Abstract
In previous work we have isolated and identified a new strain called Enterococcus faecium FL31. The active compound secreted by this strain, “BacFL31”, has been purified and characterized. In the present study, safety aspect, assessed by microbiological and molecular tests, demonstrated that Enterococcus faecium FL31 was susceptible to relevant antibiotics, free of hemolytic, gelatinase, DNase, and lipase activities. In addition, it did not harbor virulence and antibiotic resistance genes. Combined SYTOX Green dye and UV-absorbing experiments, along with released extracellular potassium and transmembrane electrical potential measurements, showed that pure BacFL31 at a concentration of 1×MIC (50 μg/mL) could damage cytoplasmic membrane of the pathogen Listeria monocytogenes ATCC19117. The same concentration causes the leakage of its intracellular constituents and leads to the destruction of this pathogenic microorganism. In summary, this work reflected characteristics of Enterococcus faecium FL31 strain and its bacteriocin in terms of functional and safety perspectives.
1. Introduction
Enterococcus is a large genus of lactic acid bacteria (LAB) and several species from this genus have been used as probiotic for humans or animals [1]. In addition, some Enterococcus faecium spp. act as protective agents against food-spoilage and pathogenic bacteria, such as Listeria monocytogenes, Salmonella typhimurium, Staphylococcus aureus, and Clostridium perfringens spores due to their ability to produce antimicrobial peptides called bacteriocins (enterocins) [2–5].
However, certain species of Enterococcus faecium can have relatively low virulence and cause nosocomial infections especially endocarditis, septicemia, urinary tract infections, meningitis, and others human infections [6]. These pathogenic strains can also carry multiple antibiotic resistances and several virulence factors like haemolysin, gelatinase, invasins, adhesins, cytolysin, and enterococcal surface proteins [7]. It should be noted that several studies have shown that enterococci possessing virulence genes are only isolated from infected patients and clinical samples, whereas the majority of Enterococcus strains isolated from foodstuffs have probiotic effects and health benefits [2]. In food storage, the application of bacteriocins of LAB as natural preservatives to control the growth of spoilage and pathogenic bacteria in food requires the safety confirmation of the producing strain and the understanding of its bacteriocin action mechanism against food-spoilage and pathogenic bacteria [8].
In previous works, a strain called FL31, isolated from fermented olives, was selected for its antimicrobial activity and identified a new lactic acid bacteria designated Enterococcus faecium FL31. The active compound of the strain FL31 was identified as a proteinaceous substance and named BacFL31. The N-terminal amino acid sequence of the purified BacFL31 showed the presence of hydroxyproline residues. BacFL31 exhibits a bactericidal mode of action against Listeria monocytogenes ATCC19117 and was proved to be useful for the inhibition of the growth of this pathogen during storage at 4°C of minced beef meat [9].
Taking into account the attractive characteristic of the Enterococcus faecium FL31 strain and its original bacteriocin BacFL31, we propose in the present paper, to define the probiotic properties and the safety of this strain as well as the elucidation of the bactericidal mechanism of BacFL31 against L. monocytogenes ATCC19117.
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
E. faecium FL31, BacFL31 producer strain [9], was grown in a De Man, Rogosa, and Sharpe (MRS) broth medium at 37°C for 18 h [10]. Enterococcus faecalis ATCC 29212 was grown overnight at 37°C in Brain Heart Infusion (BHI). The genomic DNA of this strain was extracted using molecular biology kit (Bio Basic Canada Inc.) and used as a positive control for the evaluation of the pathogenicity of E. faecium FL31. Staphylococcus aureus ATCC 6538 was cultured in LB medium overnight at 37°C and used as a positive control to study DNase and lipase activities. To measure the BacFL31 activity and to study its mechanism of action, we used the food-borne pathogen L. monocytogenes ATCC 19117 as target strain. This microorganism was cultured in Luria-Bertani (LB) medium overnight at 30°C.
2.2. Safety Evaluation of Bacteriocinogenic E. faecium FL31
2.2.1. Antibiotic Resistance
The susceptibility of the strain E. faecium FL31 to a range of relevant clinically most used antibiotics (μg/disc): ampicillin (30), streptomycin (10), kanamycin (30), chloramphenicol (30), tetracycline (30), spectinomycin (100), penicillin G (30), vancomycin (30), oxacillin (5), and amoxicillin (25) were tested by disk diffusion method on MRS solid media. Antibiotics discs were applied on MRS plates containing 107 CFU of E. faecium FL31 spread uniformly across the surface and then plates were incubated at 37°C for 24 h. Inhibition zones around the discs were measured in mm and the results were interpreted following the criteria of the Antibiogram Committee of the French Microbiology Society CA-SFM [11].
2.2.2. Hemolytic Activity, Gelatinase, DNase, and Lipase Tests
For hemolytic activity, fresh culture of E. faecium FL31 was streaked on Columbia agar plates containing 5% (w/v) sheep blood and incubated for 48 h at 37°C. Blood agar plates were examined for signs of β-hemolysis (clear zones around colonies). E. faecalis ATCC 29212 were used as a positive control for β–hemolysis assay.
Gelatinase activity of the strain E. faecium FL31 was assessed according to Su et al., 1991 [12]. GelE-positive colonies on gelatine medium were surrounded by a turbid halo after 2 days of incubation at 37°C. To measure the hydrolyzed gelatine in the agar plates, 0.5-1.0 mL of Frazier solution (mercuric chloride, 15.0 g; hydrochloric acid (37%), 20 mL; distilled water, 100 mL) was poured on the surface of the medium to precipitate the unhydrolyzed gelatine. E. faecalis ATCC 29212 was used as a positive control.
DNase activity was tested using DNase agar medium [13]. The plate was inoculated with the appropriate strain by streaking a thick line of inoculum across the plate. After incubation at 37°C for 24-48 hours, the surface of the DNase test agar plate was flooded with Toluidine Blue solution. DNase activity is indicated by a pink zone surrounding growth. The color of the medium remains unchanged if the test is negative. S. aureus ATCC 6538 was used as a positive control.
Lipase activity was performed according to Tiago et al., 2004 [14]. The appropriate strain was inoculated in MLB (tryptone 1%; 0.5% yeast extract; 0.5% NaCl) agar supplemented with 2.0 g/L of CaCl2 and 10 g/L of Tween-80. Plate was incubated at 37°C for 24-48 hours. A positive reaction was indicated by a clear halo around the colonies. S. aureus ATCC 6538 was used as a positive control.
2.2.3. Detection of Virulence Genes in E. faecium FL31 Strain
The absence or the presence of different virulence genes in E. faecium FL31 was evaluated by PCR using specific primers (Table 1). The tested genes were asa1 (aggregation substance), ace (adhesin of collagen protein), esp (enterococcal surface protein), efaAfm (cell wall adhesin), and cylB (activation and expression of cytolysin). The oligonucleotide primers were purchased from Bio Basic Canada Inc.
Table 1.
Primers used for the screening of virulence genes.
| Target gene | Primer (5′-3′) | Ann.temp (°C) | Product size (bp) | References |
|---|---|---|---|---|
| asa1 |
ASA11: GCA CGC TAT TACGAA CTA TGA ASA12: TAA GAA AGA ACA TCA CCA CGA |
56 | 375 | [15] |
|
| ||||
| ace |
ACE-F: GAA TTG AGC AAA AGT TCA ATC G ACE-R: GTC TGT CTT TTC ACT TGT TTC |
56 | 1008 | [16] |
|
| ||||
| efaAfm |
TE5: GAC AGA CCC TCA CGA ATA TE6: AGT TCA TCA TGC TGT AGT A |
54 | 705 | [17] |
|
| ||||
| esp |
ESP14F: AGA TTT CAT CTT TGA TTC TTG G ESP12R: AAT TGA TTC TTT AGC ATC TGG |
56 | 510 | [15] |
|
| ||||
| cylB |
cylB1: AAG TAC ACT AGT AGA ACT AAG GGA cylB2: ACA GTG AAC GAT ATA ACT CGC TAT T |
52 | 2020 | [18] |
PCR amplifications were performed on MultiGene™ OptiMax Thermal Cycler with a final volume of 25 μL reaction mixtures containing 5x Dream-Taq reaction buffer, 50 ng of bacterial DNA (2μL of the stock), 100 μM DNTP, 25 pM of each primer, and 1U of Taq DNA polymerase (Dream-Taq). PCR amplification of target genes was carried out using a program consisting of the initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at an appropriate temperature for 1 min, elongation at 72°C for 1 min, and a final extension step of 7 min at 72°C. The DNA from strain E. faecalis ATCC 29212 (clinical pathogen) was used as positive control. PCR products were resolved by electrophoresis in 1% agarose (Mupid EXU Japan) gels and digitized by the UVP VisiDoc-It™ Imaging System, Upland, CA, USA.
2.3. Targeting Bacteriocin Genes
Total genomic DNA of E. faecium FL31 was used as template for the detection of some enterocins genes (Ent B, Ent A, Ent P, Ent L50A, and Ent L50B) encoding known Enterococcus bacteriocins. The latter's specific primers and PCR conditions were described in Table 2.
Table 2.
Primers used for the detection of bacteriocin genes.
| Target gene | Primer (5′-3′) | Ann.temp (°C) | Product size (bp) | References |
|---|---|---|---|---|
| Ent B | F: GAA AAT GAT CAC AGA ATG CCT A R: GTT GCA TTT AGA GTA TAC ATT TG |
41 | 159 | [19] |
|
| ||||
| Ent A | F: GAG ATT TAT CTC CAT AAT CT R: GTA CCA CTC ATA GTG GAA |
45 | 542 | [20] |
|
| ||||
| Ent P | F: ATG AGA AAA AAA TTA TTT AGT TT R: TTA ATG TCC CAT ACC TGC CAA ACC |
41 | 216 | [21] |
|
| ||||
| Ent L50 A | F: CCA TGG GAG CAA TCG CAA AA R: AAG CTT AAT GTT TTT TAA TCC ACT CAA T |
50 | 135 | [22] |
|
| ||||
| Ent L50 B | F: ATG GGA GCA ATC GCA AAA TTA R: TAG CCA TTT TTC AAT TTG ATC |
49 | 252 | [23] |
PCR amplifications were performed in a final volume of 50 μL containing 50 ng of bacterial DNA, 5x Dream-Taq reaction buffer, 200 μM DNTP, 50 pM of each primer, and 1U of Taq DNA polymerase (Dream-Taq).
The following PCR conditions were used: denaturation at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 1 min; annealing at an appropriate temperature depending on Tm of each primer pair for 1 min; elongation at 72°C for 1 min and a final extension step of 7 min at 72°C. The PCR generated fragments were analyzed by electrophoresis in 1. 2% agarose gels.
2.4. BacFL31 Purification and Determination of Minimal Inhibitory Concentration (MIC)
BacFL31 was purified as described by Chakchouk-Mtibaa et al., 2014 [9]. Strain FL31 was inoculated in 1% v/v into 900 ml of MRS broth and incubated without agitation at 37°C until early stationary phase (18 h) corresponding to a maximum of bacteriocin production level. Four steps were used to purify BacFL31 from the obtained active supernatant. Briefly, the first step involved the heat treatment of the supernatant for 15 min at 90°C and then cooling at room temperature before pelleting the denatured proteins by centrifugation at 4500 g for 30 min. The active supernatant was applied (second step) to ammonium sulphate precipitation at 60%. After refrigerated centrifugation at 9000 g for 30 min, the precipitate was dissolved in 10 mL of 20 mmol/L sodium phosphate buffer (pH 7.0) and loaded on a column (128 X 2 cm) of gel filtration Sephadex G-25 (the third step) equilibrated with 20 mmol/L sodium phosphate buffer (pH 7). Ninety fractions (F1-F90) with 5 mL each were eluted from the Sephadex G-25 column. These fractions, detected at 280 nm, were fractioned into seven samples (S1-S7) and only the sample S2 (F38-F42) was noted to exhibit antibacterial activity against L. monocytogenes ATCC19117. S2 was submitted to HPLC purification (fourth step) and BacFL31 was eluted from the column with two mobile phases: A (99.9% water, 0.1% trifluoroacetic acid “TFA”) and B (99.9% acetonitrile, 0.1% TFA). The pooled biological active fraction was concentrated and stored at −20°C. BacFL31 concentration was measured using bovine serum albumin (BSA) as reference as described by Bradford in 1976 [24].
Minimal inhibitory concentration (MIC) of the pure BacFL31 against L. monocytogenes ATCC 19117 was determined in Mueller–Hinton broth. The test was performed in sterile 96-well microplates with a final volume in each microplate well of 100 μL. A stock solution of 1.2 mg/mL of BacFL31 was twofold serially diluted in LB medium. To each test well, 10 μL of L. monocytogenes ATCC 19117 cell suspension at 106 CFU/mL was seeded. The plates were then incubated overnight at 37°C. The MIC was defined as the lowest pure BacFL31 concentration at which the microorganism does not demonstrate visible growth after incubation. Twenty-five μl of Thiazolyl Blue Tetrazolium Bromide (MTT) at 0.5 mg/mL was added to the wells and incubated at room temperature for 30 min. The colorless tetrazolium salt acts as an electron acceptor and was reduced to a red-colored formazan product by the indicator microorganism. When microbial growth was inhibited, the solution in the well remained clear after incubation with MTT. Ampicillin was used as standard and experiments were performed in triplicate.
2.5. Membrane Permeabilization Assay
The membrane permeabilization assay of BacFL31 towards the cytoplasmic membrane of the targeted bacterial cells was carried out using SYTOX Green fluorescent dye (Thermo Fisher scientific). This molecular probe with a high affinity to DNA is unable to enter into an intact cell unless the membrane integrity is compromised by the addition of membrane-disrupting compounds.
The indicator strain L. monocytogenes ATCC 19117 was grown in LB broth overnight at 37°C. After centrifugation at 7000 g for 10 min, the bacterial pellets were washed 3 times with 10 mM sodium phosphate buffer (pH 7.2) before resuspending in the same buffer to reach an optical density of 0.5 at 600 nm. Five μM of SYTOX Green was added to the cells suspension. Then, 100 μL of this mixture was transferred to a 96 well PCR plate. The purified bacteriocin BacFL31 (1 X MIC) was added to the mixture (bacterial cells + SYTOX® Green). The positive control contains 70% of ethanol solution instead of BacFL31. Two negative controls were used containing bacterial cells and SYTOX Green without BacFL31 or added by a solution of tetracycline at a concentration of 1.5μg/mL. Tetracycline is an antibiotic that does not act on the cytoplasmic membrane. Then the 96 well PCR plate was immediately placed into a Varioskan Flash Spectral Scanning Multimode Reader (Thermo Fisher Scientific). The fluorescence signal produced by binding of SYTOX Green dye with the nucleic acid of the dead bacterial cells was detected at 520 nm for a period of 60 min. The experiment was performed in triplicate and raw data was analyzed using Microsoft Excel software.
2.6. Measurement of UV-Absorbing Materials
Release of UV-absorbing materials is an index of cell lysis and nonselective pore formation [25]. Absorbances were measured by UV spectrophotometer (UV-1600 PC spectrophotometer VWR) as follows: L. monocytogenes ATCC 19117 cells were resuspended in phosphate buffer 10 mM (pH 7.0) to reach a cell concentration of 107 CFU/mL. After that, BacFL31 was added to the cell suspensions (10 mL) to obtain final concentrations of 1 X MIC. A control sample (10 mL of suspensions cells without bacteriocin) was used for comparison. Aliquots of 1 mL from control and BacFL31-treated cells suspensions were taken every 30 min. Cells were harvested by centrifugation (8000 g, 10 min) and the supernatants were filtered by sterile nitrate cellulose membrane (0.22 μM). The UV-absorbing material was measured at 260 and 280 nm.
2.7. Scanning Fluorescent Microscopy Analysis
In order to determine membrane damage of L. monocytogenes ATCC 19117 cells caused by BacFL31 treatment, we carried out fluorescence studies using the SYTOX Green probe (ThermoFisher scientific). The SYTOX Green was unable to enter into cell unless if the membrane integrity was compromised [26]. Two hundred μL of cells suspension (107 CFU/ml of L. monocytogenes ATCC 19117) was treated with BacFL31 at a final concentration of 1 X MIC. As control, 200 μL of cells suspension without BacFL31 was used. The two mixtures, control and treated sample, were incubated with 2 μL SYTOX Green (10 μM final concentration) for 15 min, 30 min, and 1 h at room temperature in the dark. For each incubation time, an aliquot of cells mixture was washed with 1 mL sterile phosphate buffer (pH 7.0) and centrifuged twice at 8000 g for 10 min. The cells fixed between a slide and quartz cover-slip were examined under a fluorescent microscope OLYMPUS DP70 digital camera connected to a TV adapter.
2.8. Measurement of the Released Potassium Ions
To determine the BacFL31 impact on cells membrane permeability, the released extracellular ions K+ were measured. L. monocytogenes ATCC 19117 cells were centrifuged and resuspended in sterile water to reach 107 CFU/mL of cell concentration. BacFL31 was added at a final concentration of 1 X MIC to the cells suspensions. After various time intervals (15, 30, 45, 60, and 75 min), the control (without BacFL31 addition) and the treated samples (2 mL for each essay) were centrifuged at 7000 g for 10 min. The supernatants were subjected to measurement of the released potassium ions by an Analyst 200 atomic absorption spectrometer (Perkin Elmer).
2.9. Determination of the Transmembrane Electrical Potential (Δψ)
The effect of BacFL31 on the proton motive force (PMF) of cytoplasmic membrane was studied by ΔΨ measurement. ΔΨ was monitored with the fluorescent probe 3,3-dipropylthia-dicarbocyanine iodide “DISC3(5) Sigma–Aldrich” [27]. This probe measures the electrical potential gradient disruption across the cytoplasmic membrane cells. The L. monocytogenes ATCC 19117 cells suspension (107 CFU/mL) was prepared and mixed with 5μM DISC3(5) and then supplemented with nigericin (Sigma–Aldrich) at 1μM (negative control) or with valinomycin (Sigma–Aldrich) at 1μM (positive control) or with BacFL31 at 1 X MIC. A L. monocytogenes ATCC 19117 cells suspension (107 CFU/mL) without any addition was used as control. Fluorescence value measurements were determined with a Varioskan Flash Spectral Scanning Multimode Reader (Thermo Fisher Scientific) and the excitation and emission wavelengths were set at 622 and 670 nm, respectively.
3. Results and Discussions
3.1. Safety Evaluation of the Strain E. faecium FL31
3.1.1. Antibiotic Susceptibility Testing
E. faecium FL31 was susceptible to ampicillin, streptomycin, kanamycin, chloramphenicol, tetracycline, spectinomycin, penicillin G, vancomycin, oxacillin, and amoxicillin (Table 3). The strain E. faecium MMZ01 isolated from artisanal Tunisian fermented meat [28] has a similar antibiotic susceptibility profile to E. faecium FL31. In the last decade, vancomycin-resistant Enterococci (VRE) became a major hospital acquired pathogen which has emerged as a frequent cause of nosocomial infections. The studied strain E. faecium FL31, isolated from food, was vancomycin-sensitive bacteria. This result is in agreement with many studies demonstrating that the majority of enterococci isolated from food were sensitive to this antibiotic [4].
Table 3.
Antibiotics susceptibility of E. faecium FL31.
| Antibiotics | Concentration (μg/disc) | Sensitivity |
|---|---|---|
| Ampicillin | 30 | S |
| Streptomycin | 10 | S |
| Kanamycin | 30 | S |
| Chloramphenicol | 30 | S |
| Tetracycline | 30 | S |
| Spectinomycin | 100 | S |
| Penicillin G | 30 | S |
| vancomycin | 30 | S |
| oxacillin | 5 | S |
| amoxicillin | 25 | S |
S- Sensible
3.1.2. Physiological Test
(1) Hemolytic Activity. The hemolytic activity is associated with haemolysin production. It is an extracellular cytotoxic protein implicated in enterococcal virulence. The production of this protein can increase the severity of the infection and the frequent virulence factor [29]. In our case, no hemolytic activity was observed for the strain E. faecium FL31. This finding is in accordance with previous studies indicating that Enterococcus strains isolated from fermented food products exhibited no hemolytic activity [30].
(2) Gelatinase Activity. Gelatinase activity was not detected in E. faecium FL31. The same result was reported for E. faecium Y31 [31]. Gelatinase is a zinc metalloprotease, encoded by gelE that is capable of hydrolyzing gelatine, collagen, casein, haemoglobin, and other peptides [32]. As encoded by a plasmid gene, gelatinase could mediate binding to the host epithelium and it appears that it plays also an important role in promoting bacterial aggregation during conjugation, facilitating plasmid exchange [33].
(3) DNase Activity. Deoxyribonucleases, enzymes that hydrolyze nucleic acids to yield oligonucleotides, are involved in bacterial virulence. The strain E. faecium FL31 is devoid of DNase activity. Similarly, it has been reported that E. faecalis and E. faecium isolates do not produce DNase activity [34].
(4) Lipase Activity. The biological role of lipases might be considered the most important step in many bacterial infections [35]. The production of this enzyme enhances adhesion by degrading surface molecules of the host. According to our study, the strain E. faecium FL31 did not exhibit lipolytic activity.
3.1.3. Screening for the Presence of Virulence Genes
The safety evaluation of E. faecium FL31 was investigated by the screening of genes encoding different virulent factors cited in Table 1.
As shown in Figure 1, E. faecium FL31 strain did not harbor the five tested virulence genes efaAfm (705 bp), asa1 (375 bp), esp (510 bp), ace (1008 bp), and cylB (2020 bp). Amplification products of these virulent genes were observed only for the positive control strain E. faecalis ATCC 29212 (Figure 1). In E. faecium FL31, the absence of genes encoding endocarditis antigen (efaAfm), aggregation substance (asa1), adhesion of collagen (ace), enterococcal surface protein (esp), and cytolysin toxic protein (cylB) is in agreement with the results of Ahmadova et al. [4] and Liu et al. [31], who report the absence of these virulence determinants in E. faecium AQ71 and E. faecium Y31, respectively. Eaton and Gasson (2001) reported the presence of these virulent factors only in clinical E. faecium strains [17].
Figure 1.
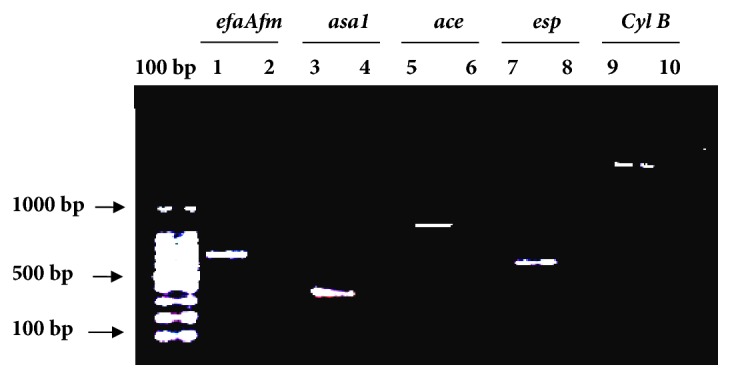
PCR screening for the presence of virulence genes in E. faecium FL31. Lanes 1, 3, 5, 7, and 9: amplification products of corresponding genes obtained from genomic DNA of the positive control E. faecalis ATCC 29212; lanes 2, 4, 6, 8, and 10: amplification products of corresponding genes obtained from genomic DNA of E. faecium FL31.
3.2. PCR Amplification of Bacteriocin Genes
PCR amplification of five well-known structural enterocin (A, B, P, Q and L050) genes was used for the screening of bacteriocin genes in chromosomal and plasmidic DNA of E. faecium FL31 strain (Table 2). Obtained PCR products showed amplification for only enterocin B from chromosomal DNA.
Gene cluster required for enterocin production may be either associated with a plasmid or located in chromosomal DNA [20]. In many cases, enterocin production is a plasmid-encoded trait such as enterocin L50A and enterocin L50B which are located in the 50 kb plasmid pCIZ1 and enterocin Q located in the 7.4 kb plasmid pCIZ2 [36]. However enterocin genes have been also found on the chromosome such as enterocin A, enterocin B, and enterocin P [37].
Enterocin produced by enterococci, including E. faecium and E. faecalis, generally belongs to Class II bacteriocins [38]. Enterocin that comes under Class IIa contains a pediocin-like structure with a YGNGVXC amino acid motif at their N-terminus and shows strong antilisterial activity. They include enterocin A and enterocin P [20]. Enterocin B is not pediocin-like but is similar to Class IIa bacteriocins with respect to its chemical characteristics, heat stability, and anti-Listeria activity [39]. Several studies showed that the enterococcal strains possess one or more enterocin structural gene(s). In the study of Gutiérrez et al., E. faecium P13 possess genes coding enterocins P, enterocin A, and L50A/B [21].
3.3. Membrane Permeabilization Test
We reported previously that BacFL31 is the first bacteriocin described as encompassing hydroxyproline residues [9]. This particular and original characteristic prompted us to study the mode of action of BacFL31 against the pathogenic L. monocytogenes ATCC19117. BacFL31 was purified to homogeneity from a cell-free culture supernatant of E. faecium FL31 strain as described by Chakchouk-Mtibaa et al. [9], and minimal inhibitory concentration (MIC) of the pure BacFL31 against L. monocytogenes ATCC19117 has been determined and is equal to 50 μg/mL.
The ability of the pure peptide BacFL31 to penetrate the cytoplasmic membrane of the pathogen L. monocytogenes ATCC 19117 was tested with SYTOX Green dye. This molecular probe is a high affinity nucleic acid stain that easily penetrates cells with compromised plasma membranes and will not cross the membranes of intact cells [26]. As shown in Figure 2, the fluorescence intensity evolution was very similar for sample treated with BacFL31 at 50 μg/mL and sample treatment with a solution of ethanol at 70% (ethanol is known to destroy cell walls membranes). This fluorescence intensity increased rapidly for the first ten minutes from 100.000 to 250.000 units. Then, the fluorescence continues to increase moderately to reach approximately 420.000 units after 60 min. Concerning the two negative controls (bacterial cells and SYTOX Green without BacFL31or added by a solution of tetracycline at 1.5μg/mL) no evolution of fluorescence was observed from the beginning to the end of the experiment. It should be noted that tetracycline is an antibiotic acting on protein synthesis by inhibiting translation. It binds to the 16S part of the 30S ribosomal subunit and prevents the amino-acyl tRNA from binding to the A site of the ribosome.
Figure 2.
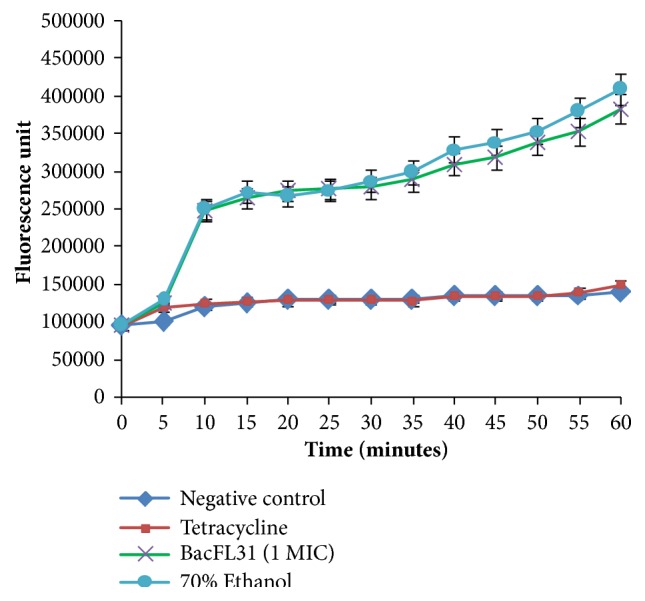
Membrane permeabilization assay of Bacteriocin Bac FL31 towards cytoplasmic membrane of L. monocytogenes ATCC 19117. Negative control without BacFL31 addition; positive control ethanol at 70%. Tetracycline did not show any permeability activity in this test.
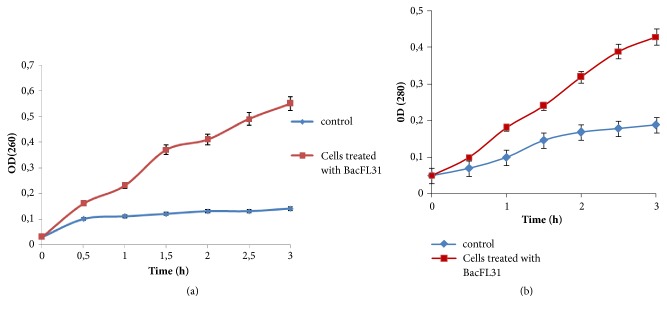
3.4. Measurement of UV-Absorbing Materials
The UV-absorbing materials were measured as an index of cell lysis and nonselective pore formation. As shown in Figure 3, the absorbance value of extracellular nucleic acids (OD260 nm) of L. monocytogenes ATCC 19117 cells treated with BacFL31 increased significantly (p<0.05) from 0.03 to 0.55 whereas, for the negative control sample, the absorbance remains constant until the end of the experience. Absorbance value of proteins (OD280 nm) of BacFL31 treated cells reaches 0.42 after 3 h of incubation. A slight increase of this absorbance was noted for the negative control to reach a value of 0.15 at the end of the experiment (Figure 3). These results supported the idea that BacFL31 damages cytoplasmic membrane and causes subsequent leakage of intracellular constituents. Thus, the crucial effect of BacFL31 on L. monocytogenes ATCC19117 cells could be the formation of nonselective pores in the plasma membrane. Similar results have been observed with other bacteriocins such as Bifidocin A against E. coli 1.90 [25], Bac C1 against B. cereus [40], and sakacin C2 against E. coli [8].
Figure 3.
Extracellular UV-absorbing materials from L. monocytogenes ATCC 19117 cells detected at 260 nm (a) and 280 nm (b).
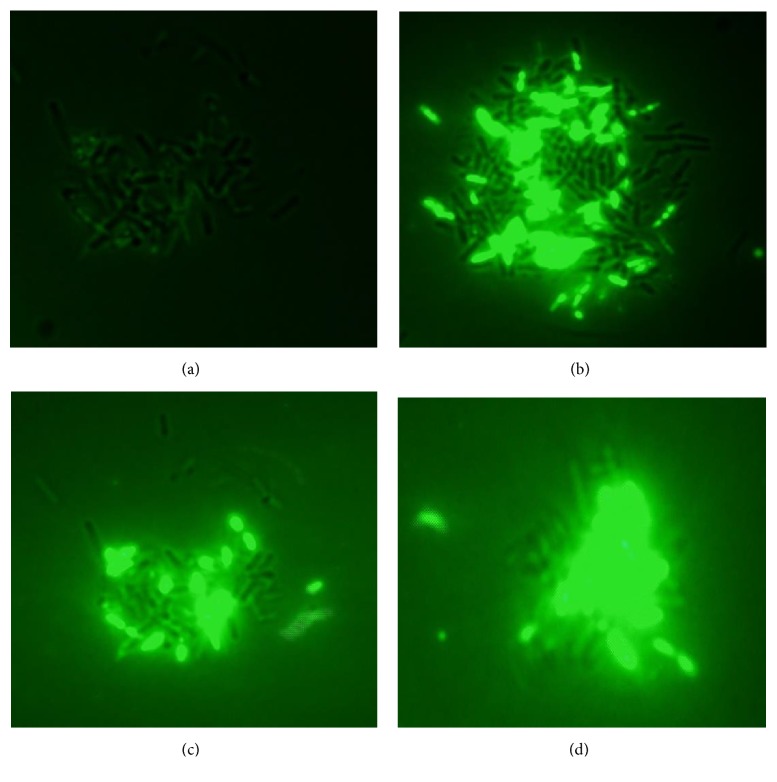
3.5. Scanning Fluorescent Microscopy Analysis
In presence of BacFL31, the alteration of L. monocytogenes ATCC19117 cells wall permeability and membrane pores formation was confirmed by scanning fluorescent microscopy using as fluorescent probe the SYTOX Green. This molecular probe can only permeate depolarized membrane cells and presents a high affinity for DNA stain [40]. As shown in Figure 4(a), there is no fluorescent signal in the untreated L. monocytogenes ATCC19117 cells (negative control) after 1 h of incubation. In presence of BacFL31 at 1 X MIC, the fluorescence intensity increased throughout the incubation time (Figures 4(b)–4(d)). For example, after 15 min of treatment with BacFL31 (Figure 4(b)), almost half of cells exposed to the SYTOX Green dye emit green fluorescence which indicate the damage of approximately 50% of cells. A treatment of 1 h leads to complete damage of cells accompanied by an increase of the fluorescence intensity (Figure 4(d)). Liu et al. [25] reported that destruction of the outer membrane permeability barrier leads to a total or partial dissipation of the proton motive force, causing energy exhaustion and cells death.
Figure 4.
Fluorescence scanning microscopy analysis of L. monocytogenes ATCC 19117 cells without (a) and treated with bacteriocin BacFL31 for 15 min (b), 30 min (c), and 1 h (d).
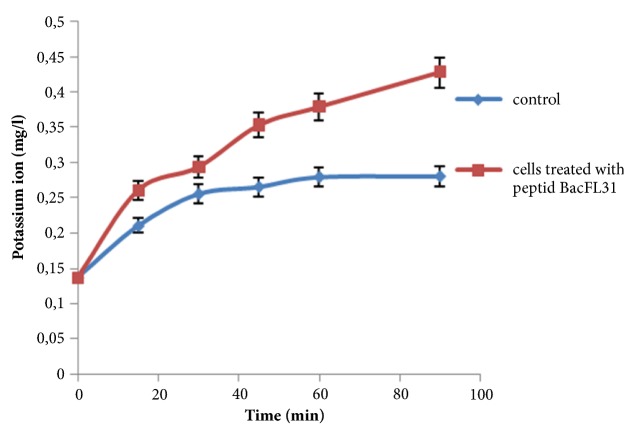
3.6. Measurement of the Released Extracellular Potassium
To determine BacFL31 effect on the integrity of L. monocytogenes ATCC19117 cells, the extracellular K+ concentration was measured in the culture supernatant of the control and the treated cells. As indicated in Figure 5, BacFL31 caused a significant increase (P<0.05) of the extracellular K+ concentration reaching 0.45 mg/L after 60 min of incubation. In contrast, for the control, the levels of the extracellular K+ were relatively stable during the test time period (Figure 5). This proves that Bac FL31 increased the permeability of L. monocytogenes ATCC19117 membrane cells by pore formation causing leakage of the extracellular potassium ions. It has been reported in previous studies that the peptide F1 causes leakage of potassium ions in Staphylococcus aureus [41] and the peptide P7 caused the disruption of Salmonella typhimurium cells membrane and the leakage of the extracellular K+ ions [42].
Figure 5.
The extracellular levels of potassium ions released by L. monocytogenes ATCC19117 cells in presence and in absence of BacFL31.
3.7. Measurement of the Transmembrane Electrical Potential (Δψ)
In order to assess the effect of BacFL31 on the proton motive force (PMF) of the cytoplasmic membrane of L. monocytogenes ATCC19117 cells, we have examined Δψ. The latter was measured qualitatively with the fluorescent probe 3,3-dipropylthia-dicarbocyanine iodide DISC3(5). The addition of the mobile ion carrier valinomycin (positive control) causes immediately a complete dissipation of the transmembrane electrical potential. In contrast, the cells maintained their Δψ in the presence of the nigericin (negative control). The addition of BacFL31 causes the depolarization of the cytoplasmic membrane of L. monocytogenes ATCC19117 cells like the valinomycin. After 09 min of the addition of BacFL31 (1×MIC) to L. monocytogenes ATCC19117 cells, the fluorescence values increased from 60 to 800 (Figure 6). These results proved the role of BacFL31 in the dissipation of the transmembrane electrical potential and membrane depolarization. Probably, BacFL31 could induce permeabilization of the target membrane cells, by forming ion-selective pores which cause dissipation of the proton motive force and depletion of intracellular ATP.
Figure 6.
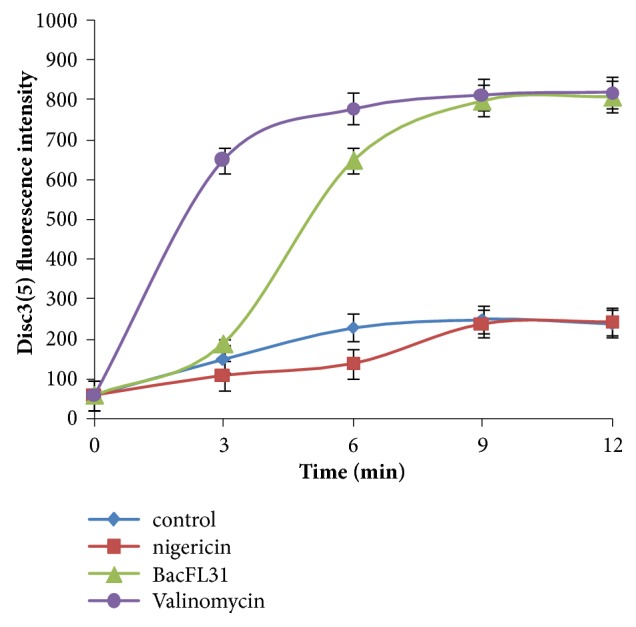
Transmembrane electrical potential (Δψ) in L. monocytogenes ATCC19117 cells. Valinomycin (positive control), nigericin (negative control), and without any addition (control).
In this regard, several studies reported that Class II bacteriocins induce permeabilization of the target membrane cells by forming ion-selective pores which cause dissipation of the proton motive force and depletion of intracellular ATP such as pentocin 31-1 [43].
4. Conclusions
In this study, we showed that the lactic acid strain E. faecium FL31 was susceptible to all tested antibiotics, free of hemolytic, gelatinase, DNase, and lipase activities and did not harbor virulence genes. PCR amplification using specific primers indicated that E. faecium FL31 possess gene encoding enterocin B. At MIC value, 50 μg/mL, the pure bacteriocin BacFL31 damages cytoplasmic membrane of L. monocytogenes ATCC19117, causes the leakage of its intracellular constituents, and leads to the destruction of this pathogenic microorganism. E. faecium FL31 and its bacteriocin BacFL31 is a real candidate as natural preservative tool in the food industry for the preservation of food-borne pathogens growth, particularly L. monocytogenes, during manipulation and storage of food derivatives.
Acknowledgments
This research was funded by the Tunisian Ministry of Higher Education and Scientific Research (Program Contract 2015-2018 of the Laboratory of Microorganisms and Biomolecules of the Center of Biotechnology of Sfax, Tunisia).
Data Availability
The Enterococcus faecium FL31 strain, the bacteriocin BacFL31 genes detection, the BacFL31 purification and its mode of action against Listeria monocytogenes data used to support the findings of this study are included within the article.
Disclosure
An earlier version of this manuscript was presented as a poster presentation in 29th International Congress of Biological Sciences and Biotechnology, 26-29 March 2018, Sousse, Tunisia.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Franz C. M. A. P., Huch M., Abriouel H., Holzapfel W., Gálvez A. Enterococci as probiotics and their implications in food safety. International Journal of Food Microbiology. 2011;151(2):125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Hadji-Sfaxi I., El-Ghaish S., Ahmadova A., et al. Antimicrobial activity and safety of use of Enterococcus faecium PC4.1 isolated from Mongol yogurt. Food Control. 2011;22(12):2020–2027. doi: 10.1016/j.foodcont.2011.05.023. [DOI] [Google Scholar]
- 3.Khan H., Flint S., Yu P.-L. Enterocins in food preservation. International Journal of Food Microbiology. 2010;141(1-2):1–10. doi: 10.1016/j.ijfoodmicro.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Ahmadova A., Todorov S. D., Choiset Y., et al. Evaluation of antimicrobial activity, probiotic properties and safety of wild strain Enterococcus faecium AQ71 isolated from Azerbaijani Motal cheese. Food Control. 2013;30(2):631–641. doi: 10.1016/j.foodcont.2012.08.009. [DOI] [Google Scholar]
- 5.Huang Y., Ye K., Yu K., Wang K., Zhou G. The potential influence of two Enterococcus faecium on the growth of Listeria monocytogenes. Food Control. 2016;67:18–24. doi: 10.1016/j.foodcont.2016.02.009. [DOI] [Google Scholar]
- 6.Muñoz-Atienza E., Araújo C., Campo R. D., Hernández P. E., Herranz C., Cintas L. M. Safety assessment and molecular genetic profiling by pulsed-field gel electrophoresis (PFGE) and PCR-based techniques of Enterococcus faecium strains of food origin. LWT- Food Science and Technology. 2016;65:357–362. doi: 10.1016/j.lwt.2015.08.038. [DOI] [Google Scholar]
- 7.Leavis H. L., Willems R. J. L., Van Wamel W. J. B., Schuren F. H., Caspers M. P. M., Bonten M. J. M. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathogens. 2007;3(1):75–96. doi: 10.1371/journal.ppat.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Li D., Sheng Y., Liu X. Mode of action of sakacin C2 against Escherichia coli. Food Control. 2011;22(5):657–661. doi: 10.1016/j.foodcont.2010.07.010. [DOI] [Google Scholar]
- 9.Chakchouk-Mtibaa A., Elleuch L., Smaoui S., et al. An antilisterial bacteriocin BacFL31 produced by Enterococcus faecium FL31 with a novel structure containing hydroxyproline residues. Anaerobe. 2014;27:1–6. doi: 10.1016/j.anaerobe.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 10.de Man J. C., Rogosa M., Sharpe M. E. A medium for the cultivation of Lactobacilli. Journal of Applied Bacteriology. 1960;23(1):130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 11. Antibiogram Committee of the French Microbiology Society CA-SFM, 2013. http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM2013vjuin.pdf.
- 12.Su Y. A., Sulavik M. C., He P., et al. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infection and Immunity. 1991;59(1):415–420. doi: 10.1128/iai.59.1.415-420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith P. B., Hancock G. A., Rhoden D. L. Improved medium for detecting deoxyribonuclease-producing bacteria. Journal of Applied Microbiology. 1969;18(6):991–993. doi: 10.1128/am.18.6.991-993.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiago I., Teixeira I., Silva S., Chung P., Veríssimo A., Manaia C. M. Metabolic and genetic diversity of mesophilic and thermophilic bacteria isolated from composted municipal sludge on poly-ε-caprolactones. Current Microbiology. 2004;49(6):407–414. doi: 10.1007/s00284-004-4353-0. [DOI] [PubMed] [Google Scholar]
- 15.Vankerckhoven V., Van Autgaerden T., Vael C., et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among european hospital isolates of Enterococcus faecium. Journal of Clinical Microbiology. 2004;42(10):4473–4479. doi: 10.1128/jcm.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben Omar N., Castro A., Lucas R., et al. Functional and Safety Aspects of Enterococci Isolated from Different Spanish Foods. Systematic and Applied Microbiology. 2004;27(1):118–130. doi: 10.1078/0723-2020-00248. [DOI] [PubMed] [Google Scholar]
- 17.Eaton T. J., Gasson M. J. Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Applied and Environmental Microbiology. 2001;67(4):1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semedo T., Santos M. A., Martins P., et al. Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. Journal of Clinical Microbiology. 2003;41(6):2569–2576. doi: 10.1128/JCM.41.6.2569-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toit M. D., Franz C. M. A. P., Dicks L. M. T., Holzapfel W. H. Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. Journal of Applied Microbiology. 2000;88(3):482–494. doi: 10.1046/j.1365-2672.2000.00986.x. [DOI] [PubMed] [Google Scholar]
- 20.Aymerich T., Holo H., Håvarstein L. S., Hugas M., Garriga M., Nes I. F. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Applied and Environmental Microbiology. 1996;62(5):1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutiérrez J., Criado R., Citti R., et al. Cloning, production and functional expression of enterocin P, a sec-dependent bacteriocin produced by Enterococcus faecium P13, in Escherichia coli. International Journal of Food Microbiology. 2005;103(3):239–250. doi: 10.1016/j.ijfoodmicro.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 22.Batdorj B., Dalgalarrondo M., Choiset Y., et al. Purification and characterization of two bacteriocins produced by lactic acid bacteria isolated from Mongolian airag. Journal of Applied Microbiology. 2006;101(4):837–848. doi: 10.1111/j.1365-2672.2006.02966.x. [DOI] [PubMed] [Google Scholar]
- 23.Cintas L. M., Casaus P., Holo H., Hernandez P. E., Nes I. F., Håvarstein L. S. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. Journal of Bacteriology. 1998;180(8):1988–1994. doi: 10.1128/jb.180.8.1988-1994.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Liu G., Song Z., Yang X., Gao Y., Wang C., Sun B. Antibacterial mechanism of bifidocin A, a novel broad-spectrum bacteriocin produced by Bifidobacterium animalis BB04. Food Control. 2016;62:309–316. doi: 10.1016/j.foodcont.2015.10.033. [DOI] [Google Scholar]
- 26.Tashyreva D., Elster J., Billi D. A Novel Staining Protocol for Multiparameter Assessment of Cell Heterogeneity in Phormidium Populations (Cyanobacteria) Employing Fluorescent Dyes. PLoS ONE. 2013;8(2):p. 55283. doi: 10.1371/journal.pone.0055283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castellano P., Raya R., Vignolo G. Mode of action of lactocin 705, a two-component bacteriocin from Lactobacillus casei CRL705. International Journal of Food Microbiology. 2003;85(1-2):35–43. doi: 10.1016/S0168-1605(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 28.Belgacem Z. B., Abriouel H., Omar N. B., et al. Antimicrobial activity, safety aspects, and some technological properties of bacteriocinogenic Enterococcus faecium from artisanal Tunisian fermented meat. Food Control. 2010;21(4):462–470. doi: 10.1016/j.foodcont.2009.07.007. [DOI] [Google Scholar]
- 29.Arias C. A., Murray B. E. The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H., Hu Y., Jiang L., Zhou H., Ma J., Liu C. Antilisterial Activity of Bacteriocin HY07 from Enterococcus faecium HY07 Isolated from Chinese Sausages. Food Biotechnology. 2015;29(1):51–68. doi: 10.1080/08905436.2014.996893. [DOI] [Google Scholar]
- 31.Liu W., Zhang L., Shi J., et al. Assessment of the safety and applications of bacteriocinogenic Enterococcus faecium Y31 as an adjunct culture in North-eastern Chinese traditional fermentation paocai. Food Control. 2015;50:637–644. doi: 10.1016/j.foodcont.2014.10.004. [DOI] [Google Scholar]
- 32.Upadhyaya G. P. M., Ravikumar K. L., Umapathy B. L. Review of virulence factors of enterococcus: an emerging nosocomial pathogen. Indian Journal of Medical Microbiology. 2009;27(4):301–305. doi: 10.4103/0255-0857.55437. [DOI] [PubMed] [Google Scholar]
- 33.Comerlato C. B., de Resende M. C. C., Caierão J., d'Azevedo P. A. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Memórias do Instituto Oswaldo Cruz. 2013;108(5):590–595. doi: 10.1590/0074-0276108052013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elsner H.-A., Sobottka I., Mack D., Claussen M., Laufs R., Wirth R. Virulence factors of Enterococcus faecalis and Enterococcus faecium blood culture isolates. European Journal of Clinical Microbiology & Infectious Diseases. 2000;19(1):39–42. doi: 10.1007/s100960050007. [DOI] [PubMed] [Google Scholar]
- 35.Travis J., Potempa J. Bacterial proteinases as targets for the development of second-generation antibiotics. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2000;1477(1-2):35–50. doi: 10.1016/S0167-4838(99)00278-2. [DOI] [PubMed] [Google Scholar]
- 36.Criado R., Diep D. B., Aakra Å., et al. Complete sequence of the enterocin Q-encoding plasmid pCIZ2 from the multiple bacteriocin producer Enterococcus faecium L50 and genetic characterization of enterocin Q production and immunity. Applied and Environmental Microbiology. 2006;72(10):6653–6666. doi: 10.1128/AEM.00859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S. H., Itoh K., Fujisawa T. Characteristics and identification of enterocins produced by Enterococcus faecium JCM 5804T. Journal of Applied Microbiology. 2003;95(2):294–300. doi: 10.1046/j.1365-2672.2003.01975.x. [DOI] [PubMed] [Google Scholar]
- 38.Franz C. M. A. P., Worobo R. W., Quadri L. E. N., et al. Atypical genetic locus associated with constitutive production of enterocin B by Enterococcus faecium BFE 900. Applied and Environmental Microbiology. 1999;65(5):2170–2178. doi: 10.1128/aem.65.5.2170-2178.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casaus P., Nilsen T., Cintas L. M., Nes I. F., Hernández P. E., Holo H. Enterocin B, a new bacteriocin from enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology. 1997;143(7):2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- 40.Goh H. F., Philip K. Isolation and mode of action of bacteriocin BacC1 produced by nonpathogenic Enterococcus faecium C1. Journal of Dairy Science. 2015;98(8):5080–5090. doi: 10.3168/jds.2014-9240. [DOI] [PubMed] [Google Scholar]
- 41.Miao J., Zhou J., Liu G., et al. Membrane disruption and DNA binding of Staphylococcus aureus cell induced by a novel antimicrobial peptide produced by Lactobacillus paracasei subsp. tolerans FX-6. Food Control. 2016;59:609–613. doi: 10.1016/j.foodcont.2015.06.044. [DOI] [Google Scholar]
- 42.Li L., Shi Y., Su G., Le G. Selectivity for and destruction of Salmonella typhimurium via a membrane damage mechanism of a cell-penetrating peptide ppTG20 analogue. International Journal of Antimicrobial Agents. 2012;40(4):337–343. doi: 10.1016/j.ijantimicag.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 43.Zhou K., Zhou W., Li P., Liu G., Zhang J., Dai Y. Mode of action of pentocin 31-1: an antilisteria bacteriocin produced by Lactobacillus pentosus from Chinese traditional ham. Food Control. 2008;19(8):817–822. doi: 10.1016/j.foodcont.2007.08.008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Enterococcus faecium FL31 strain, the bacteriocin BacFL31 genes detection, the BacFL31 purification and its mode of action against Listeria monocytogenes data used to support the findings of this study are included within the article.