Abstract
Escherichia coli is a major udder pathogen causing clinical mastitis in dairy cattle and its heat stable endotoxin in powdered infant formula milk is a potential risk factor in neonatal infections. Cephalosporins are frequently used for treatment of mastitis caused by mastitis; however, use of these antimicrobials may induce antimicrobial resistance in E. coli. The objective of this study was to explore the in vitro effect of subminimum inhibitory concentrations (sub-MIC) of cefalotin (CF) and ceftazidime (CAZ) on the morphology, antimicrobial resistance, and endotoxin releasing characteristics of 3 E. coli isolates recovered from bovine clinical mastitis. The parent E. coli isolates, which were susceptible to CF and CAZ, were exposed to CF or CAZ separately at sub-MIC levels to produce 9 generations of induced isolates. Colonies of the CAZ-induced isolates from all 3 parent E. coli were smaller on blood agar and the bacteria became filamentous, whereas the CF-induced isolates did not demonstrate prominent morphological changes. After induction by CF or CAZ, many induced isolates showed resistance to cefoxitin, CAZ, CF, kanamycin, ampicillin, and amoxicillin/clavulanic acid while their parent isolates were susceptible to these antimicrobials. Notably, 5 CAZ-induced isolates from the same parent isolate were found to produce extended-spectrum beta-lactamase (ESBL) though none of the tested ESBL related genes could be detected. All CAZ-induced isolates released more endotoxin with a higher release rate, whereas endotoxin release of CF-induced E. coli isolates was not different from parent isolates. The exposure of cephalosporins at sub-MIC levels induced resistant Escherichia coli. We inferred that cephalosporins, especially CAZ, should be used prudently for treatment of clinical E. coli mastitis.
1. Introduction
Mastitis is one of the most common and costly diseases for the dairy industry worldwide. Mastitis also affects animal welfare and is a frequent reason that cows are culled [1]. Treatment of mastitis accounts for the majority of antimicrobials administered to dairy cows [2, 3]. Gram-negative bacteria, mostly coliforms, including Escherichia coli, Klebsiella spp., and Enterobacter spp., cause a high proportion of all clinical mastitis (CM) cases [4–6]. Escherichia coli is the most common Gram-negative species causing CM in dairy cattle [4, 6, 7]. The endotoxin of E. coli, lipopolysaccharide (LPS), can remain biologically active in powdered infant formula milk because it is heat stable at 100°C and, therefore, may pose a potential risk to formula fed neonates [8].
The efficacy and necessity of antimicrobial therapy for treatment of E. coli mastitis are still a topic of debate [9]. However, fluoroquinolones and cephalosporins, particularly 3rd and 4th generation products, are the only antimicrobials for which there is supporting evidence of beneficial effects in treatment of E. coli mastitis [10, 11]. It is noteworthy that these 2 classes of antimicrobial agents are also important drugs for human health. The prevalence of resistance against these important antibiotics, particularly in E. coli, is increasing worldwide in veterinary and human medicine [12–14]. The 2 main factors involved in development of antimicrobial resistance in bacteria are the presence of resistance genes and selective pressure of antimicrobial agents, especially if suboptimal doses are administered [15]. Currently, very limited studies have been conducted to elucidate the influence of cephalosporin dosage on induction of antimicrobial resistance in E. coli isolated from CM. First-generation cephalosporins are less critical for human health, although they primarily have a Gram-positive spectrum, with limited activity against Gram-negative bacteria [16, 17].
Induction of antimicrobial resistance in E. coli has been characterized by changes in morphology, including filamentation, which is likely to protect the bacteria from deleterious effects of antimicrobials [18, 19]. There is also strong evidence that antimicrobials may enhance endotoxin release from E. coli, which could exacerbate symptoms of CM [18].
The main objective of this study was, therefore, to determine in vitro effects of sub-MIC exposure of cefalotin (CF) or ceftazidime (CAZ) on 3 E. coli isolates recovered from bovine CM cases. Characteristics such as colony morphology, antimicrobial susceptibility profile, and release of endotoxin from E. coli isolates recovered after induction were investigated.
2. Materials and Methods
2.1. Escherichia coli Isolates
Quarter milk samples (n=1252) were collected from July 2015 to May 2016 from dairy cows with CM from various dairy herds located in 16 Chinese provinces [7]. In total, 153 E. coli isolates were recovered, of which 36 produced extended-spectrum beta-lactamase (ESBL). More details on the origin and characteristics of the E. coli isolates are described [12]. All isolates were tested for resistance to CF (30 μg) and CAZ (30 μg) using the Kirby-Bauer disk diffusion method, with clinical breakpoints following recommendations of the Clinical and Laboratory Standards Institute [20]. Three E. coli isolates, subsequently referred to as E4, E11, and E21, were selected from 3 provinces (Beijing, Shanghai, and Gansu) and used as parent E. coli isolates in this study. The 3 isolates were susceptible to CF and CAZ, did not produce ESBL, and did not carry any of the tested ESBL encoding genes (data not shown).
2.2. Experimental Design
All steps of the experiment were carried out in triplicate. A single colony was inoculated into 20 mL trypticase soy broth (TSB; BD, Franklin Lake, NJ) and incubated for 12 h at 37°C. Then, 200 μL of the bacterial suspension (0.5 McFarland) was inoculated into 20 mL Mueller-Hinton broth (Luqiao, Beijing, China) to which 1/2 of the MIC was added for CF and CAZ separately and incubated for 24 h at 37°C. Subsequently, the bacterial suspension was centrifuged (6000 × g, 25°C, 5 min) (Anting, Shanghai, China) and the pellet resuspended in a solution containing 1.4 mL TSB and 0.6 mL glycerol (Luqiao, Beijing, China) and stored pending further processing. These induced isolates (i.e., first generation of growth) are further referred to as CF-1 groups (E4-CF-1, E11-CF-1, and E21-CF-1). The MIC values for CF versus CF-1 groups were determined, and 1/2 of the MIC of CF was again dissolved in Mueller-Hinton broth (Luqiao, Beijing, China) containing 1.5 x 106 CFU/mL of CF-1 groups. After 24 h of incubation at 37°C, the 2nd generation induced mutant was obtained and identified as CF-2 group. The same steps were repeated another 7 times. Induced E. coli isolates recovered after subsequent induction were designated CF-3, CF-4, CF-5, CF-6, CF-7, CF-8, and CF-9. Escherichia coli isolates were recovered after induction with CAZ, using procedures identical to those mentioned above for CF. Isolates recovered after induction were identified as CAZ-1, CAZ-2, CAZ-3, CAZ-4, CAZ-5, CAZ-6, CAZ-7, CAZ-8, and CAZ-9 (E4-CAZ, E11-CAZ, and E21-CAZ). Parent isolates and all induced isolates recovered after induction were stored in 30% glycerol at -80°C pending further processing. When MICs of the parent E. coli and induced isolates were determined, E. coli ATCC 25922 was used as a control strain. The CLSI breakpoints for CF were ≤ 8 μg/mL (susceptible), 16 μg/mL (intermediate), and ≥ 32 μg/mL (resistant). In case of CAZ, breakpoints for ceftiofur (≤ 2 μg/mL = susceptible, 4 μg/mL = intermediate, and ≥ 8 μg/mL = resistant) were used [20].
2.3. Morphological Observations
Stored isolates were recovered by streaking 100 μL of suspension on tryptone soya agar (TSA; Luqiao, Beijing, China) supplemented with 5% sheep blood and incubated for 24 h at 37°C. A pure growth was picked and streaked on a new TSA plate. Shape, size, and color of colonies were recorded. A single colony from agar was stained using the Gram-staining method and morphology examined with an optical microscope (Olympus, Tokyo, Japan).
2.4. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of the parent E. coli and isolates recovered after induction was determined using Mueller-Hinton agar (BD, Franklin Lakes, NJ) against 9 antimicrobial agents using the standard Kirby-Bauer disk diffusion method according to CLSI recommendations [20]. Inhibition zone diameter (mm) was measured using a ruler. The panel of antimicrobial agents consisted of ampicillin (10 μg), amoxicillin/clavulanic acid (20/10 μg), CAZ (30 μg), CF (30 μg), cefepime (30 μg), cefoxitin (30 μg), gentamicin (10 μg), kanamycin (30 μg), and amikacin (30 μg). For these antimicrobial agents, breakpoints for cefepime and cefoxitin referred to E. coli isolates from humans. Escherichia coli ATCC 25922 and Enterobacter cloacae CMCC45301 were used as quality control strains.
2.5. Detection of ESBL Production
Parent isolates and all isolates recovered after induction were screened on MacConkey agar containing cefotaxime (1 mg/L) for ESBL-production. Presumptive ESBL-producing isolates, if any, were further confirmed by the double-disc synergy test, following recommendations of the CLSI [20], using antimicrobial discs of cefotaxime (30 μg), cefotaxime plus clavulanic acid (30/10 μg), CAZ (30 μg), and CAZ plus clavulanic acid (30/10 μg). Production of ESBL was considered positive if the inhibition zone of cefotaxime plus clavulanic acid or CAZ plus clavulanic acid was ≥ 5 mm larger than their respective single discs [20]. Escherichia coli ATCC 25922 (ESBL-negative strain) and Klebsiella pneumoniae ATCC 700603 (ESBL-positive strain) were used as reference strains.
2.6. Detection of ESBL-Related Genes
Bacterial DNA from E. coli was isolated using the Bacterial DNA Extraction Kit (Transgen, Beijing, China) according to the manufacturer's instructions. A PCR assay was conducted to detect the presence of blaCTX-M, blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, blaCTX-M-15, blaSHV, blaTEM, and blaOXA genes, as described [21, 22] with minor modifications. The reaction mixture (20 μL) consisted of 10 μL of TaqMix (Transgen, Beijing, China), 1 μL of template DNA, 0.5 μL of each primer (10 μM; Sunbiotech, Beijing, China), and 8 μL of ultra-pure distilled water. Initial denaturation at 94°C for 5 min was followed by 35 cycles of amplification at 94°C for 45 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s, and a final step with extension at 72°C for 10 min. The PCR products were separated on a 2% agarose gel. Subsequently, PCR products were purified by TIANquick Midi Purification Kit (TIANGEN, Beijing, China) and then bidirectionally sequenced using the same primers by ABI 3730 sequencer (Applied Biosystems, Foster City, CA). Gene sequences were aligned with BLASTN software (http://www.ncbi.nlm.nih.gov/BLAST/) and compared to sequences available in GenBank. Klebsiella pneumoniae ATCC 700603 (ESBL-positive strain) and ddH2O, without template DNA, were used as positive and negative controls, respectively, in all PCR assays. Primers used in this study are presented in Table 1.
Table 1.
Primers used to detect extended spectrum beta-lactamase encoding genes.
| Genes | Primer sequence (5′ to 3′) | Amplicon size (bp) |
|---|---|---|
| blaCTX-M | CGC TTT GCG ATG TGC AG | 550 |
| ACC GCG ATA TCG TTG GT | ||
| bla CTX-M-1 | GTT ACA ATG TGT GAG AAG CAG | 1041 |
| CCG TTT CCG CTA TTA CAA AC | ||
| bla CTX-M-2 | ATG ATG ACT CAG AGC ATT CGC CGC | 876 |
| TCA GAA ACC GTG GGT TAC GAT TTT | ||
| bla CTX-M-9 | GTG ACA AAG AGA GTG CAA CGG | 857 |
| ATG ATT CTC GCC GCT GAA GCC | ||
| bla CTX-M-15 | CAC ACG TGG AAT TTA GGG ACT | 995 |
| GCC GTC TAA GGC GAT AAA CA | ||
| bla TEM | TTC TTG AAG ACG AAA GGG C | 1150 |
| ACG CTC AGT GGA ACG AAA AC | ||
| bla SHV | CAC TCA AGG ATG TAT TGT G | 885 |
| TTA GCG TTG CCA GTG CTC G | ||
| bla OXA | ACA CAA TAC ATA TCA ACT TCG C | 813 |
| AGT GTG TTT AGA ATG GTG ATC |
2.7. Detection of Endotoxin Release
Isolates recovered after induction with CF or CAZ, as well as parent isolates, were used for this part of the study. Frozen bacteria were thawed at 37°C for 60 min. Ten μL of suspension was streaked onto TSA supplemented with 5% sheep blood and incubated for 24 h at 37°C. A single colony was picked from the agar, inoculated into 10 mL of TSB, and incubated for 18 h at 37°C. Bacterial counts were measured using the 10-fold dilution method. Subsequently, bacterial solutions were diluted to 106 CFU/mL using TSB and bacteria were cultured aerobically at 37°C. At various time points (0, 2, 4, 6, 8, and 10 h), aliquots were collected and endotoxin release capability quantified. Briefly, aliquots were collected and centrifuged at 6,000 × g for 15 min. The supernatant of each aliquot was collected and stored at -20°C until further processing. Endotoxin concentration was measured using an enzyme-linked immunosorbent assay (ELISA) commercial kit (Sigma, St. Louis, MO). Each step was performed following the manufacturer's instructions. All experiments were performed in triplicate.
2.8. Statistical Analyses
Associations between endotoxin release and exposure to sub-MICs of CF and CAZ were determined by fitting a mixed effects linear regression model using replicate (n = 3) as the random effect with the lme4 package (Bates et al., 2017) in R version 3.3.0 (R Core Team). A P-value < 0.05 was considered statistically significant. Data were first visually examined to understand associations between time and endotoxin release. Due to the sigmoidal nature of the association, data were normalized by taking the natural logarithm of endotoxin release, which resulted in a more clearly parabolic association between incubation time and endotoxin release. Higher degree terms for time were included to more accurately adjust for this parabolic relationship and isolate effects of treatment and generation of isolate on endotoxin release. The final model included treatment (CF, CAZ, or control) and an interaction with generation (ranging from 1 to 9) as independent variables, with an adjustment for incubation time modelled with cubic and quadratic terms, as higher degree polynomial terms did not increase model fit as determined by P-value and change in restricted maximum likelihood criterion.
3. Results
3.1. MICs of Cefalotin and Ceftazidime
The MICs of CF and CAZ for the parent E. coli isolates were 0.5-4 μg/mL and 0.125-1 μg/mL, respectively; therefore, parent isolates were susceptible to these 2 antimicrobial agents. The MIC values of CF and CAZ against induced isolates are shown (Table 2). After exposure to sub-MIC of CF, 8, 7, and 6 isolates that were derived from E4, E11, and E21, respectively, were resistant to CF. After exposure to sub-MIC of CAZ, 6, 8, and 6 isolates derived from E4, E11, and E21, respectively, were resistant to CAZ.
Table 2.
Minimal inhibitory concentrations of cefalotin and ceftazidime against Escherichia coli isolates.
| MIC of E. coli isolates (μg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Antimicrobial | Parent type | -1 ∗ | -2 | -3 | -4 | -5 | -6 | -7 | -8 | -9 |
| E4 | Cefalotin† | 4 | 8 | 32 # | 80 # | 240 # | 240 # | 320 # | 320 # | 320 # | 320 # |
| Ceftazidime‡ | 0.125 | 0.25 | 2 | 4 | 16 # | 16 # | 32 # | 32 # | 64 # | 128 # | |
| E11 | Cefalotin | 0.5 | 2 | 16 | 32 # | # | # | # | # | # | # |
| Ceftazidime | 1 | 4 | 8 # | 8 # | 16 # | 32 # | 32 # | 64 | 128 # | 128 # | |
| E21 | Cefalotin | 2 | 8 | 16 | 16 | # | # | # | # | # | # |
| Ceftazidime | 0.5 | 1 | 4 | 4 | 8 # | 16 # | 16 # | 32 # | 32 # | 64 # | |
No mark: susceptible or intermediate to cefalotin or ceftazidime; #: resistant to cefalotin or ceftazidime. Breakpoints for cefalotin were as follows: susceptible ⩽ 8 μg/mL, intermediate 16 μg/mL, and resistant ⩾ 32 μg/mL. Breakpoints for ceftazidime were as follows: susceptible ⩽ 2 μg/mL, intermediate 4 μg/mL, and resistant ⩾ 8 μg/mL.
∗Escherichia coli isolates at various generations that were induced by cefalotin or ceftazidime.
†MIC values of cefalotin were determined for E. coli isolates induced by cefalotin.
‡MIC values of ceftazidime were determined for E. coli isolates induced by ceftazidime.
3.2. Morphological Characteristics
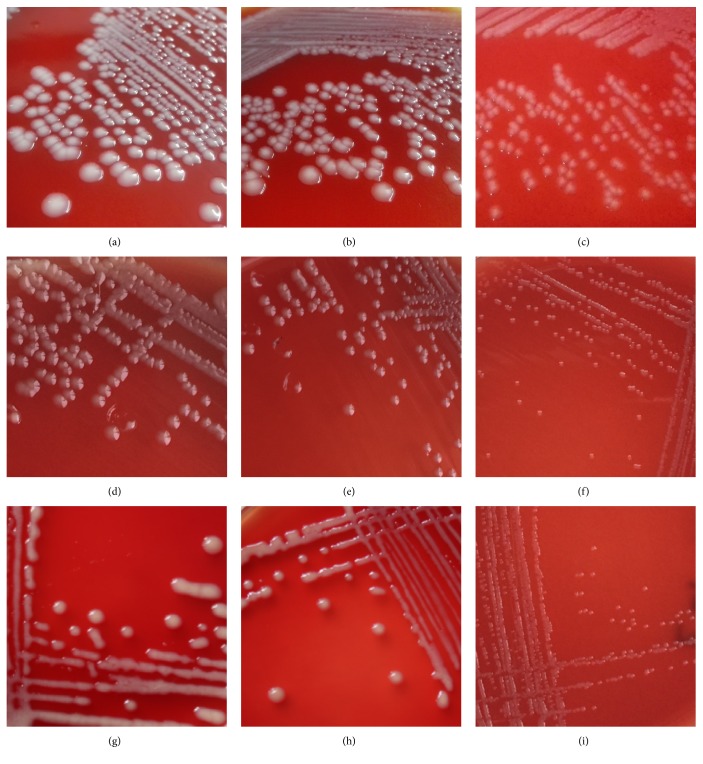
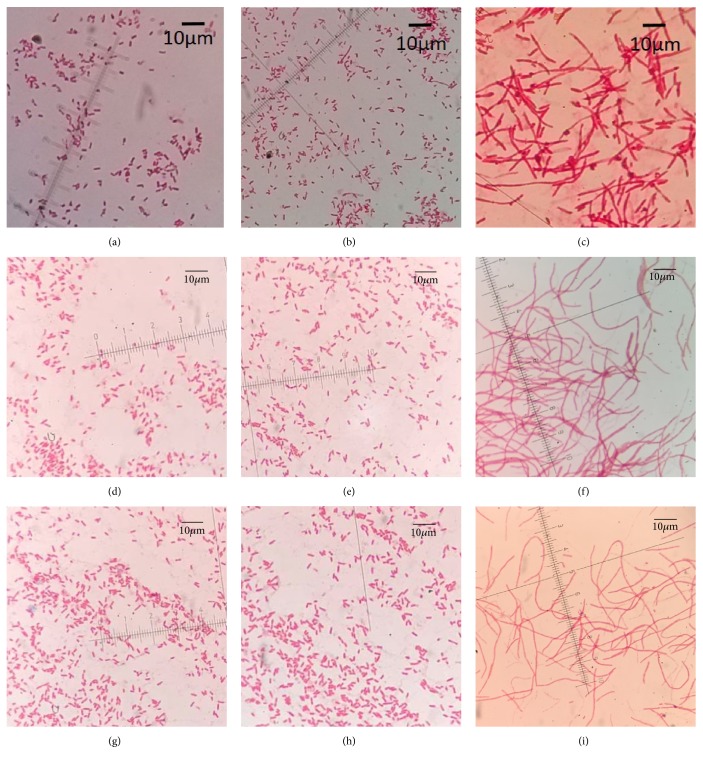
Morphological characteristics of E. coli isolates recovered after induction with CF were similar to those of their parent isolates (Figures 1 and 2). However, morphological changes on blood agar were observed in 17 isolates recovered after induction with CAZ compared to their parent isolates. Of those isolates, 6, 5, and 6 were derived from E4, E11, and E 21, respectively. Colonies of those isolates were smaller after 24 h incubation at 37°C and became sticky and were therefore difficult to retrieve (Figure 1). Also, cells of all isolates recovered after induction with CAZ were elongated, with a filamentous shape (Figure 2).
Figure 1.
Colony morphology of parent Escherichia coli and antibiotic-induced isolates recovered after induction (24 h). (a), (d), and (g) were parent strains; (b), (e), and (h) were cefalotin-induced isolates; and (c), (f), and (i) were ceftazidime-induced isolates.
Figure 2.
Cell morphology of parent Escherichia coli and isolates recovered after induction with antibiotics. Cells were subjected to Gram-staining and examined with an optical microscope (1000×). (a), (d), and (g) were parent strains; (b), (e), and (h) were cefalotin-induced isolates; and (c), (f), and (i) were ceftazidime-induced isolates.
3.3. Antimicrobial Susceptibility Profiles
Among CF-induced isolates, 17, 13, and 13 developed intermediate resistance to cefoxitin, ceftazidime, and kanamycin, respectively, whereas no isolate was resistant to these 3 antimicrobial agents (Table 3). Of isolates with intermediate resistance to cefoxitin, 7, 5, and 5 were derived from E4, E11, and E21, respectively. Of isolates with intermediate resistance to kanamycin, 6, 3, and 4 were derived from E4, E11, and E21, respectively. In addition, 6 and 4 isolates derived from E4 and E21 were resistant to ampicillin and amoxicillin/clavulanic acid, respectively, whereas 7 and 6 isolates derived from E11 became resistant to ampicillin and amoxicillin/clavulanic acid. All CF-induced isolates were susceptible to cefepime, gentamicin, and amikacin. Parent isolates remained susceptible to all tested antimicrobial agents.
Table 3.
Antimicrobial susceptibility profiles of the Escherichia coli isolates recovered after induction with cefalotin.
| Resistant E. coli isolates | |||
|---|---|---|---|
| Antimicrobial | Derived from E4 | Derived from E11 | Derived from E21 |
| Cefoxitin | CF-3/-4/-5/-6/-7/-8/-9 | CF-5/-6/-7/-8/-9 | CF-5/-6/-7/-8/-9 |
| Ceftazidime | CF-5/-6/-7/-8/-9 | CF-5/-6/-7/-8/-9 | CF-7/-8/-9 |
| Ampicillin | CF-4/-5/-6/-7/-8/-9 | CF-3/-4/-5/-6/-7/-8/-9 | CF-6/-7/-8/-9 |
| Amoxicillin/clavulanic acid | CF-4/-5/-6/-7/-8/-9 | CF-4/-5/-6/-7/-8/-9 | CF-6/-7/-8/-9 |
| Kanamycin ∗ | CF-4/-5/-6/-7/-8/-9 | CF-7/-8/-9 | CF-6/-7/-8/-9 |
∗All E. coli isolates had intermediate resistance to cefoxitin, ceftazidime, and kanamycin.
Among CAZ-induced isolates, all were resistant to CF. Nineteen isolates became resistant to cefoxitin, of which 6, 7, and 6 were derived from E4, E11, and E21, respectively. Only 1 CAZ-induced isolate (CAZ-1 derived from E4) was susceptible to ampicillin and amoxicillin/clavulanic acid. All isolates derived from E4 and E21 became intermediate to kanamycin, whereas CAZ-1 from E11 remained susceptible (Table 4). All CAZ-induced isolates were susceptible to cefepime, gentamicin, and amikacin. Parent isolates were susceptible to all tested antimicrobial agents.
Table 4.
Antimicrobial susceptibility profiles of the Escherichia coli isolates recovered after induction with ceftazidime.
| Resistant E. coli isolates | |||
|---|---|---|---|
| Antimicrobial | Derived from E4 | Derived from E11 | Derived from E21 |
| Cefalotin | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 |
| Cefoxitin | CAZ-4/-5/-6/-7/-8/-9 | CAZ-3/-4/-5/-6/-7/-8/-9 | CAZ-4/-5/-6/-7/-8/-9 |
| Ampicillin | CAZ-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 |
| Amoxicillin/ clavulanic acid |
CAZ-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 |
| Kanamycin ∗ | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-2/-3/-4/-5/-6/-7/-8/-9 | CAZ-1/-2/-3/-4/-5/-6/-7/-8/-9 |
∗All E. coli isolates had intermediate resistance to kanamycin.
3.4. ESBL Production and ESBL-Related Genes
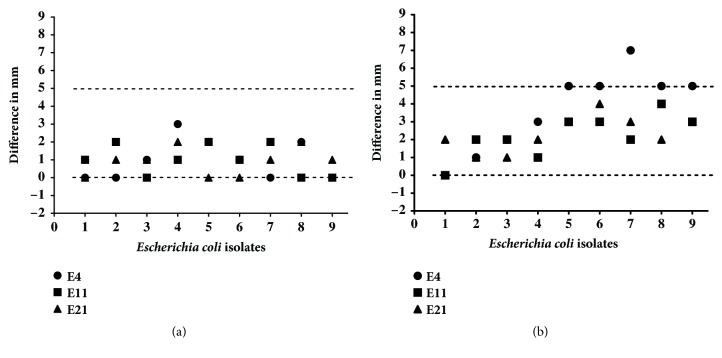
The difference in diameters between the inhibition zone of cefotaxime and cefotaxime plus clavulanic acid was < 5 mm for all induced isolates, except 5 derived from E4 (CAZ-5 to CAZ-9). The difference of the zone diameters for those 5 CAZ-induced isolates was ≥ 5 mm (Figure 3), which was an indication for ESBL production by those isolates. However, none of the tested ESBL-encoding genes were detected in ESBL-producing isolates.
Figure 3.
Extended spectrum beta-lactamase (ESBL) production of isolates recovered after induction by cefalotin (a) or ceftazidime (b). Isolates at various generations are listed on the X-axis. Differences were calculated as diameter of the inhibition zone of ceftazidime plus clavulanic acid subtracted from their respective single discs. An isolate was designated an ESBL producer if the difference was > 5 mm.
3.5. Endotoxin Release
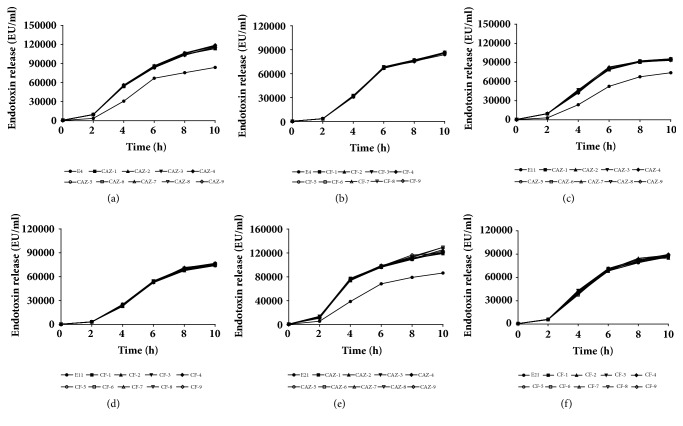
Endotoxin release increased with time (2, 4, 6, 8, and 10 h) for 2 treatment types and control (P < 0.0001). The amount of endotoxin released by CAZ-induced mutants differed from the parent E. coli isolates (P < 0.001). Therefore, CAZ at sub-MIC increased release of endotoxin from induced E. coli mutants when compared to parent strains (Figure 4). However, the amount of endotoxin released by CAZ mutants was not associated with generation of the isolate (P = 0.94). There was no difference in amount of endotoxin released at the same time point for CF-induced mutants compared to parent E. coli isolates (P = 0.70), nor did endotoxin release by CF-mutants vary by generation (P = 0.79). Therefore, CF at sub-MIC did not increase endotoxin release, compared to parent isolates, at any time point.
Figure 4.
Endotoxin release of Escherichia coli isolates included in the study at various time points (0, 2, 4, 6, 8, and 10 h).
Both antibiotic type and incubation time were associated with endotoxin release (Table 5; Figure 4). All treatments followed similar release patterns over time, starting with a high rate of endotoxin release that plateaued after 6 h.
Table 5.
Results of a linear mixed regression model for log endotoxin release (log of EU/mL) by Escherichia coli after exposure to cefalotin (CF) or ceftazidime (CAZ).
| Parameter | Estimate | SEM | P-value |
|---|---|---|---|
| Parent isolates (Ref.) ∗ | 6.07 | 0.11 | <0.001 |
| CAZ | 0.50 | 5.01 x 10−3 | <0.001 |
| CF | 0.02 | 5.01 x 10−3 | 0.699 |
| Interactions† | |||
| CAZ x generation | 3.78 x 10−4 | 5.23 x 10−3 | 0.942 |
| CF x generation | 1.39 x 10−3 | 5.23 x 10−3 | 0.791 |
| Time (h) | 1.60 | 0.02 | <0.001 |
| Time2 (h2) | -0.16 | 5.39 x 10−3 | <0.001 |
| Time3 (h3) | 0.01 | 3.54 x 10−4 | <0.001 |
|
| |||
| Between-replicate variance | 0.03 | 0.16 | |
Endotoxin release was measured at various time points (2, 4, 6, 8, and 10 h) and resistance was induced over varying numbers of generations.
∗Baseline to which other samples were compared was a parent isolate (generation 0)
†Interactions were not created with the parent isolate as there was only 1 generation
4. Discussion
This was apparently the first study to characterize changes in morphology, antimicrobial resistance, and endotoxin releasing of E. coli isolated from bovine CM under persistent exposure to CF and CAZ at sub-MIC. In this study, sub-MIC in vitro exposure of CF and CAZ resulted not only in failure of killing E. coli, but also in increased antimicrobial resistance. In addition, exposure to CAZ led to generation of more virulent strains of E. coli. Choice of antibiotic, administration route, and treatment duration are determined by many factors [23], which makes the choice of treatment strategy relatively subjective or arbitrary. Based on our findings, we inferred that prudent usage of antibiotics should be proposed when treating CM, particularly if caused by E. coli.
After continuous sub-MIC exposure to CF or CAZ, isolates became increasingly resistant to CF or CAZ, respectively. Bacteria can quickly develop resistant phenotypes under persistent cephalosporin stress [24]. In addition, several isolates became resistant to β-lactams (CF, cefoxitin, CAZ, ampicillin, and amoxicillin/clavulanic acid) after exposure to either CF or CAZ. Similarly, bovine E. coli strains isolated from cows treated with ceftiofur became resistant to cefazoline, whereas strains not previously exposed to ceftiofur remained susceptible [25]. Resistance of E. coli strains to multiple antimicrobials could result in failure of mastitis therapy and is a serious threat to human and animal health. Mutants of Pseudomonas aeruginosa isolated from humans became resistant to various antimicrobials after induction with ceftazidime [26]. Resistance to vancomycin increased in Staphylococcus aureus isolates, accompanied with morphologic alteration, after continuous exposure to ceftazidime [27]. Results of these 2 studies were in accordance with our findings.
Colonies of E. coli isolates recovered after induction with CAZ were smaller and stickier than the parent isolate. This was in accordance with a previous study that growth of Bacteroides thetaiotaomicron isolates was slower after exposure to cefoxitin [28]. In our study, bacterial cells became extensively elongated (filamentation), consistent with several E. coli strains that become filamentous after exposure to various antimicrobial agents [18, 19, 29]. Escherichia coli readily became filamentous in shape when exposed to cephalosporins [29]. Effects of antibiotics on bacterial morphology or survival depend on both type and dosage [19, 30]. Antibiotics can kill bacteria by acting on penicillin-binding proteins (PBPs) [31]. There are 3 kinds of PBPs: PBP1, PBP2, and PBP3. Various types and dosages of antibiotics could bind to different PBPs, which may result in a morphology change of bacteria [30]. A high concentration of CAZ can destroy bacteria quickly by inhibiting PBP1, whereas the same antibiotic administered at lower concentrations can lead to filamentation of bacteria [32]. In the present study, filamentation of isolates at 6th and later generations of growth was weaker than during earlier generations; perhaps induced isolates were already adapted to antimicrobial exposure. It has been reported that the filamentous bacteria return to a typical bacillus shape after antibiotic removal, so filamentation was temporary [33]. Interestingly, isolates recovered after induction with CF did not undergo obvious changes in colonial and bacterial morphology. Presumably, CF bound to different PBPs, which did not make the cell filamentous.
In this study, 5 isolates recovered after induction with CAZ produced ESBL phenotype, while no ESBL-associated genes were detected in these isolates. One potential reason for this discrepancy might be that we were unable to detect the genes that were responsible for ESBL production. In the present study, we only tried to detect common ESBL-encoding genes, as described [21, 22]. Nevertheless, other ESBL-encoding genes not targetted in this study may have been responsible for production of ESBL [34]. Moreover, there might be a misinterpretation of data, since the difference of 4 of 5 isolates was just at the threshold (5 mm) to detect ESBL phenotypes.
Release of endotoxins from the disrupted E. coli cell walls could cause a serious inflammatory reaction in an infected udder [35]. Besides, high endotoxin in ingested infant formula milk might cause neonatal bacteraemia and endotoxemia, especially in neonates with immature innate immune systems [8]. In our study, all parent and induced isolates released more endotoxin when incubation was prolonged, which was in accordance with reports that bacteria released more endotoxin in induction period by antibiotics due to overexpression or shedding at higher speed from cell wall [36, 37]. Increased release of endotoxin from isolates recovered after induction with CAZ may have been associated with filamentation of E. coli after exposure to CAZ [19, 30]. Filamentation of cells could increase the surface of the cell wall with a higher endotoxin release from cell wall and, therefore, a higher releasing speed. In contrast, isolates recovered after induction with CF did not produce more endotoxin than parent isolates, which may be associated with less morphological changes in these isolates.
Although a series of phenotypic changes (morphology, antimicrobial resistance, and endotoxin releasing capability) were observed in the current study, mechanism of bacteria's acquired resistance after exposure to the antibiotics should be further clarified. Therefore, a study including whole-genome sequencing and analysis of acquired mutations should be conducted, to provide insights regarding the origin of observed resistance.
5. Conclusions
After exposure to subminimum concentrations of CF or CAZ, parent E. coli CM isolates underwent a series of changes related to morphology, antimicrobial resistance pattern, and endotoxin release. Several isolates recovered after induction had higher resistance to CF, CAZ, and some other commonly used antimicrobials. Moreover, several isolates recovered after induction with CAZ had slower growth on blood agar, became filamentous, and released more endotoxin. Therefore, we inferred that there shoud be prudent use of cephalosporins, especially those that belong to the 3rd generation, for treatment of bovine mastitis caused by E. coli. Such treatment would increase potential risk to formula fed neonates if the milk with endotoxin was processed into powdered infant formula milk.
Acknowledgments
This research was financially supported by the Chinese 13 “Five-year” National Science and Technology Support Project (no. 2016YFD0501203), High-end Foreign Experts Recruitment Program (no. GDT20141100043 and no. GDT20171100013), and the National Natural Science Foundation of China (no. 31572587 and no. 31550110200). The authors thank Dr. John Kastelic, Faculty of Veterinary Medicine, University of Calgary, for editing the manuscript.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Gang Liu and Laidi Ding contributed equally to this work.
References
- 1.Bradley A. J. Bovine mastitis: an evolving disease. The Veterinary Journal. 2002;164(2):116–128. doi: 10.1053/tvjl.2002.0724. [DOI] [PubMed] [Google Scholar]
- 2.Kuipers A., Koops W. J., Wemmenhove H. Antibiotic use in dairy herds in the Netherlands from 2005 to 2012. Journal of Dairy Science. 2016;99(2):1632–1648. doi: 10.3168/jds.2014-8428. [DOI] [PubMed] [Google Scholar]
- 3.Stevens M., Piepers S., De Vliegher S. Mastitis prevention and control practices and mastitis treatment strategies associated with the consumption of (critically important) antimicrobials on dairy herds in Flanders, Belgium. Journal of Dairy Science. 2016;99(4):2896–2903. doi: 10.3168/jds.2015-10496. [DOI] [PubMed] [Google Scholar]
- 4.Barkema H. W., Schukken Y. H., Lam T. J. G. M., et al. Incidence of clinical mastitis in dairy herds grouped in three categories by bulk milk somatic cell count. Journal of Dairy Science. 1998;81(2):411–419. doi: 10.3168/jds.S0022-0302(98)75591-2. [DOI] [PubMed] [Google Scholar]
- 5.Breen J. E., Green M. J., Bradley A. J. Quarter and cow risk factors associated with the occurrence of clinical mastitis in dairy cows in the United Kingdom. Journal of Dairy Science. 2009;92(6):2551–2561. doi: 10.3168/jds.2008-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verbeke J., Piepers S., Supré K., De Vliegher S. Pathogen-specific incidence rate of clinical mastitis in Flemish dairy herds, severity, and association with herd hygiene. Journal of Dairy Science. 2014;97(11):6926–6934. doi: 10.3168/jds.2014-8173. [DOI] [PubMed] [Google Scholar]
- 7.Gao J., Barkema H. W., Zhang L., et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. Journal of Dairy Science. 2017;100(6):4797–4806. doi: 10.3168/jds.2016-12334. [DOI] [PubMed] [Google Scholar]
- 8.Townsend S., Caubilla Barron J., Loc-Carrillo C., Forsythe S. The presence of endotoxin in powdered infant formula milk and the influence of endotoxin and Enterobacter sakazakii on bacterial translocation in the infant rat. Food Microbiology. 2007;24(1):67–74. doi: 10.1016/j.fm.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Suojala L., Pohjanvirta T., Simojoki H., et al. Phylogeny, virulence factors and antimicrobial susceptibility of Escherichia coli isolated in clinical bovine mastitis. Veterinary Microbiology. 2011;147(3-4):383–388. doi: 10.1016/j.vetmic.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Poutrel B., Stegemann M. R., Roy O., Pothier F., Tilt N., Payne-Johnson M. Evaluation of the efficacy of systemic danofloxacin in the treatment of induced acute Escherichia coli bovine mastitis. Journal of Dairy Research. 2008;75(3):310–318. doi: 10.1017/S0022029908003348. [DOI] [PubMed] [Google Scholar]
- 11.Schukken Y. H., Bennett G. J., Zurakowski M. J., et al. Randomized clinical trial to evaluate the efficacy of a 5-day ceftiofur hydrochloride intramammary treatment on nonsevere gram-negative clinical mastitis. Journal of Dairy Science. 2011;94(12):6203–6215. doi: 10.3168/jds.2011-4290. [DOI] [PubMed] [Google Scholar]
- 12.Ali T., Ur Rahman S., Zhang L., et al. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Frontiers in Microbiology. 2016;7, article no. 1931 doi: 10.3389/fmicb.2016.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oteo J., Pérez-Vázquez M., Campos J. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Current Opinion in Infectious Diseases. 2010;23(4):320–326. doi: 10.1097/qco.0b013e3283398dc1. [DOI] [PubMed] [Google Scholar]
- 14.Ali T., Rahman S. U., Zhang L., et al. Characteristics and genetic diversity of multi-drug resistant extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from bovine mastitis. Oncotarget . 2017;8(52):90144–90163. doi: 10.18632/oncotarget.21496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silbergeld E. K., Graham J., Price L. B. Industrial food animal production, antimicrobial resistance, and human health. Annual Review of Public Health. 2008;29:151–169. doi: 10.1146/annurev.publhealth.29.020907.090904. [DOI] [PubMed] [Google Scholar]
- 16.Guérin-Faublée V., Carret G., Houffschmitt P. In vitro activity of 10 antimicrobial agents against bacteria isolated from cows with clinical mastitis. Veterinary Record. 2003;152(15):466–471. doi: 10.1136/vr.152.15.466. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer Y., Cullik A., Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. International Journal of Medical Microbiology. 2010;300(6):371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Buijs J., Dofferhoff A. S., Mouton J. W., van der Meer J. W. Continuous administration of PBP-2- and PBP-3-specific β-lactams causes higher cytokine responses in murine Pseudomonas aeruginosa and Escherichia coli sepsis. Journal of Antimicrobial Chemotherapy. 2007;59(5):926–933. doi: 10.1093/jac/dkm073. [DOI] [PubMed] [Google Scholar]
- 19.Jackson J. J., Kropp H., Hurley J. C. Influence of antibiotic class and concentration on the percentage of release of lipopolysaccharide from escherichia coli. The Journal of Infectious Diseases. 1994;169(2):471–472. doi: 10.1093/infdis/169.2.471. [DOI] [PubMed] [Google Scholar]
- 20.CLSI. Clinical and Laboratory Standards Institute, Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals—Third Edition: Approved Standard Vet01S, 2015 Wayne, PA, USA. 2015.
- 21.Jouini A., Vinue L., Slama K. B., et al. Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia colistrains of food samples in Tunisia. Journal of Antimicrobial Chemotherapy. 2007;60(5):1137–1141. doi: 10.1093/jac/dkm316. [DOI] [PubMed] [Google Scholar]
- 22.Kanamori H., Navarro R. B., Yano H., et al. Molecular characteristics of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae from the Philippines. Acta Tropica. 2011;120(1-2):140–145. doi: 10.1016/j.actatropica.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Swinkels J. M., Hilkens A., Zoche-Golob V., et al. Social influences on the duration of antibiotic treatment of clinical mastitis in dairy cows. Journal of Dairy Science. 2015;98(4):2369–2380. doi: 10.3168/jds.2014-8488. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki S., Shibata N., Yamane K., Wachino J., Ito K., Arakawa Y. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. Journal of Antimicrobial Chemotherapy. 2008;63(1):72–79. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 25.Sato T., Okubo T., Usui M., et al. Association of Veterinary Third-Generation Cephalosporin Use with the Risk of Emergence of Extended-Spectrum-Cephalosporin Resistance in Escherichia coli from Dairy Cattle in Japan. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0096101.e96101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabot G., Zamorano L., Moyà B., et al. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrobial Agents and Chemotherapy. 2016;60(3):1767–1778. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roch M., Clair P., Renzoni A., et al. Exposure of Staphylococcus aureus to Subinhibitory Concentrations of β-Lactam Antibiotics Induces Heterogeneous Vancomycin-Intermediate Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2014;58(9):5306–5314. doi: 10.1128/AAC.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang H., Edlund C., Hultenby K., Hedberg M. Effects of cefoxitin on the growth and morphology of Bacteroides thetaiotaomicron strains with different cefoxitin susceptibility. Anaerobe. 2002;8(2):55–61. doi: 10.1006/anae.2002.0417. [DOI] [Google Scholar]
- 29.Goscinski G., Tano E., Löwdin E., Sjölin J. Propensity to release endotoxin after two repeated doses of cefuroxime in an in vitro kinetic model: Higher release after the second dose. Journal of Antimicrobial Chemotherapy. 2007;60(2):328–333. doi: 10.1093/jac/dkm190. [DOI] [PubMed] [Google Scholar]
- 30.Dofferhoff A. S. M., Buys J. The influence of antibiotic-induced filament formation on the release of endotoxin from Gram-negative bacteria. Journal of Endotoxin Research. 1996;3(3):187–194. doi: 10.1177/096805199600300304. [DOI] [Google Scholar]
- 31.Sakoulas G., Kumaraswamy M., Nonejuie P., et al. Differential effects of penicillin binding protein deletion on the susceptibility of Enterococcus faecium to cationic peptide antibiotics. Antimicrobial Agents and Chemotherapy. 2015;59(10):6132–6139. doi: 10.1128/AAC.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buijs J., Dofferhoff A. S. M., Mouton J. W., Wagenvoort J. H., van der Meer J. A quantitative assessment of dose-dependent β-lactam-induced filament-formation in Gram-negative bacteria. Clinical Microbiology and Infection. 2003;9(4):88–91. doi: 10.1111/j.1469-0691.2007.01940.x. [DOI] [PubMed] [Google Scholar]
- 33.Chen K., Sun G. W., Chua K. L., Gan Y.-H. Modified virulence of antibiotic-induced Burkholderia pseudomallei filaments. Antimicrobial Agents and Chemotherapy. 2005;49(3):1002–1009. doi: 10.1128/AAC.49.3.1002-1009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Day M. J., Rodríguez I., van Essen-Zandbergen A., et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. Journal of Antimicrobial Chemotherapy. 2016;71(5):1178–1182. doi: 10.1093/jac/dkv485. [DOI] [PubMed] [Google Scholar]
- 35.Schukken Y. H., Günther J., Fitzpatrick J., et al. Host-response patterns of intramammary infections in dairy cows. Veterinary Immunology and Immunopathology. 2011;144(3-4):270–289. doi: 10.1016/j.vetimm.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Holzheimer R. G. Antibiotic induced endotoxin release and clinical sepsis: a review. Journal of Chemotherapy. 2001;13(1) suppl 2:159–172. doi: 10.1179/joc.2001.13.supplement-2.159. [DOI] [PubMed] [Google Scholar]
- 37.Flynn P. M., Shenep J. L., Gigliotti F., Davis D. S., Hildner W. K. Immunolabeling of lipopolysaccharide liberated from antibiotic-treated Escherichia coli. Infection and Immunity. 1988;56(10):2760–2762. doi: 10.1128/iai.56.10.2760-2762.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.