), and we collected the material from at least 10 siliques deprived of seeds. Briefly, the normalized expression of a gene corresponds to 2−ΔCt, which is calculated as the difference between the cycle threshold of the gene and the cycle threshold of the internal standard.
RNAseq Analysis and Identification of Differentially Expressed Genes
Raw reads were preprocessed for quality using FastQC (Andrews, 2010) version 0.11.5, cleaned, and trimmed with Trimmomatic version 0.36 (Bolger et al., 2014). The resulting reads were aligned to the Arabidopsis genome (TAIR10) using STAR version 2.5.1bb software (Dobin et al., 2013). Raw read counts were calculated using the featureCounts (http://bioinf.wehi.edu.au/featureCounts/) software (Liao et al., 2014). Read count data were used to identify differentially expressed genes by comparisons among different time points using both edgeR (Robinson et al., 2010) and the Limma empirical Bayes analysis pipeline (Smyth, 2005) and voom (Law et al., 2014), which estimates the mean variance trend of the log counts to predict the variance and to generate a precision weight to be incorporated in the linear model. To show that consistency holds for our data set, we performed differentially expressed gene analysis with edgeR using the same thresholds used for Limma/voom to rank genes (see below) and plotted the results using Venn diagrams. As expected, the results between the two tools were consistent, as most genes detected by Limma/voom also were detected by edgeR. edgeR usually reports more differentially expressed genes with respect to Limma/voom (∼1,000 in every contrast). A double cutoff on both P value and fold change was used to select differentially expressed genes; a maximum P value of 0.001 and a fold change of 2.5 were set. Contrasts were defined by considering all six possible combinations of the four time points, and genes were grouped based on their expression profiles as up-regulated (i.e. genes that were found to be significantly up-regulated between any two consecutive time points), down-regulated (i.e. genes that were significantly down-regulated between any two consecutive time points), and alternative behavior (i.e. all the other genes that exhibited various other expression profiles and that were clustered to further assess the different behaviors). Gene clustering was performed employing the hierarchical clustering method using the Pearson correlation coefficient as the distance metric to calculate pairwise distances and the unweighted pair group method with arithmetic mean to calculate the distances between the clusters thus formed. After visual inspection of the dendogram, a cutoff of 0.6 was selected and four clusters were defined. All RNAseq files are available from the European Nucleotide Archive database (accession no. PRJEB25745).
GO Enrichment Analysis
All six groups (up-regulated genes, down-regulated genes, and the four clusters generated from the genes marked as alternative behavior) were used to perform a GO enrichment analysis with agriGO (Du et al., 2010), using a hypergeometric statistical test, with the Benjamini-Hochberg (false discovery rate) correction for multiple tests, a significance level of 0.01, and the plant GOslim as parameters.
Immunoblot Analyses
For immunoblot analyses, developing siliques were collected at 3, 6, 9, and 12 DAP and total protein extracts were prepared as described by Martínez-García et al. (1999). Protein extracts, corresponding to 2 mg of silique fresh weight, were fractionated by SDS-PAGE (12% [w/v] polyacrylamide; Schägger and von Jagow, 1987) and transferred to polyvinylidene difluoride membranes (Ihnatowicz et al., 2004). Replicate filters were immunodecorated using antibodies specific for LhcA2, LhcB3, PsbO, PRPS1, PRPL4, PAO, and Histone H3 proteins. LhcA2, LhcB3, PsbO, and PAO antibodies were obtained from Agrisera. PRPS1 and PRPL4 were obtained from Uniplastomic. Histone H3 antibody was purchased from Sigma-Aldrich.
Silique Measurements
The length and width of the mature siliques were evaluated by measuring 40 siliques for each genotype in two different biological replicates. For each biological replicate, we grew at the same time 10 different plants (Col-0 or mutant), and we collected the material from 40 siliques for each genotype. Samples were first photographed using a Leica MZ6 stereomicroscope, and silique images were measured using ImageJ software (Schneider et al., 2012). Measurements were statistically analyzed by one-way ANOVA combined with Dunnett’s comparison test using StatSoft software.
GUS Assays and Trypan Blue Staining
The GUS assay was carried out as reported by Resentini et al. (2017). Marker lines were kindly provided by Takashi Araki (pFT; Takada and Goto, 2003), Jan U. Lohmann (pARR15; Zhao et al., 2010), and Ian Graham (pABI5::GUS; Penfield et al., 2006). pLOX2::GUS, pLOX3::GUS, and pLOX4::GUS were generated by Massimo Galbiati. Primers are listed in Supplemental Table S13, and fragments were cloned by Gateway cloning into pBGWFS7.
For Trypan Blue staining, the Cold Spring Harbor protocol was adopted (van Wees, 2008). For the measurement of the stained area, we used a modified version of the Phenotype Quant plugin for ImageJ (Abd-El-Haliem, 2012), while for the total area of the silique, we developed an ImageJ plugin that calculates the area of the silique through threshold levels. Measurements were analyzed statistically by two-way ANOVA combined with Dunnett’s and Duncan’s comparison test using StatSoft software.
Chlorophyll Fluorescence of Col-0 and Mutant Siliques
In vivo chlorophyll a fluorescence of Arabidopsis siliques was measured at 3, 6, 9, and 12 DPA using the pulse-modulated fluorometer Dual-PAM 100 (Walz; Tadini et al., 2012). Five siliques for each developmental stage were placed on a 24- × 24-mm coverslip and used for a single measurement. Three measurements for each stage (i.e. 15 siliques in total) were performed, and three biological replicates were used. Siliques were first dark adapted for 30 min, and minimal fluorescence (F0) was measured. Then, pulses (0.8 s) of saturating light (5,000 μmol photons m−2 s−1) were employed to determine the maximum fluorescence (Fm), and the ratio (Fm – F0)/Fm = Fv/Fm (maximum quantum yield of PSII) was calculated. A 2-min exposure to actinic red light (37 μmol photons m−2 s−1) served to drive electron transport between PSII and PSI. Then, the steady-state fluorescence (Fs) and the maximum fluorescence after light adaptation (Fm′) were determined. Finally, the ratio (Fm′ − Fs)/Fm′, representing the effective quantum yield of PSII, was calculated.
Chlorophyll Contents of Col-0 and nac058 Siliques
Pigments were extracted using 90% (v/v) acetone from valves at 12 DPA, after manual seed removal. The chlorophyll (a and b) content was measured, at 663- and 645-nm wavelength, using a spectrophotometer. Chlorophyll (a + b) values were determined as described previously by Arnon (1949) and normalized relative to tissue fresh weight.
Accession Numbers
Sequence data from this article can be found in the European Nucleotide Archive database under accession number PRJEB25745.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Number of reads obtained from each replicate by Illumina sequencing and principal component analysis.
Supplemental Figure S2. Comparison of the outputs obtained by analyzing the RNAseq datasets with the Limma and edgeR packages.
Supplemental Figure S3. Heat map of differentially expressed genes that belong to the alternative behavior group.
Supplemental Figure S4. Singular enrichment analysis of GO annotation terms overrepresented in the list of the up-regulated genes.
Supplemental Figure S5. Singular enrichment analysis of GO annotation terms overrepresented in the list of the down-regulated genes.
Supplemental Figure S6. Singular enrichment analysis of GO annotation terms overrepresented in the list of the genes showing alternative behaviors.
Supplemental Figure S7. Expression pattern of genes involved in chloroplast-nucleus communication.
Supplemental Figure S8. Detection of cell death using the Trypan Blue dye-exclusion method.
Supplemental Figure S9. Quantification of the Trypan Blue dye-exclusion method.
Supplemental Figure S10. The photosynthetic performance of the siliques is not influenced by green embryos.
Supplemental Table S1. Average expression levels, log fold change, and P values across all comparisons of the up-regulated genes, according to the criteria described in the text.
Supplemental Table S2. Average expression levels, log fold change, and P values across all comparisons of the down-regulated genes, according to the criteria described in the text.
Supplemental Table S3. Average expression levels, log fold change, and P values across all comparisons of the genes in the first cluster of the alternative behavior group, according to the criteria described in the text.
Supplemental Table S4. Average expression levels, log fold change, and P values across all comparisons of the genes in the second cluster of the alternative behavior group, according to the criteria described in the text.
Supplemental Table S5. Average expression levels, log fold change, and P values across all comparisons of the genes in the third cluster of the alternative behavior group, according to the criteria described in the text.
Supplemental Table S6. Average expression levels, log fold change and P values across all comparisons of the genes in the fourth cluster of the alternative behavior group, according to the criteria described in the text.
Supplemental Table S7. Expression levels, at the indicated time points, of genes involved in hormonal pathways.
Supplemental Table S8. List of transcription factors present in our differentially expressed gene list.
Supplemental Table S9. Expression levels, at the indicated time points, of genes involved in chloroplast-related functions.
Supplemental Table S10. Expression levels of genes analyzed by GUS assay.
Supplemental Table S11. Expression levels of commonly used housekeeping genes.
Supplemental Table S12. List of Arabidopsis T-DNA insertional mutants used in this study.
Supplemental Table S13. Primer sequences used in this work.
Acknowledgments
We thank Takashi Araki, Jan U. Lohmann, and Ian Graham for providing seeds of pFT::GUS, pARR15::GUS, and pABI5::GUS. We thank Luca Mizzi and Giorgio Binelli for help with the Trypan Blue area measurement and the statistical analysis, respectively. We also thank Carolina Cozzi for help with chlorophyll content assays, Valerio Parravicini for excellent technical assistance, and Paul Hardy for critical reading of the article.
Footnotes
The work has been supported by Cariplo Foundation [grant number 2011-2257 to C.M. and S.M.], by MIUR PRIN [grant number 2015BPM9H3_005 to S.M.] and by post doc and PhD fellowships from the Università degli Studi di Milano [to C.M., L.T., and B.G.].
References
- Abd-El-Haliem A. (2012) An unbiased method for the quantitation of disease phenotypes using a custom-built macro plugin for the program ImageJ. Methods Mol Biol 835: 635–644 [DOI] [PubMed] [Google Scholar]
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) (Suppl 1) 10: 63–75 [DOI] [PubMed] [Google Scholar]
- Barry CS, Aldridge GM, Herzog G, Ma Q, McQuinn RP, Hirschberg J, Giovannoni JJ (2012) Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol 159: 1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke MM, Lenz F (1989) Fruit photosynthesis. Plant Cell Environ 12: 31–46 [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks MD, Niyogi KK (2011) Use of a pulse-amplitude modulated chlorophyll fluorometer to study the efficiency of photosynthesis in Arabidopsis plants. Methods Mol Biol 775: 299–310 [DOI] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA (2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhu X, Ren J, Qiu K, Li Z, Xie Z, Gao J, Zhou X, Kuai B (2017) Suppressor of Overexpression of CO 1 negatively regulates dark-induced leaf degreening and senescence by directly repressing pheophytinase and other senescence-associated genes in Arabidopsis. Plant Physiol 173: 1881–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lu L, Mayer KS, Scalf M, Qian S, Lomax A, Smith LM, Zhong X (2016) POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5: e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen MW, Gregersen PL (2014) Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J Exp Bot 65: 4009–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coego A, Brizuela E, Castillejo P, Ruíz S, Koncz C, del Pozo JC, Piñeiro M, Jarillo JA, Paz-Ares J, León J; TRANSPLANTA Consortium (2014) The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. Plant J 77: 944–953 [DOI] [PubMed] [Google Scholar]
- Coombe BG. (1975) The development of fleshy fruits. Annu Rev Plant Physiol 27: 507–528 [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Busscher J, Colombo L, Losa A, Angenent GC (2004) Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol Biol 56: 351–366 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF (2005) A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132: 4687–4696 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Yu Y, Li S, Wang J, Tang S, Huang R (2016) Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol Plant 9: 126–135 [DOI] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blázquez MA, Carbonell J, Perez-Amador MA (2009) Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J 58: 318–332 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SC, Fernandez DE (2002) Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol 130: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC (2000) The embryo MADS domain factor AGL15 acts postembryonically: inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12: 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Wang CT, Zheng Y, Adamczyk BJ, Singhal R, Hall PK, Perry SE (2014) The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol 165: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C. (2002) Regulation of fruit dehiscence in Arabidopsis. J Exp Bot 53: 2031–2038 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Ghandchi FP, Caetano-Anolles G, Clough SJ, Ort DR (2016) Investigating the control of chlorophyll degradation by genomic correlation mining. PLoS ONE 11: e0162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi FM, Del Fabbro C, Licausi F (2013) Comparative study of RNA-seq- and microarray-derived coexpression networks in Arabidopsis thaliana. Bioinformatics 29: 717–724 [DOI] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LHM, Whelan J (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnan S, Marsh T, Kover PX (2017) Inflorescence photosynthetic contribution to fitness releases Arabidopsis thaliana plants from trade-off constraints on early flowering. PLoS ONE 12: e0185835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Guyer L, Hofstetter SS, Christ B, Lira BS, Rossi M, Hörtensteiner S (2014) Different mechanisms are responsible for chlorophyll dephytylation during fruit ripening and leaf senescence in tomato. Plant Physiol 166: 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Johannesson H, Engström P (2001) Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol Biol 45: 247–262 [DOI] [PubMed] [Google Scholar]
- He H, Bai M, Tong P, Hu Y, Yang M, Wu H (2018) CELLULASE6 and MANNANASE7 affect cell differentiation and silique dehiscence. Plant Physiol 176: 2186–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ (2004) N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279: 47822–47832 [DOI] [PubMed] [Google Scholar]
- Hua W, Li RJ, Zhan GM, Liu J, Li J, Wang XF, Liu GH, Wang HZ (2012) Maternal control of seed oil content in Brassica napus: the role of silique wall photosynthesis. Plant J 69: 432–444 [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D (2004) Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Jaradat MR, Ruegger M, Bowling A, Butler H, Cutler AJ (2014) A comprehensive transcriptome analysis of silique development and dehiscence in Arabidopsis and Brassica integrating genotypic, interspecies and developmental comparisons. GM Crops Food 5: 302–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo PJ, Kang SK, Park CM (2011) miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol Biol 76: 35–45 [DOI] [PubMed] [Google Scholar]
- Kalantidis K, Schumacher HT, Alexiadis T, Helm JM (2008) RNA silencing movement in plants. Biol Cell 100: 13–26 [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Saito Y, Uchimiya H (1999) Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci USA 96: 9433–9437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H (2002) The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J 21: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41: 710–721 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hong SH, Kim YW, Lee IH, Jun JH, Phee BK, Rupak T, Jeong H, Lee Y, Hong BS, et al. (2014) Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot 65: 4023–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO (2016) Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol 33: 48–56 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Kleindt CK, Stracke R, Mehrtens F, Weisshaar B (2010) Expression analysis of flavonoid biosynthesis genes during Arabidopsis thaliana silique and seed development with a primary focus on the proanthocyanidin biosynthetic pathway. BMC Res Notes 3: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Kou X, Watkins CB, Gan SS (2012) Arabidopsis AtNAP regulates fruit senescence. J Exp Bot 63: 6139–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65: 4561–4575 [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, Chung WI (2006) LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133: 4305–4314 [DOI] [PubMed] [Google Scholar]
- León P, Gregorio J, Cordoba E (2013) ABI4 and its role in chloroplast retrograde communication. Front Plant Sci 3: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lindsey K. (2001) Plant peptide hormones: the long and the short of it. Curr Biol 11: R741–R743 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Martínez-García JF, Monte E, Quail PH (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2011) The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla AJ. (2007) How is the silique fruit dismantled over its maturation? Funct Plant Sci Biotechnol 1: 85–93 [Google Scholar]
- Mazzucato A, Taddei AR, Soressi GP (1998) The parthenocarpic fruit (pat) mutant of tomato (Lycopersicon esculentum Mill.) sets seedless fruits and has aberrant anther and ovule development. Development 125: 107–114 [DOI] [PubMed] [Google Scholar]
- McAtee P, Karim S, Schaffer R, David K (2013) A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci 4: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzotti C, Mendes MA, Caporali E, Schnittger A, Kater MM, Battaglia R, Colombo L (2012) The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development. Plant J 70: 409–420 [DOI] [PubMed] [Google Scholar]
- Mizzotti C, Ezquer I, Paolo D, Rueda-Romero P, Guerra RF, Battaglia R, Rogachev I, Aharoni A, Kater MM, Caporali E, et al. (2014) SEEDSTICK is a master regulator of development and metabolism in the Arabidopsis seed coat. PLoS Genet 10: e1004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzotti C, Galliani BM, Dreni L, Sommer H, Bombarely A, Masiero S (2017) ERAMOSA controls lateral branching in snapdragon. Sci Rep 7: 41319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntz K, Rudolph A, Schlesier G, Silhengst P (1978) The function of the pericarp in fruits of crop legumes. Die Kult 26: 37–67 [Google Scholar]
- Nitsch JP. (1952) Plant hormones in the development of fruits. Q Rev Biol 27: 33–57 [DOI] [PubMed] [Google Scholar]
- O’Neill SD. (1997) POLLINATION REGULATION OF FLOWER DEVELOPMENT. Annu Rev Plant Physiol Plant Mol Biol 48: 547–574 [DOI] [PubMed] [Google Scholar]
- O’Neill SD, Nadeau JA (2010) Postpollination flower development. Hortic Rev 19: 1–58 [Google Scholar]
- Ougham H, Hörtensteiner S, Armstead I, Donnison I, King I, Thomas H, Mur L (2008) The control of chlorophyll catabolism and the status of yellowing as a biomarker of leaf senescence. Plant Biol (Stuttg) (Suppl 1) 10: 4–14 [DOI] [PubMed] [Google Scholar]
- Pandolfini T. (2009) Seedless fruit production by hormonal regulation of fruit set. Nutrients 1: 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechan PA, Morgan DG (1985) Defoliation and its effects on pod and seed development in oil seed rape (Brassica napus L.). J Exp Bot 36: 458–468 [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Mizzotti C, Colombo M, Masiero S (2014) Genetic regulation and structural changes during tomato fruit development and ripening. Front Plant Sci 5: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla B, Imlay KR, Gruissem W (1985) Plastid gene expression during fruit ripening in tomato. Plant Mol Biol 5: 373–384 [DOI] [PubMed] [Google Scholar]
- Piechulla B, Glick RE, Bahl H, Melis A, Gruissem W (1987) Changes in photosynthetic capacity and photosynthetic protein pattern during tomato fruit ripening. Plant Physiol 84: 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Prasad K, Zhang X, Tobón E, Ambrose BA (2010) The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J 62: 203–214 [DOI] [PubMed] [Google Scholar]
- Raghavan V. (2003) Some reflections on double fertilization, from its discovery to the present. New Phytol 159: 565–583 [DOI] [PubMed] [Google Scholar]
- Resentini F, Cyprys P, Steffen JG, Alter S, Morandini P, Mizzotti C, Lloyd A, Drews GN, Dresselhaus T, Colombo L, et al. (2017) SUPPRESSOR OF FRIGIDA (SUF4) supports gamete fusion via regulating Arabidopsis EC1 gene expression. Plant Physiol 173: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CK, Hill SA (1999) Altered resource allocation during seed development in Arabidopsis caused by the abi3 mutation. Plant Cell Environ 22: 117–123 [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Gracia P, Roque E, Medina M, Rochina M, Hamza R, Angarita-Díaz MP, Moreno V, Pérez-Martín F, Lozano R, Cañas L, et al. (2017) The parthenocarpic hydra mutant reveals a new function for a SPOROCYTELESS-like gene in the control of fruit set in tomato. New Phytol 214: 1198–1212 [DOI] [PubMed] [Google Scholar]
- Rollins RC. (1993) The Cruciferae of Continental North America: Systematics of the Mustard Family from the Arctic to Panama. Stanford University Press, Stanford, CA [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J 72: 922–934 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Scarpeci TE, Frea VS, Zanor MI, Valle EM (2017) Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J Exp Bot 68: 673–685 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Petreikov M (1997) Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol 113: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyednasrollah F, Laiho A, Elo LL (2015) Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform 16: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C (2013) Fruit development and ripening. Annu Rev Plant Biol 64: 219–241 [DOI] [PubMed] [Google Scholar]
- Shao H, Wang H, Tang X (2015) NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci 6: 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. (2005) Limma: linear models for microarray data. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Steinhauser MC, Steinhauser D, Koehl K, Carrari F, Gibon Y, Fernie AR, Stitt M (2010) Enzyme activity profiles during fruit development in tomato cultivars and Solanum pennellii. Plant Physiol 153: 80–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M, Rossi F, Tadini L, Labs M, Colombo M, Jahns P, Kater MM, Leister D, Finazzi G, Aro EM, et al. (2016) PGR5-PGRL1-dependent cyclic electron transport modulates linear electron transport rate in Arabidopsis thaliana. Mol Plant 9: 271–288 [DOI] [PubMed] [Google Scholar]
- Tadini L, Romani I, Pribil M, Jahns P, Leister D, Pesaresi P (2012) Thylakoid redox signals are integrated into organellar-gene-expression-dependent retrograde signaling in the prors1-1 mutant. Front Plant Sci 3: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A (2009) A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183: 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Van der Pijl L. (1982) Principles of Dispersal in Higher Plants. Springer-Verlag, New York [Google Scholar]
- van Wees S. (2008) Phenotypic analysis of Arabidopsis mutants: trypan blue stain for fungi, oomycetes, and dead plant cells. CSH Protoc 2008: pdb.prot4982 [DOI] [PubMed]
- Varoquaux F, Blanvillain R, Delseny M, Gallois P (2000) Less is better: new approaches for seedless fruit production. Trends Biotechnol 18: 233–242 [DOI] [PubMed] [Google Scholar]
- Vivian-Smith A, Koltunow AM (1999) Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol 121: 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A, Luo M, Chaudhury A, Koltunow A (2001) Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development 128: 2321–2331 [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Yang TJW, Stead AD, Buchanan-Wollaston V, Roberts JA (2009) A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 57: 690–705 [DOI] [PubMed] [Google Scholar]
- Wang X. (2010) The Dawn Angiosperms: Uncovering the Origin of Flowering Plants. Springer-Verlag, Berlin [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang W, Jiang Q, Lian J, Sun J, Xu H, Zhao H, Liu Z (2013) Lower levels of expression of FATA2 gene promote longer siliques with modified seed oil content in Arabidopsis thaliana. Plant Mol Biol Rep 31: 1368–1375 [Google Scholar]
- Wanner LA, Gruissem W (1991) Expression dynamics of the tomato rbcS gene family during development. Plant Cell 3: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Marès M, Pourtau N (2004) Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol 161: 781–789 [DOI] [PubMed] [Google Scholar]
- Woo HR, Koo HJ, Kim J, Jeong H, Yang JO, Lee IH, Jun JH, Choi SH, Park SJ, Kang B, et al. (2016) Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol 171: 452–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Tian L, Malik K, Brown D, Miki B (2000) Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J 22: 19–27 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu T, Ren G, Hörtensteiner S, Zhou Y, Cahoon EB, Zhang C (2014) Chlorophyll degradation: the tocopherol biosynthesis-related phytol hydrolase in Arabidopsis seeds is still missing. Plant Physiol 166: 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schwarz S, Saedler H, Huijser P (2007) SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol 63: 429–439 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]