Abstract
Objective
To identify differences in the expression of peroxisome proliferator-activated receptor alpha (PPARα) target genes in human peripheral blood mononuclear cells (PBMCs) associated with non-alcoholic fatty liver disease (NAFLD) among Chinese individuals.
Methods
Thirty healthy subjects were selected as the control group (CN), and 43 patients newly diagnosed with NAFLD were subdivided into two groups, non-obese group (NF, n = 21) and obese group (OF, n = 22). Expression of PPARα and its target genes was determined in PBMCs. The levels of liver cell damage markers, total cholesterol (TC), triglyceride (TG), free fatty acids (FFA), glucose, and insulin were determined in serum.
Results
Compared to the CN group, the blood pressure and homeostasis model assessment for insulin resistance (HOMA-IR) were increased in the other groups (P < 0.05), while the systolic blood pressure (SBP) and liver cell damage markers were significantly increased in the OF group (P < 0.05). In the OF group, PPARα target gene expression was 2.03–3.31 times higher than that in the CN group, and a negative correlation was found between PPARα target gene expression and abdominal circumference (AC), body mass index (BMI), diastolic blood pressure (DBP). Additionally, solute carrier family 25 (carnitine/acylcarnitine translocase) member 20 (SLC25A20) and acyl-coenzyme A dehydrogenase 2 long chain (ACADVL) were negatively correlated with HOMA-IR; PPARα, acetyl-coenzyme A dehydrogenase 2 (ACAA2), and carnitine palmitoyltransferase 1A (CPT1A) were positively correlated with HOMA-IR.
Conclusion
There is an up-expression of PPARα target genes in the PBMCs of NAFLD patients, possibly leading to changes in β-oxidation and insulin resistance.
Keywords: NAFLD, Obesity, Insulin resistance, PBMC, PPARα
Background
Non-alcoholic fatty liver disease (NAFLD) is a chronic liver lipid metabolism syndrome caused by factors other than alcohol, hepatotoxic drug consumption or liver viruses. It exhibits pathological features such as liver cell steatosis and deposition [1]. The high global incidence of NAFLD is due to its association with obesity and diabetes. The incidence of NAFLD is 20–30% among the general population, 40–70% among obese individuals, 30–50% among diabetic patients, and 90% among hyperlipidemic patients [2–6]. Based on the degree of lipid steatosis, inflammation, and fibrosis within diseased liver tissue, NAFLD is categorized into five stages: hepatic steatosis, nonalcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and cancer [7], with exact pathogenesis being unknown.
Peroxisome proliferator-activated receptors (PPARs) belong to the superfamily of nuclear hormone receptors that include PPARα, PPARβ/δ, and PPARγ, which can be used as indicators of fatty acids and fatty derivatives, playing important roles in lipid and energy processes [8]. In particular, PPARα is highly expressed in metabolically active tissues, such as the liver, heart, and muscle and brown adipose tissues [9]. In the liver, PPARα regulates peroxisomal β-oxidation, mitochondrial β-oxidation, and microsomal ω-oxidation and thus plays a significant role in the occurrence and development of NAFLD [10].
Additionally, studies have shown that the PPARα ligand can effectively lower blood lipid levels, increase insulin sensitivity, and reduce hepatic steatosis [11–13]. In contrast, the loss of PPARα results in starvation-induced accumulation of fat in the liver that cannot be degraded, increasing fatty liver conditions [14, 15]. In addition, PPARα activation can reduce NASH formation from NAFLD by suppressing inflammatory responses [16]. NASH seriousness, visceral adiposity, and insulin resistance were negatively correlated with human liver PPARα gene expression while adiponectin was positively related. A significant correlation has been found between increased expression of PPARα, but not PPARβ/δ or PPARγ, and histological improvement in the obese patients with NAFLD [17]. Based on these data, PPARα is a potential therapeutic target for NASH.
Several reports have studied the expression of human PPARα target genes; however, due to the difficulty of collecting biological samples from healthy subjects, these studies have been performed on small scales that are not representative. Blood is used as a sample source for gene extraction due to its accessibility and minimal harm to volunteers during the collection process. In humans, 20,000–25,000 genes are protein-coding genes, and 66–82% of which are expressed in human blood cells [18]. According to Cruieiras et al., assessment of gene expression profiles of peripheral blood mononuclear cells (PBMCs) is an important diagnostic tool for developing therapeutic strategies for obese patients [19]. The present study focused on studying the function of PPARα in the formation of NAFLD through the observation of changes in the expression of PPARα target genes in PBMCs.
Patients and methods
Patients
Permission of this study was granted by the Ethics Committee of Human Research at the Medical Examination Center, Shandong University Qilu Hospital, China. All subjects gave their written informed consent to participate in this study, which was approved by the Ethics Committee of Qilu Hosptial of Shandong University (Qingdao), (KYLL-2017031).
The present study consisted of 73 Chinese Han volunteers between 22 and 68 years of age. The selected individuals were divided into three groups according to liver ultrasound and body mass index (BMI), including the control group (CN, n = 30; BMI < 30.0 kg/m2; normal liver ultrasound), the fatty liver without obesity group (NF, n = 21; BMI < 30.0 kg/m2; fatty liver tested by ultrasound), and the fatty liver with obesity group (OF, n = 22; BMI ≥ 30 kg/m2; fatty liver tested by ultrasound). All three groups had absent alcohol consumption (< 20 g/d).
For the initial assessment, individuals answered a standard questionnaire that included personal information about their diets, physical activity, medication used, smoking habits, and family history of chronic diseases. Then, weight, height, abdominal circumference, and blood pressure measurements were recorded. During this evaluation, venous blood samples were collected after 10-h overnight fasting, and BMIs were calculated based on the height and weight of each individual.
Inclusion criteria: patients newly diagnosed with non-alcoholic fatty liver disease (NAFLD).
Exclusion criteria: subjects with tobacco use habits, chronic use of medications for diabetes mellitus or dyslipidemia, and the diagnosis of chronic medical conditions (e.g., diabetes, hemophilia, heart disease, anemia, and gastrointestinal disease).
Biochemical blood measurements
Peripheral venous blood (5 mL) was collected following an overnight fasting to measure aspartate aminotransferase (AST), alanine aminotransferase (ALT), r-glutamyl transpeptidase (r-GT), alkaline phosphatase (ALP), total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDLc) and low-density lipoprotein cholesterol (LDLc), free fatty acids (FFA), glucose (GLU), and insulin (INS). These substances were examined using an automatic biochemistry analyzer (Roche Diagnostics GmbH, Germany), and homeostasis model assessment for insulin resistance (HOMA-IR) was calculated from fasting insulin and glucose levels [20].
Isolation of peripheral blood mononuclear cells (PBMCs)
Immediately after blood collection, the PBMCs were isolated by gradient centrifugation with the Histopaque (density: 1.077, Sigma-Aldrich, Inc.) and washed with a 1× PBS-buffer.
Total RNA extraction
Total PBMCs RNA was isolated from all samples with Trizol LS® (cat. 15,596–026, Invitrogen), according to the manufacturer’s protocol. The isolated RNA samples were quantified by a spectrophotometer (Titertek-Berthold Colibri, Germany).
cDNA synthesis
cDNA was synthesized using the UEIris reverse transcription real-time polymerase chain reaction (RT-PCR) system. Total RNA (500 ng/ml in a final volume of 2 μl) was added to a mixture (10 μl) containing RNase-free water, 10 × DNase I buffer, and DNase I. This mixture was placed into a thermal cycler (Bioer Life Express, China) at 37 °C for 10 min, before 1 μl of 50 mM EDTA was added to the mix. This mixture was heated to 65 °C for 10 min to deactivate DNase I; then, UEIris RT MasterMix (5×) RNase-free water was added to the mix (20 μl). This mix was placed into the thermal cycler, and a reverse transcription reaction was facilitated to produce cDNA at 55 °C for 30 min, with a final step conducted for 10 s at 85 °C.
Conventional PCR
After cDNA was synthesized, a conventional PCR was conducted to evaluate its quality, and β-actin was found to be stably expressed under the described laboratory conditions in PBMCs. After quantification using a spectrophotometer (Titertek-Berthold Colibri, Germany) and confirmation of cDNA quality by electrophoresis (40 V, 30 mA for 60 min) on a 2% agarose gel (UltraPure, cat. 15,510,019, Invitrogen), a second PCR reaction was performed to assess the efficiencies of the forward and reverse primers of the following genes: pyruvate dehydrogenase kinase isoform 4 (PDK4), carnitine palmitoyltransferase 1A (CPT1A), carnitine palmitoyltransferase 1B (CPT1B), acetyl-coenzyme A dehydrogenase 2 (ACAA2), solute carrier family 25 (carnitine/acylcarnitine translocase) member 20 (SLC25A20), and acyl-coenzyme A dehydrogenase long chain (ACADVL). Primers used in this study are shown in Table 1.
Table 1.
Primer sequences for the RT-PCR
| Gene | Primers | Sequence (5’-3’) | Size (bp) |
|---|---|---|---|
| β-actin | Sense | GGCTGTATTCCCCTCCATCG | 154 |
| Antisense | CCAGTTGGTAACAATGCCATGT | ||
| Solute carrier family 25 (carnitine/acylcarnitine translocase) member 20 (SLC25A20) | Sense | GGGGTCACTCCCATGTTTG | 135 |
| Antisense | TGTGGTGAATACGCCAGATAAC | ||
| Pyruvate dehydrogenase kinase isoform 4 (PDK4) | Sense | GGAGCATTTCTCGCGCTACA | 117 |
| Antisense | ACAGGCAATTCTTGTCGCAAA | ||
| Peroxisome proliferators activated receptors α (PPARα) | Sense | ATGGTGGACACGGAAAGCC | 124 |
| Antisense | CGATGGATTGCGAAATCTCTTGG | ||
| Carnitine palmitoyltransferase 1B (CPT1B) | Sense | GCGCCCCTTGTTGGATGAT | 112 |
| Antisense | CCACCATGACTTGAGCACCAG | ||
| acetyl-coenzyme A dehydrogenase 2 (ACAA2) | Sense | CTGCTCCGAGGTGTGTTTGTA | 105 |
| Antisense | GGCAGCAAATTCAGACAAGTCA | ||
| Acyl-coenzyme A dehydrogenase, long chain (ACADVL) | Sense | ACAGATCAGGTGTTCCCATACC | 114 |
| Antisense | CTTGGCGGGATCGTTCACTT | ||
| Carnitine palmitoyltransferase 1A (CPT1A) | Sense | TCCAGTTGGCTTATCGTGGTG | 98 |
| Antisense | TCCAGAGTCCGATTGATTTTTGC |
Quantitative real-time polymerase chain reaction (qPCR)
A Q-PCR was utilized to assess the mRNA expression levels of target genes. Reactions were performed in the SLAN-96S using the Chromo 4 Detection System (MJ Research Inc.) and the Platinum SYBR Green qPCR Supermix-UDG (cat. S-2008, US Everbright Inc.). PCR amplification conditions were as follows: one cycle at 95 °C for 2 min, 40 cycles at 95 °C for 5 s, one cycle at 60 °C for 15 s, and elongation at 72 °C for 25 s. Each cDNA sample was analyzed in triplicate for each gene. The relative expression ratio of mRNA was calculated using the equation 2−ΔΔCt, where −ΔCt refers to the difference between the numbers of cycles (Ct) of the target genes and the endogenous control.
Statistical analysis
The analysis was performed using the statistical software SPSS 17.0. Values were expressed as the mean ± standard deviation (SD) whenever applicable. The differences between the groups were verified using the one-way ANOVA. The correlation of PPARα target genes with main clinical parameters in the OF group was determined using the Spearman rank test. A P-value of less than 0.05 was considered statistically significant.
Results
Clinical and biochemical characteristics
The characteristics of the studied individuals are presented in the Table 2. The patients in all three sample groups were close in age (P > 0.05). In the OF group, BMI, waist, and systolic blood pressure (SBP) were all significantly higher than those in the CN group and NF group (P < 0.01). Diastolic blood pressure (DBP) in both NF and OF groups was higher than that in the CN group (P < 0.05). Based on these data, fatty liver patients have increased blood pressure, while obese patients have higher chances of having increased SBP.
Table 2.
Clinical and biochemical characteristics of the CN, NF and OF groups
| CN group | NF group | OF group | F | P | |
|---|---|---|---|---|---|
| Age, years | 38.63 ± 12.68 | 46.31 ± 11.61 | 39.05 ± 11.14 | 2.59 | 0.08 |
| BMI,† Kg/m2 | 24.31 ± 4.14 | 25.45 ± 2.82 | 31.78 ± 2.17**ΔΔ | 30.95 | 0.00 |
| Waist,† cm | 85.45 ± 14.67 | 91.26 ± 10.78 | 112.89 ± 6.76**ΔΔ | 31.81 | 0.00 |
| SBP,† mm Hg | 120.59 ± 15.74 | 138.79 ± 18.58** | 141.37 ± 18.90**ΔΔ | 8.55 | 0.001 |
| DBP,† mm Hg | 73.36 ± 12.21 | 80.74 ± 8.52* | 87.26 ± 13.48** | 7.32 | 0.001 |
| ALT,† U/L | 24.09 ± 11.05 | 29.10 ± 13.34 | 49.74 ± 36.95**ΔΔ | 6.87 | 0.002 |
| AST,† U/L | 18.77 ± 4.30 | 20.63 ± 6.90 | 27.95 ± 14.61**Δ | 5.23 | 0.008 |
| ALP,† U/L | 66.63 ± 14.53 | 79.26 ± 28.35* | 74.68 ± 13.65* | 2.15 | 0.126 |
| rGT,† U/L | 22.64 ± 10.52 | 38.37 ± 28.40 | 83.58 ± 61.12* | 2.35 | 0.105 |
| TC,† mmol/L | 4.66 ± 0.88 | 5.52 ± 1.00** | 5.02 ± 0.91 | 4.38 | 0.017 |
| TG,† mmol/L | 1.42 ± 0.68 | 2.21 ± 1.55* | 1.73 ± 0.66 | 3.05 | 0.055 |
| HDLc,† mmol/L | 1.18 ± 0.20 | 1.15 ± 0.17 | 2.05 ± 2.64 | 2.31 | 0.109 |
| LDLc,† mmol/L | 1.93 ± 0.46 | 2.53 ± 0.56** | 2.18 ± 0.48Δ | 7.51 | 0.001 |
| FFA,† mEq/L | 0.94 ± 0.24 | 1.15 ± 0.60 | 1.06 ± 0.31 | 1.42 | 0.25 |
| Glu,† mmol/L | 4.86 ± 0.55 | 5.72 ± 1.07** | 5.59 ± 1.01* | 4.97 | 0.01 |
| INS,† uIU/ml | 8.11 ± 6.17 | 13.25 ± 8.02* | 14.86 ± 8.01** | 4.76 | 0.012 |
| HOMA-IR,† | 1.78 ± 1.41 | 3.52 ± 2.48** | 3.69 ± 2.16** | 5.52 | 0.006 |
†Data were expressed as the mean ± SD of 30 people in the CN group, 21 people in the NF group, and 22 people in the OF group. Statistically significant differences between groups were determined using ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001 vs the CN group; Δp < 0.05, ΔΔp < 0.01 vs the NF group)
ALP Alkaline phosphatase, ALT Alanine transaminase, AST Aspartate transaminase, BMI body mass index, DBP diastolic blood press, FFA free fatty acids, Glu Glucose, HDLc High-density lipoprotein cholesterol, HOMA-IR Homeostasis model assessment of insulin resistance, INS Insulin, LDLc Low-density lipoprotein cholesterol, SBP Systolic blood press, SD standard deviation, TC Total cholesterol, TG Triacylglycerol, rGT gamma-glutamyl transpeptidase
The four hepatic metabolic enzymes, ALT, AST, ALP, and r-GT were higher in the OF group compared with the CN group (P < 0.05), and the ALT and AST were significantly higher in the OF group when comparing to the NF group (P < 0.05). Increased serum AST and ALT levels are used as markers for liver damage, and here, it was proved that OF patients had increased liver function damage. Besides, NF patients had significantly higher levels of TC, TG, and LDLc than the CN patients. In contrast, there was an upward trend in blood lipids of the OF group relative to the CN group without statistical difference, indicating that the disturbances in lipid metabolism were more serious in NF patients, while the fatty liver formation in the OF group might result from other factors, such as adipocytokines. Further, the fasting blood glucose levels, insulin levels, and HOMA-IR were significantly higher in the NF and OF groups than those in the CN group (P < 0.05); there was no significant difference between the NF and OF groups; therefore, fatty liver patients have significant IR that does not depend on obesity.
Gene expression
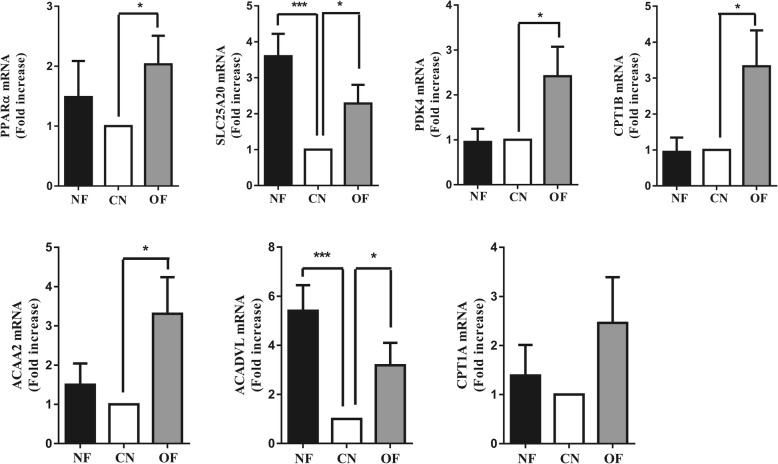
The PPARα expression was significantly higher in the OF group than that in the CN group (P < 0.05); the expression of PPARα target genes (SLC25A20, PDK4, CPT1B, ACAA2, and ACADVL) in the OF group was 2.03–3.31 times more than that in the CN group (P < 0.05), while CPT1A expression was similar in the CN group and OF group (P > 0.05) (Fig. 1). In the NF group, the SLC25A20 and ACADVL expression were significantly higher than that in the CN group (P < 0.05) (Fig. 1).
Fig. 1.
Gene expression of PPARα target genes in PBMCs. Data were expressed as the mean ± SD of 30 people in the CN group, 21 people in the NF group, and 22 people in the OF group. Statistically significant differences between groups were determined using ANOVA; *p < 0.05, **p < 0.01, ***p < 0.001 vs the CN group
The correlation of PPARα and its target genes with primary clinical parameters in the OF group is presented in the Table 3. There was a negative correlation between PPARα target gene expression and abdominal circumference (AC), BMI, DBP. SLC25A20 was negatively correlated with insulin and HOMA-IR; ACADVL was negatively correlated with blood glucose and HOMA-IR; PPARα was positively correlated to blood glucose, insulin, and HOMA-IR; ACAA2 was positively correlated with insulin and HOMA-IR; CPT1A was positively correlated with HOMA-IR.
Table 3.
Correlation of PPARα and its target genes with main clinical parameters in OF group
| OF group | SLC | PDK4 | PPARα | CPT1B | ACAA2 | ACADVL | CPT1A | |
|---|---|---|---|---|---|---|---|---|
| AC,† | Pearson Correlation | .112 | −.436* | −.383 | −.411* | −.131 | .303 | −.403* |
| Significant (unilateral) | .324 | .031 | .053 | .040 | .297 | .104 | .044 | |
| BMI,† | Pearson Correlation | .360 | −.532** | −.577** | −.512* | −.499* | .450* | −.549** |
| Significant (unilateral) | .065 | .010 | .005 | .012 | .015 | .027 | .007 | |
| SBP,† | Pearson Correlation | −.065 | −.204 | −.175 | −.233 | −.035 | −.051 | −.285 |
| Significant (unilateral) | .396 | .201 | .236 | .168 | .443 | .417 | .118 | |
| DBP,† | Pearson Correlation | .245 | −.573** | −.561** | −.628** | −.395* | .270 | −.677** |
| Significant (unilateral) | .156 | .005 | .006 | .002 | .047 | .132 | .001 | |
| ALT,† | Pearson Correlation | −.373 | −.486* | −.365 | −.387 | −.228 | −.177 | −.382 |
| Significant (unilateral) | .058 | .018 | .062 | .051 | .173 | .235 | .053 | |
| GLU,† | Pearson Correlation | −.264 | .425* | .484* | .388 | .382 | −.530** | .385 |
| Significant (unilateral) | .137 | .035 | .018 | .050 | .053 | .010 | .052 | |
| INS,† | Pearson Correlation | −.432* | .224 | .422* | .291 | .441* | −.349 | .326 |
| Significant (unilateral) | .032 | .178 | .036 | .114 | .029 | .071 | .086 | |
| HOMA -IR,† |
Pearson Correlation | −.444* | .342 | .546** | .377 | .538** | −.445* | .424* |
| Significant (unilateral) | .028 | .076 | .008 | .056 | .009 | .028 | .035 |
†In the OF group, the correlation between PPARα target genes and primary clinical parameters were determined using the Spearman rank test (*p < 0.05,**p < 0.01)
AC Abdominal circumference, ALT Alanine transaminase, BMI body mass index, DBP diastolic blood press, Glu Glucose, HOMA-IR Homeostasis model assessment of insulin resistance, INS Insulin, SBP Systolic blood press
Discussion
PPARα is a key factor involved in the cellular metabolic response, and its gene expression was examined through liver biopsy among NAFLD patients, showing significantly lower levels of expression for increasing severity of NASH [17]. In this study, high blood pressure and IR were found in all NAFLD patients, with fatty liver, obesity, and increased liver damage. The PPARα gene expression was found to be increased in the PBMCs of the OF group, having a negative correlation to BMI, DBP and a positive correlation to blood glucose, insulin, HOMA-IR. Thus, these findings suggest that PPARα gene expression follows the same trend as IR in human liver tissue.
Increasing evidence suggests that PPARα plays a main role in the management of obesity, especially central obesity associated with IR syndromes, given its involvement in the regulation of lipid metabolism and inflammation. Stimulation of β-oxidation by PPARα activation is a suggested mechanism for decreasing tissue lipid content to prevent lipid accumulation [21]. In an animal model, PPARα activation was shown to have a beneficial effect on insulin sensitivity by reducing plasma TG levels and adiposity [16]. In humans, fibrate activation of PPARα has been shown to increase the circulating levels of atheroprotective HDLc, to lower plasma levels of TG and FFA, and to improve overall atherogenic plasma lipid profiles while exerting beneficial effects on inflammation, IR, and metabolic syndrome [22–26]. In our study, NF patients had significantly higher levels of TC, TG, and LDLc than the CN patients. It has been reported that nutraceuticals and functional food ingredients that are beneficial to vascular health reduce the dyslipidemia-induced cardiovascular risk possibly by decreasing 7α-hydroxylase, increasing fecal excretion of cholesterol or decreasing the secretion of very low-density lipoprotein [27]. These findings suggest that nutraceuticals and functional food ingredients that are beneficial to liver health might reduce the incidence of NAFLD.
Through testing PPARα target gene expression in PBMC samples and conducting a correlation analysis, we found that the OF group had a 2.46-time increase in CPT1A gene expression compared to the CN group, which was positively related to HOMA-IR. Blood glucose levels decreased concomitantly with a decrease in cellular glycolysis, and CPT1A was activated, permitting the mitochondrial entry of FFA [28, 29]. However, as CPT1A is a crucial enzyme involved in FFA catabolism within mitochondria in metabolically active tissue, the up-regulation of CPT1A gene expression in PBMCs may indicate an increased β-oxidation rate to meet energy demands [21]. During fasting, increased CPT1A gene expression is an essential metabolic response to maintain cellular lipid balance in liver tissues, which increases FFA absorption and utilization [30]. In the present study, CPT1B was found to be increased and have a negative correlation with AC and BMI, however, the exact mechanism remains unclear.
PDK4 is a gatekeeper regulator for glucose metabolism [31]. According to the results in animal experiments, the mRNA and protein levels of PDK4 are elevated in livers of diabetic, fasted, and IR animals [32, 33]. Furthermore, PDK4 knock-out mice have been found to have lower blood glucose levels, better glucose tolerance, and greater insulin sensitivity compared to wild-type mice [34]. In differentiated cultured human myotubes, agonist (GW7647)-mediated activation of PPAR-α results in increased fatty acid oxidation [35, 36], diminished accumulation of TG [35], and upregulation of PDK4 [37]. In a recent study, the mRNA and protein levels of PDK4 were upregulated in NASH livers, indicating that PDK4 potentially contributes to the hepatic steatosis of NASH via regulation of several signaling pathways [38]. PDK4 phosphorylates and inactivates the pyruvate dehydrogenase complex during fasting, thereby decreasing glucose oxidation [39, 40]. We for the first time confirm that PDK4 up-expression in the PBMCs of the OF group negatively correlates to AC, BMI and positively correlates to blood glucose. Thus, PDK4 may be a new therapeutic agent for NAFLD.
Several studies have reported SLC25A20, ACADVL, and ACAA2 genes in fatty liver PBMCs; in our study, an increase in the mean fold change of the ACADVL gene among obese individuals was observed. This gene codes for an enzyme that controls a critical point in the supply of electrons to the respiratory chain and is responsible for catalyzing the first β-oxidation step [41]. CPT1A and ACADVL are lipid oxidation markers and were observed to be decreased in newborn puppies compared to obese dams [42], while another report showed that PPARα and ACAA2 are closely associated with NASH [43]. In this study, it was found that blood glucose, insulin level, and HOMA-IR were negatively related to SLC25A20, ACADVL but positively related to ACAA2 in the OF group, although further experiments are required to confirm these results.
There have been several limitations of this study. The sample size is relatively small. A further study with larger number of samples is required in the future work. Besides, a multivariate regression analysis should be performed in order to evaluate the role of confounding factors on final results.
Conclusion
In summary, the effects of PPARα gene were studied by analyzing its target gene expression in the PBMCs of NAFLD patients. PPARα target genes were found to be negatively correlated with AC, BMI, and DBP in OF patients, while SLC25A20 and ACADVL were negatively correlated with blood glucose, insulin level, and HOMA-IR, and PPARα, ACAA2, and CPT1A were positively related to the same variables. These results reveal the up-expression of PPARα target genes in NAFLD patients’ PBMCs, possibly affecting β-oxidation and IR.
Acknowledgments
We thank the staff of the Department of Medical Experimental Center, Clinical Lab and the Department of Qingdao Key Lab of Mitochondrial medicine of our hospital for their support of this study. We would also like to express our sincere gratitude to the patients who participated in this study.
Funding
This study was supported by shandong medical and health science and technology development plan (2014ws0130).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Abbreviations
- AC
Abdominal circumference
- ACAA2
Acetyl-coenzyme A dehydrogenase 2
- ACADVL
Acyl-coenzyme A dehydrogenase 2 long chain
- ALP
Alkaline phosphatase
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- BMI
Body mass index
- CN
Control group
- CPT1A
Carnitine palmitoyltransferase 1A
- CPT1B
Carnitine palmitoyltransferase 1B
- Ct
Cycles
- DBP
Diastolic blood pressure
- FFA
Free fatty acids
- GLU
Glucose
- HDLc
High-density lipoprotein cholesterol
- HOMA-IR
Homeostasis model assessment for insulin resistance
- INS
Insulin
- LDLc
Low-density lipoprotein cholesterol
- NAFLD
Non-alcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NF
Non-obese group
- OF
Obese group
- PBMCs
Peripheral blood mononuclear cells
- PDK4
Pyruvate dehydrogenase kinase isoform 4
- PPARs
Peroxisome proliferator-activated receptors
- PPARα
Peroxisome proliferator-activated receptor alpha
- qPCR
Quantitative real-time polymerase chain reaction
- r-GT
r-glutamyl transpeptidase
- RT-PCR
Reverse transcription real-time polymerase chain reaction
- SBP
Systolic blood pressure
- SD
Standard deviation
- SLC25A20
Solute carrier family 25 (carnitine/acylcarnitine translocase) member 20
- TC
Total cholesterol
- TG
Triglyceride
Authors’ contributions
TTL performed quantitative PCR and evaluated the results. TBT analyzed the results and edited English language. HQH prepared samples and measured serum biochemical parameters. XYZ participated in the planning of the study, data analysis, and manuscript preparation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Permission of this study was granted by the Ethics Committee of Human Research at the Medical Examination Center, Shandong University Qilu Hospital, China (KYLL-2017031).
Consent for publication
All subjects gave their written informed consent to participate in this study.
Competing interest
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tian Tian Li, Email: 18561810828@126.com.
Xiao Yun Zhao, Phone: +86 18561810828, Email: 18561810828@126.com.
References
- 1.Arab JP, Candia R, Zapata R, et al. Management of nonalcoholic fatty liver disease: an evidence-based clinical practice review. World J Gastroenterol. 2014;20:12182–12201. doi: 10.3748/wjg.v20.i34.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellentani S, Scaglioni F, Marino M, Bedogni G. Epidemiology of non-alcoholic fatty liver disease. Dig Dis. 2010;28:155–161. doi: 10.1159/000282080. [DOI] [PubMed] [Google Scholar]
- 3.Fazel Y, Koenig AB, Sayiner M, Goodman ZD, Younossi ZM. Epidemiology and natural history of non-alcoholic fatty liver disease. Metabolism. 2016;65:1017–1025. doi: 10.1016/j.metabol.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Milic S, Stimac D. Nonalcoholic fatty liver disease/steatohepatitis: epidemiology, pathogenesis, clinical presentation and treatment. Dig Dis. 2012;30:158–162. doi: 10.1159/000336669. [DOI] [PubMed] [Google Scholar]
- 5.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastro Hepat. 2013;10:686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 6.Reccia I, Kumar J, Akladios C, et al. Non-alcoholic fatty liver disease: a sign of systemic disease. Metabolism. 2017;72:94–108. doi: 10.1016/j.metabol.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Kanuri G, Bergheim I. In vitro and in vivo models of non-alcoholic fatty liver disease (NAFLD) Int J Mol Sci. 2013;14:11963–11980. doi: 10.3390/ijms140611963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patsouris D, Mandard S, Voshol PJ, et al. PPARα governs glycerol metabolism. J Clin Invest. 2004;114:94–103. doi: 10.1172/JCI200420468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakhshandehroo M, Hooiveld G, Müller M, Kersten S. Comparative analysis of gene regulation by the transcription factor PPARα between mouse and human. PLoS One. 2009;4:e6796. doi: 10.1371/journal.pone.0006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pyper SR, Viswakarma N, Yu S, Reddy JK. PPARα: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal. 2010;8:e002. doi: 10.1621/nrs.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cariou B, Hanf R, Lambert-Porcheron S, et al. Dual peroxisome proliferator-activated receptor α/δ agonist GFT505 improves hepatic and peripheral insulin sensitivity in abdominally obese subjects. Diabetes Care. 2013;36:2923–2930. doi: 10.2337/dc12-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fruchart JC. Selective peroxisome proliferator-activated receptor alpha modulators (SPPARMα): the next generation of peroxisome proliferator-activated receptor α-agonists. Cardiovasc Diabetol. 2013;12:82–97. doi: 10.1186/1475-2840-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiri-Sverdlov R, Wouters K, van Gorp PJ, et al. Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. J Hepatol. 2006;44:732–741. doi: 10.1016/j.jhep.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koteish A, Diehl AM. Animal models of steatosis. Semin Liver Dis. 2001;21:89–104. doi: 10.1055/s-2001-12932. [DOI] [PubMed] [Google Scholar]
- 16.Pawlak M, Bauge E, Bourguet W, et al. The transrepressive activity of peroxisome proliferator-activated receptor alpha is necessary and sufficient to prevent liver fibrosis in mice. Hepatology. 2014;60:1593–1606. doi: 10.1002/hep.27297. [DOI] [PubMed] [Google Scholar]
- 17.Francque S, Verrijken A, Caron S, et al. PPARα gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol. 2015;63:164–173. doi: 10.1016/j.jhep.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Liew Choong-Chin, Ma Jun, Tang Hong-Chang, Zheng Run, Dempsey Adam A. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. Journal of Laboratory and Clinical Medicine. 2006;147(3):126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Crujeiras AB, Parra D, Milagro FI, et al. Differential expression of oxidative stress and inflammation related genes in peripheral blood mononuclear cells in response to a low-calorie diet: a nutrigenomics study. Omics. 2008;12:251–261. doi: 10.1089/omi.2008.0001. [DOI] [PubMed] [Google Scholar]
- 20.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Teacher DF, Tumer RC. Homeostasis model assessment:insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 21.Fruchart JC. Peroxisome proliferator-activated receptor-alpha (PPARα): at the crossroads of obesity, diabetes and cardiovascular disease. Atherosclerosis. 2009;205:1–8. doi: 10.1016/j.atherosclerosis.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Guerre-Millo M, Gervois P, Raspé E, et al. Peroxisome proliferator-activated receptor α activators improve insulin sensitivity and reduce adiposity. J Biol Chem. 2000;275:16638–16642. doi: 10.1074/jbc.275.22.16638. [DOI] [PubMed] [Google Scholar]
- 23.Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC. Mechanism of action of fibrtaes on lipid and lipoprotein metabolism. Circulation. 1998;98:2088–2093. doi: 10.1161/01.CIR.98.19.2088. [DOI] [PubMed] [Google Scholar]
- 24.Katsiki N, Nikolic D, Montalto G, Banach M, Mikhilidis DP, Rizzo M. The role of fibrate treatment in dyslipidemia: an overview. Curr Pharm Des. 2013;19:3124–3131. doi: 10.2174/1381612811319170020. [DOI] [PubMed] [Google Scholar]
- 25.Patsouris D, Müller M, Kersten S. Peroxisome proliferator activated receptor ligands for the treatment of insulin resistance. Curr Opin Investig Drugs. 2004;5:1045–1050. [PubMed] [Google Scholar]
- 26.Mirza MS. Obesity, visceral fat, and NAFLD: querying the role of adipokines in the progression of nonalcoholic fatty liver disease. ISRN Gastroenterol. 2011;592404. [DOI] [PMC free article] [PubMed]
- 27.Scicchitano P, Cameli M, Maiello M, et al. Nutraceuticals and dyslipidaemia: beyond the common therapeutics. J Funct Foods. 2014;6:11–32. doi: 10.1016/j.jff.2013.12.006. [DOI] [Google Scholar]
- 28.Zandbergen F, Plutzky J. PPAR-α in atherosclerosis and inflammation. Biochim Biophys Acta. 2007;1771:972–982. doi: 10.1016/j.bbalip.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shipman KE, Strange RC, Ramachandran S. Use of fibrates in the metabolic syndrome: a review. World J Diabetes. 2016;7:74–88. doi: 10.4239/wjd.v7.i5.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenenbaum A, Fisman EZ. Fibrates are an essential part of modern anti-dyslipidemic arsenal: spotlight on atherogenic dyslipidemia and residual risk reduction. Cardiovasc Diabetol. 2012;11:125. doi: 10.1186/1475-2840-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Zuo R, Bao Y, Qu X, Sun K, Ying H. Down-regulation of PDK4 is critical for the switch of carbohydrate catabolism during syncytialization of human placental trophoblasts. Sci Rep. 2017;7:8474. doi: 10.1038/s41598-017-09163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu P, Blair PV, Sato J, Jaskiewicz J, Popov KM, Harris RA. Starvation in-creases the amount of pyruvate dehydrogenase kinase in several mammalian tissues. Arch Biochem Biophys. 2000;381:1–7. doi: 10.1006/abbi.2000.1946. [DOI] [PubMed] [Google Scholar]
- 33.Harris RA, Huang B, Wu P. Control of pyruvate dehydrogenase kinase gene expression. Adv Enzym Regul. 2001;41:269–288. doi: 10.1016/S0065-2571(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 34.Jeoung NH, Harris RA. Pyruvate dehydrogenase kinase-4 deficiency lowers blood glucose and improves glucose tolerance in diet-induced obese mice. Am J Physiol Endocrinol Metabol. 2008;295:E46–E54. doi: 10.1152/ajpendo.00536.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muoio DM, Way JM, Tanner CJ, et al. Peroxisome proliferator-activated receptor-α regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes. 2002;51:901–909. doi: 10.2337/diabetes.51.4.901. [DOI] [PubMed] [Google Scholar]
- 36.Djouadi F, Aubey F, Schlemmer D, Bastin J. Peroxisome proliferator activated receptor δ (PPARδ) agonist but not PPAR-α corrects carnitine palmitoyl transferase 2 deficiency in human muscle cells. J Clin Endocrinol Metab. 2005;90:1791–1797. doi: 10.1210/jc.2004-1936. [DOI] [PubMed] [Google Scholar]
- 37.Abbot EL, McCormack JG, Reynet C, Hassall DG, Buchan KW, Yeaman SJ. Diverging regulation of pyruvate dehydrogenase kinase isoform gene expression in cultured human muscle cells. FEBS J. 2005;272:3004–3014. doi: 10.1111/j.1742-4658.2005.04713.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Zhao Y, Li Z, Wang C. Pyruvate dehydrogenase kinase 4 mediates lipogenesis and contributes to the pathogenesis of nonalcoholic steatohepatitis. Biochem Biophys Res Commun. 2018;495:582–558. doi: 10.1016/j.bbrc.2017.11.054. [DOI] [PubMed] [Google Scholar]
- 39.Bouwens M, Afman LA, Müller M. Fasting induces changes in peripheral blood mononuclear cell gene expression profiles related to increase in fatty acid β-oxidation: functional role of peroxisome proliferator activated receptor α in human peripheral blood mononuclear cells. Am J Clin Nutr. 2007;86:1515–1523. doi: 10.1093/ajcn/86.5.1515. [DOI] [PubMed] [Google Scholar]
- 40.Houten SM, Chegary M, Te Brinke H, Wijnen WJ, Glatz JFC, Luiken JJFP. Pyruvate dehydrogenase kinase 4 expression is synergistically induced by AMP-activated protein kinase and fatty acids. Cell Mol Life Sci. 2009;66:1283–1294. doi: 10.1007/s00018-009-9066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAndrew RP, Wang Y, Mohsen AW, He M, Vockley J, Kim JP. Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase. J Biol Chem. 2008;283:9435–9443. doi: 10.1074/jbc.M709135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simino LA, Fante TD, Fontana MF, et al. Lipid overload during gestation and lactation can independently alter lipid homeostasis in offspring and promote metabolic impairment after new challenge to high-fat diet. Nutr Metab (Lond) 2017;14:16. doi: 10.1186/s12986-017-0168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia J, Xin L, Zhu W, et al. Characterization of long non-coding RNA transcriptome in high-energy diet induced nonalcoholic steatohepatitis minipigs. Sci Rep. 2016;6:307–309. doi: 10.1038/srep30709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.