Two missing enzyme activities in the amygdalan pathway result in sweet versus formation of bitter almonds.
Abstract
Almond (Prunus dulcis) is the principal Prunus species in which the consumed and thus commercially important part of the fruit is the kernel. As a result of continued selection, the vast majority of almonds have a nonbitter kernel. However, in the field, there are trees carrying bitter kernels, which are toxic to humans and, consequently, need to be removed. The toxicity of bitter almonds is caused by the accumulation of the cyanogenic diglucoside amygdalin, which releases toxic hydrogen cyanide upon hydrolysis. In this study, we identified and characterized the enzymes involved in the amygdalin biosynthetic pathway: PdCYP79D16 and PdCYP71AN24 as the cytochrome P450 (CYP) enzymes catalyzing phenylalanine-to-mandelonitrile conversion, PdUGT94AF3 as an additional monoglucosyl transferase (UGT) catalyzing prunasin formation, and PdUGT94AF1 and PdUGT94AF2 as the two enzymes catalyzing amygdalin formation from prunasin. This was accomplished by constructing a sequence database containing UGTs known, or predicted, to catalyze a β(1→6)-O-glycosylation reaction and a Basic Local Alignment Search Tool search of the draft version of the almond genome versus these sequences. Functional characterization of candidate genes was achieved by transient expression in Nicotiana benthamiana. Reverse transcription quantitative polymerase chain reaction demonstrated that the expression of PdCYP79D16 and PdCYP71AN24 was not detectable or only reached minute levels in the sweet almond genotype during fruit development, while it was high and consistent in the bitter genotype. Therefore, the basis for the sweet kernel phenotype is a lack of expression of the genes encoding the two CYPs catalyzing the first steps in amygdalin biosynthesis.
In 1802, the Berlin pharmacist Bohm discovered that the toxic principle released from bitter almonds (Prunus dulcis syn. Prunus amygdalus) upon distillation was hydrogen cyanide (Lechtenberg and Nahrstedt, 1999). It took 28 more years before the chemists Robiquet and Bourton-Chalard identified the hydrogen cyanide-releasing compound in bitter almonds as a cyanogenic glucoside (Robiquet and Boutron-Chalard, 1830). The compound was named amygdalin, reflecting its isolation from bitter almonds. Since then, cyanogenic glucosides have been established as a class of bioactive natural products distributed widely within the plant kingdom. In the detective novel Sparkling Cyanide, published in 1945 by Agatha Christie, a beautiful heiress is fatally poisoned by cyanide added to her glass of champagne. The popularity of this masterpiece detective novel greatly contributed to the general awareness of hydrogen cyanide as a poison, used to commit murders or suicides. This practice was already established in ancient Egypt in the second millennium before Christ, where traitorous priests in the capital cities of Memphis and Thebes were poisoned to death with pits of peaches, referenced in hieroglyphics as “death by peach” (Davis, 1991; Blum, 2010). The domestication of almonds occurred ∼5,000 years ago in the Fertile Crescent, with global dissemination beginning before 1300 BCE (Velasco et al., 2016). Almonds were found in the Franchthi Cave in Greece and in the tomb of Tutankhamen (Hansen and Renfrew, 1978; Zohary et al., 2012). Caius Plinius Secundus (Pliny the Elder) wrote in his 37-volume encyclopedia entitled Naturalis Historiae that the Romans were proud of knowing how to remove the bitterness and toxicity from bitter almond kernels (Pliny the Elder, 77AD). It remains unclear when and where sweet almond genotypes were first developed.
Amygdalin is a common constituent in plant species within the Rosaceae family and is present in stone fruits such as apricot (Prunus armeniaca), peach (Prunus persica), cherry (Prunus avium), and Japanese apricot (Prunus mume) and in pomaceous fruits like apples (Malus domestica). Amygdalin has a bitter taste. In stone fruits like plums (Prunus domestica), cherries, peaches, nectarines (Prunus persica), and apricots, the outer fleshy part of the fruit (mesocarp) is consumed and the pit with the kernel frequently is discarded. In these Prunus species, a high amygdalin content in the kernels does not have a negative effect on product quality.
The situation is different in almonds, where the kernel is the edible part of the fruit. The content of amygdalin may be up to 1,000-fold higher in bitter compared with sweet almond kernels (Dicenta et al., 2002; Sánchez-Pérez et al., 2008). The toxicity issues related to high kernel bitterness renders the sweet kernel trait a prime breeding trait in almonds. At present, the selection of almond trees with a sweet genotype is based on checking the bitter/sweet kernel taste. This is a slow and tardy process that can delay breeding, as breeders must wait out a long juvenile period (2–3 years) before the first flowering and fruit set occurs. The elucidation of amygdalin biosynthesis and its regulatory components will greatly enhance the understanding of the genetic differences between sweet and bitter almond genotypes. This, in turn, will guide the development and setup of marker-assisted selection to enable the differentiation between sweet/bitter phenotypes at the early seedling stage. In 2010, two molecular markers bordering the Sweet kernel (Sk) locus in linkage group 5 were characterized. These markers are effective only in a limited number of almond offspring, where the progenitors have been studied in detail (Sánchez-Pérez et al., 2010). Today, we know that the Sk locus is located in a genomic interval in scaffold 5, flanked by the two markers ppa006282/HpyCH4CV and CPDCT028, which, in peach, corresponds to a physical region of 150 kb (Ricciardi et al., 2018).
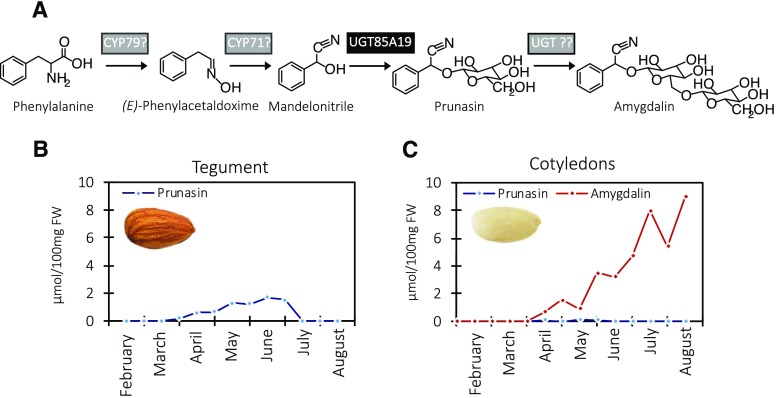
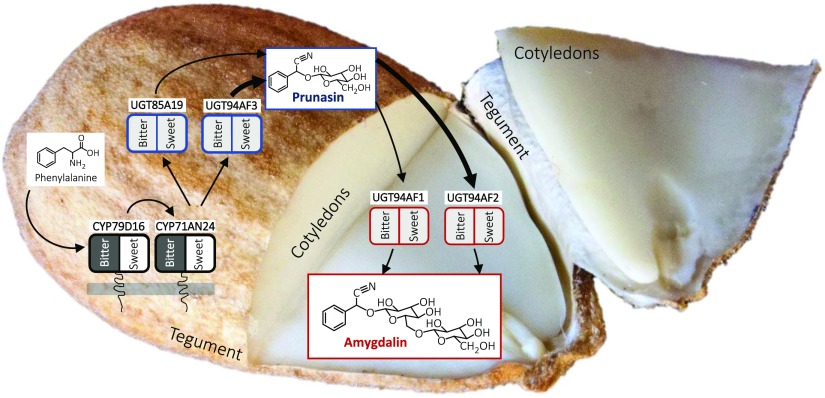
Amygdalin is a cyanogenic diglucoside. As stated above, it accumulates in high amounts in bitter almond kernels (Dicenta et al., 2002; Sánchez-Pérez et al., 2008). Prunasin is a cyanogenic monoglucoside present in all vegetative parts of the almond tree, such as leaf, stem, shoot, and root, as well as in the developing kernels. The prunasin levels in these tissues are independent of the amygdalin content and, therefore, cannot be used to predict kernel bitterness (Dicenta et al., 2002; Sánchez-Pérez et al., 2008). Prunasin, as well as amygdalin, are produced from the amino acid Phe (Fig. 1). Cyanogenic glucosides function as a two-component herbivore defense system. Upon mechanical disruption of the plant tissue, the cyanogenic glucosides are hydrolyzed by the sequential action of β-glucosidases and α-hydroxynitrilases, resulting in the formation of Glc, aldehyde or ketones, and toxic hydrogen cyanide (Morant et al., 2008; Gleadow and Møller, 2014). In almond, the hydrolytic pathway is catalyzed by amygdalin hydrolase, prunasin hydrolase, and mandelonitrile lyase (Gleadow and Møller, 2014; Del Cueto et al., 2018). The localization of the biosynthetic and hydrolytic enzymes within the tegument and cotyledon is based on earlier studies in almond and black cherry (Prunus serotina) documenting that, in almond, prunasin biosynthesis takes place in the tegument and amygdalin is formed in the cotyledons (Fig. 1; Kuroki and Poulton, 1987; Sánchez-Pérez et al., 2008, 2009, 2012).
Figure 1.
The intermediates in the biosynthesis of prunasin and amygdalin in bitter almonds (A), with product formation in the tegument (B) and cotyledons (C) shown during fruit development. In the first reaction, Phe is converted to mandelonitrile by two CYPs belonging to the CYP79 and CYP71 families. The mandelonitrile is glucosylated by the previously known UGT85A19 enzyme (Franks et al., 2008). Amygdalin is glucosylated further by an unknown UGT. The enzymes identified in this study are shown on a gray background, whereas the previously described UGT85A19 is shown on a black background. Prunasin and amygdalin contents were analyzed by liquid chromatography-mass spectrometry (LC-MS) in tegument (B) and cotyledons (C) during fruit development (February to August, analyzed every second week), which represents the period from flowering to ripening (modified from Sánchez-Pérez et al. [2008]). FW, Fresh weight.
The intermediates in the biosynthetic pathway of cyanogenic monoglucosides are conserved from arthropods to higher plants (Jensen et al., 2011; Gleadow and Møller, 2014). Specifically, in the higher plant species studied, the pathway is typically catalyzed by two cytochrome P450s (CYPs) and one or more UDP glycosyltransferases (UGTs; Fig. 1). The two CYPs are members of the CYP71 clan. The first P450 belongs to the CYP79 family and catalyzes two sequential N-hydroxylations of the parent amino acid and a decarboxylation and dehydration reaction resulting in the formation of the corresponding E-oxime (Fig. 1; Koch et al., 1995; Sibbesen et al., 1995; Andersen et al., 2000; Forslund et al., 2004; Clausen et al., 2015; Knoch et al., 2016). The second P450 belongs to the CYP71 or CYP736 family and catalyzes the rearrangement of the E-oxime into the Z-oxime, its dehydration, and a final C-hydroxylation reaction to produce a cyanohydrin (α-hydroxynitrile; Kahn et al., 1997, 1999; Bak et al., 1998a; Kannangara et al., 2011; Takos et al., 2011; Clausen et al., 2015; Knoch et al., 2016). Some eucalypt species also produce prunasin and prunasin-derived diglucosides (Neilson et al., 2011). In Eucalyptus cladocalyx, the conversion of the oxime to the cyanohydrin was shown recently to be catalyzed by two separate P450 enzymes (EcCYP706C55 and EcCYP71B103; Hansen et al., 2018). This represents yet another example of repeated evolution of this chemical defense pathway (Takos et al., 2011). In Japanese apricot, PmCYP79D16 and PmCYP71AN24 have been shown to catalyze the conversion of Phe to mandelonitrile in prunasin biosynthesis (Yamaguchi et al., 2014).
The cyanohydrin is chemically labile and toxic and is stabilized by glucosylation catalyzed by a UGT belonging to the UGT85 family. The UGT executing this reaction are as follows: in sorghum (Sorghum bicolor) it is SbUGT85B1, in barley (Hordeum vulgare) it is HvUGT85F22, in cassava (Manihot esculenta) they are MeUGT85K4 and MeUGT85K5, in lotus (Lotus japonicus) it is LjUGT85K3, and in almond it is PdUGT85A19, and their encoding genes all have been identified (Jones et al., 1999; Franks et al., 2008; Jørgensen et al., 2011; Kannangara et al., 2011; Takos et al., 2011; Knoch et al., 2016). As a member of the UGT85 family, UGT85A19 bears sequence resemblance to the other known UGT85s catalyzing the last step in cyanogenic monoglucoside formation. In sorghum, the two CYPs and the UGT catalyzing dhurrin formation are organized within a dynamic metabolon to ensure efficient substrate channeling and to avoid undesired metabolic cross talk and the leakage of labile and toxic intermediates (Møller and Conn, 1979; Laursen et al., 2016; Bassard et al., 2017).
In some plant species, the cyanogenic monoglucosides are glycosylated further into diglycosides, resulting in the formation of cyanogenic diglycosides like linustatin, neolinustatin, amygdalin, dhurrin-6-glucoside, eucalyptosin A, B, and C, and vicianin (Neilson et al., 2011; Gleadow and Møller, 2014; Pičmanová et al., 2015). In the majority of cases, the second sugar is incorporated via a β-1,6 linkage. Notably, the different eucalyptosins in Eucalyptus camphora are characterized by β-1,2, β-1,3, and β-1,4 linkages (Neilson et al., 2011). Different sugars also can be incorporated into cyanogenic diglycosides. For example, the second sugar in vicianin is arabinopyranose, whereas it is a glucopyranose unit in the other listed examples. The occurrence of apiosylated cyanogenic glycosides also has been reported (Pičmanová and Møller, 2016). A UGT enzyme catalyzing the formation of a cyanogenic diglycoside had not been identified previously. Although UGTs catalyzing the formation of cyanogenic diglycosides have remained elusive, members of several UGT families, namely UGT94, UGT79, and UGT91, have been documented or hypothesized to catalyze β(1→6)-O-glycosylation of other monoglucoside classes (Noguchi et al., 2008; Yonekura-Sakakibara and Hanada, 2011; Olsson et al., 2016), the equivalent reaction required to convert prunasin into amygdalin. For example, in furofuran lignin glucoside biosynthesis in sesame (Sesamum indicum), UGT94D1 produced (+)-sesaminol 2-O-β-d-glucosyl-(1→6)-O-β-d-glucoside.
In this study, we used the draft almond genome (Koepke et al., 2013) to find genes encoding the enzymes catalyzing the entire pathways for prunasin and amygdalin synthesis in almond. Reverse transcription quantitative PCR (RT-qPCR) analyses highlighted a dramatic difference in the expression of the two CYP biosynthetic genes between sweet and bitter almond genotypes.
RESULTS
Selection of Putative Genes Encoding the Enzymes Catalyzing Prunasin Biosynthesis in Almonds
CYP79 and CYP71 Candidate Genes
Relatedness and collinearity among the genomes of Prunus species such as almond, peach, and Japanese apricot (Dirlewanger et al., 2004; Jung et al., 2009) were exploited to characterize putative sequences and the chromosomal locations of candidate CYP genes involved in amygdalin biosynthesis in almond. Specifically, sequences of PmCYP79D16 and PmCYP71AN24, functionally shown to be involved in prunasin and amygdalin biosynthesis in Japanese apricot (Yamaguchi et al., 2014), were used for BLASTN searches against the draft almond genome sequence (Koepke et al., 2013). BLASTN of PmCYP79D16 resulted in the identification of a best-hit contig of 367,707 bp, including a greatest high-scoring segment pairs length of 976 bp with 99% identity. This 976-bp fragment was used to BLASTN against the peach genome version 1 (Verde et al., 2013) in Phytozome, resulting, as a best hit, in the annotated gene ppa021326m, located on chromosome 6 (Supplemental Table S1; Supplemental Fig. S1).
Similarly, BLASTN of PmCYP71AN24 resulted in the identification of a best-hit contig of 287,918 bp, including a greatest high-scoring segment pairs length of 876 bp with 100% identity. When BLASTed on the peach genome, this 876-bp sequence identified the annotated gene ppa004152m, located on chromosome 5, as the best hit (Supplemental Table S1; Supplemental Fig. S1). Based on the CYP nomenclature rules (http://drnelson.uthsc.edu/cytochromeP450.html), the two almond CYPs were assigned the same protein name as in Japanese apricot: PdCYP79D16 and PdCYP71AN24, respectively. A search in the National Center for Biotechnology Information (NCBI) database provided highly similar sequences within the Prunus genus and also within the Rosaceae family (Supplemental Table S2). An almond protein accession, named CYP79D16 (ADM34983.1), was found for which no functional assignment was provided. This sequence showed 99% identity to the PdCYP79D16 sequence characterized here. The encoded proteins were identical except for eight amino acid substitutions (N33K, E83Q, I95T, E184D, R187S, P317T, G510D, and L520F) when compared with Japanese apricot. The almond gene that encoded protein PdCYP71AN24 showed six single amino acid substitutions (L7F, Q52R, L343S, L371I, T399A, and A412E) compared with the Japanese apricot protein sequence.
UGT Candidate Genes
UGT85A19 was identified previously as the gene sequence encoding the UDPG-mandelonitrile glucosyltransferase catalyzing prunasin formation from mandelonitrile (Franks et al., 2008). This UGT-encoding gene is positioned on chromosome 1, according to the synteny between different Prunus species (Supplemental Table S1; Supplemental Fig. S1).
These data demonstrate that the genes encoding the three biosynthetic enzymes catalyzing prunasin formation do not cluster in Prunus species. Accordingly, it was not possible to exploit gene clusters as a tool for identifying the second UGT responsible for amygdalin formation (Takos et al., 2011; Knoch et al., 2016).
Selection of Putative Genes Encoding the Enzyme Catalyzing Amygdalin Formation from Prunasin in Almonds
Based on the aforementioned data, it was hypothesized that the UGT(s) catalyzing amygdalin formation in almond would belong to the UGT families 76, 79, 91, and 94. Therefore, to identify putative gene candidates, a UGT database was built based on gene sequences derived from these families (Noguchi et al., 2008; Yonekura-Sakakibara and Hanada, 2011). UGT73E1 from Stevia rebaudiana was used as an outgroup (Richman et al., 2005). A BLASTX search of the sweet and bitter genome assemblies (Koepke et al., 2013) against the UGT database resulted in the identification of 13 partial gene sequences encoding potentially interesting UGTs (Supplemental Table S3). Full-length gene sequences were obtained by RACE and demonstrated that six of the partial sequences could be assigned to three full-length UGT sequences. When probed against a cDNA library from cotyledons of the bitter genotype S3067 (Sc) or D05187 (Dc), five full-length UGT gene sequences were obtained as the favored candidates for encoding the UGT converting prunasin to amygdalin. Based on their sequences, the encoded UGTs were named PdUGT94AF1(Sc), PdUGT94AF2(Sc), PdUGT94AF3(Sc), PdUGT91R7(Sc), and PdUGT91C4(Dc) using the standardized nomenclature system (Mackenzie et al., 1997). The corresponding orthologs in peach were localized in the peach genome on chromosome 1 (PdUGT91C4 and PdUGT85A19), chromosome 2 (PdUGT94AF1, PdUGT94AF2, and PdUGT94AF3), and chromosome 6 (PdUGT91R7; Supplemental Table S1; Supplemental Fig. S1).
Functional Characterization of the Selected Gene Candidates in Planta
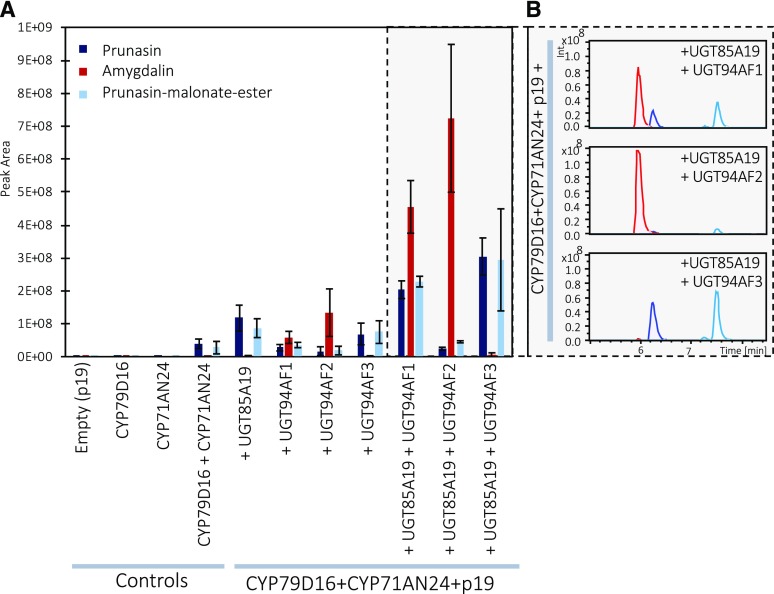
Functional characterization of the gene candidates PdCYP79D16, PdCYP71AN24, PdUGT85A19, PdUGT94AF1, PdUGT94AF2, PdUGT94AF3, PdUGT91R7, and PdUGT91C4 was performed by transient expression in Nicotiana benthamiana (Fig. 2). To avoid posttranscriptional gene silencing and obtain higher protein levels, agrobacteria harboring a vector containing the p19 gene were always coinfiltrated (Voinnet et al., 1999; Lakatos et al., 2004; Miller et al., 2004). The agroinfiltrated leaves were harvested 5 d after agroinfiltration. The products formed were extracted into methanol and identified by LC-MS analyses based on the use of prunasin (retention time [RT] 6.4 min, mass-to-charge ratio [m/z] 318) and amygdalin (RT 6 min, m/z 480) as reference compounds (Table 1).
Figure 2.
A, Functional characterization of the gene candidates PdCYP79D16, PdCYP71AN24, PdUGT85A19, PdUGT94AF1, PdUGT94AF2, and PdUGT94AF3 by transient expression in N. benthamiana and monitoring of product formation by LC-MS analysis and selected ion monitoring. When PdCYP79D16 and PdCYP71AN24 are coexpressed, small amounts of prunasin and prunasin-malonate ester can be detected, as compared with the control (p19). Amygdalin is detected only when at least three genes are coexpressed. B, Representative LC-MS recordings of the experiments shown on the gray background in A. Data are presented as means of three independent agroinfiltrated leaves ± sd.
Table 1. Substrate specificity of PdUGT94F1 and PdUGT94F2 with respect to their ability to glucosylate other cyanogenic glucosides.
The RT and m/z of infiltrated compounds and products formed are shown. n.d., Not detected.
| Infiltrated Substrate (100 µm) | m/za | RT | Expected Product Formed | m/za | RT |
|---|---|---|---|---|---|
| min | min | ||||
| Mandelonitrile | 133 | n.d. | Prunasin | 318 | 6.4 |
| Prunasin | 318 | 6.4 | Amygdalin | 480 | 6.0 |
| Amygdalin | 480 | 6.0 | Amygdalin-6-glucoside | 642 | n.d. |
| Dhurrin | 334 | 4.8 | Dhurrin-6-glucoside | 496 | 5.1 |
| Lotaustralin | 284 | 3.7, 4.3 | Lotaustralin-6-glucosideb | 446 | 4.7 |
| Linamarin | 270 | 1.8 | Linamarin-6-glucosidec | 432 | 1.6 |
The m/z value equals the mass of the sodium adduct.
Also known as neolinustatin.
Also known as linustatin.
In combination, PdCYP79D16 and PdCYP71AN24 are expected to catalyze the conversion of Phe to mandelonitrile, the aglycone of prunasin (Fig. 1). Coexpression of PdCYP79D16 and PdCYP71AN24 with PdUGT85A19 (Fig. 2) resulted in the formation of prunasin and a second component (RT 7.5 min, m/z 404) identified as a prunasin-malonate ester based on its MS and MS/MS spectra (Supplemental Fig. S2). This experiment demonstrated that PdCYP79D16 and PdCYP71AN24 catalyze the conversion of Phe to mandelonitrile, which, by the action of PdUGT85A19, was converted into prunasin (Franks et al., 2008). The expression of PdCYP79D16 and PdCYP71AN24 in the absence of any of the UGT candidate genes (Fig. 2) resulted in the formation of small amounts of prunasin. This demonstrated that the N. benthamiana leaves harbored an endogenous UGT activity able to glycosylate the mandelonitrile in a background reaction. The malonate ester formation also is catalyzed by an endogenous enzyme activity present in the N. benthamiana leaves. Separate expression of PdCYP79D16 would be expected to result in the production of (E)-phenylacetaldoxime (Clausen et al., 2015; Sørensen et al., 2018). In N. benthamiana, the free E-aldoxime is glucosylated rapidly, resulting in the accumulation of (E)-phenylacetaldoxime glucoside (Supplemental Fig. S3). Separate expression of PdCYP71AN24 did not result in any detectable products. The coexpression of PdCYP79D16 and PdCYP71AN24 resulted in the accumulation of minute levels of a mixture of (Z)- and (E)-phenylacetaldoxime glucosides (Supplemental Fig. S3). The formation of phenylacetonitrile was not detected.
The transient expression of PdUGT94AF1, PdUGT94AF2, or PdUGT94AF3 with the two P450s (Fig. 2) resulted in three distinct product profiles. The profile obtained with PdUGT94AF3 was nearly identical to that obtained with PdUGT85A19 (Fig. 2). PdUGT94AF1 and PdUGT94AF2 catalyzed the formation of amygdalin when coinfiltrated with PdCYP79D16 and PdCYP71AN24 alone (Fig. 2). The levels of amygdalin formed in these experiments were low and most likely dependent on the observed endogenous N. benthamiana UGT activity (Fig. 2). Upon simultaneous coinfiltration with PdUGT85A19, higher levels of amygdalin were formed (Fig. 2). This indicated that prunasin, and not mandelonitrile, was the preferred substrate for PdUGT94AF1 and PdUGT94AF2. Under the experimental conditions used based on transient expression in N. benthamiana, PdUGT94AF2 appeared to catalyze the formation of higher amounts of amygdalin compared with PdUGT94AF1 (Fig. 2). Introduction of PdUGT94AF3 with the two CYPs resulted in the formation of prunasin and a prunasin-malonate ester and only minute amounts of amygdalin (Fig. 2). This demonstrated that PdUGT94AF3 is a mandelonitrile glucosyltransferase. No catalytic activities were detected for PdUGT91C4 and PdUGT91R7 in any of the experiments (Supplemental Fig. S4). All compound assignments were verified by MS and MS/MS data as reported previously (Pičmanová et al., 2015). We conclude that PdCYP79D16, PdCYP71AN24, and PdUGT85A19/PdUGT94AF3 catalyze the conversion of Phe into prunasin in almond and that PdUGT94F1/PdUGT94F2 serve as diglucosyltransferases catalyzing the conversion of prunasin into amygdalin.
Gene Expression Levels in Relation to Amygdalin Formation in Sweet and Bitter Almond Genotypes throughout Fruit Development
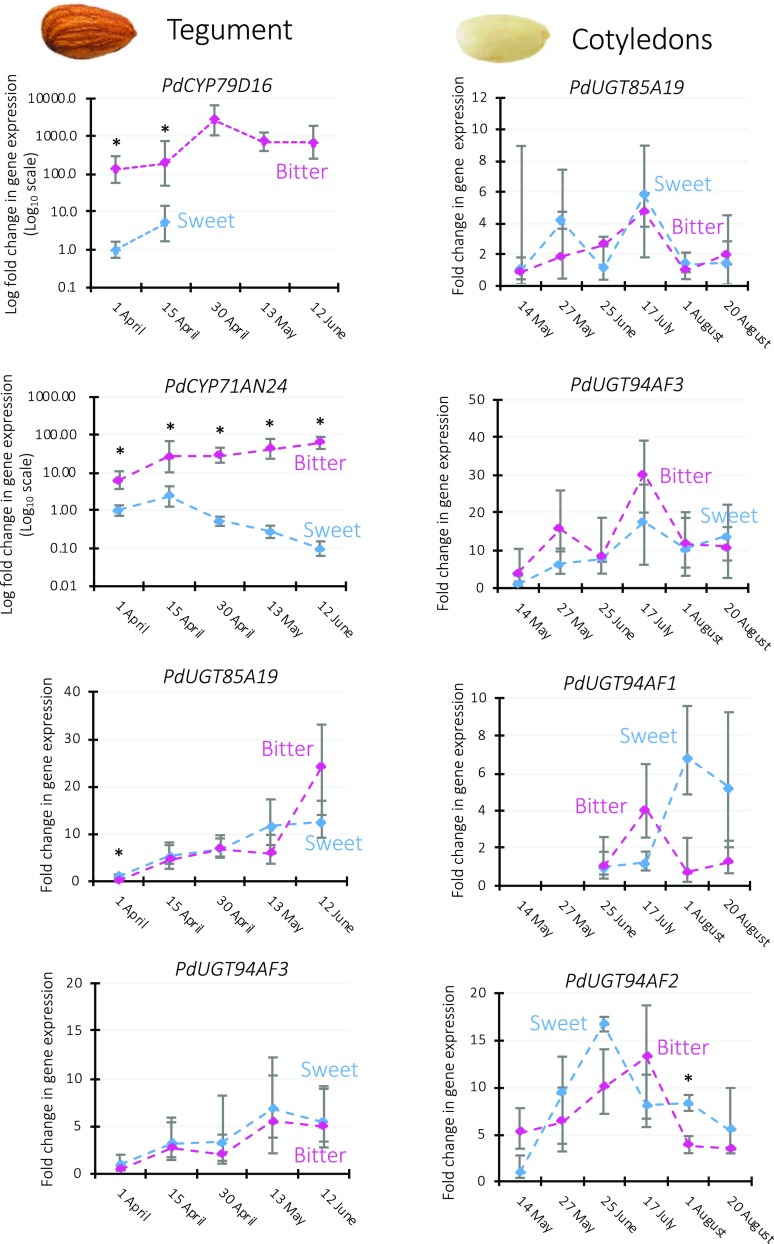
The difference between sweet and bitter almond genotypes resides in the accumulation of prunasin and amygdalin in seed tissues, including the tegument (mother tissue) and the cotyledons (also referred to as kernel [mother and father tissue]; Sánchez-Pérez et al., 2008, 2010). Identification of the entire pathway for prunasin and amygdalin enabled the examination of the expression levels of the amygdalin biosynthetic genes in the tegument and cotyledons of sweet and bitter genotypes at selected time points during fruit development using RT-qPCR (Fig. 3).
Figure 3.
Relative transcript levels of prunasin and amygdalin biosynthetic genes in the tegument and cotyledons of the sweet genotype Lauranne (blue traces) and the bitter genotype S3067 (purple traces) during fruit development. For each gene, expression levels were normalized to the one displayed by the sweet genotype at the earliest time point. TEFII, RPTII, and UBQ10 were used as housekeeping reference genes. Data are presented as means ± sd of three biological replicates. Note the logarithmic scale applied in the two graphs showing PdCYP79D16 and PdCYP71AN24 in tegument. Asterisks indicate statistically significant genotypic differences at the same time point (Student’s t test, P < 0.01). The expression of PdCYP79D16 was not detected in the sweet tegument at the last three time points, while the expression of PdUGT94AF1 was not detected in the bitter cotyledons at the first two time points. No expression of PdUGT94AF1 and PdUGT94AF2 in tegument and of PdCYP79D16 and PdCYP71AN24 in cotyledons was detectable. For relative expression level comparisons among the enzymes in the same tissue, see Supplemental Figure S5.
Strong and consistent expression of PdCYP79D16 was observed in the tegument of the bitter almond throughout fruit development. In contrast, PdCYP79D16 expression was not detectable in the tegument of the sweet genotype, except for the very early stages of fruit development, in which it was more than 100-fold lower than in the bitter genotype (Fig. 3). ANOVA showed that, besides the genotype, the fruit developmental stage also represented a significant source of variation (Supplemental Table S4).
Major genotypic differences also were observed for PdCYP71AN24, with its expression in the sweet genotype being 6- to 600-fold lower than in the bitter one throughout fruit development (Fig. 3).
Expression levels of PdUGT85A19 and PdUGT94AF3 increased significantly during fruit development, although no genotypic difference was detected between sweet and bitter (Supplemental Table S4). Interestingly, PdUGT94AF3 expression was higher than that of PdUGT85A19, up to 200-fold at the first time point (Supplemental Fig. S5).
The main site of amygdalin accumulation is the cotyledons (Sánchez-Pérez et al., 2008). No expression of PdCYP79D16 and PdCYP71AN24 could be detected in the cotyledons of sweet and bitter genotypes. PdUGT85A19, PdUGT94AF3, PdUGT94AF1, and PdUGT94AF2 all were expressed in cotyledons; however, no significant differences were observed between the sweet and bitter genotypes (Fig. 3). Pairwise comparisons between UGTs with the same enzymatic activity (PdUGT85A19/PdUGT94AF3 and PdUGT94AF1/PdUGT94AF2) revealed higher expression of PdUGT94AF3 and PdUGT94AF2 (Supplemental Fig. S5).
Based on these results, we propose the compartmentalization of the enzymes involved in the biosynthesis of cyanogenic glucosides in an almond seed as shown in Figure 3. Prunasin, which is synthesized in the tegument, needs the expression of PdCYP79D16, PdCYP71AN24, and PdUGT85A19/PdUGT94AF3. Once synthesized in the tegument, prunasin might be transported to the cotyledons and converted into the diglucoside amygdalin by PdUGT94AF1/AF2 and stored. The observed different transcriptional regulation of both PdCYP79D16 and PdCYP71AN24 may cause different amygdalin accumulation in sweet and bitter genotypes.
Glucoside Formation Catalyzed by PdUGT94AF1 and PdUGT94AF2
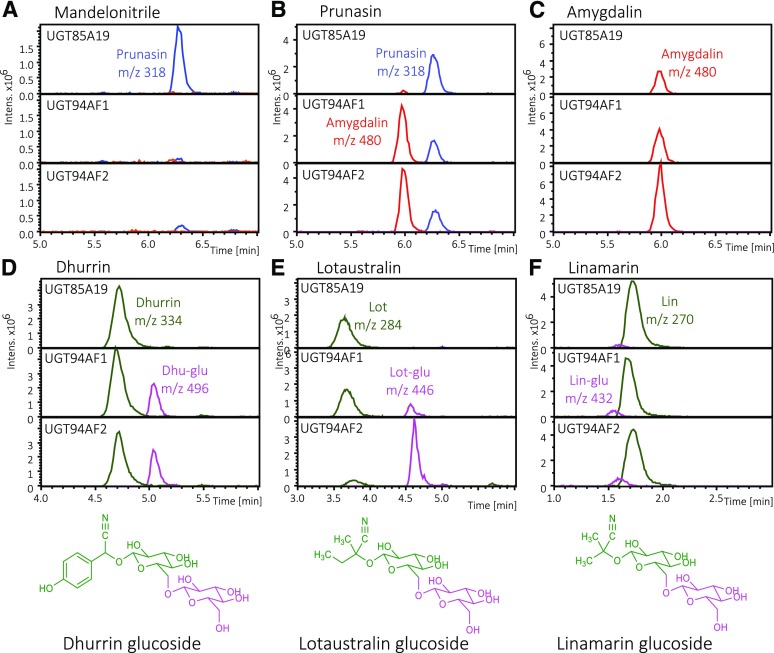
To contribute to the understanding of the functional characteristics of the two novel β(1→6)-O-diglucosyltransferases, PdUGT94AF1 and PdUGT94AF2 were transiently expressed in N. benthamiana leaves. In reference experiments, PdUGT85A19 was expressed as a representative of a previously published monoglucosyltransferase able to catalyze prunasin formation from mandelonitrile (Franks et al., 2008). On day 2 after agroinfiltration, the leaves were infiltrated with 100 µm solutions of different cyanogenic monoglucosides and diglucosides to investigate the substrate specificity of the two UGT94AFs. The infiltrated leaf material was extracted into methanol, and the products formed were identified by LC-MS analyses (Fig. 4). Agroinfiltration with p19 was used to prevent gene silencing and as a control to monitor background reactions mediated by endogenous UGT activities in the N. benthamiana leaves. The substrates administered and products formed are listed in Table 1.
Figure 4.
Substrate specificity of PdUGT85A19, PdUGT94AF1, and PdUGT94AF2 as measured by the administration of different substrates to the individual UGTs transiently produced in N. benthamiana, as monitored by LC-MS and extracted ion chromatograms. A to C, Amygdalin, m/z 480, red traces; prunasin, m/z 318, blue traces. D to F, Monoglucosides shown in green traces: dhurrin, m/z 334; lotaustralin, m/z 284; linamarin, m/z 270. Diglucosides shown in purple traces: dhurrin glucoside, m/z 496; lotaustralin glucoside, m/z 446; linamarin glucoside, m/z 432 (Supplemental Fig. S3).
In experiments where mandelonitrile was provided as a substrate, PdUGT85A19 converted mandelonitrile into prunasin, part of which was transformed into a prunasin-malonate ester (Supplemental Fig. S2). Neither PdUGT94AF1 nor PdUGT94AF2 was able to glucosylate mandelonitrile (Fig. 4A). With prunasin as a substrate, PdUGT94AF1 and PdUGT94AF2 catalyzed amygdalin formation (Fig. 4B). PdUGT94AF1 and PdUGT94AF2 also catalyzed the conversion of dhurrin, lotaustralin, and linamarin into the corresponding diglucosides, with decreasing rates of glucosylating activity observed using prunasin, dhurrin, and lotaustralin as substrates (Fig. 4, B, D, E, and F). This agrees with the amygdalin formation observed upon coexpression of PdCYP79D16, PdCYP71AN24, PdUGT85A19, and PdUGT94AF1 or PdUGT94AF2 (Fig. 2). None of these experiments resulted in the formation of triglucosides (Fig. 4C). The presence of amygdalin glucosides had been reported previously (Pičmanová et al., 2015). The experiment shows that PdUGT94AF1 and PdUGT94AF2 do not catalyze this conversion. It is notable that PdUGT85A19 did not catalyze diglucoside formation from any of the tested monoglucosides. The MS/MS spectra for the diglucosides formed from dhurrin and lotaustralin are presented in Supplemental Figure S6 and match the spectra for authentic standards.
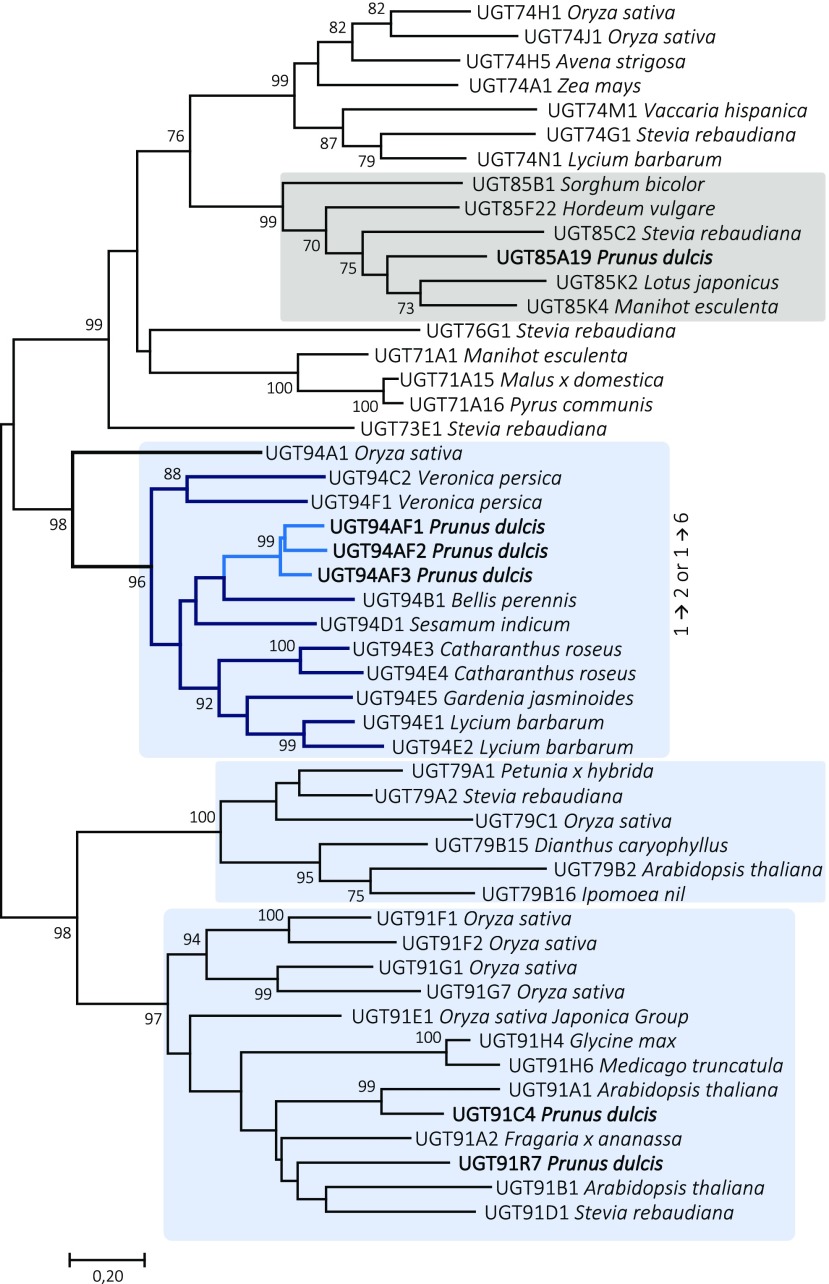
Phylogenetic Relationship of the Almond Diglucosyltransferases
The amino acid sequences of PdUGT94AF1 and PdUGT94AF2 are 83% identical, and both share about 75% sequence identity to PdUGT94AF3. A multiple sequence alignment of the three almond UGT94AF homologs is presented in Supplemental Figure S7. All three sequences harbor the plant secondary product glucosyltransferase (PSPG) motif (Hughes and Hughes, 1994). Six amino acids within this 44-amino acid motif differ among the three PdUGT94AF homologs. Compared with the PSPG consensus motif within the UGT94 enzymes characterized here, two out of the 44 amino acid residues are unique to PdUGT94AF1 and PdUGT94AF2. Within the UGTs, the relationship between the degree of amino acid sequence identity and substrate specificity is highly complex. Thus, UGTs catalyzing the same reactions may be placed in different families, and a few key amino acid substitutions may drastically alter the substrate specificity (Osmani et al., 2009). In this study, the latter is demonstrated by the difference in substrate specificity between PdUGT94AF3 compared with PdUGT94AF1 and PdUGT94AF2. Despite the low sequence conservation between UGTs belonging to different families, they show highly conserved secondary and tertiary structures (Osmani et al., 2009; Yu et al., 2017).
To investigate the phylogenetic relationship of the almond UGTs, a neighbor-joining tree was constructed (Fig. 5). The alignment included PdUGT94AF1, PdUGT94AF2, and PdUGT94AF3, functionally characterized in this study, and 27 plant UGTs from the families 76, 91, and 94, which are known to possess UGTs catalyzing the glycosylation of glucosides associated with stevioside or flavonoid metabolism (Noguchi et al., 2008; Yonekura-Sakakibara and Hanada, 2011). The member of the UGT76 family was included as a representative of UGTs catalyzing triglucoside formation (Olsson et al., 2016), and UGT73 and UGT74 sequences were used as references. The three almond UGT94 paralogs were found to cluster with UGT94B1 from Bellis perennis and UGT94D1 from sesame. Remarkably, the UGT94B1 from B. perennis is the only UGT94 known to catalyze a glucuronosylation reaction (Sawada et al., 2005; Osmani et al., 2008). The UGT94 branch is involved in making two types of linkages, β1→2 and β1→6.
Figure 5.
Phylogenetic neighbor-joining tree of 50 UGTs known to catalyze glycosylation of the Glc moiety in different classes of monoglucosides from different plant species. Based on the UGT gene data set, the derived UGT protein sequences were used to implement a multiple sequence alignment using the program ClustalV in MEGA7. The output files were used to construct the evolutionary history using the neighbor-joining method with 1,000 replications, where bootstrap values at the nodes are listed in percentage of the replications. UGT families known to harbor members catalyzing β1→2 or β1→6 bond formation are boxed in light blue. The UGT85 family catalyzing mandelonitrile glycosylation UGTs is shown in gray. The tree is a representation of the database utilized in the initial UGT search. A complete table including accession numbers is found in Supplemental Table S6.
As members of the UGT91 family, PdUGT91C4 and PdUGT91R7, studied in this work, group with UGTs involved, for example, in saponin and flavonoid metabolism. The UGT91 family catalyzes the formation of 1→6 glucosidic linkages but is known to transfer sugars other than Glc (e.g. rhamnose). This observation could serve as a hint for further studies of the UGT91s from almond.
DISCUSSION
Complete Conservation of CYPs in the Cyanogenic Pathway among Prunus Species
Cyanogenic glucosides are widespread amino acid-derived bioactive natural products found in more than 2,650 species (Gleadow and Møller, 2014). The biosynthesis of cyanogenic monoglucosides includes oximes and α-hydroxynitriles as key intermediates and is catalyzed by CYPs and a glucosyltransferase (Fig. 1). Based on the high amino acid sequence identity to Japanese apricot CYPs (Yamaguchi et al., 2014), the two almond CYPs (PdCYP79D16 and PdCYP71AN24) involved in the synthesis of prunasin and amygdalin were identified and characterized (Fig. 2). All functionally characterized members of the CYP79 family from monocots and eudicots catalyze the conversion of specific parent amino acids into the corresponding oximes (Sibbesen et al., 1995; Bak et al., 1998b; Kahn et al., 1999; Andersen et al., 2000; Takos et al., 2011; Irmisch et al., 2015; Knoch et al., 2016). The cyanogenic glucoside biosynthetic genes are clustered in the genomes in sorghum, cassava, lotus, and barley (Takos et al., 2011; Knoch et al., 2016). In almond, however, the biosynthetic genes are positioned on different chromosomes: PdCYP79D16 on chromosome 6, PdCYP71AN24 on chromosome 5, and the four UGTs characterized here on chromosomes 1, 2, and 6. PdUGT94AF1 and PdUGT94AF3 are the only genes that are in close proximity and are positioned on chromosome 2 (Supplemental Fig. S1). Based on synteny to the peach genome sequence, the Sk locus was located recently to a 150-kb region in the almond genome (Ricciardi et al., 2018). None of the almond genes encoding the biosynthetic enzymes in prunasin and amygdalin synthesis are localized within this region.
A protein BLAST search using the NCBI database with PdCYP79D16 identified the presence of a single CYP79D16 orthologous sequence in peach, sweet cherry, and Japanese apricot. All these sequences share 99% sequence identity to PdCYP79D16. A single CYP79 with high sequence identity to PdCYP79D16 also was found present in all hitherto examined other species from the Rosaceae family (Supplemental Table S2). In Japanese apricot, a second CYP79-encoding gene sequence assigned as PmCYP79A68 has been reported (Yamaguchi et al., 2014). This gene is situated on chromosome 7 and was expressed exclusively in the bud in Japanese apricot. An orthologous sequence (98% amino acid sequence identity) was found in almond and accordingly assigned as PdCYP79A68 (Supplemental Fig. S1). No evidence for the localization of a cyanogenic glucoside biosynthetic cluster around PdCYP79A68 was found, and the gene is expressed neither in the tegument nor in the cotyledons. Recently, a potential signaling role of prunasin and of endogenous prunasin turnover products in endodormancy release in almond and sweet cherry was reported (Del Cueto et al., 2017; Ionescu et al., 2017a, 2017b). In sweet cherry cultivars, PaCYP79A68 displayed a high level of expression during dormancy release. This was echoed by an observed accumulation of prunasin in the flower buds shortly after dormancy release, and the levels dropped again just before flowering time (Del Cueto et al., 2017). In sweet cherry and other Prunus species, treatment with the agrochemical hydrogen cyanamide has been used routinely to induce the breaking of dormancy to advance flowering time (Ionescu et al., 2017a, 2017b). A direct common link between prunasin formation and turnover and flower induction with hydrogen cyanamide has not been established but warrants further investigation.
In most other plant families, the species examined each contained two or more CYP79 homologs (Andersen et al., 2000; Nielsen and Møller, 2000; Takos et al., 2011; Irmisch et al., 2014). One or more of these additional CYP79 copies may play a role in the production of volatile oximes and nitriles (Irmisch et al., 2013; Sørensen et al., 2018). Most recently, a CYP79A118 in the European yew (Taxus baccata, Pinaceae family) was reported to catalyze the synthesis of phenylacetaldoxime from Phe in the biosynthesis of taxiphyllin (Luck et al., 2017), expanding the range of involvement of CYP79s in cyanogenic glucoside synthesis to the gymnosperms. The CYP79-catalyzed reaction sequence is highly complex, involving two sequential N-hydroxylations, a decarboxylation reaction, and a dehydration (Halkier et al., 1989, 1991; Vazquez-Albacete et al., 2017). The further conversion of the aldoxime to a cyanohydrin also is mediated by the action of a CYP belonging to the CYP71 clan. As in Japanese apricot, a PdCYP71AN24 was found to catalyze this process in almond. Even though CYP71s are considered more promiscuous in nature (Kahn et al., 1999), we see a clear case of strict conservation of the functional homologs within the Prunus species (Supplemental Table S2). In contrast, the conversion of the oxime to the cyanohydrin was shown recently to be catalyzed by two separate P450 enzymes in E. cladocalyx (Hansen et al., 2018). This represents yet another example showing that cyanogenic glucoside biosynthesis has evolved independently in several plant lineages (Takos et al., 2011). It was surprising to find that CYP706C55 catalyzes the dehydration of phenylacetaldoxime, because the three CYP706 members from other plant species that have been functionally characterized previously all catalyze classical oxygenation reactions.
The step of cyanohydrin glucosylation to produce cyanogenic monoglucosides has been characterized in sorghum, barley, cassava, almond, and lotus. In all cases, the glucosyltransferases involved belong to the UGT85 family (Jones et al., 1999; Franks et al., 2008; Kannangara et al., 2011; Takos et al., 2011; Knoch et al., 2016). In sorghum, the totally cyanide deficient2 mutant has a premature stop codon in the N-terminal region of UGT85B1. The mutant plants do not produce dhurrin and are characterized by reduced vigor and dwarfism, with poor root development and low fertility. LC-MS analysis shows the accumulation of numerous p-hydroxymandelonitrile-derived metabolites (Blomstedt et al., 2016). Some of these are detoxification products identical to those observed in transgenic Arabidopsis (Arabidopsis thaliana) expressing the CYP79A1 and CYP71E1 genes (Bak et al., 2000). This demonstrates that the activities of endogenous unspecific UGTs present in sorghum or Arabidopsis could not functionally replace UGT985B1. On this basis, we conclude that PdUGT85A19 has a similar essential function in almonds.
In nature, N. benthamiana does not produce cyanogenic glucosides. The prunasin formation observed upon transient expression of PdCYP79D16, PdCYP71AN24, and PdUGT85A19 in N. benthamiana demonstrated that these genes encode a set of enzymes able to catalyze the synthesis of this cyanogenic monoglucoside from Phe (Fig. 2). In addition, a malonate ester of prunasin was detected. This conjugate has been observed previously to be formed in agroinfiltrated N. benthamiana plants with CYPs from either Eucalyptus species or lotus (Tian and Dixon, 2006; Franzmayr et al., 2012; Ting et al., 2013). The conjugate may serve to facilitate the transport and/or storage of prunasin or prevent its hydrolysis by endogenous β-glucosidases.
Characterization of Three Novel UGT94s
The transient expression studies in N. benthamiana demonstrated that PdUGT94AF1 and PdUGT9AF2 are functional homologs catalyzing the conversion of the monoglucoside prunasin into the cyanogenic diglucoside amygdalin in planta (Fig. 2). To our knowledge, PdUGT94AF1 and PdUGT9AF2 represent the first UGTs identified converting a cyanogenic monoglucoside into a cyanogenic diglucoside. In a phylogenetic analysis, the UGT94 family is localized within cluster IV, characterized by harboring UGTs that are regioselective instead of substrate selective (Fig. 5). All UGT94s characterized catalyze either a β1→2 or β1→6 glycosylation of a range of glucosylated phytochemicals (Noguchi et al., 2008). It is notable that, in contrast to the reported clustering of the genes encoding the enzymes catalyzing the biosynthesis of cyanogenic glucosides in sorghum, barley, and lotus, the genes encoding the enzymes catalyzing amygdalin synthesis in almonds are not clustered (e.g. with PdCYP79D16, PdCYP71AN24, and PdUGT85A19 localized on chromosomes 6, 5, and 1, respectively). The identification of the UGT catalyzing the β(1→6)-O-glycosylation of prunasin was based on screening a UGT library generated based on UGT families previously documented or hypothesized to harbor UGTs able to catalyze β(1→6)-O-glycosylation reactions of other classes of monoglucosides. If additional UGT families turn out to harbor UGTs able to catalyze diglucoside formation, such putative candidates would have been missed.
PdUGT94AF3 was not able to catalyze the efficient conversion of prunasin to amygdalin but showed an unexpected capability to glucosylate mandelonitrile (Fig. 2). The PdUGT94AF3 reaction consisting of the conversion of an aglucon to a cyanogenic monoglucoside is not in line with the listed catalytic properties of other functionally characterized cluster IV UGTs. In studies on the functional characterization of flavonoid UGTs from sweet orange (Citrus sinensis) involved in flavonoid biosynthesis, incongruencies between tight phylogenetic relationships and experimentally determined catalytic properties also were observed (Liu et al., 2018). In addition to the demonstrated ability of PdUGT94AF3 to catalyze prunasin formation with mandelonitrile as the substrate, PdUGT94AF3 may possess transglucosylation activity and, with prunasin as the substrate, give rise to the formation of amygdalin and mandelonitrile. This could possibly supplement the activity of PdUGT94AF1 and PdUGT94AF2 with respect to amygdalin formation in specific cell types at some stage of cotyledon ontogeny. Alternatively, it could serve to trigger the initiation of endogenous recycling processes of prunasin (Pičmanová et al., 2015; Bjarnholt et al., 2018) or prunasin hydrolysis.
UGT94s Can Glycosylate Additional Cyanogenic Monoglucosides
As members of the UGT94 family, PdUGT94AF1 and PdUGT94AF2 might be expected to be catalytically active toward a wide range of acceptors. The test of mandelonitrile, prunasin, and amygdalin as putative substrates demonstrated that PdUGT94AF1 and PdUGT94AF2 glucosylate the cyanogenic monoglucoside prunasin, while no glucosylation of mandelonitrile and amygdalin was observed (Fig. 4, A–C). Because mandelonitrile slowly dissociates into benzaldehyde and hydrogen cyanide (Møller et al., 2016), a control assay was carried out with PdUGT85A19 that showed the conversion of mandelonitrile into prunasin (Fig. 4A). This control also demonstrated that PdUGT85A19 was not able to use monoglucosides and diglucosides as substrates (Fig. 4, B and C).
Upon the administration of dhurrin (Fig. 4D), lotaustralin (Fig. 4E), or linamarin (Fig. 4F) as putative substrates, PdUGT94AF1 and PdUGT94AF2 catalyzed diglucoside formation, as documented by the MS and MS / MS spectra obtained, the latter spectra with the diglucoside moiety at m/z 347, as a prominent fragment mass (Supplemental Fig. S6). In addition to amygdalin, E. camphora contains three additional cyanogenic diglucosides assigned as eucalyptosin A, B, and C, in which the second Glc residue is attached via a β1-2, β1-4, and β1-3 linkage, respectively. The presence of these diglucosides differed depending on plant ontogeny and tissue type (Neilson et al., 2011). In sorghum, the presence of three structural isomers also has been demonstrated (Pičmanová et al., 2015). In our study here, the regiospecificity of the new glucosidic linkage introduced by the action of PdUGT94AF1 and PdUGT94AF2 was not determined. According to the RT observed, we suggest that the dhurrin glucoside formed is the dhurrin-6-glucoside. The dhurrin glucosides are well separated by the LC system used, implying that PdUGT94AF1 and PdUGT94AF2 are regiospecific, catalyzing the formation of a single diglucoside. Despite the close structural resemblance between lotaustralin and linamarin, PdUGT94AF1 and PdUGT94AF2 appear to be less active with linamarin as the substrate (Fig. 4F). In our experiment, the substrate specificity of PdUGT94AF1 and PdUGT94AF2 was studied in planta. When the substrate specificity of UGTs is studied in vitro, a broader substrate specificity may be recorded (Jones et al., 1999; Kristensen et al., 2005). In this context, PdUGT94AF1 and PdUGT94AF2 may be valuable modules in synthetic biology-based approaches to produce high-value diglucosides in yeast or photosynthetic systems (Hansen et al., 2009; Lassen et al., 2014). Additional almond UGTs are predicted to catalyze the formation of prunasin-pentosides and prunasin-glucosides with different linkage types, as observed by metabolite profiling of different almond tissues (Pičmanová et al., 2015).
Bitterness in Almond Is Regulated at the CYP Expression Level
The genes encoding the enzymes required for the biosynthesis of prunasin and amygdalin are present in the genomes of both sweet and bitter almonds (Sánchez-Pérez et al., 2008). The key to unraveling the molecular basis for the sweet kernel trait, therefore, may be linked to a lack of expression of the biosynthetic genes or the complete endogenous remobilization of the amygdalin as a source of sugar and reduced nitrogen during seed development (Pičmanová et al., 2015; Bjarnholt et al., 2018). The expression analyses of the biosynthetic genes presented here documents that it is the lack of expression of PdCYP79D16 in the sweet genotype, echoed by the strongly reduced expression of PdCYP71AN24, that results in the sweet kernel trait (Fig. 6). In sorghum, the SbCYP79A1-catalyzed steps in dhurrin synthesis are rate limiting (Busk and Møller, 2002). The stoichiometric ratio between SbCYP79A1 and SbCYP71E1 in the isolated dhurrin metabolon is around 1:5 (Laursen et al., 2016; Bassard et al., 2017), which corroborates these findings. This is an efficient way of avoiding the accumulation of toxic intermediates (Sørensen et al., 2018) and would point to the transcriptional control of PdCYP79D16 expression by a transcription factor as the molecular mechanism responsible for the sweet kernel trait in almond. Identification of the transcription factor(s) responsible for the lack of PdCYP79D16 expression will offer breeders and growers a molecular marker to accelerate the selection for the sweet kernel phenotype already in juvenile almond plants.
Figure 6.
The amygdalin biosynthetic pathway and its compartmentalization in almond. In the tegument, Phe is transformed into prunasin by the action of two P450s and UGTs. The expression levels of the two P450s PdCYP79D16 and PdCYP71AN24 are different between sweet (white background, no expression) and bitter (black background, very high expression). The expression of the UGT-encoding genes is not significantly different in the sweet and bitter phenotypes. Following transport to the cotyledons, prunasin is converted into amygdalin by the action of diglucosyltransferases encoded by PdUGT94AF1 or PdUGT94AF2. These two genes also are not differentially expressed in the cotyledons. The expression levels of PdUGT94AF3 and PdUGT94AF2 are higher than those of PdUGT85A19 and PdUGT94AF1, respectively (Supplemental Fig. S5), which is represented by thicker arrows. Thus, the sweet kernel trait reflects the lack of expression of PdCYP79D16 and PdCYP71AN24.
MATERIALS AND METHODS
Plant Material
The almonds (Prunus dulcis) used in this study were obtained from the genotypes Lauranne (SkSk, sweet) and S3067 (sksk, bitter). Lauranne is a French cultivar obtained by the Institut National de la Recherche Agronomique, and S3067 is a genotype obtained by the Almond Breeding Program of Centro de Edafología y Biología Aplicada del Segura-Consejo Superior de Investigaciones Científicas (CEBAS-CSIC). Almond samples were collected every 2 to 3 weeks during the fruit development.
Isolation, Cloning, and Heterologous Expression of P450 and UGT Candidate Genes
Tegument (also called seed coat; 100 mg) and cotyledons (100 mg) tissue from the bitter genotype S3067 were collected halfway through the seed development stage (Julian Day 130; Del Cueto et al., 2018) and used for the extraction of total RNA with the Ultra Clean Plant RNA Isolation Kit (MOBIO). cDNA was synthesized from 500 ng of total RNA using the IScript cDNA Synthesis Kit (Bio-Rad). A BLAST search of CYP79D16 and CYP71AN24 from Japanese apricot (Prunus mume) versus the peach (Prunus persica) genome version 1 (Verde et al., 2013) provided the accession ppa021236m, with the highest sequence similarity to CYP79D16, and the two accessions ppa004152m and ppa017339m, with high sequence similarity to CYP71AN24, of which only the accession ppa004152m could be amplified from the almond cDNA.
Primers were designed based on peach sequences, adding CACC on the 5′ end of the forward primer, as necessary for directional cloning according to the protocol (pENTR Directional TOPO Cloning Kits) from Invitrogen (Supplemental Table S1). PCR was set up using Phusion High-Fidelity polymerase (Thermo Scientific) and the following conditions: 2 min at 98°C; 35 cycles of 20 s at 98°C, 20 s at 57°C to 65°C, and 1 min at 72°C; and one cycle of 5 min at 72°C (Supplemental Table S1). The PCR products obtained were analyzed by agarose (1% [w/v]) gel electrophoresis in Tris-acetate EDTA. Products of the expected size were cloned into the pENTR vector and transformed into Escherichia coli DH5α chemically competent cells (Invitrogen). Plasmids isolated from positive colonies were resequenced and recombined in vitro into pJAM1502 as the destination vector using the LR Clonase II Enzyme Mix (Invitrogen) reaction (Luo et al., 2007).
The PdCYP79D16 (MH992767) and PdUGT85A19 (EU015987.1) mRNAs, flanked by the attB1 and attB2 Gateway cloning sites (Supplemental Table S1), were acquired from GenScript, inserted in the pUC19 vector, and used to perform the LR reaction as described above.
To identify the UDPG-prunasin glucosyltransferase, a database was generated containing selected UGT protein sequences from the UGT76, UGT79, UGT91, and UGT94 families belonging to a common cluster (Noguchi et al., 2008) or orthologous group (Yonekura-Sakakibara and Hanada, 2011) known to harbor member UGTs catalyzing β-(1-6)-O-glucosylations. Members of the UGT71, UGT73, UGT74, and UGT88 families were included as negative controls. A BLASTX of the genome assemblies available from the sweet and bitter almond genotypes (Koepke et al., 2013) versus this protein database using the CLC Genomics Workbench provided 13 putative gene sequences belonging to the UGT94, UGT79, and UGT91 families. Full-length sequences of five of these 13 UGT-encoding genes were obtained by RACE using the SMARTer RACE cDNA Amplification Kit (Clontech; Supplemental Table S3). Each sequence was introduced into the pENTR vector by adding CACC in the full-length primers (Supplemental Table S1) to finally clone into the pJAM1502 vector.
Electroshock-based transformation of Agrobacterium tumefaciens AGL1 cells with genes cloned in pJAM1502 in DH5α E. coli cells was carried out as described previously (Del Cueto et al., 2018). Overnight cultures of A. tumefaciens-containing genes in pJAM1502 and the gene encoding the gene-silencing inhibitor protein p19 were grown in Luria-Bertani medium containing kanamycin (50 µg mL−1) and rifampicin (25 µg mL−1), while A. tumefaciens with the gene encoding p19 was grown with kanamycin, rifampicin, and tetracycline (10 µg mL−1). Cultures were harvested by centrifugation (3,000 rpm, 10 min) and resuspended to an OD600 of 2 in 1 mL of acetosyringone solution (50 mL of water, 0.5 mL of 10 mm MES, 0.5 mL of 10 mm MgCl2, and 5 µL of 100 µm acetosyringone). After incubation (2 h, room temperature), an equal volume of the A. tumefaciens cultures containing p19 and each of the different genes to be expressed was used for the agroinfiltration of Nicotiana benthamiana leaves. Leaf discs were excised and collected on day 5 after agroinfiltration and immediately frozen in liquid nitrogen until analyzed.
LC-MS Analysis of Cyanogenic Glucoside Content
Frozen leaf material was ground in liquid nitrogen in an Eppendorf tube using a pestle, and the cyanogenic glucosides were extracted in cold 85% aqueous methanol (300 μL, 5 min). The extracts were diluted five to 100 times in water and filtered through a membrane filter (0.45 μm; Merck Millipore) by centrifugation (3,000g, 5 min).
Analytical LC-MS/MS was carried out using an Agilent 1100 Series liquid chromatograph (Agilent Technologies) connected to a Bruker HCT-Ultra ion-trap mass spectrometer (Bruker Daltonics). The mobile phases were as follows: A, water with 0.1% (v/v) HCOOH and 50 mm NaCl; B, acetonitrile with 0.1% (v/v) HCOOH. The gradient program was as follows: A, 0 to 7.5 min, linear gradient 6% to 19% (v/v); and B, 7.5 to 10 min, linear gradient 19% to 100%. Electrospray ionization-MS/MS was run in the positive mode. Extracted ion chromatograms for specific [M+Na]+ adduct ions were used to search for the presence of specific compounds, and their MS, MS/MS, and RT were used to verify the identity of the compounds by comparison with authentic standards (Møller et al., 2016; Supplemental Figs. S2 and S4). Samples were analyzed with the Bruker Daltonics program Data Analysis 4.0.
Gene Expression Analysis
Tegument and cotyledon tissues of the almond genotypes Lauranne and S3067 were sampled (three biological replicates) at different time points during fruit development (April 1, April 15, April 30, May 13, and June 12 for tegument and May 14, May 27, June 25, July 17, August 1, and August 20 for cotyledons). Extraction of RNA and preparation of cDNA libraries were carried out as described above. RT-qPCR assays were carried out using the iQ SYBR Green master mix and the CFX 96 Real-Time PCR detecting system (Bio-Rad). Relative expression levels were calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001) and the geometric mean of three housekeeping genes TEF2, RPTII, and UBQ10 (Tong et al., 2009). RT-qPCR results were analyzed by Student’s t test and by two-way ANOVA using the SAS software package version 9.0 (SAS Institute). The primer pairs used for gene expression analyses are listed in Supplemental Table S5.
UGT Phylogenetic Tree
Phylogenetic analyses were conducted based on the UGT sequences available from selected UGT families. The amino acid sequences were aligned using Clustal and default parameters. Maximum-likelihood phylogenies were constructed with bootstrap n = 1,000. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. The analysis involved 57 UGT amino acid sequences (Supplemental Table S6). Evolutionary analyses were conducted using the program ClustalV in MEGA7.
Substrate Administration Assays
The substrate specificity of the individual UGTs studied was determined by A. tumefaciens-mediated transient expression in N. benthamiana of the individual UGT constructs. Two days after agroinfiltration, a 100 μm solution of putative substrates was injected into the same agroinfiltrated leaves. The tested substrates were mandelonitrile, prunasin, amygdalin, linamarin, lotaustralin, and dhurrin. Water was injected as a control. All substrates except mandelonitrile and amygdalin (Sigma) were chemically synthesized (Møller et al., 2016). On day 5 following substrate injection, leaves were harvested and analyzed as described above.
Accession Numbers
Accession numbers are as follows: PdCYP79D16, MH992767; PdUGT91C4, MH992768; PdUGT94AF1, MH992769; PdUGT94AF3, MH969427; PdUGT94AF2, MH969428; PdUGT91R7, MH969429; and PdCYP71AN24, MK014211.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Positions of the orthologous almond and peach genes in the peach genome version 2.0.
Supplemental Figure S2. Formation of a prunasin-malonate ester upon transient expression of prunasin pathway genes in N. benthamiana as verified by LC-MS analysis.
Supplemental Figure S3. Accumulation of (Z)- and (E)-phenylacetaldoxime glucosides as a result of transient expression in N. benthamiana of PdCYP79D16 in the absence or presence of coexpressed PdCYP71AN24.
Supplemental Figure S4. Functional characterization of the gene candidates PdUGT91C4 and PdUGT91R7.
Supplemental Figure S5. Relative expression of UGT genes during fruit development.
Supplemental Figure S6. Structural verification of products formed using LC-MS and MS/MS based on spectra from reference compounds.
Supplemental Figure S7. Multiple alignment of the PdUGT94AF1, PdUGT94AF2, and PdUGT94AF3 sequences.
Supplemental Table S1. Genes in almond and the primer pairs designed to obtain the full-length coding sequences.
Supplemental Table S2. The top six CYP sequences within the Rosaceae family showing high sequence identity to the almond PdCYP79D16 and PdCYP71AN24.
Supplemental Table S3. Primer pairs designed for RACE on the partial gene sequences encoding potentially interesting UGTs situated within the 13 contigs selected in the UGT database.
Supplemental Table S4. ANOVA performed on six amygdalin biosynthetic genes in tegument and cotyledon tissues of sweet and bitter genotypes at different time points.
Supplemental Table S5. Primer pairs used to study the expression of amygdalin biosynthetic genes.
Supplemental Table S6. Accession numbers of the NCBI sequences used to make the UGT database by CLC Genomics Workbench software.
Footnotes
This work was supported by the VILLUM Foundation by a grant to the VILLUM Center for Plant Plasticity (VKR023054) (B.L.M., R.S.-P., E.H.J.N.); the University of Copenhagen Excellence Program for Interdisciplinary Research to the Center for Synthetic Biology (B.L.M.); by a European Research Council Advanced Grant (ERC-2012-ADG_20120314) (B.L.M.); by two VILLUM Young Investor Program grants to R.S.-P. (VKR023124) and E.H.J.N. (VKR013167); E.H.J.N. was also funded by a Danish Independent Research Council Sapere Aude Research Talent Post-Doctoral Stipend (Grant No. 6111- 00379B); by the projects “Mejora Genética del Almendro” (MINECO-Spain) and “Breeding stone fruit species assisted by molecular tools” (Fundación Séneca-Spain) (F.D., J.D.C., and R.S.-P.).
Articles can be viewed without a subscription.
References
- Andersen MD, Busk PK, Svendsen I, Møller BL (2000) Cytochromes P-450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J Biol Chem 275: 1966–1975 [DOI] [PubMed] [Google Scholar]
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA (1998a) Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol 36: 393–405 [DOI] [PubMed] [Google Scholar]
- Bak S, Nielsen HL, Halkier BA (1998b) The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol 38: 725–734 [DOI] [PubMed] [Google Scholar]
- Bak S, Olsen CE, Halkier BA, Møller BL (2000) Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiol 123: 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard JE, Møller BL, Laursen T (2017) Assembly of dynamic P450-mediated metabolons: order versus chaos. Curr Mol Biol Rep 3: 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnholt N, Neilson EHJ, Crocoll C, Jørgensen K, Motawia MS, Olsen CE, Dixon DP, Edwards R, Møller BL (2018) Glutathione transferases catalyze recycling of auto-toxic cyanogenic glucosides in sorghum. Plant J 94: 1109–1125 [DOI] [PubMed] [Google Scholar]
- Blomstedt CK, O’Donnell NH, Bjarnholt N, Neale AD, Hamill JD, Møller BL, Gleadow RM (2016) Metabolic consequences of knocking out UGT85B1, the gene encoding the glucosyltransferase required for synthesis of dhurrin in Sorghum bicolor (L. Moench). Plant Cell Physiol 57: 373–386 [DOI] [PubMed] [Google Scholar]
- Blum D. (2010) The Poisoner’s Handbook. Penguin Press, England [Google Scholar]
- Busk PK, Møller BL (2002) Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol 129: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen M, Kannangara RM, Olsen CE, Blomstedt CK, Gleadow RM, Jørgensen K, Bak S, Motawie MS, Møller BL (2015) The bifurcation of the cyanogenic glucoside and glucosinolate biosynthetic pathways. Plant J 84: 558–573 [DOI] [PubMed] [Google Scholar]
- Davis R. (1991) Cyanogens: Toxic Substances in Crop Plants. Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- Del Cueto J, Ionescu IA, Pičmanová M, Gericke O, Motawia MS, Olsen CE, Campoy JA, Dicenta F, Møller BL, Sánchez-Pérez R (2017) Cyanogenic glucosides and derivatives in almond and sweet cherry flower buds from dormancy to flowering. Front Plant Sci 8: 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Cueto J, Møller BL, Dicenta F, Sánchez-Pérez R (2018) β-Glucosidase activity in almond seeds. Plant Physiol Biochem 126: 163–172 [DOI] [PubMed] [Google Scholar]
- Dicenta F, Martínez-Gómez P, Grané N, Martín ML, León A, Cánovas JA, Berenguer V (2002) Relationship between cyanogenic compounds in kernels, leaves, and roots of sweet and bitter kernelled almonds. J Agric Food Chem 50: 2149–2152 [DOI] [PubMed] [Google Scholar]
- Dirlewanger E, Graziano E, Joobeur T, Garriga-Calderé F, Cosson P, Howad W, Arús P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101: 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Morant M, Jørgensen B, Olsen CE, Asamizu E, Sato S, Tabata S, Bak S (2004) Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol 135: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TK, Yadollahi A, Wirthensohn MG, Guerin JR, Kaiser BN, Sedgley M, Ford CM (2008) A seed coat cyanohydrin glucosyltransferase is associated with bitterness in almond (Prunus dulcis) kernels. Funct Plant Biol 35: 236–246 [DOI] [PubMed] [Google Scholar]
- Franzmayr BK, Rasmussen S, Fraser KM, Jameson PE (2012) Expression and functional characterization of a white clover isoflavone synthase in tobacco. Ann Bot 110: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleadow RM, Møller BL (2014) Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity. Annu Rev Plant Biol 65: 155–185 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Olsen CE, Møller BL (1989) The biosynthesis of cyanogenic glucosides in higher plants: the (E)- and (Z)-isomers of p-hydroxyphenylacetaldehyde oxime as intermediates in the biosynthesis of dhurrin in Sorghum bicolor (L.) Moench. J Biol Chem 264: 19487–19494 [PubMed] [Google Scholar]
- Halkier BA, Lykkesfeldt J, Møller BL (1991) 2-Nitro-3-(p-hydroxyphenyl)propionate and aci-1-nitro-2-(p-hydroxyphenyl)ethane, two intermediates in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Proc Natl Acad Sci USA 88: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Renfrew JM (1978) Palaeolithic-Neolithic seed remains at Franchthi Cave, Greece. Nature 271: 349–352 [Google Scholar]
- Hansen CIC, Sørensen M, Veiga TAM, Zibrandtsen JFS, Heskes AM, Olsen CE, Boughton BA, Møller BL, Neilson EHJ (2018) Reconfigured cyanogenic glucoside biosynthesis involving a CYP706 in Eucalyptus cladocalyx. Plant Physiol 178: xxx–xxx [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EH, Møller BL, Kock GR, Bünner CM, Kristensen C, Jensen OR, Okkels FT, Olsen CE, Motawia MS, Hansen J (2009) De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl Environ Microbiol 75: 2765–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Hughes MA (1994) Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq 5: 41–49 [DOI] [PubMed] [Google Scholar]
- Ionescu IA, López-Ortega G, Burow M, Bayo-Canha A, Junge A, Gericke O, Møller BL, Sánchez-Pérez R (2017a) Transcriptome and metabolite changes during hydrogen cyanamide-induced floral bud break in sweet cherry. Front Plant Sci 8: 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu IA, Møller BL, Sánchez-Pérez R (2017b) Chemical control of flowering time. J Exp Bot 68: 369–382 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Unsicker SB, Gershenzon J, Köllner TG (2013) Identification and characterization of CYP79D6v4, a cytochrome P450 enzyme producing aldoximes in black poplar (Populus nigra). Plant Signal Behav 8: e27640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Clavijo McCormick A, Günther J, Schmidt A, Boeckler GA, Gershenzon J, Unsicker SB, Köllner TG (2014) Herbivore-induced poplar cytochrome P450 enzymes of the CYP71 family convert aldoximes to nitriles which repel a generalist caterpillar. Plant J 80: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Zeltner P, Handrick V, Gershenzon J, Köllner TG (2015) The maize cytochrome P450 CYP79A61 produces phenylacetaldoxime and indole-3-acetaldoxime in heterologous systems and might contribute to plant defense and auxin formation. BMC Plant Biol 15: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen NB, Zagrobelny M, Hjernø K, Olsen CE, Houghton-Larsen J, Borch J, Møller BL, Bak S (2011) Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nat Commun 2: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PR, Møller BL, Høj PB (1999) The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor: isolation, cloning, heterologous expression, and substrate specificity. J Biol Chem 274: 35483–35491 [DOI] [PubMed] [Google Scholar]
- Jørgensen K, Morant AV, Morant M, Jensen NB, Olsen CE, Kannangara R, Motawia MS, Møller BL, Bak S (2011) Biosynthesis of the cyanogenic glucosides linamarin and lotaustralin in cassava: isolation, biochemical characterization, and expression pattern of CYP71E7, the oxime-metabolizing cytochrome P450 enzyme. Plant Physiol 155: 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Jiwan D, Cho I, Lee T, Abbott A, Sosinski B, Main D (2009) Synteny of Prunus and other model plant species. BMC Genomics 10: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bak S, Svendsen I, Halkier BA, Møller BL (1997) Isolation and reconstitution of cytochrome P450ox and in vitro reconstitution of the entire biosynthetic pathway of the cyanogenic glucoside dhurrin from sorghum. Plant Physiol 115: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Fahrendorf T, Halkier BA, Møller BL (1999) Substrate specificity of the cytochrome P450 enzymes CYP79A1 and CYP71E1 involved in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys 363: 9–18 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Motawia MS, Hansen NKK, Paquette SM, Olsen CE, Møller BL, Jørgensen K (2011) Characterization and expression profile of two UDP-glucosyltransferases, UGT85K4 and UGT85K5, catalyzing the last step in cyanogenic glucoside biosynthesis in cassava. Plant J 68: 287–301 [DOI] [PubMed] [Google Scholar]
- Knoch E, Motawie MS, Olsen CE, Møller BL, Lyngkjaer MF (2016) Biosynthesis of the leucine derived α-, β- and γ-hydroxynitrile glucosides in barley (Hordeum vulgare L.). Plant J 88: 247–256 [DOI] [PubMed] [Google Scholar]
- Koch BM, Sibbesen O, Halkier BA, Svendsen I, Møller BL (1995) The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys 323: 177–186 [DOI] [PubMed] [Google Scholar]
- Koepke T, Schaeffer S, Harper A, Dicenta F, Edwards M, Henry RJ, Møller BL, Meisel L, Oraguzie N, Silva H, et al. (2013) Comparative genomics analysis in Prunoideae to identify biologically relevant polymorphisms. Plant Biotechnol J 11: 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen C, Morant M, Olsen CE, Ekstrøm CT, Galbraith DW, Møller BL, Bak S (2005) Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci USA 102: 1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki GW, Poulton JE (1987) Isolation and characterization of multiple forms of prunasin hydrolase from black cherry (Prunus serotina Ehrh.) seeds. Arch Biochem Biophys 255: 19–26 [DOI] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyán J (2004) Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J 23: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen LM, Nielsen AZ, Ziersen B, Gnanasekaran T, Møller BL, Jensen PE (2014) Redirecting photosynthetic electron flow into light-driven synthesis of alternative products including high-value bioactive natural compounds. ACS Synth Biol 3: 1–12 [DOI] [PubMed] [Google Scholar]
- Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, Silvestro D, Hatzakis NS, Wenk MR, Dafforn TR, et al. (2016) Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 354: 890–893 [DOI] [PubMed] [Google Scholar]
- Lechtenberg M, Nahrstedt A (1999) Cyanogenic glucosides. In Ikan R, ed, Naturally Occurring Glycosides. Wiley, UK [Google Scholar]
- Liu X, Lin C, Ma X, Tan Y, Wang J, Zeng M (2018) Functional characterization of a flavonoid glycosyltransferase in sweet orange (Citrus sinensis). Front Plant Sci 9: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luck K, Jia Q, Huber M, Handrick V, Wong GKS, Nelson DR, Chen F, Gershenzon J, Köllner TG (2017) CYP79 P450 monooxygenases in gymnosperms: CYP79A118 is associated with the formation of taxiphyllin in Taxus baccata. Plant Mol Biol 95: 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al. (2007) Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J 50: 678–695 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Bélanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, et al. (1997) The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7: 255–269 [DOI] [PubMed] [Google Scholar]
- Miller RE, Gleadow RM, Woodrow IE (2004) Cyanogenesis in tropical Prunus turneriana: characterisation, variation and response to low light. Funct Plant Biol 31: 491–503 [DOI] [PubMed] [Google Scholar]
- Møller BL, Conn EE (1979) The biosynthesis of cyanogenic glucosides in higher plants: N-hydroxytyrosine as an intermediate in the biosynthesis of dhurrin by Sorghum bicolor (Linn) Moench. J Biol Chem 254: 8575–8583 [PubMed] [Google Scholar]
- Møller BL, Olsen CE, Motawia MS (2016) General and stereocontrolled approach to the chemical synthesis of naturally occurring cyanogenic glucosides. J Nat Prod 79: 1198–1202 [DOI] [PubMed] [Google Scholar]
- Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchez-Pérez R, Møller BL, Bak S (2008) β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Neilson EH, Goodger JQD, Motawia MS, Bjarnholt N, Frisch T, Olsen CE, Møller BL, Woodrow IE (2011) Phenylalanine derived cyanogenic diglucosides from Eucalyptus camphora and their abundances in relation to ontogeny and tissue type. Phytochemistry 72: 2325–2334 [DOI] [PubMed] [Google Scholar]
- Nielsen JS, Møller BL (2000) Cloning and expression of cytochrome P450 enzymes catalyzing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of cyanogenic glucosides in Triglochin maritima. Plant Physiol 122: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A, Fukui Y, Iuchi-Okada A, Kakutani S, Satake H, Iwashita T, Nakao M, Umezawa T, Ono E (2008) Sequential glucosylation of a furofuran lignan, (+)-sesaminol, by Sesamum indicum UGT71A9 and UGT94D1 glucosyltransferases. Plant J 54: 415–427 [DOI] [PubMed] [Google Scholar]
- Olsson K, Carlsen S, Semmler A, Simón E, Mikkelsen MD, Møller BL (2016) Microbial production of next-generation stevia sweeteners. Microb Cell Fact 15: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Imberty A, Olsen CE, Møller BL (2008) Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol 148: 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Møller BL (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70: 325–347 [DOI] [PubMed] [Google Scholar]
- Pičmanová M, Møller BL (2016) Apiose: one of nature’s witty games. Glycobiology 26: 430–442 [DOI] [PubMed] [Google Scholar]
- Pičmanová M, Neilson EH, Motawia MS, Olsen CE, Agerbirk N, Gray CJ, Flitsch S, Meier S, Silvestro D, Jørgensen K, et al. (2015) A recycling pathway for cyanogenic glycosides evidenced by the comparative metabolic profiling in three cyanogenic plant species. Biochem J 469: 375–389 [DOI] [PubMed] [Google Scholar]
- Pliny the Elder (77AD) Naturalis Historiae, Liber XV. p 26
- Ricciardi F, Del Cueto J, Bardaro N, Mazzeo R, Ricciardi L, Dicenta F, Sánchez-Pérez R, Pavan S, Lotti C (2018) Synteny-based development of CAPS markers linked to the sweet kernel LOCUS, controlling amygdalin accumulation in almond (Prunus dulcis (Mill.) D.A.Webb). Genes (Basel) 9: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman A, Swanson A, Humphrey T, Chapman R, McGarvey B, Pocs R, Brandle J (2005) Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J 41: 56–67 [DOI] [PubMed] [Google Scholar]
- Robiquet PJ, Boutron-Chalard AF (1830) Nouvelles expériences sur les amandes amères et sur l’huile volatile qu’elles fournissent. Ann Chim Phys 44: 352–382 [Google Scholar]
- Sánchez-Pérez R, Jørgensen K, Olsen CE, Dicenta F, Møller BL (2008) Bitterness in almonds. Plant Physiol 146: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Jørgensen K, Motawia MS, Dicenta F, Møller BL (2009) Tissue and cellular localization of individual β-glycosidases using a substrate-specific sugar reducing assay. Plant J 60: 894–906 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Howad W, Garcia-Mas J, Arús P, Martínez-Gómez P, Dicenta F (2010) Molecular markers for kernel bitterness in almond. Tree Genet Genomes 6: 237–245 [Google Scholar]
- Sánchez-Pérez R, Belmonte FS, Borch J, Dicenta F, Møller BL, Jørgensen K (2012) Prunasin hydrolases during fruit development in sweet and bitter almonds. Plant Physiol 158: 1916–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T (2005) UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers: enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem 280: 899–906 [DOI] [PubMed] [Google Scholar]
- Sibbesen O, Koch B, Halkier BA, Møller BL (1995) Cytochrome P-450TYR is a multifunctional heme-thiolate enzyme catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J Biol Chem 270: 3506–3511 [DOI] [PubMed] [Google Scholar]
- Sørensen M, Neilson EHJ, Møller BL (2018) Oximes: unrecognized chameleons in general and specialized plant metabolism. Mol Plant 11: 95–117 [DOI] [PubMed] [Google Scholar]
- Takos AM, Knudsen C, Lai D, Kannangara R, Mikkelsen L, Motawia MS, Olsen CE, Sato S, Tabata S, Jørgensen K, et al. (2011) Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J 68: 273–286 [DOI] [PubMed] [Google Scholar]
- Tian L, Dixon RA (2006) Engineering isoflavone metabolism with an artificial bifunctional enzyme. Planta 224: 496–507 [DOI] [PubMed] [Google Scholar]
- Ting HM, Wang B, Rydén AM, Woittiez L, van Herpen T, Verstappen FWA, Ruyter-Spira C, Beekwilder J, Bouwmeester HJ, van der Krol A (2013) The metabolite chemotype of Nicotiana benthamiana transiently expressing artemisinin biosynthetic pathway genes is a function of CYP71AV1 type and relative gene dosage. New Phytol 199: 352–366 [DOI] [PubMed] [Google Scholar]
- Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Albacete D, Montefiori M, Kol S, Motawia MS, Møller BL, Olsen L, Nørholm MHH (2017) The CYP79A1 catalyzed conversion of tyrosine to (E)-p-hydroxyphenylacetaldoxime unravelled using an improved method for homology modeling. Phytochemistry 135: 8–17 [DOI] [PubMed] [Google Scholar]
- Velasco D, Hough J, Aradhya M, Ross-Ibarra J (2016) Evolutionary genomics of peach and almond domestication. G3 (Bethesda) 6: 3985–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F, et al. (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45: 487–494 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Yamamoto K, Asano Y (2014) Identification and characterization of CYP79D16 and CYP71AN24 catalyzing the first and second steps in L-phenylalanine-derived cyanogenic glycoside biosynthesis in the Japanese apricot, Prunus mume Sieb. et Zucc. Plant Mol Biol 86: 215–223 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J 66: 182–193 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu F, Dossa K, Wang Z, Ke T (2017) Genome-wide analysis of UDP-glycosyltransferase super family in Brassica rapa and Brassica oleracea reveals its evolutionary history and functional characterization. BMC Genomics 18: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D, Hopf M, Weiss E (2012) Domestication of Plants in the Old World : The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin. Oxford University Press, Oxford [Google Scholar]