Abstract
Context: Ma Huang Tang (MHT) has been used to treat influenza, fever, bronchial asthma, etc. as a traditional Chinese medication. However, the anti-inflammation mechanism of MHT remains unclear.
Objective: The study identifies the possible mechanisms of MHT on ovalbumin (OVA)-induced acute bronchial asthma in mice.
Materials and methods: First, an asthma-related protein-protein interaction (PPI) network was constructed. And then, the acute bronchial asthma mice models were established by exposing to aerosolized 1% ovalbumin for 30 min/day for 1 week, and the mice were administered 2.0, 4.0, or 8.0 g/kg of MHT daily. To evaluate therapeutic effect, sensitization time, abdominal breathing time, eosinophils in bronchoalveolar lavage fluid, and tissue and trachea pathology were examined. Related genes were measured using RNA sequencing (RNA-seq). The expression levels of TLR9 in lung and trachea tissues were determined by immunohistochemical staining.
Results: MHT had a LD50 = 19.2 g/kg against asthma, while MHT at high doses (8 g/kg) effectively extended the sensitization time and abdominal breathing time and alleviated OVA-induced eosinophilic airway inflammation and mitigated pathological changes. The RNA-seq assay showed that the high-dose MHT resulted in a significant decrease in the levels of TLR9, TRAF6, TAB2, etc. in the lung tissue. Immunohistochemical assay confirmed the down-regulated of TLR9. Molecular docking revealed that six MHT compounds potentially mediated the TLR9 signaling pathway.
Discussion and conclusions: MHT could mitigate the pathological changes of acute asthma-like syndrome through inhibition of the TLR9 pathway. Results of this study may provide a reference for the development of a novel therapy for patients with allergic asthma.
Keywords: MHT, bioinformatics, targets, immunohistochemical assay
Introduction
Asthma is a chronic inflammatory disease that often occurs in the airway. Many cells of the innate immune system and acquired immune system (such as eosinophils, lymphocytes, and neutrophils) can interact with epithelial cells and cause bronchial hyper-responsiveness (BHR), excess mucus generation, airway remodeling, and airway stenosis (Lambrecht and Hammad 2015). The pathological characteristics of asthma are mainly referred to chronic airway inflammation and airway remodeling. Although humans are able to produce their own immune response, statistics from all over the world show that asthma diagnosis technology is still lacking (Marklund et al. 1999; Van Schayck et al. 2000; Lindensmith et al. 2004) and that the rate of treatment is extremely low (Beasley 1998; Rabe et al. 2000; Pearce et al. 2007; Carroll et al. 2012; Levy 2014). Therefore, further research on the pathogenesis of asthma is imperative.
The therapy effect of MHT on human respiratory diseases has been highly praised by Chinese people for centuries. MHT comprises four different kinds of medicines: Ephedra sinica Stapf. (Ephedraceae), Cinnamomum cassia Presl. (Lauraceae), Prunus armeniaca L. var. ansu Maxim. (Rosaceae) and Glycyrrhiza uralensis Fisch. (Fabaceae). This formula has been widely used in the clinic to treat colds, bronchial asthma, etc. Previous studies (Xiao et al. 2017) found that the components in MHT can relieve bronchial smooth muscle spasm, reduce inflammation factor release and inhibit the occurrence of asthma symptoms. Although MHT has displayed a significant curative effect for asthma, the mechanism of action is not completely understood at present, and there is no clarity as to the exact targets regulated by specific compounds within MHT. Therefore, determining the anti-asthmatic active ingredients in MHT is necessary.
As the development of bioinformatics progresses, the fast screening of drug targets and precise predictions of drug therapy mechanisms become more realistic. The similarity ensemble approach (SEA) is an online target prediction program based on the concept that compounds sharing high structural similarity may have relatively similar target association profiles (Gong et al. 2013). By using this tool, the structures of selected target proteins were picked out or constructed to screen for potential target compounds. In addition, we constructed protein–protein interaction (PPI) networks associated with selected proteins. Functions related to gene expression of proteins were annotated by using the DAVID database (Huang et al. 2009), and the asthma-related sub-network of the global human PPI network was extracted to explore the interrelations. Potential ligands of MHT were further investigated using a molecular docking analysis. The bioinformatics results indicated that 20 compounds in MHT targeted 32 kinds of proteins in the asthma network. An OVA-induced acute bronchial asthma model was successfully established. After treatment with MHT, the potential asthma-associated target was confirmed by RNA-seq. We selected the Toll-like receptor 9 (TLR9) pathway to further verify the expression level of proteins.
Materials and methods
Reagents
Herba Ephedra (Ephedra), Ramulus Cinnamomi (Cassia twig), Semen Armeniacae Amarum (Bitterapricot kernel) and Radix Glycyrrhizae Praeparata (Prepared licorice) were purchased from Nanta Drug Store (Shenyang, China) and authenticated according to the standards documented in the Chinese Pharmacopoeia. Dexamethasone was obtained from the Guoda Drug Store (Shenyang, China). Ovalbumin (OVA) was obtained from Solarbio (Lot. NO. 326A0512), and aluminum hydroxide was obtained from Damao Chemical Reagent Factory (Tianjin). Wright's stain was obtained from Solarbio (Lot.NO.20150803). Hematoxylin stain was obtained from Nanjing Jiancheng (Nanjing, China). Anti-TLR9 antibody (ab12121, Abcam) and RNAwait were obtained from Solarbio (Lot. NO. 20150824).
Preparation of MHT
As mentioned, MHT comprised four traditional Chinese herbs. Samples of Ephedra sinica (288 g), Cinnamomum cassia (196 g), Prunus armeniaca var. ansu (196 g) and Glycyrrhiza uralensis (96 g) were accurately weighed and mixed, and then these herbs were immersed for 30 min in three times their volume of distilled water. After 4 h of extraction via an essential oil extractor, the remaining medicinal materials were added to 500 mL of distilled water and decocted by boiling for 30 min. This was done twice, and the liquids mixed, yielding 1000 mL of decocted liquids. The liquids were concentrated and dried under vacuum at 65 °C; the final concentration of MHT was 8 g/mL, while the primary extracted essential oils were embedded into hydroxypropyl-β-cyclodextrinat a 1:2 ratio, yielding 0.8687 g of powder. The embedded essential oils and the concentrated suspension liquids were dissolved according to their corresponding proportions. The sample was stored at 4 °C. The doses of MHT in distilled water were expressed as grams of the original Ma Huang dry materials per kilogram body weight. Doses of 2, 4 and 8 g/kg of the distilled water extract of MHT were given to animals according to the human dose.
Experimental animals
A total of 120 female BALB/c mice (6–8 weeks, weight 18–20 g) were obtained from the Hua Fu Kang Biotechnology Co. (Certificate No. of SCXK [Liao] 2014-0004, Beijing, China). Mice were maintained in an animal facility under specific pathogen-free conditions for 7 d prior to experiments. All animal experiments were strictly conducted in accordance with regulations of Shenyang Pharmaceutical University (Shenyang, China) and were approved by the Animal Ethics Committee for Animal Studies (Ethical Review No. of SYPU-IACUC-S20150917-05).
Screening the key targets
Prediction of targets
The MHT compounds obtained were identified by consulting a large number of databases, such as SciFinder, NCBI, Web of Science, database of traditional Chinese medicine and chemical composition, etc. We then utilized the SEA database (Zheng et al. 2015) for determining targets of the MHT compounds. Here, to predict potential targets of MHT, we used a similarity-based method, the core idea of which is to rank potential compound-target interactions based on their similarity to known compound-target interactions. Furthermore, the predicted targets were evaluated by E-value to distinguish true and false compound-target interactions. A smaller E-value would lead to a smaller incidence of accidental error. The function of the targets can be determined by querying a database such as the Universal Protein Resource (UniProt).
Construction of the target PPI network
A primary global human PPI network of the determined proteins was constructed with diverse PPIs from online databases. These protein interaction data were collected from Human Protein Reference Database (HPRD) and PrePPI database (Keshava Prasad et al. 2009; Zhang et al. 2013). As different data sources can predict protein interactions from different aspects, we applied a naive Bayesian model to integrate diverse data and make the final interaction predictions (Li et al. 2010). Bronchial asthma-related proteins were filtered by the DAVID database, in which the functional determined protein in each pair is included. The unified conceptual framework of the PPI network was finally integrated by Cytoscape (Politano et al. 2014).
Homology modeling
Based on the protein–protein interaction network, we found that TLR9 was closely related to a known inflammatory response (Chen et al. 2014; Hu et al. 2015; Burgueño et al. 2016; Suresh et al. 2016), so TLR9 was selected to verify our prediction. First, sequences of the human TLR9 protein were downloaded from the UniProt database. SWISS-MODEL was used to search the modeling templates, of which 3WPC shared 83.69% similarity in comparison with TLR9; hence, we established the model of TLR9 with 3WPC. Finally, by comparing the similarity of our modelled TLR9 protein with TLR8 (PDB ID: 3w3j), we found that TLR9 has a high degree of sequence similarity with the TLR8 protein in its structural and active sites, so we used the TLR8 structure to define the active site of TLR9.
Molecular docking
In total, 238 compounds from MHT were confirmed and collected by using SciFinder, and their 3 D structures were modelled with Chem 3 D Ultra 12.0 (Cambridge Soft Corporation, Cambridge MA, USA. http://www.cambridgesoft.com.). Before docking, the fully minimized energy conformation for each compound was generated by the MMFF94 energy minimization protocol in Chem 3 D Ultra 12.0, and the crystallization of key proteins and their cocrystallized ligands equipped with spatial coordinates was downloaded from the Protein Data Bank (PDB). The docking study was processed by CDOCKER (Tiwari and Lee 2010), a CHARMM and simulated annealing-based method for ligand docking in DS 3.5. In the beginning, 20 random conformations for each ligand were generated by 5000 steps at a target temperature of 1000 K. The grid extension was set to 4 Å and 50 random orientations of each ligand conformation were generated. These conformations were then translated into receptors and moved into the binding sphere to search their appropriate binding modes. A final full potential minimization was then automatically called to refine the ligand poses. According to the CDOCKER scores, the top ten-ranked conformations for each ligand were obtained.
Experimental design
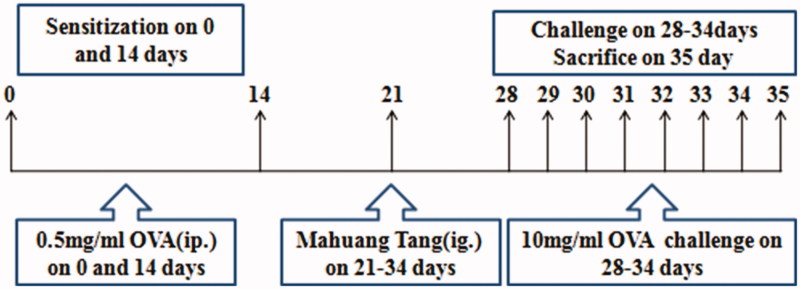
A total of 120 mice were divided into six groups (n = 20) as follows: blank group, model group, dexamethasone group (2.0 mg/kg) and three MHT-treatment groups (low dose: 2.0 g/kg, medium dose: 4.0 g/kg, high dose: 8.0 g/kg). Experimental processes are summarized in Figure 1. For the asthma model, mice were sensitized on day 0 and day 14 by intraperitoneal (i.p.) injection of 100 µg of OVA and 2 mg of aluminum hydroxide in 0.2 mL of normal saline. At 14 d, after the second sensitization, the mice were challenged with 1.0% OVA-NS or NS for the blank group for 30 min for 7 d. From 21 d to 34 d, 1 h before each challenge, the MHT groups received treatment (2.0, 4.0 and 8.0 g/kg) through the intragastric (i.g.) route and the dexamethasone group received 2.0 mg/kg through i.p. injection.
Figure 1.
Time line representation of the acute asthma model and pharmacological intervention.
Behavioral indexes
As sensitization, challenge and treatment procedures for different groups were conducted, we obtained the influence of drugs on mice by daily monitoring of the body weight change in mice during the experiment. During the atomization challenge stage, each six mice were placed in an airtight container one time connected to an ultrasonic atomizer for 30 min, at a rate of 1% OVA-atomized excitation, for 7 d. To chart symptoms exhibited by the mice, such as intense nose scratching, cyanosis of lips, dysphoria and shortness of breath, we kept a record of abdominal breathing times and sensitization times.
Collection of BALF and wright's stain
Mice were anesthetized using 0.7% paraformaldehyde after the last challenge and sacrificed immediately, then tracheotomy was performed. Normal saline (3.0 mL) was perfused into the lung four times to collect a total of 3 mL of bronchoalveolar lavage fluid (BALF), which was then resuspended for counting. After cell counting, BALF was centrifuged at 3000 rpm at 4 °C for 10 min to isolate the inflammatory cells. Centrifugal sediment was resuspended in 1 mL of normal saline and 20 μL of cell suspension was dislodged. Then, Wright’s dye was used to stain the eosinophils cells in cell suspension according to the manufacturer’s instructions.
Histological assessment of lung and trachea tissue
Lung and trachea specimens were fixed with 10% formalin and embedded in paraffin. The paraffin-embedded specimens were sectioned into 5 μm thick slices, deparaffinized in xylene for 15 min, dehydrated in a gradient alcohol series and rinsed three times with 1% PBS. The slices were stained with hematoxylin and eosin (H&E) according to the manufacturer’s instructions. The sections were examined under a Nikon 80 i plus confocal laser-scanning microscope (Nikon, Tokyo, Japan) at ×100 magnification.
RNA-seq analysis of lung
Total RNA was extracted using Trizol reagent (Invitrogen, Pleasanton, CA). The quality and quantity were first determined with a Bioanalyzer 2100 (Agilent, Santa Clara, CA) prior to any further processing. The relative expression of microRNAs was assessed by Affymetrix Gene Chip miRNA 2.0 arrays (Affymetrix, Santa Clara, CA) in accordance with manufacturer’s instruction. Briefly, total RNA was poly A-tailed and labelled with biotin with the Flash Tag Biotin HSR RNA Labeling Kit (Genisphere, Hatfield, PA). The hybridization was performed under rotation at 48 °C overnight, and the array was scanned on the Affymetrix 3000 Gene Scanner. Data were analysed with Affymetrix Gene Chip Command Console software, and the relative signal intensity and normalization were generated by the Robust Multichip Analysis (RMA) algorithm. The miRNA candidates with >2-fold change and p < 0.01 were considered biologically significant.
Immunohistochemical detection of TLR9 in lung and trachea tissue
Toll like receptor 9 (TLR9) expression levels in the lung and trachea were measured by diaminobenzidine (DAB) according to the manufacturer’s instructions. Each sample was tested in triplicate. The number of positively stained cells in each tissue section was calculated from a 50 μm magnified field under a light microscope. For immunohistochemical analyses of TLR9, staining density was determined using Image Pro plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA) in one field of each section with a prominent DAB reaction.
Analysis of HPLC fingerprints
Preparation of MHT: Ephedra sinica (28.8 g), Cinnamomum cassia (19.2 g), Prunus armeniaca L. var. ansu (19.2 g) and Glycyrrhiza uralensis (9.6 g) powder were mixed with distilled water in an amount three times that of the medicinal material and soaked for 30 min. After 4 h of extraction with a volatile oil extractor, the material was filtered through four layers of gauze to obtain 60 mL of primary water extract, then 50 mL of water was added for another boiling for 30 min. After gauze filtering, the two aqueous extracts were mixed to yield 100 mL extracts finally. The final concentration of MHT was 8 g/mL (calculated as Ephedra herb content).
The separate preparation of Ephedra sinica, Cinnamomum cassia, Prunus armeniaca L. var. ansu and Glycyrrhiza uralensis: 1 g of Ephedra sinica was crushed and sonicated in 25 mL of methanol for 30 min, and then the supernatant was obtained after centrifugation. Cinnamomum cassia, Prunus armeniaca L. var. ansu and Glycyrrhiza uralensis were prepared in the same way as Ephedra sinica.
For HPLC, a sunFire C18 (4.6 mm ×150 mm, 5 μm) chromatographic column was used with 0.1% H3PO4 water solution and acetonitrile as the mobile phase for gradient elution. The gradient elution program includes 0 min with 5% acetonitrile, 20 min with 35% acetonitrile, 50 min with 45% acetonitrile, 55 min with 50% acetonitrile and 65 min with 80% acetonitrile. The detection wavelength was 260 nm.
Statistical analysis
The data are expressed as the means ± SDs. Statistical comparisons were made by one-way ANOVA using SPSS software, version 16.0. p < 0.05 was considered statistically significant.
Results and discussion
MHT extended sensitization time and abdominal breathing time in OVA-induced asthma mice
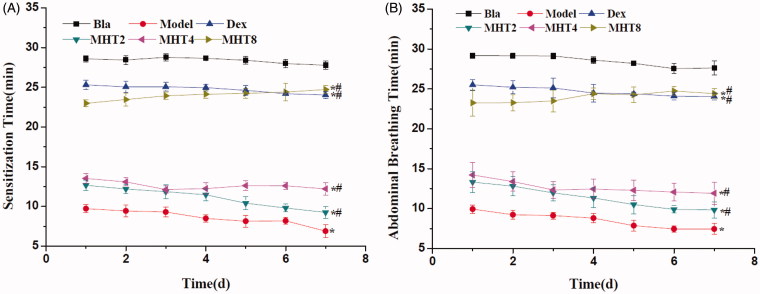
Sensitization time is the duration when asthma symptoms occur in mice, including vigorous scratching of ear and nose. Abdominal breathing is as the duration when mice have deep breathing. As shown in Figure 2(A,B), the sensitization time and abdominal breathing time in the model groups were faster than those in the blank groups. In addition, the above measures in the MHT group were significantly different from those in the model group, especially the high-dose group which showed effects similar to those in the dexamethasone group.
Figure 2.
The effect of MHT on OVA-induced asthma in Balb/c. (A) The effect of MHT on daily sensitization time of OVA-induced asthma in Balb/c. (B)The effect of MHT on daily Abdominal Breathing time of OVA-induced asthma in Balb/c. *p < 0.01, different from the sensitization time of the blank group, n = 6, mean þ SD; #p < 0.01, different from the sensitization time of the model group, n = 6, mean þ SD. Bla: blank group; Dex: dexamethasone group; MHT2: Mahuang Tang group of 2 g/kg; MHT4: Mahuang Tang group of 4 g/kg; MHT8: Mahuang Tang group of 8 g/Kg; Model: model group.
MHT alleviated OVA-induced eosinophilic airway inflammation
Table 1 lists the total number of cells and percentage of eosinophils (EOS%) in BLFA. Commonly, leucocytes can be detected in BLFA in low amounts. During induced bronchial asthma, the mice showed significantly increased leucocyte levels, especially eosinophils, which directly reflected the severity of asthma. The total leucocyte number and EOS% in the model group were significantly increased from the blank group. Increasing the treatment dosage of MHT decreased the total leucocyte number and EOS% of BLFA, especially in the high-dose group, where the total number of leucocytes and EOS% were significantly decreased in comparison with the model group. The results indicated that a high dose of MHT inhibited the generation and infiltration of inflammatory cells, thus significantly relieving asthma symptoms.
Table 1.
Total cell of BLFA and EOS% of BLFA (values are mean ± S.D., n = 20).
| Groups | Total cells (×104/mL) | EOS (%) |
|---|---|---|
| A | 8.42 ± 0.95 | 0.67 ± 0.29 |
| B | 24.08 ± 1.76* | 21.50 ± 1.00* |
| C | 9.92 ± 1.01# | 3.33 ± 0.29*# |
| D | 20.17 ± 0.52*# | 19.50 ± 0.50*# |
| E | 19.17 ± 0.52*# | 19.00 ± 0.50*# |
| F | 14.67 ± 1.01*# | 10.50 ± 0.50*# |
*p < 0.05, different from the total cells of BLFA of the blank group; #p < 0.05, different from the total cells of BLFA of the model group; *p < 0.05, different from the EOS% of BLFA of the blank group; #p < 0.05, different from the EOS% of BLFA of the model group; A: blank group, B: model group, C: dexamethasone group, D: Mahuang Tang group of 2 g/kg, E: Mahuang Tang group of 4 g/kg and F: Mahuang Tang group of 8 g/kg. BLFA: bronchoalveolar lavage fluid. EOS: eosinophils.
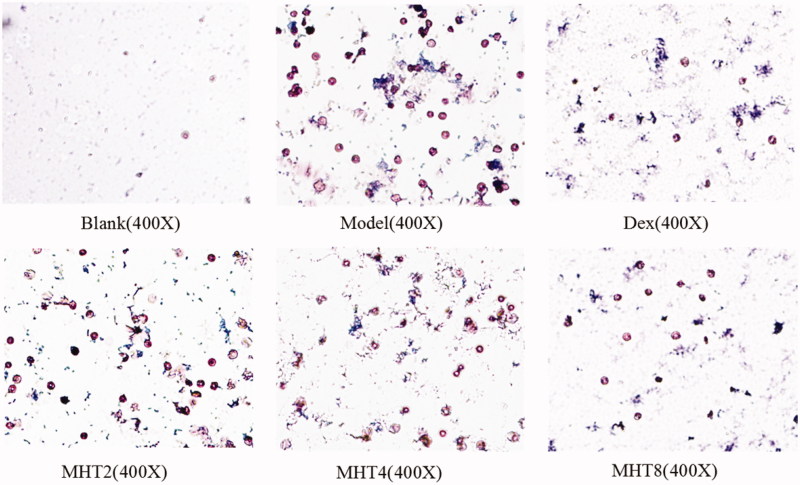
In a detailed analysis of eosinophils with Wright’s stain, cell nuclei were dyed pink, and the cytoplasm was dyed blue. In Figure 3, the numbers of eosinophils in the model group were significantly greater than those in the blank group. Cell numbers in the treatment group were significantly decreased in comparison with the model group, especially in the high dose group. These results further illustrated that a high dose of MHT can reduce the generation of inflammatory cells and therefore achieve the purpose of the treatment of asthma.
Figure 3.
Eosinophils determined by wright's stain. The number of cells reflects the severity of the inflammation. Eosinophil decreased significantly in the high dose group of MHT. Scale bar, 50 μm. Blank: blank group, Model: model group, Dex: dexamethasone group; MHT2: Mahuang Tang group of 2 g/kg; MHT4: Mahuang Tang group of 4 g/kg; MHT8: Mahuang Tang group of 8 g/kg.
MHT mitigated pathological changes
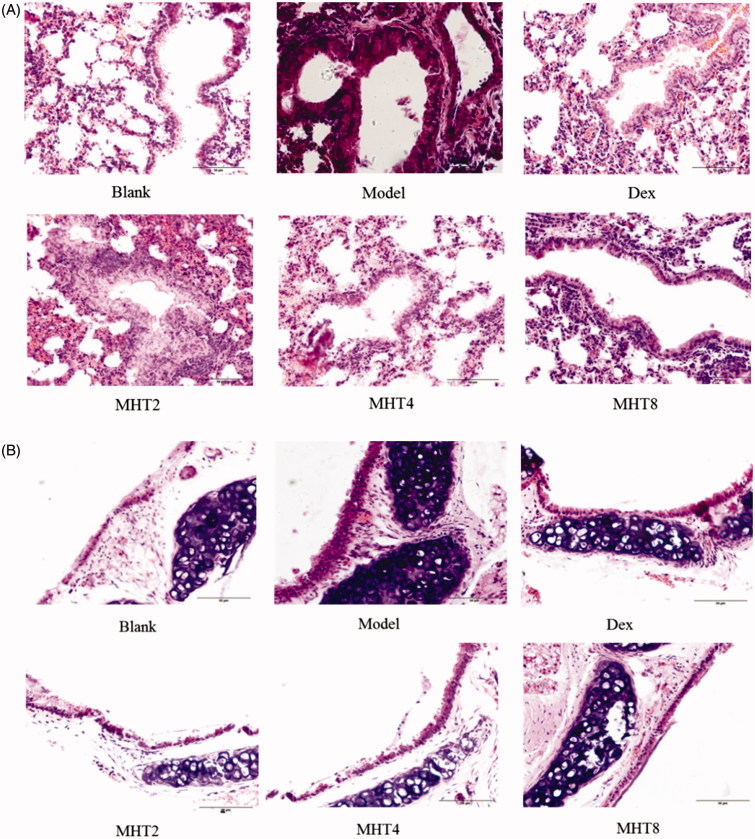
We next examined the lung and trachea histopathology stained with H&E. As shown in Figure 4, the lung blood vessel walls and bronchial walls in the model group tended to be thickened, and the simple columnar epithelium cell of bronchioles fell off. The other pathological changes observed were summarized as follows: simple columnar epithelium cells changed into tall columnar epithelial cells with a disordered arrangement; cell degeneration was observed, specifically necrosis of the nucleus and infiltration of inflammation cells was observed. Finally, visible emphysema occurred. Treatment with MHT alleviated the pulmonary inflammatory infiltration issue to some extent, In particular, in the high dose group, an outline of the lung bronchioles was visualized, and the arrangement of the cells tended to the norm. In summary, a high dose of MHT exhibited significant relief of asthma symptoms.
Figure 4.
Hematoxylin-eosin stained (A–B). (A) Representative H&E-stained sections of lung. (B) Representative H&E-stained sections of trachea. Scale bar, 50 µm. Blank: blank group; Model: model group; Dex: dexamethasone group; MHT2: Mahuang Tang group of 2 g/kg; MHT4: Mahuang Tang group of 4 g/kg; MHT8: Mahuang Tang group of 8 g/kg.
MHT regulated signal pathways by RNA-seq were analyzed
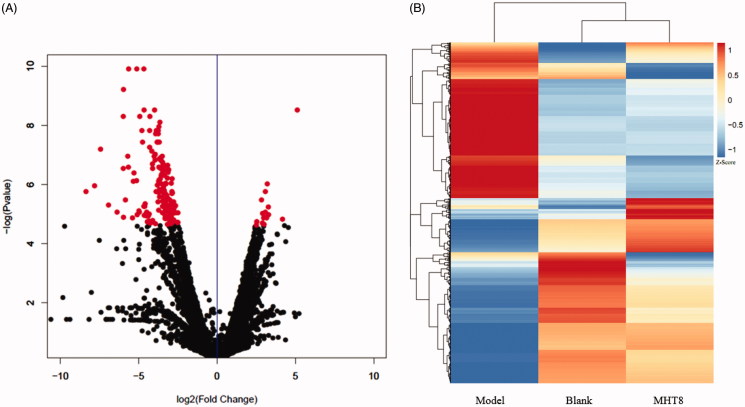
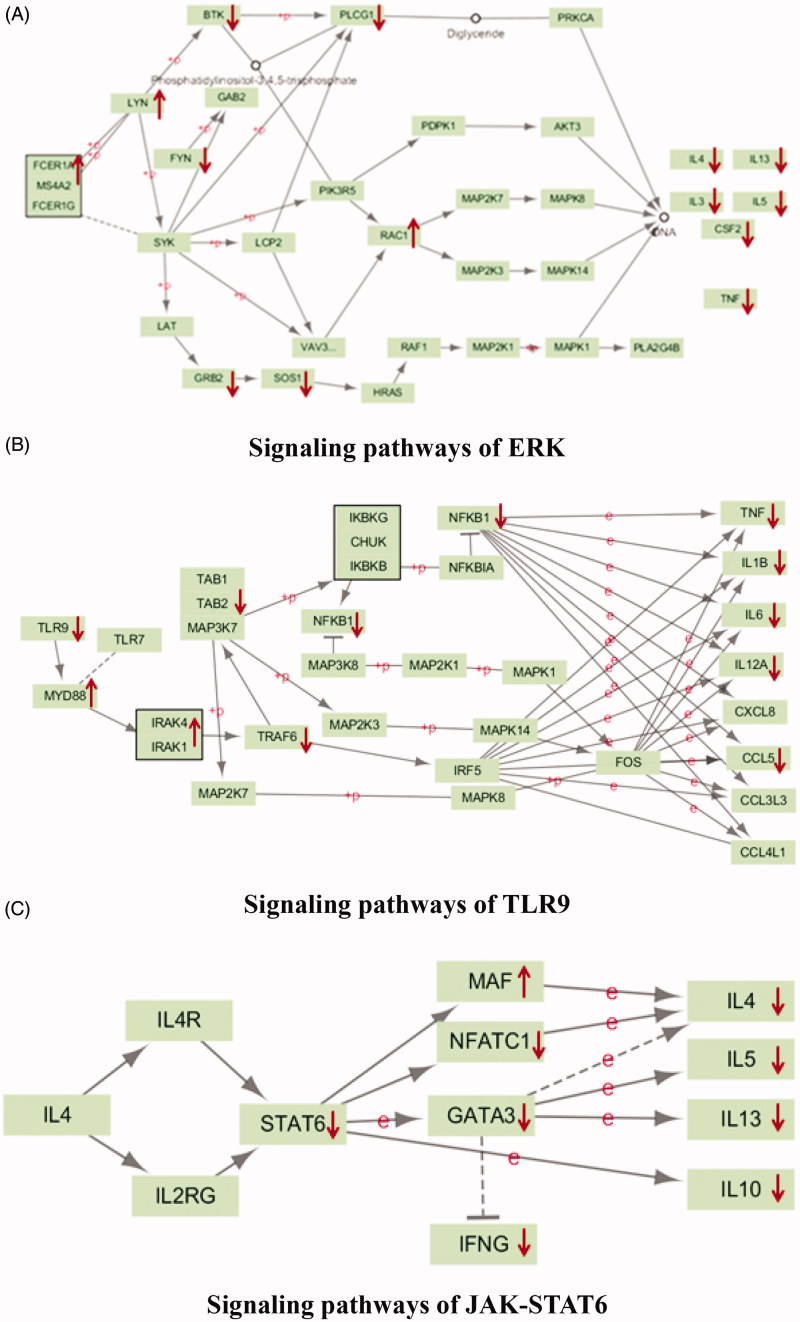
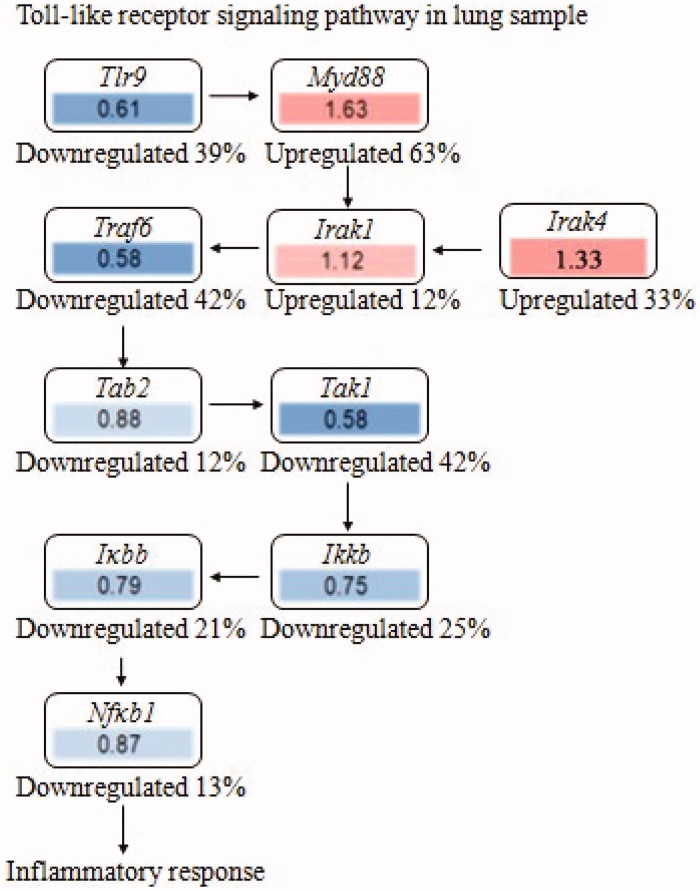
To further validate interrelation between the 20 kinds of components in MHT and proteins, the RNA-seq method was applied to detect of the transcriptome genes of lung tissues. Astringent threshold (fold change >2 and p < 0.01) was applied for specific identification of potential candidate RNAs. In total, MHT treatment upregulated 115 RNAs and downregulated 185 (Figure 5); these data are detailed in Supplement Table S3. Results of RNA-seq mainly involve the changes in the ERK, FcεRI, PI3K-Akt, TLRs and JAK-STAT6 signaling pathways (Figure 6). It is speculated that MHT regulates these pathways to exert anti-inflammatory and anti-asthmatic effects. Among these, TLR9 showed the most marked decrease (Figure 7); therefore, we chose the TLR9 pathway to further analyse. Combined with the preliminary results of bioinformatics, the 32 types of key proteins in the asthma pathway were further explored (Table 2).
Figure 5.
The results of RNA-seq. (A) volcano plot. After 24 h, the RNAs of the lung were isolated and were analyzed by microarray, and the different expression genes (DEGs) were displayed as volcano plot. Data were presented as mean þ SD from three independent experiments with triple replicatesper experiment.*p < 0.01 compared to blank group. (B) Heatmap analysis of gene expression profiles. The heatmap shows unsupervised hierarchical clustering of blank, MHT8 and model group based on the expression levels of all genes. Blank: blank group; Model: model group; MHT8: Mahuang Tang group of 8 g/kg.
Figure 6.
The result of RNA-seq mainly involves the signaling pathway. (A) signaling pathways of ERK; (B) signaling pathways of TLR9; (C) signaling pathways of JAK-STAT6.
Figure 7.
The effect of fold change with RNA-seq analysis for toll-like receptor nine signaling pathway in lung. Fold change: MHT8/Model, upregulated or downregulated percentage: (MHT8-Model)/Model. Toll-like receptor 9 signaling pathway in lung sample. MHT8: Mahuang Tang group of 8 g/kg; Model: model group.
Table 2.
RNA-seq analysis of lung for targeted protein in asthma signaling pathway.
| Protein | Gene_id | Model/Blank | MHT8/Model |
|---|---|---|---|
| IL-4 | Il4 | 7.85 | 0.13 |
| STAT6 | Stat6 | 1.10 | 0.95 |
| C-MAF(MAF) | Maf | 0.60 | 1.32 |
| CD40L(Cd40lg) | Cd40lg | 9.85 | 0.34 |
| IFN-γ(IFNG) | Ifng | 9.02 | 0.42 |
| SOS1 | Sos1 | 1.14 | 0.89 |
| GRB2 | Grb2 | 1.81 | 0.63 |
| TNFα(TNFA) | Tnf | 5.80 | 0.28 |
| RAC(RAC1) | Rac1 | 0.86 | 1.10 |
| LYN | Lyn | 0.61 | 1.58 |
| AKT(Akt1) | Akt1 | 1.07 | 0.96 |
| PDK1 | Pdk1 | 1.18 | 0.85 |
| BTK | Btk | 1.65 | 0.98 |
| PI3K(PIK3CA) | Pik3ca | 0.93 | 1.03 |
| FYN | Fyn | 1.69 | 0.63 |
| FcεRI(FCER1A) | Fcer1a | 0.60 | 1.46 |
| PLCγ(PLCG1) | Plcg1 | 1.10 | 0.93 |
| CHRNA4 | Chrna4 | 0.45 | 1.14 |
| PTPRC | Ptprc | 7.75 | 0.22 |
| F2R | F2r | 0.38 | 2.35 |
| PTK2B | Ptk2b | 3.76 | 0.34 |
| TLR9 | Tlr9 | 3.62 | 0.61 |
| RARA | Rara | 0.84 | 1.17 |
| BCL2 | Bcl2 | 2.17 | 0.47 |
| PRKCD | Prkcd | 1.15 | 0.88 |
| SHH | Shh | 0.51 | 1.75 |
| FLT3 | Flt3 | 2.24 | 0.61 |
| PLA2G4A | Pla2g4a | 0.42 | 2.01 |
| LCK | Lck | 57.90 | 0.04 |
| ITGAL | Itgal | 7.02 | 0.22 |
| FKBP1A | Fkbp1a | 0.71 | 1.30 |
| ERBB2 | Erbb2 | 0.32 | 2.95 |
Combined with the KEGG database, 32 key proteins were involved in the ERK, FcεRI, PI3K-Akt, TLRs and JAK-STAT6 signaling pathways. Based on the decrease in inflammatory factor expression after treatment, the anti-inflammatory effect of MHT might be associated with the above signaling pathways. Combining these finding with the literature, we selected toll-like pathways and the key protein TLR9 for launching a detailed investigation. Results of gene sequencing in the lung and changes in the candidate key target of TLR9 in the subnet are shown below.
According to related research (Li et al. 2011; Cho and Hsieh 2016; Pan et al. 2016), we learned that the TLR9 ligand is a kind of CpG motif or synthetic CpG-ODN in bacterial and viral DNA. Through swallowing, CpG DNA enters the body, then binds with TLR9 and promotes dimerization; based on the TIR domain, dimerized TLR9 raises adapter molecules with myeloid differentiation factor 88 (MyD88) (Martin and Wesche 2002). MyD88 belongs to the family of Toll/IL-1R; in essence, MyD88 is a kind of soluble protein. The structure of this protein comprises three functional segment which are an N-terminal dead zone (death domain, DD), a center area and a C-terminal Toll region. Similar to IL-1 receptor cytoplasmic domains, the Toll zone possesses approximately 130 amino acids and transport signals through the recruitment connexin. Toll-interacting protein (Tollip, MyD88 regulatory protein) is commonly associated with IL-1R-associated kinase (IRAK, downstream kinase of MyD88). When binding of the Toll receptor with its ligand occurred, recruiting of IRAK will be accompanied by the falling of Tollip from the dimer. Thus, we accomplish the auto-phosphorylation of IRAK and signal transportation downstream. IRAK1 and IRAK4 are the primarily involved IRAK proteins (Takeda and Akira 2004). The auto-phosphorylation of IRAK4 (which interacted with the protein complex) promotes auto-phosphorylation of IRAK4 as well, leading to the introduction of a large amount of charge. Finally, IRAK1 dissociates from the protein complex. Moreover, based on phosphorylation of IRAK1 to TRAF6 (TNF-receptor-associated factor 6), a new protein complex of IRAK1-TRAF6 forms and dissociates from the TLR9-MyD88-IRAK complex (Heit et al. 2003). IRAK1-TRAF6 continued to bind with TAB2 (the adapter protein of TAB1), and the IRAK1-TRAF6-TAB2 complex will interact with TAB1 to cause degradation of IRAK1 in cell membranes. Then, the newly generated protein complex TRAF6-TAB2-TAK1is transported to the cytoplasm, and TRAF6 and E2 ligase together catalyze the generation of poly-ubiquitinated protein chains that can eventually activate TAK1 and the IKK signal pathway. The activation of I-κB facilitates the release and transport of NF-κB into the nucleus. Finally, a series of pro-inflammatory cytokines (Brentano et al. 2005) are generated.
The RNA-seq results showed that the TLR9 regulatory effect of MHT might function by inhibiting TLR9, thus blocking the activation of theTLR9-MyD88-IRAK complex and disabling the dissociation of IRAK1 (Figure 7). A series of subsequent steps, including the recruiting of TRAF6, the transport of TAB2 and TAK1 into the cytoplasm and the release of NF-κB and pro-inflammatory cytokines were also inhibited, thus achieving the therapeutic goal of MHT against asthma. The other results, including the high expression of MyD88 and IRAK, might be mediated by some other members of the TLR family. Future detailed research into these mechanisms is needed.
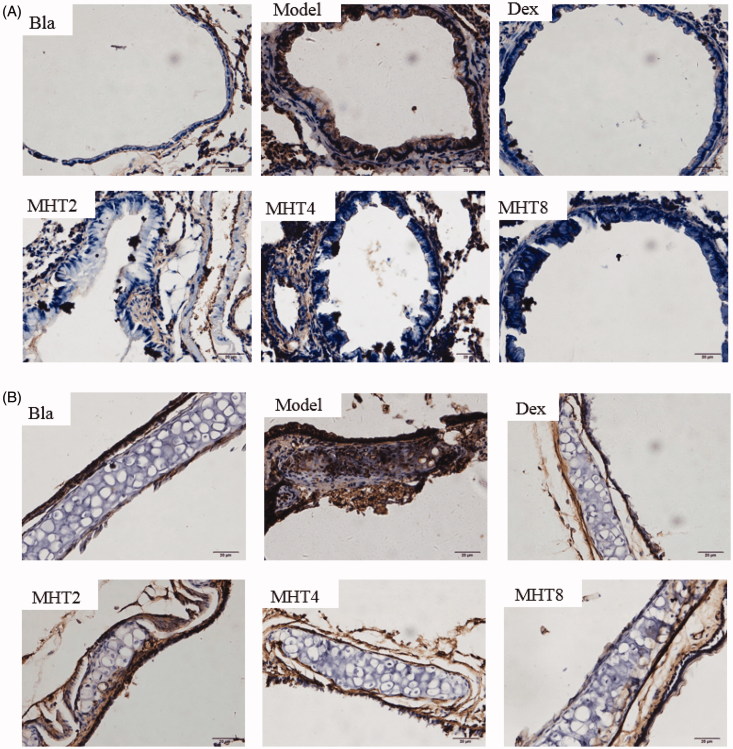
MHT attenuated TLR9 expression in OVA-induced mice
In the lung, TLR9 was significantly increased by OVA challenge and was remarkably decreased in the MHT-treated groups (Figure 8(A)). In tracheal epithelial cells, TLR9 was also induced in the OVA group and showed a significant reduction in the MHT groups as well (Figure 8(B)). In particular, TLR9 was attenuated by a high dose of MHT. These results were consistent with the TLR9 expression by RNA-seq in the lungs. In brief, the finding that a high dose of MHT had an inhibitory effect on TLR9 was certified again, thus achieving the desired asthma therapy effect.
Figure 8.
MHT decreases the level of TLR9 in lung and trachea tissues. (A) Lung determined by histology staining for TLR9 (brown). Quantification of staining intensity and representative images scale bars, 20 µm. (B) Trachea determined by histology staining for TLR9 (brown). Quantification of staining intensity and representative images. Scale bars, 20 µm. Bla: blank group; Model: model group; Dex: dexamethasone group; MHT2: Mahuang Tang group of 2 g/kg, MHT4: Mahuang Tang group of 4 g/kg; MHT8: Mahuang Tang group of 8 g/kg.
Further analysis of compounds from MHT with TLR9 interaction by molecular docking
To further explore the molecular mechanism of MHT in inhibiting TLR9, according to a previous study (Dai et al. 2015), VD3 can downregulate TLR9 expression levels. Accordingly, molecular docking was used to analyze interactions between VD3 and TLR9. We also analyzed the binding site between MHT and TLR9. As seen in Table 3, compounds in MHT that showed similar binding energies to VD3 included flavonoids in licorice and phenyl propanoid compounds in almonds, specifically ganS002, ganS003, ganS005, ganS027, xingS001 and xingS008. These compounds showed strong hydrophobicity and could fit well with the hydrophobic binding pocket in the TLR9 active site. In a detailed analysis, the H-bond-forming sites of VD3 and TLR9 showed high similarities, especially with ganS002, ganS003, ganS005, ganS027, xingS001 and xingS008 (Table 4), and all these compounds can interact with Arg426 through H-bonding. Combined with results of RNA-seq, immunohistochemistry and bioinformatics with computational simulation, we reasonably predicted that six compounds, namely, ganS002, ganS003, ganS005, ganS027, xingS001 and xingS008 (Table 5), may possessTLR9inhibitory activity and may mediate the TLR9 signal pathway, thus achieving the desired asthma therapy effect.
Table 3.
Compared VD3 and TLR9 protein docking with the 19 compounds.
 |
 |
 |
Table 4.
H-bond interactions of compounds and 1,25-dihydroxy vitamin D3 with active sites of TLR9.
| H-bond interactions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compounds | Arg426 | Ser350 | Gly563 | Tyr345 | Lys347 | Gly565 | Ala511 | Val349 | Gln346 | Asp534 |
| 1,25-dihydroxy vitamin D3 | + | – | + | – | – | – | – | – | – | – |
| maS010 | – | – | – | – | – | – | – | – | – | – |
| maS040 | – | – | – | + | – | + | – | – | – | – |
| ganS002 | + | – | – | – | + | – | – | – | – | – |
| ganS003 | + | – | – | – | + | – | + | – | – | – |
| ganS005 | + | – | – | – | + | – | + | – | – | – |
| ganS027 | + | – | – | – | + | – | – | + | + | – |
| xingS001 | + | + | – | – | – | + | – | – | – | – |
| xingS002 | + | – | – | – | – | – | – | – | – | – |
| xingS003 | + | – | – | – | + | – | – | – | – | – |
| xingS007 | + | – | – | – | – | – | – | – | – | – |
| xingS008 | + | – | – | + | – | – | – | – | – | – |
| xingS022 | + | – | – | – | – | – | – | – | – | – |
| xingS042 | – | – | – | – | – | – | – | – | – | – |
| ximgS043 | – | – | – | – | – | – | – | – | – | – |
| guiS001 | + | + | – | – | – | – | – | – | – | – |
| guiS014 | + | – | – | – | – | – | – | – | – | – |
| guiS021 | + | – | – | – | – | – | – | – | – | – |
| guiS041 | + | + | – | – | – | – | – | – | – | – |
| guiS077 | – | – | – | – | – | – | – | – | – | – |
“+” referred to formed H-bond interactions; “–” referred to have no H-bond interactions.
Table 5.
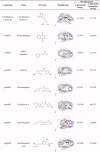
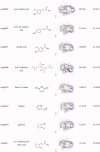
The compounds with the effect of inhibiting TLR9 from MHT.
| Compounds | Name | Structure |
|---|---|---|
| ganS002 | echinatin | 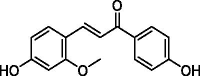 |
| ganS003 | isoliquiritigenin | 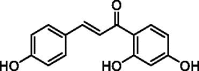 |
| ganS005 | isoliquiritigenin |  |
| ganS027 | Neoisoliquiritin |  |
| xingS001 | neochlorogenic acid |  |
| xingS008 | 8-O,4'-Diferulic acid |  |
Identify fingerprints for MHT
The MHT and single-herb samples were analysed by HPLC under the same conditions to determine the source of each peak. The HPLC fingerprint of MHT was established to exhibit 69 peaks. Through comparative analysis, it was determined that 50 common peaks were derived from Ephedra sinica; 16 common peaks were derived from Cinnamomum cassia; 10 common peaks were derived from Prunus armeniaca L. var. ansu and 29 common peaks were derived from Glycyrrhiza uralensis (Supplement Figure 2). The established HPLC fingerprint has better precision, reproducibility and stability and can be applied in the quality control of MHT.
Conclusions
Asthma, a major chronic inflammatory disease of the airways, occurs in people of all ages (Szefler et al. 2014). Current treatment is limited. Hence, we aimed to determine whether MHT, which shows efficacy against inflammation, has a potential application in treating asthma. According to results of behavioural indicators, Wright’s staining and H&E staining, we investigated three doses of MHT that might be effective in asthma mice in this experiment. Our study clearly demonstrated that MHT in high doses effectively prevented inflammation and airway remodelling in a mouse model of acute asthma. Moreover, there were appreciable differences between the protective effects of the two lower doses of MHT used throughout the study, suggesting that MHT in 8 g/kg doses might be effectively used for treating asthma.
Eosinophils are a critical component in the pathogenesis of asthma. The release of eosinophilic granulomatous proteins leads to pathological changes characteristic of asthma and airway hyper responsiveness (Cardet and Israel 2015). Therefore, using the eosinophil count to judge the occurrence of bronchus asthma is a great help. In our study, there was a decreased release of eosinophils in the high-dose group compared with the low- and middle-dose groups, which might result from a therapeutic dose-dependent response to MHT. Moreover, MHT mitigated pathological changes, especially in the high-dose group.
The use of bioinformatics was prevalent at the moment (Dolled-Filhart et al. 2013; Magana et al. 2014). Combining the results of bioinformatics and RNA-seq, it was found that MHT can regulate multiple signaling pathways to treat asthma. We selected the TLR9 pathway for further analysis. We assumed that lignans in MHT played a role as the main ligand of TLR9. TLR9 is mainly expressed in a variety of immune cells such as B cells, mononuclear cells and dendritic cells and so on; this was determined mainly through the identification of unmethylated CpG fragments on the relevant biological DNA (Xu et al. 2015). In asthmatic patients, more IL-25 was released in respiratory epithelial cells than in normal individuals. IL-25-induced expression of TLR9 on dendritic cells was increased and enhanced the response to TLR9 activation (Tworek et al. 2016). Duechs et al. (2014) found that activation of TLR9 significantly reduced airway eosinophil infiltration and IgE levels in patients with asthma and that TLR9 receptor activation can increase the Th1-type immune response and reduce Th2 type cytokines IL- 4, IL-5, IL-1β and IL-12, suggesting that enhancing the Th1-type immune response may be the mechanism by which TLR9 activation inhibits Th2-type inflammation. TLR9 has been shown to suppress airway remodelling in the respiratory tract (Jain et al. 2002). In vivo experiments also confirmed that TLR9 activation reduces allergen-induced respiratory allergic inflammation (Ashino et al. 2008). These findings suggest that TLR9 has potential as a prophylactic and therapeutic asthma target. The RNA-seq and immunohistochemical data demonstrated that MHT effectively attenuated TLR9 expression in lung and bronchia, particularly in the high-dose MHT group. It is noteworthy that the Myd88 and Irak-related cytokines Irak1 and Irak4 were not significantly reduced in our model or affected by MHT. Although it was not clear why the levels of these cytokines were not decreased by OVA exposure, our data suggested that the TLR9-producing inflammatory response could play an important role. Therefore, the inhibition of TLR9 by MHT could eventually prevent subsequent airway inflammation.
In summary, the current study demonstrated that the traditional Chinese medicine MHT could effectively decrease OVA-induced sensitization time, abdominal breathing time, eosinophilic airway inflammation and pathological changes, most likely through downregulation of TLR9 by the MHT constituent compounds ganS002, ganS003, ganS005, ganS027, xingS001 and xingS008. However, none of the compounds regulatory TLR9 pathways are from the key component herb of Herba Ephedrae, the key compounds may regulate bronchial asthma through other pathways, such as ERK, FcεRI, PI3K-Akt, TLRs and JAK-STAT6 signaling pathways, these conclusions require us to further verify other pathways involved in asthma treatment of MHT. In any case, our findings support a possible application ofTLR9 as a therapeutic target in MHT for patients with acute asthma.
Supplementary Material
Funding Statement
This work was supported by National Science and Technology Major Projects for New Drug Development [grant number: 2017ZX09305005].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ashino S, Wakita D, Zhang Y, Chamoto K, Kitamura H, Nishimura T. 2008. CpG-ODN inhibits airway inflammation at effector phase through down-regulation of antigen-specific Th2-cell migration into lung. Int Immunol. 20:259–266. [DOI] [PubMed] [Google Scholar]
- Beasley R. 1998. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet. 351:1225–1232. [PubMed] [Google Scholar]
- Brentano F, Kyburz D, Schorr O, Gay R, Gay S. 2005. The role of Toll-like receptor signalling in the pathogenesis of arthritis. Cell Immunol. 233:90–96. [DOI] [PubMed] [Google Scholar]
- Burgueño JF, Barba A, Eyre E, Romero C, Neunlist M, Fernández E. 2016. TLR2 and TLR9 modulate enteric nervous system inflammatory responses to lipopolysaccharide. J Neuroinflammation. 13:187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardet JC, Israel E. 2015. Update on reslizumab for eosinophilic asthma. Expert Opin Biol Ther. 15:1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll WD, Wildhaber J, Brand PL. 2012. Parent misperception of control in childhood/adolescent asthma: the Room to Breathe survey. Eur Respir J. 39:90–96. [DOI] [PubMed] [Google Scholar]
- Chen YC, Liu CM, Jeng JH, Ku CC. 2014. Association of pocket epithelial cell proliferation in periodontitis with TLR9 expression and inflammatory response. J Formos Med Assoc. 113:549–556. [DOI] [PubMed] [Google Scholar]
- Cho KO, Hsieh J. 2016. Microglial TLR9: Guardians of homeostatic hippocampal neurogenesis. Epilepsy Currents. 16:39–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai ZH, Tan B, Yang H, Wang O, Qian JM, Lv H. 2015. 1,25-Hydroxyvitamin D relieves colitis in rats via down-regulation of toll-like receptor 9 expression. Croat Med J. 56:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolled-Filhart MP, Lee M Jr, Ou-Yang CW, Haraksingh RR, Lin JC. 2013. Computational and bioinformatics frameworks for next-generation whole exome and genome sequencing. Scientific World J. 2013:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duechs MJ, Tilp C, Tomsic C, Gantner F, Erb KJ. 2014. Development of a novel severe triple allergen asthma model in mice which is resistant to dexamethasone and partially resistant to TLR7 and TLR9 agonist treatment. PLoS One. 9:e91223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Cai C, Liu X, Ku X, Jiang H, Gao D, Li H. 2013. ChemMapper: a versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics. 29:1827–1829. [DOI] [PubMed] [Google Scholar]
- Heit A, Maurer T, Hochrein H, Bauer S, Huster KM, Busch DH, Wagner H. 2003. Cutting edge: Toll-like receptor 9 expression is not required for CpG DNA-aided cross-presentation of DNA-conjugated antigens but essential for cross-priming of CD8 T cells. J Immunol. 170:2802–2805. [DOI] [PubMed] [Google Scholar]
- Hu D, Yang X, Xiang Y, Li H, Yan H, Zhou J, Caudle Y, Zhang X, Yin D. 2015. Inhibition of Toll-like receptor 9 attenuates sepsis-induced mortality through suppressing excessive inflammatory response. Cell Immunol. 295:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Jain VV, Kitagaki K, Businga T, Hussain I, George C, O'Shaughnessy P, Kline JN. 2002. CpG-oligodeoxynucleotides inhibit airway remodeling in a murine model of chronic asthma. J Allergy Clin Immun. 110:867–872. [DOI] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. . 2009. Human protein reference database-2009 update. Nucleic Acids Res. 37:D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN, Hammad H. 2015. The immunology of asthma. Nat Immunol. 16:45–56. [DOI] [PubMed] [Google Scholar]
- Levy ML. 2014. Asthma still kills: little change over five decades. NPJ prim Care Resp M. 24:14029–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai H, Xu J, Ying S, Zhang Y. 2010. A mouse protein interactome through combined literature mining with multiple sources of interaction evidence. Amino Acids. 38:1237–1252. [DOI] [PubMed] [Google Scholar]
- Li Y, Cao H, Wang N, Xiang Y, Lu Y, Zhao K, Zheng J, Zhou H. 2011. A novel antagonist of TLR9 blocking all classes of immunostimulatory CpG-ODNs. Vaccine. 29:2193–2198. [DOI] [PubMed] [Google Scholar]
- Lindensmith J, Morrison D, Deveau C, Hernandez P. 2004. Over diagnosis of asthma in the community. Can Respir J. 11:111–116. [DOI] [PubMed] [Google Scholar]
- Magana AJ, Taleyarkhan M, Alvarado DR, Kane M, Springer J, Clase K. 2014. A survey of scholarly literature describing the field of bioinformatics education and bioinformatics educational research. CBE Life Sci Educ. 13:607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund B, Tunsäter A, Bengtsson C. 1999. How often is the diagnosis bronchial asthma correct? Fam Pract. 16:112–116. [DOI] [PubMed] [Google Scholar]
- Martin MU, Wesche H. 2002. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochim Biophys Acta. 1592:265–280. [DOI] [PubMed] [Google Scholar]
- Pan X, Li B, Kuang M, Liu X, Cen Y, Qin R, Ding G, Zheng J, Zhou H. 2016. Synthetic human TLR9-LRR11 peptide attenuates TLR9 signaling by binding to and thus decreasing internalization of CpG oligodeoxynucleotides. Int J Mol Sci. 17:242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce N, Aït-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, Robertson C. 2007. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax. 62:758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politano G, Benso A, Savino A, Di Carlo S. 2014. ReNE: a cytoscape plugin for regulatory network enhancement. PLoS One. 9:e115585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe KF, Vermeire PA, Soriano JB, Maier WC. 2000. Clinical management of asthma in 1999: the Asthma Insights and Reality in Europe (AIRE) study. Eur Respir J. 16:802–807. [DOI] [PubMed] [Google Scholar]
- Suresh MV, Thomas B, Dolgachev VA, Sherman MA, Goldberg R, Johnson M, Chowdhury A, Machado-Aranda D, Raghavendran K. 2016. Toll-like receptor-9 (TLR9) is requisite for acute inflammatory response and injury following lung contusion. Shock. 46:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szefler SJ, Chmiel JF, Fitzpatrick AM, Giacoia G, Green TP, Jackson DJ, Nielsen HC, Phipatanakul W, Raissy HH. 2014. Asthma across the ages: knowledge gaps in childhood asthma. J Allergy Clin Immun. 133:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Akira S. 2004. TLR signaling pathways. Semin Immunol. 16:3–9. [DOI] [PubMed] [Google Scholar]
- Tiwari M, Lee JK. 2010. Molecular modeling studies of l-arabinitol 4-dehydrogenase of Hypocrea jecorina: its binding interactions with substrate and cofactor. J Mol Graph Model. 28:707–713. [DOI] [PubMed] [Google Scholar]
- Tworek D, Smith SG, Salter BM, Baatjes AJ, Scime T, Watson R, Obminski C, Gauvreau GM, O'Byrne PM. 2016. IL-25 receptor expression on airway dendritic cells after allergen challenge in subjects with asthma. am J RespirCrit Care Med. 193:957–964. [DOI] [PubMed] [Google Scholar]
- Van Schayck CP, Van Der Heijden FM, Van Den Boom G, Tirimanna PR, Van Herwaarden CL. 2000. Underdiagnosis of asthma: is the doctor or the patient to blame? The DIMCA project. Thorax. 55:562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao MM, Pan CS, Liu YY, Ma LQ, Yan L, Fan JY, Wang CS, Huang R, Han JY. 2017. Post-treatment with Ma-Huang-Tang ameliorates cold-warm-cycles induced rat lung injury. Sci Rep. 7:312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Wong EB, Rubio D, Roscoe F, Ma X, Nair S, Remakus S, Schwendener R, John S, Shlomchik M, et al. . 2015. Sequential activation of two pathogen-sensing pathways required for Type I interferon expression and resistance to an acute DNA virus infection. Immunity. 43:1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Garzón JI, Deng L, Honig B. 2013. PrePPI: a structure-informed database of protein-protein interactions. Nucleic Acids Res. 41:D828–D833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YX, Wu JM, Feng XS, Jia Y, Huang J, Hao ZH, Zhao SY, Wang JH. 2015. In silico analysis and experimental validation of lignan extracts from Kadsura longipedunculata for potential 5-HT1AR agonists. PLoS One. 10:e0130055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.