Abstract
Background:
Elevated tumor infiltrating lymphocytes (TIL) within the tumor microenvironment is a known positive prognostic factor in colorectal cancer (CRC). We hypothesized that since cytotoxic T cells release cytolytic proteins such as Perforin (PRF1) and pro-apoptotic Granzymes (GZMA) to attack cancer cells, Cytolytic Activity Score (CAS) would be a useful tool to assess anticancer immunity.
Methods:
Genomic expression data was obtained from 456 patients from The Cancer Genome Atlas (TCGA). CAS was defined by GZMA and PRF1 expression. CIBERSORT was used to evaluate intra-tumoral immune cell composition.
Results:
High CAS was associated with high microsatellite instability (MSI-H), as well as high levels of activated memory CD4+ T cells, gamma delta T cells and M1 macrophages. CAS-high CRC patients had improved OS (p=0.019) and DFS (p=0.016) compared to CAS-low, especially in TIL-positive tumors. Multivariate analysis demonstrated that CAS-high associates with improved survival independently after controlling for age, lymphovascular invasion, colonic location, microsatellite instability and TIL positivity. The levels of immune checkpoint molecules (ICM)- PD-1, PD-L1, CTLA4, LAG-3, TIM3 and IDO1 correlated significantly with CAS (p<0.0001); with improved survival in CAS-high, ICM-low patients and poorer survival in ICM-high patients.
Conclusions:
High CAS within CRC associated with improved survival likely due to increased immunity and cytolytic activity of T cells and M1 macrophages. High CAS also associated with high expression of immune checkpoint molecules, thus further studies to elucidate the role of CAS as a predictive biomarker of the efficacy of immune checkpoint blockade are warranted.
Keywords: Colon cancer, Immune cytolytic activity, Immunity, Prognosis
Introduction
Colorectal cancer (CRC) is the fourth most common cause of cancer related death 1,2. Impaired DNA repair mechanisms and genomic instability are key factors in carcinogenesis via the accumulation of somatic mutations within DNA 2,3. This accrual of DNA mutations, can in turn, promote neoantigens (immunogenic tumor mutated peptides), which attract increased numbers of lymphocytes and other immune cells into the tumor microenvironment 3-5. A high degree of tumor infiltrating lymphocytes (TILs) has been identified as a positive prognostic factor for CRC with decreased tumor invasiveness, less nodal involvement, earlier stage tumors and improvement in survival 6-10. High levels of CD8+ (cytotoxic) TILs have also been noted to be good markers of response to neoadjuvant chemotherapy 4. Thus, the current dogma is that cancer biology and patient outcomes are at least partly determined by immune cells within the tumor microenvironment. We hypothesized that it is not merely the existence of the immune cells, but rather their anti-tumoral cytolytic activity which translate to patient outcomes.
The granzyme-perforin pathway is a primary method by which cytotoxic lymphocytes destroy cancer cells 11. Perforin creates pores within the target cell membrane and mediates the entry of granzymes, which are tryptases that cleave caspases and induce apoptosis 11,12. Rooney etal. devised a quantitative measure of immune cytolytic activity based on transcript levels of granzyme A (GZMA) and perforin (PRF1), which are upregulated with cytotoxic T cell activation and can be used to calculate a Cytolytic Activity Score (CAS) 13.
We hypothesized that tumors with higher mutation load and a greater degree of intratumoral immunogenicity would be associated with higher CAS, which would in turn, result in improved patient survival.
Materials and Methods
Patient cohort and genomic data processing
Both the clinical and the genomic data were acquired from The Cancer Genome Atlas (TCGA) CRC cohort within the Genomic Data Common (GDC) data portal through R/Bioconductor package “TCGAbiolinks” in September 2017 14. Samples without gene expression data were excluded. The data used included 456 tumor samples with 39 matched normal samples. Somatic mutation data of 417 tumor samples were also retrieved in the Mutation Annotation Format (MAF) aligned against hg19 15,16.
Gene expression data were obtained in RSEM format from GDC and converted to Transcripts Per Million (TPM) by a given gene’s estimated fraction of transcripts and multiplying with 106. The immune cytolytic activity score (CAS) was defined as the geometric mean of GZMA and PRF1 expression values in TPM. The threshold of dichotomization of CAS high and low groups was determined by comparing differences in the overall survival between the two groups at multiple candidate cutoff points within the range of CAS, and the optimal cutoff point gave the most significant results was chosen. The classification of high and low groups for immune-related genes was also determined by this running Cox proportional hazard model 17.
CIBERSORT 18 deconvolution algorithm was used to estimate the fraction of twenty-two immune cell types in each tumor tissue to evaluate intra-tumor immune cell composition. The twenty-two cell fractions were calculated via their online calculator (https://cibersort.stanford.edu/).
Microsatellite instability (MSI) status: MSI-high (MSI-H) and microsatellite stable (MSS) for 291 CRC subjects were obtained from a genomic classifier including >200,000 microsatellite loci created by Hause et al. 19. This study was deemed exempt from Institutional Review Board evaluation because all information within TCGA is publicly accessible and de-identified 20,21,22,23.
Statistical analysis
All statistical analyses were performed using R software (https://www.r-project.org/) and Bioconductor (https://www.bioconductor.org/). Overall survival (OS) was defined as the time from date of diagnosis to death, while disease-free survival (DFS) was defined as the time from date of diagnosis to date of first recurrence or date of death when no recurrence. Kaplan-Meier method with log-rank test and Cox proportional hazard models were used to compare survival curves between groups. Hazard ratios (HR) and 95% confidence intervals (CI) were provided when fitting the multivariate Cox models of CAS and other clinicopathological features. Through literature review, we identified six clinical factors (Age, Gender, Lymphatic invasion, Venous invasion, Tumor location, Stage) and two molecular factors (MSI and TIL status) which we used to perform a univariate analysis based on the cox proportional hazard model for overall survival with each clinical variable as covariates. Clinical variables which showed significance were then selected as covariates for multivariate analysis.
For continuous variables, the differences between two groups were assessed by Mann–Whitney test; for discrete variables, chi-square tests were used to evaluate the association between factors. Spearman correlation was used to describe the relationship between gene expressions and CAS. Unadjusted p-values were determined to be appropriate for this exploratory study. In all analyses, a two-sided p<0.05 was considered statistically significant.
Results
CAS was Low in Cancer Tissue and High in Microsatellite Unstable CRC with High Mutation Load
Of the 456 patient TCGA cohort, 221 patients (48.5%) were CAS-low and 235 patients (51.5%) were CAS-high. There was a normal distribution of cytolytic molecules throughout the cohort of CRC patients in TCGA (Supplementary Figure S1).
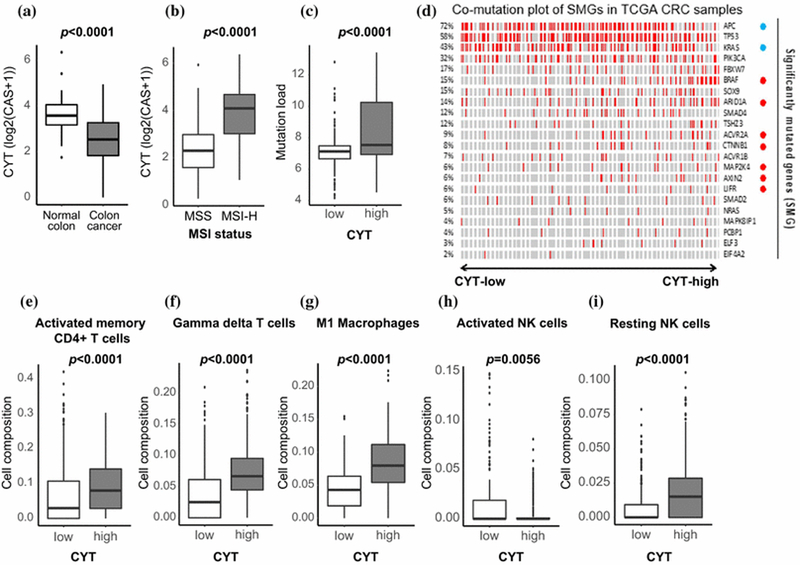
Since CAS is a quantitative measurement of immune cytolytic activity, it is expected to be high in tissues where cytotoxic T cells infiltrate and are activated. Indeed, CRCs were found to have lower CAS than normal colon (p<0.0001, Figure 1a). It was noted that MSI-H tumors associated with high CAS compared with MSS (p<0.0001, Figure 1b). We found that CAS-high tumors were associated with a higher mutational load than CAS-low (p<0.0001, Figure 1c). We further examined the association between CAS and significantly mutated genes (SMG) 24 in CRC. Interestingly, of the SMGs, frequently mutated genes (APC and KRAS) showed significantly negative association with CAS-high status, while several SMGs which less-frequent mutational events had showed significantly positive association with high CAS (Figure 1d).
Figure 1:
Association between CAS and mutation load, MSI status, and TILs. a) CAS expression in 39 matched samples of Colon cancer vs. Normal colonic tissue. b) CAS expression in Microsatellite Instability-High (MSI-H) vs. Microsatellite stable (MSS). c) Assessment of mutational load in CAS-high vs. CAS-low tumors. d) Association between CAS and mutational events within SMGs in CRC; blue circles (APC and KRAS), the mutations significantly correlated with CAS-low (p<0.05); red circles (BRAF, ARID1A, ACVR2A, CTNNB1, MAP2K4, AXIN2, and LIFR), mutations significantly correlated with CAS-high (p<0.05); red bar, each mutational event (left column, mutation rate %).
CAS-high vs. low in e) Activated memory CD4+ T-cells, f) Gamma-Delta T-cells, g) M1 Macrophages, h) Activated Natural Killer (NK) Cells and i) Resting NK Cells
CAS-high associates with Tumor Infiltrating Lymphocytes and Macrophages
Tumors with high CAS associated significantly with high levels of TIL including, activated memory CD4+ T cells, gamma-delta T-cells as well as M1 macrophages (p<0.0001). Interestingly, we did not note any elevation in CAS in CD8+ T cells (p=0.335). We also found that resting Natural Killer (NK) cells were significantly associated with being CAS-high (p<0.0001) and activated NK cells had greater association with CAS-low status (p=0.0056, Figure 1e-i).
Clinicopathologic Features of CAS-high versus CAS-low Patients
There was no difference in mean age, gender, and lymphovascular invasion between CAS-high and CAS-low patients (Table 1). More CAS-high patients had right sided CRC than left (p=0.01). CAS-high patients also had early stage (I and II) cancers compared with the CAS- low group (p=0.005). Univariate analysis (Table 2) determined that male gender, lymphatic and vascular invasion and stage IV disease adversely impacted survival. Whereas CAS-high status was a significant positive prognostic factor. In subsequent multivariate cox regression analysis (Table 2), only female gender and CAS-high status remained as factors associating with improved survival independently after controlling for age, lymphovascular invasion, colonic location, microsatellite instability and TIL positivity.
Table 1.
Demographic characteristics of two groups stratified by CAS status
| Variable | CAS low (n=220) |
CAS high (n=235) | p-value 1 |
|---|---|---|---|
| Age (mean) | 66.83 | 68.04 | 0.3348 |
| Gender (female/male) | 101/120 | 115/120 | 0.5501 |
| Lymphatic invasion (no/yes) | 114/81 | 134/83 | 0.5617 |
| Venous invasion (no/yes) | 144/47 | 155/49 | 0.9851 |
| Location (left or other/right) | 121/95 | 97/127 | 0.0101 |
| Stage (I/II/III/IV) | 31/78/65/44 | 44/99/63/21 | 0.0052 |
| MSI status (MSS/MSI-H) | 132/9 | 103/47 | <0.0001 |
| TIL (positive/negative) | 107/113 | 107/128 | 0.5693 |
Bold, significant difference
Table 2.
CAS-high is a positive prognostic factor independent of age, lymphovascular invasion, colonic location, microsatellite instability and TIL positivity
| Variable | Univariate prognostic analysis (Log rank) |
Multivariate prognostic analysis (cox regression) |
||||
|---|---|---|---|---|---|---|
| HR1 | 95% CI2 | p-value | HR1 | 95% CI2 | p-value | |
| Age >60 (vs. <60) | 1.528 | (0.7737, 3.0176) | 0.2221 | |||
| Gender male (vs. female) | 2.0026 | (1.0844, 3.6980) | 0.0265 | 1.9088 | (1.0247, 3.5556) | 0.0417 |
| Lymphatic invasion positive (vs. negative) | 2.2936 | (1.2748, 4.1265) | 0.0056 | 1.2999 | (0.5669, 2.9807) | 0.5356 |
| Venous invasion positive (vs. negative) | 2.237 | (1.2214, 4.0972) | 0.0091 | 1.6397 | (0.7215, 3.7268) | 0.2378 |
| Location right (vs. center or other) | 1.5044 | (0.8276, 2.7349) | 0.1805 | |||
| Stage II (vs. stage I) | 1.6351 | (0.4644, 5.7564) | 0.4439 | 1.5327 | (0.4335, 5.4183) | 0.5075 |
| Stage III (vs. stage I) | 2.9149 | (0.8476, 10.0235) | 0.0896 | 2.2915 | (0.6476, 8.1076) | 0.1984 |
| Stage IV (vs. stage I) | 6.6306 | (1.8840, 23.3353) | 0.0032 | 3.7305 | (0.9911, 14.0420) | 0.0516 |
| MSI-H (vs. MSS) | 0.8513 | (0.3958, 1.8311) | 0.6804 | |||
| TIL positive (vs. negative) | 1.0619 | (0.5908, 1.9085) | 0.8409 | |||
| CAS high (vs. low) | 0.4626 | (0.2457, 0.8710) | 0.017 | 0.513 | (0.2634, 0.9993) | 0.0498 |
HR, hazard ratio;
95% confidence interval; Bold, significant difference
CAS-high Patients have Improved Survival
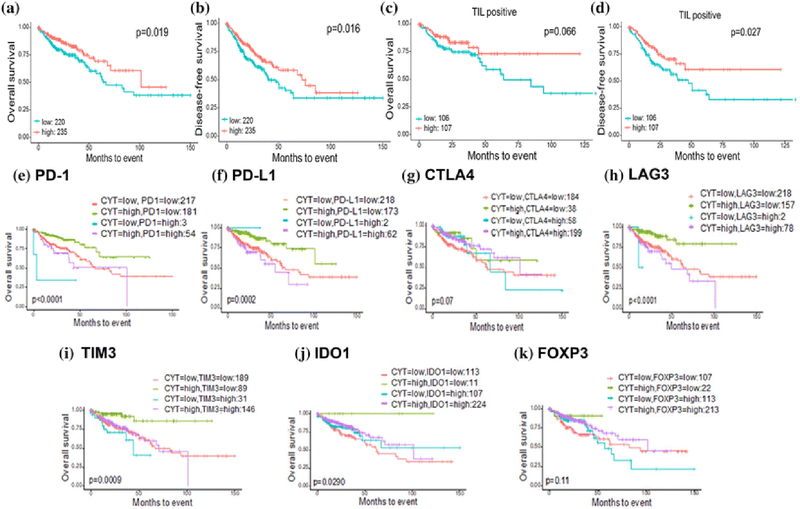
We found that CAS-high patients have significantly improved median and 5-year OS and DFS compared from CAS-low patients (Figure 2a-b). Median OS for the CAS-high cohort was 101.4 months compared to 66.7 months in CAS-low. 5-year OS was also significantly better in CAS-high compared with CAS-low patients (69.1% vs. 57.9%, p=0.019). A significant difference in median DFS was also seen between CAS-high and CAS-low patients (72.5 vs. 40.5 months). 5-year DFS was 58.4% for CAS-high patients compared to 40.8% in CAS-low (p=0.016).
Figure 2:
Survival analysis of CAS in CRC patients. Kaplan-Meier (KM) curve of a) Overall Survival and b) Disease-Free Survival in CAS-high vs. CAS-low. KM curves of c) Overall Survival and d) Disease-Free survival stratified by CAS-high and CAS-low in TIL positive patients. “TIL positivity” is determined by higher CD8+ T-cells composition than the median value within the TCGA CRC cohort. CAS and its association with immune checkpoint molecules: e) PD-1, f) PD-L1, g) CTLA4, h) LAG3, i) TIM3, and j) IDO1 and k) Treg marker FOXP3 and their effect on patient survival. High, high expression of each gene; low, low expression of each gene.
We especially noted an improvement in survival in the CAS-high patients who were also found to be TIL positive, as determined by higher CD8+ T cell composition than the mean value within the TCGA CRC cohort (Figure 2c-d). Median OS was not reached at 125 months in the CAS-high group compared to 63.7 months in the CAS–low group even when all included patients had TIL positive tumors (p=0.066). Similarly, median DFS in TIL positive CAS-high patients was also not reached in this time period compared to 47.4 months in the CAS-low group (p=0.027). Conversely, patients who were TIL negative did not show significant improvement in OS and DFS with CAS-high (Supplementary Figure S2).
The effect of CAS and its association with Immune Checkpoint Molecules on Patient Survival
We found that high expression of several immune checkpoint molecules (ICM) directly associates with high CAS with differences in survival based on their expression; these were PD- 1, PD-L1, CTLA4, LAG3, TIM3 and IDO1 (Figure 2e-k). We found that patients with high CAS and low ICM expression had significantly superior survival to other patients in the cohort. None of these patients had median OS reached at 150 months- PD-1 (p<0.0001), PD-L1 (p=0.0002), CTLA4 (p=0.07), LAG3 (p<0.0001), TIM3 (p=0.0009) and IDO1 (p=0.029). Poorer prognosis was seen in the CAS-low groups with several groups demonstrating the poorest survival in patients with low CAS and high ICM. However, it is difficult to draw conclusions given the low number of patients in this group. Patients who were CAS-high and ICM-high had variable median OS (in months); PD-1 (43.5), PD-L1 (55.4), CTLA4 (101.4), LAG3 (50.1), TIM3 (71.1) and IDO1 (101.4). A similar association was seen with regulatory T-cell (Treg) marker FOXP3-median OS not reached at 150 months in CAS-high, FOXP3-low and 57 months in CAS-low, FOXP3- high (p=0.11, Figure 2i).
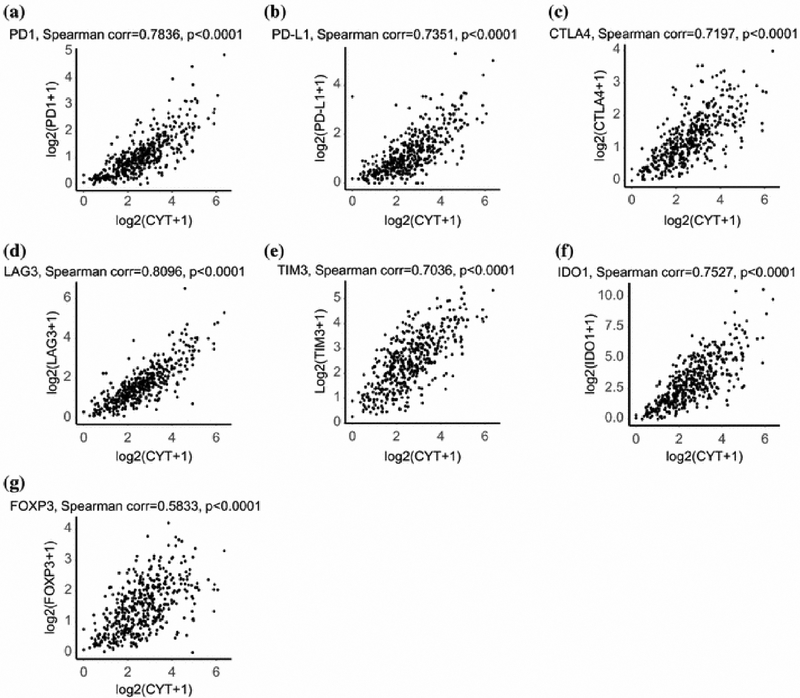
CAS Correlates with expression of Immune Checkpoint Molecules
We found that high expression of the most common ICM (PD-1, PD-L1, CTLA4, LAG3, TIM3 and IDO1) directly correlated with high CAS (Spearman corr. 0.70-0.81, p<0.0001, Figure 3a-f). A similar correlation was seen between high CAS and high expression of FOXP3 (Spearman corr. 0.58, p<0.0001, Figure 3g). This same trend was noted with lesser known checkpoint molecules IDO2, TIGIT, A2AR, VISTA, and an inhibitory checkpoint index (described by Balli etal.) 12 created to assess the expression of key checkpoint molecules across CRC patients, as well as Treg markers CCR4, CCR5 and IL2RA (p<0.0001, Supplementary Figure S3).
Figure 3:
Correlation between CAS and immune checkpoint molecules and Treg marker. Scatter plot by Spearman’s correlation test between CAS and a) PD-1, b) PD-L1, c) CTLA4, d) LAG3, e) TIM3, f) IDO1, and g) FOXP3. X-axis, log2(CAS+1); y-axis, log2(each gene+1).
Discussion
The concept of immunoediting has become increasingly important in understanding the immune system’s ability to control tumor growth and spread in a variety of cancer types 25. Higher infiltration of lymphocytes and macrophages into the tumor microenvironment is known to associate with favorable outcomes in several cancers including CRC 25. We hypothesized that improvement in patient outcomes with TILs are related to increased anticancer cytolytic activity, which we are able to quantitate by CAS, rather than TIL numbers. CAS associated with MSI-H tumors, mutational load and TIL infiltration, but not with mutations in KRAS or APC, likely due to their lack of association with MSI or TIL. We also found that CAS-high patients demonstrated significant improvement in OS and DFS compared to CAS-low patients (especially in TIL- positive patients), an association never before reported in CRC.
We found that MSI-H tumors associated significantly with high CAS compared to MSS tumors. It is known that MSI-H CRC has 10 to 100 times more somatic mutations than MSS, with prominent lymphocytic infiltrate into these tumors due to increased neoantigen load 4,5,13,26,27. Since CAS measures intra-tumoral immune cytolytic activity, it makes sense to see high CAS associating with high TIL conditions such as MSI-H CRC. Patients with MSI-H tumors have also been found in several studies to have improved survival compared to their MSS counterparts 1,5. This may be attributed to the elevation in cytolytic activity within their cancer cells secondary to increased TIL infiltration.
We also found associations between certain immune cells and elevated CAS likely due to their status as cytolytic effectors. CAS-high tumors have more infiltration of activated memory CD4+ T cells whereas they did not for resting memory CD4+ T-cells or naïve CD4+ T-cells. This is likely secondary to re-activation of these CD4+ memory T-cells by tumor antigens resulting in stimulation of effector cytotoxic T cells and elevated CAS 28,29. Gamma-delta T-cells are a subset of cytotoxic T-cells which produce TNF-α resulting in cell mediated lysis of tumor cells and had more infiltration into CAS-high tumors within our analysis 30. M1 macrophages were also identified as being associated with CAS-high tumors; likely due to their known role in inflammatory response and anti-tumor immunity via the release of pro-inflammatory cytokines such as TNF-α 31. Conversely, no increase in M2 macrophages was seen in CAS-high tumors, probably secondary to their function in tumor progression 31. We also found an association between CAS-high tumors and resting NK cells as compared to activated NK cells. The reason for this is difficult to elucidate with CIBERSORT alone given that NK cells are known to be a highly diverse lymphocyte population with unique regulation and activation processes 32.
ICMs are an immune-inhibitory mechanism by which cancer cells may evade anti-tumor immune cells 33. PD-1 expressed on T cells is initially bound by its ligand (PD-L1), resulting in negative regulation of T cell anti-tumoral activity, deemed “T cell exhaustion” 6,33. Other ICMs include CTLA-4 which has been targeted by Ipilimumab in the treatment of metastatic melanoma, LAG3, TIM3 and IDO1 34-36. In our analyses, higher expression of ICMs: PD-1, PD- L1, CTLA4, LAG3, TIM3 and IDO1 were directly correlated with higher CAS. Balli etal. In their evaluation of CAS in pancreatic adenocarcinoma found that CAS-high tumors were concurrently enriched with activated CD8+, PD1-high T-cells 12. These findings indicate that elevation of cytotoxic TILs with higher CAS result in a contemporaneous up-regulation of ICMs that function as “brakes”.
Higher levels of Treg marker FOXP3 were also directly related to high CAS. Treg are CD4+ T-cells which inhibit the function of tumor specific T-cells by expressing immunosuppressive cytokines such as IL-10 and TGF-ß as well as ICMs and have been determined to be independent predictors of poorer survival when found in the tumor immune micro-environment 37-39. A positive correlation has also been noted between CD8+ and FOXP3+ cell counts, indicating that tumors utilize Treg to counteract elevated cytotoxic T cells 40,41. It has been reported that poorer prognosis of tumors with a high density of Treg is likely a result of an unfavorable ratio to cytotoxic T lymphocytes rather than the number of Tregs alone 41. We similarly identified poorer survival with higher expression of ICMs and Treg which diminished the improvement in survival demonstrated in CAS-high patients.
The developments of immune checkpoint inhibitors have become increasingly popular in the treatment of a variety of cancer types 6,34,42,43. Le et al. in their phase II trial examining anti- PD-1 blockade found a significantly improved objective response rate and survival in MSI-H CRC compared to MSS with associated TIL elevation 44. The efficacy of these checkpoint inhibitors is likely correlated to their ability to obtain a cytolytic response as denoted by CAS. Given CAS’s correlation with ICM expression in this analysis, it is expected that CAS-high tumors may have a superior response to immune checkpoint inhibition. Thus, measurement of CAS could possibly be used as a predictive biomarker of the efficacy of immune checkpoint blockade or of a tumor’s possible responsiveness to immunotherapy. Interestingly, we found that CAS-high patients with high expression of ICM showed poorer survival. This may reflect the fact that these ICMs are also known as T-cell exhaustion markers which may determine survival despite concurrent cytolytic activity. This allows us to speculate that this population may benefit the most from immune checkpoint inhibition.
Some limitations in the present study included our inability to obtain precise information regarding patients’ co-morbid conditions and therapeutic interventions from TCGA. We also could not assess underlying molecular mechanisms given our bioinformatics approach.
Conclusions
CAS- high CRC is associated with improved OS and DFS likely due to enhanced anti- cancer immunity. This is especially true of tumors with increased mutation load and MSI which have increased cytolytic T cell and M1 macrophage burden. High CAS also associated with high expression of ICMs and Tregs likely due to concurrent elevation of cytolytic effectors. Thus, further studies to elucidate the role of CAS as a predictive biomarker of the efficacy of immune checkpoint blockade are warranted.
Supplementary Material
Supplementary Figure S1 Distribution of cytolytic molecules (GZMA and PFR1) throughout the cohort of CRC patients in TCGA. Gene expression was defined as log2(TPM + 1).
Supplementary Figure S2 Kaplan-Meier (KM) survival curves of a) Overall Survival and b) Disease-Free survival stratified by CAS-high and CAS-low in TIL negative patients. “TIL negativity” is determined by lower CD8+ T-cells composition than the median value within the TCGA CRC cohort.
Supplementary Figure S3 a-e) Correlation between CAS and immune checkpoint molecules; a) IDO2, b) TIGIT, c) A2AR, and d) VISTA, and e) inhibitory checkpoint index. The immune checkpoint index was generated by taking the log- average expression in TPM of the following molecules: PD1, PD-L1, CTLA4, A2AR TIM3, IDO1, IDO2, PD-L2, TIGIT, VISTA, and VTCN1. f-h) Correlation between CAS and Treg markers; f) CCR4, g) CCR5, and h) IL2RA.
Synopsis.
Cytolytic activity score associates with increased infiltration of lymphocytes and macrophages into the tumor immune microenvironment with improvement in survival.
Acknowledgement
Kazuaki Takabe is supported by NIH/NCI grant R01CA160688 and Susan G. Komen Investigator Initiated Research Grant IIR12222224, and also supported by Institutional Startup Grant 71-4085-01. This work was also supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Cancer Institute’s Bioinformatics and Biostatistics Shared Resources.
Footnotes
Disclosure
S. Narayanan, T. Kawaguchi, L. Yan, X. Peng, Q. Qi, K. Takabe have no potential conflicts of interest to declare.
References
- 1.Li P, Xiao Z, et al. A relationship to survival is seen by combining the factors of mismatch repair status, tumor location and age of onset in colorectal cancer patients. PLoS One 2017; 12:e0172799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marmol I, Sanchez-de-Diego C, et al. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci 2017; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chae YK, Anker JF, et al. Genomic landscape of DNA repair genes in cancer. Oncotarget 2016; 7:23312–23321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green AR, Aleskandarany MA, et al. Clinical Impact of Tumor DNA Repair Expression and T- cell Infiltration in Breast Cancers. Cancer Immunol Res 2017; 5:292–299. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, Powell AG, et al. Mismatch repair status in patients with primary operable colorectal cancer: associations with the local and systemic tumour environment. Br J Cancer 2016; 114:562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prall F, Huhns M. The PD-1 expressing immune phenotype of T cell exhaustion is prominent in the ‘immunoreactive’ microenvironment of colorectal carcinoma. Histopathology 2017; 71:366–374. [DOI] [PubMed] [Google Scholar]
- 7.Salama P, Phillips M, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol 2009; 27:186–192. [DOI] [PubMed] [Google Scholar]
- 8.Governa V, Trella E, et al. The Interplay Between Neutrophils and CD8(+) T Cells Improves Survival in Human Colorectal Cancer. Clin Cancer Res 2017; 23:3847–3858. [DOI] [PubMed] [Google Scholar]
- 9.Daster S, Eppenberger-Castori S, et al. High frequency of CD8 positive lymphocyte infiltration correlates with lack of lymph node involvement in early rectal cancer. Dis Markers 2014; 2014:792183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berntsson J, Nodin B, et al. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int J Cancer 2016; 139:1129–1139. [DOI] [PubMed] [Google Scholar]
- 11.Woodsworth DJ, Dreolini L, et al. Targeted Cell-to-Cell Delivery of Protein Payloads via the Granzyme-Perforin Pathway. Mol Ther Methods Clin Dev 2017; 7:132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balli D, Rech AJ, et al. Immune Cytolytic Activity Stratifies Molecular Subsets of Human Pancreatic Cancer. Clin Cancer Res 2017; 23:3129–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rooney MS, Shukla SA, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015; 160:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colaprico A, Silva TC, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016; 44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N, Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowley JLM, Jacobson J, Salmon S. Some exploratory tools for survival analysis. Lecture Notes on Statistics Proceedings of the First Seattle Symposium in Biostatistics: Survival Analysis. New York: Springer; 1997:199–229. [Google Scholar]
- 18.Newman AM, Liu CL, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hause RJ, Pritchard CC, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016; 22:1342–1350. [DOI] [PubMed] [Google Scholar]
- 20.Ramanathan R, Olex AL, et al. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat 2017; 162:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young J, Kawaguchi T, et al. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget 2017; 8:99978–99989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi T, Yan L, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Sci Rep 2017; 7:15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SY, Kawaguchi T, et al. Clinical Relevance of microRNA Expressions in Breast Cancer Validated Using the Cancer Genome Atlas (TCGA). Ann Surg Oncol 2017; 24:2943–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandoth C, McLellan MD, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markl B, Wieberneit J, et al. Number of Intratumoral T Lymphocytes Is Associated With Lymph Node Size, Lymph Node Harvest, and Outcome in Node-Negative Colon Cancer. Am J Clin Pathol 2016; 145:826–836. [DOI] [PubMed] [Google Scholar]
- 26.Giannakis M, Mu XJ, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 2016; 17:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peltomaki P DNA mismatch repair gene mutations in human cancer. Environ Health Perspect 1997; 105 Suppl 4:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLeod MK, Kappler JW, et al. Memory CD4 T cells: generation, reactivation and re- assignment. Immunology 2010; 130:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H, Zhang P, et al. Primary Tr1 cells from metastatic melanoma eliminate tumor-promoting macrophages through granzyme B- and perforin-dependent mechanisms. Tumour Biol 2017; 39:1010428317697554. [DOI] [PubMed] [Google Scholar]
- 30.Kiladjian JJ, Visentin G, et al. Activation of cytotoxic T-cell receptor gammadelta T lymphocytes in response to specific stimulation in myelodysplastic syndromes. Haematologica 2008; 93:381–389. [DOI] [PubMed] [Google Scholar]
- 31.Burmeister K, Quagliata L, et al. Vascular endothelial growth factor A amplification in colorectal cancer is associated with reduced M1 and M2 macrophages and diminished PD-1- expressing lymphocytes. PLoS One 2017; 12:e0175563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holder KA, Comeau EM, et al. Origins of natural killer cell memory: special creation or adaptive evolution. Immunology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee LH, Cavalcanti MS, et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol 2016; 29:1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pennock GK, Chow LQ. The Evolving Role of Immune Checkpoint Inhibitors in Cancer Treatment. Oncologist 2015; 20:812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toh JW, de Souza P, et al. The Potential Value of Immunotherapy in Colorectal Cancers: Review of the Evidence for Programmed Death-1 Inhibitor Therapy. Clin Colorectal Cancer 2016; 15:285–291. [DOI] [PubMed] [Google Scholar]
- 36.Prado-Garcia H, Romero-Garcia S, et al. The PD-L1/PD-1 pathway promotes dysfunction, but not “exhaustion”, in tumor-responding T cells from pleural effusions in lung cancer patients. Cancer Immunol Immunother 2017; 66:765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber RD, Old LJ, et al. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011; 331:1565–1570. [DOI] [PubMed] [Google Scholar]
- 38.Ali HR, Chlon L, et al. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med 2016; 13:e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katz SC, Bamboat ZM, et al. Regulatory T cell infiltration predicts outcome following resection of colorectal cancer liver metastases. Ann Surg Oncol 2013; 20:946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saigusa S, Toiyama Y, et al. Implication of programmed cell death ligand 1 expression in tumor recurrence and prognosis in rectal cancer with neoadjuvant chemoradiotherapy. Int J Clin Oncol 2016; 21:946–952. [DOI] [PubMed] [Google Scholar]
- 41.Gao Q, Qiu SJ, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol 2007; 25:2586–2593. [DOI] [PubMed] [Google Scholar]
- 42.Anderson AC. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol Res 2014; 2:393–398. [DOI] [PubMed] [Google Scholar]
- 43.Brahmer JR, Tykodi SS, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366:2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le DT, Uram JN, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015; 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Distribution of cytolytic molecules (GZMA and PFR1) throughout the cohort of CRC patients in TCGA. Gene expression was defined as log2(TPM + 1).
Supplementary Figure S2 Kaplan-Meier (KM) survival curves of a) Overall Survival and b) Disease-Free survival stratified by CAS-high and CAS-low in TIL negative patients. “TIL negativity” is determined by lower CD8+ T-cells composition than the median value within the TCGA CRC cohort.
Supplementary Figure S3 a-e) Correlation between CAS and immune checkpoint molecules; a) IDO2, b) TIGIT, c) A2AR, and d) VISTA, and e) inhibitory checkpoint index. The immune checkpoint index was generated by taking the log- average expression in TPM of the following molecules: PD1, PD-L1, CTLA4, A2AR TIM3, IDO1, IDO2, PD-L2, TIGIT, VISTA, and VTCN1. f-h) Correlation between CAS and Treg markers; f) CCR4, g) CCR5, and h) IL2RA.