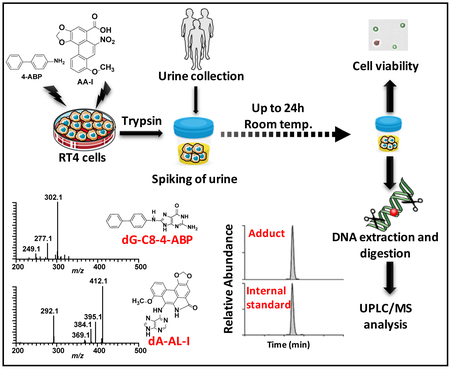
Abstract
Tobacco smoking contributes to about 50% of the bladder cancer (BC) cases in the United States. Some aromatic amines in tobacco smoke are bladder carcinogens; however, other causal agents of BC are uncertain. Exfoliated urinary cells (EUCs) are a promising noninvasive biospecimen to screen for DNA adducts of chemicals that damage the bladder genome. Though, the analysis of DNA adducts in EUCs is technically challenging because of the low number of EUCs and limiting quantity of cellular DNA. Moreover, EUCs and their DNA adducts must remain viable during the time of collection and storage of urine, to develop robust screening methods. We employed RT4 cells, well-differentiated transitional epithelial bladder cell line, as a cell model system in urine to investigate cell viability and the chemical stability of DNA adducts of two prototypical bladder carcinogens: 4-aminobiphenyl (4-ABP), an aromatic amine found in tobacco smoke, and aristolochic acid (AA-I) a nitrophenanthrene found in Aristolochia herbaceous plants used for medicinal purposes world-wide. The cell viability of RT4 cells pretreated with 4-ABP or AA-I in urine exceeded 80%, and the major DNA adducts of 4-ABP and AA-I, quantified by liquid chromatography-mass spectrometry, were stable for 24 hours. Thereafter, we successfully screened EUCs of mice treated with AA-I to measure DNA adducts of AA-I, which were still detected 25 days following treatment of the carcinogen. EUCs are promising biospecimens that can be employed for the screening of DNA adducts of environmental and dietary genotoxicants that may contribute to the development of BC.
Graphical Abstract
INTRODUCTION
Bladder cancer (BC) is the sixth most common cancer in the United States and almost 80,000 new cases are reported each year.1 Smoking is estimated to account for 50% of all bladder cancer cases.2 Some aromatic amines present in tobacco smoke, including 4-aminobiphenyl (4-ABP) and 2-naphthylamine, are believed to contribute to BC risk based on elevated BC among factory workers in the textile dye and rubber tire industries who were exposed to these chemicals.2–4 Aristolochic acids (AA-I) are potent upper urinary tract carcinogens found in Aristolochia plant species, which have been used in traditional Chinese herbal medicines world-wide.5,6 Recently, AA-I has also been implicated in BC.7 However, other major causative agents of BC remain undefined.
The measurement of biomarkers in urine is a useful approach to obtain important information about tobacco, diet and occupational exposures in BC risk.8 For example, the urine of smokers and meat-eaters have elevated levels of mutagenicity, which has been attributed to aromatic amines or structurally related heterocyclic aromatic amines (HAAs).9–12 Several aromatic amines and HAAs have been identified in urine of smokers and omnivores.13,14 These chemicals bathe the urothelium and can undergo bioactivation by cytochrome P450s expressed in the bladder and damage DNA.15
DNA adducts of genotoxicants are a measure an internal exposure to chemicals and can provide clues about exposures that can contribute to cancer risk.16 A DNA adduct of 4-ABP was detected in human bladder by the 32P-postlabeling method.17 More recently, DNA adducts of acrolein, a by-product of lipid peroxidation and a toxicant present in tobacco smoke, were also detected in bladder, by 32P-postlabeling.18 However, 32P-postlabeling methods do not provide spectral data and the chemical identities of these lesions were not confirmed by unambiguous mass spectrometry methods. Moreover, other putative DNA adducts were detected by 32P-postlabeling in bladder, but the identities of these presumed lesions remain unknown.
The employment of exfoliated urinary cells (EUCs) is a promising non-invasive biospecimen to screen for DNA adducts and to identify chemicals that may contribute to BC. The exfoliation of cells is a main mechanism involved in the homeostatic control of epithelium cell population size.19 EUCs are a heterogeneous mixture of cell types that include urothelial cells shed from the upper and lower urinary tracts, tubular epithelial cells from the kidney, squamous cells from the vagina and urethra; and inflammatory cells (with infection).20,21 The urothelium has a very low rate of cell turnover, but a high regenerative capacity, showing rapid cell proliferation during development and in response to damage or injury.22 Thus, EUCs can serve as specimens to screen for DNA damage of the urinary system. For example, micronuclei formation in EUCs are elevated in smokers compared to non-smokers,23 factory workers exposed to nitrite and N-phenyl-1-naphthylamine,24 and for individuals who chronically drink water contaminated with arsenic.25
Relatively few studies have attempted to measure DNA adducts in human EUCs.26 Talaska and co-workers screened EUC of smokers and factory workers exposed to aromatic amines and detected putative DNA adducts of 4-ABP, benzidine, and 4,4′-methylenebis-2-chloroaniline, by 32P-postlabeling.27–29 The Vouros laboratory used liquid chromatography-mass spectrometry (LC-MS) to screen for DNA adducts of 4-ABP in human EUCs.30 Even though a very low limit of quantification (LOQ) of 2 adducts per 108 bases was achieved, 4-ABP adducts were not detected in EUCs of nonsmokers. In a pilot study, we showed that EUCs obtained from subjects with compromised renal function who were exposed to AA-I through ingestion of Chinese herbal medicines could be used to screen for DNA adducts of AA-I by ultraperformance liquid chromatography–electrospray ionization/multistage mass spectrometry (UPLC-ESI/MS3).31 Our pilot study demonstrated the feasibility of employing human EUCs to screen for DNA adducts by LC-MS methods. Nevertheless, systematic studies examining the viability of EUCs during collection and storage of urine, and the stability of DNA adducts have not been reported. Such studies are required to validate the use of EUCs in biomonitoring of DNA adducts in humans. In this investigation, we employed RT4 cells, a well-differentiated human urinary bladder cell line,32 which bioactivates AA-I and 4-ABP to reactive intermediates that bind to DNA (vide infra), as an in vitro cell model in urine. We pretreated RT4 cells with AA-I and 4-ABP and then examined cell viability, DNA recovery, and the stability of DNA adducts of RT4 cells incubated in human urine for 24 h. Thereafter, we demonstrated that DNA adducts of AA-I can be screened in EUCs of mice four weeks following treatment with AA-I.
EXPERIMENTAL SECTION
Caution:
AA-I and 4-ABP are human carcinogens. These chemicals should be handled with caution in a well-ventilated fume hood with appropriate personal protective equipment.
Materials
4-ABP, calf thymus DNA (CT-DNA), Dimethyl sulfoxide (DMSO), Proteinase K (Tritirachium album), DNase I (type IV, bovine pancreas), alkaline phosphatase (Escherichia coli), nuclease P1 (Penicillium citrinum, RNase A (bovine pancreas), and RNase T1 (Aspergillus oryzae) were purchased from Sigma-Aldrich (St. Louis, MO). Phosphodiesterase I (Crotalus adamanteus venom) was purchased from Worthington Biochemical Corp. (Newark, NJ). Isopore™ membrane filter (10 µm) and Sterifil® Aseptic filter holder (500 mL) were purchased from Millipore-Sigma (Bedford, MA). 8-Methoxy-6-nitrophenanthro-[3,4-d]-1,3-dioxole-5-carboxylic (aristolochic acid, AA-I) was provided by Dr. H. Priestap, Department of Biological Sciences, Florida International University. The DNA adducts 7-(2′-deoxyadenosin-N6-yl)aristolactam-I (dA-AL-I) and [15N5]-dA-AL-I,33 N-(2′-deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8–4-ABP), and [13C10]-dG-C8–4-ABP were synthesized as described.34 DNeasy blood/tissue kit was purchased from Qiagen (Germantown, MD). Microliter CapLC vials with silanized inserts were purchased from Wheaton (Millville, NJ).
Methods.
Cell culture, treatment with 4-ABP and AA-I, cell harvesting, and cell viability.
The human bladder epithelial cell line RT4 (American Type Culture Collection, Manassas, VA), was maintained in McCoy’s 5A (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich) and antibiotics (100 units/mL penicillin and 100 mg/mL streptomycin) (Gibco, ThermoFisher Scientific, Carlsbad, CA) at 37 ºC in a humidified atmosphere of 5% CO2. For DNA adducts experiments, cells were seeded at 10 × 106 cells/plate on 10 cm diameter plates in in McCoy’s 5A 10% FBS and allowed to reach 90% confluence. The cells were washed with pre-warmed phosphate-buffered saline (PBS) (Gibco) at 37 °C, and the media was renewed with fresh media containing 4-ABP (1 µM or 10 µM) or AA-I (0.1 µM or 0.5 μM) or DMSO (0.1% v/v) as a solvent control. After 24 h of treatment, the cells were washed 3 times with pre-warmed PBS and detached from the cell culture dish using 2 mL of 0.25% Trypsin-EDTA (Gibco) for 3 min at 37 °C, followed by dilution with 7 mL of pre-warmed culture media. After centrifugation at 200 × g for 7 min, the media was removed and the cell pellets were resuspended in 10 mL PBS. The cell viability and density were evaluated using the trypan blue exclusion assay with a TC20™ automated cell counter (BioRad, Hercules, CA). The recovery of viable RT4 cells following treatment of carcinogens exceeded 90%.
The effect of urine and duration of collection on the viability of RT4 cells.
De-identified fresh urine specimens were collected over 6 h from three healthy male volunteers (200 to 600 mL and stored at 4 ºC). The pH values of urine specimens were measured with pH strip indicators and the values ranged between pH 6 to 7. The RT4 cells (one million cells per mL PBS) were spiked into 5 mL urine to achieve a count of 2 × 105 cells/mL and incubated for 24 h at 4 ºC or room temperature. Cell viability was measured over time (T=0, 3, 6, and 24 h), by mixing 10 µL of the urine with 10 µL of trypan blue (0.4%).
Optimization of conditions for collection of urinary cells: filtration versus centrifugation.
Studies on urinary cell collection and recovery of DNA were conducted with urine (40 mL) containing 1 million non-treated RT4 cells. The efficacy of cell recovery was based on the yield of DNA. The RT4 cells in urine were collected using a two-step centrifugation procedure or by vacuum filtration. The centrifugation of urine was performed at 4 ºC unless stated otherwise. The urine specimens containing RT4 cells were centrifuged at three different centrifugal forces (600, 1500, or 3000 × g) for 10 min. The cell pellets were reconstituted in 1.8 mL of cold PBS buffer and transferred into a new 2 mL tube for high-speed centrifugation at 14,000 × g for 10 min. The cell pellets were stored at −80 ºC until analyzed. Non-spiked urine samples (control urine) containing endogenous EUCs were collected by the two-step centrifugation. In the vacuum filtration procedure, a polycarbonate membrane filter (10 µm pore size) was placed on the filter holder (500 mL) that was connected to a 1-liter vacuum filter flask. The RT4 cells were collected on the membrane filter by gentle vacuum pressure (~500 mm Hg) to preserve the morphology of cells. The urinary cells collected on the membrane filter were recovered by repeatedly washing the membrane with 1.8 mL chilled PBS using a transfer pipette. Thereafter, the PBS containing RT4 cells was transferred to a new 2 mL tube for high-speed centrifugation. The cell pellets were stored at −80 ºC until analyzed.
Isolation of DNA.
Cell pellets were resuspended in 200 µL of 50 mM TE lysis buffer containing 10 mM ßME and lysed in an ultrasonic water bath (40 kHz, Bransonic CPX3800H, Branson Ultrasonics Corp, Danbury, CT) for 15 min at ambient temperature. The urinary cell DNA was isolated by DNeasy blood/tissue kit with minor modifications as reported in supporting information (Protocol S-1). The quantities of urinary cell DNA d were determined by UV spectroscopy.
The effect of the urine matrix and duration of urine collection on the stability of DNA adducts in RT4 cells.
The effects of urine and duration of collection on the stability of DNA adducts in RT4 cells were examined using urine (40 mL) or PBS buffer (40 mL) containing one million RT4 cells pretreated with 4-ABP or AA-I. After 24 h, the treated RT4 cells were trypsinized. The RT4 cells were recovered and incubated at room temperature with urine or PBS for up to 24 h. Cells were collected by the two-step centrifugation step (vide supra). DNA (5 µg) modified with 4-ABP or AA-I was spiked with respective internal standards and incubated with nucleases (vide infra).
Effect of cell population on the stability of DNA adducts of RT4 cells in urine.
The stability of DNA adducts of 4-ABP and AA-I in RT4 cells spiked in urine was tested at three different concentrations of RT4 cells (1 × 104, 2 × 104, and 4 × 104 cells per 40 mL of urine). The samples were incubated at room temperature for up to 24 h, and the levels of DNA adducts were measured over time (T 0, 10, and 24 h). The control samples of RT4 cells pretreated with carcinogens were harvested and stored at – 80 °C prior to DNA workup and adduct measurements.
Analysis of dA-AL-I adduct in the urinary exfoliated cells of mice dosed with AA-I.
The animal studies followed the guidelines of the National Institutes of Health Office of Laboratory Animal Welfare. Five male C57BL/6J mice aged 8–10 weeks old (Jackson Laboratories, Bar Harbor, ME) were dosed (i.p.) with AA-I (1 mg/kg in PBS). The mice were placed in metabolic cages for overnight collection of urine at room temperature during the following days: day 0, 4, 11, and day 25 post-dosing. The volume of urine collected per mouse was approximately 1 mL at each timepoint. The exfoliated urinary cells were centrifuged at 1500 x g and cellular pellets were washed once with 1 mL of cold PBS. The cells were lysed with TE buffer (vide supra), and the DNA was isolated with the DNeasy blood/tissue kit. The concentration of mouse urine DNA was measured by Qubit fluorometric quantitation employing Quant-iT™ dsDNA HS assay kit, which is specific for double-stranded DNA measurements (Invitrogen, Waltham, MA). The amount of DNA ranged from 9 to 122 ng after pooling DNA per collection of each time point. Then, DNA samples were mixed 20 µg of carrier CT DNA, and precipitated with isopropanol, to remove residual salts and buffer that interfered with the UPLC-ESI/MS3 assay (B-H Yun, unpublished observation) The carrier CT DNA was added to minimize losses of DNA from EUCs during the precipitation step and for nuclease digestion of AA-I-modified DNA, which had been optimized with 10 to 20 μg DNA.33 Due to limited quantity of DNA, the dA-AL-I adduct measurements were conducted in singlicate by UPLC-MS3 (vide infra).
Enzymatic digestion of carcinogen modified DNA.
The protocol of DNA digestion was reported previously.33,35 In brief, RT4 cell DNA (2 – 5 µg) was spiked with isotopically labeled internal standards, [15N5]-dA-AL-I or [13C10]-dG-C8–4-ABP, at a level of 1 adduct per 107 bases prior to DNA digestion. For mouse EUC DNA, 0.42 pg of [15N5]-dA-AL-I was added to the EUC DNA, corresponding to a level of internal standard ranging from 2 to 28 adducts per 106 bases. DNA samples were incubated with DNase I, nuclease P1, phosphodiesterase I, and alkaline phosphatase for 18 h at 37 ºC. The DNA digests were dried by vacuum centrifugation and resuspended in 40 µL (for RT4 cells) and 20 µL (for mouse EUCs) of 1:1 DMSO:H2O, and centrifuged at 21,000 × g for 10 min. The supernatants were carefully transferred to a silanized vial insert for subsequent analyses. Calf thymus (CT) DNA was spiked with internal standards and served as a negative control. The completion of enzymatic digestion and the purity of DNA were assessed by HPLC analysis of 2′-deoxynucleosides.33,35
Measurement of dG-C8–4-ABP and dA-AL-I by UPLC-ESI-Ion Trap-MS3.
Analyses were conducted with a nanoAcquity UPLC system (Waters Corp., Milford, MA) or Dionex Ultimate™ 3000 RSLCnano system (Thermo Fisher Scientific, Waltham, MA) interfaced with an Advanced CaptiveSpray source (Michrom Bioresources, Auburn, CA) and Velos Pro Ion Trap Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA). An Acclaim PepMap C18 µ-precolumn (0.3 × 5 mm, 5 µm, 100 Å) was used for online sample enrichment of DNA adducts. A Magic C18 AQ column (Michrom Bioresources, 0.3 × 150 mm, 5 µm, 100 Å) was employed for chromatographic analyses. The LC mobile phases were (A) 0.01% formic acid and (B) 95% acetonitrile containing 0.01% formic acid. The DNA digest was injected onto the trapping column and washed with mobile phase A for 4 min at a flow rate of 12 µL/min. Then the adducts were back-flushed onto the Magic C18 AQ column at a flow rate of 5 µL/min. A linear gradient of 1–99% B over 10 min was applied for the separation, followed by 3 min of washing at 99% B.
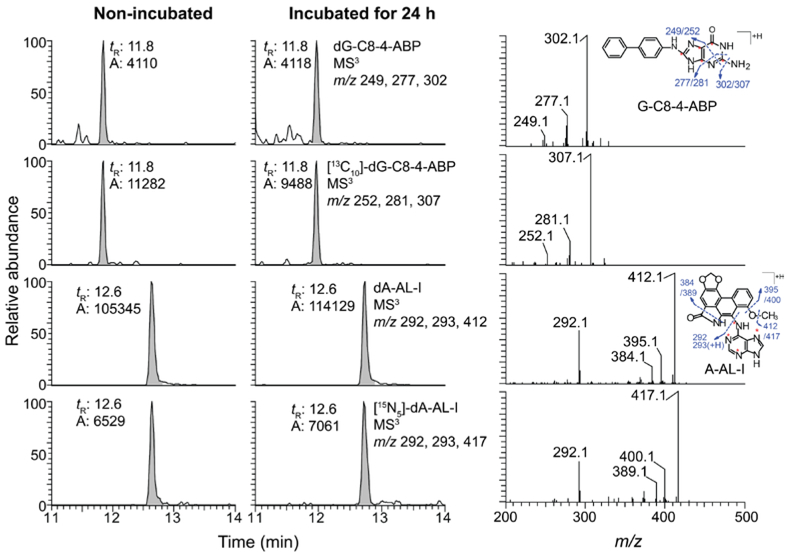
DNA adducts were measured in positive-ion mode at the MS3 scan stage. The MS parameters were described previously.33,35 The following transitions employed were: dA-AL-I at m/z 543.3 → 427.2 → 292.1, 293.1, and 412.1; [15N5]-dA-AL-I at m/z 548.3 → 432.2 → 292.1, 293.1, and 417.1; dG-C8–4-ABP at m/z 435.2 → 319.1 → 277.1 and 302.1; [13C10]-dG-C8–4-ABP at m/z 445.2 → 324.1 → 281.1 and 307.1. Mass spectrometry data acquisition and analysis were conducted with Xcalibur version 3.0.63.
RESULTS AND DISCUSSION
We examined the feasibility of employing urinary cells as a non-invasive biospecimen to monitor DNA adducts by LC-MS methods. We selected 4-ABP and AA-I as prototypes of bladder carcinogens for study (Scheme 1).3,4,36 However, the limiting number of EUCs and low quantity of DNA in urine of healthy individuals37 precludes multiple analyses on the kinetics of cell viability and DNA adduct stability in human urine. Therefore, we employed RT4 cells, a well-differentiated human urinary bladder cell line,32 pretreated with these carcinogens, as a urinary cell model. RT4 cells retain many morphological and metabolic features of bladder epithelial cells, including membrane rigidity, expression of xenobiotic metabolism enzymes, and biological responses to carcinogens.32,38–41 Moreover, RT4 cells carry the normal p53,42 a key gene in the repair of ABP-induced genomic DNA damage in human bladder cells.40 We employed our validated UPLC-ESI/MS3 methods to measure DNA adducts of 4-ABP and AA-I.33,43
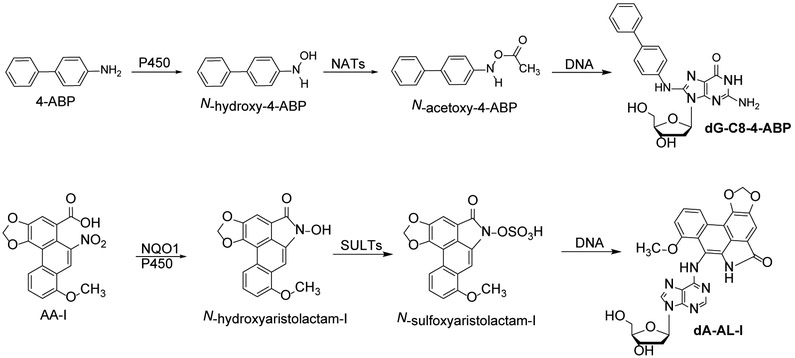
Scheme 1. Metabolic activation of 4-aminobiphenyl and aristolochic acid I and DNA adduct formation.
The bioactivation of 4-ABP by cytochrome P450 (P450) and the bioactivation of AA-I occurs by several enzymes, including P450 or NAD(P)H dehydrogenase [quinone] 1. N-acetylransferases (NATs) or sulfotransferases activate the hydroxylamine metabolites to reactive intermediates that bind to DNA.
The DNA recovery from RT4 cells: centrifugation versus filtration.
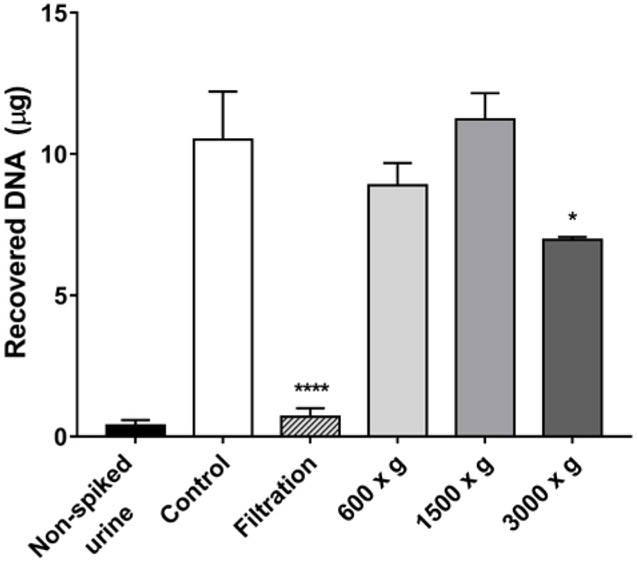
We examined the recovery of urinary cells by comparing the yield of DNA recovered from RT4 cells using centrifugation or vacuum filtration (Figure 1). The Isopore™ membrane filter is made of polycarbonate and it is track-etched screen filter with pore size 10 µm. The reported diameters of cells in human urine are: erythrocytes (~ 7 µm), leukocytes (10 −12 µm), and epithelial cells (20– 65 µm).44,45 Thus, filtration is a rapid means to concentrate EUCs from urine and remove many other cell types. However, the filtration of urine resulted in nearly a 90% loss of RT4 cells compared to the control sample containing RT4 cells processed by centrifugation. Most of the RT4 cells adhered to the surface of the membrane filter and were not efficiently recovered by washing the membrane with PBS buffer. The optimum recovery of DNA from RT4 cells in urine was obtained when the urine was centrifuged at 1500 × g, and the yield of DNA was similar to that of the control sample. However, the centrifugation of RT4 cells (or EUCs) at the higher 3000 × g can rupture the cell membrane and cause cell lysis, resulting in the loss of some cellular DNA.46
Figure 1.
The recovery of DNA from RT4 cells spiked in urine of a subject by centrifugation (g-forces indicated) or vacuum filtration with a membrane filter (pore size 10 µm). The yield of DNA was obtained from 40 mL of urine in the absence and presence of one million RT4 cells. The RT4 cell control sample was processed in PBS and centrifuged at 1500 g. All measurements were reported as the mean ± SD. One-way analysis of variation with Sidak’s multiple comparisons test was performed against the control sample (Prism 6, San Diego, CA); *p <0.05 and ****p<0.0001.
The effect of urine matrix and duration of urine collection on the viability of RT4 cells.
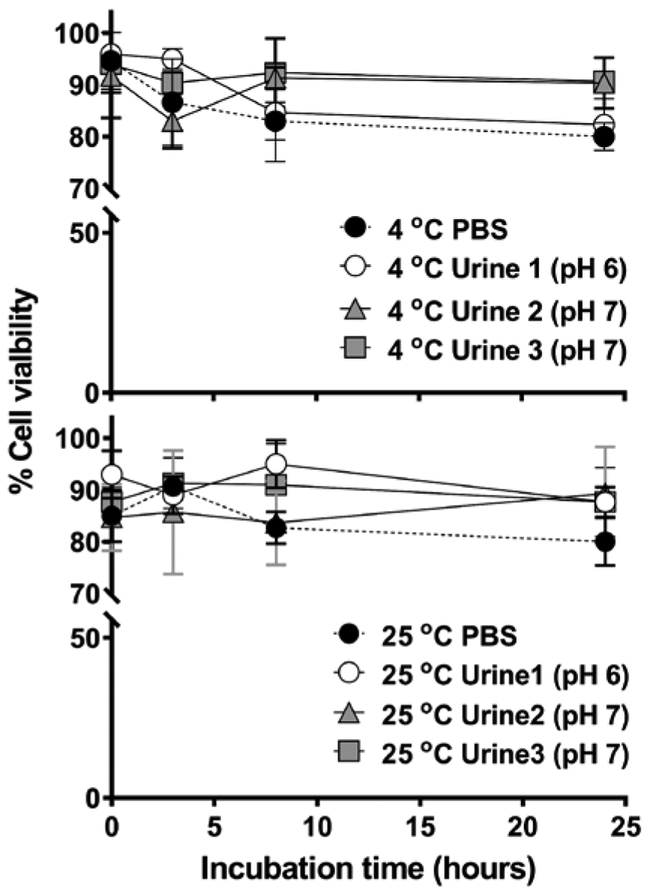
A time interval of 12 to 24 h may be required to obtain a sufficient amount of EUCs and cellular DNA for MS analysis of DNA adducts (Figure 2). Depending upon the diet and water consumption, urine can be isotonic or otherwise hypertonic or hypotonic, which can deform and rupture the cell membrane of EUCs.44 The viability of RT4 cells in urine stored at room temperature or 4 ºC was examined with the trypan blue exclusion test. Our findings show that the viability of RT4 cells in urine stored for up to 24 h exceeded 80%, regardless of the urine donor or the temperature of urine storage (Figure 2).
Figure 2.
The viability of RT4 cells incubated in urine of three donors at 4 ºC or 25 ºC over 24 h. Cell viability was determined by the trypan blue exclusion assay. All measurements were performed in triplicate and reported as the mean ± SD.
The effect of urine and duration of urine collection on the stability of DNA adducts in RT4 cells.
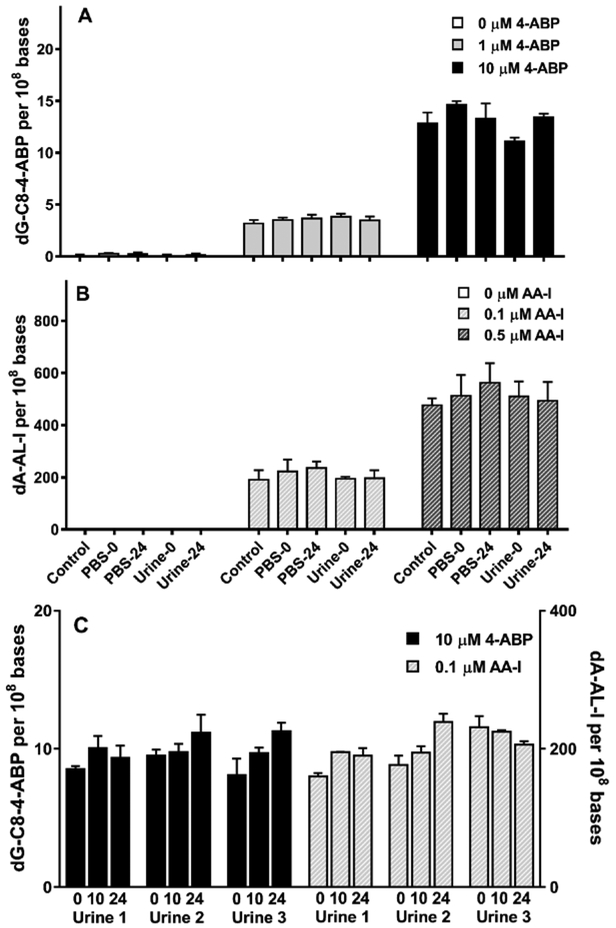
The stabilities of DNA adducts of 4-ABP and AA-I in RT4 cells pretreated with both carcinogens were examined by adding the pretreated cells to urine of three donors and incubated at room temperature for 24 h (Figure 3). The DNA adducts of RT4 cells diluted in urine or PBS were stable over time and comparable to those values measured at T0. Representative extracted- ion chromatogram at MS3 scan stage and the product ion spectra of dA-AL-I and dG-C8–4-ABP in pretreated RT4 cells with and without incubation in urine are shown in Figure 4.
Figure 3.
Effect of urine matrix and duration of urine collection on the levels of DNA adducts in RT4 cells. (A and B) Urine from a single subject or PBS (40 mL) were spiked with 1 million RT4 cells pretreated with 4-ABP or AA-I at variable concentrations. (C) Urine from three subjects were incubated with 4-ABP (10 µM) or AA-I (0.1 µM) over T0, 10 or 24 h. N = 3 or 4 independent analyses for each subject. All measurements were reported as the mean ± SD.
Figure 4.
Extracted ion chromatograms at the MS3 scan stage and product ion spectra of dA-AL-I of DNA adducts of 4-ABP and AA-I isolated from RT4 cells incubated in urine of a donor at room temperature for 24 h. [13C10]-dG-C8–4-ABP and [15N5]-dA-AL-I were spiked into 5 µg DNA at a level of one adduct per 107 bases. The amount of DNA injected was ~1.6 µg.
The effect of the cell number on the stability of DNA adducts in RT4 cells in urine over time.
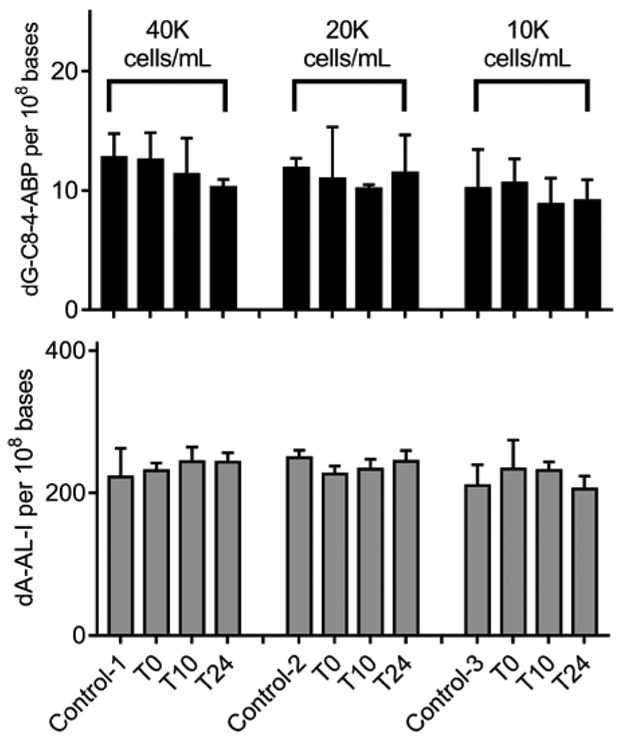
The levels of EUCs in urine of healthy male subjects can range from < 5,000 up to 600,000 cells per 40 mL urine.37 The population of EUCs in urine can be influenced by the diet and consumption of water, urine osmality, the pH of urine, and urination frequency.44,47 We observed that the levels of dG-C8–4-ABP and dA-AL-I adducts were comparable for all populations of RT4 cells studied in urine (1 × 104 - 4 × 104 cells per 40 mL of urine). (Figure 5).
Figure 5. The effect of cell number on the stability of DNA adducts of RT4 cells in urine.
RT4 cells pretreated with 4-ABP and AA-I were spiked in urine of a single donor at cell counts ranging between 1 × 104 and 4 × 104 cells per mL of urine. The pretreated RT4 cells were incubated at room temperature and DNA adduct levels were measured at T 0, 10, and 24 h. All measurements were reported as the mean ± SD.
Monitoring DNA adducts of AA in EUCs of the mouse model.
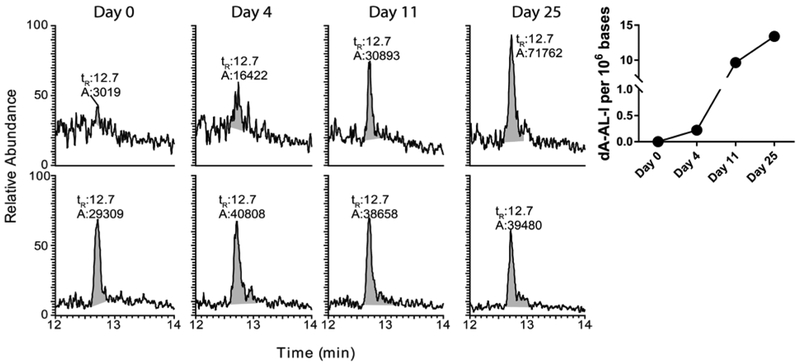
The UPLC/MS3 chromatograms and the kinetics of dA-AL-I elimination in EUCs of mice following a single dose of AA-I (1 mg/kg) are shown in Figure 6. dA-AL-I in EUCs was successfully measured with <5 mL of pooled mouse urine. The product ion spectrum of dA-AL-I acquired from DNA of EUCs at day 25 post-dosing is shown in supporting information (Figure S-1). The dA-AL-I adduct was detected in EUCs of mice four days post-treatment of AA-I, and the levels of dA-AL-I in EUCs continued to increase over the 25 days post-dosing period. The dA-AL-I adduct is likely derived from urothelial cells shed from the upper and lower urinary tracts and tubular epithelial cells from the kidney.20,21 The kinetics and the time course of appearance of EUCs harboring dA-AL-I adducts are consistent with the kinetics of urinary biomarkers of kidney injury following a single dose of AA-I in mice.48 Urinary biomarkers of kidney injury and dysfunction have been examined in AA-I-sensitive and AA-I resistant mouse strains.48 In the AA-sensitive mouse strain, such markers appear in urine two to four days after treatment with a single dose of AA-I and return to pre-treatment levels over the following week. However, for the C57Bl/6J mouse, a resistant strain, renal injury biomarkers are difficult to detect.48 The appearance of dA-AL-I adducts in the urine of the C57Bl/6J mouse four days post-treatment is consistent with the appearance of renal-specific injury markers, but dA-AL-I adducts levels continue to rise and remain detectable four weeks after treatment with AA-I (Figure 6). Thus, the dA-AL-I in EUCs is a superior biomarker revealing exposure, even for the C57Bl/6J mouse normally resistant to AA-induced injury, for a much longer time interval than conventional renal injury biomarkers. These findings recapitulate our pilot study in humans, where the dA-AL-I was detected in EUCs up to 3 months after the discontinued use of herbal medicines.31 Our analytical method and level of sensitivity are vastly superior to that previously reported, where 3 liters of pooled urine from rats dosed daily at 10 mg of AA-I /kg/day for a month was required to detect dA-AL-I in EUCs.36
Figure 6. The level of dA-AL-I recovered from exfoliated urinary cells of mice treated with AA-I.
The UPLC/MS33 total ion chromatogram and the kinetics of dA-AL-I recovered in EUCs of the mouse following a single dose of AA-I (1 mg/kg). Urine was collected on days 0, 4, 11, and 25 after dosing.
CONCLUSIONS
The employment of EUCs as biospecimens to noninvasively screen for DNA adducts formed by renal and bladder carcinogens in rodents and humans using specific and quantitative LC-MS methods is highly promising. Our data show that RT4 cells collected in urine are viable and that DNA adducts of two important urinary carcinogens in RT4 cells are stable for 24 hours. Two recent studies have reported that EUCs of healthy adults are viable in urine stored at room temperature for at least 24 hours49,50 Thus, urine collected for 8 to 24 hours can be employed to recover sufficient number of EUCs for DNA adduct measurements. We previously showed in a pilot study that dA-AL-I could be measured in EUCs of patients with renal disease, who had ingested traditional Chinese herbs containing AA-I.31 The LOQ value of dA-AL-I approached 3 adducts per 109 bases when assaying 2 µg of DNA per injection.31,33 UPLC-ESI/MS3 was performed with a capillary column at a flow rate of 5 μL/min and employed a CaptiveSpray ion source. The same UPLC-MS configuration was used in this current study. If necessary, our level of sensitivity can be further improved by employing nano-flow chromatography and a nanoESI source as described by the Vouros laboratory.30 With improved sensitivity, the time period of urine collection can be reduced and lower amounts of EUC37 may be used for screening DNA adducts in humans. As a first approach, we will extend our studies to EUCs of mice exposed to 4-ABP and other aromatic amine bladder carcinogens found in tobacco smoke, and then conduct analyses of EUCs in urine of smokers to determine if such DNA adducts are detected by UPLC-ESI/MS3. The success of these measurements can pave the way to screen EUCs for DNA adducts of other chemicals that may contribute to BC.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by R01ES019564 (R.J.T) from National Institute of Environmental Health Sciences, and R01CA220367 (R.J.T and T.A.R.) from the National Cancer Institute, National Institutes of Health Sciences. Mass spectrometry was carried out in the Analytical Biochemistry Share Resources of the Masonic Cancer Center, University of Minnesota, funded in part by Cancer Center Support Grant CA-077598. We thank Ms. Lihua Yao and Mr. Sesha Krishnamachari, University of Minnesota, for their technical assistance.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: the method of DNA isolation from RT4 cells in urine (Protocol S-1) and the product ion spectra of dA-AL-I recovered from mouse urinary cells (Figure S-1)
CONFLICT OF INTEREST DISCLOSURE
The authors have no conflict of interest to disclose.
REFERENCES
- (1).American Cancer Society, Cancer Facts & Figures 2017; American Cancer Society: Atlanta, 2017. [Google Scholar]
- (2).Yu MC; Skipper PL; Tannenbaum SR; Chan KK; Ross RK Mutat.Res 2002, 506–507, 21–28. [DOI] [PubMed] [Google Scholar]
- (3).International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Tobacco smoking Lyon, France, 1986; Vol. 38. [Google Scholar]
- (4).International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Personal habits and indoor combustions: Lyon, France, 2012; Vol. 100E. [PMC free article] [PubMed] [Google Scholar]
- (5).Grollman AP; Shibutani S; Moriya M; Miller F; Wu L; Moll U; Suzuki N; Fernandes A; Rosenquist T; Medverec Z; Jakovina K; Brdar B; Slade N; Turesky RJ; Goodenough AK; Rieger R; Vukelic M; Jelakovic B Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).International Agency for Research on Cancer. IARC Monographs on the evaluation of carcinogenic risks to humans: Some traditional herbal medicines, some mycotoxins, naphthalene and styrene.: Lyon, France, 2002; Vol. 82. [PMC free article] [PubMed] [Google Scholar]
- (7).Poon SL; Huang MN; Choo Y; McPherson JR; Yu W; Heng HL; Gan A; Myint SS; Siew EY; Ler LD; Ng LG; Weng WH; Chuang CK; Yuen JS; Pang ST; Tan P; Teh BT; Rozen SG Genome medicine 2015, 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hecht SS Carcinogenesis 2002, 23, 907–922. [DOI] [PubMed] [Google Scholar]
- (9).Doolittle DJ; Rahn CA; Burger GT; Lee CK; Reed B; Riccio E; Howard G; Passananti GT; Vesell ES; Hayes AW Food Chem. Toxicol 1989, 27, 657–666. [DOI] [PubMed] [Google Scholar]
- (10).Smith CJ; McKarns SC; Davis RA; Livingston SD; Bombick BR; Avalos JT; Morgan WT; Doolittle DJ Mutat. Res 1996, 361, 1–9. [DOI] [PubMed] [Google Scholar]
- (11).DeMarini DM; Hastings SB; Brooks LR; Eischen BT; Bell DA; Watson MA; Felton JS; Sandler R; Kohlmeier L Mutat.Res 1997, 381, 83–96. [DOI] [PubMed] [Google Scholar]
- (12).Peters U; Sinha R; Bell DA; Rothman N; Grant DJ; Watson MA; Kulldorff M; Brooks LR; Warren SH; DeMarini DM Environ.Mol.Mutagen 2004, 43, 53–74. [DOI] [PubMed] [Google Scholar]
- (13).Grimmer G; Dettbarn G; Seidel A; Jacob J Sci.Total Environ 2000, 247, 81–90. [DOI] [PubMed] [Google Scholar]
- (14).Turesky RJ; Le Marchand L Chem. Res. Toxicol 2011, 24, 1169–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nakajima M; Itoh M; Sakai H; Fukami T; Katoh M; Yamazaki H; Kadlubar FF; Imaoka S; Funae Y; Yokoi T Int. J. Cancer 2006, 119, 2520–2526. [DOI] [PubMed] [Google Scholar]
- (16).Jarabek AM; Pottenger LH; Andrews LS; Casciano D; Embry MR; Kim JH; Preston RJ; Reddy MV; Schoeny R; Shuker D; Skare J; Swenberg J; Williams GM; Zeiger E Crit. Rev. Toxicol 2009, 39, 659–678. [DOI] [PubMed] [Google Scholar]
- (17).Talaska G; al Juburi AZ; Kadlubar FF Proc. Natl. Acad. Sci. U.S.A 1991, 88, 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lee HW; Wang HT; Weng MW; Hu Y; Chen WS; Chou D; Liu Y; Donin N; Huang WC; Lepor H; Wu XR; Wang H; Beland FA; Tang MS Oncotarget 2014, 5, 3526–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Loktionov A In Encyclopedia of Cancer, Schwab M, Ed.; Springer Berlin; Heidelberg: Berlin, Heidelberg, 2011, pp 1357–1360. [Google Scholar]
- (20).Dorrenhaus A; Muller JI; Golka K; Jedrusik P; Schulze H; Follmann W Arch. Toxicol 2000, 74, 618–626. [DOI] [PubMed] [Google Scholar]
- (21).Ringsrud KM Lab. Med 2001, 32, 153–155. [Google Scholar]
- (22).Crallan RA; Georgopoulos NT; Southgate J Carcinogenesis 2006, 27, 374–381. [DOI] [PubMed] [Google Scholar]
- (23).Reali D; Dimarino F; Bahramandpour S; Carducci A; Barale R; Loprieno N Mutat. Res 1987, 192, 145–149. [DOI] [PubMed] [Google Scholar]
- (24).Fontana L; Lasfargues G; Ughetto S; Rogier S; Masdieu E; Lafaure M; Aublet-Cuvelier B; Catilina P Mutagenesis 2001, 16, 449–452. [DOI] [PubMed] [Google Scholar]
- (25).Tian D; Ma H; Feng Z; Xia Y; Le XC; Ni Z; Allen J; Collins B; Schreinemachers D; Mumford JL J. Toxicol. Environ. Health A 2001, 64, 473–484. [DOI] [PubMed] [Google Scholar]
- (26).Talaska G; Schamer M; Skipper P; Tannenbaum S; Caporaso N; Kadlubar F; Bartsch H; Vineis P Environ. Health Perspect 1993, 99, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Talaska G; Schamer M; Skipper P; Tannenbaum S; Caporaso N; Unruh L; Kadlubar FF; Bartsch H; Malaveille C; Vineis P Cancer Epidemiol. Biomarkers Prev 1991, 1, 61–66. [PubMed] [Google Scholar]
- (28).Rothman N; Bhatnagar VK; Hayes RB; Zenser TV; Kashyap SK; Butler MA; Bell DA; Lakshmi V; Jaeger M; Kashyap R; Hirvonen A; Schulte PA; Dosemeci M; Hsu F; Parikh DJ; Davis BB; Talaska G Proc. Natl. Acad. Sci. U.S.A 1996, 93, 5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kaderlik KR; Talaska G; DeBord DG; Osorio AM; Kadlubar FF Cancer Epidemiol. Biomarkers Prev 1993, 2, 63–69. [PubMed] [Google Scholar]
- (30).Randall KL; Argoti D; Paonessa JD; Ding Y; Oaks Z; Zhang Y; Vouros P J Chromatogr A 2010, 1217, 4135–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yun BH; Sidorenko V; Rosenquist TA; Dickman KG; Grollman AP; Turesky RJ Toxicol. Res 2015, 4 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Reshetnikova G; Sidorenko VS; Whyard T; Lukin M; Waltzer W; Takamura-Enye T; Romanov V Exp Cell Res 2016, 349, 101–108. [DOI] [PubMed] [Google Scholar]
- (33).Yun BH; Rosenquist TA; Sidorenko V; Iden CR; Chen CH; Pu YS; Bonala R; Johnson F; Dickman KG; Grollman AP; Turesky RJ Chem. Res. Toxicol 2012, 25, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Jones CR; Sabbioni G Chem.Res.Toxicol 2003, 16, 1251–1263. [DOI] [PubMed] [Google Scholar]
- (35).Xiao S; Guo J; Yun BH; Villalta PW; Krishna S; Tejpaul R; Murugan P; Weight CJ; Turesky RJ Anal. Chem 2016, 88, 12508–12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Guo L; Wu H; Yue H; Lin S; Lai Y; Cai ZJ Chromatogr.B Analyt. Technol .Biomed. Life Sci 2011, 879, 153–158. [DOI] [PubMed] [Google Scholar]
- (37).Wiggins R; Horner PJ; Whittington K; Holmes CH BMC research notes 2009, 2, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Baker SC; Arlt VM; Indra R; Joel M; Stiborova M; Eardley I; Ahmad N; Otto W; Burger M; Rubenwolf P; Phillips DH; Southgate J Mol Carcinog 2018, 57, 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liu S; Wang Y Chem Soc Rev 2015, 44, 7829–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Swaminathan S; Torino JL; Burger MS Mutat Res 2002, 499, 103–117. [DOI] [PubMed] [Google Scholar]
- (41).Plottner S; Bastian LA; Kafferlein HU; Bruning T J Toxicol Environ Health A 2016, 79, 1106–1117. [DOI] [PubMed] [Google Scholar]
- (42).Sanchez-Carbayo M; Socci ND; Charytonowicz E; Lu M; Prystowsky M; Childs G; Cordon-Cardo C Cancer Res 2002, 62, 6973–6980. [PubMed] [Google Scholar]
- (43).Bessette EE; Spivack SD; Goodenough AK; Wang T; Pinto S; Kadlubar FF; Turesky RJ Chem. Res. Toxicol. 2010, 23, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Mundt LA; Shanahan K Graff’s Textbook of Urinalysis and Body Fluids, 3rd ed.; Lippincott Williams & Wilkins: Philadelphia, 2015, p 352. [Google Scholar]
- (45).Liu H; Tan Q; Geddie WR; Jewett MA; Phillips N; Ke D; Simmons CA; Sun Y Cell Biochem. Biophys 2014, 68, 241–246. [DOI] [PubMed] [Google Scholar]
- (46).Corry WD; Meiselman HJ Biophys. J 1978, 21, 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Belik R; Follmann W; Degen GH; Roos PH; Blaszkewicz M; Knopf HJ; Golka K J Toxicol Environ Health A 2008, 71, 923–929. [DOI] [PubMed] [Google Scholar]
- (48).Rosenquist TA Am. J. Physiol. Renal Physiol 2011, 300, F1360–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ercan M; Akbulut ED; Abusoglu S; Yilmaz FM; Oguz EF; Topcuoglu C; Oztekin V; Bogdaycioglu N Clin Biochem 2015, 48, 919–922. [DOI] [PubMed] [Google Scholar]
- (50).Eksioglu MK; Madenci OC; Yucel N; Elci A; Turhan B; Orhan G; Orcun A Biochem Med (Zagreb) 2016, 26, 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.