Abstract
Four species of true crocodile (genus Crocodylus) have been described from the Americas. Three of these crocodile species exhibit non-overlapping distributions—Crocodylus intermedius in South America, C. moreletii along the Caribbean coast of Mesoamerica, and C. rhombifer confined to Cuba. The fourth, C. acutus, is narrowly sympatric with each of the other three species. In this study, we sampled 113 crocodiles across Crocodylus populations in Cuba, as well as exemplar populations in Belize and Florida (USA), and sequenced three regions of the mitochondrial genome (D-loop, cytochrome b, cytochrome oxidase I; 3,626 base pair long dataset) that overlapped with published data previously collected from Colombia, Jamaica, and the Cayman Islands. Phylogenetic analyses of these data revealed two, paraphyletic lineages of C. acutus. One lineage, found in the continental Americas, is the sister taxon to C. intermedius, while the Greater Antillean lineage is most closely related to C. rhombifer. In addition to the paraphyly of the two C. acutus lineages, we recovered a 5.4% estimate of Tamura-Nei genetic divergence between the Antillean and continental clades. The reconstructed paraphyly, distinct phylogenetic affinities and high genetic divergence between Antillean and continental C. acutus populations are consistent with interspecific differentiation within the genus and suggest that the current taxon recognized as C. acutus is more likely a complex of cryptic species warranting a reassessment of current taxonomy. Moreover, the inclusion, for the first time, of samples from the western population of the American crocodile in Cuba revealed evidence for continental mtDNA haplotypes in the Antilles, suggesting this area may constitute a transition zone between distinct lineages of C. acutus. Further study using nuclear character data is warranted to more fully characterize this cryptic diversity, resolve taxonomic uncertainty, and inform conservation planning in this system.
Keywords: Crocodylus acutus, American crocodile, mtDNA, Phylogeny, Cryptic species
Introduction
The order Crocodylia is currently comprised of 27 recognized or proposed living species distributed among three families (Alligatoridae, Crocodylidae, and Gavialidae) and nine genera (Alligator, Caiman, Melanosuchus, Paleosuchus, Crocodylus, Mecistops, Osteolaemus, Tomistoma, and Gavialis) representing, along with birds, the unique surviving members of the Archosauria (Grigg & Kirshner, 2015). The genus Crocodylus alone comprises almost 50% (12 out of 27) of all extant crocodylian species and is unique for its global distribution in the tropics and recent diversification (Oaks, 2011). Yet, ambiguity remains regarding the phylogenetic relationships within the genus Crocodylus (Meganathan et al., 2010; Man et al., 2011; Meredith et al., 2011; Hekkala et al., 2011; Oaks, 2011; Shirley et al., 2014). For example, the sister relationships of the four New World Crocodylus species have not been consistent and significantly vary depending on the origin of the species sampled (Milián-García et al., 2015, 2018).
Cuba represents an important area in the distribution of New World Crocodylus, given its geographic proximity to mainland North America (∼150 km from Florida, USA) and Central America (∼210 km from the Yucatán peninsula, Mexico) and the fact that it hosts two Crocodylus species. The Cuban crocodile (Crocodylus rhombifer), endemic to the main island of Cuba, is largely restricted to Zapata Swamp (Matanzas Province) where it is sympatric with C. acutus. In most other coastal areas in Cuba, including Birama Swamp in the southeast and the Guanahacabibes´ peninsula (Pinar del Río province) on the western tip, C. acutus is found in allopatry with no evidence of historical overlap with C. rhombifer (Tabet et al., 2014). Previous studies have indicated that the phylogenetic placement of C. acutus is sensitive to sampling location, with most studies relying solely on individuals sampled from continental populations in Central and/or South America (Meredith et al., 2011; Milián-García et al., 2011, 2015; Oaks, 2011). In these cases, a sister taxon relationship between C. acutus and C. intermedius has been consistently recovered with high support (Brochu & McEachran, 2000; Meganathan et al., 2010; Meredith et al., 2011; Hekkala et al., 2011; Oaks, 2011). However, studies that included Antillean C. acutus sampled in Cuba have revealed a well-supported sister relationship with C. rhombifer, to the exclusion of continental C. acutus that continued to cluster with C. intermedius (Milián-García et al., 2011, 2015, 2018). The observed inconsistency in phylogenetic placement of C. acutus is likely a consequence of excluding intraspecific genetic variation when trying to resolve higher order relationships.
Although Cuban and American crocodiles have been included in previous phylogenetic studies, insufficient sampling has precluded a definitive investigation of cryptic diversity in C. acutus and what impact that might have on phylogenetic reconstruction, taxonomic status, and conservation management. To fill this knowledge gap, here we collected mitochondrial DNA character data for C. acutus sampled throughout Cuba to characterize patterns of variation and lineage diversity across the island. We combined these data with exemplar sampling and previously collected data from Central America (Colombia, Belize), North America (Florida, USA) and elsewhere in the Greater Antilles (Jamaica and Cayman Islands) to reconstruct phylogenetic relationships across the species distribution and relative to all living Crocodylus species. Reconstructed relationships using newly combined data sets allowed us to test a hypothesis of monophyly of C. acutus as currently described and explore implications for current taxonomic and conservation status in Cuba and elsewhere.
Materials and Methods
Sampling
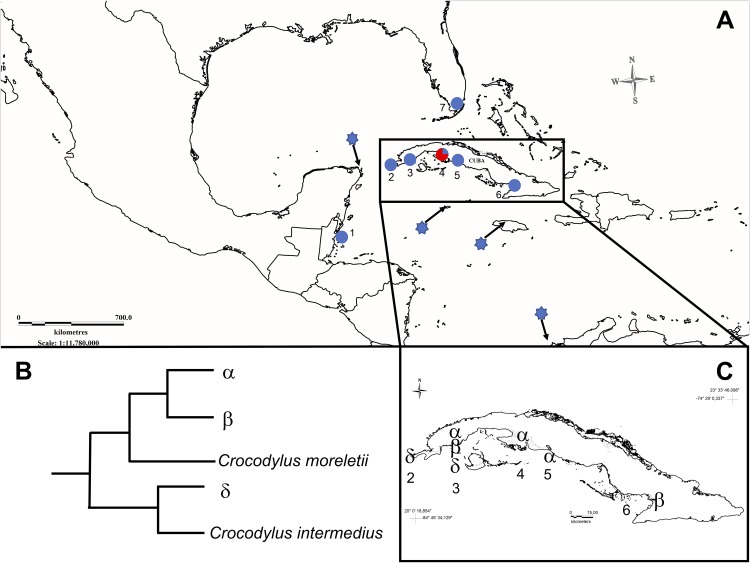
We sampled a total of 65 Crocodylus individuals between the years 2007 and 2014 from four main areas across the Cuban archipelago (Fig. 1). These included three areas where C. acutus is found in allopatry to C. rhombifer (Birama Swamp (n = 27); Province of Cienfuegos (n = 1); Province of Pinar del Río along the Guanahacabibes peninsula (n = 2)) and Zapata Swamp, where the two species are sympatric and known to hybridize (C. rhombifer (n = 31); C. acutus (n = 2); suspected hybrids (n = 2); Milián-García et al., 2015). In addition, we sampled the most western ex situ population for C. acutus at the Sabanalamar captive breeding facility (n = 24), also located in the province of Pinar del Río. The facility was founded in 1986 with a small group of adult breeders caught in the surrounded areas of Pinar del Río, up to five individuals from Lanier Swamp, and a group of neonates from Birama Swamp (R. Rodríguez-Soberón, 2018, personal communication).
Figure 1. Map showing sampled populations of Crocodylus species in North-America, Central-America, and the Caribbean.
(A) 1, Belize; 2, Guanahacabibes, Cuba; 3, Sabanalamar, Cuba; 4, Zapata Swamp, Cuba; 5, Cienfuegos, Cuba; 6, Birama Swamp, Cuba; 7, Florida, USA. Blue dots indicate C. acutus populations. The red and blue dot indicates the only population where C. rhombifer (red) and C. acutus (blue) were both relative proportions of samples (not exactly to scale for display purposes). The blue stars represent the reported location of the GenBank reference sequences of C. acutus analyzed in this study. (B) Representation of the relationships for the main mitochondrial haplotypes. (C) Distribution of main mitochondrial haplotypes across the Cuban archipelago.
All individuals were classified as C. rhombifer, C. acutus, or as suspected hybrids based on 26 external morphological characters (Targarona, 2013) and a piece of a caudal scale was clipped from each animal’s tail for genetic analysis. We collected and transported all samples in accordance with an agreement established between the Faculty of Biology at the University of Havana and the National Enterprise for the Protection of Flora and Fauna in Cuba, and CITES export and import permits, C002135 and E-00134/14, respectively.
We also included tissue samples from continental C. acutus collected from the Florida Everglades, USA (n = 12) and Belize (n = 12). All of these individuals have been previously classified as C. acutus based on morphology and genetics (Hekkala et al., 2015; Rossi, 2016).
Mitochondrial DNA sequencing
Total DNA was isolated for the samples from tail scale tissue preserved in 95% ethanol, using the NucleoSpin® kit for DNA extraction (Macherey-Nagel, Düren, Germany) following manufacturer’s instructions. A total of 716 base pair (bp) segment of mtDNA including the tRNAPro-tRNAPhe-D-loop region (hereafter referred to as D-loop) was amplified as a single fragment for all individuals using primers drL15459 (5′-AGGAAAGCGCTGGCCTTGTAA-3′) and CR2HA (5′-GGGGCCACTAAAAACTGGGGGGA-3′) (Weaver et al., 2008) except for individuals from the Florida Everglades and Belize. Two overlapping fragments of cytochrome b (cytb) covering a total of 1,320 bp were amplified by polymerase chain reactions (PCR) using the primers L14212 (5′-TTG GGC TTT AGACCA AGA CC-3′) with CB3H (5′-GGC AAA TAG GAA RTATCA-3′) (Palumbi, 1996), and L14849 (5′-TCCTCCACGAACGCGGAR C-3′) with H15453 (5-CCKTCCAYYTCTGTCTTACAAG-3′) (Milián-García et al., 2011). We also amplified a 665 bp fragment from the 3′ end of cytochrome oxidase I (COI) using the primer pair COIa (5′-AGT ATA AGC GTC TGGGTA GTC-3′) with COIf (5′-CCT GCA GGA GGA GGA GAY CC-3′) (Kessing et al., 1989).
We carried out the PCR on either a BioRad T100™ or an Eppendorf thermal cycler. Each PCR was prepared in a 15 μL total volume, containing: six μL of VWR® Taq DNA-polymerase (5 U/μL) Master Mix 2X, seven μL of distilled water, 0.5 μL of each primer (10 μmol), and 20–50 ng of DNA. Cycling conditions for the D-loop were as follows: 94 °C (2 min), 33 cycles of 94 °C (30 s), 58 °C (30 s), 72 °C (45 s), and a final extension of 72 °C (7 min). For cytb and COI, cycling conditions were: 94 °C (2 min), 35 cycles of 94 °C (45 s), 48 °C (45 s), 72 °C (90 s), and a final extension of 72 °C (10 min). All PCR products were purified by ExoSAP-IT (USB® Products) and sequenced using Big Dye terminators on an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The regions sequenced per sample are shown in Table S1.
Data analysis
We edited and aligned mitochondrial DNA sequences in Sequencher 5.2.4 (Gene Codes, Ann Arbor, MI, USA). We also calculated haplotypic (h) and nucleotide (π) diversity (Nei, 1987) estimates based on mtDNA sequences after excluding gaps and missing data, as executed in DNASP v5 (Librado & Rozas, 2009). We then generated a haplotype network using statistical parsimony as implemented in TCS (Clement, Posada & Crandall 2000). Haplotype networks were visualized and edited using the web-based program tcsBU (Múrias dos Santos et al., 2016). In addition, we calculated Tamura-Nei genetic distances in MEGA v 7 (Kumar, Stecher & Tamura, 2016).
To examine observed haplotype variation recovered in Cuban Crocodylus within the context of all extant Crocodylus, we analyzed our sequences relative to previously published data at overlapping mtDNA fragments for all living Crocodylus spp. (Table 1). We tested for significant geographic structure among populations using analysis of molecular variance (AMOVA) (Excoffier, Smouse & Quattro, 1992) as implemented in ARLEQUIN v3.5 (Excoffier & Lischer, 2010). We unambiguously aligned sequences using MAFFT as implemented in GENEIOUS 10.2.4 (Biomatters Ltd., Auckland, New Zealand; Kearse et al., 2012) with default settings and concatenated the regions following their clockwise order in the mitochondrial genome (cytb-D-loop-COI). We conducted a Bayesian phylogenetic analysis for the combined dataset using MrBayes 3.0b4 (Ronquist & Huelsenbeck, 2003), assuming a mixed-model of nucleotide substitution using the best-fit model inferred for each partition separately according to the Bayesian information criterion as implemented in jModelTest 2 (Posada & Crandall, 1998). We used the African Slender-snouted crocodile (Mecistops cataphractus; EF551000.1) as an outgroup to root the tree. We ran four simultaneous chains for a chain length of 2 × 106, each using a random tree as a starting point, the default heating scheme, and a subsampling frequency of 100. The first 2,000 trees were discarded as burn-in samples and the remaining trees were used to construct a majority-rule consensus tree and derive posterior probability values.
Table 1. Origin of previously published reference mtDNA sequences used per Crocodylus spp.
| Species | GenBank accession number | Mitochondrial sequences | Authors |
|---|---|---|---|
| C. rhombifer | JF502247.1 | Partial mitochondrial genome | Meredith et al. (2011) |
| C. acutus | HM636894.1, HQ595003, HQ595041, HQ595005, HQ595043, KF273834, KF273842, KF273841, KF273849, AY568314 | Complete mitochondrial genome, Cyt b, COI, D-loop | Ray et al. (2004); Rodriguez et al., 2011; Man et al. (2011); Milián-García et al. (2011); Bloor, Ibáñez & Viloria-Lagares, (2015) |
| C. moreletii | HQ585889.1, HQ595007, HQ595045 | Complete mitochondrial genome, Cyt b, COI | Meganathan et al. (2010); Milián-García et al. (2011) |
| C. intermedius | JF502242.1 | Partial mitochondrial genome | Meredith et al. (2011) |
| C. niloticus | JF502245.1 | Partial mitochondrial genome | Meredith et al. (2011) |
| C. suchus | JF502244.1 | Partial mitochondrial genome | Meredith et al. (2011) |
| C. johnsoni | HM488008.2 | Complete mitochondrial genome | Meganathan et al. (2010) |
| C. porosus | DQ273698.1 | Complete mitochondrial genome | Li et al. (2007) |
| C. palustris | HM488007.1 | Complete mitochondrial genome | Meganathan et al. (2010) |
| C. mindorensis | GU144287.1 | Complete mitochondrial genome | Feng et al. (2010) |
| C. novaeguineae | HM636896.1 | Complete mitochondrial genome | Man et al. (2011) |
| C. siamensis | EF581859.1 | Complete mitochondrial genome | Srikulnath et al. (2012) |
Results
Haplotypic variation and population differentiation
D-loop
The number of identified mtDNA D-loop haplotypes within Cuban Crocodylus ranged from one (Birama Swamp and Zapata Swamp) to four (Pinar del Río). Haplotypic and nucleotide diversities ranged from 0.000–0.634 to 0.000–0.01791, respectively, with the highest levels found in Pinar del Río, particularly the captive population (Table 2).
Table 2. Genetic variation within Crocodylus populations inferred from mitochondrial DNA markers.
| Population | Mitochondrial DNA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D-loop | COI | Cyt b | |||||||||||
| N | n | No. haplotypes | Haplotype diversity | Nucleotide diversity (π) | n | No. haplotypes | Haplotype diversity | Nucleotide diversity (π) | n | No. haplotypes | Haplotype diversity | Nucleotide diversity (π) | |
| Captive C. acutus-Pinar del Río | 24 | 24 | 4 | 0.634 (0.00580) | 0.01791 | 22 | 3 | 0.558 (0.0033) | 0.02420 | 24 | 3 | 0.583 (0.00373) | 0.02948 |
| Wild C. acutus-Pinar del Río | 2 | 2 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | 0 | 0 |
| C. acutus-Birama | 27 | 19 | 1 | 0 | 0 | 7 | 2 | 0.286 (0.03856) | 0.00055 | 26 | 1 | 0 | 0 |
| C. acutus-Florida | 12 | – | – | – | – | 11 | 1 | 0 | 0 | 12 | 1 | 0 | 0 |
| C. acutus-Belize | 12 | – | – | – | – | 12 | 1 | 0 | 0 | 12 | 1 | 0 | 0 |
| C. rhombifer-Zapata | 31 | 30 | 1 | 0 | 0 | 25 | 1 | 0 | 0 | 31 | 1 | 0 | 0 |
Note:
N, Total number of individuals per population; n, number of sequences used. The variance of the haplotype diversity is indicated between parentheses.
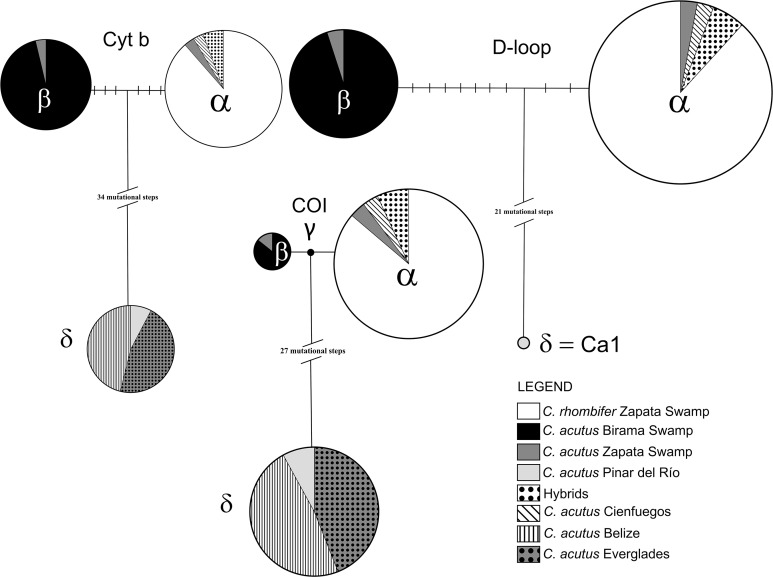
Among the wild caught individuals, the most common haplotype was identical to the α haplotype (Weaver et al., 2008; Milián-García et al., 2015), which is shared by C. rhombifer (n = 30) and hybrids (n = 2) from Zapata Swamp, as well as for C. acutus from Cienfuegos (n = 1) and Zapata Swamp (n = 1) (Fig. 2). The second most common haplotype was identical to the β haplotype (Weaver et al., 2008; Milián-García et al., 2015), differing by nine steps from the α haplotype, and was found in C. acutus from Zapata Swamp (n = 1) and Birama Swamp (n = 19). The third most common haplotype was δ (identical to the Ca1 haplotype in Rodriguez et al. (2011); GenBank accession no. MH753361), which has been considered the most frequent haplotype in North and Central America (Fig. 2). It differs by 23 steps from the α haplotype and was found in all wild C. acutus from Pinar del Río (n = 2).
Figure 2. Haplotype networks generated for each mitochondrial DNA region for the Crocodylus populations included in the present study.
Mutational steps between haplotypes are indicated by slashes. Greek letters correspond to the identifier of each sampled haplotype, following the order suggested in previous studies (Weaver et al., 2008).
Cytochrome oxidase I
The COI fragment resolved into four haplotypes (Fig. 2) with an overall haplotype and nucleotide diversity per population ranging from 0.000–0.558 to 0.000–0.02420, respectively (Table 2). Similar to patterns at the D-loop, the most common haplotype was identical to the α haplotype and was shared among C. rhombifer (n = 25), C. acutus (n = 1), and hybrids (n = 2) from Zapata Swamp as well as for C. acutus from Cienfuegos (n = 1) (Fig. 2). The second most common haplotype δ (GenBank accession no. MH758763) was shared for C. acutus individuals from Pinar del Río (n = 2), Belize (n = 12), and Florida (n = 11). The third most common haplotype was identical to the β haplotype and was shared among C. acutus from Birama (n = 7), and Zapata Swamp (n = 1). The fourth mtDNA COI haplotype was unique to C. acutus from Birama (n = 1) and differed by just one step from the α and β haplotypes (GenBank accession no. MH758764).
Cytochrome b
Three mtDNA cytb haplotypes were recovered among the wild individuals sampled from Pinar del Río, Birama Swamp, Zapata Swamp, Belize, and Florida. Following the same pattern as for the D-loop, the most common haplotype was identical to the α haplotype and was shared among C. rhombifer (n = 31) and hybrids (n = 2) from Zapata Swamp, as well as for C. acutus from Cienfuegos (n = 1) and Zapata Swamp (n = 1) (Fig. 2). The second most common haplotype, δ, was shared for C. acutus from Pinar del Río (n = 2), Belize (n = 12), and Florida (n = 12) differing by 37 steps from the α haplotype (GenBank accession no. MH758762). The third haplotype was identical to the β haplotype and was shared among C. acutus from Birama (n = 26) and Zapata Swamps (n = 1), differing by six steps from the α haplotype. Overall haplotypic and nucleotide diversities ranged from 0.000–0.583 to 0.000–0.02948, respectively (Table 2).
The fully concatenated sequences revealed unique haplotypes for all Cuban samples for which all regions were successfully sequenced (Table S1). The AMOVA using the concatenated mtDNA sequences recovered significant levels of genetic structure, with a highest fraction distributed among (>99%), rather than within (<1%), the populations of Cuban and American crocodiles studied (P < 0.0001).
Captive population
Thirteen individuals with the D-loop α haplotype were identified in captivity, while two possessed the D-loop β haplotype and seven presented the D-loop δ haplotype. The other two individuals presented a fourth new haplotype differing by only one mutational step away from the δ haplotype (GenBank accession no. MH719199). When analyzing the COI fragment, captive individuals were distributed into three haplotypes. Twelve individuals presented the COI α haplotype, one the COI β haplotype and nine individuals shared the continental haplotype (δ). When analyzing the cytb, three haplotypes were recovered for the 24 captive individuals. A total of 13 individuals presented the cytb α haplotype, two exhibited the cytb β haplotype, while nine shared the same haplotype (δ) showed by the continental individuals.
Phylogenetic analysis
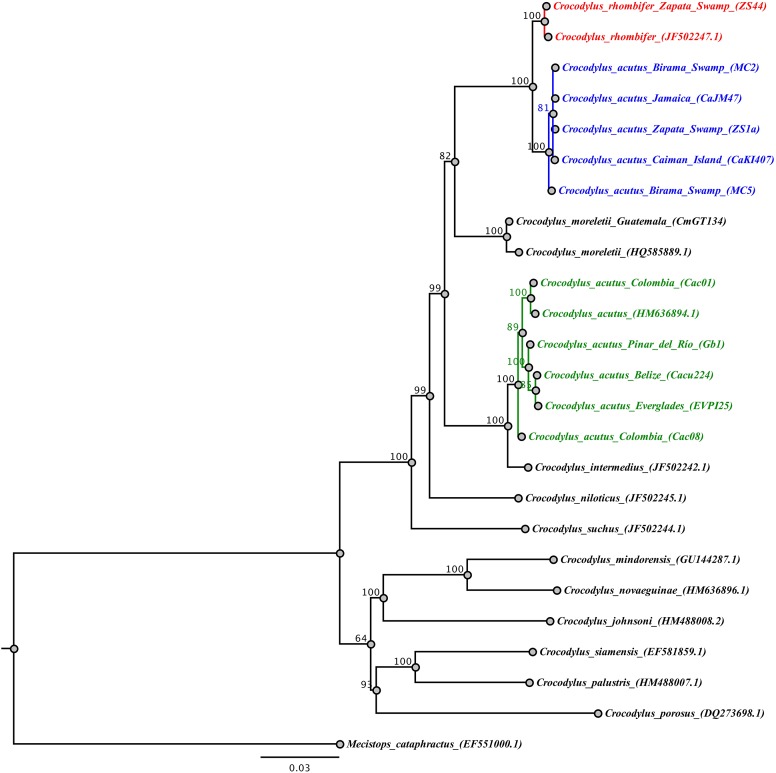
The analysis of the concatenated mtDNA regions for the wild-caught samples revealed that Cuban Crocodylus are split into three independent clades. Two of these correspond to the previous clades recovered for Antillean C. acutus (central and eastern Cuba, Jamaica, Cayman Islands) and C. rhombifer (Fig. 3) revealing a well-supported split (posterior probability = 1), and a sister relationship with C. moreletti. The third clade grouped wild C. acutus from western Cuba with C. acutus from continental South, Central, and North America, sister to C. intermedius. Tamura-Nei genetic distances revealed shallow divergence between Antillean C. acutus and C. rhombifer (0.4%), substantially lower than the value resolved between western and eastern C. acutus populations within Cuba (5.4%).
Figure 3. Phylogenetic tree based on Bayesian inference and the concatenated mtDNA dataset.
The Cuban crocodile (Crococylus rhombifer) clade is highlighted in red, while the two clades for the American crocodile (Crocodylus acutus) are indicated in blue and green, respectively. The numbers on the nodes indicate posterior probability values expressed in percentage.
Discussion
Recent studies have uncovered cryptic diversity across a diverse group of crocodylians (e.g., C. niloticus, C. suchus, M. cataphractus, Osteolaemus; McAliley et al., 2006; Eaton et al., 2009; Hekkala et al., 2011; Oaks, 2011; Shirley et al., 2014; Cunningham, Shirley & Hekkala, 2016). Consistent with previous findings (Milián-García et al., 2011, 2015, 2018), our results revealed a high degree of genetic differentiation within the American crocodile, C. acutus. Based on the most expansive geographic and mtDNA character sampling to date, we reconstructed C. acutus as paraphyletic, exhibiting two distinct lineages composed of Antillean and continental populations with high genetic divergence (5.4%). Moreover, these two lineages are more closely related to other species of crocodiles than they are to each other. Antillean C. acutus exhibits a close affiliation with C. rhombifer in a clade sister to C. moreletii. Conversely, continental C. acutus is most closely related to C. intermedius.
At a finer-scale, we found significant differentiation between haplotypes from western Cuba and the rest of the sampled populations, including Zapata Swamp and Birama Swamp. Interestingly, C. acutus haplotypes recovered from Pinar del Río province do not exhibit significant differentiation from continental C. acutus, and have individuals placed in the same clade (Fig. 3). This result further suggests that the western tip of Cuba—which is closer to the Yucatan peninsula than it is to Zapata Swamp—may either be a transition zone or an area of recent colonization from continental C. acutus. The fact that individuals sampled within the most western on-island C. acutus captive population share the continental haplotype is consistent with the hypothesis of a transition zone in the area. Most founders at the Sabanalamar captive population were captured locally back in the 1980s, while others were transported from Birama and Lanier Swamps (R. Rodríguez-Soberón, 2018, personal communication). The latter helps account for the admixture exhibited within the captive population. Our results also demonstrate that human-mediated translocation should no longer be a supported practice as it produces a mix of highly divergent genetic lineages.
The close genetic relationship between C. rhombifer and Antillean C. acutus based on mtDNA has been previously hypothesized to be a consequence of mitochondrial capture through hybridization, and C. rhombifer and C. acutus have been documented to hybridize in Zapata Swamp (Milián-García et al., 2015). Although karyological differences exist between C. rhombifer (2n = 30) and C. acutus (2n = 32) (Cohen & Gans, 1970), there is evidence of fertile hybrids (at least F1) between the species (E. Pérez-Fleitas, 2018, personal communication). However, most C. acutus populations in the Antilles have never been recorded to overlap in distribution with the Cuban crocodile (Tabet, 2010; Tabet et al., 2014; Brochu & Jiménez-Vázquez, 2014). For example, the American crocodile population in Birama Swamp, recognized as the second most extensive wetland in the insular Caribbean, is considered to be the largest and most protected population for the species in the Antilles (Tabet, 2010; Tabet et al., 2014). As such, mitochondrial capture through hybridization would be a feasible hypothesis to explain genetic similarity between C. rhombifer and Antillean C. acutus if the event occurred before the dispersion of C. acutus around the Cuban archipelago and the rest of the Greater Antilles. It would also imply that Antillean C. acutus with C. rhombifer captured-mitochondria would have higher fitness than the continental C. acutus, given the absence of the latter across the Antilles.
An alternative explanation for this pattern could be one of very recent divergence of the Antillean lineages from continental congeners coupled with the slow mtDNA mutation rate in crocodylians (Hekkala et al., 2011; Oaks, 2011). This hypothesis is consistent with the observation of low levels of genetic differentiation between C. rhombifer and Antillean C. acutus (Milián-García et al., 2011, 2015, 2018), albeit at a lower level than the average genetic distance between conspecifics within Crocodylus (∼1%; Hekkala et al., 2011). These findings, in tandem with observed morphological, ecological, and ethological characteristics (Cuvier, 1807; Targarona, 2013), suggest that C. rhombifer constitutes a unique lineage, but further study is warranted (see below).
These patterns reconstructed from mtDNA character data represent clear challenges for the current taxonomy of C. acutus. Combined with the results of previous studies (Weaver et al., 2008; Milián-García et al., 2011, 2015, 2018), we found evidence for a cryptic lineage of American crocodile currently found in Cuba and other localities in the Antilles. Moreover, the paraphyletic linages of C. acutus show a genetic distance (5.4%) similar to interspecific comparisons within Crocodylus and exhibit closer phylogenetic affinities with other species than each other, suggesting a formal taxonomic reassessment may be in order. Yet it is imperative to highlight that this work was limited to insights from mitochondrial DNA character data; additional analyses using nuclear data are essential for resolving taxonomic uncertainty in this system and to more directly test phylogeographic hypotheses. That said, our results do have immediate relevance for conservation, as the cryptic diversity uncovered here and elsewhere is not currently accounted for in status assessments and management plans for new world Crocodylus.
Conclusions
Overall, the reconstructed paraphyly, distinct phylogenetic affinities and high genetic divergence between Antillean and continental C. acutus populations reinforce the need for a formal taxonomic reassessment. Such an endeavor would benefit from a nuclear genome-wide study of Crocodylus, as has been demonstrated in other systems that exhibit a complex history and high prevalence of cryptic diversity (e.g., Galapagos tortoise, Chelonoidis sp.; Miller et al., 2018). This work benefited from the most comprehensive geographic and character data sampling conducted to date for C. acutus and other congenerics. Importantly, the inclusion, for the first time, of samples from the western population of the American crocodile in Cuba revealed evidence for continental mtDNA haplotypes in the Antilles, suggesting this area may constitute a transition zone between distinct lineages of C. acutus. More broadly, detection of cryptic diversity has important implications for conservation. Although C. acutus population sizes have been increasing since CITES protections have been in place (Platt, Rainwater & Nichols, 2004), assessments to date have not evaluated the status of Antillean populations as potentially constituting an independently evolving lineage of New World crocodiles. Likewise, cryptic diversity may be reflective of ongoing evolutionary processes as closely related species and populations adapt to changing environments (Bickford et al., 2007), punctuating the need for formal characterization to resolve taxonomic uncertainty and inform conservation planning (Struck et al., 2018), in this case for the American and Cuban crocodiles.
Supplemental Information
Acknowledgments
We would like to dedicate this study to the memory of Rafael Crespo for his valuable contribution to the project in Florida. We thank the Cuban crocodile field work specialists Etiam Pérez Fleitas, Gustavo Sosa Rodríguez and Leiter Guerra Manchena. We are especially thankful to Vicente Berovides Álvarez and specialists of the Sabanalamar captive breeding facility for their help during sample collection. We are also deeply grateful to Roberto Ramos Targarona and Manuel Alonso Tabet for providing samples to complete the present study. We thank Dr. Joseph J Gillespie, Dr. Kornsorn Srikulnath, and the two other anonymous reviewers for their constructive suggestions and comments that significantly improved the manuscript. We acknowledge the assistance of all members of the Biodiversity and Climate Research Centre at Goethe University and the Sackler Institute for Comparative Genomics at the American Museum of Natural History, New York.
Funding Statement
The study was supported by the Leibniz Association, the Student Research Assistance Scheme (SRAS) and the IUCN-SSC Crocodile Specialist Group’s Student Research Assistance Scheme (SRAS) which provided funding that supported collaborations and student exchange between Goethe University and the University of Havana (YMG). Research funding was provided to YMG by the International Foundation for Science (no. D 5354), Idea Wild Organization, a Theodore Roosevelt Grant from the American Museum of Natural History (NY), and the Rufford Foundation for Nature Conservation (Booster Grant for Nature Conservation no. 19318-B). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Yoamel Milián-García conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Michael A. Russello conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Jessica Castellanos-Labarcena analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Martin Cichon performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Vikas Kumar analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Georgina Espinosa contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Natalia Rossi contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Frank Mazzotti contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Evon Hekkala analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
George Amato analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Axel Janke conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
All samples were collected and transported in accordance with an agreement established between the Faculty of Biology at the University of Havana and the National Enterprise for the Protection of Flora and Fauna in Cuba, and CITES export and import permits, C002135 and E-00134/14, respectively.
Data Availability
References
- Bickford et al. (2007).Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends in Ecology & Evolution. 2007;22(3):148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bloor, Ibáñez & Viloria-Lagares (2015).Bloor P, Ibáñez C, Viloria-Lagares TA. Mitochondrial DNA analysis reveals hidden genetic diversity in captive populations of the threatened American crocodile (Crocodylus acutus) in Colombia. Ecology and Evolution. 2015;5:130–140. doi: 10.1002/ece3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu & Jiménez-Vázquez (2014).Brochu CA, Jiménez-Vázquez O. Enigmatic crocodyliforms from the early miocene of Cuba. Journal of Vertebrate Paleontology. 2014;34(5):1094–1101. doi: 10.1080/02724634.2014.855225. [DOI] [Google Scholar]
- Brochu & McEachran (2000).Brochu CA, McEachran JD. Phylogenetic relationships and divergence timing of crocodylus based on morphology and the fossil record. Copeia. 2000;2000(3):657–673. doi: 10.1643/0045-8511(2000)000[0657:PRADTO]2.0.CO;2. [DOI] [Google Scholar]
- Clement, Posada & Crandall (2000).Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cohen & Gans (1970).Cohen MM, Gans C. The chromosomes of the order crocodilia. Cytogenetic and Genome Research. 1970;9(2):81–105. doi: 10.1159/000130080. [DOI] [PubMed] [Google Scholar]
- Cunningham, Shirley & Hekkala (2016).Cunningham SW, Shirley MH, Hekkala ER. Fine scale patterns of genetic partitioning in the rediscovered African crocodile, Crocodylus suchus (Saint-Hilaire 1807) PeerJ. 2016;4:e1901. doi: 10.7717/peerj.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier (1807).Cuvier G. Sur les differentes especes de crocodiles vivantes et sur leurs caracteres distinctifs. Annales du Muséum d’histoire Naturelle. 1807;10:8–66. [Google Scholar]
- Eaton et al. (2009).Eaton MJ, Martin A, Thorbjarnarson J, Amato G. Species-level diversification of African dwarf crocodiles (Genus Osteolaemus): a geographic and phylogenetic perspective. Molecular Phylogenetics and Evolution. 2009;50(3):496–506. doi: 10.1016/j.ympev.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Excoffier & Lischer (2010).Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier, Smouse & Quattro (1992).Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng et al. (2010).Feng G, Wu X, Yan P, Li X. Two complete mitochondrial genomes of Crocodylus and implications for crocodilians phylogeny. Amphibia-Reptilia. 2010;31:299–309. doi: 10.1163/156853810791769464. [DOI] [Google Scholar]
- Grigg & Kirshner (2015).Grigg G, Kirshner D. Biology and evolution of Crocodylians. Canberra: Csiro Publishing; 2015. [Google Scholar]
- Hekkala et al. (2015).Hekkala ER, Platt SG, Thorbjarnarson JB, Rainwater TR, Tessler M, Cunningham SW, Twomey C, Amato G. Integrating molecular, phenotypic and environmental data to elucidate patterns of crocodile hybridization in Belize. Royal Society Open Science. 2015;2(9):150409. doi: 10.1098/rsos.150409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekkala et al. (2011).Hekkala E, Shirley MH, Amato G, Austin JD, Charter S, Thorbjarnarson J, Vliet KA, Houck ML, Desalle R, Blum MJ. An ancient icon reveals new mysteries: mummy DNA resurrects a cryptic species within the Nile crocodile. Molecular Ecology. 2011;20(20):4199–4215. doi: 10.1111/j.1365-294X.2011.05245.x. [DOI] [PubMed] [Google Scholar]
- Kearse et al. (2012).Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessing et al. (1989).Kessing B, Croom H, Martin A, McIntosh C, Mcmillan WO, Palumbi S. The Simple Fool’s Guide to PCR. Honolulu: University of Hawaii; 1989. [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2007).Li Y, Wu X, Ji X, Yan P, Amato G. The complete mitochondrial genome of salt-water crocodile (Crocodylus porosus) and phylogeny of crocodilians. Journal of Genetics and Genomics. 2007;34:119–128. doi: 10.1016/S1673-8527(07)60013-7. [DOI] [PubMed] [Google Scholar]
- Librado & Rozas (2009).Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Man et al. (2011).Man Z, Yishu W, Peng Y, Xiaobing W. Crocodilian phylogeny inferred from twelve mitochondrial protein-coding genes, with new complete mitochondrial genomic sequences for Crocodylus acutus and Crocodylus novaeguineae. Molecular Phylogenetics and Evolution. 2011;60(1):62–67. doi: 10.1016/j.ympev.2011.03.029. [DOI] [PubMed] [Google Scholar]
- McAliley et al. (2006).McAliley LR, Willis RE, Ray DA, White PS, Brochu CA, Densmore LD. Are crocodiles really monophyletic?—Evidence for subdivisions from sequence and morphological data. Molecular Phylogenetics and Evolution. 2006;39(1):16–32. doi: 10.1016/j.ympev.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Meganathan et al. (2010).Meganathan PR, Dubey B, Batzer MA, Ray DA, Haque I. Molecular phylogenetic analyses of genus Crocodylus (Eusuchia, Crocodylia, Crocodylidae) and the taxonomic position of Crocodylus porosus. Molecular Phylogenetics and Evolution. 2010;57(1):393–402. doi: 10.1016/j.ympev.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Meredith et al. (2011).Meredith RW, Hekkala ER, Amato G, Gatesy J. A phylogenetic hypothesis for Crocodylus (Crocodylia) based on mitochondrial DNA: evidence for a trans-Atlantic voyage from Africa to the New World. Molecular Phylogenetics and Evolution. 2011;60(1):183–191. doi: 10.1016/j.ympev.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Milián-García et al. (2018).Milián-García Y, Castellanos-Labarcena J, Russello MA, Amato G. Mitogenomic investigation reveals a cryptic lineage of Crocodylus in Cuba. Bulletin of Marine Science. 2018;94(2):329–343. doi: 10.5343/bms.2016.1134. [DOI] [Google Scholar]
- Milián-García et al. (2015).Milián-García Y, Ramos-Targarona R, Pérez-Fleitas E, Sosa-Rodríguez G, Guerra-Manchena L, Alonso-Tabet M, Espinosa-López G, Russello MA. Genetic evidence of hybridization between the critically endangered Cuban crocodile and the American crocodile: implications for population history and in situ/ex situ conservation. Heredity. 2015;114(3):272–280. doi: 10.1038/hdy.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milián-García et al. (2011).Milián-García Y, Venegas-Anaya M, Frias-Soler R, Crawford AJ, Ramos-Targarona R, Rodríguez-Soberón R, Alonso-Tabet M, Thorbjarnarson J, Sanjur OI, Espinosa-López G, Bermingham E. Evolutionary history of Cuban crocodiles Crocodylus rhombifer and Crocodylus acutus inferred from multilocus markers. Journal of Experimental Zoology. Part A: Ecological Genetics and Physiology. 2011;315A(6):358–375. doi: 10.1002/jez.683. [DOI] [PubMed] [Google Scholar]
- Miller et al. (2018).Miller JM, Quinzin MC, Edwards DL, Eaton DAR, Jensen EL, Russello MA, Gibbs JP, Tapia W, Rueda D, Caccone A. Genome-wide assessment of diversity and divergence among extant galapagos giant tortoise species. Journal of Heredity. 2018;109(6):611–619. doi: 10.1093/jhered/esy031. [DOI] [PubMed] [Google Scholar]
- Múrias dos Santos et al. (2016).Múrias dos Santos A, Cabezas MP, Tavares AI, Xavier R, Branco M. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics. 2016;32(4):627–628. doi: 10.1093/bioinformatics/btv636. [DOI] [PubMed] [Google Scholar]
- Nei (1987).Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Oaks (2011).Oaks JR. A time-calibrated species tree of crocodylia reveals a recent radiation of the true crocodiles. Evolution. 2011;65(11):3285–3297. doi: 10.1111/j.1558-5646.2011.01373.x. [DOI] [PubMed] [Google Scholar]
- Palumbi (1996).Palumbi S. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sunderland: Sinauer Associates; 1996. pp. 205–247. [Google Scholar]
- Platt, Rainwater & Nichols (2004).Platt SG, Rainwater TR, Nichols S. A recent population assessment of the American crocodile (Crocodylus acutus) in Turneffe Atoll, Belize. Herpetological Bulletin. 2004;89:26–32. [Google Scholar]
- Posada & Crandall (1998).Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ray et al. (2004).Ray DA, Dever JA, Platt SG, Rainwater TR, Finger AG, McMurry ST, Batzer MA, Barr B, Stafford PJ, McKnight J, Densmore LD. Low levels of nucleotide diversity in Crocodylus moreletii and evidence of hybridization with C. acutus. Conservation Genetics. 2004;5(4):449–462. doi: 10.1023/B:COGE.0000041024.96928.fe. [DOI] [Google Scholar]
- Rodriguez et al. (2011).Rodriguez D, Forstner MRJ, Moler PE, Wasilewski JA, Cherkiss MS, Iii LDD. Effect of human-mediated migration and hybridization on the recovery of the American crocodile in Florida (USA) Conservation Genetics. 2011;12(2):449–459. doi: 10.1007/s10592-010-0153-1. [DOI] [Google Scholar]
- Ronquist & Huelsenbeck (2003).Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rossi (2016).Rossi NA. Population genetic structure and reproductive ecology of Crocodylus across local and regional scales. 2016. PhD thesis.
- Shirley et al. (2014).Shirley MH, Vliet KA, Carr AN, Austin JD. Rigorous approaches to species delimitation have significant implications for African crocodilian systematics and conservation. Proceedings of the Royal Society of London B: Biological Sciences. 2014;281(1776):20132483. doi: 10.1098/rspb.2013.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath et al. (2012).Srikulnath K, Thongpan A, Suputtitada S, Apisitwanich S. New haplotype of the complete mitochondrial genome of Crocodylus siamensis and its species-specific DNA markers: distinguishing C. siamensis from C. porosus in Thailand. Molecular Biology Reports. 2012;39:4709–4717. doi: 10.1007/s11033-011-1263-7. [DOI] [PubMed] [Google Scholar]
- Struck et al. (2018).Struck TH, Feder JL, Bendiksby M, Birkeland S, Cerca J, Gusarov VI, Kistenich S, Larsson K-H, Liow LH, Nowak MD, Stedje B, Bachmann L, Dimitrov D. Finding evolutionary processes hidden in cryptic species. Trends in Ecology & Evolution. 2018;33(3):153–163. doi: 10.1016/j.tree.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Tabet (2010).Tabet MA. Comportamiento del cocodrilo americano (Crocodylus acutus) en el refugio de fauna monte Cabaniguan Cuba. 2010. http://purl.org/dc/dcmitype/Text. http://purl.org/dc/dcmitype/Text thesis.
- Tabet et al. (2014).Tabet MA, Targarona RR, Soberón RR, Thorbjarnarson J, Ferrer JB, Álvarez VB. Los Crocodylia de Cuba. Sant Vicent del Raspeig: Universidad de Alicante; 2014. [Google Scholar]
- Targarona (2013).Targarona RR. Ecologia y conservación del cocodrilo cubano (Crocodylus rhombifer) en la “Ciénaga de Zapata,” Cuba. 2013. http://purl.org/dc/dcmitype/Text. http://purl.org/dc/dcmitype/Text thesis.
- Weaver et al. (2008).Weaver JP, Rodriguez D, Venegas-Anaya M, Cedeño-Vázquez JR, Forstner MRJ, Densmore LD. Genetic characterization of captive Cuban crocodiles (Crocodylus rhombifer) and evidence of hybridization with the American crocodile (Crocodylus acutus) Journal of Experimental Zoology Part A: Ecological Genetics and Physiology. 2008;309A(10):649–660. doi: 10.1002/jez.471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
GenBank accession no. MH719199, MH753361, MH758762–MH758764.