Abstract
Objectives
To evaluate the influence of MRI scanning parameters on texture analysis features.
Methods
Publicly available data from the Reference Image Database to Evaluate Therapy Response (RIDER) project sponsored by The Cancer Imaging Archive included MRIs on a phantom comprised of 18 25‐mm doped, gel‐filled tubes, and 1 20‐mm tube containing 0.25 mM Gd‐DTPA (EuroSpinII Test Object5, Diagnostic Sonar, Ltd, West Lothian, Scotland). MRIs performed on a 1.5 T GE HD, 1.5 T Siemens Espree (VB13), or 3.0 T GE HD with TwinSpeed gradients with an eight‐channel head coil included T1WIs with multiple flip angles (flip‐angle = 2,5,10,15,20,25,30), TR/TE = 4.09–5.47/0.90–1.35 ms, NEX = 1 and DCE with 30° flip‐angle, TR/TE=4.09–5.47/0.90–1.35, and NEX = 1,4. DICOM data were imported into an in‐house developed texture analysis program which extracted 41‐texture features including histogram, gray‐level co‐occurrence matrix (GLCM), and gray‐level run‐length (GLRL). Two‐tailed t tests, corrected for multiple comparisons (Q values) were calculated to compare changes in texture features with variations in MRI scanning parameters (magnet strength, flip‐angle, number of excitations (NEX), scanner platform).
Results
Significant differences were seen in histogram features (mean, median, standard deviation, range) with variations in NEX (Q = 0.003–0.045) and scanner platform (Q < 0.0001), GLCM features (entropy, contrast, energy, and homogeneity) with NEX (Q = 0.001–0.018) and scanner platform (Q < 0.0001), GLRL features (long‐run emphasis, high gray‐level run emphasis, high gray‐level emphasis) with magnet strength (Q = 0.0003), NEX (Q = 0.003–0.022) and scanner platform (Q < 0.0001).
Conclusion
Significant differences were seen in many texture features with variations in MRI acquisition emphasizing the need for standardized MRI technique.
Keywords: phantom, quantitative MRI, texture analysis
Abbreviations
- GLCM
gray‐level co‐occurrence matrix
- GLGM
gray‐level gradient matrix
- GLN
gray‐level nonuniformity
- GLRL
gray‐level run‐length
- NEX
number of excitations
- RLN
run‐length nonuniformity
- SRE
short‐run emphasis
- SRLGE
short‐run low gray‐level emphasis
1. INTRODUCTION
Increasing radiology research efforts have been focused on the investigation of potential genotype‐phenotype relationships of tumor biology and behavior, often referred to as radiomics. Radiomics has been employed in an effort to identify distinct behavioral subtypes of tumors based on their imaging appearance, with the ultimate hope of predicting tumor prognosis and treatment response. This field of radiomics often uses quantitative post‐processing techniques, known as a texture analysis, to identify unique pixel intensity patterns, or textures, within a tumor lesion based on its imaging appearance (e.g., CT, MRI, ultrasound, etc.).
Texture analysis describes the patterns of pixel intensity variations within an image calculated by a series of mathematical algorithms.1 Numerous texture analysis features have been described in the literature and defined in the work of Haralick et al.1 The use of a texture analysis applied to imaging studies including CT and MRI have been previously performed for the evaluation of multiple nonneoplastic disorders including the evaluation for mesial temporal sclerosis on MRI,2 evaluation of intervertebral disc disease on MRI,3 evaluation of hepatic fibrosis on both CT and MRI,4, 5, 6, 7, 8 evaluation of subchondral bone on MRI.9 Prior oncologic studies have also employed texture analyses to evaluate specific tumor features including the assessment of HPV status of oropharyngeal squamous cell carcinomas,8 prognosis of head and neck neoplasms,10, 11, 12 classification of gastric and colorectal tumors on CT,13, 14, 15 genomic mapping and predictive marker identification of gliomas on MRI,16, 17, 18, 19 the identification of potentially prognostic predictors in lung cancer,20, 21 evaluation of genitourinary neoplasms on both CT and MRI,22, 23, 24, 25 and for the radiomic classifications of breast carcinoma subtypes.26, 27, 28
In an effort to study specific patterns of tumor biology correlating with different imaging appearances, multi‐institutional centers have worked toward pooling resources to make publicly available cancer imaging databases, such as The Cancer Imaging Archive (TCIA) and The Cancer Genomic Atlas (TCGA), to help facilitate research efforts in the arena of tumor genotype‐phenotype analyses.16, 26, 28 Prior research studies have used a radiomics approach for investigating prostate cancer radiotherapy responses,25 responsiveness of neoadjuvant chemotherapy in breast cancer,28 and prognostic predictions of advanced nasopharyngeal carcinoma.12 However, larger studies and systematic reviews on radiomics have noted methodological variations as a source of difficulty precluding an accurate and collective interpretation of data.11, 29, 30
Based on our knowledge of how changes in the CT scanning parameters varies texture analysis features30, as well as preliminary studies investigating the sensitivity of texture features to variations in MRI technique,29, 31, 32, 33 we could similarly deduce that changes in MRI scanning parameters such as differences in magnet strength and scanner platform could also influence texture analysis features. Thus, the purpose of this study was to evaluate and quantify changes in MRI sequence parameters may have on texture analysis features using a simple, nonanatomic phantom model.
2. MATERIALS AND METHODS
This study employed the use of a phantom for all image acquisitions, precluding the requirement for IRB approval.
2.A. Phantom development and MR imaging techniques
The construction of the phantom, and scan data of serial MRI scans of this phantom are publicly available as part of the Reference Image Database to Evaluate Therapy Response (RIDER) at The Cancer Imaging Archive (TCIA).34 The original DICOM datasets and scan data on the RIDER phantom are available for public use in an effort to generate an initial consensus on how to harmonize the data collection and analysis for quantitative imaging methods applied to the measurement of drug and/or radiation treatment response.35
The nonanatomic phantom used in the RIDER database was comprised of 18 25‐mm doped gel‐filled tubes, and a single 20‐mm tube containing 0.25 mM GdDTPA (EuroSpin II Test Object 5, Diagnostic Sonar, Ltd, West Lothian, Scotland),34 as shown in Fig 1.
Figure 1.
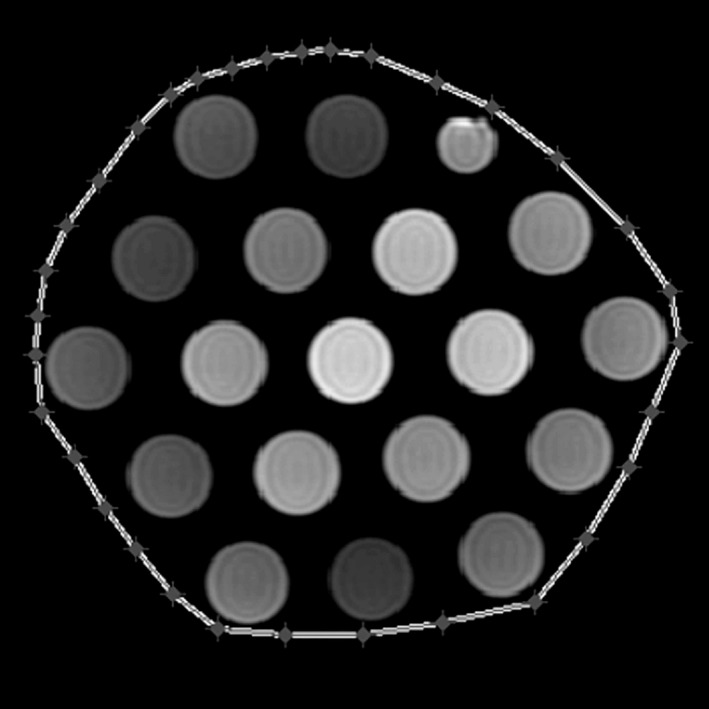
A cross‐sectional T1 weighted DICOM image through the nonanatomic phantom with a peripheral contour created within our in‐house developed MatLab platform. The phantom was composed of 18 dope‐filled gel tubes and a single tube filled with 0.25 mM of Gd‐DTPA.
All MRI examinations were performed at MD Anderson on either a 1.5 T GE HD, a 1.5 T Siemens Espree (VB13), or a 3.0 T GE HD with TwinSpeed gradients. An eight‐channel head coil was used for all scans.
Acquired scans included a T1‐weighted image using multiple flip angles, three‐dimensional Fast Spoiled Gradient Recalled Echo Sequence with flip angles = 2,5,10,15,20,25,30, a TR/TE = 4.09–6.469/0.90–1.35 ms, receiver bandwidth = ±31.25 kHz, 256 × 192 matrix, NEX = 1, slice thickness = 5 mm, and a 24 × 19 cm field of view.
A DCE acquisition was also performed using a three‐dimensional Fast Spoiled Gradient Recalled Echo Sequence with 30‐degree flip angle, a TR/TE = 4.09–5.47/0.90–1.35, receiver bandwidth = ±31.25 kHz, and NEX = 1,4, slice thickness = 5 mm, 256 × 160 matrix, and a 24 × 19 cm field of view.
2.B. DICOM segmentation and texture analysis
Original DICOM data sets were downloaded and then imported into in‐house developed MATLAB (MathWorks, Natick, MA) texture analysis software to calculate texture analysis features. The texture analysis software was developed by the co‐author (BL) and the use of this texture analysis program has been previously reported in the literature.7, 8, 30 Image segmentation of phantom was performed manually by an experienced radiologist (co‐author HK), using the same geometric boundaries and a uniform contour volume for each dataset in an effort to reduce potential variation related to the manual segmentation process. The entirety of the phantom was contoured including each of the doped gel‐filled tubes, the gadolinium filled tube, as well as the negative space between in the inserts. A correction for spatial inhomogeneity was not applied. Prior to the texture analysis, the contoured images were preprocessed (or corrected) which consisted of the following steps: (a) partial volume artifact correction, and (b) global grayscale normalization. These steps are described in the work by Li et al.7 In brief, to correct for partial volume artifact, an optimal thresholding algorithm was applied using an iterative optimal thresholding algorithm.36 This method assumes all image pixels are from two probability distributions (e.g., structure of interest and the dark background) and attempts to find the gray‐level threshold corresponding to the minimum probability between the maxima of the two distributions, which results in minimal segmentation error. To find the optimal threshold, this algorithm was applied iteratively (usually four to ten iterations were sufficient), updating the threshold in each iteration from the weighted sum of the two distributions. For global grayscale normalization, the images were corrected by the mean and standard deviation to minimize the overall grayscale variation across images, similar to that described in the work of Collewet et al.33 The correction was applied to the entire image. The mean gray value of each corrected image was set to 250 and the standard deviation to 30.
In total, 41 texture features, including 12 histogram features, five gray‐level co‐occurrence matrix (GLCM) features, 11 gray‐level run‐length (GLRL) features, four gray‐level gradient matrix (GLGM) features, and nine Laws features, were calculated and averaged over the contoured images of each dataset. Numerous texture analysis equations have been defined and developed. Only a subset of 41 texture features were employed in this study based on our prior work, and based on the popularity of reported texture features in the radiomics literature.7, 8, 30
The use of our in‐house developed MATLAB program and the specific details of the texture analysis features calculated by this program have been previously published.7, 8, 30 A full description of the mathematical equations is described in the work by Haralick et al.1 and Tang el al.37 GLCM features, in contrast to histogram features, are highly spatially dependent. In this study, the GLCM texture features were calculated using only directly adjacent pixels for simplicity. Horizontal, 45°, vertical, and 135° directions were averaged together to eliminate any directional dependence. The following GLCM features proposed by Haralick et al.1 were tested:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where (i, j) represents the (i, j) value of the GLCM.
GLRL matrices were used as these texture features provide additional insights into spatial dependence18. The same directions considered for the calculation of the GLCM features, were averaged for the GLRL matrix features. The features explored included equations utilizing short‐run emphasis (SRE), long‐run emphasis (LRE), gray‐level nonuniformity (GLN), run‐length nonuniformity (RLN), run percentage (RP), low gray‐level run emphasis (LGRE), high gray‐level run emphasis (HGRE), short‐run low gray‐level emphasis (SRLGE), short‐run high gray‐level emphasis (SRHGE), long‐run low gray‐level emphasis (LRLGE), and long‐run high gray‐level emphasis (LRHGE), defined as follows:
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
where p(i, j) represents the (i, j) value of the GLRL matrix, nr is the total number of runs, and np is the total number of pixels.
GLGM features were also investigated to provide the histogram of the absolute gradient values in the interrogated region of interest. As a preprocessing step, the gradient of each pixel within the ROI was computed using a 3 × 3 neighborhood. The GLGM features mathematically summarize the gradient values of the pixels in the ROI and include mean, variance, skewness, and kurtosis.
2.C. Statistical analysis
Serial MRI scans on the phantom were performed with variations in specific MRI scanning parameters. Multiple scan sequences are available in the RIDER dataset where a specific MRI scanning parameter is varied i.e., flip angle, while the remaining scanning parameters are held constant. We collated such scans where individual scanning parameters were sequentially varied in order to make the following assessments:
Assessment of Magnet Strength: T1‐weighted images performed on a 1.5 T GE Signa Excite compared to a T1‐weighted image performed on a 3 T GE Signa Excite with flip angle, TR/TE, number of excitations (NEX), echo train length, slice thickness, and matrix held constant
Assessment of Flip Angle: T1‐weighted images performed on a 1.5 T Siemens Espree with flip angle varying from 2, 5, 10, 15, 20, 25, and 30 degrees with the TRTE, NEX, echo train length, slice thickness, and matrix held constant.
Assessment of NEX: DCE images performed on a 1.5 T Siemens Espree with NEX either 1, or 4. The flip angle, TR/TE, slice thickness, and matrix were held constant
Assessment of Scanner Platform: DCE images were performed on a 1.5 T GE Signal Excite compared to a 1.5 T Siemens Espree with the flip angle, TR/TE, NEX, echo train length, slice thickness and matrix held constant.
For each of the four comparisons, a student's t test for assessing independent samples was used to evaluate variations in the 41 texture features based and was reported as a P value. To adjust for multiple comparisons, a false discovery rate (FDR) correction was performed and the FDR correction of the P values (termed Q values) were calculated in addition to raw P values using Benjamini and Hochberg method described in the literature.38 Statistical computations were performed using SAS 9.1.3 software (SAS Institute, Cary, NC).
3. RESULTS
Changes in texture analysis features based on variations in MR scanning parameters are shown in Tables 1, 2, 3, 4, and Table S1.
Table 1.
Texture parameters: 1.5T vs 3T
| 1.5T (n = 80) | 3T (n = 61) | P value | Q value | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Histogram | ||||||
| Mean | 247.2 | 2.2 | 246.8 | 2.4 | 0.225 | 0.298 |
| Median | 244.7 | 4.8 | 243.4 | 5.7 | 0.170 | 0.239 |
| STD | 30.6 | 4.5 | 31.8 | 5.7 | 0.184 | 0.251 |
| Range | 8.3 | 2.8 | 7.9 | 2.4 | 0.465 | 0.554 |
| Geometric mean | 248.2 | 0.49 | 248.0 | 0.61 | 0.142 | 0.220 |
| Harmonic mean | 246.4 | 0.90 | 246.2 | 1.1 | 0.164 | 0.238 |
| 2nd STD | 2.9 | 1.0 | 2.7 | 0.84 | 0.336 | 0.432 |
| STD5 | 3.1 | 1.1 | 2.9 | 0.96 | 0.159 | 0.238 |
| STD9 | 3.2 | 1.0 | 3.2 | 0.93 | 0.892 | 0.934 |
| 4th moment | 3069652.7 | 2076989.3 | 4128044.7 | 2905848.7 | 0.018 | 0.045 |
| IQR | 39.6 | 4.3 | 39.1 | 3.8 | 0.503 | 0.580 |
| Entropy | 7.2 | 0.31 | 7.2 | 0.25 | 0.702 | 0.790 |
| GLCM | ||||||
| Entropy | 2.1 | 0.45 | 2.1 | 0.43 | 0.745 | 0.818 |
| Contrast | 16.9 | 6.5 | 18.9 | 6.6 | 0.075 | 0.143 |
| Correlation | 0.91 | 0.08 | 0.90 | 0.08 | 0.360 | 0.450 |
| Energy | 0.01 | 0.003 | 0.01 | 0.003 | 0.817 | 0.875 |
| Homogeneity | 0.60 | 0.05 | 0.59 | 0.04 | 0.940 | 0.940 |
| GLRL | ||||||
| SRE | 0.09 | 0.03 | 0.10 | 0.04 | 0.054 | 0.128 |
| LRE | 0.09 | 0.04 | 0.10 | 0.04 | 0.065 | 0.133 |
| GLN | 0.09 | 0.03 | 0.10 | 0.04 | 0.090 | 0.153 |
| RLN | 0.09 | 0.04 | 0.10 | 0.04 | 0.063 | 0.133 |
| RP | 162.1 | 42.3 | 149.4 | 41.2 | 0.076 | 0.143 |
| LGRE | 159.5 | 43.3 | 147.1 | 42.5 | 0.092 | 0.153 |
| HGRE | 157.9 | 42.6 | 146.4 | 41.7 | 0.113 | 0.182 |
| SRLGE | 159.8 | 43.4 | 147.2 | 42.5 | 0.087 | 0.153 |
| SRHGE | 3052.4 | 1691.4 | 1833.1 | 626.2 | <0.0001 | 0.0003 |
| LRLGE | 3661.3 | 2077.6 | 2100.8 | 755.5 | <0.0001 | 0.0003 |
| LRHGE | 2647.5 | 1455.7 | 1568.1 | 532.6 | <0.0001 | 0.0003 |
| Law's features | ||||||
| L1 | 162232.6 | 83374.1 | 252842.7 | 54759.9 | <0.0001 | 0.0003 |
| L2 | 18049.8 | 12630.7 | 30827.7 | 11995.7 | <0.0001 | 0.0003 |
| L3 | 6438.0 | 3814.3 | 10443.2 | 2874.5 | <0.0001 | 0.0003 |
| L4 | 33648.4 | 14046.5 | 49080.4 | 7890.0 | <0.0001 | 0.0003 |
| L5 | 6830.8 | 7083.6 | 13929.7 | 6922.4 | <0.0001 | 0.0003 |
| L6 | 5044.4 | 4893.9 | 9891.0 | 4395.4 | <0.0001 | 0.0003 |
| L7 | 3793.4 | 3264.1 | 7070.4 | 2912.7 | <0.0001 | 0.0003 |
| L8 | 10919.5 | 9724.9 | 20981.7 | 8112.5 | <0.0001 | 0.0003 |
| L9 | 15398.6 | 6675.3 | 22754.1 | 3790.9 | <0.0001 | 0.0003 |
| GLGM | ||||||
| MGR | 20.7 | 10.2 | 29.1 | 13.0 | <0.0001 | 0.0003 |
| VGR | 16410.5 | 8816.7 | 24038.9 | 11063.9 | <0.0001 | 0.0003 |
| Skewness | 7.7 | 2.0 | 6.5 | 1.9 | 0.001 | 0.003 |
| Kurtosis | 67.6 | 34.4 | 50.2 | 29.0 | 0.002 | 0.005 |
| Mean skewness | 0.64 | 0.59 | 0.72 | 0.59 | 0.468 | 0.554 |
| Mean kurtosis | 3.0 | 0.5 | 3.2 | 0.6 | 0.063 | 0.133 |
| Mean laws | 270470.4 | 141778.4 | 423628.7 | 100109.0 | <0.0001 | 0.0003 |
Mean texture analysis features on a 1.5 T vs a 3.0 T scanner. n: number of contoured slices; STD: standard deviation; STD5: 5‐neighborhood standard deviation; STD9: 9‐neighborhood standard deviation; IQR: indicates interquartile range; GLCM: gray‐level co‐occurrence matrix; GLRL: gray‐level run length; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; GLGM: gray‐level gradient matrix; SRE: short‐run emphasis; LRE: long‐run emphasis; GLN: gray‐level nonuniformity; RLN: run‐length nonuniformity; RP: run percentage; LGRE: low gray‐level run emphasis; HGRE: high gray‐level run emphasis; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; LRLGE: long‐run low gray‐level emphasis; LRHGE: long‐run high gray‐level emphasis; MGR: mean gradients; VGR: variance of gradients.
Bold indicates statistically significant as determined with the two‐tailed t test and false detection analyses (Q < 0.05).
Table 2.
Texture parameters: mean by flip angle
| 2 (n = 36) | 5 (n = 36) | 10 (n = 36) | 15 (n = 36) | 20 (n = 36) | 25 (n = 36) | 30 (n = 44) | P‐value | Q‐value | |
|---|---|---|---|---|---|---|---|---|---|
| Histogram | |||||||||
| Mean | 248.0 | 246.6 | 246.4 | 247.3 | 247.1 | 247.3 | 247.5 | 0.106 | 0.227 |
| Median | 247.2 | 243.8 | 241.4 | 245.6 | 247.4 | 246.5 | 244.8 | <0.0001 | 0.002 |
| STD | 27.0 | 29.5 | 30.3 | 28.5 | 29.2 | 28.2 | 27.7 | 0.081 | 0.220 |
| Range | 10.2 | 9.3 | 9.5 | 9.7 | 10.2 | 10.1 | 10.0 | 0.693 | 0.958 |
| Geometric mean | 248.6 | 248.3 | 248.2 | 248.4 | 248.3 | 248.4 | 248.5 | 0.104 | 0.227 |
| Harmonic mean | 247.2 | 246.7 | 246.6 | 246.9 | 246.8 | 247.0 | 247.1 | 0.102 | 0.227 |
| 2nd STD | 3.4 | 3.1 | 3.2 | 3.3 | 3.5 | 3.4 | 3.4 | 0.697 | 0.958 |
| STD5 | 4.3 | 4.0 | 4.1 | 4.1 | 4.3 | 4.3 | 4.2 | 0.826 | 0.958 |
| STD9 | 5.1 | 4.9 | 5.1 | 5.0 | 5.3 | 5.2 | 5.2 | 0.946 | 0.992 |
| 4th moment | 2166675.3 | 2993396.8 | 3359004.4 | 2519117.0 | 3053199.6 | 2557749.2 | 2269343.1 | 0.298 | 0.559 |
| IQR | 39.0 | 41.3 | 41.5 | 40.2 | 40.2 | 39.9 | 39.4 | 0.362 | 0.603 |
| Entropy | 7.5 | 7.4 | 7.3 | 7.4 | 7.0 | 7.0 | 7.1 | <0.0001 | 0.002 |
| GLCM | |||||||||
| Entropy | 2.7 | 2.3 | 2.1 | 2.4 | 2.3 | 2.4 | 2.4 | 0.021 | 0.118 |
| Contrast | 18.0 | 14.3 | 12.7 | 16.4 | 17.5 | 17.5 | 18.3 | 0.001 | 0.011 |
| Correlation | 0.92 | 0.94 | 0.95 | 0.93 | 0.92 | 0.92 | 0.92 | 0.019 | 0.118 |
| Energy | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.087 | 0.220 |
| Homogeneity | 0.53 | 0.56 | 0.57 | 0.55 | 0.54 | 0.54 | 0.54 | 0.036 | 0.135 |
| GLRL | |||||||||
| SRE | 0.09 | 0.11 | 0.13 | 0.11 | 0.11 | 0.11 | 0.11 | 0.756 | 0.958 |
| LRE | 0.09 | 0.11 | 0.13 | 0.11 | 0.11 | 0.11 | 0.11 | 0.813 | 0.958 |
| GLN | 0.09 | 0.11 | 0.13 | 0.11 | 0.11 | 0.11 | 0.10 | 0.861 | 0.969 |
| RLN | 0.09 | 0.11 | 0.13 | 0.11 | 0.11 | 0.11 | 0.11 | 0.801 | 0.958 |
| RP | 174.7 | 149.3 | 134.2 | 164.2 | 172.6 | 172.8 | 180.9 | 0.988 | 0.992 |
| LGRE | 174.7 | 149.0 | 133.5 | 163.9 | 172.1 | 171.7 | 179.6 | 0.986 | 0.992 |
| HGRE | 178.1 | 153.7 | 137.9 | 167.8 | 176.3 | 175.6 | 184.2 | 0.989 | 0.992 |
| SRLGE | 173.5 | 147.8 | 132.6 | 162.7 | 171.1 | 171.2 | 179.2 | 0.992 | 0.992 |
| SRHGE | 2917.7 | 2167.3 | 1952.6 | 2589.0 | 2569.0 | 2592.7 | 2549.7 | 0.830 | 0.958 |
| LRLGE | 3424.4 | 2631.8 | 2429.1 | 3029.8 | 3003.8 | 3049.7 | 2988.9 | 0.806 | 0.958 |
| LRHGE | 2910.0 | 2195.3 | 2006.0 | 2568.6 | 2571.9 | 2606.3 | 2569.7 | 0.656 | 0.958 |
| Law's features | |||||||||
| L1 | 192705.6 | 192761.2 | 192439.2 | 193311.5 | 195378.8 | 194563.7 | 193770.3 | 0.733 | 0.958 |
| L2 | 16489.5 | 16373.4 | 16323.5 | 16576.5 | 17381.4 | 17285.1 | 16808.4 | 0.001 | 0.011 |
| L3 | 6162.2 | 6141.1 | 6255.8 | 6155.8 | 6387.6 | 6363.1 | 6304.5 | 0.073 | 0.219 |
| L4 | 38082.9 | 37979.5 | 37990.6 | 38104.2 | 38882.5 | 38666.9 | 38384.9 | 0.424 | 0.681 |
| L5 | 4816.5 | 4812.1 | 4769.3 | 4869.8 | 5128.8 | 5112.0 | 4929.7 | 0.034 | 0.135 |
| L6 | 4024.3 | 4032.9 | 3989.2 | 4066.0 | 4269.3 | 4269.7 | 4111.7 | 0.027 | 0.135 |
| L7 | 3002.0 | 3000.4 | 3081.4 | 3010.1 | 3136.7 | 3145.4 | 3091.9 | 0.035 | 0.135 |
| L8 | 9396.0 | 9481.2 | 9217.9 | 9434.4 | 10117.2 | 10063.3 | 9574.5 | 0.010 | 0.090 |
| L9 | 18404.7 | 18234.2 | 18211.3 | 18321.4 | 18878.1 | 18750.6 | 18650.3 | 0.199 | 0.407 |
| GLGM | |||||||||
| MGR | 14.5 | 13.4 | 13.5 | 14.1 | 13.1 | 13.4 | 12.7 | 0.329 | 0.569 |
| VGR | 11028.3 | 10337.9 | 10427.8 | 10781.4 | 9984.6 | 10181.8 | 9640.4 | 0.213 | 0.417 |
| Skewness | 8.5 | 8.8 | 8.7 | 8.7 | 9.1 | 9.0 | 9.3 | 0.088 | 0.220 |
| Kurtosis | 78.1 | 83.2 | 82.1 | 81.4 | 89.2 | 88.6 | 94.6 | 0.045 | 0.151 |
| Mean skewness | 0.46 | 0.80 | 0.92 | 0.62 | 0.60 | 0.58 | 0.54 | 0.020 | 0.118 |
| Mean kurtosis | 2.8 | 3.1 | 3.4 | 3.0 | 3.2 | 3.1 | 3.1 | 0.047 | 0.151 |
| Mean laws | 300502.0 | 300676.9 | 300393.7 | 301260.7 | 307050.4 | 305634.3 | 303027.0 | 0.320 | 0.569 |
Mean texture analysis features variation with changes in flip angle. n: number of contoured slices; STD: standard deviation; STD5: 5‐neighborhood standard deviation; STD9: 9‐neighborhood standard deviation; IQR: indicates interquartile range; GLCM: gray‐level co‐occurrence matrix; GLRL: gray‐level run length; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; GLGM: gray‐level gradient matrix; SRE: short‐run emphasis; LRE: long‐run emphasis; GLN: gray‐level nonuniformity; RLN: run‐length nonuniformity; RP: run percentage; LGRE: low gray‐level run emphasis; HGRE: high gray‐level run emphasis; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; LRLGE: long‐run low gray‐level emphasis; LRHGE: long‐run high gray‐level emphasis; MGR: mean gradients; VGR: variance of gradients.
Bold indicates statistically significant as determined with the two‐tailed t‐test and false detection analyses (Q < 0.05).
Table 3.
Texture parameters: number of excitations 1 vs 4
| 1 (n = 36) | 4 (n = 44) | P‐value | Q‐value | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Histogram | ||||||
| Mean | 249.6 | 2.3 | 247.5 | 2.8 | 0.001 | 0.003 |
| Median | 249.5 | 6.0 | 244.8 | 6.8 | 0.002 | 0.005 |
| STD | 24.3 | 4.2 | 27.7 | 4.9 | 0.002 | 0.005 |
| Range | 11.4 | 2.3 | 10.0 | 2.7 | 0.013 | 0.022 |
| Geometric mean | 248.8 | 0.38 | 248.5 | 0.45 | 0.002 | 0.005 |
| Harmonic mean | 247.6 | 0.69 | 247.1 | 0.83 | 0.003 | 0.006 |
| 2nd STD | 3.9 | 0.78 | 3.4 | 0.93 | 0.013 | 0.022 |
| STD5 | 4.8 | 1.0 | 4.2 | 1.1 | 0.028 | 0.045 |
| STD9 | 5.7 | 1.3 | 5.2 | 1.4 | 0.107 | 0.146 |
| 4th moment | 1257285.5 | 1220668.6 | 2269343.1 | 1656035.4 | 0.003 | 0.006 |
| IQR | 35.1 | 5.0 | 39.4 | 6.2 | 0.001 | 0.003 |
| Entropy | 7.2 | 0.48 | 7.1 | 0.29 | 0.413 | 0.502 |
| GLCM | ||||||
| Entropy | 3.0 | 0.61 | 2.4 | 0.69 | 0.0001 | 0.001 |
| Contrast | 23.1 | 7.7 | 18.3 | 8.5 | 0.010 | 0.018 |
| Correlation | 0.89 | 0.04 | 0.92 | 0.05 | 0.005 | 0.009 |
| Energy | 0.005 | 0.002 | 0.008 | 0.004 | <0.0001 | 0.001 |
| Homogeneity | 0.49 | 0.06 | 0.54 | 0.07 | 0.002 | 0.005 |
| GLRL | ||||||
| SRE | 0.07 | 0.04 | 0.11 | 0.05 | 0.001 | 0.003 |
| LRE | 0.07 | 0.04 | 0.11 | 0.05 | 0.0004 | 0.002 |
| GLN | 0.07 | 0.04 | 0.10 | 0.05 | 0.001 | 0.003 |
| RLN | 0.07 | 0.04 | 0.11 | 0.05 | 0.001 | 0.003 |
| RP | 211.7 | 63.2 | 180.9 | 66.2 | 0.038 | 0.055 |
| LGRE | 212.3 | 63.5 | 179.6 | 66.9 | 0.029 | 0.045 |
| HGRE | 214.4 | 62.7 | 184.2 | 65.3 | 0.040 | 0.056 |
| SRLGE | 212.1 | 64.1 | 179.2 | 67.8 | 0.030 | 0.045 |
| SRHGE | 3842.0 | 1316.4 | 2549.7 | 1735.8 | 0.0004 | 0.002 |
| LRLGE | 4431.0 | 1453.1 | 2988.9 | 1914.6 | 0.0004 | 0.002 |
| LRHGE | 3845.2 | 1318.9 | 2569.7 | 1738.3 | 0.001 | 0.003 |
| Law's features | ||||||
| L1 | 194138.1 | 8172.2 | 193770.3 | 8258.3 | 0.843 | 0.843 |
| L2 | 17098.3 | 1319.8 | 16808.4 | 1211.3 | 0.310 | 0.388 |
| L3 | 6323.2 | 406.3 | 6304.5 | 380.0 | 0.832 | 0.843 |
| L4 | 38727.6 | 2029.3 | 38384.9 | 2147.6 | 0.469 | 0.555 |
| L5 | 5002.1 | 517.8 | 4929.7 | 427.0 | 0.495 | 0.571 |
| L6 | 4142.0 | 409.5 | 4111.7 | 308.3 | 0.707 | 0.758 |
| L7 | 3072.6 | 210.6 | 3091.9 | 170.2 | 0.651 | 0.715 |
| L8 | 9678.2 | 1135.5 | 9574.5 | 872.1 | 0.645 | 0.715 |
| L9 | 18972.1 | 1287.3 | 18650.3 | 1373.0 | 0.287 | 0.369 |
| GLGM | ||||||
| MGR | 16.2 | 1.9 | 12.7 | 4.0 | <0.0001 | 0.001 |
| VGR | 12135.7 | 1227.1 | 9640.4 | 2842.2 | <0.0001 | 0.001 |
| Skewness | 7.9 | 0.5 | 9.3 | 1.6 | <0.0001 | 0.001 |
| Kurtosis | 67.2 | 9.0 | 94.6 | 32.2 | <0.0001 | 0.001 |
| Mean skewness | 0.03 | 0.64 | 0.54 | 0.75 | 0.002 | 0.005 |
| Mean kurtosis | 2.7 | 0.63 | 3.1 | 0.58 | 0.004 | 0.008 |
| Mean laws | 303992.7 | 14617.7 | 303027.0 | 14279.6 | 0.767 | 0.803 |
Mean texture analysis features with variations in number of excitations (NEX). n: number of contoured slices; STD: standard deviation; STD5: 5‐neighborhood standard deviation; STD9: 9‐neighborhood standard deviation; IQR: indicates interquartile range; GLCM: gray‐level co‐occurrence matrix; GLRL: gray‐level run length; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; GLGM: gray‐level gradient matrix; SRE: short‐run emphasis; LRE: long‐run emphasis; GLN: gray‐level nonuniformity; RLN: run‐length nonuniformity; RP: run percentage; LGRE: low gray‐level run emphasis; HGRE: high gray‐level run emphasis; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; LRLGE: long‐run low gray‐level emphasis; LRHGE: long‐run high gray‐level emphasis; MGR: mean gradients; VGR: variance of gradients.
Bold indicates statistically significant as determined with the two‐tailed t‐test and false detection analyses (Q < 0.05).
Table 4.
Texture parameters: GE vs Siemens
| GE (n = 83) | Siemens (n = 36) | P‐value | Q‐value | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Histogram | ||||||
| Mean | 245.9 | 1.8 | 249.6 | 2.3 | <0.0001 | 0.0001 |
| Median | 242.0 | 4.5 | 249.5 | 6.0 | <0.0001 | 0.0001 |
| STD | 32.5 | 5.2 | 24.3 | 4.2 | <0.0001 | 0.0001 |
| Range | 9.2 | 1.4 | 11.4 | 2.3 | <0.0001 | 0.0001 |
| Geometric mean | 248.0 | 0.56 | 248.8 | 0.38 | <0.0001 | 0.0001 |
| Harmonic mean | 246.1 | 1.04 | 247.6 | 0.70 | <0.0001 | 0.0001 |
| 2nd STD | 3.2 | 0.48 | 3.9 | 0.78 | <0.0001 | 0.0001 |
| STD5 | 3.6 | 0.56 | 4.8 | 0.96 | <0.0001 | 0.0001 |
| STD9 | 3.5 | 0.54 | 5.7 | 1.3 | <0.0001 | 0.0001 |
| 4th moment | 4384199.1 | 2820540.2 | 1259420.0 | 1216812.6 | <0.0001 | 0.0001 |
| I QR | 41.0 | 3.1 | 35.1 | 5.1 | <0.0001 | 0.0001 |
| Entropy | 7.3 | 0.20 | 7.2 | 0.48 | 0.516 | 0.554 |
| GLCM | ||||||
| Entropy | 2.2 | 0.39 | 3.0 | 0.61 | <0.0001 | 0.0001 |
| Contrast | 16.7 | 4.5 | 23.2 | 7.7 | <0.0001 | 0.0001 |
| Correlation | 0.93 | 0.04 | 0.89 | 0.04 | <0.0001 | 0.0001 |
| Energy | 0.009 | 0.002 | 0.005 | 0.002 | <0.0001 | 0.0001 |
| Homogeneity | 0.57 | 0.02 | 0.49 | 0.06 | <0.0001 | 0.0001 |
| GLRL | ||||||
| SRE | 0.11 | 0.03 | 0.07 | 0.04 | <0.0001 | 0.0001 |
| LRE | 0.11 | 0.03 | 0.07 | 0.04 | <0.0001 | 0.0001 |
| GLN | 0.12 | 0.03 | 0.07 | 0.04 | <0.0001 | 0.0001 |
| RLN | 0.11 | 0.03 | 0.07 | 0.04 | <0.0001 | 0.0001 |
| RP | 144.5 | 19.2 | 212.0 | 63.3 | <0.0001 | 0.0001 |
| LGRE | 141.0 | 20.6 | 212.6 | 63.6 | <0.0001 | 0.0001 |
| HGRE | 139.5 | 20.4 | 214.8 | 62.7 | <0.0001 | 0.0001 |
| SRLGE | 140.6 | 20.5 | 212.5 | 64.2 | <0.0001 | 0.0001 |
| SRHGE | 2037.7 | 592.1 | 3844.4 | 1321.0 | <0.0001 | 0.0001 |
| LRLGE | 2421.5 | 660.9 | 4429.5 | 1460.6 | <0.0001 | 0.0001 |
| LRHGE | 1753.3 | 466.9 | 3845.8 | 1323.4 | <0.0001 | 0.0001 |
| Law's features | ||||||
| L1 | 186793.4 | 33134.5 | 194202.9 | 8099.2 | 0.189 | 0.236 |
| L2 | 17662.7 | 8333.2 | 17104.7 | 1293.8 | 0.691 | 0.707 |
| L3 | 7092.6 | 2150.3 | 6323.8 | 399.1 | 0.036 | 0.049 |
| L4 | 39166.7 | 4904.5 | 38746.2 | 2016.0 | 0.621 | 0.650 |
| L5 | 5836.4 | 4462.7 | 5002.2 | 524.1 | 0.267 | 0.316 |
| L6 | 4712.7 | 3135.9 | 4141.0 | 407.6 | 0.279 | 0.322 |
| L7 | 3724.9 | 2070.2 | 3072.0 | 206.5 | 0.062 | 0.082 |
| L8 | 10812.2 | 5484.5 | 9691.9 | 1184.0 | 0.229 | 0.279 |
| L9 | 17868.8 | 2408.8 | 18978.0 | 1267.0 | 0.010 | 0.014 |
| GLGM | ||||||
| MGR | 14.1 | 8.9 | 16.2 | 1.9 | 0.166 | 0.213 |
| VGR | 11304.8 | 7622.5 | 12137.4 | 1222.2 | 0.517 | 0.554 |
| Skewness | 9.2 | 1.7 | 7.9 | 0.52 | <0.0001 | 0.0001 |
| Kurtosis | 94.3 | 30.4 | 67.2 | 9.0 | <0.0001 | 0.0001 |
| Mean skewness | 0.95 | 0.42 | 0.03 | 0.64 | <0.0001 | 0.0001 |
| Mean kurtosis | 3.2 | 0.47 | 2.7 | 0.67 | <0.0001 | 0.0001 |
| Mean laws | 301700.2 | 63339.7 | 304095.9 | 14624.5 | 0.823 | 0.823 |
Mean texture analysis features based on scanner platform: GE vs Siemens. STD: standard deviation; STD5: 5‐neighborhood standard deviation; STD9: 9‐neighborhood standard deviation; IQR: indicates interquartile range; GLCM: gray‐level co‐occurrence matrix; GLRL: gray‐level run length; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; GLGM: gray‐level gradient matrix; SRE: short‐run emphasis; LRE: long‐run emphasis; GLN: gray‐level nonuniformity; RLN: run‐length nonuniformity; RP: run percentage; LGRE: low gray‐level run emphasis; HGRE: high gray‐level run emphasis; SRLGE: short‐run low gray‐level emphasis; SRHGE: short‐run high gray‐level emphasis; LRLGE: long‐run low gray‐level emphasis; LRHGE: long‐run high gray‐level emphasis; MGR: mean gradients; VGR: variance of gradients.
Bold indicates statistically significant as determined with the two‐tailed t‐test and false detection analyses (Q < 0.05).
3.A. Assessment of magnetic strength
Variations in magnetic strength (1.5 T vs 3 T) resulting in changes in texture features are displayed in Table 1. No statistically significant differences were noticed in the histogram, or GLCM texture features. A few of the GLRL texture features including the short‐run high gray‐level emphasis (SRHGE), long‐run low gray‐level emphasis (LRLGE), and long‐run high gray‐level emphasis (LRHGE) demonstrated statistically significant differences (Q = 0.0003), however, the remaining GLRL features did not demonstrate a significant difference. All Law's features demonstrated statistically significant differences (Q = 0.0003), and all of the GLGM features, with the exception of mean skewness and mean kurtosis (Q = 0.554, and 0.133, respectively), demonstrated statistically significant differences (Q = 0.0003–0.005).
3.B. Assessment of flip angle
Variations in flip angles produced variations in texture analysis features, as shown in Table 2 and Table S1. Only two histogram features, median and entropy, demonstrated statistically significant differences with changes in flip angle (Q = 0.002, each). Similarly, only the GLCM feature, contrast, demonstrated a statistically significant difference related to changes in the flip angle (Q = 0.011). No statistically significant difference in the GLRL features, Law's features, or GLGM features with variations in flip angle.
3.C. Assessment of NEX
Changes in NEX (1 vs 4) produced variations in texture analysis features as shown in Table 3. All histogram texture features, with the exception of neighborhood standard deviation (STD9) (Q = 0.146) and entropy (Q = 0.502), demonstrated statistically significant differences (Q = 0.003–0.045). All GLCM texture features demonstrated statistically significant differences with changes in NEX (Q = 0.001–0.018). All GLRL texture features, with the exception of run percentage (RP) (Q = 0.055) and high gray‐level run emphasis (HGRE) (Q = 0.056), demonstrated statistically significant differences with changes in NEX (Q = 0.002–0.045). None of the Law's features demonstrated statistically significant differences. All of the GLGM texture features demonstrated statistically significant differences with changes in NEX (Q = 0.001–0.008).
3.D. Assessment of scanner platform
Differences in scanner platform (GE vs Siemens) produced differences in the texture analysis features as shown in Table 4. All histogram features, except for entropy (P = 0.554), demonstrated statistically significant differences (Q = 0.0001). All GLCM and GLRL texture features demonstrated statistically significant differences with different scanner platforms (Q = 0.0001). Only the Laws feature, L9, demonstrated a statistically significant difference (Q = 0.014). All GLGM texture features demonstrated statistically significant differences (Q = 0.0001), with the exception of mean gradients (MGR) (Q = 0.236), variance of gradients (VGR) (Q = 0.554), and mean Laws features (Q = 0.823).
4. DISCUSSION
The results of this study demonstrate statistically significant differences in multiple texture analysis features (histogram, GLCM, GLRL, and GLGM) related to changes in several, specific MRI scan parameters such as magnet strength, flip angle, NEX, and scanner platform.
While multiple prior research studies have investigated the use of a texture analysis applied to MR images2, 3, 4, 5, 6, 7, 9, 10, 12, 15, 16, 17, 18, 19, 39 the underlying influence of MRI scan parameters on texture analysis features are not entirely understood. Furthermore, despite the increasing use of texture analysis in the field of radiology, a fundamental understanding of the histopathologic and biologic correlation between tissue and texture analysis features remains in its infancy.
In this study, we demonstrated statistically significant differences in the Law's features and several GLGM features with differences in magnet strength, while histogram, GLCM, and GLRL features were invariant of these changes in magnet strength. Differences in flip angle significantly influenced GLCM texture features and changes in NEX significantly influenced histogram, many GLRL, and GLGM texture features. The most substantial changes in texture analysis features were encountered with differences in MRI scanner platform (GE vs Siemens). Differences in the MRI scanner platform generated statistically significant differences in all categories of texture analysis features, except for the Laws features. Histogram features by in large measure the image signal‐to‐noise ratio (SNR), which is mostly a low frequency signal. In contradistinction, Laws features measure distinct features within an image such as edges and lines, which are predominately high frequency signals. Changes in both NEX and flip angle would only affect the SNR, but not the spatial resolution. We postulate that different scanner platforms (i.e., GE vs Siemens and 3 T vs 1.5 T) employ dramatically different image processing algorithms and this is the primary reason there is a statistical significance observed in Laws features. On the contrary, histogram features are less sensitive to changes in spatial resolution and are more sensitive to changes in SNR (e.g., NEX, flip angle, etc.)
Changes in NEX and flip angle may only affect image contrast and would not affect spatial resolution. We postulate that different scanner vendors, i.e., GE and Siemens, employ dramatically different image processing algorithms and this is the primary reason there is a statistical significance observed in Laws features. On the contrary, histogram features are less sensitive to changes in spatial resolution and are more sensitive to changes in contrast (e.g., NEX, flip angle, etc.).
Multiple prior studies have highlighted the potential promise and importance of using a texture analysis as a quantitative, post‐processing technique to evaluate subtle changes in pixel intensity which may not be evident to the human eye.7, 8, 14 These subtle patterns of pixel variation could potentially serve as a biomarker for lesion characterization, early disease detection, and prediction of lesional behavior.6, 8, 20 A recent prior study demonstrated a dependency of texture analysis features on variations in CT scanning parameters.30 The results of this study, build off those from the prior study examining how texture analysis features are influenced by MRI, in addition to CT acquisition parameters. This work highlights the importance of using standardized and rigorously controlled scanning protocol when conducting research utilizing a texture analysis. This current study expands upon prior studies published in the literature which previously investigated a limited set of MRI acquisition parameters and their influence on texture features.31, 32 The study performed by Mayerofer et al.31 investigated changes in TR/TE, sampling bandwidth, and number of acquisitions and the influence of these parameters on texture analysis features. Mayerofer et al., noted that changes in these features had a substantial impact on the sensitivity of the texture analysis features,31 however, this study examined a limited set of MRI scanning parameters which did not include a study of NEX, flip angle, magnet strength, and scanner platform (GE vs Siemens). This current study seeks to bridge the gap in knowledge investigating the influence these additional MRI scanning parameters have on certain texture analysis features.
The results of this study underscore the importance of understanding how texture analysis features are influenced by imaging acquisition parameters. The ability to distinguish changes in texture analysis features related to tissue biology and pathology vs effects related to technical differences in MRI scanning protocol is of paramount importance for designing future research investigations which will use a texture analysis.
There are several limitations to the current study. The first is that this was a study using a nonanatomic phantom with basic architecture variations in internal structure. The use of this phantom and associated scanner data was advantageous as an initial pilot investigation into the dependency texture features on MRI scanning parameters as the raw scanning data are publicly available for research efforts. The phantom used in this study has a well‐defined, well‐characterized, and simple internal geometric structure. We recognize that the simplicity of this phantom is a far reach from a phantom with anatomically relevant internal structure, but we feel that the simplicity of this nonanatomic phantom initially helps us to understand the results of this study and the effects the changes in MRI scanning parameters has on the texture features. Future research efforts will need to be conducted using a phantom with more anatomically relevant internal structure and with more complex internal components, perhaps with an internal composition mimicking that of fat, muscle, and bone. Additionally, a robustness analysis on real test‐retest data should be also performed, similar to the work of van Timmeren et al. for CT.40 A second limitation of this study is that only a discrete subset of MRI scanning parameters was investigated. This study was limited based on the information available in the RIDER dataset. We would have liked to investigate the influence additional scanning parameters such as slice thicknesses, matrix size, and differences in TR/TE have on these texture analysis features, however, this information was not available in the RIDER dataset. Future investigations on this topic will also be to examine a broader set of MRI scan parameters and evaluate the influence these parameters have on texture analysis features. Thirdly, this study investigated a limited set of 41 defined texture analysis features. There are hundreds of defined texture analysis features described in the literature. We sought to investigate a subset of 41 texture features which we have investigated in our previous works, and which we feel are most frequently reported in the radiomics literature.7, 8, 30 We recognize that this subset may have excluded additional texture features of interest. The inclusion of additional texture features in our in‐house developed texture analysis program will be addressed in future research endeavors. Lastly a limitation of this study, is the investigation of only GRE‐based MRI sequenced. Again, our investigation was limited to the information available in the RIDER dataset. We do recognize that investigating how texture analysis features are influenced by MRI scanning parameters on non‐GRE based sequences would be of great interest. Future investigations in this subject matter with an expanded analysis of additional texture analysis features are warranted.
5. CONCLUSION
Texture analysis represents an increasingly popular, post‐processing, quantitative evaluation technique that can potentially be used as an adjunct in diagnostic imaging, and as a possible imaging biomarker. The results of this study demonstrate that MRI acquisition parameters have a significant influence on specific texture analysis features. This work serves as a pilot study highlighting the importance of using a standardized and controlled MRI scanning protocol when using a texture analysis. Multi‐institutional research endeavors, or single institution endeavors using different MRI scanning platforms and scanning protocols should exercise caution when using texture analysis.
CONFLICT OF INTEREST
No conflicts of interest.
Supporting information
Table S1. Texture parameters: Standard deviation by flip angle
ACKNOWLEDGMENTS
None.
Advances in Knowledge
This is the first study specifically investigating the influence of specific MRI parameters on texture analysis features underscoring the importance of using uniform and standardized MRI scanning protocols when employing a texture analysis.
REFERENCES
- 1. Haralick R, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern A Syst Hum. 1973;SMC‐3:610–621. [Google Scholar]
- 2. de Carvalho Alegro M, Valotta Silva A, Yumi Bando S, et al. Texture analysis of high resolution MRI allows discrimination between febrile and afebrile initial precipitating injury in mesial temporal sclerosis. Magn Reson Med. 2012;68:1647–1653. [DOI] [PubMed] [Google Scholar]
- 3. Mayerhoefer ME, Stelzeneder D, Bachbauer W, Welsch GH, Mamisch TC, Szczypinski P. Quantitative analysis of lumbar intervertebral disc abnormalities at 3.0 Tesla: value of T(2) texture features and geometric parameters. NMR Biomed. 2012;25:866–872. [DOI] [PubMed] [Google Scholar]
- 4. Risse F, Pesic J, Young S, Olsson LE. A texture analysis approach to quantify ventilation changes in hyperpolarised ³He MRI of the rat lung in an asthma model. NMR Biomed. 2012;25:131–141. [DOI] [PubMed] [Google Scholar]
- 5. Fujimoto K, Tonan T, Azuma S, Kage M, Nakashima O, Johkoh T. Evaluation of the mean and entropy of apparent diffusion coefficient values in chronic hepatitis C: correlation with pathologic fibrosis stage and inflammatory activity grade. Radiology. 2011;258:739–748. [DOI] [PubMed] [Google Scholar]
- 6. Jirák D, Dezortová M, Taimr P, Hajek M. Texture analysis of human liver. J Magn Reson Imaging. 2002;15:68–74. [DOI] [PubMed] [Google Scholar]
- 7. Li B, Jara H, Yu H, O'Brien M, Soto J, Anderson SW. Enhanced laws textures: a potential MRI surrogate marker of hepatic fibrosis in a murine model. Magn Reson Imaging. 2017;30:33–40. [DOI] [PubMed] [Google Scholar]
- 8. Buch K, Fujita A, Li B, Kawashima Y, Qureshi MM, Sakai O. Using texture analysis to determine human papillomavirus status of oropharyngeal squamous cell carcinomas on CT. AJNR Am J Neuroradiol. 2015;36:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daginawala N, Li B, Buch K, et al. Using texture analyses of contrast enhanced CT to assess hepatic fibrosis. Eur J Radiol. 2016;85:511–517. [DOI] [PubMed] [Google Scholar]
- 10. Ramkumar S, Ranjbar S, Ning S, et al. MRI‐based texture analysis to differentiate sinonasal squamous cell carcinoma from inverted papilloma. AJNR Am J Neuroradiol. 2017;38:1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jethanandani A, Lin T, Volpe S, et al. Exploring applications of radiomics in magnetic resonance imaging of head and neck cancer: a systematic review. Front Oncol. 2018;8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang G, He L, Yuan C, Huang Y, Liu Z, Liang C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur J Radiol. 2018;98:100–106. [DOI] [PubMed] [Google Scholar]
- 13. Ba‐Ssalamah A, Muin D, Schernthaner R, et al. Texture‐based classification of different gastric tumors at contrast‐enhanced CT. Eur J Radiol. 2013;82:e537–e543. [DOI] [PubMed] [Google Scholar]
- 14. Miles KA, Ganeshan B, Griffiths MR, Young RC, Chatwin CR. Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology. 2009;250:444–452. [DOI] [PubMed] [Google Scholar]
- 15. Lubner MG, Stabo N, Lubner SJ, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre‐treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40:2331–2337. [DOI] [PubMed] [Google Scholar]
- 16. Colen RR, Vangel M, Wang J, et al. TCGA Glioma Phenotype Research Group, Zinn PO . Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project. BMC Med Genomics. 2014;2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kickingereder P, Götz M, Muschelli J, et al. Large‐scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti‐angiogenic treatment response. Clin Cancer Res. 2016;22:5765–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ingrisch M, Schneider MJ, Nörenberg D, et al. Radiomic analysis reveals prognostic information in T1‐weighted baseline magnetic resonance imaging in patients with glioblastoma. Invest Radiol. 2017;52:360–366. [DOI] [PubMed] [Google Scholar]
- 19. Jakola AS, Zhang YH, Skjulsvik AJ, et al. Quantitative texture analysis in the prediction of IDH status in low‐grade gliomas. Clin Neurol Neurosurg. 2018;164:114–120. [DOI] [PubMed] [Google Scholar]
- 20. Cunliffe AR, Armato SG 3rd, Straus C, Malik R, Al‐Hallaq HA. Lung texture in serial thoracic CT scans: correlation with radiologist‐defined severity of acute changes following radiation therapy. Phys Med Biol. 2014;59:5387–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahn SY, Park CM, Park SJ, et al. Prognostic value of computed tomography texture features in non‐small cell lung cancers treated with definitive concomitant chemoradiotherapy. Invest Radiol. 2015;50:719–725. [DOI] [PubMed] [Google Scholar]
- 22. Schieda N, Thornhill RE, Al‐Subhi M, et al. Diagnosis of sarcomatoid renal cell carcinoma with CT: evaluation by qualitative imaging features and texture analysis. AJR Am J Roentgenol. 2015;204:1013–1023. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X, Xu X, Tian Q, et al. Radiomics assessment of bladder cancer grade using texture features from diffusion‐weighted imaging. J Magn Reson Imaging. 2017;46:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shahedi M, Halicek M, Guo R, Zhang G, Schuster DM, Fei B. A semiautomatic segmentation method for prostate in CT images using local texture classification and statistical shape modeling. Med Phys. 2018;45:2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shiradkar R, Ghose S, Jambor I, et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J Magn Reson Imaging. [Epub ahead of print]. 10.1002/jmri.26178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saha A, Yu X, Sahoo D, Mazurowski MA. Effects of MRI scanner parameters on breast cancer radiomics. Expert Syst Appl. 2017;87:384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun X, He B, Luo X, et al. Preliminary study on molecular subtypes of breast cancer based on magnetic resonance imaging texture analysis. J Comput Assist Tomogr. 2018;42:531–535. [DOI] [PubMed] [Google Scholar]
- 28. Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE‐MRI. Breast Cancer Res. 2017;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application‐oriented study. Med Phys. 36:1236–1243. [DOI] [PubMed] [Google Scholar]
- 30. Buch K, Li B, Qureshi MM, Kuno H, Anderson SW, Sakai O. Quantitative assessment of variation in CT parameters on texture features: Pilot study using a nonanatomic phantom. AJNR Am J Neuroradiol. 2017;38:981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brynolfsson P, Nilsson D, Torheim T, et al. Haralick texture features from apparent diffusion coefficient (ADC) MRI images depend on imaging and pre‐processing parameters. Sci Rep. 2017;7:4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fave X, Cook M, Frederick A, Zhang L, Yang J, Fried D. Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput Med Imaging Graph. 2015;44:54–61. [DOI] [PubMed] [Google Scholar]
- 33. Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22:81–91. [DOI] [PubMed] [Google Scholar]
- 34. The Cancer Imaging Archive: RIDER NEURO MRI . https://wiki.cancerimagingarchive.net/display/Public/RIDER+NEURO+MRI Accessed Jan 21, 2017.
- 35. Prior F, Smith K, Sharma A. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci Data. 2017;4:170124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sonka M, Hlavac V, Boyle R. Image processing, analysis and machine vision. London: Chapman & Hall; 2008. [Google Scholar]
- 37. Tang X. Texture information in run‐length matrices. IEEE Trans Image Process. 1998;7:1602–1609. [DOI] [PubMed] [Google Scholar]
- 38. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 39. MacKay JW, Murray PJ, Low SB, et al. Quantitative analysis of tibial subchondral bone: texture analysis outperforms conventional trabecular microarchitecture analysis. J Magn Reson Imaging. 2016;43:1159–1170. [DOI] [PubMed] [Google Scholar]
- 40. van Timmeren JE, Leijenaar R, van Elmpt W, et al. Test‐retest data for radiomics feature stability analysis: generalizable or study‐specific? Tomography. 2016;2:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Texture parameters: Standard deviation by flip angle


