Abstract
Cardiac Nav1.5 and Kir2.1–2.3 channels generate Na (INa) and inward rectifier K (IK1) currents, respectively. The functional INa and IK1 interplay is reinforced by the positive and reciprocal modulation between Nav15 and Kir2.1/2.2 channels to strengthen the control of ventricular excitability. Loss-of-function mutations in the SCN5A gene, which encodes Nav1.5 channels, underlie several inherited arrhythmogenic syndromes, including Brugada syndrome (BrS). We investigated whether the presence of BrS-associated mutations alters IK1 density concomitantly with INa density. Results obtained using mouse models of SCN5A haploinsufficiency, and the overexpression of native and mutated Nav1.5 channels in expression systems — rat ventricular cardiomyocytes and human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) — demonstrated that endoplasmic reticulum (ER) trafficking–defective Nav1.5 channels significantly decreased IK1, since they did not positively modulate Kir2.1/2.2 channels. Moreover, Golgi trafficking–defective Nav1.5 mutants produced a dominant negative effect on Kir2.1/2.2 and thus an additional IK1 reduction. Moreover, ER trafficking–defective Nav1.5 channels can be partially rescued by Kir2.1/2.2 channels through an unconventional secretory route that involves Golgi reassembly stacking proteins (GRASPs). Therefore, cardiac excitability would be greatly affected in subjects harboring Nav1.5 mutations with Golgi trafficking defects, since these mutants can concomitantly trap Kir2.1/2.2 channels, thus unexpectedly decreasing IK1 in addition to INa.
Keywords: Cardiology, Cell Biology
Keywords: Arrhythmias, Potassium channels, Sodium channels
Patients carrying Brugada Syndrome-associated Golgi trafficking-defective Nav1.5 mutations exhibit a greater depression of cardiac excitability than those carrying other type of loss-of-function mutations.
Introduction
Cardiac Na+ channels (Nav1.5) generate inward Na+ current (INa), which is responsible for the depolarization of atrial and ventricular action potentials (APs) (1, 2). Inward rectifier current (IK1) is generated by homo- and/or heterotetramers of Kir2.1, Kir2.2, and Kir2.3 channels. IK1 is critical for the setting resting membrane potential (RMP) and, to a lesser extent, the duration of AP (APD) during the final phase of repolarization (3). In the 1950s Weidmann (4) established that the encompassed voltage-dependent activity of INa and IK1 is a major determinant of cardiac excitability and conduction velocity. Furthermore, we demonstrated that the functional expression of Nav1.5 and Kir2.1/Kir2.2 channels is reciprocally regulated (5–8), a mechanism that probably strengthens control of ventricular excitability.
There are many inherited cardiac rhythm disturbances (i.e., BrS, progressive cardiac conduction defect, idiopathic ventricular fibrillation, sick sinus node syndrome, atrial fibrillation, and sudden infant death syndrome) (2, 9, 10) that result from loss-of-function mutations in the SCN5A gene, which encodes human Nav1.5 channels. Many Nav1.5 mutants fail to properly traffic to the myocyte membrane or generate channels with gating defects, both resulting in reduced INa density (1, 2, 9). Some such mutations also produce a dominant negative effect (DNE); i.e., they decrease the expression/function of native (WT) subunits. A DNE, although unexpected, considering that each Nav1.5 channel is functional by itself, has been explained on the basis of possible WT and mutated subunit interactions (1, 11). Since Nav1.5 also interacts with Kir2.1/2.2 (5, 6), it seems reasonable to investigate whether patients carrying SCN5A mutations that encode loss-of-function channels also have altered expression of Kir2.1–2.2 channels and consequently reduced ventricular IK1 density. Another key question is whether Kir2.1–2.2 channels are able to restore the membrane expression of mutated Nav1.5 channels.
To answer the above questions, we systematically assessed the changes in INa and IK1 generated in the presence of Kir2.x channels alone or together with WT or BrS-associated mutated Nav1.5 channels. We used an expression system, rat ventricular cardiomyocytes (CMs) and human induced pluripotent stem cell–derived CMs (hiPSC-CMs), as well as mouse models of SCN5A haploinsufficiency and Kir2.1 overexpression. Our results demonstrated that endoplasmic reticulum (ER) trafficking–defective Nav1.5 channels significantly decreased IK1, since they did not positively modulate Kir2.1/2.2 channels. Moreover, Golgi trafficking–defective Nav1.5 mutants produced a DNE on Kir2.1/2.2 and, thus, an additional IK1 reduction. On the other hand, ER trafficking–defective Nav1.5 channels can be partially rescued by Kir2.1/2.2 channels through an unconventional secretory route that involves Golgi reassembly stacking proteins (GRASPs). Therefore, cardiac excitability would be greatly affected in subjects harboring Nav1.5 mutations with Golgi trafficking defects, since these mutants can concomitantly trap Kir2.1/2.2 channels, thus unexpectedly decreasing IK1 in addition to INa.
Results
Kir2.1 overexpression rescues Nav1.5 current in transgenic mice.
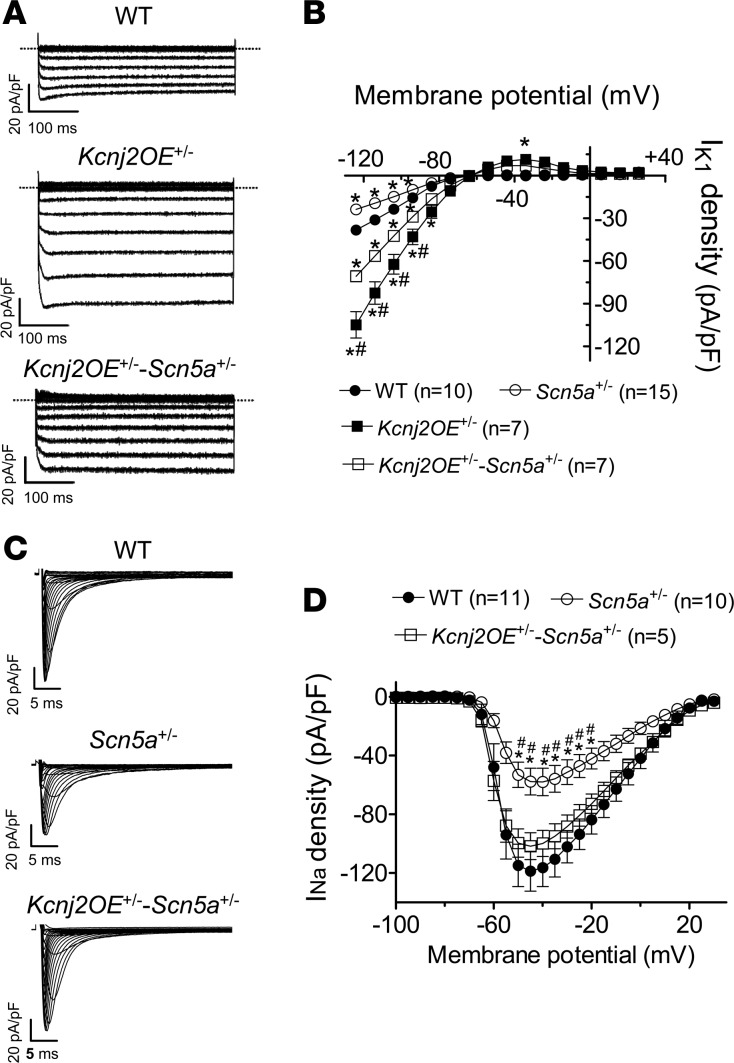
To test whether an INa decrease occurs concomitantly with an IK1 decrease (reciprocal modulation), we first used the transgenic mouse models Kcnj2OE+/– and Scn5a+/–, which had been previously created and functionally characterized (12, 13). Kcnj2OE+/– mice overexpress the KCNJ2 gene, which encodes for Kir2.1 channels (12). Scn5a+/– mice present a heterozygous targeted disruption of the SCN5A gene (see Supplemental Methods) and recapitulate most of the features of BrS (13). Additionally, we successfully obtained the new Kcnj2OE+/–-Scn5a+/– mouse line, whose genetic distribution was that of Mendelian inheritance (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96291DS1). As expected, IK1 was significantly larger in Kcnj2OE+/– than WT CMs at all voltages tested (Figure 1, A and B) (n ≥ 7, P < 0.05), and in Scn5a+/– CMs INa density was halved compared with WT at all voltages tested (Figure 1, C and D) (n ≥ 10, P < 0.05). Importantly, Figure 1B shows that in Scn5a+/– mouse hearts, IK1 density was actually significantly lower than in WT and Kcnj2OE+/– mouse hearts. On the other hand, the INa density recorded in Kcnj2OE+/–-Scn5a+/– CMs was significantly greater than in Scn5a+/– CMs and indistinguishable from that in WT (Figure 1, C and D). Thus, INa was completely rescued to normal in Kcnj2OE+/–-Scn5a+/– mouse hearts. However, the effects of the heterozygous decrease in Nav1.5 expression on IK1 density might be different than those produced by the presence of Nav1.5 loss-of-function mutants.
Figure 1. Heterozygous deletion of the SCN5A gene in mice concomitantly decreased the inward rectifier current, and Kir2.1 overexpression rescued the sodium current.
(A) Inward rectifier current (IK1) traces recorded in ventricular cardiomyocytes (CMs) from WT, Kcnj2OE+/–, and Kcnj2OE+/–-Scn5a+/– mice. (B) Mean current density-voltage curves for IK1 recorded in ventricular CMs from 3 WT (black circles), 4 Scn5a+/– (white circles), 3 Kcnj2OE+/– (black squares), and 3 Kcnj2OE+/–-Scn5a+/– (white squares) mice. (C) Sodium current (INa) traces recorded in ventricular CMs from WT, Scn5a+/-, and Kcnj2OE+/–-Scn5a+/– mice. (D) Mean current density-voltage curves for INa recorded in ventricular CMs from 3 WT (black circles), 3 Scn5a+/–, and 3 Kcnj2OE+/–-Scn5a+/– mice. Each point represents the mean ± SEM of n cells. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons. *P < 0.05 vs. WT; #P < 0.05 vs. Kcnj2OE+/–-Scn5a+/–.
Cotransfection of Kir2.x may rescue Nav1.5 current generated by trafficking-defective mutants.
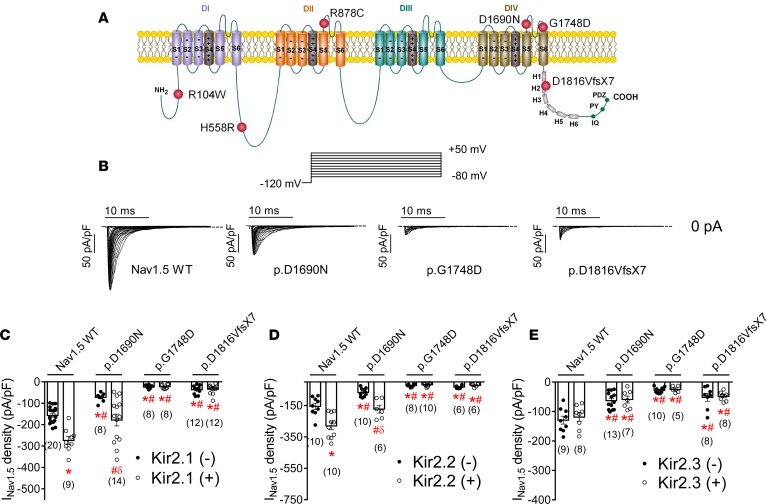
Figure 2A shows a topological diagram of the Nav1.5 channel (locus NM_198056.2) depicting the location of residues whose loss-of-function mutations or polymorphisms were tested here. Figure 2B shows Nav1.5 current (INav1.5) traces generated in CHO cells transfected with WT or some of the mutated human Nav1.5 channels (1.6 μg) together with the ancillary hNavβ1 subunit (1.6 μg). Indeed, Nav1.5 channels, both WT and mutated, were always transfected together with the hNavβ1 subunit. p.D1690N and p.G1748D mutations, located at the pore region in the fourth domain of the channel (Figure 2A), have been found in compound heterozygosity in a BrS patient (14). Moreover, the p.H558R polymorphism was present in one allele of this proband, in addition to the p.D1690N mutation (14). p.D1816VfsX7 is a frameshift mutation that suppresses 201 amino acids of the Nav1.5 C-terminal domain, which was responsible for an overlapping phenotype (including idiopathic ventricular fibrillation, bradycardia, intracardiac conduction delay, atrial fibrillation, and BrS) in a large Spanish family (15). All 3 mutations, located at the fourth domain or the C-terminus, reduced channel trafficking toward the plasma membrane, which together with channel gating abnormalities (very intense for the p.G1748D mutant) accounted for the reduction in INav1.5 density (Figure 2B) (14, 15). Interestingly, both p.D1690N and p.G1748D (which resulted in the most marked INav1.5 reduction) mutants produced a DNE when coexpressed with WT channels (14). Conversely, the p.D1816VfsX7 mutation did not produce DNE, since cotransfection with WT channels (0.5:0.5 ratio) resulted in “haploinsufficiency,” i.e., it reduced the current density by just ≈50% (15).
Figure 2. In CHO cells, cotransfection of Kir2.x may rescue Nav1.5 current generated by trafficking-defective Brugada syndrome–associated mutants in CHO cells.
(A) Topological diagram of the Nav1.5 channel showing the location of the mutations studied. The residue numbering corresponds to that of the transcript NM_198056.2. (B) Nav1.5 current (INav1.5) traces obtained by applying the protocol shown at the top in cells transfected with WT, p.D1690N, p.G1748D, and p.D1816VfsX7 Nav1.5 channels. Dashed lines represent the zero current level. (C–E) Peak INav1.5 density recorded in cells expressing WT or mutated Nav1.5 channels alone (black circles) or together (white circles) with Kir2.1 (C), Kir2.2 (D), or Kir2.3 (E) channels. Each bar represents the mean ± SEM of n cells from at least 3 different batches, and each dot represents 1 experiment. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons. *P < 0.05 vs. Nav1.5 WT alone; #P < 0.05 vs. Nav1.5WT+Kir2.x; δP < 0.05 vs. p.D1690N alone.
Figure 2, C–E, depicts the maximum INav1.5 densities generated by WT, p.D1690N, p.G1748D, and p.D1816VfsX7 channels in the absence and presence of human cardiac Kir2.1 (Figure 2C), Kir2.2 (Figure 2D), and Kir2.3 (Figure 2E) channels in transiently transfected CHO cells. Coexpression of Nav1.5 and Kir2.x channels was tested electrophysiologically and by fluorescence detection, and only cells coexpressing both channels were patched. Coexpression with Kir2.1 (1.6 μg) or Kir2.2 (1.6 μg) significantly (n ≥ 9, P < 0.05) increased the maximum INav1.5 density generated by WT (Figure 2, C and D, and Supplemental Figure 2). Similarly, coexpression with Kir2.1 and Kir2.2 also positively modulated the current density generated by trafficking-defective Nav1.5 mutant channels such as p.D1690N (P < 0.05) (Figure 2, C and D). However, current generated by p.D1690N when coexpressed with Kir2.1 or Kir2.2 channels was still significantly lower than that generated by WT Nav1.5 coexpressed with Kir2.x (P < 0.05). Conversely, cotransfection with Kir2.1 or Kir2.2 failed to increase the INav1.5 density generated by p.G1748D or p.D1816VfsX7; i.e., the positive reciprocal modulation was absent for these 2 Nav1.5 mutants (Figure 2, C and D). Furthermore, the results demonstrated that cotransfection with Kir2.3 (1.6 μg) did not positively modulate INav1.5 at any of the voltages tested (Supplemental Figure 2), and thus the INav1.5 generated when Kir2.3 and Nav1.5 channels were coexpressed was similar to that generated by Nav1.5 channels alone (Figure 2E) (n ≥ 8, P > 0.05). Consistent with this finding, the presence of Kir2.3 channels did not modify the INav1.5 density generated by p.D1690N, p.G1748D, and p.D1816VfsX7 channels (Figure 2E).
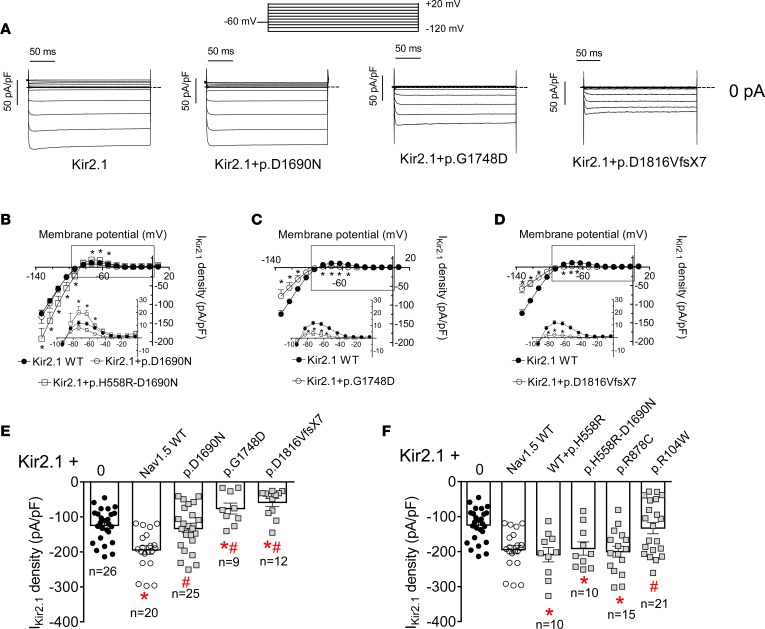
Kir2.x currents are reduced in the presence of trafficking-defective Nav1.5 channels.
Figure 3A shows Kir2.1 current (IKir2.1) traces generated in CHO cells transfected with Kir2.1 alone or together with p.D1690N, p.G1748D, or p.D1816VfsX7. Figure 3, B–D, shows IKir2.1 density-voltage curves, and the insets show the current density generated at physiologically relevant membrane potentials recorded in cells cotransfected or not with mutant Nav1.5 channels. Figure 3E depicts IKir2.1 density at –120 mV to better appreciate the differences under each condition. IKir2.1 density generated in cells cotransfected with p.D1690N Nav1.5 was similar to that generated by Kir2.1 alone (Figure 3, A and E) at all voltages tested (Figure 3B) (P > 0.05). Therefore, p.D1690N Nav1.5 did not positively modulate Kir2.1 channels. Figure 3A shows that cotransfection with either p.G1748D or p.D1816VfsX7 Nav1.5 channels not only abolished the positive modulation of IKir2.1 density at all voltages tested (Figure 3, C and D), but also significantly reduced IKir2.1 density compared with that generated by Kir2.1 alone (Figure 3E) (P < 0.05).
Figure 3. In CHO cells Kir2.1 current is reduced in the presence of trafficking-defective Brugada syndrome–associated mutated Nav1.5 channels.
(A) Kir2.1 current (IKir2.1) traces obtained by applying 250-ms pulses from –60 mV to potentials ranging between –120 and +20 mV in CHO cells transfected with Kir2.1 channels alone or together with p.D1690N, p.G1748D, and p.D1816VfsX7 Nav1.5 channels. Dashed lines represent the zero current level. (B–D) Mean current density-voltage curves for IKir2.1 recorded in cells expressing Kir2.1 alone (black symbols) or together (white symbols) with p.D1690N (B), p.H558R-D1690N (B), p.G1748D (C), and p.D1816VfsX7 (D) Nav1.5 channels. Insets in B–D: data at potentials positive to potassium equilibrium potential (EK) on an expanded scale. (E and F) IKir2.1 density at –120 mV generated in cells transfected with Kir2.1 channels alone or together with WT or mutated Nav1.5 channels. For comparison, data from cells cotransfected or not with WT Nav1.5 channels are also presented in F. Each data point/bar represents the mean ± SEM of n cells from at least 3 different batches, and each dot (in E and F) represents 1 experiment. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons. *P < 0.05 vs. Kir2.1 alone; #P < 0.05 vs. Kir2.1+Nav1.5 WT.
For comparison, we also tested the effects of the cotransfection with other loss-of-function Nav1.5 channels on IKir2.1 density (Figure 3F). The p.R878C mutant did not generate current at all as a consequence of a gating defect, even when it properly trafficked toward the plasma membrane (16). Figure 3F shows that cotransfection of p.R878C with Kir2.1 also positively modulated IKir2.1 density (P < 0.05). We previously demonstrated that currents generated by p.H558R-D1690N Nav1.5 channels were indistinguishable from those generated by native channels, since p.H558R-D1690N properly traffics toward the membrane (14). Consistent with this finding, cotransfection with p.H558R-D1690N Nav1.5 channels also increased IKir2.1 density (Figure 3, B and F). The same occurred with the cotransfection of native and p.H558R Nav1.5 channels with Kir2.1 channels (Figure 3F). That is an interesting result considering that the heterozygous presence of the polymorphism is very common in the general population. Clatot and coworkers (17) described a Nav1.5 point mutation (p.R104W) in the N-terminus that generated channels that are barely expressed in the cell membrane. Figure 3F shows that p.R104W channels failed to positively modulate the expression of Kir2.1 channels.
As described previously (6), cotransfection with Nav1.5 channels significantly increased the current generated by Kir2.2 (IKir2.2) but not that generated by Kir2.3 (IKir2.3) channels (Supplemental Figure 3). Consistent with this finding, IKir2.3 density measured at –120 mV was not affected when Kir2.3 was cotransfected with any of the 3 trafficking-defective Nav1.5 mutants tested (Supplemental Figure 3). Conversely, IKir2.2 density was significantly decreased when Kir2.2 channels were cotransfected with p.G1748D and p.D1816VfsX7 Nav1.5 channels compared with the current generated by Kir2.2 channels alone (n ≥ 5, P < 0.05). p.D1690N, on the other hand, was unable to positively modulate IKir2.2 (Supplemental Figure 3). Thus, cotransfection with the loss-of-function Nav1.5 mutants similarly affected IKir2.2 and IKir2.1.
In summary, the foregoing results suggest that cotransfection with Nav1.5 loss-of-function mutant channels that properly traffic toward the membrane positively modulates IKir2.1/2.2. Conversely, cotransfection with Nav1.5 mutant channels that do not properly traffic either abolishes the positive modulation or produces an additional IKir2.1/2.2 decrease, i.e., a DNE on Kir2.1/2.2 channels. Hereafter, we mainly focus on the interactions of Kir2.x with p.D1690N, p.G1748D, and p.D1816VfsX7 Nav1.5 channels, which were selected as models of Nav1.5 loss-of-function trafficking-defective mutants that do or do not produce DNEs on INav1.5 and/or IKir2.1/2.2.
Nav1.5 mutants produce DNEs on IK1 in adult rat ventricular CMs.
To test for the effects of Nav1.5 mutants on cardiac IK1 and INa, we overexpressed WT (Ad-Nav1.5) and trafficking-defective Nav1.5 channels in rat ventricular CMs using adenoviral constructs. Control CMs were infected with an adenoviral construct encoding GFP (Ad-GFP) alone. INa density almost doubled in CMs infected with Ad-Nav1.5 (Supplemental Figure 4, A and B) (n ≥ 35, P < 0.05). INa recorded in Ad-p.D1690N– and Ad-p.G1748D–infected CMs significantly decreased compared with that in Ad-Nav1.5–infected CMs. Furthermore, Ad-p.D1690N– and Ad-p.G1748D–infected CMs also exhibited decreased INa compared with Ad-GFP–infected CMs (Supplemental Figure 4, A and B), a result that can be explained by the fact that both mutants produce DNEs on WT Nav1.5 channels (14). Infection of CMs with Ad-p.D1816VfsX7 did not significantly modify INa density relative to Ad-GFP, consistent with the fact that this mutant does not produce a DNE on WT channels (15). However, Ad-p.D1816VfsX7 infection of CMs significantly decreased INa relative to Ad-Nav1.5 (n ≥ 15, P < 0.05) (Supplemental Figure 4, A and B). Additionally we tested the effects of overexpression of each mutant on the voltage dependence of INa activation and inactivation (Supplemental Figure 4, C and D). Overexpression of p.G1748D and p.D1816VfsX7 Nav1.5 channels significantly shifted the midpoint of the activation and inactivation curves toward more depolarized potentials (Supplemental Table 1), while overexpression of WT or p.D1690N did not modify either the activation or inactivation of the INa. These results are in perfect agreement with the effects described for these mutants on voltage-dependent gating of INa (14, 15).
Supplemental Figure 5, A and B, demonstrates that the effects of overexpression of the Nav1.5 mutants on IK1 recorded in adult rat ventricular CMs were identical to those obtained in CHO cells. Indeed, the IK1 density recorded in CMs infected with Ad-p.D1690N was not different from that generated in Ad-GFP–infected CMs, but it was smaller than that generated in those infected with Ad-Nav1.5 (n ≥ 18, P < 0.05). Conversely, infection with either Ad-p.G1748D or Ad-p.D1816VfsX7 significantly decreased IK1 compared with Ad-GFP–infected CMs. These results confirmed that p.G1748D and p.D1816VfsX7 produce a DNE on Kir2.1/2.2 channels that therefore produces a significant IK1 reduction.
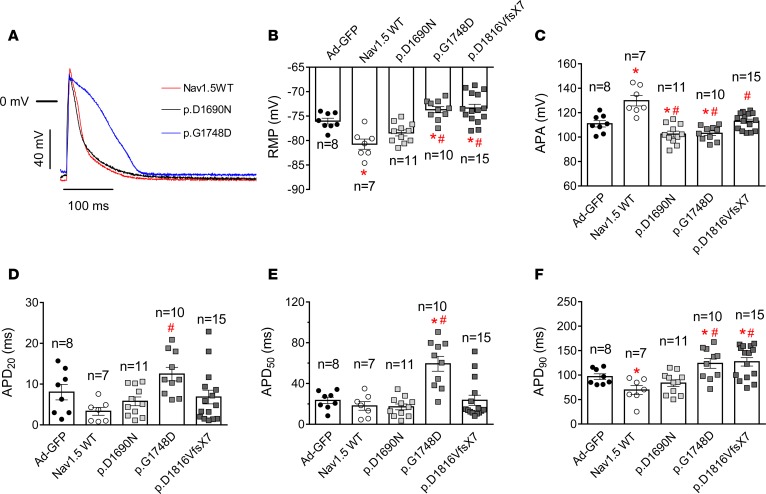
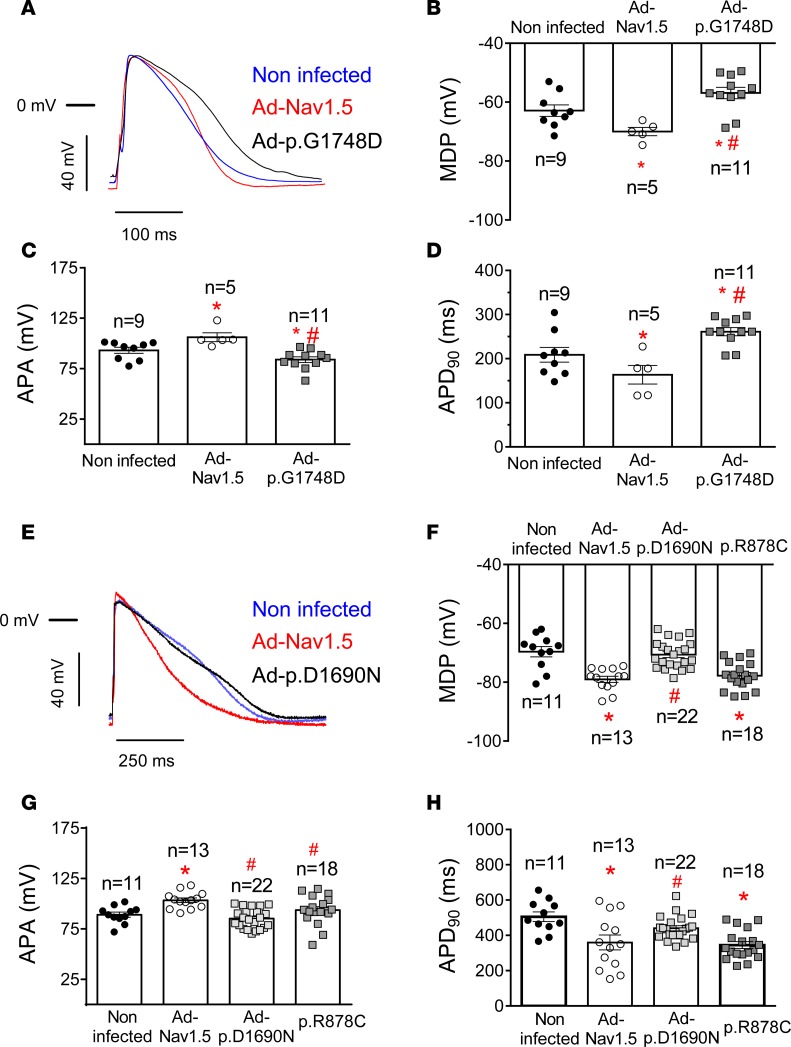
We also recorded APs in rat ventricular CMs overexpressing or not WT and mutated Nav1.5 channels. Figure 4A shows illustrative superimposed AP traces recorded in 3 CMs infected with Ad-Nav1.5, Ad-p.D1690N, and Ad-p.G1748D particles, respectively. Infection with Ad-Nav1.5 hyperpolarized, while infection with either Ad-p.G1748D or Ad-p.D1816VfsX7 depolarized, RMP compared with that of CMs infected with Ad-GFP (Figure 4B). Indeed, only overexpression of p.D1690N channels did not modify RMP significantly (n ≥ 8, P > 0.05). AP amplitude (APA) was significantly increased by the overexpression of WT Nav1.5 channels (Figure 4C, n ≥ 7, P < 0.05), while it was decreased by overexpression of those mutants that produce a DNE on INa, i.e., p.D1690N and p.G1748D (n ≥ 8, P < 0.05). AP duration (APD) was measured at 20% (APD20), 50% (APD50), and 90% (APD90) of repolarization (Figure 4, D–F). Infection with Ad-Nav1.5 significantly shortened APD90 (Figure 4F), an effect that was attributable to the reciprocal increase in IK1. Interestingly, infection with Ad-p.G1748D significantly lengthened APD50 and APD90, while infection with Ad-p.D1816VfsX7 only lengthened APD90 (Figure 4, D–F, n ≥ 8, P < 0.05).
Figure 4. Overexpression of trafficking-defective Nav1.5 mutants modifies action potential characteristics in rat ventricular cardiomyocytes.
(A) Action potential (AP) traces recorded in isolated ventricular cardiomyocytes (CMs) driven at 1 Hz and infected with the adenoviral (Ad) constructions encoding for WT, p.D1690N, or p.G1748D Nav1.5 channels. The horizontal bar represents the 0 mV level. (B–F) Resting membrane potential (RMP) (B), AP amplitude (APA) (C), and AP duration measured at 20% (APD20) (D), 50% (APD50) (E), and 90% (APD90) (F) of repolarization obtained in ventricular CMs obtained from 8 rats infected with Ad-GFP (black circles) or with adenovirus encoding WT (white circles) or mutated Nav1.5 channels (gray squares). Each bar represents mean ± SEM of n cells, and each dot represents 1 experiment. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons. *P < 0.05 vs. Ad-GFP: #P < 0.05 vs. Ad-Nav1.5 WT.
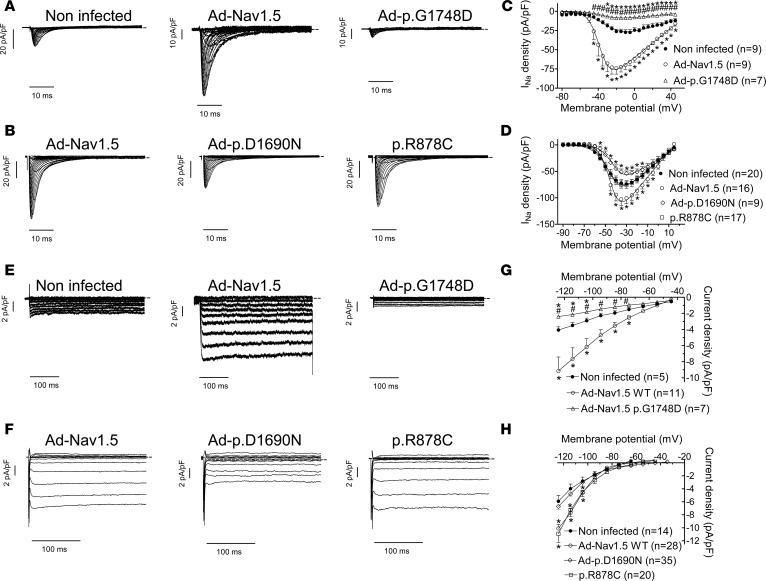
p.G1748D, but not p.D1690N, exerts a DNE on Kir2.1 in hiPSC-CMs.
We used adenoviral gene transfer in hiPSC-CMs to confirm in human CMs the results obtained in CHO cells and in rat CMs. We selected the p.G1748D mutant because of its significant DNE on IK1 (similar to that produced by p.D1816VfsX7 channels); the p.D1690N mutant, which produced DNE on INa but not IK1; and the p.R878C mutant, which positively modulated IKir2.1/2.2. We used mature hiPSC-CMs from 2 different origins, i.e., custom made in our laboratory (DF19-9-11T) (18) (infected with Ad-p.G1748D) and commercial (iCell2) (infected with Ad-p.D1690N and transfected with p.R878C) (see Supplemental Methods). Since the densities of INa and IK1 recorded in noninfected hiPSC-CMs of the 2 groups were not identical, the results are presented separately.
Figure 5A shows INa traces recorded in hiPSC-CMs infected or not with adenoviral constructs encoding Nav1.5 WT or p.G1748D; and Figure 5B, INa traces recorded in hiPSC-CMs infected with adenoviruses encoding either WT or p.D1690N and transfected with p.R878C Nav1.5 channels. As expected, infection with Ad-Nav1.5 produced a significant increase in INa (Figure 5, A, C, and D), while infection with Ad-p.G1748D and Ad-p.D1690N markedly decreased INa density (n ≥ 7, P < 0.05) (Figure 5, A–D) compared with that generated by noninfected hiPSC-CMs as a result of the DNE produced by p.G1748D and p.D1690N on Nav1.5 WT channels (14). Conversely, liposomal transfection with p.R878C significantly decreased the INa generated by hiPSC-CMs compared with that generated by CMs overexpressing Nav1.5 channels, but not compared with that generated by noninfected cells (Figure 5, B and D). Infection with Ad-Nav1.5 also markedly increased the total IK1 density, demonstrating that the reciprocal modulation between INa and IK1 is also observed in hiPSC-CMs (Figure 5, E, G, and H). Interestingly, infection with Ad-p.G1748D (Figure 5, E and G), but not with Ad-p.D1690N (Figure 5, F and H), significantly decreased (by ≈32%) the density of total inward potassium current compared with that generated in noninfected hiPSC-CMs (n ≥ 5, P < 0.05) (Figure 5, G and H). On the other hand, the total inward potassium current generated in hiPSC-CMs transfected with p.R878C channels was indistinguishable from that generated in human CMs overexpressing Nav1.5 channels (Figure 5, F and H).
Figure 5. Effects of trafficking-defective Nav1.5 mutants on sodium current and inward rectifier current generated in human induced pluripotent stem cell–derived cardiomyocytes.
(A and B) Sodium current (INa) traces recorded in DF19-9-11T (A) or in iCell2 (B) human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) expressing or not Ad-Nav1.5 WT or Nav1.5 mutants by applying 100-ms pulses from –160 mV to potentials ranging between –80 and +45 mV in 5-mV steps (A) or 50-ms pulses from –120 mV to potentials ranging between –90 and +40 mV in 5-mV steps (B). (C and D) Mean peak current density-voltage curves for INa recorded in DF19-9-11T (C) or in iCell2 (D) hiPSC-CMs expressing or not (black circles) Ad-Nav1.5 WT (white circles) or p.G1748D (white triangles), p.D1690N (white diamonds), and p.R878C (white squares). (E and F) Inward rectifier current traces recorded in DF19-9-11T (E) or in iCell2 (F) hiPSC-CMs expressing or not Ad-Nav1.5 WT or Nav1.5 mutants by applying 400-ms (E) or 250-ms (F) pulses from –40 mV to potentials ranging between –120 and –40 mV in 10-mV steps. (G and H) Mean current density-voltage curves for the inward rectifier current recorded in DF19-9-11T (G) or iCell2 (H) hiPSC-CMs expressing or not Ad-Nav1.5 WT or the indicated Nav1.5 mutants. In A, B, E, and F, dashed lines represent the zero current level. In C, D, G, and H, each point represents mean ± SEM of n experiments from at least 5 different dishes. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons.*P < 0.05 vs. noninfected; #P < 0.05 vs. Ad-Nav1.5.
APs were also recorded in hiPSC-CMs, which exhibited spontaneous automatic activity (Supplemental Figure 6) (18). Infection with Ad-Nav1.5 significantly reduced automatic firing frequency (n ≥ 6, P < 0.05), while infection with Ad-p.G1748D significantly increased it (n ≥ 7, P < 0.05) (Supplemental Figure 6). On the other hand, transfection with p.R878C Nav1.5 channels and infection with Ad-p.D1690N, respectively, decreased (n ≥ 12, P < 0.05) and did not significantly modify automatic activity compared with that of noninfected cells (Supplemental Figure 6). Figure 6A shows superimposed APs recorded at 37°C in 3 hiPSC-CMs cells driven at 1 Hz and infected or not with Ad-Nav1.5 and Ad-p.G1748D. Infection with Ad-Nav1.5 significantly hyperpolarized maximum diastolic potential (MDP), increased APA, and shortened APD90 (Figure 6, B–D). Conversely, infection with Ad-p.G1748D significantly depolarized MDP, decreased APA, and prolonged APD90 (P < 0.05 vs. Nav1.5 WT and noninfected cells) (Figure 6, B–D). Figure 6E shows superimposed APs recorded at room temperature in 3 hiPSC-CMs cells driven at 1 Hz and infected or not with Ad-Nav1.5 and Ad-p.D1690N. Compared with Ad-Nav1.5–infected hiPSC-CMs, Ad-p.D1690N depolarized MDP, diminished APA, and lengthened APD90 (Figure 6, F–H, n ≥ 11, P < 0.05). Conversely, and also compared with Ad-Nav1.5–infected hiPSC-CMs, p.R878C channels did not significantly modify either MDP or APD90 (Figure 6, F and H, n ≥ 13, P < 0.05)
Figure 6. Effects of trafficking-defective Nav1.5 mutants on action potentials recorded in human induced pluripotent stem cell–derived cardiomyocytes.
(A) Superimposed action potential (AP) traces recorded in noninfected and Ad-Nav1.5– or Ad-p.G1748D–infected DF19-9-11T human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) driven at 1 Hz. (B–D) Mean maximum diastolic potential (MDP) (B), AP amplitude (APA) (C), and AP duration measured at 90% (APD90) (D) for APs recorded at 1 Hz in noninfected hiPSC-CMs (black circles) and hiPSC-CMs infected with Ad-Nav1.5 (white circles) or Ad-p.G1748D (gray squares). (E) Superimposed AP traces recorded in noninfected and Ad-Nav1.5– or Ad-p.D1690N–infected iCell2 hiPSC-CMs driven at 1 Hz. (F–H) Mean MDP (F), APA (G), and APD90 (H) for APs recorded at 1 Hz in noninfected hiPSC-CMs (black circles) and hiPSC-CMs infected with Ad-Nav1.5 (white circles) or Ad-p.D1690N (light gray squares) or transfected with the cDNA encoding p.R878C Nav1.5 (dark gray squares). In B–D and F–H, each bar represents mean ± SEM of n experiments from at least 5 different dishes, and each dot represents 1 experiment. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons. *P < 0.05 vs. noninfected; #P < 0.05 vs. Ad-Nav1.5.
Mechanistic insight.
The results suggested that trafficking-defective Nav1.5 mutants either do not reciprocally modulate (p.D1690N) or produce a DNE (p.G1748D and p.D1816VfsX7) on IK1. We hypothesized that such different behaviors may depend on the intracellular compartment in which the individual trafficking-defective channel is “trapped.” Thus, it might be that mutants retained in the ER or in the Golgi are those that produce a DNE on IK1.
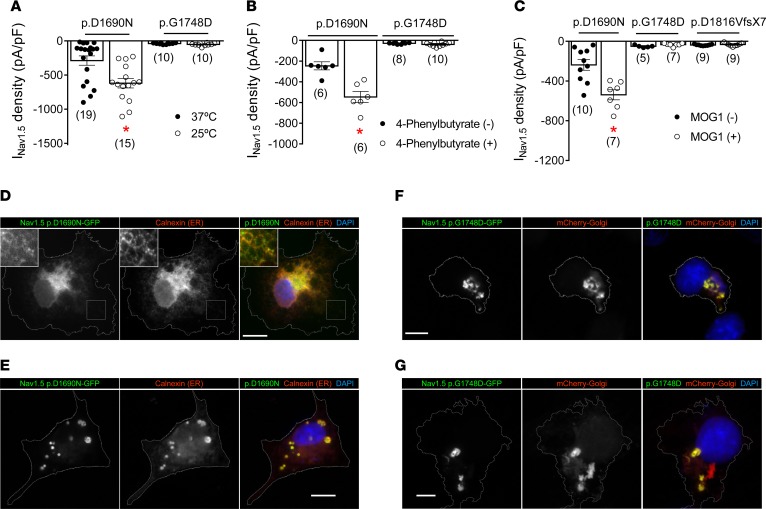
Trafficking-defective mutants that are retained at the ER can be rescued by incubation either at room temperature or with a chemical “chaperone” such as 4-phenylbutyrate (1). We previously demonstrated that expression of p.D1816VfsX7 Nav1.5 channels is not modified by either maneuver (15). Here, we functionally tested whether p.D1690N and p.G1748D present trafficking defects at either the ER or the Golgi. As illustrated in Figure 7, A and B, CHO cells transfected with p.D1690N, but not p.G1748D, showed significantly increased INav1.5 density when incubated at 25°C (Figure 7A) or in the presence of 4-phenylbutyrate (5 mmol/l) (Figure 7B). We also studied the effects of MOG1, a guanine nucleotide release factor for Ras-related nuclear protein (Ran), which interacts with Nav1.5 channels and increases their expression at the cell membrane (19). Recently it has been described that MOG1 restores the defective trafficking of the Nav1.5 mutants that are retained at the ER (20). Figure 7C demonstrates that cotransfection with MOG1 significantly and selectively increased INav1.5 density generated by p.D1690N but not INav1.5 generated by either the p.G1748D or p.D1816VfsX7 channel.
Figure 7. p.D1690N and p.G1748D are endoplasmic reticulum and Golgi trafficking–defective channels, respectively.
(A and B) Maximum Nav1.5 current (INav1.5) density generated by p.D1690N and p.G1748D channels in CHO cells incubated at 37°C (black circles) or 25°C (white circles) (A) or in the presence (white circles) or absence (black circles) of 4-phenylbutyrate (B). (C) Maximum INav1.5 density generated by WT or mutated Nav1.5 channels cotransfected (white circles) or not (black circles) with MOG1. *P < 0.05 vs. p.D1690N without treatment. Each bar represents mean ± SEM of n cells from at least 3 different batches, and each dot represents 1 experiment. Unpaired t test and multilevel mixed-effects model were used for comparisons. (D and E) Colocalization of Nav1.5 p.D1690N-GFP channels and the endoplasmic reticulum (ER) marker calnexin in 2 different COS cells representative of 3 different dishes. In D, inset images are included to make low-contrast highlighted regions visible. (F and G) Colocalization between Nav1.5 p.G1748D-GFP channels and the Golgi marker mCherry-Golgi in 2 different COS cells representative of 3 different dishes. Cell contours are delineated in gray for enhanced visualization. Scale bars: 25 μm.
Together with functional analysis, we determined the intracellular localization of p.D1690N and p.G1748D channels by immunofluorescence. Figure 7, D and E, shows the intracellular distribution of p.D1690N in 2 different cells. A high degree of colocalization with the resident ER chaperone calnexin was observed, suggesting that p.D1690N is retained and, in most cells, accumulated (see the accumulations in the cell shown in Figure 7E), in the ER. As a consequence, p.D1690N channels did not colocalize with the Golgi marker (Supplemental Figure 7). On the other hand, transient expression of p.G1748D channels resulted in local intracellular accumulations (Figure 7, F and G) that colocalized with the Golgi marker mCherry-Golgi, suggesting that these channels transit through Golgi and from there are sorted and retained in the Golgi itself, in cargos possibly targeted for degradation (Figure 7G). p.G1748D did not colocalize with the ER marker (Supplemental Figure 7). Interestingly, it would seem that p.G1748D induced stress at the ER, considering the unusual ER distribution (in local clusters) observed with calnexin (Supplemental Figure 7). Overall, these results strongly suggest that p.D1690N channels exhibit a traffic defect at the ER, whereas p.G1748D (and probably p.D1816VfsX7) at the Golgi. Together the results support the hypothesis that Golgi trafficking–defective Nav1.5 mutants are those that produce a DNE on Kir2.1 channels.
Why do Nav1.5 mutants with trafficking defects at the Golgi produce a DNE on Kir2.1 channels?
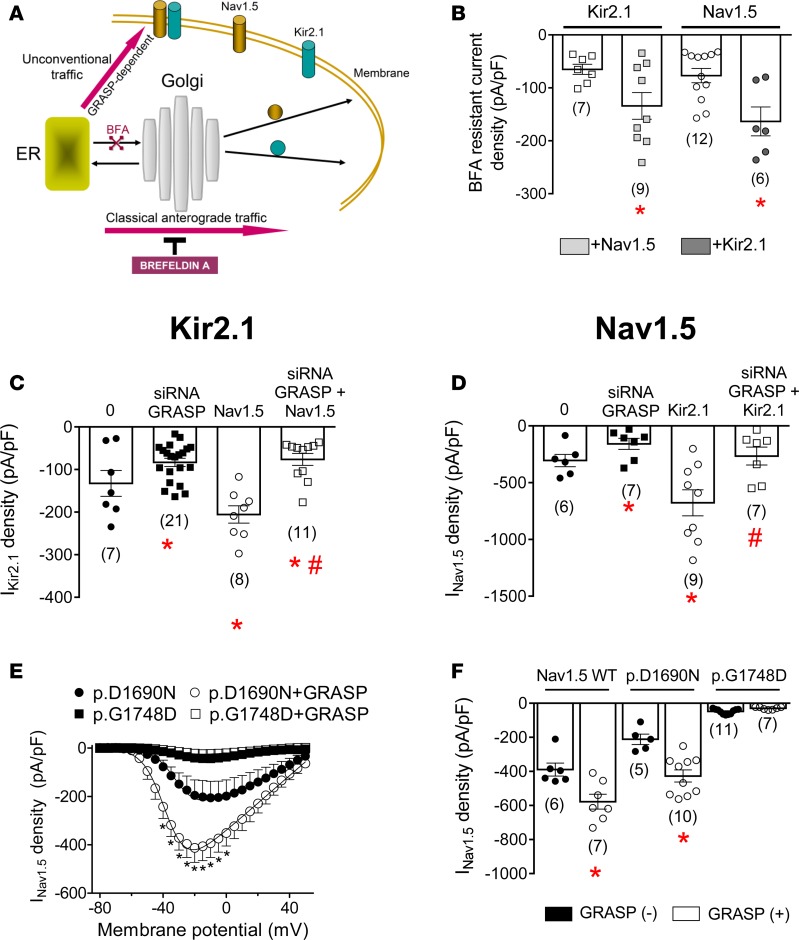
As an initial step to answering the above question, we explored the trafficking of Nav1.5 and Kir2.1 channels when they were transfected either alone or together. First we treated Nav1.5-, Kir2.1-, and Nav1.5-Kir2.1–transfected cells with brefeldin A (BFA) (50 ng/ml for 24 hours), a fungal metabolite that specifically blocks the forward movement of newly synthesized membrane proteins from the ER to the Golgi (Figure 8A) (21). Before BFA treatment, IKir2.1 generated by either Kir2.1 alone or cotransfected with Nav1.5 channels averaged –129.8 ± 18.1 and –199.6 ± 17.2 pA/pF, respectively. INav1.5 generated by Nav1.5 alone or cotransfected with Kir2.1 channels was –214.9 ± 26.1 and –466.3 ± 42.1 pA/pF, respectively. Figure 8B depicts IKir2.1 and INav1.5 densities recorded after 24-hour incubation with BFA (BFA-resistant current). After incubation, IKir2.1 and INav1.5 generated by Kir2.1 and Nav1.5 channels alone significantly decreased (n ≥ 6, P < 0.05). Importantly, after BFA treatment, cotransfection with Nav1.5 channels significantly increased IKir2.1 density (n ≥ 7, P < 0.05) and vice versa (n ≥ 6, P < 0.05). Therefore, incubation with BFA did not abolish the positive reciprocal modulation between Kir2.1 and Nav1.5 channels. Since BFA blocks classical membrane protein trafficking without interfering with unconventional secretory routes (Figure 8A) (21, 22), the BFA-resistant current would represent that generated by those channels that could reach the plasma membrane through an unconventional Golgi-independent pathway.
Figure 8. GRASP proteins can rescue endoplasmic reticulum trafficking–defective Nav1.5 mutants.
(A) Hypothetical diagram of anterograde trafficking of Nav1.5 and Kir2.1 channels through conventional or unconventional routes. (B) Brefeldin A–resistant (BFA-resistant) Kir2.1 (IKir2.1) and Nav1.5 (INav1.5) currents recorded in CHO cells transfected with Kir2.1, Nav1.5, or Kir2.1+Nav1.5 channels after incubation with BFA for 24 hours. (C) IKir2.1 density at –120 mV recorded in cells in which GRASP55 was silenced or not, transfected with either Kir2.1 or Kir2.1+Nav1.5 channels. (D) Maximum INav1.5 density recorded in cells in which GRASP55 was silenced or not, transfected with either Nav1.5 or Kir2.1+Nav1.5 channels. In B–D, *P < 0.05 vs. Kir2.1 or Nav1.5 in control conditions; #P < 0.05 vs. Nav1.5+Kir2.1. (E) Mean current density-voltage curves for INav1.5 recorded in cells expressing p.D1690N or p.G1748D cotransfected or not with GRASP55 (GRASP). (F) Maximum INav1.5 density generated by WT, p.D1690N, and p.G1748D Nav1.5 channels cotransfected or not with GRASP55. In E and F, *P < 0.05 vs. WT or p.D1690N in the absence of GRASP55. Each bar/point represents mean ± SEM of n cells from at least 3 different batches, and each dot (B–D and F) represents 1 experiment. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons.
Nav1.5 channels can reach the plasma membrane via both the classical and an unconventional pathway (23). Otherwise, it has been demonstrated that the cAMP-activated cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel can reach the plasma membrane through an unconventional mechanism that is dependent of GRASPs (22). Thus, we explored whether GRASPs could be involved in the trafficking of the channels responsible for the positive reciprocal modulation. We used siRNAs to reduce the expression of GRASP55, the isoform that is present in CHO cells (they lack the GRASP65 isoform) (22). Expression of GRASP was markedly reduced in cells transfected with siRNA1 (~65%), and its specific effects were confirmed by using a scrambled siRNA (Supplemental Figure 8). IKir2.1 (n = 21, P < 0.05) and INav1.5 (n = 7, P < 0.05) densities were significantly reduced in GRASP-silenced cells (Figure 8, C and D, respectively). Importantly, Figure 8C demonstrates that in GRASP-silenced cells, transfection with Nav1.5 did not increase but rather decreased IKir2.1 density (n = 11, P < 0.05). Moreover, in GRASP-silenced cells, transfection with Kir2.1 did not increase INav1.5 density (Figure 8D). Therefore, GRASP silencing abolished positive reciprocal modulation, suggesting that GRASP-dependent unconventional trafficking of Kir2.1 and Nav1.5 channels is involved in this process. Should this occur, it might be possible that those trafficking-defective mutants that are retained at the ER can be rescued by GRASP overexpression, while those with trafficking defects at the Golgi cannot. Thus, in the next group of experiments, GRASP55 was overexpressed together with either p.D1690N or p.G1748D channels, respectively. Indeed, Figure 8, E and F, shows that overexpression of GRASP55 significantly increased the INav1.5 density generated by WT and p.D1690N channels but not by the p.G1748D mutant. Overall, the results suggested that GRASP proteins can rescue trafficking-defective Nav1.5 channels when they are retained at the ER.
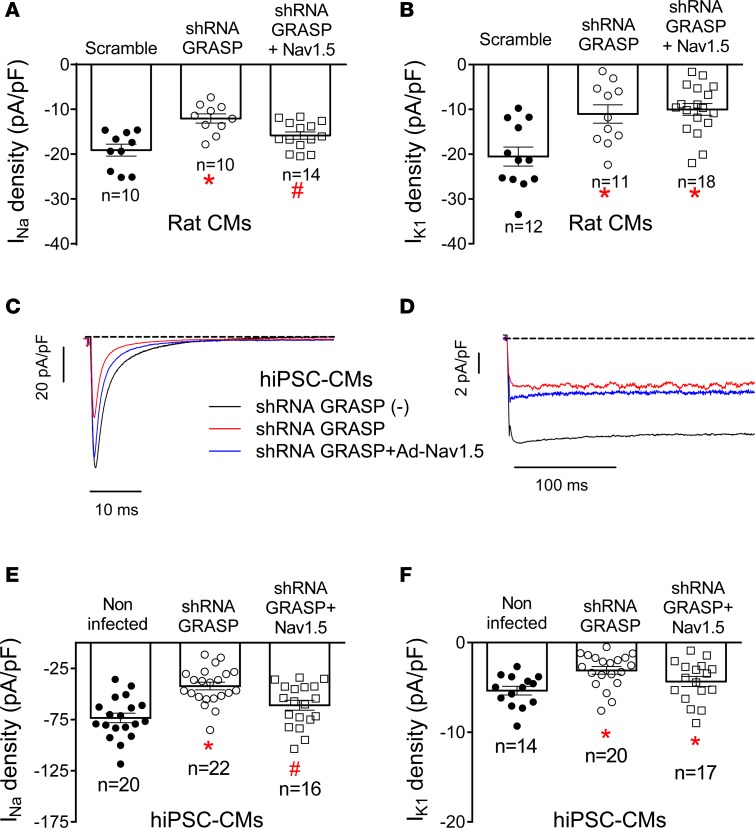
Finally, we also tested the role of GRASP55 and GRASP65 in adult rat ventricular (Figure 9, A and B) and in hiPSC-derived CMs (Figure 9, C–F). In CMs the expression of GRASP55 and GRASP65 was significantly and specifically reduced (Supplemental Figure 8) with shRNAs that were included in lentiviral particles. Figure 9, C and D, shows superimposed INa and IK1 traces recorded in hiPSC-CMs in which GRASP expression was silenced or not. In other groups of experiments, CMs were infected with Ad-Nav1.5 after GRASP silencing (see Supplemental Methods). Examples of the INa and IK1 recorded under this condition are shown in Figure 9, C and D, respectively. The results demonstrated that both INa and IK1 densities were decreased in rat and hiPSC-derived CMs infected with shRNA-GRASP55/65 (Figure 9, A–F). Furthermore, in GRASP-silenced rat ventricular and hiPSC-derived CMs, the overexpression of Nav1.5 channels did not concomitantly increase IK1 density, confirming the role of GRASP in the positive reciprocal modulation between Nav1.5 and Kir2.1 channels.
Figure 9. Silencing of GRASP abolished the positive reciprocal modulation in rat ventricular cardiomyocytes and human induced pluripotent stem cell–derived cardiomyocytes.
(A and B) Maximum sodium current (INa) density (A) and inward rectifier K current (IK1) density at –120 mV (B) recorded in rat ventricular cardiomyocytes (CMs) infected with lentivirus encoding shRNA-GRASP55/65 (white symbols) or scrambled shRNA (black circles) together or not with Ad-Nav1.5. (C and D) INa traces recorded by applying 50-ms pulses from –120 to –20 mV (C) and IK1 traces recorded by applying 250-ms pulses from –40 to –120 mV (D) in noninfected human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) (black lines), hiPSC-CMs infected with lentivirus encoding shRNA-GRASP55/65 together (blue lines) or not (red lines) with Ad-Nav1.5. (E and F) Maximum INa density (E) and IK1 density at –120 mV (F) recorded in noninfected hiPSC-CMs (black circles), hiPSC-CMs infected with lentivirus encoding shRNA-GRASP55/65 together (white squares) or not (white circles) with Ad-Nav1.5. In A, B, E, and F, each bar represents mean ± SEM of n cells from at least 5 different batches, and each dot represents 1 experiment. One-way ANOVA followed by Newman-Keuls and multilevel mixed-effects model were used for comparisons. *P < 0.05 vs. scramble or noninfected; #P < 0.05 vs. shRNA GRASP.
Discussion
Results presented here suggest that BrS patients harboring Golgi trafficking–defective Nav1.5 mutations should show greater reduction in cardiac excitability than patients whose Nav1.5-mutated channels are retained at the ER. Indeed, while all trafficking-defective Nav1.5 mutants decrease IK1 because they do not positively modulate IKir2.1/2.2, Golgi trafficking–defective Nav1.5 mutants produce an extra IK1 decrease because they exert a DNE on Kir2.1/2.2 channels. Interestingly, an unconventional GRASP-dependent secretory route seems to be responsible for the positive reciprocal modulation of Nav1.5/Kir2.1/2.2 channels as long as none of these channels have trafficking defects at the Golgi.
We investigated whether loss-of-function Nav1.5 mutants also positively modulate IKir2.1/2.2. The initial answer to this question came from a transgenic mouse model of BrS in which heterozygous deletion of the SCN5A gene decreased both INa and IK1. However, the mouse model raised the question of whether a 50% reduction in Nav1.5 expression could have consequences different from those produced by loss-of-function mutants.
The results demonstrate that proper trafficking toward the plasma membrane of Nav1.5 channels is a sine qua non condition for the positive reciprocal modulation to occur. In other words, Nav1.5 mutants that do not traffic toward the membrane (such as p.R104W, p.D1690N, p.G1748D, and p.D1816VfsX7) do not positively modulate IKir2.1/2.2. Conversely, p.R878C, a loss-of-function Nav1.5 mutant with a gating defect that is expressed in the membrane (16), positively modulated IK1 in hiPSC-CMs. Consistent with this finding, when p.D1690N trafficking was normalized by the presence of the H558R polymorphism (p.H558R-D1690N) (14), the positive modulation over IKir2.1 was also restored. Therefore, in CMs from patients carrying any trafficking-defective Nav1.5 mutation, IK1 generated by Kir2.1/2.2 channels is diminished. Indeed, in hiPSC-CMs expression of p.D1690N did not increase IK1 density as WT channels did, and, as a consequence, the mutant did not significantly hyperpolarize MDP, decrease automatic firing frequency, increase APA, or shorten APD90.
However, it was unexpected that some trafficking-defective Nav1.5 mutants, specifically p.G1748D and p.D1816VfsX7, produced a DNE on Kir2.1/2.2 channels, and thus further decreased IKir2.1/2.2. In fact, expression of p.G1748D Nav1.5 channels in hiPSC-CMs driven at 1 Hz led to, along with an APA decrease, a depolarization of the MDP, a significant increase in automatic firing frequency, and a prolongation of APD — effects attributable to the marked IK1 decrease. Therefore, as mentioned, the results presented here provide an example of DNEs between channels encoded by two different genes (SCN5A and KCJN2).
DNE implies that the protein encoded by the mutated allele (“poisoning protein”) reduces the activity/expression of the protein encoded by the WT gene (24). Therefore, the results would indicate that, at least at some stages (biosynthesis and/or forward and retrograde trafficking), Nav1.5 and Kir2.1 proteins do interact, forming complexes. Indeed, previous results from our group demonstrated that at least a pool of Nav1.5 and Kir2.1 channels form complexes early after their synthesis at the ER (8) and that these complexes, whose cellular biology is regulated differently than that of Nav1.5 and Kir2.1 channels alone (7), exhibit preferential forward trafficking toward the membrane (7, 8). Interestingly, when a Nav1.5 mutant produces a DNE on Nav1.5, this does not necessarily imply that it concomitantly produces a DNE on Kir2.1/2.2 channels (p.D1690N) and vice versa (p.D1816VfsX7). This reinforces the contention that the biology of Nav1.5-Kir2.1 complexes differs somehow from that of each channel alone, even though the anterograde and retrograde trafficking routes of the complexes are similar to those of Nav1.5 channels alone (7).
It is accepted that 2 main pools of Nav1.5 channels associated to different scaffolding proteins can be distinguished in cardiomyocytes, which are highly differentiated cells (25). Results from our group demonstrated that even when both Nav1.5 and Kir2.1 channels bind to SAP97 (5), it is binding to α1-syntrophin that is critical for positive reciprocal modulation to occur (6). This led us to the hypothesis that Nav1.5-Kir2.1 complexes are preferentially located at lateral membranes, i.e., the membrane domain at which both Nav1.5 and Kir2.1 can be coimmunolocalized with α1-syntrophin (6). Here, however, we did not analyze whether the pools of Nav1.5 channels or Nav1.5-Kir2.1 complexes that are reduced are those that are localized at the lateral membranes or at the intercalated discs.
It has been suggested that some proteins reach the extracellular medium or plasma membrane by unconventional secretory pathways (26, 27). As mentioned, a pool of Nav1.5 channels can reach the membrane of the cardiomyocyte by means of the unconventional secretory pathway (23). The functionality of the pool of Nav1.5 channels not exported through the ER-Golgi membrane route is a matter of debate (28, 29), since the fully glycosylated forms of the channel are generated at the Golgi (23). Interestingly, a fraction of Nav1.5 channels associated to Kir2.1 are sensitive to endoglycosidase H (8), which removes only high mannose carbohydrates that are incorporated in the ER (22, 30), thus suggesting that a pool of Nav1.5 channels that form part of the Nav1.5-Kir2.1 complexes also bypasses the Golgi. The unconventional secretory route seems to be mediated by GRASP65 and GRASP55 (26, 27), which contain 2 PDZ domains at the N-terminus (27) and, besides the Golgi, are located at ER exit sites (31). The unconventional GRASP-dependent secretory pathway is insensitive to BFA, which selectively inhibits the ER to Golgi trafficking (21). In our hands, blockade of the ER-Golgi membrane secretory route with BFA reduced both INav1.5 and IKir2.1, but did not abolish the positive reciprocal modulation of Kir2.1 and Nav1.5. These results led us to hypothesize that the positive reciprocal modulation is mediated by GRASP in such a way that the joint presence of Nav1.5 and Kir2.1 channels might enhance the export of both through the unconventional route (see Figure 8A). The results demonstrate that when expression of GRASP is silenced, positive reciprocal modulation is abolished in CHO cells and also in rat and hiPSC-derived CMs. On the other hand, the current densities generated by the WT and p.D1690N, but not p.G1748D, Nav1.5 channels can be significantly increased by GRASP overexpression. Our functional and immunolabeling analysis indicated that p.D1690N Nav1.5 channels are retained at the ER, while p.G1748D and p.D1816VfsX7 present trafficking defects at the Golgi. Thus, we hypothesize that those ER trafficking–defective Nav1.5 mutants may be delivered to the membrane via an unconventional GRASP-dependent secretory pathway. Similarly, the trafficking-defective p.Δ508-CFTR anion channel, which is retained at the ER, can reach the plasma membrane through an unconventional mechanism that is dependent of GRASPs (22). Furthermore, recent data demonstrated that Golgi trafficking–defective Δ314-315 Kir2.1 channels, which are associated to the Andersen-Tawil syndrome because they generate almost no IK1 (32), produced a DNE on INa (8). Gathering all the results, we propose that Nav1.5 channels that are retained early in the secretory route (mainly the ER and/or the cis-Golgi) can be exported together with Kir2.1 channels via GRASP through the unconventional route. Conversely, when Nav1.5 mutant channels do not properly traffic through the Golgi, they concomitantly retain Kir2.1 WT channels, thus producing the observed DNE because they cannot benefit from the GRASP-dependent export route.
Clinical implications.
Unlike in ventricles, the presence of Kir2.3 subunits in the human atria precludes the interaction that is seen between Kir2.1/2.2 and Nav1.5 channels (6). Since Kir2.3 channels are more abundant in atria than in ventricles (33), we surmise that in humans Nav1.5 trafficking–defective mutations would decrease INa in both atria and ventricles, whereas they would decrease IK1 only in ventricles. Unfortunately, this hypothesis cannot be tested using transgenic mouse models, since the mouse heart does not express Kir2.3 channels (34).
Altogether, the results presented here suggest that patients carrying mutations such as p.G1748D or p.D1816VfsX7 would experience, at least under certain circumstances, a more marked reduction in ventricular excitability because of the combined decrease in INa and IK1. It is accepted that the more reduced the excitability, the greater the risk of ventricular arrhythmias. Should this occur, patients carrying Golgi trafficking–defective Nav1.5 mutations will be more prone to experience ventricular arrhythmias than those carrying mutations that do not produce DNE on Kir2.1/2.2 channels. Recently it has been demonstrated that BrS patients with SCN5A mutations exhibit more conduction abnormalities on electrocardiogram and have a higher risk for cardiac events (35). Furthermore, it has been suggested that depolarization abnormalities can be detected more often in the electrocardiogram of BrS patients with prior ventricular fibrillation/sudden cardiac death (VF/SCD) than in those without VF/SCD (36, 37). Indeed, the presence of fragmented QRS (f-QRS), which is a marker of depolarization abnormalities and consists of the presence of multiple spikes within the QRS complex, exists more frequently in BrS patients with prior VF than in those asymptomatic or with syncope (36). More recently it has been suggested that the assessment of the Tpeak-Tend (Tp-Te) interval, a marker of the transmural dispersion of ventricular repolarization (38) or of combined depolarization and repolarization parameters on electrocardiogram (39), may effectively predict risk in BrS patients. Whether patients carrying Nav1.5 mutations that produce DNE on Kir2.1 channels are more prone to present depolarization and/or repolarization abnormalities than those carrying other kinds of loss-of-function mutations merits further analysis.
Some data suggested that there might be a discrepancy between the functional consequences of a mutation in vitro and the phenotype of the affected patients. In fact, patients carrying apparently “benign” mutations sometimes present marked phenotypes and vice versa (40). Importantly, ER, but not Golgi, trafficking–defective Nav1.5 mutants could be partially “rescued” by Kir2.1/2.2 channels, an effect that can ameliorate the INa reduction produced by the former. Therefore, Golgi trafficking–defective Nav1.5 mutants would produce a greater reduction in INa, and as a consequence of cardiac excitability, than ER trafficking–defective mutants.
Limitations of the study.
Our study has some potential limitations. First, BrS is mainly due to SCN5A mutations that commonly appear in heterozygosis. Conversely, the results presented here were obtained in heterologous transfection systems, and in rat ventricular and hiPSC-derived CMs in which mutated Nav1.5 channels were overexpressed. However, data from BrS transgenic mice in which one SCN5A allele was deleted also pointed in the same direction. Second, a limited number of SCN5A mutations (5 plus the association of a mutation with the common p.H558R polymorphism) were studied. However, we believe that they are fairly representative of different types of BrS-associated mutations because they exhibit a broad spectrum of phenotypic manifestations (i.e, only traffic or gating defects and traffic defects combined with strong gating defects; mutations producing or not DNE on Nav1.5 channels) and are not located at the same Nav1.5 domain. Third, results obtained at the cellular level should be confirmed in a multicellular setting, since the strong electrotonic interactions among cells could partially compensate the APD prolongation and the resting membrane depolarization produced by the IK1 decrease. In summary, our results suggest that ER trafficking–defective Nav1.5 mutants diminish IK1, because these mutants are not able to reciprocally increase IKir2.1/2.2. Moreover, Golgi trafficking–defective Nav1.5 mutants produce a DNE on Kir2.1/2.2 channels that would lead to a further IK1 decrease. Moreover, Nav1.5 mutants that are retained at the ER can be partially rescued by Kir2.1/2.2 channels via an unconventional GRASP-dependent secretory route. The correlation between, on one hand, the functional effects produced by SCN5A mutations on INa and IK1 and, on the other, the clinical features in a large cohort of BrS patients is beyond our scope, but would be very important for confirmation of our observations.
Methods
Further information can be found in Supplemental Methods, available online with this article.
Cell culture and transfection.
CHO cells (ATCC) were transiently transfected with WT and mutant Nav1.5 or Kir2.x channels by using FUGENE X-tremeGENE (Roche Diagnostics) and cultured as described previously (6, 14, 15, 18). In all experiments, Nav1.5 channels, both WT and mutated, were transfected together with the hNavβ1 subunit.
Rat and mouse ventricular myocyte isolation.
Ventricular CMs isolated from male Sprague-Dawley rats by enzymatic dissociation were infected with adenoviral constructs encoding for human WT and mutant Nav1.5 channels (6). Ventricular CMs were also isolated from male heterozygous Kcnj2-overexpressing (Kcnj2OE+/–) (12), heterozygous Scn5a-knockout (Scn5a+/–) mice generated using previously described methods (41) or a double mutant resulting from cross-breeding both mice (Kcnj2OE+/–-Scn5a+/–).
Patch clamping.
Currents were recorded using the whole-cell patch-clamp technique (6, 14, 15, 18). Series resistance was compensated manually using the compensation unit of the Axopatch amplifier (Molecular Devices); ≥80% compensation was achieved. No significant voltage errors (<5 mV) due to series resistance were expected with the micropipettes used.
INa, IK1, and AP recordings in hiPSC-CMs.
Enriched and mature DF19-9-11T hiPSC-CMs generated as described elsewhere (18) and commercial hiPSC-CMs (iCell Cardiomyocytes2, Cellular Dynamics) were used and infected with the adenoviral constructs coding WT or mutant Nav1.5 channels. Currents (recorded at 21–23°C) and APs were recorded using the whole-cell patch-clamp technique.
Immunofluorescence and imaging.
COS7 cells (ATCC) were cotransfected with Nav1.5-GFP mutants and ER or Golgi markers and thereafter (48 hours) fixed and processed for immunofluorescence (42).
GRASP silencing.
To analyze the consequences of GRASP silencing, CHO cells or rat ventricular and hiPSC-derived CMs were transfected with siRNA GRASP55 or infected with lentiviral particles encoding shRNA GRASP55/65, respectively. Control cells/CMs were transfected/infected with scrambled siRNA or shRNA (6). Silencing was confirmed by Western blotting using a GRASP55-specific antibody.
Statistics.
Results are expressed as mean ± SEM. Unpaired t test or 1-way ANOVA followed by Newman-Keuls test was used where appropriate. In small-size samples (n < 15), statistical significance was confirmed by using nonparametric tests. To take into account repeated sample assessments, data were analyzed with multilevel mixed-effects models. A P value less than 0.05 was considered significant.
Study approval.
Animal studies were approved by the Committees on the Use and Care of Animals at Complutense University and the University of Michigan and conformed to the guidelines of European Union Directive 2010/63/EU on the protection of animals used for scientific purposes.
Author contributions
MPH, MM, SA, PNM, RGU, DT, RdA, TC, DPB, BCW, ENJV, GGS, AMdR, FJDG, and RC conducted experiments, acquired data, and analyzed data. KC and TJH generated and maturated hiPSC-CMs. JT, JJ, RC, and ED designed the research, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Sandra Sacristán, Paloma Vaquero, Lorena Ondo, Luis Tamargo, and Yan Chen for their invaluable technical assistance. We also thank Dan Roden of Vanderbilt University for generously providing the Scn5a+/– mouse model and Ignacio Sandoval and the Optical Microscopy Core Facility, Centro de Biología Molecular Severo Ochoa (CSIC-UAM), for assistance with imaging studies. This work was supported by Fondos Europeos de Desarrollo Regional; Ministerio de Economía y Competitividad (SAF2017-88116P and SAF2014-55686-R [FJDG]); Instituto de Salud Carlos III (PI16/00398); Comunidad Autónoma de Madrid (B2017/BMD-3738); and grants from ERA-Net for Research on Rare Diseases (AC14/00029), Banco Bilbao Vizcaya Argentaria Foundation, the Spanish Society of Cardiology, the National Heart, Lung, and Blood Institute (R01 HL122352 to JJ), and the Leducq Foundation (JJ).
Version 1. 09/20/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(18):e96291. https://doi.org/10.1172/jci.insight.96291.
Contributor Information
Marcos Matamoros, Email: marcosmatamoros@ucm.es.
Silvia Alfayate, Email: silvalfa@ucm.es.
Paloma Nieto-Marín, Email: paloma.nieto.marin@ucm.es.
Raquel G. Utrilla, Email: ragarc12@ucm.es.
David Tinaquero, Email: dtinaque@ucm.es.
Teresa Crespo, Email: tcresp01@ucm.es.
Daniela Ponce-Balbuena, Email: dponceba@med.umich.edu.
Guadalupe Guerrero-Serna, Email: gpegros@med.umich.edu.
Todd J. Herron, Email: toddherr@umich.edu.
Juan Tamargo, Email: jtamargo@med.ucm.es.
Ricardo Caballero, Email: rcaballero@med.ucm.es.
References
- 1.Chen-Izu Y, et al. Na+ channel function, regulation, structure, trafficking and sequestration. J Physiol (Lond) 2015;593(6):1347–1360. doi: 10.1113/jphysiol.2014.281428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veerman CC, Wilde AA, Lodder EM. The cardiac sodium channel gene SCN5A and its gene product NaV1.5: role in physiology and pathophysiology. Gene. 2015;573(2):177–187. doi: 10.1016/j.gene.2015.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anumonwo JM, Lopatin AN. Cardiac strong inward rectifier potassium channels. J Mol Cell Cardiol. 2010;48(1):45–54. doi: 10.1016/j.yjmcc.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidmann S. The effect of the cardiac membrane potential on the rapid availability of the sodium-carrying system. J Physiol (Lond) 1955;127(1):213–224. doi: 10.1113/jphysiol.1955.sp005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milstein ML, et al. Dynamic reciprocity of sodium and potassium channel expression in a macromolecular complex controls cardiac excitability and arrhythmia. Proc Natl Acad Sci USA. 2012;109(31):E2134–E2143. doi: 10.1073/pnas.1109370109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matamoros M, et al. Nav1.5 N-terminal domain binding to α1-syntrophin increases membrane density of human Kir2.1, Kir2.2 and Nav1.5 channels. Cardiovasc Res. 2016;110(2):279–290. doi: 10.1093/cvr/cvw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Utrilla RG, et al. Kir2.1-Nav1.5 channel complexes are differently regulated than Kir2.1 and Nav1.5 channels alone. Front Physiol. 2017;8:903. doi: 10.3389/fphys.2017.00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponce-Balbuena D, et al. Cardiac Kir2.1 and NaV1.5 channels traffic together to the sarcolemma to control excitability. Circ Res. 2018;122(11):1501–1516. doi: 10.1161/CIRCRESAHA.117.311872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remme CA. Cardiac sodium channelopathy associated with SCN5A mutations:electrophysiological, molecular and genetic aspects. J Physiol. 2013;591(17):4099–4116. doi: 10.1113/jphysiol.2013.256461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shryock JC, Song Y, Rajamani S, Antzelevitch C, Belardinelli L. The arrhythmogenic consequences of increasing late INa in the cardiomyocyte. Cardiovasc Res. 2013;99(4):600–611. doi: 10.1093/cvr/cvt145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balse E, Steele DF, Abriel H, Coulombe A, Fedida D, Hatem SN. Dynamic of ion channel expression at the plasma membrane of cardiomyocytes. Physiol Rev. 2012;92(3):1317–1358. doi: 10.1152/physrev.00041.2011. [DOI] [PubMed] [Google Scholar]
- 12.Piao L, Li J, McLerie M, Lopatin AN. Transgenic upregulation of IK1 in the mouse heart is proarrhythmic. Basic Res Cardiol. 2007;102(5):416–428. doi: 10.1007/s00395-007-0659-y. [DOI] [PubMed] [Google Scholar]
- 13.Papadatos GA, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci USA. 2002;99(9):6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Núñez L, et al. p.D1690N Nav1.5 rescues p.G1748D mutation gating defects in a compound heterozygous Brugada syndrome patient. Heart Rhythm. 2013;10(2):264–272. doi: 10.1016/j.hrthm.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 15.Dolz-Gaitón P, et al. Functional characterization of a novel frameshift mutation in the C-terminus of the Nav1.5 channel underlying a Brugada syndrome with variable expression in a Spanish family. PLoS ONE. 2013;8(11):e81493. doi: 10.1371/journal.pone.0081493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Correlations between clinical and physiological consequences of the novel mutation R878C in a highly conserved pore residue in the cardiac Na+ channel. Acta Physiol (Oxf) 2008;194(4):311–323. doi: 10.1111/j.1748-1716.2008.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clatot J, et al. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Na(v)1.5 α-subunits. Cardiovasc Res. 2012;96(1):53–63. doi: 10.1093/cvr/cvs211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caballero R, et al. Tbx20 controls the expression of the KCNH2 gene and of hERG channels. Proc Natl Acad Sci USA. 2017;114(3):E416–E425. doi: 10.1073/pnas.1612383114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, et al. Identification of a new co-factor, MOG1, required for the full function of cardiac sodium channel Nav1.5. J Biol Chem. 2008;283(11):6968–6978. doi: 10.1074/jbc.M709721200. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarti S, et al. MOG1 rescues defective trafficking of Na(v)1.5 mutations in Brugada syndrome and sick sinus syndrome. Circ Arrhythm Electrophysiol. 2013;6(2):392–401. doi: 10.1161/CIRCEP.111.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56(5):801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gee HY, Noh SH, Tang BL, Kim KH, Lee MG. Rescue of ΔF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011;146(5):746–760. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Mercier A, Clément R, Harnois T, Bourmeyster N, Bois P, Chatelier A. Nav1.5 channels can reach the plasma membrane through distinct N-glycosylation states. Biochim Biophys Acta. 2015;1850(6):1215–1223. doi: 10.1016/j.bbagen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Sottas V, Abriel H. Negative-dominance phenomenon with genetic variants of the cardiac sodium channel Nav1.5. Biochim Biophys Acta. 2016;1863(7 Pt B):1791–1798. doi: 10.1016/j.bbamcr.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Petitprez S, et al. SAP97 and dystrophin macromolecular complexes determine two pools of cardiac sodium channels Nav1.5 in cardiomyocytes. Circ Res. 2011;108(3):294–304. doi: 10.1161/CIRCRESAHA.110.228312. [DOI] [PubMed] [Google Scholar]
- 26.Giuliani F, Grieve A, Rabouille C. Unconventional secretion: a stress on GRASP. Curr Opin Cell Biol. 2011;23(4):498–504. doi: 10.1016/j.ceb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Vinke FP, Grieve AG, Rabouille C. The multiple facets of the Golgi reassembly stacking proteins. Biochem J. 2011;433(3):423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- 28.Waechter CJ, Schmidt JW, Catterall WA. Glycosylation is required for maintenance of functional sodium channels in neuroblastoma cells. J Biol Chem. 1983;258(8):5117–5123. [PubMed] [Google Scholar]
- 29.Zhang Y, Hartmann HA, Satin J. Glycosylation influences voltage-dependent gating of cardiac and skeletal muscle sodium channels. J Membr Biol. 1999;171(3):195–207. doi: 10.1007/s002329900571. [DOI] [PubMed] [Google Scholar]
- 30.Gee HY, Kim J, Lee MG. Unconventional secretion of transmembrane proteins. Semin Cell Dev Biol. doi: 10.1016/j.semcdb.2018.03. https://doi.org/10.1016/j.semcdb.2018.03.016 [published online ahead of print March 28, 2018]. [DOI] [PubMed] [Google Scholar]
- 31.Marra P, et al. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat Cell Biol. 2001;3(12):1101–1113. doi: 10.1038/ncb1201-1101. [DOI] [PubMed] [Google Scholar]
- 32.Ma D, et al. Golgi export of the Kir2.1 channel is driven by a trafficking signal located within its tertiary structure. Cell. 2011;145(7):1102–1115. doi: 10.1016/j.cell.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaborit N, et al. Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol (Lond) 2007;582(Pt 2):675–693. doi: 10.1113/jphysiol.2006.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panama BK, McLerie M, Lopatin AN. Heterogeneity of IK1 in the mouse heart. Am J Physiol Heart Circ Physiol. 2007;293(6):H3558–H3567. doi: 10.1152/ajpheart.00419.2007. [DOI] [PubMed] [Google Scholar]
- 35.Yamagata K, et al. Genotype-phenotype correlation of SCN5A mutation for the clinical and electrocardiographic characteristics of probands with Brugada syndrome: a Japanese multicenter registry. Circulation. 2017;135(23):2255–2270. doi: 10.1161/CIRCULATIONAHA.117.027983. [DOI] [PubMed] [Google Scholar]
- 36.Morita H, et al. Fragmented QRS as a marker of conduction abnormality and a predictor of prognosis of Brugada syndrome. Circulation. 2008;118(17):1697–1704. doi: 10.1161/CIRCULATIONAHA.108.770917. [DOI] [PubMed] [Google Scholar]
- 37.Takagi M, Yokoyama Y, Aonuma K, Aihara N, Hiraoka M, Japan Idiopathic Ventricular Fibrillation Study (J-IVFS) Investigators Clinical characteristics and risk stratification in symptomatic and asymptomatic patients with Brugada syndrome: multicenter study in Japan. J Cardiovasc Electrophysiol. 2007;18(12):1244–1251. doi: 10.1111/j.1540-8167.2007.00971.x. [DOI] [PubMed] [Google Scholar]
- 38.Zumhagen S, Zeidler EM, Stallmeyer B, Ernsting M, Eckardt L, Schulze-Bahr E. Tpeak-Tend interval and Tpeak-Tend/QT ratio in patients with Brugada syndrome. Europace. 2016;18(12):1866–1872. doi: 10.1093/europace/euw033. [DOI] [PubMed] [Google Scholar]
- 39.Kawazoe H, et al. Risk stratification of ventricular fibrillation in Brugada syndrome using noninvasive scoring methods. Heart Rhythm. 2016;13(10):1947–1954. doi: 10.1016/j.hrthm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Hoshi M, et al. Brugada syndrome disease phenotype explained in apparently benign sodium channel mutations. Circ Cardiovasc Genet. 2014;7(2):123–131. doi: 10.1161/CIRCGENETICS.113.000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, et al. Recombinase-mediated cassette exchange to rapidly and efficiently generate mice with human cardiac sodium channels. Genesis. 2006;44(11):556–564. doi: 10.1002/dvg.20247. [DOI] [PubMed] [Google Scholar]
- 42.Garrido-García A, Andrés-Pans B, Durán-Trío L, Díez-Guerra FJ. Activity-dependent translocation of neurogranin to neuronal nuclei. Biochem J. 2009;424(3):419–429. doi: 10.1042/BJ20091071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.