Abstract
Background
Lead (Pb) and mercury (Hg) are persistent hazardous metals in industrially polluted soils which can be toxic in low quantities. Metal toxicity can cause changes at cellular and molecular level which should be studied for better understanding of tolerance mechanism in plants. Soybean (Glycine max L.) is an important oilseed crop of the world including India. Indian soils growing soybean are often contaminated by Pb and Hg. The aim of this study was to explore how soybean root nodule responds to Pb and Hg through proteomic and ecophysiological alterations in order to enhance tolerance to metal stress.
Results
Soybean plants were exposed to Pb (30 ppm PbCl2) and Hg (0.5 ppm HgCl2) to study histological, histochemical, biochemical and molecular response of N2-fixing symbiotic nodules. Both Pb and Hg treatment increased the level of oxidative stress in leaves and nodules. Chlorosis in leaves and morphological/anatomical changes in nodules were observed. Activities of ascorbate peroxidase, glutathione reductase and catalase were also modulated. Significant changes were observed in abundance of 76 proteins by Pb and Hg. Pb and Hg influenced abundance of 33 proteins (17 up and 16 down) and 43 proteins (33 up and 10 down), respectively. MS/MS ion search identified 55 proteins which were functionally associated with numerous cellular functions. Six crucial proteins namely catalase (CAT), allene oxide synthase (AOS), glutathione S-transferase (GST), calcineurin B like (CBL), calmodulin like (CML) and rapid alkalinisation factor (RAF) were selected for transcript abundance estimation. The qRT-PCR based real time expression exhibited a positive correlation with proteomics expression except for GST and RAF.
Conclusion
Soybean root nodule responds to metal stress by increased abundance of defence, development and repair related proteins. An efficient proteomic modulation might lead to metal-induced stress tolerance in N2-fixing nodules. Although concentrations of Pb and Hg used in the study cannot be considered equimolar, yet Hg seems to induce more changes in nodule proteomic profile, and higher damage to both bacteroides and root anatomy.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1499-7) contains supplementary material, which is available to authorized users.
Keywords: Soybean, Root nodules, Metal stress, Antioxidants, Proteomics, Real time expression
Background
Plant roots take up nutrient ions from the soil and while doing so they can pick toxic ions of unwanted elements inadvertently. Such elements include Hg, Pb, Cd, As, etc. which are deleterious for plant health. The US Agency for Toxic Substances and Disease Registry (ATSDR) 2013 ranked such elements in top four priority list of hazardous substances [1].
Soybean is an important oilseed crop due to its high nutritional value. It is a major source of food in terms of protein and oil. Being a member of Leguminosae, soybean is characterised by the presence of nitrogen-fixing nodules in the roots [2]. These nodules home rhizobium for a symbiotic relation which makes soybean far more important apart from source of nutrition. Soybean production improves soil nitrogen content, thus helps in reduced application of nitrogen fertilizer for not only soybean itself but also for subsequent crop on the same soil [3]. Thus, symbiotic nitrogen fixation greatly contributes to sustainable agriculture through its contribution to plant growth, enrichment of nitrogen-deficient soils and limiting the use of nitrogen fertilizers [4].
India is the fifth largest producer (approximately 10.5 million metric tonnes) of soybean in the world. Around half of soybean produce in India comes from Madhya Pradesh [5]. Interestingly, soils of Madhya Pradesh are contaminated with many toxic metals including Hg, As, Pb, etc. [6]. According to Central Pollution Control Board (CPCB), Madhya Pradesh (MP) is accountable for producing more than 85% of hazardous waste which includes high levels of toxic metals including Pb and Hg.
Our understanding for symbiotic interactions has improved well with recent developments in field of multiomics (e.g., transcriptome, proteome and metabolome) and instrumentations (e.g., Real Time PCR and mass spectrometry). There are only few reports on impact of toxic metals on symbiotic nodules [7]. However, there are no reports on proteomic and molecular response of soybean nodules to Pb and Hg. Improving our understanding about the impact of such stress on nodules would help to assess the crop losses, designing strategies for stress management and improvement of stress tolerance against toxic metals.
It has been reported that toxic metals cause reduction in nodulation and nitrogen fixation through decreased cellular water potential, reduced nutrient uptake and induction of oxidative stress [8]. At molecular level, toxic metals can alter expression of key enzymes of metabolic pathways [9]. Metal stress can cause changes in gene expression and protein profile [10]. There are no reports on impact of nodule proteome and real time expression of important genes in soybean under Pb and Hg stress. Therefore, present study was conducted to assess the impact of Pb and Hg on soybean plant with major focus on proteome and transcripts of stress responsive proteins in symbiotic nodules.
Methods
Bradyrhizobium japonicum culture
B. japonicum strain KAS-1 was obtained from Department of Microbiology, Indian Agricultural Research Institute, Pusa, New Delhi. Bacterial culture was grown at 28 °C for 7 days at 150×g on orbital shaker in yeast extract mannitol broth medium. Bradyrhizobium japonicum KAS-1 is a slow growing strain which was identified at IARI, New Delhi. Slow growing rhizobia have better nitrogen fixing efficiency as compared to fast growing rhizobia. The slow growing rhizobia were under log phase at the time of inoculation.
Plant culture and stress treatment
Soybean (Glycine max L. var. JS-335) seeds were obtained from ICAR- Indian Institute of Soybean Research, Indore, Madhya Pradesh (MP), India. Seeds were thoroughly washed with detergent and surface sterilized using 1% sodium hypochlorite for 10 min and rinsed 10 times with autoclaved double distilled water (DDW). Seeds were placed on wet Whatman filter paper in regular petri-dishes and kept overnight in dark for uniform germination. Forty five plastic pots (7″ × 7″; w × d) were divided into three sets viz. control, Lead (Pb) and Mercury (Hg), each pot filled with 1 kg Soilrite® a soil free system having peat, perlite, and vermiculite (1:1:1). Germinated healthy seedlings were transferred to pots. For nodulation, three-days-old seedlings were inoculated with Bradyrhizobium japonicum culture. This was done by dispensing 2 mL of bacterial culture (nearly 108 cells mL− 1) in a small pit made in Soilrite® in which seedlings were placed by transferring from petri-dish. In soil, plants were watered with nitrogen free nutrient media via wicking system.
For stress induction, 5 pots per set in triplicate were supplied with 30 ppm PbCl2 and another set of 5 pots in triplicate were supplied with 0.5 ppm HgCl2 dissolved in nutrient solution. One set of 5 pots in triplicates was used as Control, without metal treatment. The concentrations of Pb and Hg were selected as reported in soils of MP. The pots were kept inside a growth chamber under 28 °C temperature and 16 h light of 600 μmol photons m− 2 s− 1 and 8 h dark cycle. After sixty days (Fig. 1a-c), the plants were carefully up-rooted and the roots were washed with DDW, dried with soft lint-free tissue paper (Kimwipes, Kimberly-Clark, USA). The nodules were harvested from all the sets, immediately frozen in liquid nitrogen, and stored at − 80 °C until used for further study.
Fig. 1.

(a-c). a. View of experimental plants at the age of 60 days. b. A complete root with mature nodules. c. A close up of nodules in the root
Protein extraction from nodules
Nodule proteins were extracted using ReadyPrep™ total protein extraction kit (Bio-Rad, USA) as per instruction manual. One gram of nodules was pulverized to a fine powder using mortar-pestle in liquid nitrogen. Powder was transferred to Nalgene™ Oak Ridge high-speed centrifuge tubes (Thermo Scientific, USA). 2 mL of 2D rehydration buffer (7 M urea, 2 M thiourea, 1% w/v amidosulfobetaine-14 detergent, 40 mM tris base and 0.001% bromophenol blue) was added. The extract was centrifuged at maximum speed (15,339×g) for 20 min at 18 °C to pellet bacteria and cell debris. The supernatant was removed and transferred to a clean tube containing cold acetone and kept overnight at − 20 °C for nodule protein precipitation, then centrifuged at 6440×g and supernatant was discarded. Protein pellet was re-suspended in rehydration buffer followed by protein estimation using Bradford reagent [11]. BSA was used as standard.
2D (IEF SDS-page)
Following quantification, 200 μg of nodule protein in a total volume of 250 μL in solubilisation buffer was loaded onto 11 cm, non-linear, pH 4–7 IPG (Immobilized pH gradient) strips (Ready Strip™, Bio-Rad, USA) in a rehydration tray. The strips were covered with mineral oil and kept overnight for passive rehydration of proteins. The strips were then placed in isoelectric focusing tray (Protean II IEF™ cell, Bio-Rad, USA) with pre-moistened paper wicks covering each electrode. Protein focusing was performed at 20 °C with total 65,000 Vh current supply in a multi-step programme (Additional file 1: Table S1). After IEF completion, IPG strips having immobilized proteins were equilibrated with reduction buffer (50 mM Tris-HCl, pH 8.8, 8 M urea, 20% (v/v) glycerine, 2% (w/v) SDS and 130 mM DTT) and with alkylation buffer (50 mM Tris-HCl, pH 8.8, 8 M urea, 20% (v/v) glycerine, 2% (w/v) SDS and 135 mM iodoacetamide) for 20 min each, then rinsed out with ultrapure water. The strips were placed on 12% polyacrylamide gels in a Protean II XL Cell™ (Bio-Rad, USA) for second dimensional run with Precision Plus Protein™ Standards (Bio-Rad, USA) loaded on one side of gels for accurate protein molecular mass determination. Two mL of 0.5% (w/v) agarose added with some crystals of bromophenol blue was placed on top of glass plates to serve as a sealing agent and tracking dye, respectively. Electrophoretic run (2nd dimensional) was performed with a current flow of 40 mA per gel at 10 °C. After completion, the gels were stained and de-stained by the method of Candiano at al. [12] for protein visualization. The gels were digitized using gel documentation system (Gel Doc™, Bio-Rad, USA). Protein spot densities were analysed using PDQuest™ software (Bio-Rad, USA). The 2-D gel images of control and stressed plants were thoroughly compared for upregulated and downregulated proteins.
In-gel tryptic digestion of differentially expressed proteins
The selected protein spots were trypsin-digested according to Bagheri et al. [13]. Spots were picked in 0.2 mL PCR tubes and 100 μL ultrapure water was added, then centrifuged at 101×g, followed by a double wash. To de-stain the gel pieces, 100 μL of 50% (v/v) acetonitrile (ACN) was added and tubes were centrifuged at 101×g for 20 min at 22–24 °C. The process was repeated with 100 μL of absolute ACN. 5 μL of 1 M DTT and 45 μL of 100 mM ammonium bicarbonate were added to the tubes and incubated at 56 °C for 45 min. The supernatant was discarded and 100 μL of 0.1 M Iodoacetamide (IAA) in 50 mM ammonium bicarbonate was added to the tubes, then centrifuged at 101×g for 30 min at 22–24 °C. IAA was removed, then 100 μL of 50% (v/v) ACN was added to the tubes and centrifuged at 101×g for 20 min at 22–24 °C. ACN was discarded and the step was repeated with absolute ACN. The pellet was dried and incubated for 45 min with 30 μL trypsin (Promega, USA) which was suspended at 1 μg/μL in 50 mM acetic acid and diluted in 50 mM NH4HCO3 at 20 μg/mL. Trypsin was discarded and 50 μL of 50 mM NH4HCO3 was added to the tubes and kept overnight (12–16 h) at 37 °C. 5 μL of 1% (v/v) TFA was added and centrifuged at 1100×g for 20 min at 22–24 °C. Supernatant was taken in another tube and to the gel slices 100 μL of 50% (v/v) ACN and 5% (v/v) TFA was added and incubated for 30–60 min at room temperature. Supernatant was added to the first extract. The step was repeated and all the extracts were pooled and freeze dried in a lyophilizer (VirTis, SP Scientific, USA) and stored at − 80 °C until used for further study.
MALDI-TOF MS/MS analysis for peptide identification
Trypsin digested peptides were dissolved in 0.1% (v/v) TFA and mixed with equal volume of matrix (50% v/v acetonitrile,0.1% v/v TFA and 20 gL− 1 α-cyano-4-hydroxycinnamic acid in ultrapure water). The peptide-matrix solution was loaded on to a MALDI plate and allowed to dry. MS/MS analysis was performed on a MALDI-TOF/TOF MS analyzer (AB-SCIEX, TOF/TOF 5800, Applied Biosystems, USA). The peptide mass spectra were detected through result-dependent analysis on Protein Pilot™ v.3.2 software (AB Sciex, MA, USA) with MS (precursor-ion) peak filtering 800–4000 m/z interval, monoisotopic, mass tolerance 50 ppm. MS/MS (fragmentation) peak filtering monoisotopic, MH+, minimum signal-to-noise ratio (S/N) 10, MS/MS fragment tolerance 0.75 Da. Protein identification was done using MASCOT database search engine (Matrix Science, London, UK) with MS/MS ion search performed against non-redundant NCBI protein database. Parameters were set as taxonomy-viridiplantae, enzyme-trypsin with one missed cleavage, fixed modification to carbamidomethyl C, peptide and MS/MS tolerance set to 1 Da and 0.8 Da respectively with 2+ peptide charge. Significant hits with MASCOT probability-based score (p < 0.05) and best matched molecular weight, pI value and percent sequence coverage were considered to evaluate protein identification.
qRT-PCR based gene expression analysis
Six proteins from among those involved in stress response were selected for further confirmation at transcriptional level through qRT-PCR expression analysis. The genes which were selected on the basis of proteomic results of soybean nodules are catalase (CAT), allene oxide synthase (AOS), gluitathione S-Transferase (GST), calcineurin B like (CBL), calmodulin like (CML) and rapid alkalinisation factor (RAF). Actin (ACTIN) was used as the standard. The gene sequences were retrieved from NCBI nucleotide database. For every gene, the corresponding forward and reverse primers were designed (Additional file 2: Table S2) using primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Total nodule RNA from control and stressed plants was isolated using RNeasy plant mini kit (Qiagen, USA). RNA was quantified using BioPhotometer plus (Eppendorf, Germany). One μg of total RNA was reverse transcribed using M-MuLV RT-PCR kit (Merck, Germany). qRT-PCR was done with three replicates on Rotor Gene™ 6000 real time rotary analyzer (Corbett Research, Australia) using SYBR green master mix, gene specific primers and cDNA in a final volume of 20 μL with actin as internal control. For data normalisation CT values of genes were subtracted from CT value of reference gene (ΔCT) and relative quantification (2-ΔΔCT) was calculated by subtracting ΔCT of interest from ΔCT of reference gene. Information about assay and run conditions has been presented in (Additional file 3: Table S3, Additional file 4: Table S4).
Nodule oxidative damage and antioxidant enzyme assays
Thiobarbituric acid reactive substances (TBARS) was estimated to measure magnitude of oxidative stress by the method of Heath and Packer [14]. 0.5 g fresh nodules were grounded to a fine powder using liquid nitrogen in a mortar and pestle. The powder was homogenized in 1% (w/w) TCA (10 mL g− 1 FW) and centrifuged for 5 min at 10062×g. To the fresh tube 1.0 mL supernatant and 4.0 mL of 0.5% (w/v) TBA were added. The homogenate was incubated at 95 °C for 30 min, then immediately cooled on ice bath and centrifuged at 4200×g for 5 min to form a clear solution. The absorbance of supernatant was read at 532 nm and the value was subtracted from absorbance value at 600 nm for correction of unspecific turbidity.
For measurement of antioxidant enzyme activities fresh nodules from each treatment were harvested, washed with distilled water, and blot dried. Enzyme extract was prepared by homogenizing 0.5 g nodules in a pre-chilled mortar and pestle with 100 mM potassium phosphate buffer (pH 7.5) containing 3 mM DL-dithiothreitol, 1 mM EDTA, 5% (w/v) insoluble polyvinyl pyrrolidone and 1 Mm ascorbic acid. The solution was centrifuged at 11,269×g for 30 min and supernatant was kept at -20 °C until enzyme assays were performed. Ascorbate peroxidase (APX) assay was performed by measuring the absorbance of supernatant at 290 nm according to Qureshi et al. [15]. A unit of enzyme is defined as the amount of APX required to oxidize 1 μmol of ascorbate per min at 25 °C (2.8 mmol− 1 cm− 1extinction coefficient of ascorbate at 290 nm). The activity of catalase (CAT) was measured by the decrease in absorbance of supernatant at 240 nm due to degradation of H2O2 as reported by Aebi et al. [16]. A unit of enzyme is defined as the amount of catalase required to decompose 1 μmol of H2O2 per min with molar extinction coefficient of 0.04 mmol− 1 cm− 1 at 240 nm absorbance. Glutathione reductase (GR) activity was performed by the method of Anderson [17].The decrease in absorbance of reaction mixture at 340 nm was recorded to access the enzyme activity. A unit of enzyme is defined as the amount of GR that catalyzes the reduction of 1 μmol of GSSG min− 1 mg− 1 protein.
Histochemical assays for detection of hydrogen peroxide and superoxide in leaf
The accumulation of hydrogen peroxide (H2O2) and superoxide (O2˙ˉ) anion in leaves was histochemically assessed by 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining of soybean leaves following the procedure described by Scarpeci et al. [18].
Visualization of hydrogen peroxide
For detecting H2O2, the leaves were immersed overnight in a solution of DAB (1 mg mL− 1, pH 3.8). Thereafter, chlorophyll was removed by boiling in ethanol for 10 min and the reddish-brown coloration denoting the H2O2 content was visualized and photographed.
Visualization of superoxides
For detection of O2˙ˉ, the leaves were floated in 50 mM sodium phosphate (pH 7.5) containing 0.2% NBT and the dark blue insoluble formazan compound formed by reaction of NBT with O2˙ˉ which was visualized in leaf and photographed.
Nodule growth related parameters
Immediately after harvesting plants were washed with DDW and blot-dried. Plants were dissected into nodules, roots, stems and leaves. Nodules were counted and expressed in per plant basis. Each organ was carefully weighed, and fresh weight (FW) was recorded as FW per plant in grams. Material was placed in hot-air oven at 65 °C for a week. Dry weight was recorded as DW per plant in grams.
Study of nodule transverse section by light microscopy
Fresh nodules from control and stressed plants were cut into 1–3 mm2 pieces and kept in solution for fixation containing 2.5% (w/v) glutaraldehyde, 2% (v/v) paraformaldehyde and 1% (v/v) formaldehyde. The fixing solution containing nodule pieces was vacuum infiltrated for 10 min and stored at 4 °C for overnight. The sections were washed with 0.1 M phosphate buffer (pH 7.4) and kept for 2 h in osmium tetraoxide solution at 4 °C followed by washing with 0.1 M phosphate buffer (pH 7.4). The samples were added with 30–90% (v/v) acetone for dehydration and placed in dry acetone (saturated with copper sulphate) at 4 °C for 1 h. Samples were then treated twice with toluene for 1 h each and kept overnight in resin and toluene (1:3) under vacuum, then impregnated in resin and toluene (2:2 and 3:1) overnight in vacuum. The samples were finally impregnated in pure resin at room temperature for 6 h. Samples were cut into 500 nm sections and stained in 1% methylene blue for 40s. The sections were observed with bright field microscope (CX31, Olympus, UK).
GO term enrichment and statistical analysis
Gene ontology enrichment analysis for protein identification was carried out using agriGO database (http://bioinfo.cau.edu.cn/agriGO/) [19]. Protein spot intensities were log2 transformed for hierarchical clustering and partial least squares discriminant analysis (PLS-DA) using Metaboanalyst 3.0 software [20]. All data are presented as the mean ± standard error (SE). The measurements were made using five biological replicates. The statistical analyses were carried out using one-way analysis of variance (ANOVA) and means were compared using Tukey’s test. NS = Non-significant, *P ≤ 0.05 and **P ≤ 0.01.
Results
Pb and hg induced oxidative stress in leaves
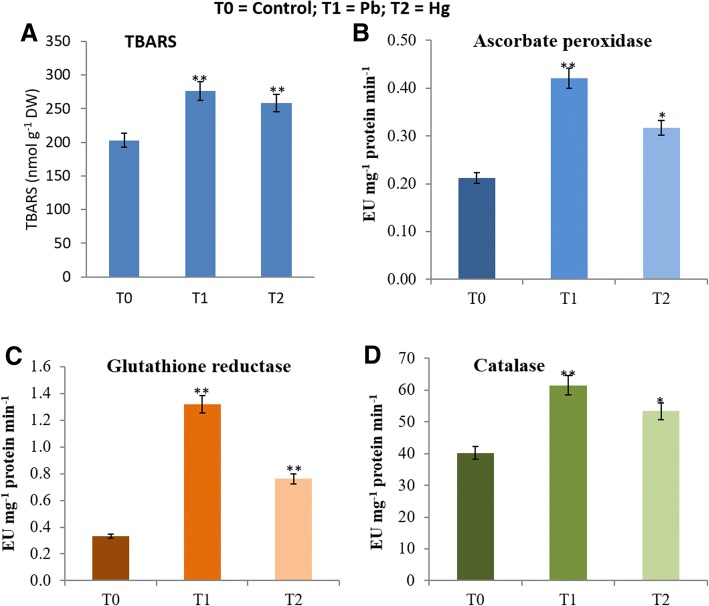
Both Pb and Hg induced oxidative stress in leaves. Control, Pb and Hg treated plants accumulated 72, 342 (~ 4 fold) and 202 (~ 2 fold) nmol TBARS g− 1 DW, respectively (Fig. 2).
Fig. 2.
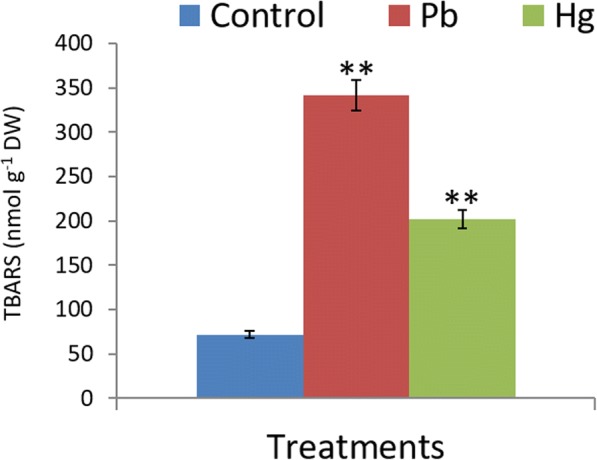
Impact of Pb and Hg on magnitude of oxidative stress in Glycine maxl. Leaves. Oxidative stress marker was estimated in terms of TBARS content. All data are presented as the mean ± standard error (SE). The measurements were made using five biological replicates. The statistical analyses were carried out using one-way analysis of variance (ANOVA) and means were compare using Tukey’s test. NS = Non-significant, *P ≤ 0.05 and **P ≤ 0.01
Histochemical assays demonstrated oxidative stress in leaves
Histochemical detection of hydrogen peroxide
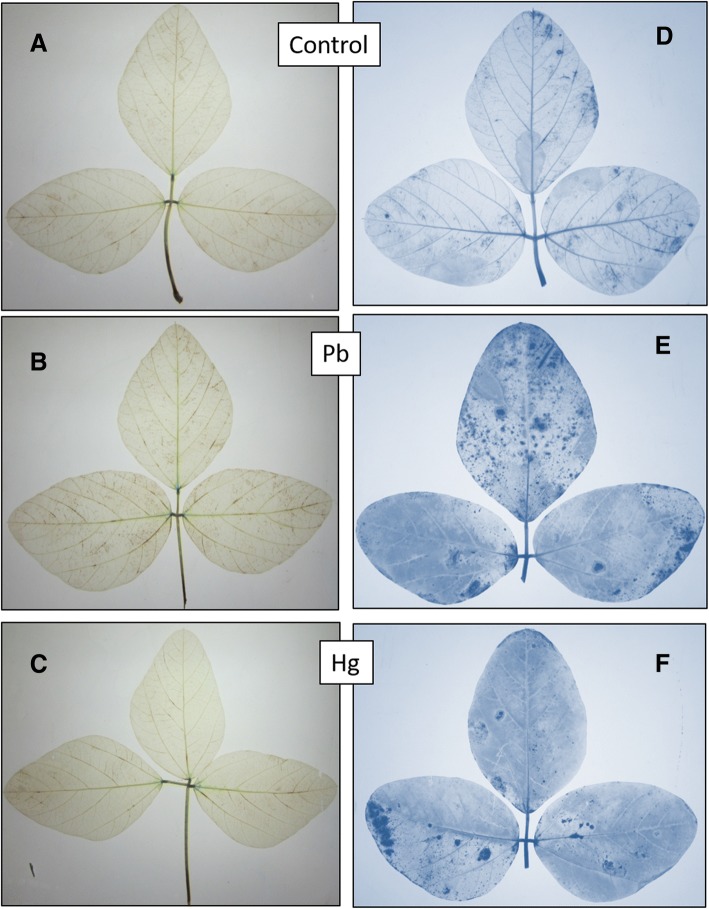
Results show that Pb increased the concentration of hydrogen peroxide in the leaves. However, Hg did not make much difference compared to control (Fig. 3a-c).
Fig. 3.
(a-f). Glycine max L. leaves showing localization of H2O2under control (a), Pb (b) and Hg (c) treatments and superoxidesunder control (d), Pb (e) and Hg (f) treatments
Histochemical detection of superoxides
Images show that both Pb and Hg elevated the levels of superoxides as compared to control (Fig. 3d-f).
Impact of Pb and hg on growth parameters and nodule anatomy
Soybean growth and yield attributes were significantly affected by Pb and Hg treatment after 60 days of treatment (DAS). Pb and Hg resulted into a decline of nodule number by 31 and 60%, nodule fresh weight by 17 and 47%, nodule dry weight by 6 and 30%, respectively. Treatment of Pb and Hg resulted into a decline of fresh weight and dry weight of root, stem and leaf. Pb and Hg treatment caused decline of root fresh weight by 19 and 43%, root dry weight by 25 and 47%, stem fresh weight by 20 and 34%, stem dry weight by 22 and 39%, leaf fresh weight by 24 and 38%, leaf dry weight by 24 and 42% respectively (Table 1).
Table 1.
Effect of lead (Pb) and mercury (Hg) on nodule number, fresh weight (FW) and dry weight (DW) of nodule, root, stem and leaf in Soybean (Glycine max L.) at 60 DAS
| Attributes per plant | Treatment | ||
|---|---|---|---|
| Control | Lead (Pb) | Mercury (Hg) | |
| Nodules | |||
| Number | 138 ± 13.3 (00) | 95 ± 7.67 (−31.16%)* | 58 ± 4.67 (−57.97%)** |
| Fresh weight (g) | 1.327 ± 0.06 (00) | 1.101 ± 0.05 (−17.03%) NS | 0.704 ± 0.04 (−46.95%)** |
| Dry weight (g) | 0.405 ± 0.03 (00) | 0.380 ± 0.01 (−6.17%) NS | 0.250 ± 0.01 (−38.27%)** |
| Root | |||
| Fresh weight (g) | 5.97 ± 0.21 (00) | 4.85 ± 0.30 (−18.8%) NS | 3.37 ± 0.18 (−43.55%)** |
| Dry weight (g) | 0.588 ± 0.03 (00) | 0.44 ± 0.03 (−25.17%)* | 0.31 ± 0.02 (−47.27%)** |
| Stem | |||
| Fresh weight (g) | 7.13 ± 0.42 (00) | 5.72 ± 0.38 (−19.77%) NS | 4.73 ± 0.25 (−33.66%)*** |
| Dry weight (g) | 1.29 ± 0.06 (00) | 1.00 ± 0.08 (−22.48%)* | 0.78 ± 0.06 (−39.53%)** |
| Leaf | |||
| Fresh weight (g) | 9.77 ± 0.85 (00) | 7.37 ± 0.65 (−24.56%)* | 6.02 ± 0.42 (−38.38%)** |
| Dry weight (g) | 1.56 ± 0.08 (00) | 1.20 ± 0.04 (−23.8%)* | 0.91 ± 0.05 (− 41.67%)** |
All data are presented as the mean ± standard error (SE). The measurements were made using five biological replicates. The statistical analyses were carried out using one-way analysis of variance (ANOVA) and means were compare using Tukey’s test. NS = Non-significant, *P ≤ 0.05 and **P ≤ 0.01
Nodule transverse section (TS) of control plants showed all the components of root vasculature besides well-formed bacteroides (Fig. 4a). However, root fibres were lesser, and population of bacterial cell was dense in nodules of Pb treated plants with non-visibility of regular vasculature (Fig. 4b). Nodule TS of Hg treated plants showed overall damage to both vasculature and bacteroides (Fig. 4c).
Fig. 4.
a-c. Impact of Pb (30 ppm), and Hg (0.5 ppm) on morphology and anatomy of Soybean roots and nodules. Nodule components are OC = outer cortex, S = sclerid layer, IC = inner cortex, V = vascular bundle, B = bacteroids
Pb- and hg-induced oxidative stress in root nodules
Nodules of Control, Pb and Hg treated plants accumulated 203, 276 (+36%) and 258 (+27%) nmol TBARS g− 1 DW, respectively (Fig. 5a).
Fig. 5.
a-d. Impact of Pb and Hg on magnitude of oxidative stress (a) and changes in activities of ascorbate peroxidase (APX, b), catalase (CAT, c) and glutathione reductase (GR, d). Enzyme actities have been expressed in enzyme units (EU) mg− 1 protein min− 1 in soybean root nodules exposed to Pb (30 ppm) and Hg (0.5 ppm). All data are presented as the mean ± standard error (SE). The measurements were made using five biological replicates. The statistical analyses were carried out using one-way analysis of variance (ANOVA) and means were compare using Tukey’s test. NS = Non-significant, *P ≤ 0.05 and **P ≤ 0.01
Pb and hg induced changes in antioxidant enzymes
Activity of enzymes of ascorbate-glutathione antioxidant system viz. ascorbate peroxidise (APX) and glutathione reductase (GR) got modulated in response to Pb as well as Hg (Fig.5b-d). Control, Pb- treated and Hg-treated plants showed 0.212 EU, 0.421 EU (+98%) and 0.317 EU (+49%) respectively, of APX activity (Fig. 5b). GR activity in control, Pb-treated and Hg-treated plants was 0.333 EU, 1.32 EU (+296%) and 0.762 EU (+128%), respectively (Fig. 5c).
Activities of another important antioxidant enzyme, catalase (CAT), were found to be around 40 EU (Control), 61 EU (Pb-treated) (+53%) and 53 EU (Hg-treated) (+33%) (Fig. 5d).
Impact of Pb and hg stress on relative abundance of nodule proteins
Untargeted comparative proteomics for differential abundance of proteins, to explore response mechanism of nodule to Pb and Hg, was taken into account.
Differential relative expression of proteins on 2D gels
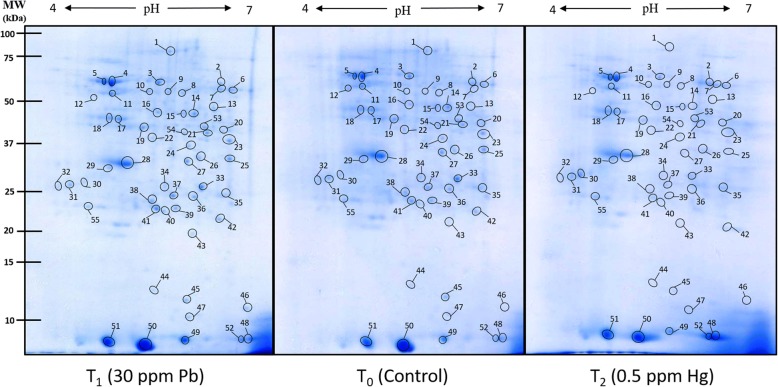
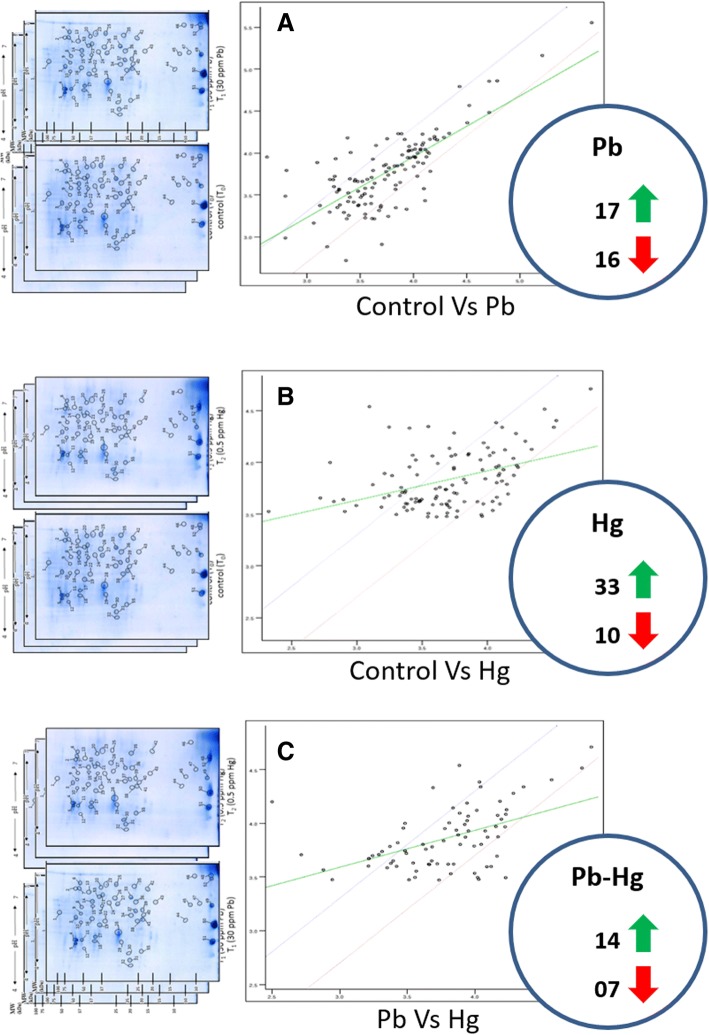
Expression of altered proteome was successfully visualised on 2D gels (Fig. 6) and digitized for comparative analysis using PDQuest™ (Bio-Rad, USA). Around 245 spots were reproducibly detected on each gel. Comparative image analysis showed that overall 76 spots were two-fold differentially expressed against control, out of which 33 were expressed in Pb treatment (17 upregulated and 16 downregulated) and 43 spots in Hg treatment (33 upregulated and 10 downregulated) (Fig. 7).
Fig. 6.
Representative 2D proteome maps of soybean root nodules of control, Pb-treated (30 ppm) and Hg-treated (0.5 ppm). 400 μg of total protein was loaded on to 11 cm/non-linear/pH 4–7 IPG strips for first dimensional run on IEF cell followed by second dimensional run on 12% SDS-PAGE. The proteins were visualised by staining of gels with blue silver stain. The encircled protein spots with numbering represents differentially expressed proteins which were considered for tryptic digestion and subsequent identification through MALDI-TOF MS/MS
Fig. 7.
(a-c). Image analyses of 2D proteome maps for computing the changes in expression of protein profile as affected by Pb and Hg in root nodules of Glycine max
Hierarchical clustering and partial least squares discriminant analysis
Hierarchical clustering was performed for analyzing correlation between protein profiles and changes between Control, Pb and Hg treatments. Protein spot intensities were normalized by log transformation for analysis by MetaboAnalyst software (Fig. 8a-c). Hierarchical clustering showed variation between differentially expressed proteins (Fig. 8a). Variable importance in projection (VIP) plot was created from PLS-DA loading plots (Fig. 8b) for identification of differentially expressed proteins with maximum abundance (Fig. 8c). Fifteen protein spots were identified with VIP score (1.5–3) which include ring finger protein (spot 55), mitochondrial transcription termination factor protein (spot 16), probable L-gulonolactone oxidase 6 (spot 7), aluminium activated malate transporter (spot 5), probable calcium binding protein CML43 (spot 45), V-ATPase related protein (spot 51), hydroperoxide lyase (spot 9), glutaredoxin C-5 like protein (spot 52), armadillo/beta-catenin repeat family protein (spot 11), catalase 3 (spot 4), MADS box transcription factor (spot 31), calcineurin B-like protein (spot 32), allene oxide synthase (spot 13), SCF ubiquitin ligase (spot 30), and bet1-like SNARE protein (spot 48).
Fig. 8.
(a-c). Multivariate data analysis of differentially expressed proteins in root nodules of Soybean performed by MetaboAnalyst software. Hierarchical clustering (a), Loadings plot (b) and VIP plot (c) displays differentially expressed proteins with maximum abundance in Control (1),Pb (2) and Hg (3) treatments
Proteins with significant changes in relative abundance and belonging to a variety of stress response (Additional file 5: Figure S1) were selected to study the quantitative real time expression by targeting at transcriptional level.
Identification and functional categorization of nodule proteins
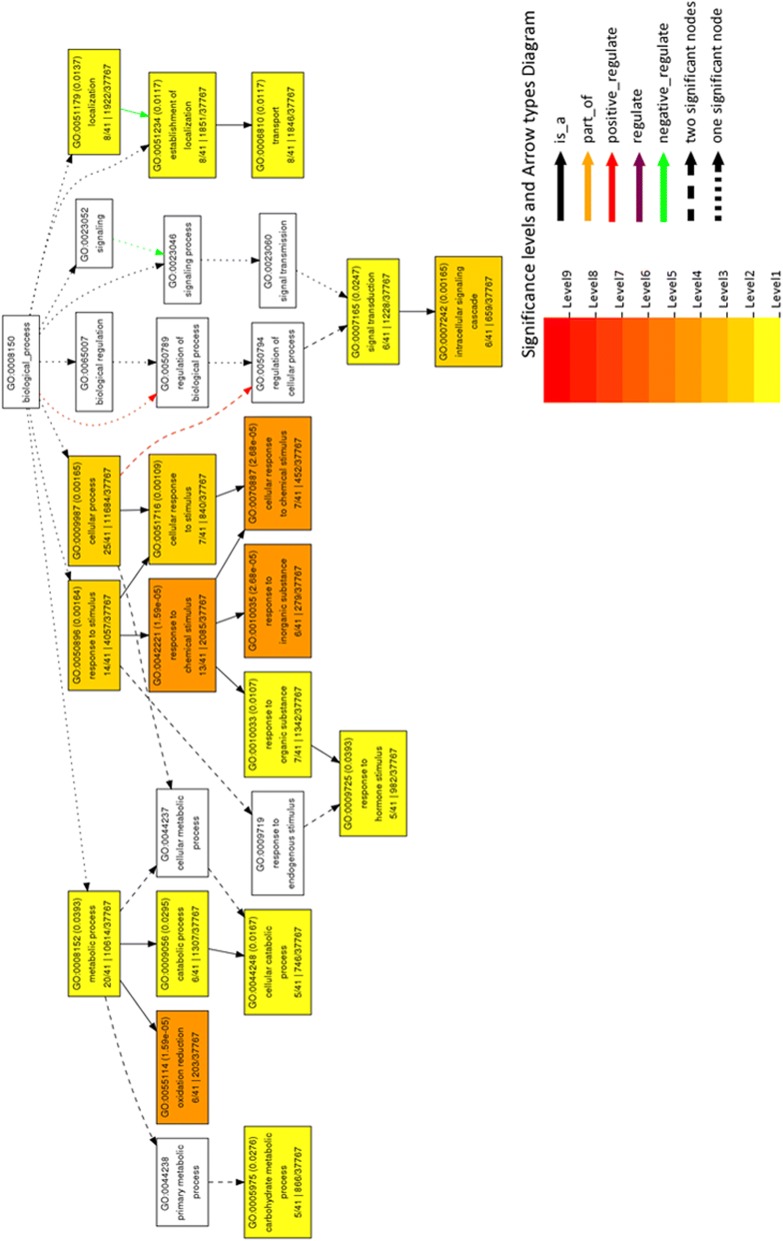
Fifty-five differentially expressed protein spots were selected for tryptic digestion and identification using MALDI TOF MS/MS followed by MS/MS ion search on MASCOT database (http://www.matrixscience.com/). The identified proteins were searched against UniProt database (http://www.uniprot.org/) for their biological function and cellular component. Homologous soybean gene ID’s were retrieved from SoyKB database (http://soykb.org/). Gene ontology enrichment analysis with agriGO database shows functional distribution of proteins (Fig. 9 and Fig. 10a-b). Biological process shows an overall increase in metabolic process, hormone signalling and signal transduction (Fig. 9) Cellular component shows decrease in membrane and organelle activity (Fig. 10a) while molecular function has increased oxidoreductase activity and protein binding with decreased calcium and metal ion binding (Fig. 10b).
Fig. 9.
GO term enrichment analysis of differentially expressed proteins using AgriGo database corresponding to biological function
Fig. 10.
(a-b). GO term enrichment analysis of differentially expressed proteins using AgriGo database corresponding to (a). Cellular components and (b). Molecular function
Additional file 6: Table S5 describes various parameters of identified proteins including theoretical/practical molecular weight and pI values, number of peptides matched, per cent sequence coverage, NCBI accession, SoyKB gene ID’s, biological function, and cellular location.
The identified proteins were found associated with diverse biological functions (Fig. 11a) with maximum contribution to protein metabolism (22%), oxidative stress response (18%) and nucleotide metabolism (13%) which might be due to Pb and Hg toxicity leading to activation of defence, cell division and repair process. Proteins with differential expression involved in various other vital cellular functions were also identified such as carbohydrate metabolism (9%), signalling (7%), hormone metabolism (7%), transport (5%), development (4%), structural proteins (4%), co-factor and vitamin metabolism (4%), lipid metabolism (3%), sulfur assimilation (2%) and polyamine metabolism (2%).
Categorization of differentially expressed proteins at sub-cellular level
The proteins were further categorized based on their cellular location (Fig. 11b) with maximum proteins located in cytosol (25%), membrane (22%), mitochondrion (15%) and nucleus (13%). Proteins located in other cell locations are less abundant such as extracellular proteins (7%), endoplasmic reticulum (5%), Golgi (5%), vacuole (4%) and peroxisomes (4%).
Fig. 11.

a-b. Functional categorization (a) and subcellular localization (b) of differentially expressed proteins in soybean root nodules exposed to Pb and Hg
Comparative study of gene transcripts with protein expression
Real time quantitative estimation of gene transcripts was achieved and compared to protein expression level (Fig. 12 a-b). A relation between relative abundance of protein (Fig. 12a) and related transcripts (Fig. 12b) reveals a comparative account of protein and related transcript population. Results showed that expression of CAT transcripts was elevated by both Pb and Hg, more under Hg stress. Both Pb and Hg could increase the expression of AOS up to a same level. Levels of GST transcripts were high under Hg stress, compared to a non-significant increase under Pb stress. The abundance of CBL transcripts was also very high under Hg stress whereas some decrease was noted under Pb stress. Abundance of CML was decreased by both stressors. Abundance of RAF transcripts was much higher under Pb stress compared to lower elevation under Hg stress.
Fig. 12.
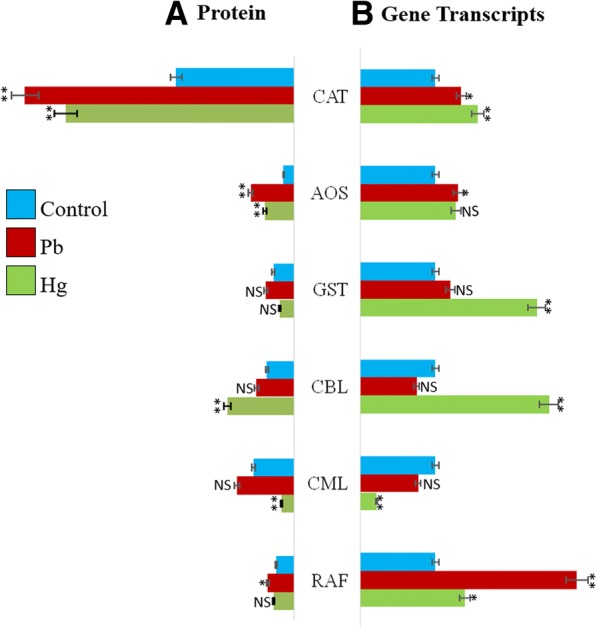
a-b. A comparative account of Pb- and Hg-induced changes in relative abundance of some important proteins and related transcripts. The data were normalised against control plants. CAT, catalase; AOS, Allene oxide synthase; GST, Gluitathione S-Transferase; CBL, Calcineurin B like; CML, Calmodulin like; RAF, Rapid alkalinisation factor. All data are presented as the mean ± standard error (SE). The measurements were made using three replicates (n = 3). The statistical analyses were carried out using one-way analysis of variance (ANOVA) and means were compare using Tukey’s test. NS = Non-significant, *P ≤ 0.05 and **P ≤ 0.01
Discussion
Abiotic stresses are denoted by an array of different stress-causing mechanism. Such mechanisms vary from plasmolysis to oxidative stress to interruption in electron flow to biomolecules, cellular degradation/dysfunction, and much more. In turn, plant responds by modulating its transcriptome, proteome and metabolome for defence and repair process, and amelioration of growth and development [21].
Soybean belongs to the legume family which is characterized by N2-fixing root nodules that harbour symbiotic bacteria and fixes atmospheric nitrogen to available nitrogenous forms [22]. Thus, their nodules, besides roots, are the primary sites at the risk of metal toxicity. In fact, rate of germination and efficiency of bacteria for nodulation might also be hugely compromised. Therefore, the present study focuses on understanding Pb and Hg induced changes in proteome, selective transcripts and antioxidant enzymes of nitrogen-fixing root nodules in soybean.
Nodule anatomy was modulated by Pb and hg
Toxic metal induced oxidative stress causes damage to cellular structures and alteration in functions which might be responsible for damage to nodule structures. Both Pb and Hg had differential damaging pattern to the nodule anatomy. It could be due to the differential mechanism of Pb and Hg toxicity; Pb affected the nodule anatomy more whereas Hg affected the bacteroides.
Both Pb and hg induces oxidative stress in the nodules and free radicals in the leaves
Induction of oxidative stress by toxic metals is a common phenomenon in plant cells [23]. The marker of oxidative stress in both nodules and leaves confirms that magnitude of oxidative damage was higher by Pb than Hg. Perhaps, Pb involves in production of reactive oxygen species more efficiently than Hg.
Pb induces higher production of hydrogen peroxide and superoxide radicals in leaves
Both Pb and Hg are known to induce oxidative stress [15] in leaves of crop plants. Histochemical localization reveals that Pb produces H2O2 more efficiently. Hg was not found to elevate the levels of H2O2. Superoxides were elevated by both Pb and Hg.
Pb and hg induced changes in protein metabolism
Toxic metals such as Pb and Hg are non-essential and among common contaminants found in polluted soils and water near industrial sites [24]. Plants respond to toxic effects of metals by altered expression of stress associated genes and proteins leading to activation of signalling networks for stress regulation [25]. Protein metabolism related proteins with altered expression due to Pb and Hg stress were identified in our study. These proteins were found to be involved in varied biological processes such as protein glycosylation, folding, ubiquitination, defence response, metal ion binding, transport, and protein synthesis (Fig. 13).
Fig. 13.
An overview of different pathways with Pb- and Hg-induced changes in abundance of identified proteins
Ubiquitin mediated protein degradation is an important cellular defence response process against environmental stresses. In our study Armadillo/beta-catenin repeat family protein and SCF ubiquitin ligase, SKP1 component were upregulated in both Pb and Hg treatment. Armadillo/beta-catenin repeat family protein belong to E3 ubiquitin ligases which functions as signalling molecule through protein-protein interaction [26]. Abscisic acid (ABA) signalling functions in various environmental perturbances. RING finger protein 141 is positively correlated with ABA signalling and its abundance was down regulated in Pb treatment while upregulated in Hg treatment. Sel1 repeat protein was down regulated in both Pb and Hg treatment. This protein is involved in ER-associated protein degradation for efficient removal of mis-folded proteins. It has been reported that stressed ER aggravate defect in ER-associated protein degradation [27].
Plants respond to environmental changes by acclimatization at various levels including cell turgor, volume, membrane surface modifications and cell expansion which are restricted within cell wall [28]. Membrane trafficking is an integral part of cell growth and development which involves specialized proteins for vesicle traffic and membrane transport. Abundance of ER-Golgi transport protein bet 1-like SNARE was found to be upregulated under Pb and Hg stress while alpha-soluble NSF attachment protein was downregulated in both stresses. FKBPs (FK506-binding proteins) are functionally diverse proteins that belong to Peptidyl-prolyl cis-trans isomerases (PPIases) involved in protein folding and a subset of these proteins are stress responsive [29]. PPIase FKBP42 was found to be upregulated under Pb stress and downregulated under Hg stress.
Proteins involved in oxidative stress and redox homeostasis
Metal toxicity lead to excessive production of reactive oxygen species (ROS) which cause damage to DNA, proteins, carbohydrates, and lipids resulting in oxidative stress. In plants accumulation of ROS such as hydrogen peroxide and superoxide radicals are indicators of oxidative stress. Response to oxidative stress include changes in gene expression related to growth, development, defence, signalling, and programme cell death (PCD) [30]. In our study, we have identified 18% proteins related to oxidative stress and redox homeostasis. Catalase (CAT), glutathione S-transferase (GST) and peroxidase are involved in detoxification of metal toxicity caused due to oxidative stress [31]. Balestrasse et al. (2008) [32] reported overexpression of CAT gene in nodules of soybean plants exposed to cadmium. In our study, CAT3 was upregulated in both stresses while GST and peroxidase were upregulated in Pb and downregulated in Hg treatment suggesting a pronounced effect of Hg over Pb. Changes in gene expression due to oxidative stress are regulated by network of regulatory transcription factors. AP2 class of transcription factors are known to involve in abiotic stress response [33]. This transcription factor was upregulated in both Pb and Hg treatment suggesting alteration in gene expression due to metal stress.
Small peptides act as signalling molecules in plant growth, development and defence response. One such peptide, rapid alkalinization factor (RAF) which induces extracellular pH elevation was overexpressed in both stresses. Alkalinization of extracellular media in plant helps in functioning of various bioactive peptides which are involved in defence response. RALF was found to arrest root growth in tomato and Arabidopsis seedlings and regulates nodulation in Medicago truncatula [34, 35]. Oxidative stress causes membrane damage due to lipid peroxidation. Overexpression of Cytochrome b5 in Pb stressed nodules indicate possible membrane repair process as this protein is involved in fatty acid synthesis of ER membrane [36]; however this protein is downregulated in Hg stress.
Dehydroascorbate reductase (DHAR) maintains reduced ascorbate levels in the cells which act as ROS scavenger and detoxify H2O2 via ascorbate glutathione cycle. Both Pb and Hg stress reduces expression of DHAR. Loscos et al. [37] reported post-translational inhibition of DHAR in nodules of bean plants exposed to stress signalling compound jasmonic acid. Inhibition of DHAR causes apoplast oxidation which is perceived as a stress signal by nodules.
Nucleotide metabolism related proteins
Nucleotide metabolism is a fundamental process for plant growth and development. Metal stress can affect nucleotide metabolism. ROS mediated damage to different cellular molecules can trigger defence and repair processes involving antioxidant system, growth and cell division which require nucleotide synthesis.
Pb and Hg influenced the abundance of various proteins involved in nucleotide metabolism. Auxin responsive protein IAA14 involved in auxin signalling pathway and which controls lateral root morphogenesis was down regulated by both Pb and Hg stress. Lateral root formation can be suppressed by inhibition of auxin transport [38]. MADS-box transcription factors play important role in plant development. Around 50 MADS-box genes are known to express in Arabidopsis roots with most of them having unknown function. ANR1 is a member of MADS-box family which control lateral root development and respond to nitrogen availability [39]. Contrary to IAA14 expression under Pb and Hg stress, MADS-box transcription factor was upregulated under both the stressors implying positive regulation towards stress response. Arabidopsis mitochondrial transcription termination factor family protein (mTERF) encoded by shot1 (suppressor of hot 1–4) which is a suppressor of heat shock protein HSP101 (hot 1–4) provides thermo-tolerance and protection against oxidative damage through altered expression of redox related genes [40].
In the present study, mTERF was up regulated under both Pb and Hg stress indicating possible role in protection against oxidative stress. Nudix hydrolases are enzymes that catalyze hydrolysis of various nucleoside diphosphate derivatives. These enzymes protect against mutagenesis owing to excessive ROS production [41]; [42]. Mitochondrial nudix hydrolase 18 was upregulated by Pb and downregulated by Hg stress. Zinc-finger proteins have diverse biological functions such as binding to DNA, RNA, lipids, and proteins as well as providing acclimation to various biotic and abiotc stresses. In Arabidopsis 14 genes encoding stress associated proteins (SAP) having A20/AN1 zinc-finger domains were identified and were grouped based on their domain structure [43].
In this study, we have identified zinc-finger A20 and AN1 domain containing stress associated protein 1 and ADP-ribosylation factor GTPase-activating protein having C4 type zinc finger domain. These proteins were up regulated by Pb and down regulated by Hg stress.
Carbohydrate metabolism related proteins
Carbohydrates serve as structural components, energy source for plant cells and maintain enzyme activities during stress. In our study, we have identified five proteins related to carbohydrate metabolism with altered expression due to Pb and Hg stress. Enzymes of glycolytic pathway pyruvate dehydrogenase E1 [44] component subunit beta and ATP-dependent 6-phosphofructokinase 3 isoform X2 were up regulated by Pb and down regulated by Hg stress while plastid enolase [45] was downregulated in both stresses. Endoglucanase 19 catalyzes cell wall organization through carbohydrate binding and cellulase activity. Arabidopsis beta -glucosidase 15 (BGLU15) degrades flavonol 3-O-beta-glucoside-7-O-alpha-rhamnosides which is a major flavonol that accumulate during abiotic stresses mainly during nitrogen deficiency and low temperature [46]. The expression of both endoglucanase 19 and beta-glucosidase 15 was up regulated by Pb and down regulated by Hg stress.
Proteins related to hormone and lipid metabolism
Hormones play a crucial role in abiotic stress tolerance in plants through growth and development. Optimum hormone levels are critical for proper physiological functioning of plant cells [47]. In our study Pb and Hg induced prominent changes in expression of proteins related to hormone metabolism. Jasmonates are lipid hormones which functions as signalling molecules in many physiological processes of plants including response to abiotic stress. Allene oxide synthase (AOS) catalyzes the first committed step of jasmonic acid biosynthesis pathway by conversion of 13-hydroperoxylinolenic acid to allene oxide. Germinating soybean seeds release jasmonic acid in rhizosphere which is perceived by Bradyrhizobium japonicum for induction of nodulation (nod) genes [48]. High abundance of AOS under Pb and Hg stress indicates possible activation of jasmonic acid pathway for signalling and stress response.
Several studies demonstrated the role of auxins in stress response. The active form of IAA is converted to inactive form by conjugation with amino acids which is catalysed by IAA-amido synthetases belonging to group II GH3 protein family. GH3 genes have been shown to participate in biotic and abiotic stress resistance. The OsGH3–8, OsGH3–1 and OsGH3–13 genes belonging to group II GH3 family of rice have roles in development and stress resistance [49]. In our study, abundance of IAA-amido synthetase was increased by both stresses. Transcription factors have DNA binding domains which bind at cis-elements in promoter regions of abiotic stress responsive genes and induce their expression [50].
Plant basic leucine zipper (bZIP) transcription factors are involved in different growth and developmental processes including abiotic stress signalling. In Arabidopsis ER stress associated unfolded protein response (UPR) activates bZIP28, which interacts with CCAAT box binding factors NY-Fs for upregulation of ER-stress responsive genes [51]. Transgenic Arabidopsis over expressing soybean bZIP genes GmbZIP132, GmbZIP44, GmbZIP62, GmbZIP78 negatively regulate ABA signalling and confers stress tolerance [52].
Transcription factor TGA6 is the member of bZIP family which is expressed in emerging young roots and is involved in cellular defence against biotic and abiotic stresses. TGA6 regulate gene expression in response to bioactive compounds oxylipins under stressed conditions and GST25 activity in Arabidopsis [53]. In this study bZIP transcription factors were overexpressed in Pb and underexpressed in Hg stress. Role of ethylene in plant defence and abiotic stress tolerance is evident by alteration in expression of stress associated genes in Arabidopsis, Medicago truncatula and rice exposed to various abiotic stress conditions. 1-aminocyclopropane-1-carboxylate (ACC) oxidase catalyzes conversion of ACC to ethylene. Altered expression of ACC is known to regulate abiotic stress tolerance in Arabidopsis and rice [54]. Pb and Hg stress caused downregulation of ACC oxidase in our study which might be due to decreasing level of ethylene.
Lipids are integral part of cell membranes, impart energy for metabolism, maintain growth, development, and stress response [55]. Activation of oxylipin pathway by environmental or developmental stimuli induces synthesis of bioactive compounds called oxylipins. The oxylipin pathway has branch pathways for hydroperoxide lyase (HPL) and allene oxide synthase (AOS) which belong to cytochrome P450s and are stress responsive. HPL and AOS metabolize hydroperoxide fatty acid to different oxylipins [56].
In this study, Pb and Hg treatment caused over expression of hydroperoxide lyase which suggests overproduction of bioactive oxylipins for stress tolerance. Under Pb stress, a newly induced protein sulfotransferase was identified. Activity of several bioactive compounds such as steroids, brassinosteroids, and flavonols is controlled by sulfotransferases which catalyse sulfate group transfer from 3’-Phosphoadenosine-5′-phosphosulfate (PAPS) to hydroxyl or amine group of substrates. Several functional genomics studies of sulfotransferases in Arabidopsis characterize various sulfotransferase genes which are regulated by growth, development, and stress response [57].
Signalling and transport related proteins
The elevation in cytosolic calcium (Ca2+) levels is the first signal sensing response towards most environmental stresses. Higher plants have several calcium sensor protein families. Calmodulins (CaM), calmodulin like proteins (CML) and calcineurin B-like (CBL) proteins are well known calcium sensors having multiple calcium binding domains and are involved in abiotic stress response. AtCML43 is a root tip specific calcium sensor in Arabidopsis which was overexpressed when exposed to defence hormone salicylic acid (SA) [58]. In this study, treatment of Pb increased the abundance of CML43 and CML36 whereas Hg decreased the abundance of both proteins. In contrast, calcineurin B-like protein was found to be decreased by Pb and increased by Hg treatment. Calcineurin B-like protein is known to regulate abscisic acid responses in Arabidopsis [59]. Abscisic acid receptor PYL9 provide drought tolerance and regulates lateral root formation in Arabidopsis [60, 61]. Abundance of ABA PYL9 receptor protein was upregulated by Pb and downregulated by Hg stress.
Transport across membranes is a vital process for maintaining cell homeostasis. Vacuoles maintain cell turgor pressure by osmotic entry of water through V-ATPases which are multisubunit proton pumps localized in plant cell endomembranes. Under environmental stress, maintenance of V-ATPase activity is indispensable for cell survival [62]. Over expression of V-ATPase like protein under Pb and Hg stress suggests increased active transport across membranes for maintenance of cell homeostasis under toxic metal stress.
ROS and lipid peroxidation products can stimulate mitochondrial uncoupling protein(UCP) activity for uncoupling of mitochondria which causes reduction in electron transport chain for limiting ROS formation inside mitochondria. Over-expression of AtUCP1 gene from Arabidopsis confers multiple abiotic stress tolerance in transgenic tobacco plants [63]. The abundance of mitochondrial uncoupling protein 2 was increased in Pb stress and decreased in Hg stress. Aluminium activated malate transporter (ALMT) is a ligand gated anion transporter protein which exudate malate and other inorganic anions for tolerance against Al3+ and functions in malate homeostasis and inorganic anion transport in guard cells [64]. ALMT9 protein was overexpressed in both the stresses in our study.
Miscellaneous proteins
Proteins with diverse cellular processes such as co-factor, vitamin, polyamine metabolism, sulfur assimilation and structural proteins were identified in our study. Nicotinamide/nicotinic acid mononucleotide adenylyltransferase (NaMNAT) is involved in biosynthesis of nicotinamide adenine dinucleotide (NAD) which is a co-factor that regulates cellular redox reactions and ROS signalling in plant development and stress response [65]. NaMNAT was overexpressed in Pb and underexpressed in Hg treatment. L-gulonolactone oxidase (GLOase) is the final precursor of ascorbic acid biosynthesis via myo-inositol pathway. Overexpression of GLOase gene in transgenic potato confers increased ascorbate accumulation and provides abiotic stress tolerance [66]. GLOase was overexpressed in both Pb and Hg stress suggesting enhanced accumulation of ascorbate to cope with metal induced oxidative stress.
Myosins are motor proteins involved in cell dynamics. Myosin XI-K is the member of myosin XI family which regulate root hair elongation and organelle trafficking [67]. Downregulation of myosin XI-K under both Pb and Hg stress indicates possible loss of cell motility. Annexins are well documented for their role in environmental stress tolerance. Arabidopsis annexin ATANN1 regulates calcium mediated ROS signalling in root epidermal cells under salinity stress [68]. In our study, Annexin D8 was upregulated by Pb and downregulated by Hg stress.
Polyamines in association with brassinosteroids activate ABA and IAA pathways for tolerance against metal stress. Putrescine, spermine, spermidine and thermospermine are common polyamines found in plants. Arabidopsis ACAULIS5 (ACL5) gene encodes thermospermine synthase which converts spermidine to thermospermine [69]. In this study, abundance of thermospermine synthase ACAULIS5-like protein was up regulated by Pb and down regulated by Hg stress.
Sulfite oxidase (SOX) is an important enzyme of sulfur assimilation pathway which catalyses re-oxidation of sulfite to sulfate. SOX gene in Arabidopsis was differentially expressed under various abiotc stress conditions [70]. Abundance of SOX expression was found to be upregulated by Pb and downregulated by Hg stress in root nodules of soybean. Therefore, it was quite evident that an array of proteomic response including numerous metabolic pathways were modulated under metal stress (Fig. 13).
Correlation of gene transcription with protein expression
Expression pattern of selective key proteins due to Pb and Hg stress was further validated by transcript abundance in Pb and Hg exposed nodules using qPCR analysis. These genes included those that encode CAT, AOS, GST, CBL, CML and RAF. Expression of CAT transcripts was elevated by both Pb and Hg, more under Hg stress. Both Pb and Hg could increase the expression of AOS up to the same level indicating the possible role in defence. However, levels of GST transcripts were high under Hg stress, compared to a non-significant increase under Pb stress. The data indicates towards a special feature of plant to increase the level of GST. The levels of CBL transcripts were also high under Hg stress whereas some decrease was noted under Pb stress, indicating its role during metal stress. Expression of CML was decreased under both stressors and hence seems a major protein adversely affected by metal stress. Interestingly, quantity of RAF transcripts was much higher under Pb stress compared to lower elevation under Hg stress. The data clearly indicatethe important role of RAF under Pb stress.
Post mRNA synthesis, many regulatory processes such as post transcriptional, translational and protein degradation control protein abundances. Proteins have longer half-life as compared to mRNA [71]. The rate of mRNA transcription is much lower than protein translation. The biochemical diversity of proteins can cause variation in protein and mRNA correlation level which might be the cause of difference in expression pattern of GST, CBL and CML genes as compared to protein expression.
Conclusions
Our study provides novel insights into proteomic and ecophysiological responses to Pb and Hg induced phytotoxicity in nodules of soybean plant. Symbiotic nodules are among the primary sites that encounter and counter metal stress. For strengthening the defence, understanding the response of nodule becomes pivotal in order to design proper strategies. In our study, it is evident that toxic metals (Pb and Hg) cause differential expression of proteins and transcript abundance in soybean nodules even when present in low concentrations. Nodules responded in an array of responses at the level of cellular antioxidants, transcription regulation, modulation of protein profile and causes prominent changes in nodule anatomy and bacteroidal volume. There is an increased abundance in defence, repair and development related proteins due to Pb and Hg stress. Pb newly induced a unique protein, sulfotransferase which controls the activity of several bioactive compounds involved in stress response. Correlation between protein expression and gene transcript levels due to Pb and Hg stress provided clues about regulation at transcriptional level. Pb and Hg stress responses in soybean root nodules has increased our knowledge regarding the mechanisms of stress tolerance which can be helpful in development of stress tolerant varieties through gene transfer and marker-assisted breeding. Such study would contribute not only to fill the gap of our understanding about nodule but also raise a hope of improved yield and production of soybean.
Additional files
Table S1. Program with varied current ramp, voltage, duration and total current used to perform for isoelectric focusing of protein extracted from root nodules of Glycine max L. Merr. (DOCX 13 kb)
Table S2. Summary of genes selected for qRT-PCR analysis with their corresponding forward and reverse primers and their attributed including melting temperature (Tm), GC content (%) and product size. (DOCX 14 kb)
Table S3. Information about conditions used in quantification assay of qRT-PCR (DOCX 13 kb)
Table S4. Information about qRT-PCR run conditions (DOCX 12 kb)
Figure S1. Impact of Pb and Hg stress on some key proteins of Glycine max L. Changes in actual relative abundance, bar graphs and picture have been demonstrated. (PPTX 1266 kb)
Table S5. List of proteins identified through MALDI TOF MS/MS and their theoretical/practical molecular weight and pI values, percent sequence coverage, NCBI accession, SoyKB gene ID’s, biological function, and cellular component. (DOCX 25 kb)
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for supporting this work through research group no. RGP 231.
Authors are thankful to Dr. K. Annapurna, Principal Scientist, Division of Microbiology, Indian Agricultural Research Institute (IARI), Pusa, New Delhi, India for generously providing Bradyrhizobium japonicum strain KAS-1. Authors also acknowledge Dr. Nitin Semwal, Sequence Referral Laboratories, New Delhi for helping in qPCR related studies.
Funding
Deanship of Scientific Research at King Saud University supported this work through research group no. RGP 231.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Abbreviations
- 2DE
2-Dimensional Electrophoresis
- ABA
Abscisic Acid
- ACN
Acetrinitrile
- ALMT
Aluminium activated malate transporter
- ANOVA
Analysis of variance
- AOS
Allene oxide synthase
- APX
Ascorbate peroxidase
- As
Arsenic
- ATSDR
Agency for Toxic Substances and Disease Registry
- BSA
Bovine serum albumin
- CAT
Catalase
- CBL
Calcineurin B like
- Cd
Cadmium
- CML
Calmodulin like
- CPCB
Central Pollution Control Board
- CT
Cycle threshold
- DTT
Dithiothreitol
- EDTA
Ethylenediaminetetraacetic acid
- FAOSTAT
Food and agriculture organization corporate statistical database
- FICCI
Federation of Indian Chambers of Commerce and Industry
- GST
Glutathione S-transferase
- H2O2
Hydrogen peroxide
- Hg
Mercury
- HgCl2
Mercuric chloride
- HNE
4-hydroxy-2-nonenal
- IAA
Indole-3-acetic acid
- IAA
Iodoacetamide
- IEF
Iso Electric Focusing
- IPG
Immobilized pH Gradient
- MALDI TOF MS
Matrix-assisted laser desorption/ionization- time-of-flight mass spectrometry
- MDA
Malondialdehyde
- MP
Madhya Pradesh
- MS
Mass spectrometry
- NADPH
Nicitinamide adeninedinucleotide phosphate
- NH4HCO3
Ammonium bicarbonate
- Pb
Lead
- PbCl2
Lead chloride
- PCD
Programme cell death
- PCR
Polymerase chain reaction
- PEP
phosphoenolpyruvate
- pI
Isoelectric point
- PLS-DA
Partial least squares discriminant analysis
- PUFAs
Polyunsaturated fatty acids
- qRT-PCR
Quantitative real time polymerase chain reaction
- RAF
Rapid alkalinisation factor
- ROS
Reactive oxygen species
- RT-PCR
Reverse transcription polymerase chain reaction
- SDS
Sodium Dodecyl Sulfate
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBARS
Thiobarbituric acid reactive substances
- TFA
Trifluoroacetic acid
- UCPs
Mitochondrial uncoupling proteins
- USDA
United States Department of Agriculture
- VIP
Variable importance in projection
- YEM
Yeast Extract-Mannitol
Authors’ contributions
MIQ and MAB designed the work. MAB, JA and RB performed plant growth and sampling. MAB, JA, RB and AAA performed biochemical and antioxidant experiments and related calculations. MAB, JA and MIQ performed proteomic analysis. MAB, JA, RB, MMI and AAH retrieved proteins and correlated with functions. MAB, AAH and MMI compiled and analysed data. MAB and MIQ wrote the MS. All authors approve the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohd Affan Baig, Email: affan.mac@gmail.com.
Javed Ahmad, Email: javedahmad12@gmail.com.
Rita Bagheri, Email: rita.bagheri@gmail.com.
Arlene Asthana Ali, Email: arlene.seven@gmail.com.
Asma Abdulkareem Al-Huqail, Email: aalhuqail@ksu.edu.sa.
Mohamed Mohamed Ibrahim, Email: mibramim@gmail.com.
Mohammad Irfan Qureshi, Email: mirfanq@gmail.com, Email: miqureshi@jmi.ac.in.
References
- 1.Clemens S, Ma JF. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu Rev Plant Biol. 2016;67:489–512. doi: 10.1146/annurev-arplant-043015-112301. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki Y, Adhikari D, Itoh K, Suyama K. Effects of temperature on competition and relative dominance of Bradyrhizobium japonicum and Bradyrhizobium elkanii in the process of soybean nodulation. Plant Soil. 2014;374:915–924. doi: 10.1007/s11104-013-1924-5. [DOI] [Google Scholar]
- 3.Zhang F, Li L. Using competitive and facilitative interactions in intercropping systems enhances crop productivity and nutrient-use efficiency. Plant Soil. 2003;248:305–312. doi: 10.1023/A:1022352229863. [DOI] [Google Scholar]
- 4.Beatty PH, Good AG. Future prospects for cereals that fix nitrogen. Sci. 2011;333:416–417. doi: 10.1126/science.1209467. [DOI] [PubMed] [Google Scholar]
- 5.Hati KM, Chaudhary RS, Mandal KG, Bandyopadhyay KK, Singh RK, Sinha NK, et al. Effects of tillage, residue and fertilizer nitrogen on crop yields, and soil physical properties under soybean–wheat rotation in vertisols of Central India. Agric Res. 2015;4:48–56. doi: 10.1007/s40003-014-0141-7. [DOI] [Google Scholar]
- 6.Malik N, Biswas AK, Raju CB. Plankton as an indicator of heavy metal pollution in a freshwater reservoir of Madhya Pradesh, India. Bull Environ Contam Toxicol. 2013;90:725–729. doi: 10.1007/s00128-013-0985-8. [DOI] [PubMed] [Google Scholar]
- 7.Hao X, Taghavi S, Xie P, Orbach MJ, Alwathnani HA, Rensing C, et al. Phytoremediation of heavy and transition metals aided by legume-rhizobia symbiosis. Int J Phytoremediation. 2014;16:179–202. doi: 10.1080/15226514.2013.773273. [DOI] [PubMed] [Google Scholar]
- 8.Haddad SA, Tabatabai MA, Loynachan TE. Inhibition of nodulation and nitrogen nutrition of leguminous crops by selected heavy metals. Air Soil Water. 2015;8:1–7. [Google Scholar]
- 9.Gururani MA, Upadhyaya CP, Baskar V, Venkatesh J, Nookaraju A, Park SW. Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J Plant Growth Regul. 2013;32:245–258. doi: 10.1007/s00344-012-9292-6. [DOI] [Google Scholar]
- 10.Singh S, Parihar P, Singh R, Singh VP, Prasad SM. Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front Plant Sci. 2016;6:1–36. doi: 10.3389/fpls.2015.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 12.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 13.Bagheri R, Bashir H, Ahmad J, Iqbal M, Qureshi MI. Spinach (Spinacia oleracea L.) modulates its proteome differentially in response to salinity, cadmium and their combination stress. Plant Physiol Biochem. 2015;97:235–245. doi: 10.1016/j.plaphy.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Heath R, Packer L. Photoperoxidation in isolated chloroplasts: I Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- 15.Qureshi MI, Abdin MZ, Qadir S, Iqbal M. Lead-induced oxidative stress and metabolic alterations in Cassia angustifolia Vahl. Biol Plant. 2007;51:121–128. doi: 10.1007/s10535-007-0024-x. [DOI] [Google Scholar]
- 16.HE A, Aebi HE. Catalase. Methods Enzym Anal 1983;105:121–26.
- 17.Anderson ME. Determination of glutathione and glutathione disulphide in biological samples. Annu Rev Biochem. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- 18.Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol. Biol. 2008;66:361–378. doi: 10.1007/s11103-007-9274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Zhou, Zhou Xin, Ling Yi, Zhang Zhenhai, Su Zhen. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research. 2010;38(suppl_2):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia J., Psychogios N., Young N., Wishart D. S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Research. 2009;37(Web Server):W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad J, Bashir H, Bagheri R, Baig A, Al-Huqail A, Ibrahim MM, et al. Drought and salinity induced changes in ecophysiology and proteomic profile of Parthenium hysterophorus. PLoS One. 2017;12:1–27. doi: 10.1371/journal.pone.0185118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qureshi MI, Muneer S, Bashir H, Ahmad J, Iqbal M. Nodule physiology and proteomics of stressed legumes. In:Kader J-K, Delseny M, editors. Advances in botanical research. Elsevier Inc 2010;56: p. 1–48.
- 23.Sytar O, Kumar A, Latowski D, Kuczynska P, Strzałka K, Prasad MNV. Heavy metal-induced oxidative damage, defense reactions, and detoxification mechanisms in plants. Acta Physiol Plant. 2013;35:985–999. doi: 10.1007/s11738-012-1169-6. [DOI] [Google Scholar]
- 24.Wang Y, Xu L, Shen H, Wang J, Liu W, Zhu X, et al. Metabolomic analysis with GC-MS to reveal potential metabolites and biological pathways involved in Pb & cd stress response of radish roots. Sci Rep. 2015;5:18296. doi: 10.1038/srep18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thapa G, Sadhukhan A, Panda SK, Sahoo L. Molecular mechanistic model of plant heavy metal tolerance. Biometals. 2012;25:489–505. doi: 10.1007/s10534-012-9541-y. [DOI] [PubMed] [Google Scholar]
- 26.Samuel MA, Salt JN, Shiu SH, Goring DR. Multifunctional arm repeat domains in plants. Int Rev Cytol. 2006;253:1–26. doi: 10.1016/S0074-7696(06)53001-3. [DOI] [PubMed] [Google Scholar]
- 27.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 28.Sutter JU, Campanoni P, Blatt MR, Paneque M. Setting SNAREs in a different wood. Traffic. 2006;7:627–638. doi: 10.1111/j.1600-0854.2006.00414.x. [DOI] [PubMed] [Google Scholar]
- 29.Geisler M, Bailly A. Tête-à-tête: the function of FKBPs in plant development. Trends Plant Sci. 2007;12:465–473. doi: 10.1016/j.tplants.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Asif MH, Chakrabarty D, Tripathi RD, Dubey RS, Trivedi PK. Expression of a rice lambda class of glutathione S-transferase, OsGSTL2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J Hazard Mater. 2013;248–249:228–237. doi: 10.1016/j.jhazmat.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Balestrasse KB, Yannarelli GG, Noriega GO, Batlle A, Tomaro ML. Heme oxygenase and catalase gene expression in nodules and roots of soybean plants subjected to cadmium stress. Biometals. 2008;21:433–441. doi: 10.1007/s10534-008-9132-0. [DOI] [PubMed] [Google Scholar]
- 33.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Pearce G, Yamaguchi Y, Munske G, Ryan CA. Structure-activity studies of RALF, rapid Alkalinization factor, reveal an essential - YISY - motif. Peptides. 2010;31:1973–1977. doi: 10.1016/j.peptides.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Murphy E, De Smet I. Understanding the RALF family: a tale of many species. Trends Plant Sci. 2014;19:664–671. doi: 10.1016/j.tplants.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Kumar R, Tran LSP, Neelakandan AK, Nguyen HT. Higher plant cytochrome b5 polypeptides modulate fatty acid desaturation. PLoS One. 2012;7(2):e31370. doi: 10.1371/journal.pone.0031370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loscos J, Matamoros MA, Becana M. Ascorbate and homoglutathione metabolism in common bean nodules under stress conditions and during natural senescence. Plant Physiol. 2008;146:1282–1292. doi: 10.1104/pp.107.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the solitary-root/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu LH, Miao ZQ, Qi GF, Wu J, Cai XT, Mao JL, et al. MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol Plant. 2014;7:1653–1669. doi: 10.1093/mp/ssu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim M, Lee U, Small I, des Francs-Small CC, Vierling E. Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell. 2012;24:3349–3365. doi: 10.1105/tpc.112.101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogawa T, Ueda Y, Yoshimura K, Shigeoka S. Comprehensive analysis of cytosolic nudix hydrolases in Arabidopsis thaliana. J Biol Chem. 2005;280:25277–25283. doi: 10.1074/jbc.M503536200. [DOI] [PubMed] [Google Scholar]
- 42.Jambunathan N, Mahalingam R. Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta. 2006;224:1–11. doi: 10.1007/s00425-005-0183-y. [DOI] [PubMed] [Google Scholar]
- 43.Giri J, Vij S, Dansana PK, Tyagi AK. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011;191:721–732. doi: 10.1111/j.1469-8137.2011.03740.x. [DOI] [PubMed] [Google Scholar]
- 44.Du CX, Fan HF, Guo SR, Tezuka T, Li J. Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry. 2010;71:1450–1459. doi: 10.1016/j.phytochem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Prabhakar V, Löttgert T, Gigolashvili T, Bell K, Flügge UI, Häusler RE. Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett. 2009;583:983–991. doi: 10.1016/j.febslet.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Roepke J, Bozzo GG. Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides. Phytochemistry. 2015;109:14–24. doi: 10.1016/j.phytochem.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 47.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Costanzo ME, Andrade A, Del Carmen Tordable M, Cassán F, Abdala G. Production and function of jasmonates in nodulated roots of soybean plants inoculated with Bradyrhizobium japonicum. Arch Microbiol. 2012;194:837–845. doi: 10.1007/s00203-012-0817-y. [DOI] [PubMed] [Google Scholar]
- 49.Du H, Wu N, Fu J, Wang S, Li X, Xiao J, et al. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot. 2012;63:6467–6480. doi: 10.1093/jxb/ers300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agarwal PK, Jha B. Transcription factors in plant and ABA dependent and independent abiotic stress signaling. Biol Plant. 2010;54:201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- 51.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Zhang JS, Chen SY, Zhang WK. Role of soybean GmbZIP132 under abscisic acid and salt stresses. J Integr Plant Biol. 2008;50:221–230. doi: 10.1111/j.1744-7909.2007.00593.x. [DOI] [PubMed] [Google Scholar]
- 53.Stotz HU, Mueller S, Zoeller M, Mueller MJ, Berger S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J Exp Bot. 2013;64:963–975. doi: 10.1093/jxb/ers389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20:219–229. doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Xiao S, Chye ML. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog Lipid Res. 2011;50:141–151. doi: 10.1016/j.plipres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Mwenda Cynthia Mugo, Matsuki Atsushi, Nishimura Kohji, Koeduka Takao, Matsui Kenji. Spatial expression of the Arabidopsishydroperoxide lyasegene is controlled differently from that of theallene oxide synthasegene. Journal of Plant Interactions. 2015;10(1):1–10. doi: 10.1080/17429145.2014.999836. [DOI] [Google Scholar]
- 57.Marsolais F, Boyd J, Paredes Y, Schinas AM, Garcia M, Elzein S, et al. Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760) Planta. 2007;225:1233–1244. doi: 10.1007/s00425-006-0413-y. [DOI] [PubMed] [Google Scholar]
- 58.Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah BW. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015;6:1–12. doi: 10.3389/fpls.2015.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F, Li L, Jiao Z, Chen Y, Liu H, Chen X, et al. Characterization of the calcineurin B-like (CBL) gene family in maize and functional analysis of ZmCBL9 under abscisic acid and abiotic stress treatments. Plant Sci. 2016;253:118–129. doi: 10.1016/j.plantsci.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci. 2016;113:1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xing L, Zhao Y, Gao J, Xiang C, Zhu J-K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep. 2016;6:27177. doi: 10.1038/srep27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dietz KJ, Tavakoli N, Kluge C, Mimura T, Sharma SS, Harris GC, et al. Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J Exp Bot. 2001;52:1969–1980. doi: 10.1093/jexbot/52.363.1969. [DOI] [PubMed] [Google Scholar]
- 63.Begcy K, Mariano ED, Mattiello L, Nunes AV, Mazzafera P, Maia IG, et al. An Arabidopsis mitochondrial uncoupling protein confers tolerance to drought and salt stress in transgenic tobacco plants. PLoS One. 2011;6(8):e23776. doi: 10.1371/journal.pone.0023776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furuichi T, Sasaki T, Tsuchiya Y, Ryan PR, Delhaize E, Yamamoto Y. An extracellular hydrophilic carboxy-terminal domain regulates the activity of TaALMT1, the aluminum-activated malate transport protein of wheat. Plant J. 2010;64:47–55. doi: 10.1111/j.1365-313X.2010.04309.x. [DOI] [PubMed] [Google Scholar]
- 65.Noctor G, Queval G, Gakière B. NAD(P) Synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot. 2006;57:1603–1620. doi: 10.1093/jxb/erj202. [DOI] [PubMed] [Google Scholar]
- 66.Hemavathi UCP, Akula N, Young KE, Chun SC, Kim DH, et al. Enhanced ascorbic acid accumulation in transgenic potato confers tolerance to various abiotic stresses. Biotechnol Lett. 2010;32:321–330. doi: 10.1007/s10529-009-0140-0. [DOI] [PubMed] [Google Scholar]
- 67.Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 2008;146:1109–1116. doi: 10.1104/pp.107.113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szalonek M, Sierpien B, Rymaszewski W, Gieczewska K, Garstka M, Lichocka M, et al. Potato annexin STANN1 promotes drought tolerance and mitigates light stress in transgenic Solanum tuberosum L. plants. PLoS One. 2015;10(7):e0132683. doi: 10.1371/journal.pone.0132683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naka Y, Watanabe K, Sagor GHM, Niitsu M, Pillai MA, Kusano T, et al. Quantitative analysis of plant polyamines including thermospermine during growth and salinity stress. Plant Physiol Biochem. 2010;48:527–533. doi: 10.1016/j.plaphy.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 70.Filiz E, Vatansever R, Ozyigit II. Insights into a key sulfite scavenger enzyme sulfite oxidase (SOX) gene in plants. Physiol Mol Biol Plants. 2017;23:385–395. doi: 10.1007/s12298-017-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Program with varied current ramp, voltage, duration and total current used to perform for isoelectric focusing of protein extracted from root nodules of Glycine max L. Merr. (DOCX 13 kb)
Table S2. Summary of genes selected for qRT-PCR analysis with their corresponding forward and reverse primers and their attributed including melting temperature (Tm), GC content (%) and product size. (DOCX 14 kb)
Table S3. Information about conditions used in quantification assay of qRT-PCR (DOCX 13 kb)
Table S4. Information about qRT-PCR run conditions (DOCX 12 kb)
Figure S1. Impact of Pb and Hg stress on some key proteins of Glycine max L. Changes in actual relative abundance, bar graphs and picture have been demonstrated. (PPTX 1266 kb)
Table S5. List of proteins identified through MALDI TOF MS/MS and their theoretical/practical molecular weight and pI values, percent sequence coverage, NCBI accession, SoyKB gene ID’s, biological function, and cellular component. (DOCX 25 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].