Mutation of the coiled-coil domain of the intracellular immune receptor Rx1 led to distinct cell death and resistance phenotypes and revealed regions of Rx1 required for interactions between Rx1 and the co-factor RanGAP2.
Abstract
The intracellular immune receptor Rx1 of potato (Solanum tuberosum), which confers effector-triggered immunity to Potato virus X, consists of a central nucleotide-binding domain (NB-ARC) flanked by a carboxyl-terminal leucine-rich repeat (LRR) domain and an amino-terminal coiled-coil (CC) domain. Rx1 activity is strictly regulated by interdomain interactions between the NB-ARC and LRR, but the contribution of the CC domain in regulating Rx1 activity or immune signaling is not fully understood. Therefore, we used a structure-informed approach to investigate the role of the CC domain in Rx1 functionality. Targeted mutagenesis of CC surface residues revealed separate regions required for the intramolecular and intermolecular interaction of the CC with the NB-ARC-LRR and the cofactor Ran GTPase-activating protein2 (RanGAP2), respectively. None of the mutant Rx1 proteins was constitutively active, indicating that the CC does not contribute to the autoinhibition of Rx1 activity. Instead, the CC domain acted as a modulator of downstream responses involved in effector-triggered immunity. Systematic disruption of the hydrophobic interface between the four helices of the CC enabled the uncoupling of cell death and disease resistance responses. Moreover, a strong dominant negative effect on Rx1-mediated resistance and cell death was observed upon coexpression of the CC alone with full-length Rx1 protein, which depended on the RanGAP2-binding surface of the CC. Surprisingly, coexpression of the N-terminal half of the CC enhanced Rx1-mediated resistance, which further indicated that the CC functions as a scaffold for downstream components involved in the modulation of disease resistance or cell death signaling.
To defend themselves against pests, plants have developed a complex cell-based immune system. Resistance proteins (R proteins) mediate the recognition of pathogen-derived molecules and are able to initiate defense responses that hamper further spreading of the pathogen. Most known plant R proteins belong to the NB-LRR class, which is characterized by a central nucleotide-binding domain, the NB-ARC, and a C-terminal leucine-rich repeat (LRR) domain. The NB-ARC domain shares homology with animal immune receptors and apoptosis proteins, hence its acronym: nucleotide-binding-Apaf-1, R proteins, CED-4 (van der Biezen and Jones, 1998). By cycling between an autoinhibited ADP-bound conformation and a signaling-competent ATP-bound conformation, the NB-ARC acts like a molecular switch (Tameling et al., 2006; Williams et al., 2011; Bernoux et al., 2016). The LRR domain regulates this NB-ARC switch and often functions as the sensor for the pathogen (Takken et al., 2006). The NB-LRR class of R proteins can be separated in two main subgroups according to the domain located N-terminal to the NB-ARC domain, which in most cases is either a putative coiled-coil (CC) domain or a Toll/IL receptor-like (TIR) domain. These two subgroups represent an ancient split in the lineage of plant NB-LRR proteins, and each subgroup harbors specific conserved motifs in the NB-ARC and LRR domains (Meyers et al., 1999).
CC domains derive their name from a specific helix-helix interaction in which α-helices wind around each other in a superhelical conformation (Crick, 1953; Cohen and Parry, 1986). A signature motif for the coiled coil, the Leu-rich heptad repeat, was recognized early on in CC-NB-LRRs. The first two structures of CC domains have been solved experimentally for the barley (Hordeum vulgare) immune receptor MLA against powdery mildew (Blumeria graminis f. sp. hordei; Maekawa et al., 2011) and the potato (Solanum tuberosum) immune receptor Rx1 against Potato virus X (PVX; Hao et al., 2013). The published tertiary structures of the CC domains of MLA and Rx1 differ markedly, despite the similarity in amino acid sequence and secondary structure. The CC domain of MLA folds into an antiparallel CC structure and forms a tight antiparallel dimer with the CC domain of a second MLA protein. Correspondingly, full-length MLA occurs as a homodimer in the cell, and mutations in the CC domain that disrupt the dimerization of MLA result in a loss of functionality, indicating the importance of this self-association (Maekawa et al., 2011). The CC domain of Rx1, however, forms a compact four-helix bundle in which the hydrophobic residues of the heptad repeat form the hydrophobic core of the bundle (Hao et al., 2013). A solution structure of the CC domain (amino acids 6–120) of Sr33, a close homolog of MLA from diploid wheat (Aegilops tauschii) that confers resistance to stem rust (Puccinia graminis f. sp. tritici), revealed a monomeric structure similar to that of the CC of Rx1, but longer constructs (amino acids 1–142) dimerized in a yet unknown structure and displayed autoactive cell death signaling in planta (Casey et al., 2016). In the case of Rx1, there are no indications of homodimerization, but the CC of Rx1 forms a heteromeric complex with the WPP domain of RanGTPase-activating protein2 (RanGAP2; Sacco et al., 2007; Tameling and Baulcombe, 2007; Hao et al., 2013).
Several roles have been attributed to the CC domains of NB-LRR immune receptors. They may play a role in (1) intramolecular interactions regulating the activity of the NB-LRR, (2) interactions with downstream signaling components, (3) interactions with cofactors required for pathogen recognition, and (4) regulation of the subcellular distribution of NB-LRRs in the cell. A role in signaling is evident from the finding that CC domains of several NB-LRRs can activate cell death responses when expressed as a single domain. The CC domains of homologs of Rp1-D, a resistance protein against maize common rust (Puccinia sorghi) in maize (Zea mays) and MLA, Sr33, and Sr50 from barley, diploid wheat, and rye (Secale cereale) are able to induce cell death autonomously (Maekawa et al., 2011; Wang et al., 2015; Cesari et al., 2016). The signaling activity of the CC of Rp1 proteins is suppressed by the NB domain in cis, showing a mechanism by which an autoinhibited state is retained in the full-length protein in the absence of pathogens. The direct interaction of the CC of MLA with transcription factors, which either positively (MYB6) or negatively (WRKY1) regulate resistance, is an example of how a CC domain connects to downstream signaling components (Chang et al., 2013). Both CC domains belong to the subclass that harbor a conserved EDVID motif (Rairdan et al., 2008). For the CC of Rx1, which belongs to the same EDVID subclass, no such autonomous activity could be demonstrated. In contrast, the NB domain of Rx1 acts as the minimal part of the protein that can induce an autoactive cell death response (Rairdan et al., 2008).
The CC often plays an indirect role in recognition. It interacts with host proteins targeted by effectors, which act as bait to allow the NB-LRR to detect effector-induced modifications (Innes, 2004; Collier and Moffett, 2009). For example, the CC of RESISTANCE TO PSEUDOMONAS SYRINGAE5 (RPS5) binds the kinase AVRPPH1 Susceptible1 (PBS1), and RPS5 is activated when PBS1 is processed by the bacterial effector protease Avirulence protein Pseudomonas phaseolicolaB (AvrPphB; DeYoung et al., 2012; Qi et al., 2014). Whether the interaction of the CC domain of Rx1 with a regulator of the small GTPase Ran (Sacco et al., 2007; Tameling and Baulcombe, 2007) functions similarly in pathogen recognition is unknown. However, the enhanced activation of the highly homologous potato R protein Gpa2 against the potato cyst nematode Globera pallida, which occurs when RanGAP2 and the cognate nematode effector are artificially tethered, hints at such a role (Sacco et al., 2007, 2009; Tameling and Baulcombe, 2007).
Moreover, a complex role has emerged for the CC domain in intramolecular interactions regulating the activity of NB-LRRs (Sukarta et al., 2016) . Studies on the potato NB-LRR protein Rx1 revealed that the CC interacts with both the NB-ARC and LRR domains of the protein, probably by contacting both domains simultaneously (Moffett et al., 2002). These intramolecular interactions between the CC, NB-ARC, and LRR domains of Rx1 depend on the activation state of the protein, as mutations modifying the activation- or nucleotide-bound state of the protein result in distinct patterns of domain interactions (Moffett et al., 2002; Rairdan et al., 2008). Mutagenesis of the CC domain of Rx1 reveals that the conserved EDVID motif (EDMVD in Rx1) is required for the interaction between the CC and the NB-ARC and LRR domains and that this motif is essential for the functionality of the resistance proteins (Rairdan et al., 2008; Mazourek et al., 2009; Lukasik-Shreepaathy et al., 2012 ).
A specific localization of NB-LRR proteins in the cell is required for effector recognition and for the subsequent activation of resistance signaling pathways (Bernoux et al., 2011a). In the case of Rx1, the CC domain plays a dual role in balancing the nucleocytoplasmic partitioning required for full functionality (Slootweg et al., 2010; Tameling et al., 2010). The interaction of the Rx1 CC domain with cytoplasmic protein RanGAP2 retains the NB-LRR protein in the cytoplasm (Tameling et al., 2010), and interaction of the CC with nuclear components is linked to a nuclear accumulation of Rx1 (Slootweg et al., 2010). Recently, it was shown that the CC-NB-ARC of Rx1 can bind DNA directly and cause it to bend, which suggests a nuclear role in transcriptional regulation. This DNA binding is aspecific, and interactions of the CC with other nuclear proteins, like the Golden2-like transcription factor1 from Nicotiana benthamiana (NbGLK1), are likely required to target Rx1 to specific regions (Fenyk et al., 2015; Townsend et al., 2018).
It is still not clear how the CC of Rx1, with its relatively simple structure, can integrate multiple interactions and regulatory functions and how it contributes to resistance signaling. We used a structure-informed approach to resolve the contribution of specific regions of the CC structure in the activation of Rx-mediated cell death and disease resistance signaling. Specific mutations were introduced to disrupt the interaction of individual helices or combinations thereof with the rest of the CC. In addition, mutations were introduced in specific nonoverlapping surfaces of the first and fourth helices of the CC domain. We tested the impact of these local disruptions on the interactions of the CC with the NB-ARC-LRR of Rx1 and with RanGAP2 and on the functionality of Rx1. Our results indicate that separate parts of the CC domain of Rx1 play a role in Rx1-mediated cell death responses and disease resistance against PVX. Any mutation that disrupted the structure of the CC prevented the accumulation of Rx1 in the nucleus. Residues in the surface of the N-terminal helix 1 were involved in the interaction of the CC with the NB-ARC-LRR. Furthermore, coexpression of a CC construct with full-length Rx1 exerted a strong dominant negative effect on Rx1-mediated resistance and cell death, which depended on the RanGAP2-binding surface of the CC. From this, a functional model emerged in which the CC domain of Rx1 is a modulator of downstream signaling responses involved in effector-triggered immunity in plant cells.
RESULTS
A Hydrophobic Interface between the N- and C-Terminal Two-Helix Segments of the Rx1 CC Domain Is Required for the Reconstitution of a Functional Immune Receptor
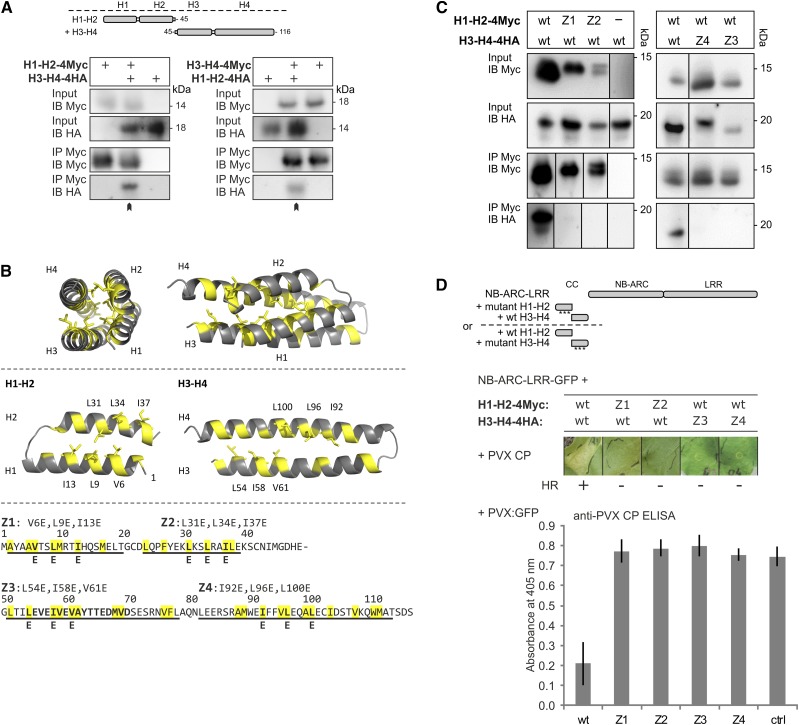
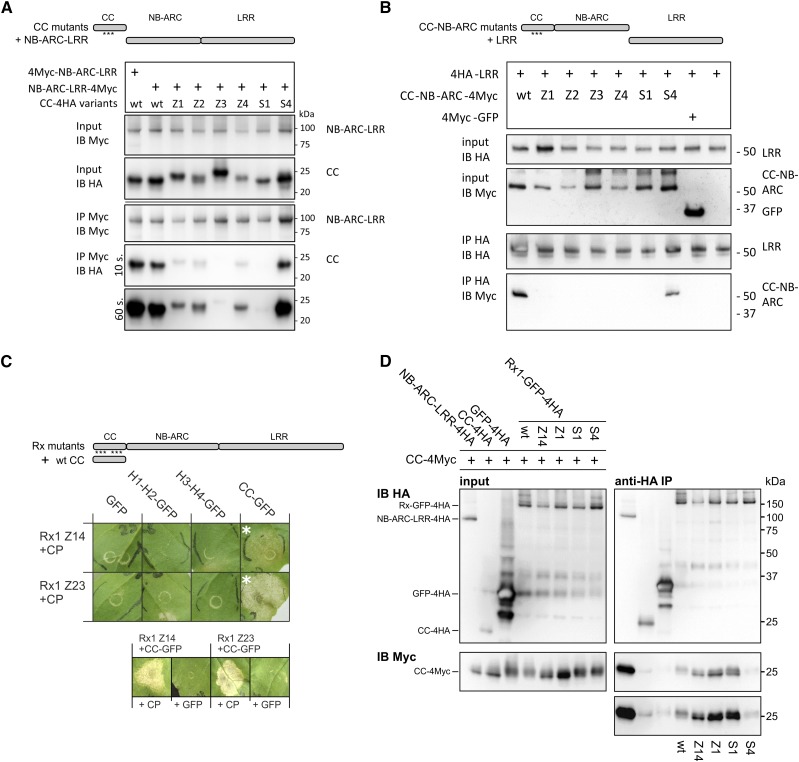
The CC domain plays a role in the nuclear and cytoplasmic localization of Rx1, but strikingly different localizations occur for the two halves of the CC domain (Slootweg et al., 2010). The N-terminal segment encompassing the first two α-helices of the CC domain (H1-H2) accumulates in the cytoplasm, whereas the C-terminal segment consisting of the third and fourth α-helices (H3-H4) accumulates in the nucleus. Moreover, the coexpression of the two fragments of the CC domain with the corresponding NB-ARC-LRR domains results in the functional reconstitution of Rx1 (Slootweg et al., 2010). Consistent with its compact four-helix bundle structure (Hao et al., 2013), we expected that the reconstitution of the functional CC from coexpressed segments requires these segments to interact. To test this, we performed a coimmunoprecipitation experiment with epitope-tagged versions of both segments following the agroinfiltration of N. benthamiana leaves. H1-H2 and H3-H4 CC segments coimmunoprecipitated, demonstrating that the separate CC segments indeed form a functional CC structure in plant cells (Fig. 1A).
Figure 1.
Assessing the role of the hydrophobic interface between the N- and C-terminal two-helix segments of the Rx1 CC domain. A, The two halves of the Rx1 CC interact in a coimmunoprecipitation assay. Combinations of coexpressed CC fragments H1-H2 (amino acids 1–45) and H3-H4 (amino acids 45–116) fused to 4xMyc or 4xHA affinity tags were subjected to anti-c-Myc immunoprecipitation. Extracts of leaves expressing the single strands were included as controls for aspecific binding and protein stability. The blots show the cell extract used as input and the result of the immunoprecipitation as detected with anti-c-Myc and anti-hemagglutinin (HA) antibodies. B, Targeted mutagenesis of hydrophobic residues in the heptad repeats. A set of constructs was made in which three apolar residues per α-helix were exchanged for Glu (E). The groups of three substitutions were named Z1 to Z4 in correspondence with the predicted helix in which they are positioned. Hydrophobic positions of the heptad repeat are indicated in yellow in the CC structure and the amino acid sequence. The substituted residues are indicated. C, Effects of mutations Z1 to Z4 on the intramolecular interaction of the Rx1 CC domain. Mutant versions of H1-H2-4Myc were coexpressed with wild-type (wt) H3-H4-4HA or vice versa. The H1-H2-4Myc constructs were pulled down by anti-Myc antibodies, and the coimmunoprecipitated H3-H4-4HA constructs were visualized by anti-HA immunoblot. Some of the lanes have been rearranged to align input and corresponding immunoprecipitation results; this is indicated by solid lines on the immunoblot. D, Mutant versions of H1-H2-4Myc and H3-H4-4HA were transiently expressed with Rx1 NB-ARC-LRR-GFP (amino acids 142–937) in the presence of the coat protein (CP106) of PVX to assess the cell death response. The combinations of the Rx1 segments are indicated in a schematic drawing. Images were taken 5 d post infiltration (dpi). To assess PVX resistance, variants of H1-H2 and H3-H4 were coinfiltrated in N. benthamiana leaves with the NB-ARC-LRR-GFP and a PVX:GFP amplicon. Virus accumulation was determined by anti-PVX CP ELISA of leaf extracts at 5 dpi with alkaline phosphatase-conjugated antibodies. p-Nitrophenol accumulation was detected via its A405. As a control (ctrl), PVX:GFP was expressed in the absence of Rx1.
Even though the NB domain is the minimal region of Rx1 that can initiate a cell death response, the CC is essential for the activity of the full-length Rx1 protein (Rairdan et al., 2008). To study the role of the interaction between the individual helices of the CC in the functioning of Rx1, we applied a targeted mutagenesis strategy aimed at disrupting the interaction of each helix with the rest of the helix bundle. The hydrophobic residues in the heptad repeats of the helices form the hydrophobic core of the helix bundle in the crystal structure (Hao et al., 2013). Hydrophobic residues in the core are packed in such a way that the hydrophobic residues from one helix fit in the spaces lined by the hydrophobic residues of the other helices: a hydrophobic zipping interaction (Fig. 1B). In a series of constructs, we replaced in each of the four helices three residues in the hydrophobic zipping interface with Glu, which carries a negative charge under physiological conditions. Glu residues are relatively well accommodated in α-helices and, thus, are less likely to disrupt the local secondary structure (Cohen and Parry, 1990). Each group of substitutions was named after the helix that harbors it: Z1 (V6E, L9E, and I13E) in helix 1, Z2 (L31E, L34E, and I37E) in helix 2, Z3 (L54E, I58E, and V61E) in helix 3, and Z4 (I92E, L96E, and L100E) in helix 4 (Fig. 1B).
The effectiveness of the mutations in disrupting the hydrophobic CC interactions of the helices in the CC structure was tested by a coimmunoprecipitation experiment combining mutant and wild-type versions of the H1-H2 and H3-H4 CC segments. Z1 and Z2 completely disrupted the ability of the H1-H2 CC segment to interact with wild-type H3-H4 (Fig. 1C). Similarly, both Z3 and Z4 disrupted the interaction of H3-H4 with the wild type H1-H2 (Fig. 1C). In accordance with the loss of interaction, the substitutions also abolished the ability of H1-H2 and H3-H4 to reconstitute Rx1-mediated elicitor-dependent cell death signaling and PVX resistance in trans when coexpressed with the NB-ARC-LRR (Fig. 1D). Thus, the hydrophobic residues of the heptad repeat in each of the four helices are required for the interaction between the two segments of the CC, and the integrity of the CC is important to enable the reconstitution of Rx1 functionality in trans.
Local Disruption of the Interaction between the Two Segments of the CC Has Distinct Effects on Cell Death and PVX Resistance Mediated by Full-Length Rx1
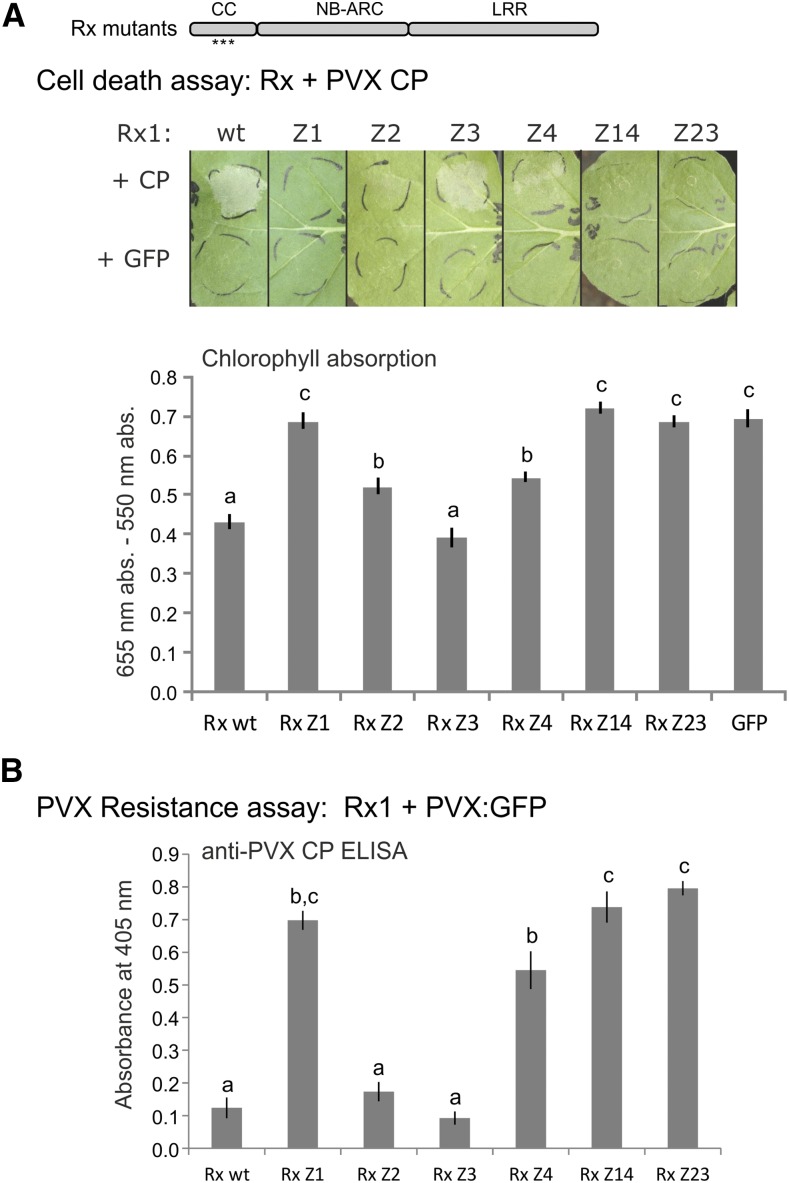
Reconstitution of a functional protein from coexpressed fragments depends primarily on the interaction between those fragments. Therefore, the disruption of the interaction between the CC fragments by the mutations probably results in a stronger effect on the functionality of Rx1 in trans than when the mutations are introduced in the full-length protein. To test the impact of the disruption of the local tertiary structure of the CC on the functioning of Rx1 in cis, we introduced the mutations Z1, Z2, Z3, and Z4 in full-length protein constructs and monitored the induction of an elicitor-dependent cell death response after coexpression of the Rx1 variants with the avirulent PVX coat protein CP106. PVX resistance was monitored upon transient coexpression of the mutant full-length Rx1 constructs in N. benthamiana leaves with an amplicon of the avirulent PVX strain. Coexpression of the full-length Rx1 constructs with GFP showed that none of the mutant Rx1 proteins (Z1–Z4) initiated a cell death response in the absence of the PVX coat protein, indicating that none of the mutations alone releases the autoinhibitory mechanisms that keep Rx1 in its resting state (Fig. 2A). Analysis of tagged versions of the proteins via SDS-PAGE and immunoblotting showed the mutant variants accumulated to similar levels to the wild-type Rx1 protein (Supplemental Fig. S1). Combinations of mutated helices in the CC, Z1 + Z4 and Z2 + Z3, resulted in a complete loss of function of Rx1, whereas each of the single CC Z variants gave rise to a distinct combination of effects on cell death and resistance levels. Mutations of the hydrophobic residues in helix 3 (Z3) did not lead to a marked decrease in cell death (Fig. 2A) or PVX resistance (Fig. 2B) mediated by Rx1. The substitution of hydrophobic zipping residues in helix 4 (Z4) resulted in a reduced elicitor-dependent cell death and a partial loss of PVX resistance (Fig. 2). The Z2 mutant of Rx1 showed a similar partial loss of elicitor-dependent cell death but, surprisingly, retained almost wild-type levels of PVX resistance in this assay. The mutations in helix 1 (Z1) resulted in a complete loss of both elicitor-dependent cell death and PVX resistance. The distinct effects of the local disruption of the CC structure on elicitor-dependent cell death and PVX resistance mediated by the mutant full-length Rx1 proteins suggest that different regions of the CC contribute to cell death and virus resistance signaling pathways.
Figure 2.
Effects of mutations Z1 to Z4 on the functionality of full-length Rx1, either as the separate groups of mutations or in the combinations of Z1 with Z4 (Rx1 Z14) and Z2 with Z3 (Rx1 Z23). A, Rx1 constructs were coexpressed with the CP of PVX to assess their ability to induce elicitor-dependent cell death. The constructs were coexpressed with GFP to detect autoactivity. Images of the cell death response at 5 dpi are shown. The level of cell death was quantified by measuring the absorption of light at 655 nm by chlorophyll in a leaf extract (see “Materials and Methods”). A stronger cell death leads to lower chlorophyll levels. The error bars represent se (n = 8). Different letters represent significant differences (one-way ANOVA with a posthoc Tukey’s test, P < 0.05). B, Rx1-mediated resistance was tested by coexpressing the Rx1 constructs with a PVX:GFP amplicon and subsequent detection of the CP of PVX with an alkaline phosphatase-conjugated antibody in an ELISA at 5 dpi. The error bars represent se (n = 8). Statistical analysis is as in A.
Aromatic and Hydrophobic Surface Residues in Helix 1 and in the RanGAP2 Interface on Helix 4 Contribute to the Compact Fold of the CC and the Reconstitution of Rx1 Functionality in Trans
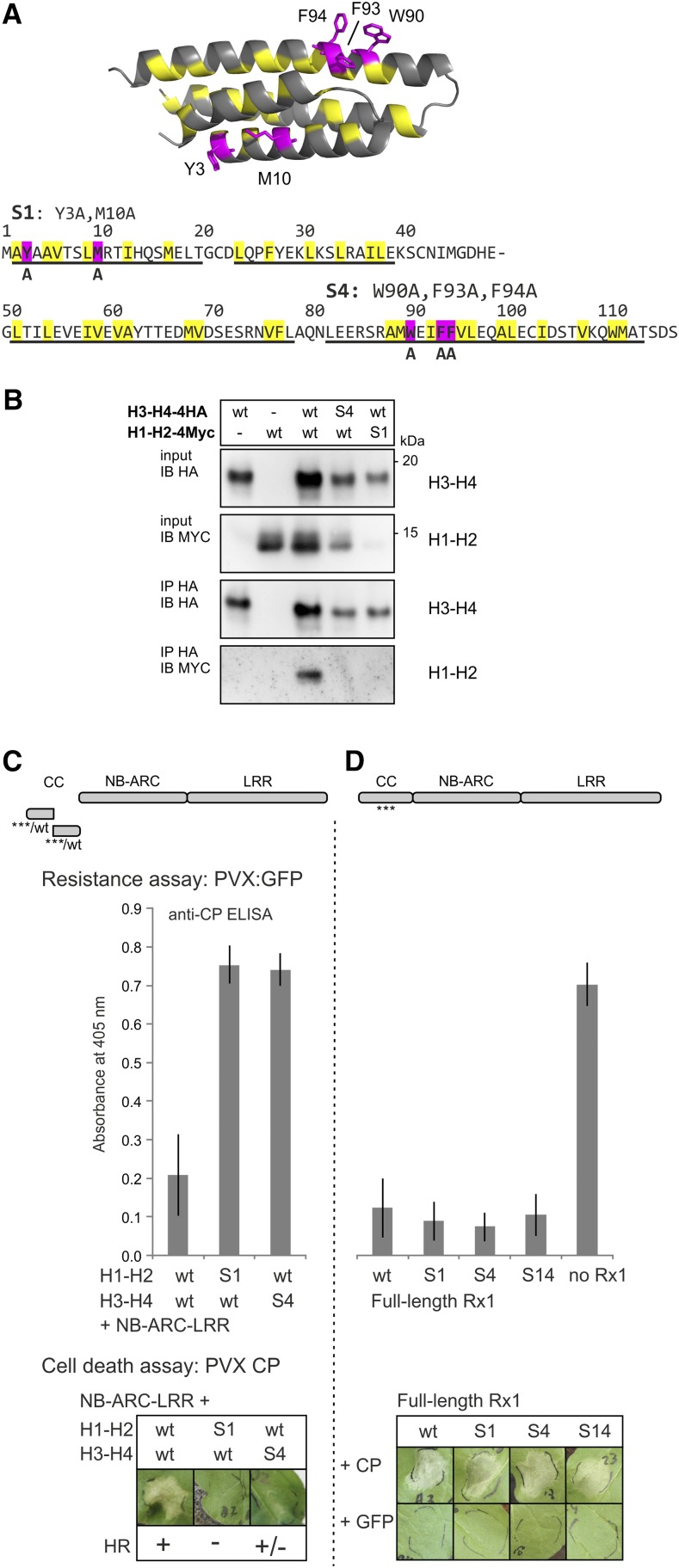
The distinct effects of the local disruption of the CC structure on cell death and PVX resistance signaling might be related to changes in either the intramolecular interactions of the CC or the interaction of Rx1 with its cofactor RanGAP2 mediated by the CC surface. In the crystal structure, a surface-exposed hydrophobic patch on helix 4 containing a cluster of aromatic residues (F93, F94, and W90; Fig. 3A) interacts with a hydrophobic groove on the WPP domain of RanGAP2, and the Trp at position 90 is required for the interaction between the CC and the WPP domain (Hao et al., 2013). Here, we replaced all three aromatic residues in helix 4 with the small and hydrophobic amino acid Ala (constructs named S4; Fig. 3A) to test if altering the intermolecular interaction via helix 4 can explain the functional effects observed after disrupting the local structure of helix 4 in Z4. In helix 1, the residues Y3 and M10 also contribute to a hydrophobic surface area separate from the RanGAP2-interacting surface (Fig. 3A). To determine if these residues are involved in interactions or the functioning of Rx1, we created constructs in which Y3 and M10 are replaced by Ala. The constructs with these substitutions were named S1 (Fig. 3A).
Figure 3.
Ala substitution of aromatic and hydrophobic surface residues in helix 1 and helix 4 of the CC. A, Two groups of mainly aromatic residues in the CC (highlighted in magenta in the structure and amino acid sequence) were substituted for Ala (A). In helix 1, Y3A and M10A were combined and named S1. In helix 4, the substitutions W90A, F93A, and F94A were introduced, and this combination was referred to as S4. Both groups of substitutions were introduced in several constructs, including the H1-H2 and H3-H4 CC strands and full-length Rx1. B, Effects of the S1 and S4 mutations on the interaction between the H1-H2 and H3-H4 strands of the CC. Anti-HA immunoprecipitation was performed to study the interaction between coexpressed wild-type (wt) and mutated (S1 and S4) versions of the H1-H2-4Myc and H3-H4-4HA constructs. Expression of only H1-H2-4Myc or H3-H4-4HA was used as a negative control. C, The effects of the S1 and S4 mutations on Rx1 functioning were tested for the transcomplementation of H1-H2 and H3-H4 with the NB-ARC-LRR, as shown by a schematic drawing. Resistance was tested by coexpressing the Rx1 constructs with the PVX:GFP amplicon in N. benthamiana followed by an anti-PVX CP ELISA with extracts of the infiltrated leaf material. Error bars indicate the sd of six samples. The ability of the constructs to induce cell death (HR) was assessed by coexpression of the complementary Rx1 fragments with the avirulent PVX elicitor CP106. D, The effects of S1 and S4 on the functioning of full-length Rx1 were tested in transient PVX resistance and cell death assays. Mutant constructs (S1, S4, or the combination S14) and wild-type Rx1 were coexpressed with a PVX:GFP amplicon to test for resistance. A leaf infiltrated with PVX:GFP, but not Rx1, was included as a control. Error bars indicate the sd (n = 6). To assess the ability of the mutants to induce a cell death response, the full-length Rx1 constructs were coexpressed with the avirulent PVX CP, and images of the response were taken at 3 dpi. As a control for the autoactive cell death response, the Rx1 constructs were coexpressed with GFP.
The effect of the S1 and S4 mutations on the intradomain interaction between the helices of the Rx1 CC was investigated in a coimmunoprecipitation assay of the S1 mutant version of the H1-H2 segment of the CC and of the S4 mutant version of the H3-H4 segment with the complementary wild-type H1-H2 and H3-H4 segments. The S1 mutant version of the H1-H2 constructs was not stably expressed, which made it difficult to assess its ability to bind the wild-type H3-H4 segment of the CC domain (Fig. 3B). On the other hand, the S4 mutant of the H3-H4 segment of the CC was detected in the total protein extract but did not interact with the wild-type H1-H2 construct in the coimmunoprecipitation assay (Fig. 3B). This indicates that the residues mutated in S4, which are mostly surface exposed in the crystal structure, also play a role in the intramolecular helix-helix interactions. The S4 mutant of the H3-H4 segment of the CC could not fully reconstitute a functional Rx1 protein when coexpressed with the wild-type H1-H2 segment and the NB-ARC-LRR of Rx1 in a transcomplementation assay. This combination did not exhibit PVX resistance, and only a weak cell death was detected upon coexpression with the avirulent PVX coat protein CP106 (Fig. 3C). The lack of PVX resistance and cell death response in the combinations of the S1 mutant of the H1-H2 segment with the wild-type H3-H4 segment and the NB-ARC-LRR likely is due to the instability of the H1-H2 S1 CC segment. From this, we concluded that the aromatic residues in the surface region of helix 4, which are mutated in S4, contribute to the compact fold of the CC and the reconstitution of Rx1 functionality in trans.
To test the impact of the Ala substitutions in the hydrophobic outer surface areas of helix 1 and helix 4 on the functioning of Rx1, mutations S1 and S4 were introduced in full-length Rx1 constructs. Surprisingly, neither S1 nor S4 led to a marked decrease in elicitor-dependent cell death or resistance mediated by Rx1 in the transient assays or in an autoactive cell death response at the expression levels used here (Fig. 3D). Because cell death and virus resistance appear to be signaled via at least partially independent pathways, we also tested if any of the CC mutations led to an autoactive resistance response. For that purpose, the Rx1 variants (wild type, S1, S4, Z1–Z4) were overexpressed in N. benthamiana leaves with a TMV:GFP amplicon. Tobacco mosaic virus (TMV) is not recognized by Rx1, but the resistance response initiated by Rx1 is able to stop the replication of viruses unrelated to PVX, including TMV (Kohm et al., 1993; Bendahmane et al., 1995). Thus, an autoactive resistance response of the Rx1 variants would result in lower levels of TMV and, consequently, reduced GFP expression. The overexpression of wild-type Rx1 caused a mild cell death response, as expected (Bendahmane et al., 2002), and consistently inhibited the accumulation of TMV:GFP expression (Supplemental Fig. S2). Interestingly, the Rx1 S1 variant and the Z1, Z2, Z3, and Z4 variants lost this autoactive aspecific resistance response to TMV. Rx1 S4, however, displayed a mild autoactive cell death response and an autoactive resistance response against TMV similar to that of wild-type Rx1 (Supplemental Fig. S2). Compared with the zipping mutations Z1 to Z4, it seems that changes in the surface regions brought about by the S1 and S4 mutations had a subtler effect on the local structure of the CC and Rx1 functionality in cis, and although S1 and S4 do not affect the elicitor-dependent response against PVX in cis, at least the S1 mutations abolish the autoactive resistance response to TMV.
The Effect of the Mutations on the Functioning of the Full-Length CC Coincides with Local Disruptions of the Helix-Helix Interactions
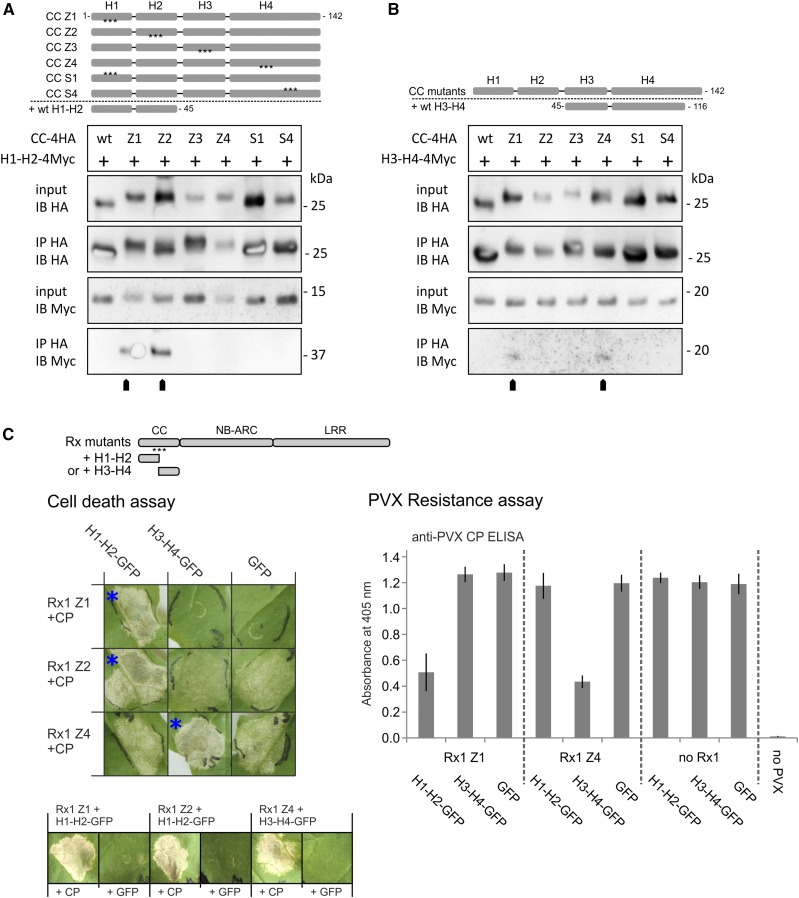
The more pronounced effects of the Z mutations on the functionality of Rx1 in comparison with the limited effect of the S mutations could be due to a more severe disruption of the local structure of the CC by the Z mutations. If mutations cause the dissociation of a helix from the rest of the helix bundle, this would partially open up the structure of the CC and, as a consequence, would make the hydrophobic core surface of the complementary wild-type segment of the CC available for binding and transcomplementation by separate H1-H2 and H3-H4 segments. To test this hypothesis, we performed a coimmunoprecipitation assay with the mutant and wild-type versions of the complete CC domain (amino acids 1–142) coexpressed with wild-type versions of either of the two-helix segments (H1-H2 or H3-H4) to test their binding in planta (Fig. 4, A and B). As expected for a closed conformation of the helix bundle, no interaction of either H1-H2 or H3-H4 with the wild-type CC could be detected. The lack of interaction between the S1 and S4 variants of the complete CC and either wild-type two-helix segment indicates that the substitution of these outer surface residues in helix 1 and 4 does not cause the helices of the CC to dissociate (Fig. 4A). The Z4 mutation in the complete CC, however, allowed the separate H3-H4 segment to bind (Fig. 4B). No such binding was detected for the Z3 mutant of the complete CC. The H1-H2 segment coimmunoprecipitated with the Z1 and Z2 version of the CC (Fig. 4A). Together, these results show that the hydrophobic surface regions, buried in the wild-type structure of the Rx1 CC domain, become exposed if the hydrophobic zipping interaction between the helices in the four-helix bundle is disrupted.
Figure 4.
A and B, Immunoprecipitation assay to test if the loss of interaction between H1-H2 and H3-H4 due to mutations (Z1–Z4, S1, and S4) causes the strands to dissociate in the complete CC and expose internal binding surfaces. HA-tagged versions of the wild-type (wt) and mutant Rx1 CC (amino acids 1–142) were coexpressed with wild-type H1-H2-4Myc (amino acids 1–45; A) or H3-H4-4Myc (amino acids 45–116; B), as indicated by schematic overviews of the constructs. The CC constructs were immunoprecipitated with antibodies against the HA tag. Coimmunoprecipitation of the interacting H1-H2 or H3-H4 strands was detected by anti-Myc immunoblotting. C, Complementation of the loss of function caused by the Z1, Z2, and Z4 mutations via coexpression of wild-type H1-H2 or H3-H4 strands. Full-length Rx1 mutant constructs displaying decreased elicitor-dependent cell death (Z1, Z2, and Z4) or decreased PVX resistance (Z1 and Z4) were coexpressed with H1-H2-GFP, H3-H4-GFP, or GFP to investigate if the presence of the wild-type strands could restore the functionality of Rx1. Combinations in which Rx1-mediated cell death was reconstituted are indicated by blue asterisks. These three combinations also were tested in the absence of the CP to determine if the coexpressed CC fragment induces an autoactive response (row of images at bottom). Resistance was assessed by the detection of PVX in an ELISA (error bars represent the sd; n = 8). The CC strands or GFP were coexpressed with PVX:GFP in the absence of Rx1 as a negative control.
The interaction of the wild-type H3-H4 segment with the CC Z1 construct was unexpected (Fig. 4B), because the Z1 mutations in helix 1 disrupt the interaction of the H1-H2 segment with H3-H4. However, we found that, in contrast to the complete CC, H3-H4 segments of the CC can homodimerize using coimmunoprecipitation and fluorescence resonance energy transfer by fluorescence lifetime imaging (FRET-FLIM) assays (Supplemental Fig. S3, A–C). The H3-H4 dimerization was sensitive to the Z3 and Z4 mutations but not the S4 mutations, indicating that the interactions depended on the hydrophobic helix-helix interface (Supplemental Fig. S3A). In the wild-type CC, the interface of the H3-H4 interaction is buried in the structure and not expected to be available for homodimerization.
The results shown in Figure 4, A and B, indicate that the Z1, Z2, and Z4 mutations open up the structure of the CC and enable wild-type segments to bind in trans to the exposed hydrophobic surface. To test if this interaction could allow transcomplementation and, thereby, the possibility to restore wild-type cell death or resistance to the full-length Rx1 mutants, we coexpressed the full-length Rx1 constructs that displayed a complete (Z1) or partial (Z2 and Z4) loss of function with the wild-type versions of either the H1-H2 or H3-H4 segment (Fig. 4C). Resistance was assessed by coexpression with the PVX amplicon and cell death induction via coexpression with the PVX CP106 (Fig. 4C). The loss of function caused by mutations Z1 and Z2 in the first half of the CC were complemented by coexpression of the wild-type segment H1-H2 (Fig. 4C). Coexpresssion of GFP instead of the CP demonstrated that the observed cell death was not an autoactive response triggered by the CC segment. The reduced cell death induction and loss of resistance of Rx1 Z4 was complemented by coexpression of wild-type H3-H4 (Fig. 4C). As expected, no complementation was observed with coexpression of the CC segments that do not encompass the helices mutated in the full-length Rx1 protein. In conclusion, the local disruption of the interaction between the helices of the CC has distinct effects on Rx1-mediated cell death and PVX resistance. It seems that the disruption and, thereby, opening up of the CC structure in the full-length mutant proteins underlies the transcomplementation by coexpressed wild-type CC segments.
Local Disruption of the CC Structure Affects the Interaction of the CC with the NB-ARC and LRR
The CC domain of Rx1 interacts with the combined NB-ARC and LRR domains but not at all or at a much lower affinity with the separate NB-ARC or LRR domains (Moffett et al., 2002; Rairdan et al., 2008). To determine the effect of the local disruption of the CC structure on the interdomain interactions within Rx1, we used affinity-tagged versions of the CC and the CC-NB-ARC harboring the hydrophobic core (Z) or surface (S) mutations in a coimmunoprecipitation experiment with the NB-ARC-LRR and the LRR, respectively. In the combination of the CC and NB-ARC-LRR constructs, only the HA-tagged CC S4 coimmunoprecipitated with the Myc-tagged NB-ARC-LRR at levels similar to that of the wild-type HA-tagged CC (Fig. 5A). The binding of all other CC variants to the Myc-tagged NB-ARC-LRR was reduced strongly (Z1, Z2, and Z4), and two CC constructs were not detected on the immunoblot after immunoprecipitation (Z3 and S1), even though all CC constructs were detected at similar levels in the input material (Fig. 5A). Similarly, from the six mutant versions of the CC-NB-ARC, only S4 still displayed a wild-type-like binding to the LRR (Fig. 5B). For all other mutant CC-NB-ARC variants, we did not detect a coimmunoprecipitation of the LRR, emphasizing the importance of the CC in the interaction of the CC-NB-ARC with the LRR (Fig. 5B).
Figure 5.
Effects of the mutations in the CC on the domain interactions of Rx1. A, Effects of the mutations Z1 to Z4, S1, and S4 on the interaction between the CC and the NB-ARC-LRR of Rx1. A Myc-tagged construct of the Rx1 NB-ARC-LRR (amino acids 144–937) was coexpressed with 4xHA-tagged constructs of the wild-type (wt) and mutant CC. The NB-ARC-LRR was pulled down using anti-Myc antibodies, and the coimmunoprecipitation of the CC constructs was detected via an anti-HA immunoblot. B, Immunoprecipitation assay testing the effect of the CC mutations on the interaction between the CC-NB-ARC and LRR of Rx1. 4Myc-tagged CC-NB-ARC constructs were coexpressed with 4HA-LRR. Immunoprecipitation was performed with anti-Myc antibodies, and the coprecipitation of the HA-tagged LRR was visualized by anti-HA immunoblot. C, Complementation of functionality for full-length Rx1 mutants by coexpression of the wild-type CC. The full-length Rx1 constructs carrying the combined Z14 or Z23 mutations were coexpressed with wild-type versions of the individual CC strands (H1-H2 and H3-H4) or the complete CC (shown in the schematic drawing at top). The PVX CP was coexpressed to test if the presence of the CC or CC strands could restore the ability of the Rx1 mutant to initiate a cell death response. The combinations in which a cell death occurred are marked with white asterisks. These combinations were tested with GFP instead of the CP to test for autoactivity (row of images at bottom). D, Interaction study to test if mutations in the CC disrupt the interaction between the CC and NB-ARC-LRR in the full-length Rx1 protein and, thereby, make the CC-binding surface on the NB-ARC-LRR accessible for coexpressed CC constructs. GFP-4HA-tagged constructs of full-length Rx1 (wild type, Z14, Z1, S1, and S4) were coexpressed with a wild-type CC-4Myc construct. The full-length Rx1 constructs were immunoprecipitated by anti-HA antibodies, and the coimmunoprecipitation of the CC was detected via anti-Myc immunoblot. The truncated NB-ARC-LRR served as a positive control for this interaction. HA-tagged CC and HA-tagged GFP constructs were used as negative controls.
In planta transcomplementation assays, where either the CC and NB-ARC-LRR or the CC-NB-ARC and LRR were coexpressed with an avirulent PVX CP106 or a PVX:GFP amplicon, yielded results that matched the interaction data of the coimmunoprecipitation experiments; loss of interaction led to a loss of function in trans. Only the combinations of the CC S4 and the NB-ARC-LRR and of the CC-NB-ARC S4 and LRR mediated a PVX resistance and cell death similar to the wild-type constructs (Supplemental Fig. S4, A and B). CC-NB-ARC S1 and the double mutant CC-NB-ARC S14 no longer conferred virus resistance but still initiated a weak cell death response (Supplemental Fig. S4B). The loss of affinity between the CC-NB-ARC and LRR caused by the S1 mutations affected the induction of a virus resistance response more than the induction of the cell death response.
The Integrity of the Overall Structure of the CC Domain Is Required to Assemble a Functional Rx1 Protein
Most of the tested CC mutations reduced the interdomain interaction of the CC, NB-ARC, and LRR in trans. We reasoned that if the CC mutations disrupted the intramolecular interaction of the CC with the NB-ARC and LRR in full-length Rx1 in cis, then the CC-binding surface on the NB-ARC and LRR would become exposed. This exposed binding surface could enable a coexpressed wild-type CC to bind in trans and functionally complement the mutations in the CC of the full-length protein (Fig. 5C). The loss of function displayed by the full-length Rx1 Z14 and Z23 versions was not functionally complemented by the coexpression of only the H1-H2 or H3-H4 CC segments. However, coexpressing the wild-type CC construct with the full-length Rx1 Z14 or Rx1 Z23 indeed restored their ability to initiate cell death in response to the PVX CP (Fig. 5C). No elicitor-independent response was observed. The restoration of Rx1 functionality by coexpression of a functional CC supports the hypothesis that the CC mutations disrupted the intramolecular interaction of the CC with the NB-ARC-LRR and, thereby, provided access to the CC-interacting surface on the NB-ARC-LRR.
We tested whether these mutations indeed expose the CC-binding surface on the NB-ARC-LRR of the full-length Rx1 via a coimmunoprecipitation experiment. The full-length Rx1 carrying the Z14, Z1, and S1 mutations was coexpressed with an affinity-tagged wild-type CC construct. Wild-type Rx1 and Rx1 S4 served as controls, because domain interactions were not affected in these proteins in trans as shown (Fig. 5A). In comparison with the wild-type Rx1 and Rx1 S4, markedly more CC-4Myc coimmunoprecipitated with Rx1 Z14, Z1, and S1 (Fig. 5D). It was interesting to observe the increased interaction, especially for Rx1 S1, as full-length Rx1 S1 was not visibly affected in its functionality in transient assays and, therefore, could not be tested in a functional complementation assay with the wild-type CC domain (Fig. 5C). A truncated Rx1 construct that lacks the CC domain (NB-ARC-LRR) coimmunoprecipitated relatively higher amounts of the CC than any of the full-length Rx1 constructs. Steric hindrance by the CC in cis might limit accessibility to the CC-binding surface by the CC expressed in trans, even in the full-length constructs in which the intramolecular interaction between the CC and the rest of the protein was disrupted. No signs of CC dimerization were seen in the assay; the low level of CC-4Myc that coimmunoprecipitated with CC-4HA did not exceed the level of background binding of CC-4Myc to GFP(HA).
H3 and H4 of the CC of Rx1 Are Required and Sufficient for the Interaction with RanGAP2
The surface residues mutated in the fourth helix (S4) are located in the interface of the CC and the RanGAP2 WPP domain. Previous interaction studies with truncated versions of the Rx1 CC domain indicate that the RanGAP2-binding surface partially overlaps with the NB-ARC-LRR-binding region of the CC (Rairdan et al., 2008). The mutations we introduced in the helices of the CC domain to investigate the effect of local structure disruptions on the intramolecular interactions and the functionality of Rx1 also might influence the interaction with RanGAP2. We tested this with immunoprecipitation assays.
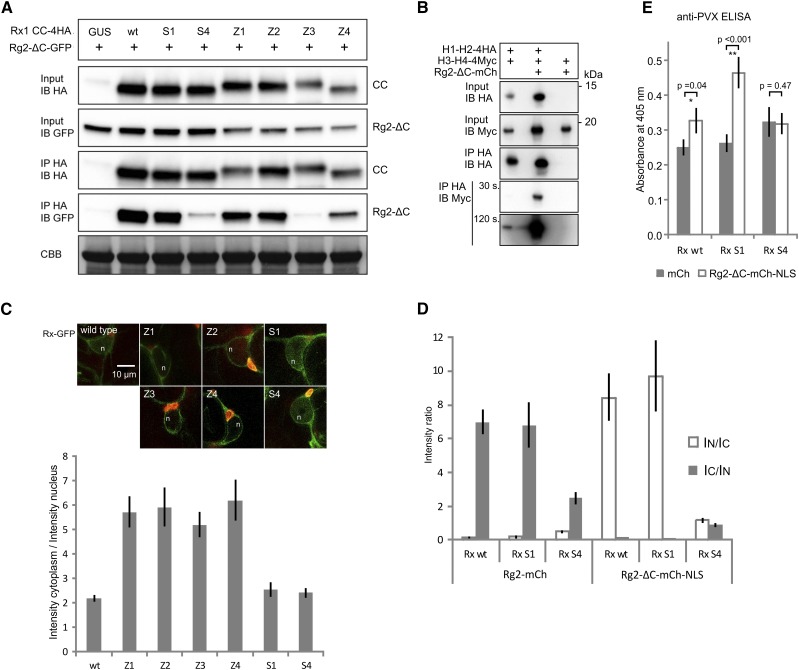
The CC constructs with mutations in the two N-terminal helices (S1, Z1, and Z2) were not affected in their interaction with a GFP-tagged RanGAP2 WPP domain (amino acids 112; NbRg2-ΔC-GFP; Fig. 6A). As expected, CC S4 showed a strong reduction in the amount of Rg2-ΔC-GFP that coimmunoprecipitated. From the hydrophobic core mutants, CC Z3 exhibited a similar loss of interaction to the WPP domain, whereas CC Z4 exhibited an intermediate binding level (Fig. 6A). The three groups of mutations that affected the interaction between the CC domain and the RanGAP2 WPP domain all were positioned in the C-terminal segment of the CC (H3-H4). It is interesting that, in contrast to the S1 substitutions, the S4 substitutions did not affect the intramolecular interactions of Rx1, and both did not have a visible effect on the elicitor-dependent activation of Rx1 in transient cell death and resistance assays. Coimmunoprecipitations of full-length Rx1 carrying either the S1 or S4 surface substitutions with the RanGAP2 WPP domain or with full-length RanGAP2 yielded similar results: the S4 substitutions abolished the interaction of Rx1 with RanGAP2, but the S1 substitutions did not affect this interaction (Supplemental Fig. S5). To exclude the possibility that additional plant proteins play a role in the changes in interaction of the mutant CC domains and RanGAP2 WPP, we tested the effect of the mutations in a yeast two-hybrid assay (Supplemental Fig. S6). In the yeast system, the wild-type CC and CC S1 interacted strongly with the WPP domain whereas the CC S4 did not interact with the WPP, corresponding to the interaction pattern in the coimmunoprecipitation experiments. From the tested Z mutations (Z1, Z3, and Z4), only Z3 appeared to interact with the WPP domain (Supplemental Fig. S6). No interaction was seen for CC Z1 and WPP, even though this CC variant interacted with the WPP domain in coimmunoprecipitation experiments.
Figure 6.
Effects of the mutations in the CC on the interaction with RanGAP2 and the subcellular localization of Rx1. A, Coimmunoprecipitation of HA-tagged versions of the Rx1 CC variants Z1 to Z4, S1, S4, and the wild type (wt) with the N-terminal WPP domain of RanGAP2 (Rg2-ΔC-GFP). Equal loading of the input material is shown by the Coomassie Brilliant Blue-stained Rubisco (CBB). B, Effects of the presence of the RanGAP2 WPP domain on the interaction between H1-H2 and H3-H4. Anti-HA immunoprecipitation of H1-H2-4HA coexpressed with H3-H4-4Myc in the presence or absence of mCherry-tagged RanGAP2 WPP domain (Rg2-ΔC-mCh) is shown. Two exposures (30 and 120 s) are shown for the anti-Myc immunoblot with the results of the anti-HA immunoprecipitation to show the two bands of different intensity. C, Full-length Rx1 constructs (wild type, Z1–Z4, S1, and S4) with a C-terminal GFP fusion were imaged using confocal microscopy after 2 d of expression in N. benthamiana leaves. The images show nuclei (n) and surrounding cytoplasm in representative cells. Chlorophyll autofluorescence is shown in red. Bar = 10 μm for all images. The ratio of GFP fluorescence intensity in the cytoplasm and nucleus was determined in seven to 12 cells for each construct. The graph shows the average cytoplasmic intensity/nuclear intensity ratio (IC/IN). The error bars represent the se. Higher values indicate a more cytoplasmic localization profile. D, Coexpression of Rx-GFP variants with RanGAP2 constructs to test the effect on the localization of Rx. Rx-GFP (wild type, S1, and S4) was coexpressed with either full-length RanGAP2-mCherry (Rg2-mCh) or with Rg2-ΔC-mCh-NLS, a construct in which the RanGAP2 WPP domain was tagged to a nuclear localization signal. The localization of wild-type Rx is affected by the coexpression of these constructs: RanGAP2 sequesters it in the cytoplasm, and Rg2-ΔC-mCh-NLS targets it to the nucleus. The GFP intensities from the Rx-GFP constructs were determined for the nucleus and cytoplasm, and the average ratios of the intensities are plotted (n = 9; error bars denote the se). E, PVX resistance assay. Full-length Rx (wild type, S1, and S4) was coexpressed with either mCherry as a control or with Rg2-ΔC-mCh-NLS and an avirulent PVX amplicon. Previously, we showed that targeting full-length Rx to the nucleus led to a partial loss of resistance (Slootweg et al., 2010). The level of virus after 5 d was determined by an anti-PVX CP ELISA. Error bars represent the se (n = 9). Student’s t test was used to determine if coexpression of Rg2-ΔC-mCh-NLS resulted in a significantly higher virus level than coexpression with free mCherry (*, P < 0.5 and **, P < 0.05).
Interestingly, we found that RanGAP2 stabilized the interaction between the two-helix segments of the CC (H1-H2 and H3-H4) when it was coexpressed with these segments in planta. In this experiment, the interaction between the wild-type H1-H2 and H3-H4 constructs was tested in the presence or absence of a coexpressed RanGAP2 WPP construct (Rg2-ΔC-mCh; Fig. 6B). The strongly increased H1-H2/H3-H4 interaction seems to suggest that the WPP domain of RanGAP2 has a stabilizing effect on the complete CC structure, even though the minimal fragment of the CC domain required for binding RanGAP2 encompasses only H3 and H4 (Supplemental Fig. S7).
Changes in the CC Structure and RanGAP2 Interaction Affect the Subcellular Localization of Rx1
Previously, we demonstrated that Rx1 requires a localization in both the nucleus and cytoplasm for full functionality and that the CC is required for nuclear localization (Slootweg et al., 2010). To test if changes in the surface and structure of the CC domain impact the subcellular distribution of Rx1, the localizations of GFP-tagged versions of the full-length Rx1 mutants (Z1–Z4, S1, and S4) were studied by confocal microscopy. Rx1 S1 and S4 displayed a nucleocytoplasmic distribution similar to wild-type Rx1, whereas all the hydrophobic zipping mutations (Z1–Z4) resulted in a clear reduction in the nuclear pool of Rx1, and fluorescence intensities in the cytoplasm were not visibly reduced (Fig. 6C). These results indicate that proper folding of the Rx1 CC domain is essential for the accumulation of Rx1 in the nucleus.
RanGAP2 can function as a cytoplasmic retention factor for Rx1; coexpression of the mostly cytoplasmic RanGAP2 leads to lower Rx1 levels in the nucleus, and virus-induced gene silencing of RanGAP2 leads to higher nuclear Rx1 levels (Tameling et al., 2010). Because of their contrasting effects on the interaction of the CC with the NB-ARC and LRR and on the interaction with RanGAP2, we tested the effect of the S1 and S4 mutations on the subcellular localization of Rx1 coexpressed with full-length RanGAP2 (Fig. 6D). Coexpression of RanGAP2 with wild-type Rx1 reduced the nuclear pool of wild-type Rx1 and Rx1 S1, consistent with its role as a cytoplasmic retention factor of Rx1. The subcellular distribution of Rx1 S4 was not affected by the coexpression of RanGAP2 (Fig. 6D). Cell fractionation of leaf material coexpressing RanGAP2 and the Rx1 variants also showed that the distribution of Rx1 S4 was not affected by the coexpression of RanGAP2 (Supplemental Fig. S8).
For full functionality, Rx1 requires a balanced nucleocytoplasmic distribution, and a change in the ratio between the cytoplasmic and nucleus-localized Rx1 results in reduced PVX resistance (Slootweg et al., 2010). The coexpression of the N-terminal WPP domain of RanGAP2 fused to a nuclear localization signal (Rg2-ΔC-mCh-NLS) directs the wild-type Rx1 and Rx1 S1 into the nucleus but does not alter the nucleocytoplasmic distribution of Rx1 S4 (Fig. 6D). To test whether the observed differences in the nucleocytoplasmic distribution patterns also would result in differences in PVX resistance, wild-type Rx1, Rx1 S1, and Rx1 S4 were coexpressed with Rg2-ΔC-mCh-NLS and PVX:GFP in a transient resistance assay. The forced nuclear accumulation caused by Rg2-ΔC-mCh-NLS led to a reduction in PVX resistance for wild-type Rx1 and Rx1 S1 in comparison with the control, where Rg2-ΔC-mCh-NLS was replaced by the fluorescent protein mCherry (Fig. 6E). No change in PVX resistance was observed for Rx1 S4 when coexpressed with Rg2-ΔC-mCh-NLS (Fig. 6E).
Together, these data show a link between RanGAP2 binding, its effect on the nucleocytoplasmic partitioning of Rx1 in the cell, and Rx1 functioning in disease resistance. Upon forced translocation to the nucleus, however, the Rx1 S1 mutant shows a stronger loss of PVX resistance compared with wild-type Rx1 (Fig. 6E), indicating that the disruption of the intramolecular interaction between the CC and the NB-ARC-LRR domains (Fig. 5) has an impact on the functioning of Rx1, even though we could not detect any in a transient virus resistance assay in which the localization of Rx1 was not altered.
The Wild-Type CC of Rx1 Is Found Predominantly in a Heteromeric Complex with RanGAP2 in the Cell, Whereas the S4 Variant of the CC Appears to Be Monomeric
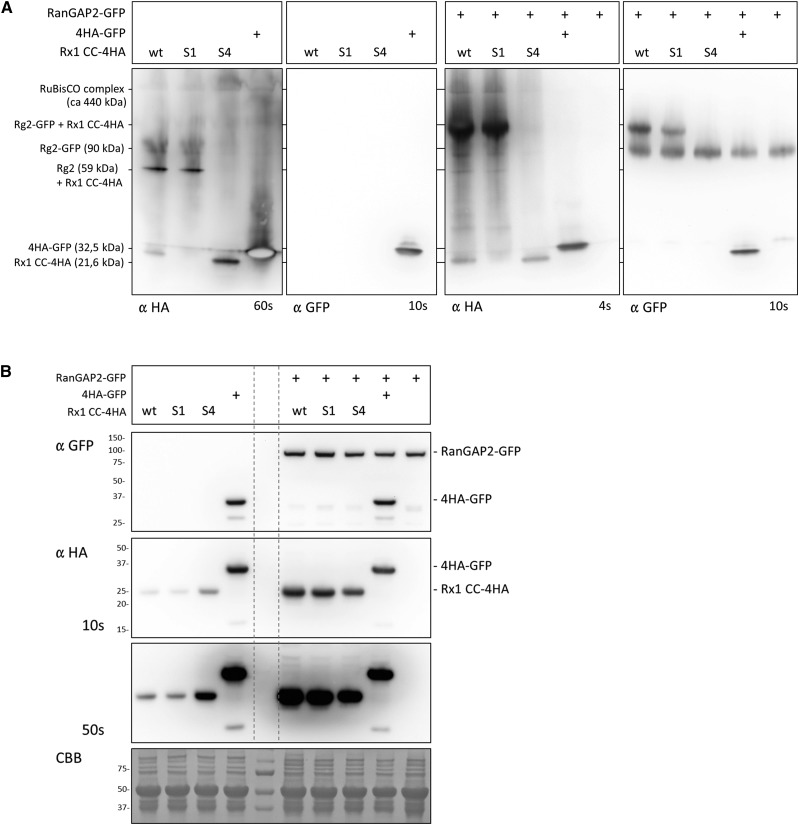
Although coimmunoprecipitation demonstrated that the S4 mutations and not the S1 mutations affected the complex formation of the CC of Rx1 and the WPP domain of RanGAP2, this method does not exclude the possible contribution of other, untagged, endogenous proteins in this complex formation. To further investigate whether the observed interaction between the CC and WPP depends on other host proteins, Blue Native PAGE was used, as it allows the analysis of proteins and protein complexes without denaturing them and, therefore, is informative on relative complex sizes (Wittig et al., 2006; Wittig and Schägger, 2009). We analyzed the behavior of HA-tagged versions of the wild-type CC and the S1 and S4 mutants without or with coexpressed GFP-tagged RanGAP2 (Fig. 7A). The wild type and S1 were found predominantly in a larger complex, although some of the wild-type CC also was detected as a band that ran below the GFP-4HA control (29 kD). The higher band was likely the complex of the 4HA-tagged CC (21.6 kD) with endogenous RanGAP2 (59 kD). The lower band was likely a monomeric form of the CC, as a dimer would be expected to run higher than the GFP control. The S4 mutant could not be detected in the higher complex and ran predominantly at a height corresponding with a monomer. Coexpression of GFP-tagged full-length RanGAP2 shifts the larger complex upward when expressed with the wild type and the S1 variant, but the lower bands containing the wild type and S4 variants remained at the same position with respect to the GFP control (Fig. 7A). On the anti-GFP blot, a corresponding pattern was seen: RanGAP2-GFP expressed alone or in combination with 4HA-GFP ran as a single band. Coexpression of the CC S4 construct did not affect this, but coexpression of the CC S1 or the wild-type CC shifted a fraction of the RanGAP2-GFP upward to the same height at which those CCs were detected on the anti-HA blot. Separating the same set of protein combinations on a denaturing gel shows all proteins as single bands, as the interactions are not retained under denaturing conditions (Fig. 7B). A similar experiment in which a GFP-tagged WPP domain of RanGAP2 was used instead of full-length RanGAP2-GFP yielded similar results (Supplemental Fig. S9). Thus, most of the wild-type CC appeared to be present in the cell as a predominant complex with RanGAP2 or its WPP domain and partially as a monomer. The S4 mutations of the residues in the CC required for RanGAP2 binding via the WPP domain resulted in a complete absence of the CC in the larger complex, further supporting this observation. However, the detection of a few additional bands indicated that a minor portion of the CC formed complexes with either modified versions of RanGAPs or other host components.
Figure 7.
Blue Native gel analysis of the complex formed by the Rx1 CC and RanGAP2 in the cell. A, Blue Native gel analysis of Rx1 CC constructs (wild type [wt], S1, and S4) coexpressed with RanGAP2-GFP (right two images) or expressed alone (left two images). HA-tagged GFP (4HA-GFP) was included as a control and was detected with the anti-HA and anti-GFP antibody. The mass given for the RanGAP2, Rx1-CC, and GFP constructs represents the mass of a monomer, and for Rubisco the approximate mass of the complex is given. The behavior of the proteins on this gel is determined by the mass of the complex they are part of and by their shape. The blots were aligned to each other using the Rubisco complex and the 4HA-GFP, which is present on each immunoblot. B, SDS-PAGE analysis of the samples used in A demonstrating that the banding patterns on the Native Blue blot are not due to protein degradation or modifications. RanGAP2-GFP and the CC-4HA constructs run as single bands on SDS-PAGE. A Coomassie Brilliant Blue (CBB)-stained blot is included as a control for equal loading. The dashed vertical lines indicate the positions of the marker lane on the immunoblots.
Coexpressed Rx1 Domains Have a Dominant Negative Effect on Rx1-Mediated Resistance, Which Is Linked with the RanGAP2-Interacting Surface for the CC
Coexpression of the CC domain complemented mutations in the CC domain in full-length Rx1 by the reconstitution of a functional protein in trans. The multidomain architecture of the protein allowed the CC domain to replace the mutated CC domain by interacting with surface-exposed areas on the NB-ARC-LRR, resulting in the reconstitution of a functional protein (Figs. 6 and 8A). However, truncated versions of proteins also are known to interfere with the functionality of their wild-type counterparts (Dinesh-Kumar et al., 2000; Du et al., 2012). This prompted us to address the question of whether this also occurs when coexpressing the CC domain in trans with a normal functional wild-type Rx1 protein, potentially interfering with regular functionality. We tested whether coexpression of the individual CC domain would affect the resistance signaling by Rx1 and, if so, how the intramolecular or intermolecular interactions of the CC contribute to this effect.
Figure 8.
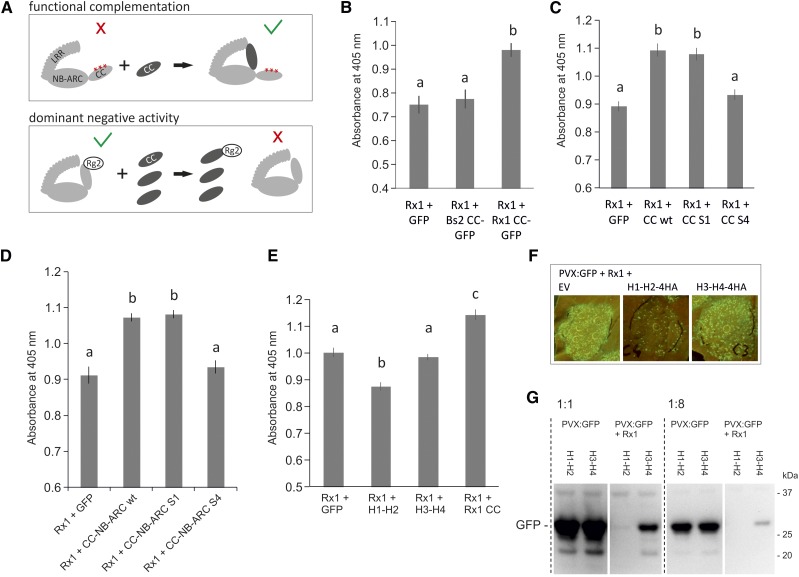
Exploring suppressive effects of the coexpressed CC domain on the functionality of wild-type Rx1. A, Schematic view of the potential mechanisms behind the functional complementation and the dominant negative phenotypes that could occur upon coexpression of the wild-type CC domain with mutant or wild-type Rx, respectively. A red cross indicates a loss of signaling of the full-length protein, and the green V indicates a state in which the protein is able to signal. B, Assay to test if coexpression of the CC of Rx1 suppresses the resistance mediated by Rx1, leading to higher PVX levels in a transient PVX resistance assay. Coexpression of GFP or the CC domain of Bs2 was used as negative controls. PVX accumulation was determined via an anti-PVX ELISA. Error bars present the se (n = 8), and letters denote significantly (P < 0.05) different groups (one-way ANOVA with posthoc Tukey’s test). C, To determine if the S1 or S4 mutation affected the suppressive effect seen for the wild-type Rx1 CC, CC constructs harboring these mutations were coexpressed with wild-type Rx1 in a transient PVX assay. Coexpression of GFP served as a negative control. Error bars present the se (n = 12), and letters denote significantly (P < 0.05) different groups (one-way ANOVA with posthoc Tukey’s test). D, Coexpression of the wild type and S1 and S4 variants of the CC-NB-ARC with wild-type Rx1 in a transient PVX assay to test if the mutations affect the suppressive effect of the CC-NB-ARC. Coexpression of GFP was used as a negative control. Error bars present the se (n = 8), and letters denote significantly (P < 0.05) different groups (one-way ANOVA with posthoc Tukey’s test). E, Coexpression of the N- and C-terminal CC segments with the full-length Rx1 in a transient PVX resistance assay to test if the suppressive effect is caused by a specific region in the CC. Error bars present the se (n = 8), and letters denote significantly (P < 0.01) different groups (one-way ANOVA with posthoc Tukey’s test). F, Coexpression of PVX:GFP, Rx1, and the CC segments. The virus accumulation was visualized via the GFP expressed from the PVX genome. Lower GFP levels are apparent on leaves in which H1-H2 is coexpressed. G, To test if the reduction of PVX levels was a direct effect of the H1-H2 segment on the virus or if it was due to an enhancement of Rx1 activity, the CC segments were coexpressed with PVX:GFP in the absence and presence of Rx1. PVX levels were compared on an anti-GFP immunoblot via the GFP expressed by PVX. All samples were loaded undiluted and 8× diluted to make a better comparison possible between the samples with high and low GFP concentrations.
In a transient PVX resistance assay, we expressed wild-type full-length Rx1 under the control of its native promoter with the avirulent PVX:GFP in combination with either the CC domain of Rx1 or, as a control, GFP or the CC domain of the related R protein Bs2, a CC-NB-LRR that confers resistance against bacterial spot disease (Xanthomonas campestris pv vesicatoria) in pepper (Capsicum annuum; Tai et al., 1999). The use of the native promoter Rx1 construct resulted in a lower expression level for Rx1 than the equivalent Cauliflower mosaic virus 35S construct, and an assay based on this combination of Rx1 and PVX constructs sensitively detects the effects of coexpressed proteins on Rx1-mediated resistance. Surprisingly, coexpression of the CC in this assay led to an increase in PVX accumulation, which indicates a decrease in Rx1-mediated resistance (Fig. 8B). Coexpression of GFP or the CC domain of the pepper R protein Bs2 did not affect Rx1-mediated resistance, demonstrating the specificity of this effect. In contrast to the functional complementation of Rx1 constructs with mutations in the CC, coexpression of the CC with wild-type Rx1 resulted in a loss of function. We hypothesized that this was the result of a dominant negative effect of the overexpression of the CC domain on Rx1 functioning by competing for available interactions normally involved in the activation of an Rx-mediated defense response and the formation of nonfunctional complexes through this interaction (Fig. 8A). Coexpression of either the NB-ARC or LRR had a similar suppressive effect on full-length Rx1 (Supplemental Fig. S10A).
To test if the dominant negative effect of the CC is linked to the intramolecular interaction of the CC with the NB-ARC and LRR or with the intermolecular interaction with RanGAP2, the S1 and S4 substitutions of surface residues in the CC were used. Coexpression of wild-type CC, CC S1, and CC S4 with full-length Rx1 and PVX demonstrated a negative effect on Rx1-mediated resistance for the wild-type CC and the CC S1 construct, which had a strongly reduced interaction with the NB-ARC-LRR (Fig. 8C). The CC S4 construct, however, lost the dominant negative effect on resistance signaling by full-length Rx1. Similarly, coexpression of the CC-NB-ARC S4 construct with full-length Rx1 did not suppress Rx1-mediated resistance, whereas both the wild-type CC-NB-ARC and the S1 version suppressed resistance by Rx1 (Fig. 8D). These results indicate that the aromatic residues in helix 4, which are required for the interaction with RanGAP2, also are required for the dominant negative suppression of Rx1 signaling.
If the dominant negative effect of the CC is linked to the interaction of the CC with RanGAP2, then the minimal interacting region, in this case the H3-H4 CC segment, also might be the minimal region needed to suppress Rx1 resistance. To test this, we coexpressed the complete CC, the H1-H2 segment, or the H3-H4 segment with full-length Rx1 and PVX CP106. To our surprise, the H3-H4 construct did not affect Rx1-mediated resistance like the CC (Fig. 8, E–G). Immunoblots demonstrated that it accumulated to lower levels in the cell than the complete CC (see Fig. 7 in Slootweg et al., 2010), which could explain the lack of an effect. Alternatively, additional surface regions in H1-H2 are required for the dominant negative effect or the H3-H4 helices adopt a different conformation than in the context of the complete CC.
Even more remarkable, coexpression of the N-terminal two helices of the Rx1 CC consistently stimulated the resistance mediated by Rx1, as seen by the reduced virus levels (Fig. 8E) and reduced levels of GFP expressed via PVX:GFP (Fig. 8, F and G). Expression of the H1-H2 segment with PVX in the absence of Rx1 did not result in reduced virus levels compared with the control (Fig. 8G).
DISCUSSION
From the data presented in this study, a picture emerges that shows that distinct surfaces are important for the intramolecular interaction of the CC with the NB-ARC-LRR domains of Rx1 on the one hand and the intermolecular interaction with the protein RanGAP2 on the other. Surface changes disrupting the RanGAP2 interaction also affect the nucleocytoplasmic partitioning of Rx1 in the cell, and the overall integrity of the CC is required for the accumulation of Rx1 in the nucleus. Finally, we demonstrated that distinct regions of the CC play a role in different Rx1-mediated responses, which uncouple defense-related cell death and virus resistance in plant cells. From these findings, a functional model can be inferred in which the CC domain of Rx1 plays a dual role in modulating effector-triggered immunity in plants.
In NB-LRR proteins, the switch from the resting state to the active state is regulated strictly by intramolecular domain interactions. For example, a slight change in the interface of the NB-ARC and LRR can trigger the activation of Rx1 and, thereby, initiate resistance and cell death responses (Bendahmane et al., 2002; Slootweg et al., 2013). The simultaneous binding of the CC to both the NB-ARC and LRR and its dependency on the nucleotide-bound state of the NB-ARC suggest that it plays a role in the regulation of the conformational switch (Moffett et al., 2002; Rairdan et al., 2008). The way the CC mutations in this study affected the domain interaction in Rx1 demonstrates their complex interdependency. Five out of the six groups of mutations, involving each of the four helices of the CC, disrupted in trans binding of the CC to the NB-ARC-LRR but also binding of the CC-NB-ARC to the LRR. It is surprising, therefore, that the S1 and Z3 mutations, which severely disrupted the interaction of the CC with the NB-ARC and LRR, have so little effect on the functionality of the full-length Rx1 protein. None of the CC mutants resulted in increased activity or autoactivity, which suggests that the intramolecular interaction of the CC with the NB-ARC and LRR is not required to retain an autoinhibited state. The overexpression-induced autoactive resistance response of Rx1 to TMV:GFP was observed only for wild-type Rx1 and the Rx1 S4 variant and was lost in all other mutants (Supplemental Fig. S2), even in those like S1 and Z3, which display wild-type-like elicitor-dependent responses.
In the TIR-NB-LRR flax resistance proteins L6 and L7, the N-terminal TIR domain cooperates with the NB in the regulation of nucleotide binding to the NB-ARC, both in elicitor-dependent and independent cell death signaling (Bernoux et al., 2011b, 2016). Variants in which combinations of residues in the TIR and NB domains negatively regulate the ability of the NB-ARC to switch from the ADP-bound state to the ATP-bound state exhibit a reduced activity and also fail to bind to the cognate effector for which the LRR is required. This can be explained by a model in which the NB-LRRs exist in an equilibrium between on and off states, and the effector shifts the equilibrium by stabilizing the on state (Bernoux et al., 2016). In Rx1, the cooperation between the ARC and LRR might be governed by a similar mechanism, but the role of the CC probably is relatively small, as some mutations, like S1, that strongly affect domain interactions have little effect on elicitor-dependent activity (Fig. 3).
Changes in the domain interactions that play a role in the conformational switch of NB-LRRs have been studied by observing the interactions of the domains when coexpressed as separate polypeptides. The ability of the domains of Rx1 to reconstitute a functional protein is one of the reasons Rx1 is such an interesting model protein. In this study, we demonstrate changes in the intramolecular interactions in the full-length protein by detecting exposed domain-binding surfaces. The interaction surface of the CC on the NB-ARC-LRR is mostly shielded by the CC in the wild-type protein. Its exposure due to mutations in the CC that disrupt this intramolecular interaction was probed successfully by testing if the full-length protein could interact with a coexpressed free wild-type CC domain (Fig. 5D). In a similar vein, the partial disruption of the CC structure could be demonstrated via the induced interaction with coexpressed CC segments (Fig. 4, A and B). These induced in trans interactions explain why coexpressed wild-type versions of domains transcomplement the loss-of-function phenotype of the full-length protein caused by these mutations (Figs. 4, C and D, and 5C). In a previous study, a truncated Rx1 protein lacking its CC domain was functionally complemented by a coexpressed full-length Gpa2 protein. No interaction could be detected between full-length Rx1 proteins, but Rx1 lacking the CC domain interacted with a full-length protein (Moffett et al., 2002). Previously, we also were unable to detect self-association of either the CC-NB-ARC or the LRR domains of Rx1 under conditions where interdomain interactions are readily detected (see Supplemental Fig. S7 in Slootweg et al., 2013).
In contrast to Rx1, self-association is required for the functioning of several other CC- and TIR-NB-LRR proteins (Mestre and Baulcombe, 2006; Ade et al., 2007 ; Bernoux et al., 2011a; Maekawa et al., 2011). In our experiments, however, we could not detect the self-association of the wild-type CC domain via Blue Native PAGE (Fig. 7; Supplemental Fig. S9), FRET-FLIM (Supplemental Fig. S3), or coimmunoprecipitation (Fig. 5D). Only a truncated CC (H3-H4) lacking the first two α-helices exhibited homotypic interactions in coimmunoprecipitation (Supplemental Fig. S3A) and FRET-FLIM assays (Supplemental Fig. S3, B and C), but these are likely an artifact caused by the exposure of residues that would be buried in the complete CC structure. Previously, sucrose density gradient centrifugation in combination with native PAGE showed that Rx1 is present either as a monomer or in a complex with the size of Rx1 plus the interacting RanGAP2 (Sacco et al., 2007), which is consistent with our Blue Native PAGE results (Fig. 7; Supplemental Fig. S9).
The autoactive cell death signaling by the CC domains of the resistance proteins MLA10, Sr33, and Sr50 coincides with their ability to self-associate (Casey et al., 2016). Interestingly, the ability to self-associate depends on a small sequence in these CC domains; constructs that are truncated upstream of a position equivalent to amino acid 142 in MLA10 (or position 160 in Rx1) do not self-associate and behave like monomers in solution. MLA10, Sr33, and Sr50 CC constructs of 142 amino acids or longer do self-associate and induce cell death in planta. The NMR spectroscopy-determined structure of Sr33 6-120 reveals a four-helix bundle similar to the structure of Rx1 1-122 determined via x-ray crystallography (Hao et al., 2013; Casey et al., 2016). The structure of self-associated Sr33 1-142 could not be resolved, so it is still unknown if it resembles the dimer found in the MLA10 5-120 crystal structure (Maekawa et al., 2011). The Rx1 CC constructs used in our study extend to amino acid 142 and do not show signs of self-association, in contrast with the behavior of corresponding CC fragments of MLA10, Sr33, and Sr50. If the CC of Rx1 forms an MLA10 CC-like dimer, then the hydrophobic residues mutated in the Z constructs would be positioned in the interface between the helices of the two protomers and the residues mutated in S1 and S4 would be located on the surface of the structure (Supplemental Fig. S11). In an in vitro size-exclusion chromatography analysis of Rx1 CC 1-142, the CC migrates like a protein close to twice its predicted size (Supplemental Fig. S12). In the study by Casey et al. (2016), a similar behavior was shown for the Rx1 1-122 and Sr33 6-120 constructs, which also were shown convincingly to be monomeric in solution via, among others, small-angle x-ray scattering (Casey et al., 2016). We conclude that Rx1 preferably associates with RanGAP2 via the CC and WPP domain and is present in plant cells as a heteromeric complex when in its resting state instead of a homomeric complex, as shown for other CC-type NLRs.
Secondary structure prediction (PSIPred) of the Rx1 CC indicates that helix 4 does not extend beyond amino acids 122 and is followed by a linker region rich in Ser and Pro. Around amino acid 160 (corresponding to amino acid 185 in MLA10), the first secondary structure motifs of the NB domain appear (Supplemental Fig. S13). Much smaller segments of the CC of Rx1 are still functional in trans with the NB-ARC-LRR, as shown by Rairdan et al. (2008). For example, Rx1 1-86, lacking most of the fourth α-helix, still allows the induction of elicitor-dependent cell death. Rx1 1-116 combined with the NB-ARC-LRR confers both cell death and resistance in a transient virus resistance test. A 158-amino acid-long construct of the CC domain of Bs2, a CC-NB-LRR from pepper related to Rx1, also does not induce a cell death response when expressed transiently in leaves (Collier et al., 2011). Alignment of MLA, Sr33, and Sr50 with Rx and Bs2 (Supplemental Fig. S13) shows that the region in MLA10, Sr33, and Sr50 required for autonomous cell death signaling is not conserved between the two groups of CCs. The CC of the resistance protein Resistance to Pseudomonas syringae pv maculicola1 from Arabidopsis (Arabidopsis thaliana) does self-associate, even when truncated at the C terminus to amino acid 135 (El Kasmi et al., 2017). In contrast to MLA10-like CCs, none of the tested CC constructs in that study displayed autonomous cell death signaling. Thus, Rx1-like CCs might have evolved different mechanisms for signaling than the MLA10-like CCs. The helper NLRs NRC2, NRC3, and NRC4 were shown recently to be required for Rx1-mediated resistance (Wu et al., 2017). It is possible that these helper NLRs have taken over a role in signaling from Rx1 that the autoactive CC domains of the MLA-like proteins still possess themselves. The possibility that the self-association of Rx1 occurs after activation cannot yet be excluded and will be the focus of future research.
Rx1 and RanGAP2, a regulator of nucleocytoplasmic trafficking, form a heteromeric complex via the interaction of their CC and WPP domains, respectively (Sacco et al., 2007; Tameling and Baulcombe, 2007; Hao et al., 2013). The crystal structure of the complex of the CC and WPP has been resolved, and the interface between the domains has been probed by targeted mutagenesis (Hao et al., 2013). In our S4 mutant, we specifically substituted the aromatic surface residues in or adjacent to the RanGAP2-binding surface of helix 4 (W90A, F93A, and F94A). Like the W90D substitution of Hao et al. (2013), the three Ala substitutions of S4 strongly disrupted the binding of the CC to RanGAP2. CC S4 was not affected in its interaction with the NB-ARC and LRR, showing that at least this part of the interface with RanGAP2 does not overlap with the binding surface for the intramolecular interactions and that the substitutions do not disrupt the overall structure of the CC. Surprisingly, S4 and W90D appear to have different effects on the functionality of Rx1. Hao et al. (2013) observed a loss of resistance, but not the cell death response, when the CC-NB-ARC W90D was coexpressed with the LRR. In our experiments, we did not observe a loss of resistance or cell death for the full-length Rx1 S4 or for the coexpressed CC S4/NB-ARC-LRR and CC-NB-ARC S4/LRR combinations (Supplemental Fig. S4). The substitution of W90 by an Asp might have a different effect on the structure of the CC than the Ala substitutions in S4, explaining the difference in the effect on Rx1 functionality.
RanGAP2 has a positive role in Rx1 activation. Experiments with RanGAP2 and elicitors of Gpa2, which is highly homologous to Rx1, suggest a role for RanGAP2 as a cofactor in effector recognition, analogous to the role of several other proteins interacting with the N termini of NB-LRRs (Collier and Moffett, 2009; Sacco et al., 2009). In that context, it is surprising that the Z3 and S4 mutant constructs, which exhibit a strong reduction in RanGAP2 binding, are not affected in their ability to induce elicitor-dependent cell death or resistance in our assays (Fig. 2). This suggests that if RanGAP2 plays a role in the recognition of PVX, it is rather an enhancer of recognition than it is an absolute requirement for recognition. Under circumstances where either Rx1 or the coat protein of PVX is present at low concentration in the cell, such enhancement of recognition could be beneficial. Beyond a certain concentration threshold of coat protein or Rx1, the recognition might take place without assistance by RanGAP2. Surprisingly, even the overexpression-induced autoactive resistance response that wild-type Rx1 displays is not visibly reduced for Rx1 S4, which indicates that the RanGAP2 interaction is not required for downstream signaling (Supplemental Fig. S2).
The overlap in helix 3 of the CC between the binding surfaces for the NB-ARC-LRR and RanGAP2 raises the question of whether both interactions modulate each other. The results from the immunoprecipitations with the S1 and S4 mutants do not indicate that the interactions are mutually exclusive. The mutant CC S4 exhibits a strong reduction in binding RanGAP2 but does not show a clear increase in its interaction with the NB-ARC-LRR (Fig. 5A), and the CC S1 does not seem to bind more RanGAP2 than the wild-type CC, even though its intramolecular interactions with the NB-ARC and LRR are reduced (Fig. 6A). On the other hand, the expression of RanGAP2 with the coexpressed segments H1-H2 and H3-H4 strongly enhances the interaction between these two segments (Fig. 6B), and if this mechanism acts in the full-length Rx1, it would stabilize the four-helix bundle structure of the CC. Previous findings support this notion: the overexpression of RanGAP2 has been shown to increase the stability of full-length Rx1, and the silencing of RanGAP2 leads to a lower level of Rx1 in the cell (see Fig. 6 in Tameling et al., 2010). One could envision that the binding of RanGAP2 to the CC alters its conformation or stability, and the RanGAP2-bound and free pools of Rx1 in the cell differ in activity or stability.
The coexpression of CC, CC-NB-ARC, NB-ARC, or LRR with full-length Rx1 leads to a dominant negative suppression of Rx1-mediated resistance and cell death (Fig. 8; Supplemental Fig. S10A). The dominant negative activities of truncated proteins usually are ascribed to the competition of the truncated protein with the full-length protein for binding partners required for signaling; the truncated protein sequesters these binding partners in a complex that is not signaling competent. Therefore, dominant negative activities can be informative on the functional mechanisms of a protein. The suppressive activity by the CC domain appears specific; coexpression of the CC domain of Bs2 does not affect the activity of Rx1 (Fig. 8B). The suppressive effect of the CC of Rx1 likely works via a specific interaction that the CC of Bs2 cannot establish. Comparing the suppressive activity of the wild-type CC with that of the S1 and S4 mutants makes clear that the suppression does not act via an intramolecular interaction. The CC S1 has no detectable interaction with the NB-ARC and LRR but still exhibits a pronounced suppressive effect, indistinguishable from the wild-type CC (Fig. 8, C and D). This sets the suppressive effect of the Rx1 CC apart from the suppression of the autoactivity of chimeric constructs of the tomato (Solanum lycopersicum) resistance protein Mi-1.2 against the root knot nematode Meloidogyne incognita and its nonfunctional homolog Mi-1.1 by the coexpressed N-terminal SD domain of Mi-1.2. The activity of these chimeric Mi constructs was suppressed only by an N-terminal domain originating from the same Mi protein as the LRR in the chimeric construct (Hwang et al., 2000; Hwang and Williamson, 2003). In contrast to the Rx1 CC S1 construct, the CC S4 and CC-NB-ARC S4 constructs lost the ability to suppress the activity of full-length Rx1. This implies that the residues in helix 4 of the CC that are required for the interaction with RanGAP2 also are required for the suppressive effect of the CC. If the suppressive effect depends on competition for an interaction partner, then RanGAP2 is a candidate, although other yet unknown interactors cannot be excluded. If they form a complex with Rx1, the NRC-like helper NLRs, which are required for signaling by Rx1 and related NLRs (Wu et al., 2017), also might be candidates for which sequestering in a nonfunctional complex would affect resistance. In this context, it is interesting that coexpression of the CC of Bs2, which shows a similar dependence on NRCs as Rx1, does not affect the resistance mediated by Rx1 (Fig. 8B). Blue Native PAGE indicates that most of the cytoplasmic wild-type CC is complexed with RanGAP2 and that the CC S4 products are predominantly monomeric (Fig. 7A). Blue Native PAGE analysis does not take the nuclear pool of the CC into account, and previous observations of a high nuclear accumulation and reduced diffusion speed of the CC inside the nucleus suggest that the CC has nuclear interactors besides RanGAP2 (Slootweg et al., 2010). The CC-NB-ARC of Rx1 has been shown to interact directly with DNA (Fenyk et al., 2015), and recently, one nuclear interactor has been identified as a Golden2-like transcription factor (Townsend et al., 2018).
Several mutants or truncated versions of NB-LRR proteins are known to have a dominant negative effect. Loss-of-function mutants of Arabidopsis RPS2 with alterations in the CC domain suppress the activity of the wild-type RPS2 expressed in the same Arabidopsis background, whereas loss-of-function mutants with changes in the nucleotide-binding site do not display a suppressive effect (Tao et al., 2000). Substitution E572K in the conserved third LRR inactivates the Arabidopsis CC-NB-LRR RPS5 (rps5-1) but, surprisingly, also suppresses a subset of other NB-LRRs in Arabidopsis not limited to CC-NB-LRR proteins (Warren et al., 1998; Bittner-Eddy et al., 2000; Sinapidou et al., 2004). A mutation in the 12th repeat of the LRR also inactivates RPS5 (rps5-2), but the expression of this mutant does not affect other resistance proteins. Similarly, autoactive variants of the tomato NB-LRR Prf are suppressed when the LRR of Prf is coexpressed. This suppression is specific: a similar autoactive mutant of the resistance protein Rpi-blb1 from Solanum bulbocastanum is not suppressed by the LRR of Prf. Interestingly, coexpression of the LRR leads to a destabilization of the full-length Prf (Du et al., 2012). There does not appear to be a difference in stability in the full-length Rx1 mutants coexpressed with a wild-type CC construct that coincides with the ability to interact (Fig. 5D). However, to conclude if coexpression of the CC affects the stability of full-length Rx1, a direct comparison of the protein levels of full-level Rx1 in the presence and absence of coexpressed CC would be necessary. Full-length CC-NB-LRR proteins encoded by Pm3 alleles in wheat (Triticum aestivum) can suppress the resistance against powdery mildew (B. graminis f. sp. tritici) mediated by the homologous protein Pm8 (Hurni et al., 2014) and by other Pm3 alleles (Stirnweis et al., 2014). The suppression appears to depend on the formation of heteromeric CC-NB-LRR/CC-NB-LRR complexes, although not all Pm3 variants that interact have a suppressive effect. The N-terminal half of the LRR is crucial in this suppression, and in contrast to what we observe for Rx1, the CC-NB-ARC of the Pm3 proteins does not suppress the activity of full-length proteins. Resistance genes often are located in clusters of homologous genes and subject to sequence exchange via unequal crossing over (Meyers et al., 2005). The dominant negative effects seen in examples as the Pm3 homologs or in the case of the truncated Rx1 proteins in our experiments show that the presence of genes encoding such proteins in these clusters poses a risk to resistance when their expression is not suppressed.
In this study, we observed that some of the mutants lost their ability to initiate PVX resistance but still responded with a wild-type-like cell death response to the PVX elicitor (CP). This is the case when CC-NB-ARC variants containing the substitutions of the aromatic residues in helix 1 (S1) only, or in combination with the substitution of aromatic residues in helix 4 (S14), are coexpressed with the LRR (Supplemental Fig. S4B). A similar loss of resistance, but not cell death, has been seen in the series of deletion constructs of the Rx1 CC tested by Rairdan et al. (2008). At least one truncation (construct C2: Rx1 1-86, lacking most of helix 4) resulted in a loss of resistance but not of elicitor-dependent cell death in trans, which could be interpreted as a quantitative difference in the immune response. However, we also observed the reverse phenotype. The full-length Rx1 Z2 construct, in which hydrophobic residues in helix 2 are replaced by Glu, conferred PVX resistance in a transient assay, which was indistinguishable from the response of the wild-type construct but responded consistently with a reduced cell death response to the PVX CP (Fig. 2A). This suggests that the pathways leading to the cell death response or the response that provides resistance against PVX are not identical and are affected differently by the Z2 mutations.
The NB domain of Rx1 induces an autoactive cell death response when expressed as a free domain (Rairdan et al., 2008). When tested for dominant negative activity, the NB did not have a suppressive effect on Rx1-mediated resistance, unlike the CC, LRR, and NB-ARC constructs (Supplemental Fig. S10, B–D). Maybe more surprisingly, it also did not enhance virus resistance like the H1-H2 segment of the CC does. If expressed transiently with PVX, a cell death response occurs, as it does in the absence of PVX, but the replication of PVX is not hampered markedly by the autoactive response of the NB domain (Supplemental Fig. S10D). Further study is required, but this lack of effect on PVX accumulation suggests that the NB-mediated response is limited to cell death and that other domains, like the CC, are required for a full resistance response.
Previous studies have indicated that the Rx1-induced virus resistance and the cell death response might indeed be distinct responses (Kohm et al., 1993; Bendahmane et al., 1999). The antiviral pathway includes the Argonaute4-dependent inhibition of the translation of viral RNAs (Bhattacharjee et al., 2009). Rx1-mediated resistance was called extreme resistance because its highly efficient inhibition of viral replication often did not include a cell death response. Only when resistance is less efficient, as for example after the silencing of RanGAP2, do necrotic spots accompany the response (Tameling and Baulcombe, 2007). It will be interesting to see whether the phenotype observed in this study, in which cell death and virus resistance are uncoupled, can be linked to different downstream signaling pathways that are initiated by distinct CC-dependent configurations of Rx1. The R protein MLA10 can no longer induce cell death if forced into the nucleus but still is able to confer resistance (Bai et al., 2012). The autoactive cell death induction by the CC domains of MLA10, Sr33, and Sr50 also requires a cytosolic localization, and exclusion from the nucleus via the fusion of a nuclear export signal even appears to enhance the cell death response for Sr50 (Cesari et al., 2016). These examples suggest that the pathways resulting in cell death and other defense responses are linked to different subcellular compartments (Heidrich et al., 2012).
Previously, we have shown that the subcellular localization of Rx1 must be balanced between the cytoplasm and the nucleus for full resistance and that the PVX coat protein is recognized in the cytoplasm and not in the nucleus (Slootweg et al., 2010). Here, we show that proper folding of the CC domain is required for the accumulation of Rx1 in the nucleus, as disruption of the CC structure by the introduction of hydrophobic zipping mutations results in a shift of Rx1 toward a more cytoplasmic localization. The overall integrity of the CC domain is required to maintain a balanced nucleocytoplasmic distribution pattern, in line with our previous finding that a truncated Rx1 lacking the CC domain is mostly absent from the nucleus. The expression level of the Rx variants in this study is higher than that of the nucleus- or cytoplasm-targeted Rx construct in our previous study, which might explain why the nuclear exclusion of the CC mutants does not always correspond with a loss of functioning. Nuclear targeting of Rx1 or Rx1 S1 via the coexpression of WPP-NLS resulted in reduced PVX resistance (Fig. 6E), further supporting the role of the cytoplasmic Rx1 protein pool in recognition and signaling.
MATERIALS AND METHODS
DNA Constructs and Mutagenesis
The potato (Solanum tuberosum) mutations were introduced into the sequence of the Rx1 CC by DNA synthesis. The following codon changes were introduced in the coding sequence of Rx to create the mutants: Z1, 16-GTT-18 to GAG (V6E), 25-CTT-27 to GAG (L9E), and 37-ATA-39 to GAA (I13E); Z2, 91-CTC-93 to GAG (L31E), 100-TTG-102 to GAG (L34E), and 109-ATT-111 to GAG (I37E); Z3, 160-TTG-162 to GAG (L54E), 172-ATC-174 to GAG (I58E), and 175-GTA-177 to GAA (V61E); Z4, 274-ATTA-276 to GAG (I92E), 286-CTG-288 to GAG (L96E), and 298-CTA-300 to GAG (L100E); S1, 7-TAT-9 to GCC (Y3A) and 28-ATG-30 to GCG (M10A); S4, 268-TGG-270 to GCG (W90A), 277-TTT-279 to GCA (F93A), and 280-TTC-282 to GCT (F94A). The altered sequences were cloned into the constructs encoding segments of Rx (amino acids 1–45, 45–116, 1–142, and 1–473) or the full-length Rx sequence, which have been described previously as GFP-tagged constructs (Slootweg et al., 2010). Here, the GFP sequence was replaced by either a 4× HA tag or a 4× c-Myc tag.
Plant Material, Transient Expression, and PVX Virus Resistance Assay
For expression in plants, the constructs were cloned into the binary vector pBINPLUS (van Engelen et al., 1995). Agrobacterium tumefaciens MOG101 containing the vector was infiltrated in the leaves of Nicotiana benthamiana, and leaf material was harvested 2 d after infiltration for further analysis of the expressed protein, as described previously (Slootweg et al., 2010). Transient PVX resistance assays were performed by coinfiltrating A. tumefaciens carrying the Rx1 constructs (OD600 = 0.05) with A. tumefaciens carrying a PVX:GFP amplicon (pGR208 [OD600 = 0.002]; Peart et al., 2002). At 5 dpi, the leaves were harvested and an extract (24 mg of leaf material in 250 μL of 50 mm phosphate buffer, pH 7) was incubated in anti-PVX CP antibody (Prime Diagnostics)-coated plates. After stringent washing, the presence of PVX was detected using alkaline phosphatase-conjugated anti-PVX antibodies. p-Nitrophenol formation was measured in a plate reader (Bio-Rad model 680 microplate reader) via its absorption at a wavelength of 405 nm. As a measure for the strengths of the cell death response, the loss of chlorophyll was determined in the leaf area transiently expressing the tested constructs (modified from Harris et al. [2013]). A 13-mm-diameter leaf disc was collected per agroinfiltration spot at 3 to 5 d after infiltration and incubated overnight in 500 μL of dimethyl sulfoxide. From this volume, 150 μL of chlorophyll extract was transferred to a clear 96-well plate, and the absorption at 655 nm was measured in a plate reader. IBM SPSS Statistics 22 & 24 software was used to statistically analyze data from the ELISA and chlorophyll assays.
Confocal Microscopy
Enhanced GFP- or the monomeric red fluorescent protein mCherry (Shaner et al., 2004)-tagged proteins were transiently expressed in N. benthamiana and imaged at 2 d after agroinfiltration using a Zeiss LSM 510 confocal microscope. The 488-nm line from an argon-ion laser was used for the excitation of GFP, and the 543-line from a HeNe laser was used for the excitation of mCherry. GFP emission was detected after a 505- to 530-nm band-pass filter. Chloroplast autofluorescence excited by the 488-nm line was detected through a 650-nm long-pass filter. The emission from mCherry was detected through a 600- to 650-nm band-pass filter. Images were analyzed with ImageJ software (Abramoff et al., 2004).
Immunoprecipitation and Protein Analysis
Protein was extracted from N. benthamiana leaves by grinding them in extraction buffer (50 mm Tris-HCl, pH 7.5, 10% [v/v] glycerol, 150 mm NaCl, 1 mm EDTA, 2% [w/v] polyclar-AT polyvinylpolypyrrolidone, 0.4 mg mL−1 Pefabloc SC plus [Roche], and 5 mm DTT) on ice. Prior to immunoprecipitation, the total protein extract was passed through a Sephadex G-25 column. The extract was precleared with rabbit-IgG agarose (40 μL slurry mL−1 protein extract). After preclearing, the extract was mixed with 25 μL of anti-Myc agarose beads (Sigma A7470) or anti-HA agarose beads (Roche 11815016001, 3F10) and incubated for 2 h at 4°C. After 6× washing in protein extraction buffer to which 0.15% (v/v) Igepal CA-630 was added, the beads were resuspended in Laemmli buffer and the bound protein was separated by SDS-PAGE and blotted. c-Myc-tagged protein was detected by Abcam 9132 rabbit anti-c-Myc (1:5,000) as the primary antibody and Jackson 705-035-147 peroxidase-conjugated donkey anti-rabbit (1:10,000) as the secondary antibody. HA-tagged proteins were detected with a peroxidase-conjugated anti-HA antibody (Roche 10836800, 1:500), GFP with Abcam AB 290 (1:2,000) or Miltenyi 130-091-833 anti-GFP-HRP (1:2,000), and mCherry with Abcam AB 34767 HRP conjugate (1:500). The peroxidase activity was visualized with the luminescent substrate SuperSignal West Dura & Femto (Techno Pierce) and imaged by the Syngene G:Box imaging system.
Blue Native Gel Electrophoresis
The protocol used for the Blue Native gel electrophoresis was adopted from the protocol provided with the NativePAGE system (Life Technologies). Protein combinations were expressed in N. benthamiana leaves via agroinfiltration and extracted by grinding 100 mg of leaf sample frozen in liquid nitrogen and resuspending in 1 mL of native sample buffer (50 mm Bis-Tris, 25 mm NaCl, 10% [w/v] glycerol, 0.001% Ponceau S, and 5 mm DTT, pH 7.2). Fifteen microliters of the soluble fraction was loaded on a cooled NativePAGE 4-16% Bis-Tris gel. The gradient gel was run at 150 to 250 V with the 1× NativePAGE anode buffer and the Light Blue NativePAGE cathode buffer. After running the gel, the protein was blotted on a polyvinylidene difluoride membrane that was incubated subsequently in 8% acetic acid, dried, and rewetted in methanol. The rewetted blot was developed further via standard immunoblotting protocols. The NativeMark unstained protein standard (Life Technologies) and Rubisco complex were visualized by staining the blots with Coomassie Brilliant Blue R250 after the tagged proteins detected by the anti-GFP or anti-HA antibodies had been visualized.
Accession Numbers
GenBank accession numbers are as follows: Rx1 (Solanum tuberosum), AJ011801; RanGAP2 (Nicotiana benthamiana), EF396237.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Stability of the full-length Rx1 mutant constructs.
Supplemental Figure S2. Effect of overexpressed Rx1 on the replication of TMV as a test for autoactive resistance responses in wild-type and mutant Rx1.
Supplemental Figure S3. Homodimerization of the H3-H4 segment of the Rx1 CC.
Supplemental Figure S4. A loss of interaction between domains in trans (CC/NB-ARC-LRR and CC-NB-ARC/LRR) results in a loss of resistance and cell death in trans.
Supplemental Figure S5. Interaction of full-length Rx1 variants with the RanGAP2 WPP domain or full-length RanGAP2.
Supplemental Figure S6. Yeast two-hybrid assay to test the interaction between the WPP domain of RanGAP2 and the variants of the Rx1 CC in the absence of additional plant proteins.
Supplemental Figure S7. Interaction of the H3-H4 segment of the Rx1 CC with the WPP domain of RanGAP2.
Supplemental Figure S8. Effect of RanGAP2 coexpression on the localization of Rx1 S1 and S4 tested via cell fractionation.
Supplemental Figure S9. Analysis of complex formation by the Rx1 CC variants (wild type, S1, and S4) and the WPP domain of RanGAP2 via Blue Native gel electrophoresis.
Supplemental Figure S10. The Rx1 NB-ARC and LRR domains exhibit a suppressive activity on Rx1-mediated resistance.
Supplemental Figure S11. CC mutations mapped on a hypothetical dimer of the Rx1 CC.
Supplemental Figure S12. Size-exclusion chromatography analysis of the purified CC domain of Rx1 (amino acids 1–144).
Supplemental Figure S13. Structural alignment of the amino acid sequences of the CC domains and P-loop motifs of the NB domain of the proteins Rx1, Bs2, MLA10, Sr33, and Sr50.
Footnotes
This work was supported by the EU-funded Integrated Project Bioexploit, the Dutch Top Technology Institute Green Genetics, the Dutch Technology Foundation STW and Earth and Life Sciences ALW, which are part of the Netherlands Organization for Scientific Research (NWO), the UEFISCDI grant PN-III-ID-PCE-2016-0650, projects 1 and 3 of IBAR from the Romanian Academy, and Biotechnology and Biological Sciences Research Council Grant BB/M007405/1. This work also benefited from interactions through the COST Action SUSTAIN FA1208.
Articles can be viewed without a subscription.
References
- Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–41 [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FLW, et al. (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Köhn BA, Dedi C, Baulcombe DC (1995) The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J 8: 933–941 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bernoux M, Ellis JG, Dodds PN (2011a) New insights in plant immunity signaling activation. Curr Opin Plant Biol 14: 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN (2011b) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Burdett H, Williams SJ, Zhang X, Chen C, Newell K, Lawrence GJ, Kobe B, Ellis JG, Anderson PA, et al. (2016) Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28: 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Zamora A, Azhar MT, Sacco MA, Lambert LH, Moffett P (2009) Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J 58: 940–951 [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J 21: 177–188 [DOI] [PubMed] [Google Scholar]
- Casey LW, Lavrencic P, Bentham AR, Cesari S, Ericsson DJ, Croll T, Turk D, Anderson PA, Mark AE, Dodds PN, et al. (2016) The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc Natl Acad Sci USA 113: 12856–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Moore J, Chen C, Webb D, Periyannan S, Mago R, Bernoux M, Lagudah ES, Dodds PN (2016) Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc Natl Acad Sci USA 113: 10204–10209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH (2013) Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25: 1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C, Parry DAD (1986) Alpha-helical coiled coils: a widespread motif in proteins. Trends Biochem Sci 11: 245–248 [Google Scholar]
- Cohen C, Parry DA (1990) Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins 7: 1–15 [DOI] [PubMed] [Google Scholar]
- Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Crick FHC. (1953) The packing of α-helices: simple coiled-coils. Acta Crystallogr 6: 689–697 [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW (2012) Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Tham WH, Baker BJ (2000) Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc Natl Acad Sci USA 97: 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Miao M, Ma X, Liu Y, Kuhl JC, Martin GB, Xiao F (2012) Plant programmed cell death caused by an autoactive form of Prf is suppressed by co-expression of the Prf LRR domain. Mol Plant 5: 1058–1067 [DOI] [PubMed] [Google Scholar]
- El Kasmi F, Chung EH, Anderson RG, Li J, Wan L, Eitas TK, Gao Z, Dangl JL (2017) Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proc Natl Acad Sci USA 114: E7385–E7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenyk S, Townsend PD, Dixon CH, Spies GB, de San Eustaquio Campillo A, Slootweg EJ, Westerhof LB, Gawehns FKK, Knight MR, Sharples GJ, et al. (2015) The potato nucleotide-binding leucine-rich repeat (NLR) immune receptor Rx1 is a pathogen-dependent DNA-deforming protein. J Biol Chem 290: 24945–24960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Collier SM, Moffett P, Chai J (2013) Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J Biol Chem 288: 35868–35876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci USA 110: 21189–21194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K, Blanvillain-Baufumé S, Parker JE (2012) Molecular and spatial constraints on NB-LRR receptor signaling. Curr Opin Plant Biol 15: 385–391 [DOI] [PubMed] [Google Scholar]
- Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA, Keller B (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79: 904–913 [DOI] [PubMed] [Google Scholar]
- Hwang CF, Williamson VM (2003) Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J 34: 585–593 [DOI] [PubMed] [Google Scholar]
- Hwang CF, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM (2000) Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12: 1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW. (2004) Guarding the goods: new insights into the central alarm system of plants. Plant Physiol 135: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohm BA, Goulden MG, Gilbert JE, Kavanagh TA, Baulcombe DC (1993) A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik-Shreepaathy E, Slootweg E, Richter H, Goverse A, Cornelissen BJ, Takken FL (2012) Dual regulatory roles of the extended N terminus for activation of the tomato MI-1.2 resistance protein. Mol Plant Microbe Interact 25: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Mazourek M, Cirulli ET, Collier SM, Landry LG, Kang BC, Quirin EA, Bradeen JM, Moffett P, Jahn MM (2009) The fractionated orthology of Bs2 and Rx/Gpa2 supports shared synteny of disease resistance in the Solanaceae. Genetics 182: 1351–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre P, Baulcombe DC (2006) Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8: 129–134 [DOI] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC (2002) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J 29: 569–579 [DOI] [PubMed] [Google Scholar]
- Qi D, Dubiella U, Kim SH, Sloss DI, Dowen RH, Dixon JE, Innes RW (2014) Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on S-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol 164: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P (2008) The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco MA, Mansoor S, Moffett P (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Sacco MA, Koropacka K, Grenier E, Jaubert MJ, Blanchard A, Goverse A, Smant G, Moffett P (2009) The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tör M, Crute I, Bittner-Eddy P, Beynon J (2004) Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J 38: 898–909 [DOI] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg EJ, Spiridon LN, Roosien J, Butterbach P, Pomp R, Westerhof L, Wilbers R, Bakker E, Bakker J, Petrescu AJ, et al. (2013) Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol 162: 1510–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnweis D, Milani SD, Brunner S, Herren G, Buchmann G, Peditto D, Jordan T, Keller B (2014) Suppression among alleles encoding nucleotide-binding-leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J 79: 893–903 [DOI] [PubMed] [Google Scholar]
- Sukarta OCA, Slootweg EJ, Goverse A (2016) Structure-informed insights for NLR functioning in plant immunity. Semin Cell Dev Biol 56: 134–149 [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Albrecht M, Tameling WIL (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Tameling WIL, Baulcombe DC (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJC, Takken FLW (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12: 2541–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend PD, Dixon CH, Slootweg EJ, Sukarta OCA, Yang AWH, Hughes TR, Sharples GJ, Pålsson LO, Takken FLW, Goverse A, et al. (2018) The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. J Biol Chem 293: 3218–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG (1998) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Wang GF, Ji J, El-Kasmi F, Dangl JL, Johal G, Balint-Kurti PJ (2015) Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog 11: e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SJ, Sornaraj P, deCourcy-Ireland E, Menz RI, Kobe B, Ellis JG, Dodds PN, Anderson PA (2011) An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact 24: 897–906 [DOI] [PubMed] [Google Scholar]
- Wittig I, Schägger H (2009) Native electrophoretic techniques to identify protein-protein interactions. Proteomics 9: 5214–5223 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H (2006) Blue native PAGE. Nat Protoc 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S (2017) NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci USA 114: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]