We designed a porous 3D Au–DNA hydrogel network for simultaneously imaging multiplex miRNAs in living cells through dual-signal amplification.
We designed a porous 3D Au–DNA hydrogel network for simultaneously imaging multiplex miRNAs in living cells through dual-signal amplification.
Abstract
Non-invasively imaging multiplex microRNAs (miRNAs) in living cells is pivotal to understanding their physiological functions and pathological development due to the key regulatory roles of miRNAs in gene expression. However, developing smart delivery systems with large gene loading capacity, biocompatibility and responsiveness remains a significant challenge. Herein, we successfully incorporated DNA-capped Au nanoparticles (NPs) and their complementary fluorescent DNA sequences into a porous 3D hydrogel network (AuDH), in which hairpin-locked DNAzyme strands and active metal ions were loaded (AuDH/Mn+/H) for simultaneously imaging multiplex miRNAs in living cells. After transfection into cells, the specific miRNAs trigger the strand-displacement reaction and sequentially activate the DNAzyme-assisted target recycling, leading to a strong increase in the corresponding fluorescence intensity for imaging. This enables simultaneous assessment of the abundance of multiplex cancer-related miRNAs, even if at a very low expression level, in different cells through the different fluorescence intensities due to the dual signal amplification, and the change in abundance of miRNAs induced by siRNA or miRNA mimics in living cells can also be efficiently monitored. The versatile and responsive DNA hydrogel system holds great potential for miRNA biomedical applications.
Introduction
MicroRNAs (miRNAs), endogenous small molecule RNAs (18–22 nt),1,2 are regarded as key regulators for an array of biological processes including cellular differentiation, proliferation and cell apoptosis.3 The deregulated expressions of miRNAs are interrelated with a variety of pathological conditions such as cancer and cardiovascular disease.4,5 Thus, they have received considerable attention as potential diagnostic and prognostic biomarkers and novel therapeutic targets.6,7 However, the gene regulation network of miRNAs exhibits a complex and orchestrated mode as a result of the imperfect complement between miRNAs and their target mRNAs,8 and single miRNA monitoring could not provide reliable and satisfactory information about the pathological processes. So far, a variety of novel fluorescence nanoprobes have been designed for multiplex detection and imaging of cancer-related mRNAs in living cells with high sensitivity and selectivity.9–12 Developing strategies for accurate and sensitive multiplex miRNA detection in living cells is significant for miRNA biomedical applications.13,14 To address this issue, advanced delivery systems assisting the transfection of DNA probes into cells15,16 and efficient intracellular signal amplification strategies for improving the detection sensitivity are urgently needed.
The rapid development of DNA nanotechnology provides an opportunity to construct smart and versatile delivery nanocarriers with high cell uptake efficiency.17–19 DNA hydrogels, a porous three-dimensional structured network using DNA as building blocks composed of complementary DNA sequences,20 have gained increasing attention for biological applications.21,22 Owing to their high biocompatibility, responsiveness, versatility and large loading capacity,23–25 DNA hydrogels have attracted immense interest in various function material, biomedicine and sensor applications.26–32 Especially, DNA hydrogels could be appended to any type of nucleic acid molecule (e.g. miRNA, siRNA or DNA aptamers) for cell-specific gene delivery in biological applications.18,29,33,34 The intrinsic responsive properties, high cell uptake efficiency and gene loading capacity of DNA hydrogels35 endow them with great potential for intracellular DNA/RNA functional molecular trigger responsive detection, which has barely been explored.
Catalytic DNA molecules (known as DNAzymes),36 in the presence of specific metal ions,37 enable cleavage at a single ribonucleotide embedded within their complementary DNA substrate without assistance from any other nicking enzyme.38 As a burgeoning enzyme-free signal amplification technique, the use of metal ion-specific DNAzymes39–41 provides great prospects for fabricating highly sensitive sensors for specific intracellular detection owing to their designability, versatility and high catalytic efficiency.42–44 A DNAzyme motor in response to a specific intracellular target operating in living cells was reported.45 A Zn2+-specific DNAzyme responsive to intracellular miRNA was designed for intracellular miRNA amplified detection.46 However, the abundance of the intracellular metal ions limits the sensitivity of the ion-dependent DNAzyme amplification efficiency and its practical application.
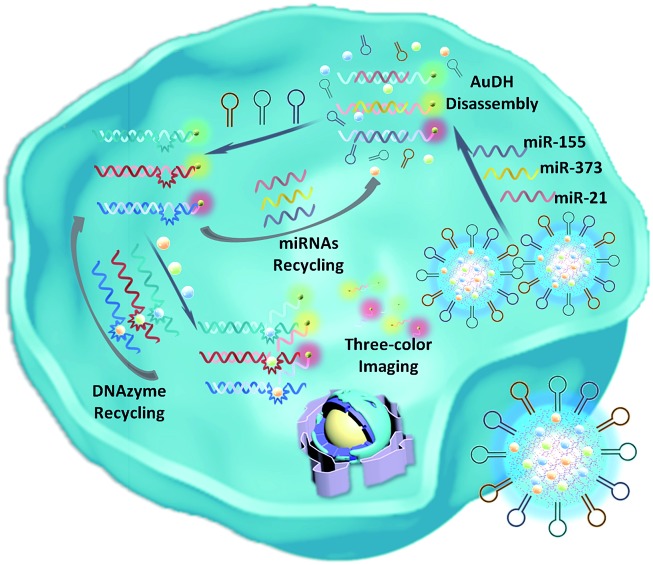
Herein, we develop an Au nanoparticle (NP) DNA hydrogel (AuDH) network constructed from three different DNA-capped Au NPs (Au-P1, Au-P2 and Au-P3) and their complementary fluorescent dye-modified DNA probes (P1, P2 and P3, embedded with ribonucleotides). Three hairpin-locked DNAzyme strands (H1, H2 and H3) and their specific metal ions (Cu2+, Mg2+ and Zn2+)47,48 are simultaneously loaded into the AuDH (AuDH/Mn+/H) to fabricate a smart and versatile system (Fig. 1A). It is specifically responsive to intracellular multiplex miRNAs (miR-21, miR-373 and miR-155), which specifically trigger the strand-displacement reaction and hairpin-locked DNAzyme-assisted target recycling dual signal amplification to simultaneously assess the abundance of multiplex cancer-related miRNAs, even if at a very low expression level, in different cells through the amplified specific fluorescence signal (Scheme 1).
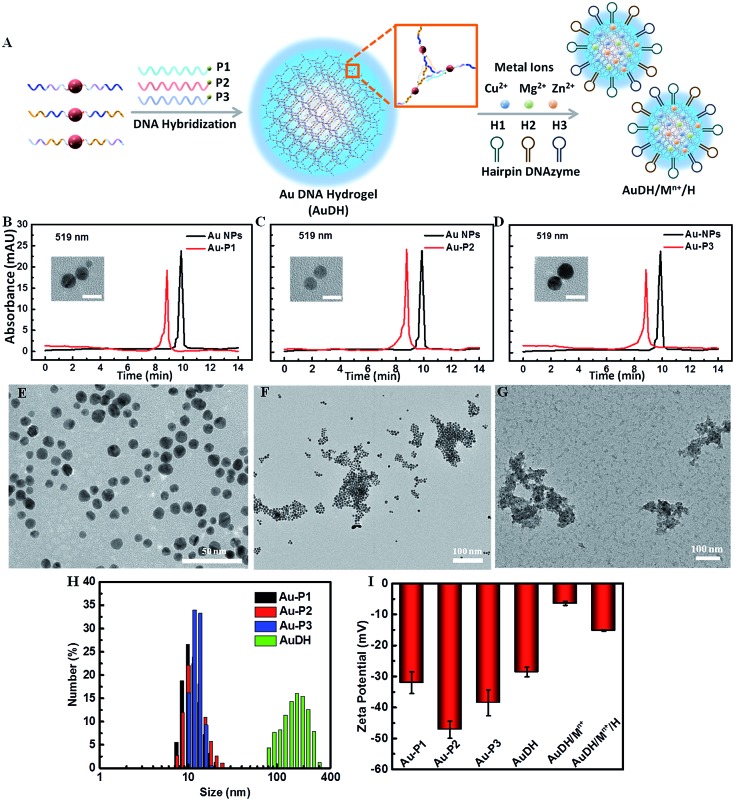
Fig. 1. (A) The self-assembly procedure of AuDH/Mn+/H. HPLC characterization of Au NPs and DNA-capped Au NPs, Au-P1 (B), Au-P2 (C) and Au-P3 (D); the insets present the TEM images of Au-P1, Au-P2 and Au-P3; the scale bar is 5 nm. TEM characterization of Au-P1, Au-P2 and Au-P3 without linkers (E), AuDH (F), and AuDH/Mn+/H (G). (H) DLS analysis of DNA-capped Au NPs and AuDH. (I) Zeta potential measurement of DNA-capped Au NPs, AuDH, AuDH/Mn+ and AuDH/Mn+/H.
Scheme 1. Schematic illustration of the imaging of multiplex miRNAs in living cells using AuDH/Mn+/H-mediated dual signal amplification.
Results and discussion
Characterization of the self-assembled DNA hydrogel
High-performance liquid chromatography (HPLC) was used to verify the synthesis of phosphorothioate domain-embedded DNA-capped Au NPs. The characteristic peaks of pure Au NPs and DNA-capped Au NPs at 519 nm (Au NP absorbance) were recorded and are shown in Fig. 1B–D. Compared to the profile of the pure Au NPs, the absorbance peak of DNA-capped Au NPs at 519 nm shifted, which demonstrated that the three DNA-capped Au NPs (Au-P1, Au-P2 and Au-P3) had been successfully synthesized. Transmission electron microscopy (TEM) revealed that the average size of the DNA-passivized Au NPs was about 4 nm (inset in Fig. 1B–D). The agarose gel electrophoresis analysis indicated the successful stepwise assembly of the designed DNA-three way junction nanostructure and feasibility for further forming Au DNA hydrogels (AuDH) (Fig. S1, ESI†). Furthermore, the assembly of AuDH was adequately characterized. The DNA-capped Au NPs dispersedly distributed in the absence of linked DNA probes (Fig. 1E). The addition of the linker DNA probes induced obvious connection and assembly of DNA-capped Au NPs to form AuDH with an average size of ≈200 nm (Fig. 1F), which was consistent with the dynamic light scattering (DLS) results (Fig. 1H). As shown in Fig. 1G, when the metal ions (Cu2+, Mg2+ and Zn2+) and hairpin-locked DNAzyme strands (H1, H2 and H3) were loaded onto AuDH, a layer of floccule was obviously observed on the surface of the AuDH, indicating the successful coating and generation of AuDH/Mn+/H. As shown in Fig. 1I, the zeta potential of the AuDH was about –28.6 mV, and the surface zeta potential changed from –28.6 mV to –6.43 mV after the AuDH was loaded with positively charged metal ions. The coating of negatively charged hairpin-locked DNAzyme strands induced a zeta potential change to –15.2 mV, further indicating the successful formation of AuDH/Mn+/H. Under optimal conditions, the AuDH showed large loading capacity for hairpin-locked DNAzyme strands and metal ions (Fig. S2, ESI†).
Feasibility of AuDH/Mn+/H for dual-signal amplification
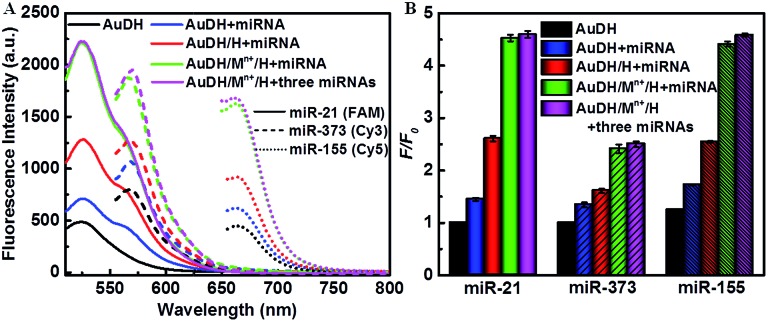
The amounts of hairpin-locked DNAzyme strands and metal ions significantly influenced the DNAzyme-assisted target miRNA recycling and strand-displacement reaction, which were firstly optimized. The optimal molar ratio of AuDH to hairpin-locked DNAzyme strands was 1 : 0.25 for all three H1, H2 and H3 probes (Fig. S3A–C, ESI†), while 1 mM Cu2+, 10 mM Mg2+ and 2.5 mM Zn2+ were selected as optimal concentrations for the DNAzyme-assisted signal amplification step (Fig. S3D–F, ESI†). Under optimized conditions, the feasibility of this strategy for ternary miRNA detection was investigated (Fig. 2). As shown in Fig. 2A, the solid lines, the short dashed lines and the short dotted lines indicate the fluorescence intensity in response to miR-21, mi-373 or miR-155. Upon excitation at the corresponding wavelength, the fluorescence intensities of FAM, Cy3 and Cy5 between the AuDH/Mn+/H exposed to the one specific target miRNA (green curves) and the mixture containing all three miRNAs (purple curves) were almost the same, which indicated negligible mutual interference for miRNA detection and good feasibility of ternary miRNA simultaneous detection. Meanwhile, polyacrylamide gel electrophoresis (PAGE) was employed to verify the feasibility of AuDH/Mn+/H for dual amplification in vitro, demonstrating the feasibility of AuDH/Mn+/H for dual-signal amplification (Fig. S4, ESI†).
Fig. 2. (A) Feasibility analysis of the AuDH/Mn+/H for detection of multiplex miRNAs (miR-21/FAM, miR-373/Cy3, and miR-155/Cy5). (B) The corresponding fluorescence intensities in (A).
We further investigated the dual-signal amplification efficiency. Compared to the control (black curves), the fluorescence intensities of the AuDH hybridized with miRNAs induced a slight increase (blue curves). The addition of hairpin-locked DNAzyme strands further caused an increase in fluorescence intensities due to the toe-hold strand-displacement recycling amplification reaction (red curves). The combined introduction of hairpin-locked DNAzyme strands and metal ions led to a sharp increase in the fluorescence values as a result of toe-hold strand-displacement and DNAzyme-assisted miRNA recycling dual-signal amplification (green curves), and it was 4.48-fold higher for miR-21, 2.65-fold higher for miR-373 and 3.49-fold higher for miR-155, respectively, compared to the control groups (Fig. 2B).
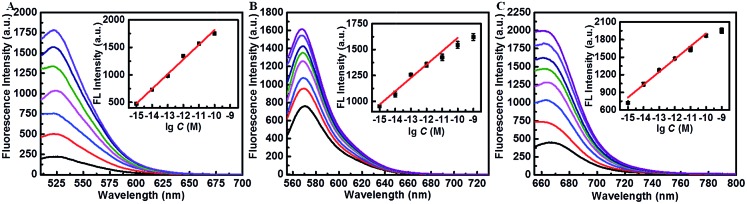
Subsequently, the fluorescence intensity of this proposed assay for ternary miRNA detection was evaluated through dual amplification steps. As shown in Fig. 3A–C, the fluorescence intensity increased with the increase of target miRNA concentration, exhibiting a wide linear relationship with the logarithm of miRNA concentration from 1 fM to 100 pM (inset in Fig. 3A–C). The regression equations between the concentration (M) of the three types of target miRNAs and their fluorescence intensities are described as follows:miR-21: F (a.u.) = 267.5 lg C (M) + 4491.6, R = 0.996miR-373: F (a.u.) = 128.8 lg C (M) + 2902.2, R = 0.984miR-155: F (a.u.) = 218.1 lg C (M) + 4084.7, R = 0.986where R is the corresponding correlation coefficient of the calibration curve. The limit of detection (LOD) of the target miRNAs was calculated by using three times the standard deviation of the control fluorescence intensity values according to previous reports,49,50 which were estimated to be 179 × 10–18 M for miR-21, 58.8 × 10–18 M for miR-373 and 24.9 × 10–18 M for miR-155, respectively, suggesting the good sensitivity of this strategy. These results suggested the high amplification efficiency of the proposed system.
Fig. 3. (A–C) Fluorescence spectral responses to the different concentrations of miR-21 (FAM, control and 1 fM to 100 pM), miR-373 (Cy3, control and 1 fM to 1 nM) and miR-155 (Cy5, control and 1 fM to 1 nM). The insets in A–C show the linear correlation between the corresponding fluorescence intensity and the logarithm of miRNA concentrations.
Multiplex miRNA imaging in living cells
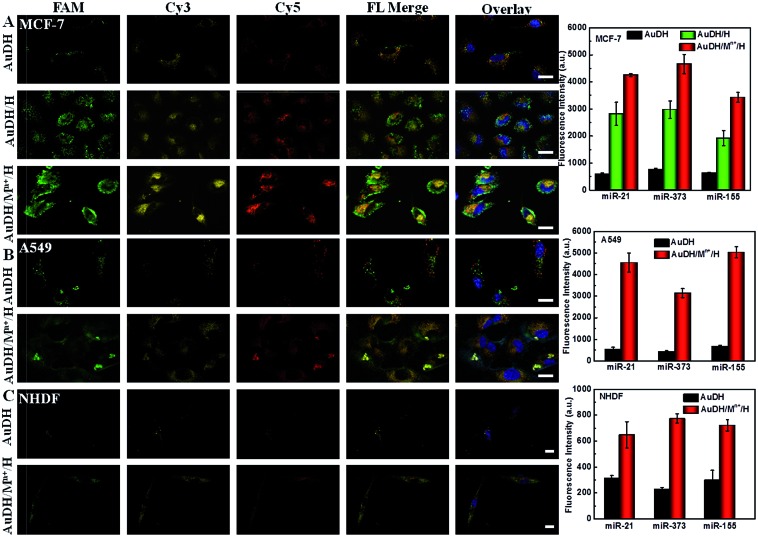
The high amplification efficiency and multiplex miRNA detection capability of the system motivated us to further investigate its performance for multiplex miRNA imaging in living cells. The investigation revealed that the prepared AuDH/Mn+/H nanoconstruct exhibited good stability in cell media containing serum (Fig. S5, ESI†). The cytotoxicity of the resultant AuDH and AuDH/Mn+/H was evaluated. The cell viability of AuDH/Mn+/H-treated MCF-7 cells was slightly lower than that of the AuDH-treated cells due to the loaded metal ions. The cell viabilities of AuDH/Mn+/H-treated groups were still high at 86.5% even upon adding 100 μL AuDH/Mn+/H into 4 mL cell media, which is similar to the conditions selected in the subsequent experiments. These results suggested the low cell cytotoxicity of AuDH/Mn+/H under these conditions (Fig. S6, ESI†). Furthermore, the cellular uptake procedure of the resultant AuDH was also explored using MCF-7 as a model. As shown in Fig. S7,† the AuDH was taken up when the cells were exposed to AuDH in one hour and the AuDH was abundant in the cells incubated for 4 hours (Fig. S7A, ESI†), which was similar to the commercial liposome-treated cells (Fig. S7C, ESI†). The weak fluorescence signal of DNA hydrogels without AuNP modification showed the low cellular uptake efficiency compared with the prepared AuDH (Fig. S7B, ESI†). The endocytosis mechanism of the prepared AuDH was also investigated. Low temperature is a well-known factor for suppressing the production of adenosine triphosphate (ATP), hindering the endocytic pathway and inhibiting the cellular activity. When the incubation temperature was chosen to be 4 °C, a very weak fluorescence signal could be observed (Fig. S8A, ESI†) and the inhibition percentage was up to 86.7% (Fig. S8C, ESI†), assuming that just 14% of AuDH entered into cells by energy-independent pathways and the rest was taken up through energy-dependent pathways. Subsequently, the endocytosis mechanism for AuDH cellular uptake as an energy-dependent pathway was explored. Chlorpromazine and colchicine were used as a clathrin-mediated endocytosis and macro-pinocytosis inhibitor, respectively. The cellular uptakes of AUDH in chlorpromazine- and colchicine-treated MCF-7 cells were decreased to 57.5% and 49.8%, respectively, indicating that the AuDH entered cells via clathrin-mediated endocytosis and/or macro-pinocytosis pathways (Fig. S8, ESI†). The expression levels of miR-21, miR-373 and miR-155 in two cancer cell lines including A549 (a lung cancer cell line) and MCF-7 cells (a human breast cancer cell line) and a normal cell line of NHDF cells (normal human dermal fibroblast cells) were explored using the AuDH/Mn+/H nanosystem.
As shown in Fig. 4A, the AuDH-treated MCF-7 cells (as control) presented three weak fluorescence signals (green FAM for miR-21, yellow Cy3 for miR-373 and red Cy5 for miR-155) derived from the direct recognition of miRNAs by AuDH probes without signal amplification. For the MCF-7 cells incubated with AuDH/H, the corresponding fluorescence intensities of the three miRNAs were increased, being 4.77-fold higher for miR-21, 3.91-fold higher for miR-373 and 3.02-fold higher for miR-155 compared with their corresponding signals in control groups. In contrast, the corresponding fluorescence intensities of the AuDH/Mn+/H-treated cells were sharply enhanced, being 7.22-fold higher for miR-21, 6.12-fold higher for miR-373 and 5.39-fold higher for miR-155 than that observed in the AuDH-treated group, respectively (Fig. 4A and S9, ESI†), indicating that the AuDH/Mn+/H mediated dual-signal amplification system could be used for simultaneously imaging miRNAs in living cells with high sensitivity. Similar results were observed in the A549 cells (Fig. 4B) and NHDF cells (Fig. 4C). Notably, although almost no visible fluorescence signals were observed in the AuDH-treated NHDF cells (Fig. 4C), an approximately 2.63-fold fluorescence enhancement and thus obvious fluorescence signals were observed in the cells treated with AuDH/Mn+/H.
Fig. 4. Confocal Laser Scanning Microscopy (CLSM) images and the corresponding fluorescence intensity derived from miRNA recognition in MCF-7 (A), A549 (B) and NHDF (C) incubated with AuDH, AuDH/H and AuDH/Mn+/H, respectively. The nucleus was stained blue by Hoechst; the FAM, Cy3 and Cy5 fluorescence signals were presented as green, yellow and red, respectively. The scale bar is 25 μm.
As expected, the differences in abundance of these miRNAs in the three types of cells were clearly and sensitively revealed, suggesting the much lower expression level of the three miRNAs in NHDF cells compared with the two types of cancer cells. Meanwhile, the higher abundance of miR-373 in MCF-7 cells and miR-155 in A549 cells than that in the other cells was also demonstrated.51 These results demonstrated that the AuDH/Mn+/H nanosystem for simultaneously and sensitively imaging intracellular miRNAs provided a facile and promising way to discriminate between different types of cells with various miRNA expression levels.
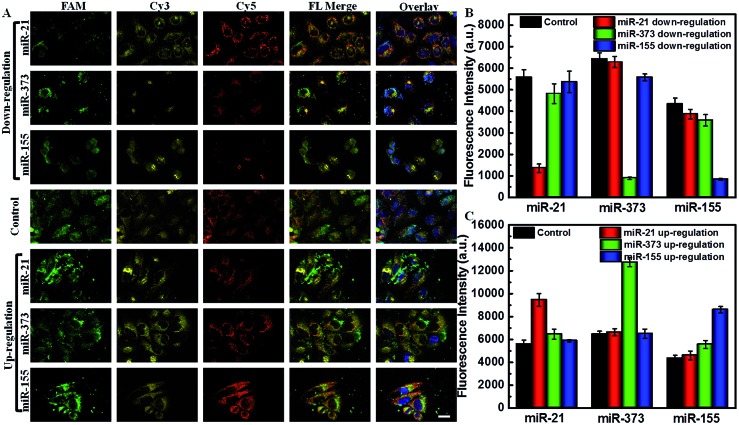
The abundance of miRNAs varied in various physiological processes and under various pathological conditions. It is important to sensitively monitor the change of miRNAs in living cells to provide useful diagnostic and prognostic information. Using siRNA and miRNA mimics as regulators, the down- or up-regulated miRNA expression levels monitored by the AuDH/Mn+/H nanosystem were further studied. As shown in Fig. 5, the green, yellow and red fluorescence signals were derived from miR-21, miR-373 and miR-155 detection, respectively. The green fluorescence intensity of cells that received miR-21 inhibitor treatment exhibited a significant decrease (Fig. 5A), which was 4.11-fold lower than that of the control group, and the yellow and red fluorescence intensities were almost the same (Fig. 5B). The cells treated with miR-373 inhibitors or miR-155 inhibitors also exhibited a sharp decrease in the corresponding fluorescence intensity compared to the control group (Fig. 5A and B). In contrast, when the MCF-7 cells were treated with specific miRNA mimics, a higher corresponding fluorescence intensity related to the high expression level of miRNA compared to that in the control group was observed (Fig. 5A and C). These results indicated that this proposed assay enabled simultaneous imaging of the change of abundance of miRNAs, even if at a low expression level, which may be useful for disease diagnosis and prognosis.
Fig. 5. (A) CLSM images of MCF-7 cells that received down-regulated (miR-21 inhibitors, miR-373 inhibitors or miR-155 inhibitors) or up-regulated treatments (miR-21 mimics, miR-373 mimics or miR-155 mimics) and were detected by the AuDH/Mn+/H nanosystem. The corresponding fluorescence intensities for the MCF-7 cells that received down-regulated (B) and up-regulated treatments (C).
Conclusions
A versatile Au–DNA hydrogel for simultaneously and sensitively imaging intracellular multiplex miRNAs was developed using a toe-hold strand-displacement reaction and hairpin-locked DNAzyme-assisted miRNA recycling dual signal amplification. A wide linear range, low limit of detection and good selectivity for simultaneous detection of multiplex miRNAs in homogeneous solution were demonstrated. The abundance of three types of intracellular cancer-related miRNAs in different types of cancer cells and normal cells was simultaneously and sensitively detected with only one step transfection. The changes regulated by siRNA or miRNA mimics in living cells were successfully and sensitively monitored, which was useful for cancer diagnosis and prognosis. This strategy enables accurate and sensitive differentiation of cancer cells by imaging intracellular multiplex miRNAs even with a very low expression level, and we hope that it will provide an opportunity for the early diagnosis of cancer with high accuracy and sensitivity.
Experimental
Materials and reagents
Copper(ii) chloride dihydrate (CuCl2·2H2O), magnesium chloride hexahydrate (MgCl2·6H2O), zinc chloride (ZnCl2) and trisodium citrate dihydrate were purchased from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Tetrachloroauric acid (HAuCl4) was provided by Toronto Research Chemicals Inc. (Toronto, Canada). All the reagents were of analytical reagent grade. Ultrapure water generated from a Millipore Milli-Q water purification system (Billerica, MA, USA) was used in all experiments. Chlorpromazine and colchicine were purchased from Energy Chemical (Shanghai, China). Phosphate-buffered saline (PBS, pH 7.4, 10 mM, 137 mM NaCl), Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were provided by Life Technologies Corporation (Los Angeles, CA, USA). The oligonucleotide strands in our experiments were obtained from Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). All of the nucleotide strands were purchased from GenePharma (Shanghai, China) and diluted in diethy pyrocarbonate (DEPC) water for further use. The DNA and RNA sequences are presented in the ESI.†
Synthesis of DNA-capped Au nanoparticles (NPs)
The distillation 10 mL-flakes and stirring bars were soaked in aqua regia solution and repeatedly rinsed thoroughly with ultrapure water before using. 20 μL of HAuCl4 (1 mM) solution was added into 1.8 mL of 1 nmol DNA-template solution, and stirred for 30 min at room temperature. Then the mixture was heated until boiling, and 2 μL of trisodium citrate dihydrate was added to the boiling solution rapidly, and the colorless mixture changed to wine red, maintaining the reaction for 5 min. The DNA-capped Au NPs were obtained, labeled as Au-P1, Au-P2 and Au-P3.
Assembly of Au-based DNA hydrogels (AuDH)
DNA-capped Au nanoparticles were purified via centrifugation at 12 000 for 10 min to remove residual DNA and inorganic salts, and the operation was repeated three times. The resultant DNA-capped Au NPs were re-dispersed in PBS (pH 7.4, 10 mM) for use. DNA-capped Au NPs (Au-P1, Au-P2 and Au-P3) and linker DNA strands (P1, P2 and P3) were mixed in a PCR-tube with a ratio of 1 : 4, and incubated at 95 °C for 2 min, and cooled down to 80 °C, 75 °C, 70 °C, 65 °C, 60 °C, 55 °C, 50 °C, 45 °C, and 40 °C (each temperature was maintained for 5 min). Afterwards, the mixture was cooled down to 37 °C and this temperature was maintained for 2 h. The assembled AuDH was purified via centrifugation at 6000 rpm for 10 min to remove the free DNA-capped Au NPs and linker DNA strands. The purified AuDH was re-suspended in PBS (pH 7.4, 10 mM) for use.
Loading of metal ions and DNA hairpins (AuDH/Mn+/H)
The assembled AuDH was mixed with different concentrations of metal ion solutions (Cu2+, Mg2+ and Zn2+) in PBS (pH 7.4, 10 mM) and incubated for 2 h at room temperature. Then the solution mixture was centrifuged at 5000 rpm for 10 min to remove the free metal ions. The resultant AuDH/Mn+ was incubated with different concentrations of hairpin-locked DNAzyme strands (H1, H2 and H3) for 2 h at room temperature, and the final AuDH/Mn+/H nanoconstruct was obtained for miRNA detection.
miRNA fluorescence detection in vitro
50 μL of AuDH/Mn+/H and different types of miRNAs with different concentrations were mixed in a 100 μL PCR-tube containing PBS (pH 7.4, 10 mM, 137 mM NaCl) and incubated for 3 h at 37 °C. Three types of fluorescence intensities were recorded at 525 nm, 568 nm and 662 nm with excitation at 490 nm, 540 nm and 640 nm, respectively, using an F-7000 fluorescence spectrometer (HITACHI, Tokyo, Japan).
Cellular uptake investigation with inhibitors
MCF-7 cells were seeded in 20 mm glass bottom cell culture dishes in DMEM with 10% FBS at 37 °C under a humidified atmosphere with 5% CO2 for 24 h. The MC-7 cells were incubated with AuDH (FAM), pure DNA hydrogels (FAM) and commercial Lipofectamine®2000 (FAM) solution in Opti-MEM medium. The fluorescence signals of the three nanomaterials were observed at intervals of one hour with a confocal laser scanning fluorescence microscope (UltraVIEW VoX, PerkinElmer, Germany).
Low temperature 4 °C (ATP inhibitor): MCF-7 cells were seeded in 20 mm glass bottom cell culture dishes in DMEM with 10% FBS at 37 °C under a humidified atmosphere with 5% CO2 for 24 h. The dish was placed in a 4 °C environment for 10 min and then the medium was removed. Subsequently, the AuDH (FAM) solution was dispersed in Opti-MEM medium, which was then maintained at 4 °C for 1 h. The fluorescence signals were observed with a confocal laser scanning fluorescence microscope (UltraVIEW VoX, PerkinElmer, Germany).
Chlorpromazine/colchicine (inhibitor for clathrin-mediated endocytosis): MCF-7 cells were pre-incubated with colchicine or chlorpromazine (5 μg mL–1) in serum-free medium. After 1 h of incubation, the medium was replaced with fresh Opti-MEM medium containing AuDH (FAM) and further incubated for 1 h. Then, the cells were washed with PBS (10 mM, pH 7.4) three times and the fluorescence intensity was observed using a confocal laser scanning fluorescence microscope (UltraVIEW VoX, PerkinElmer, Germany).
miRNA imaging in living cells
NHDF, A549 and MCF-7 cells were respectively cultured in 20 mm glass bottom cell culture dishes in DMEM with 10% FBS at 37 °C under a humidified atmosphere with 5% CO2 for 24 h. The cells were exposed to AuDH/Mn+/H solution in Opti-MEM medium. After 4 h of incubation, the cells were replaced with fresh medium and incubated for another 12 h, accomplishing the intracellular miR-21/miR-373/miR-155 recognition. The fluorescence signals for miR-21 (FAM)/miR-373 (Cy3)/miR-155 (Cy5) imaging in the cells were observed with a confocal laser scanning fluorescence microscope (UltraVIEW VoX, PerkinElmer, Germany).
Conflicts of interest
The authors declare no competing financial interests.
Supplementary Material
Acknowledgments
The work was supported by the Special Foundation for State Major Research Program of China (Grant No. 2016YFC0106602 and 2016YFC0106601); the National Natural Science Foundation of China (21475008); the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University (PRRD-2016-YB2); the Fundamental Research Funds for the Central Universities (Grant No. FRF-BD-17-016A); and the Beijing Municipal Science and Technology Commission (Grant No. z131102002813058).
Footnotes
†Electronic supplementary information (ESI) available: Additional documentation (9 figures) including agarose gel electrophoresis, zeta potential, proportional optimization, polyacrylamide gel electrophoresis, dynamic light scattering, MTT experiment, cellular uptake efficiency, cellular uptake mechanism and the ratio of fluorescence intensities. See DOI: 10.1039/c8sc02858c
References
- Ryoo S. R., Lee J., Yeo J., Na H. K., Kim Y. K., Jang H., Lee J. H., Sang W. H., Lee Y., Kim V. N. Acs Nano. 2013;7:5882–5891. doi: 10.1021/nn401183s. [DOI] [PubMed] [Google Scholar]
- Meng X., Dai W., Zhang K., Dong H., Zhang X. Chem. Sci. 2018;9:1184–1190. doi: 10.1039/c7sc04725h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Dong H., Lei J., Ding L., Wen Y., Ju H., Zhang X. Chem. Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- Zhang W., Hu F., Zhang Q., Zhang J., Mao Y., Wang P., Gu Y. Talanta. 2018;179:685–692. doi: 10.1016/j.talanta.2017.11.068. [DOI] [PubMed] [Google Scholar]
- Dong H., Lei J., Ju H., Zhi F., Wang H., Guo W., Zhu Z., Yan F. Angew. Chem., Int. Ed. 2012;51:4607–4612. doi: 10.1002/anie.201108302. [DOI] [PubMed] [Google Scholar]
- Duan R., Zuo X., Wang S., Quan X., Chen D., Chen Z., Jiang L., Fan C., Xia F. J. Am. Chem. Soc. 2013;135:4604–4607. doi: 10.1021/ja311313b. [DOI] [PubMed] [Google Scholar]
- Arenz C. Angew. Chem., Int. Ed. 2006;45:5048–5050. doi: 10.1002/anie.200601537. [DOI] [PubMed] [Google Scholar]
- Li N., Chang C., Pan W., Tang B. Angew. Chem., Int. Ed. 2012;51:7426–7430. doi: 10.1002/anie.201203767. [DOI] [PubMed] [Google Scholar]
- Pan W., Zhang T., Yang H., Diao W., Li N., Tang B. Anal. Chem. 2013;85:10581–10588. doi: 10.1021/ac402700s. [DOI] [PubMed] [Google Scholar]
- Pan W., Yang H., Li N., Yang L., Tang B. Chem. - Eur. J. 2015;21:6070–6073. doi: 10.1002/chem.201500365. [DOI] [PubMed] [Google Scholar]
- Pan W., Li Y., Wang M., Yang H., Li N., Tang B. Chem. Commun. 2016;52:4569–4572. doi: 10.1039/c5cc10147f. [DOI] [PubMed] [Google Scholar]
- Hu J., Liu M. H., Zhang C. Y. Chem. Sci. 2018;9:4258–4267. doi: 10.1039/c8sc00832a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L. P., Hu J., Liang L., Zhang C. Y. Chem. Commun. 2014;50:11883–11886. doi: 10.1039/c4cc05598e. [DOI] [PubMed] [Google Scholar]
- Kirby A. J., Camilleri P., Engberts J. B., Feiters M. C., Nolte R. J., Soderman O., Bergsma M., Bell P. C., Fielden M. L., Garcia Rodriguez C. L., Guedat P., Kremer A., McGregor C., Perrin C., Ronsin G., van Eijk M. C. Angew. Chem., Int. Ed. 2003;42:1448–1457. doi: 10.1002/anie.200201597. [DOI] [PubMed] [Google Scholar]
- Chittimalla C., Zammut-Italiano L., Zuber G., Behr J. P. J. Am. Chem. Soc. 2005;127:11436–11441. doi: 10.1021/ja0522332. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Kim H., Jo C., Jeong J., Ko J., Han S., Lee M. S., Lee H.-Y., Han J. W., Lee J., Lee J. B. Adv. Funct. Mater. 2017;27:1704213. [Google Scholar]
- Li J., Fan C., Pei H., Shi J., Huang Q. Adv. Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- Veetil A. T., Chakraborty K., Xiao K., Minter M. R., Sisodia S. S., Krishnan Y. Nat. Nanotechnol. 2017;12:1183–1189. doi: 10.1038/nnano.2017.159. [DOI] [PubMed] [Google Scholar]
- Li C., Rowland M. J., Shao Y., Cao T., Chen C., Jia H., Zhou X., Yang Z., Scherman O. A., Liu D. Adv. Mater. 2015;27:3298–3304. doi: 10.1002/adma.201501102. [DOI] [PubMed] [Google Scholar]
- Cai W., Xie S., Zhang J., Tang D., Tang Y. Biosens. Bioelectron. 2017;98:466–472. doi: 10.1016/j.bios.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Deshpande S. R., Hammink R., Das R. K., Nelissen F. H. T., Blank K. G., Rowan A. E., Heus H. A. Adv. Funct. Mater. 2016;26:9075–9082. [Google Scholar]
- Um S. H., Lee J. B., Park N., Kwon S. Y., Umbach C. C., Luo D. Nat. Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- Noll T., Wenderhold-Reeb S., Schonherr H., Noll G. Angew. Chem., Int. Ed. 2017;56:12004–12008. doi: 10.1002/anie.201705001. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jean S. R., Ahmed S., Aldridge P. M., Li X., Fan F., Sargent E. H., Kelley S. O. Nat. Commun. 2017;8:381. doi: 10.1038/s41467-017-00298-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Wu C., Liu H., Zou Y., Zhang X., Kang H., Yang C. J., Tan W. Angew. Chem., Int. Ed. 2010;49:1052–1056. doi: 10.1002/anie.200905570. [DOI] [PubMed] [Google Scholar]
- Kahn J. S., Trifonov A., Cecconello A., Guo W., Fan C., Willner I. Nano Lett. 2015;15:7773–7778. doi: 10.1021/acs.nanolett.5b04101. [DOI] [PubMed] [Google Scholar]
- Yata T., Takahashi Y., Tan M., Nakatsuji H., Ohtsuki S., Murakami T., Imahori H., Umeki Y., Shiomi T., Takakura Y., Nishikawa M. Biomaterials. 2017;146:136–145. doi: 10.1016/j.biomaterials.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Liu S., Su W., Li Y., Zhang L., Ding X. Biosens. Bioelectron. 2018;103:1–5. doi: 10.1016/j.bios.2017.12.021. [DOI] [PubMed] [Google Scholar]
- Zhao M., Wang P., Guo Y., Wang L., Luo F., Qiu B., Guo L., Su X., Lin Z., Chen G. Talanta. 2018;176:34–39. doi: 10.1016/j.talanta.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Shi Z., Gao X., Ullah M. W., Li S., Wang Q., Yang G. Biomaterials. 2016;111:40–54. doi: 10.1016/j.biomaterials.2016.09.020. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang D., Willner I., Tian Y., Jiang L. Angew. Chem., Int. Ed. 2018;57:1–6. doi: 10.1002/anie.201803222. [DOI] [PubMed] [Google Scholar]
- Angell C., Xie S., Zhang L., Chen Y. Small. 2016;12:1117–1132. doi: 10.1002/smll.201502167. [DOI] [PubMed] [Google Scholar]
- Lai J., Jiang P., Gaddes E. R., Zhao N., Abune L., Wang Y. Chem. Mater. 2017;29:5850–5857. doi: 10.1021/acs.chemmater.7b00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu Y., Hu Y., Zeng G., Zhang Y., Zhang C., Feng C. Small. 2018;14:e1703305. doi: 10.1002/smll.201703305. [DOI] [PubMed] [Google Scholar]
- Yousefi H., Ali M. M., Su H. M., Filipe C. D. M., Didar T. F. ACS Nano. 2018;12:3287–3294. doi: 10.1021/acsnano.7b08010. [DOI] [PubMed] [Google Scholar]
- Wang W., Satyavolu N. S. R., Wu Z., Zhang J. R., Zhu J. J., Lu Y. Angew. Chem., Int. Ed. 2017;56:6798–6802. doi: 10.1002/anie.201701325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Yu C., Niu Y., Chen J., Zhao Y., Zhang Y., Gao R., He J. Biosens. Bioelectron. 2018;101:297–303. doi: 10.1016/j.bios.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Tang Y., He X., Zhou Z., Tang J., Guo R., Feng X. Chem. Commun. 2016;52:13905–13908. doi: 10.1039/c6cc07719f. [DOI] [PubMed] [Google Scholar]
- Wang F., Elbaz J., Teller C., Willner I. Angew. Chem., Int. Ed. 2011;50:295–299. doi: 10.1002/anie.201005246. [DOI] [PubMed] [Google Scholar]
- Yin H. S., Li B. C., Zhou Y. L., Wang H. Y., Wang M. H., Ai S. Y. Biosens. Bioelectron. 2017;96:106–112. doi: 10.1016/j.bios.2017.04.049. [DOI] [PubMed] [Google Scholar]
- Huang P. J., Liu J. Anal. Chem. 2014;86:5999–6005. doi: 10.1021/ac501070a. [DOI] [PubMed] [Google Scholar]
- Huang Y., Fang L., Zhu Z., Ma Y., Zhou L., Chen X., Xu D., Yang C. Biosens. Bioelectron. 2016;85:496–502. doi: 10.1016/j.bios.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Liu M., Zhang Q., Chang D., Gu J., Brennan J. D., Li Y. Angew. Chem., Int. Ed. 2017;56:6142–6146. doi: 10.1002/anie.201700054. [DOI] [PubMed] [Google Scholar]
- Peng H., Li X. F., Zhang H., Le X. C. Nat. Commun. 2017;8:14378. doi: 10.1038/ncomms14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cui M., Zhou H., Yang W. ACS Sens. 2017;2:1847–1853. doi: 10.1021/acssensors.7b00710. [DOI] [PubMed] [Google Scholar]
- Yun W., Wu H., Liu X., Fu M., Jiang J., Du Y., Yang L., Huang Y. Anal. Chim. Acta. 2017;986:115–121. doi: 10.1016/j.aca.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Balogh D., Aleman Garcia M. A., Albada H. B., Willner I. Angew. Chem., Int. Ed. 2015;54:11652–11656. doi: 10.1002/anie.201501777. [DOI] [PubMed] [Google Scholar]
- Dong H., Meng X., Dai W., Cao Y., Lu H., Zhou S. F., Zhang X. Anal. Chem. 2015;87:4334–4340. doi: 10.1021/acs.analchem.5b00029. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Jin J., Wang H., Tang H., Yang R., Wang K. Anal. Chem. 2009;81:9703–9709. doi: 10.1021/ac901906w. [DOI] [PubMed] [Google Scholar]
- Mahdiannasser M., Karami Z. Biosens. Bioelectron. 2018;107:123–144. doi: 10.1016/j.bios.2018.02.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.