Abstract
The pathogenetic mechanisms underlying the pathologic fibrosis in diseases such as idiopathic pulmonary fibrosis (IPF) are poorly understood. To identify genetic factors affecting susceptibility to IPF, we analyzed a murine genetic model of IPF in which a profibrotic cytokine (TGF-β1) was expressed in the lungs of 10 different inbred mouse strains. Surprisingly, the extent of TGF-β1–induced lung fibrosis was highly strain dependent. Haplotype-based computational genetic analysis and gene expression profiling of lung tissue obtained from fibrosis-susceptible and -resistant strains identified laminin α1 (Lama1) as a genetic modifier for susceptibility to IPF. Subsequent studies demonstrated that Lama1 plays an important role in multiple processes that affect the pulmonary response to lung injury and susceptibility to fibrosis, which include: macrophage activation, fibroblast proliferation, myofibroblast transformation, and the production of extracellular matrix. Also, Lama1 mRNA expression was significantly increased in lung tissue obtained from IPF patients. These studies identify Lama1 as the genetic modifier of TGF-β1 effector responses that significantly affects the development of pulmonary fibrosis.
Keywords: Pulmonology
Keywords: Fibrosis
Haplotype genetic analysis and gene expression profiling of TGF-β transgenic mice on different genetic strains of mice identified Laminin-α1 as a genetic modifier for pulmonary fibrosis.
Introduction
Although human fibrotic lung disorders cause substantial morbidity and mortality, we know relatively little about their pathogenesis and have few highly effective treatments for them (1). In addition, although genetic factors are believed to play important roles in the regulation and progression of human fibrotic disorders, the factors that account for the variations in the development and progression of pulmonary fibrosis have not been defined (2, 3). Idiopathic pulmonary fibrosis (IPF) is a prototypical fibrotic lung disorder that is characterized by epithelial damage, fibroproliferative matrix deposition and parenchymal remodeling (4–6). TGF-β1, a multifunctional cytokine that plays pivotal roles in diverse biologic processes, is believed to play an important role in the pathogenesis of this disorder. This is based on studies that demonstrate that (i) TGF-β1 is an essential mediator of the pathologic scarring in a variety of fibrotic disorders (7–12); (ii) a high level of biologically active TGFβ-1 is expressed in IPF tissue (13–15); (iii) TGFβ-1 is a critical mediator of the bleomycin-induced pulmonary fibrosis in rodent modeling systems (10, 11); and (iv) TGF-β1 overexpression causes progressive pulmonary fibrosis in vivo (16, 17) and in vitro (15). In accordance with these observations, the juxtaposition of tissue injury, fibrosis, and exaggerated TGF-β1 expression are well documented in IPF (18–21). However, the pathways that mediate and the factors that regulate the fibrogenic effects of TGF-β1 are poorly understood. Moreover, the rate of progression, severity, and response to therapy observed in IPF patients are extremely variable, and the factors that contribute to this variability have not been defined (22).
In addition to its roles in the generation of fibrotic tissue and cellular responses, recent studies have highlighted a wide variety of other TGF-β1–induced effector responses and the fact that these responses can appear to contradict one another (7, 23–25). In keeping with its importance in fibrosis, TGF-β1 is essential for wound healing and stimulates matrix molecule deposition and angiogenesis in some settings. In contrast, in other settings TGF-β1 can also induce tissue injury and cellular apoptosis, decrease epithelialization, and inhibit wound healing (23, 25–28). Similarly, TGF-β1 has important antiinflammatory (29, 30) and proinflammatory effects (30, 31) in different settings. In accordance with this complexity and in contrast to its roles in fibrosis, TGF-β1 has also been implicated in the pathogenesis of diseases characterized by tissue destruction, such as the emphysema in chronic obstructive lung disease (COPD) (32, 33). However, the mechanisms that contribute to these complex and context-dependent effects of TGF-β1 have not been defined.
To begin to characterize effector responses of TGF-β1, we generated transgenic (Tg) mice in which bioactive TGF-β1 was inducibly targeted to the murine lung. These mice were initially on a C57BL/6J (C57) murine genetic background. As expected, the TGF-β1 transgene induced a prominent pulmonary fibrotic response in these C57 background animals (34). In contrast, when the mice were bred for more than 10 generations onto a BALB/c background, the same level of transgene expression induced a phenotype in which fibrosis was markedly diminished and tissue destruction and emphysema dominated the tissue response. These differences demonstrated that genetic modifiers play a critical role in determining the biologic responses that are induced by TGF-β1. To further define these genetic modifier(s), the TGF-β1 Tg mice were bred for more than 10 generations onto 10 different inbred murine strains. and their phenotypes were assessed and compared. These studies demonstrated that the extent of pulmonary fibrosis was highly strain dependent. Haplotype-based computational genetic mapping (HBCGM) and mRNA profiling of the lungs obtained from fibrosis-prone and -resistant mice identified laminin α1 (Lama1) as a genetic modifier of pulmonary fibrosis. Subsequent in vivo and in vitro studies highlighted strain-specific differences in Lama1 expression in key cell types and the critical role(s) of Lama1 in determining the magnitude of TGF-β1–induced pulmonary repair and or fibrotic responses. The disease relevance of these murine findings was also confirmed in studies that demonstrated that the levels of Lama1 expression are increased in lung tissues from patients with IPF versus controls.
Results
The extent of pulmonary fibrosis is strain dependent.
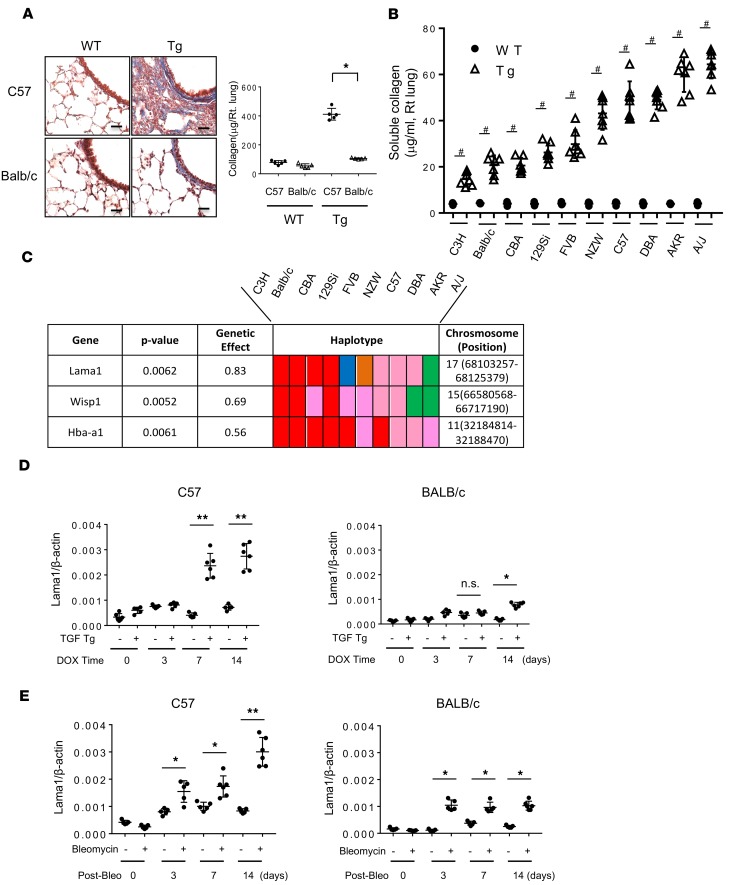
To define the mechanisms that underlie the heterogeneity of TGF-β1 effector responses, we generated Tg mice in which bioactive TGF-β1 was inducibly expressed in the lungs of C57 mice (34). Then, this transgene was bred for more than 10 generations onto 9 other strains (BALB/cJ, C3H/HeJ, CBA/J, 129Si/J, FVB/J, NZW/J, DBA/J, AKR/J, and A/J). As previously described, bioactive TGF-β1 induced epithelial apoptosis and subsequent pulmonary fibrosis in C57 mice (Figure 1A) (34). The fibrogenic response in C57 mice was much more prominent than in the lungs of Tg BALB/c mice (Figure 1A). When the pulmonary phenotypes in all 10 strains were compared, it became clear that there was an extensive amount of fibrosis in certain strains (A/J, AKR, DBA, and C57), while others (C3H, BALB/c, CBA, and 129Si) had markedly reduced levels of collagen accumulation (Figure 1B and Table 1). Across the 10 strains, the collagen concentration in the lungs of TGF-β1 Tg mice varied from 15 μg/ml (C3H) to 66 μg/ml (A/J). The observed inter-strain difference was strong and very significant (P = 4 × 10–27), suggesting that the observed variations were indeed due to genetic differences. These differences represented intrinsic properties of the genetic backgrounds of the strains. They could not be explained by differences in the level of Tg TGF-β1 expression, because comparable levels of TGF-β1 were present in bronchoalveolar lavage (BAL) fluid obtained from the 10 inbred strains after doxycycline induction (Supplemental Figure 1). These results demonstrate that the pulmonary responses to TGF-β1 depend upon genetic modifiers present in the genetic background of the different strains.
Figure 1. Identification of Lama1 as a genetic modifier of TGF-β–stimulated pulmonary fibrosis.
Lungs from 6- to 8-week-old WT and TGF-β1–Tg mice on the noted genetic backgrounds were evaluated after the mice were given doxycycline in drinking water for 14 days of to induce Tg. (A) Mallory trichrome evaluation (left panel) and total collagen content measured by Sircol collagen assay (right panel) of lungs from WT and TGF-β1 Tg mice on C57BL/6J (C57) and BALB/cJ (BALB/c) backgrounds. Scale bars: 25 μm. (B) Levels of collagen in lungs from WT and Tg mice on 10 different background strains were measured by Sircol evaluations (n = 7 for each). (C) Illustration of haplotype blocks of Lama1, Wisp1, and Hba-a1. The haplotype of each gene is represented by a colored block, and is presented in the same order as the phenotypic data shown in B. (D) Kinetic evaluation (using qRT-PCR) of the levels of mRNA encoding Lama1 in lungs from WT and TGF-β1 Tg mice on C57 and BALB/c backgrounds at various time points after transgene induction with doxycycline (DOX). (E) Expression of Lama1 mRNA in the lungs of bleomycin-challenged C57 and BALB/c mice. The histology shown in A is representative of at least 5 mice per each group. The values in A, B, D, and E represent mean ± SEM of a minimum number of 5 mice in each group. Post-Bleo, days after bleomycin challenge. *P < 0.05. **P < 0.01. #P < 0.001.
Table 1. Soluble collagen levels in the lungs from WT and TGF-β Tg mice on different background strains measured by Sircol collagen assay.
Identification of Lama1 as a genetic modifier of TGF-β1–stimulated pulmonary fibrosis. To identify genetic modifiers of TGF-β1 responses, HBCGM was used to analyze the fibrogenic responses (collagen levels) in the 10 inbred strains as previously described (35). This analysis indicated that 560 genes had allelic patterns that correlated with the extent of collagen accumulation (using a P value cutoff of 0.01; Supplemental Table 1). In order to prioritize this extensive candidate gene list, we compared the lung gene expression profiles of fibrosis-prone (C57, AKR/J, A/J) and fibrosis-resistant (C3H/J, 129 Si/J, and BALB/c) TGF-β1 Tg mice. For each strain, we determined the expression fold change of each gene transcript in Tg mice compared with controls. We identified 127 genes with at least a 2-fold difference on average in the fibrosis-prone strains but less than a 2-fold change in fibrosis-resistant strains (Supplemental Table 2). Finally, by intersecting the lists of genes identified by HBCGM and by gene expression profiling, 3 candidate genetic modifiers were identified: Lama1, Wisp1, and Hba-a1 (Figure 1C). Of these, Lama1 was selected for analysis based on its relatively high genetic effect (O.83) and consistent responsiveness to TGF-β1 stimulation in vivo and in vitro (Figure 1D and see below). A schematic illustration of the overall selection process and expression levels of Lama1 in fibrosis-prone and -resistant strains of mice is shown in Supplemental Figure 2. We identified 851 SNPs that were variable among these 10 strains, among which 15 coding SNPs induced amino acid changes. Additional evidence supporting a critical role for Lama1 was found when the baseline levels and levels of TGF-β1–stimulated expression of pulmonary Lama1 were examined. These studies demonstrated that both were significantly higher in C57 Tg mice than in BALB/c mice (Figure 1D). Similar time-dependent increases in the expression of Lama1 were noted in the lungs of bleomycin-challenged C57 mice, another animal model of pulmonary fibrosis, and the levels of Lama1 expression in C57 mice were significantly higher than in BALB/c mice (Figure 1E).
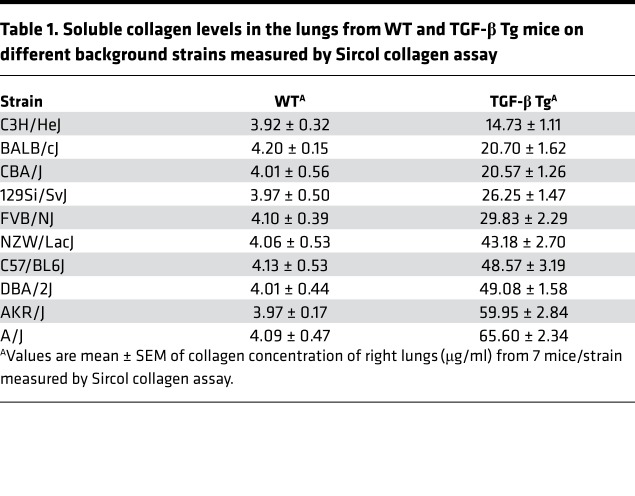
Strain-dependent localization of Lama1 expression in the lungs of TGF-β1 Tg mice. Studies were next undertaken to localize the expression of Lama1 in lungs from C57 and BALB/c TGF-β1 Tg mice. Low levels of Lama1 expression were noted in the regions near the basement membranes as well as interstitial areas of lungs from both strains of mice (Figure 2A). Interestingly, Lama1 expression was prominently induced in macrophages in lungs from C57 TGF-β1 Tg mice but not in lungs from BALB/c TGF-β1 Tg mice. This strain-specific macrophage expression was further confirmed by double-label IHC staining using CD68 macrophage-specific and Lama1 antibodies (Figure 2B). The differential induction of Lama1 by TGF-β1 was also confirmed in vitro using alveolar macrophages isolated from C57 and to BALB/c animals (Supplemental Figure 3). Similarly, TGF-β1 significantly induced the expression of Lama1 in fibroblasts from C57 mice but did not have comparable effects in cells from BALB/c animals (Figure 2C). Interestingly, the TGF-β–stimulated Smad2 activation was markedly decreased with Lama1 siRNA silencing in the fibroblasts from C57 but not BALB/c mice (Figure 2D). Collectively these studies demonstrate that TGF-β1 stimulates Lama1 expression in a strain-specific and cell-specific manner, with TGF-β1 inducing a high level of Lama1 expression in macrophages and fibroblasts of fibrogenic C57 mice, and with significantly lower levels of induction in fibrosis-resistant BALB/c mice.
Figure 2. Strain- and cell-dependent Lama1 expression in lungs from TGF-β Tg mice.
Lungs from doxycycline-treated TGF-β1 Tg mice on the noted genetic backgrounds were evaluated. (A) Representative Lama1 IHC evaluation comparing C57 and BALB/c TGF-β Tg mice. White arrow indicates Lama1 positive macrophages while blue arrow indicates Lama1 stained area of basement membrane (lightly stained compared to macrophages) (B) Double IHC using anti-Lama1 (green) and anti-CD68 (red) antibodies. Arrows indicate cells stained with, Lama1, CD68 (macrophage specific marker), or both. Scale bars in A and B: 25 μm. (C) Fibroblasts were established from lungs of C57 and BALB/c mice and treated with TGF-β1 for 2 or 24 hours. The levels of Lama1 mRNA were evaluated by qRT-PCR. (D) Western blot evaluation on Smad2 activation in fibroblasts from C57 and BALB/c mice with and without TGF-β stimulation. A, B, and D represent evaluations in a minimum of 3 mice. Values in C represent mean ± SEM of evaluations in a minimum of 5 mice in each group. **P < 0.01, ***P < 0.001 compared with controls with no TGF-β treatment.
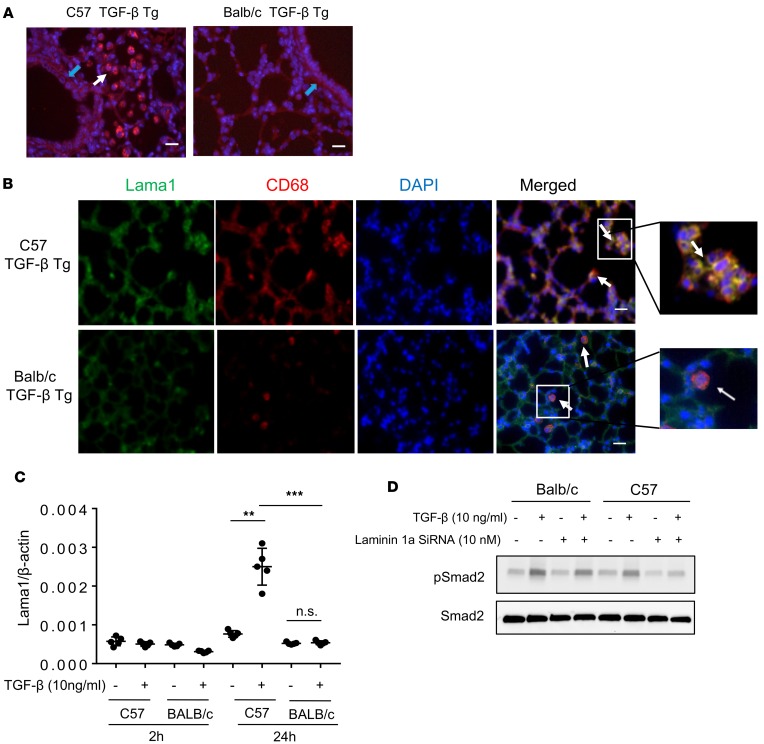
Lama1 plays a critical role in TGF-β1–stimulated pulmonary fibrosis. To investigate the role(s) of Lama1 in TGF-β1–induced pulmonary fibrosis, Lama1-specific siRNA experiments were performed. In these experiments, 6- to 8-week-old C57 WT and TGF-β1 Tg mice were treated with Lama1-specific (3 nmol/mouse/d) or control (scrambled) siRNAs every other day via nasal aspiration, and the extent of collagen accumulation in the lungs was characterized using Mallory trichrome staining and Sircol assays 2 weeks later. In these experiments, Lama1-specific siRNA decreased TGF-β1–stimulated pulmonary Lama1 mRNA expression by more than approximately 90% to levels comparable to those in WT mice (Supplemental Figure 4). This Lama1 siRNA silencing significantly decreased the collagen accumulation in lungs of TGF-β1 Tg mice compared with those receiving control siRNA (Figure 3A). Similarly, levels of expression of extracellular matrix proteins (type I and III collagens, fibronectin) were markedly decreased in lungs from Lama1-specific siRNA–treated TGF-β1 Tg mice (Figure 3B). In addition, the accumulation of α–smooth muscle actin+ (α-SMA+)/myosin– myofibroblasts surrounding airways in the lungs of C57 TGF-β1 Tg mice was also markedly reduced in the lungs of mice treated with Lama1-specific siRNA compared with controls treated with scrambled control siRNA (Figure 3C). We also noted increased α-SMA staining in the interstitial areas of TGF-β Tg mice, and Lama siRNA silencing significantly reduced the α-SMA–positive region compared with scrambled siRNA–treated mice (Figure 3D). These studies demonstrated that Lama1 plays a critical role in TGF-β1–stimulated pulmonary fibrosis, collagen accumulation, and in myofibroblast accumulation.
Figure 3. Lama1 plays an essential role in TGF-β–stimulated pulmonary fibrosis.
Lungs from 8-week-old WT and TGF-β1 Tg mice were evaluated after 2 weeks of Tg induction by doxycycline in drinking water. (A) Histologic evaluation with Mallory trichrome staining (left) and evaluation of lung collagen content with Sircol assays (right) comparing WT and Tg lungs from mice treated with control scrambled siRNA and Lama1 siRNA. (B) qRT-PCR evaluation of the levels of mRNA encoding extracellular matrix proteins (collagen type1 alpha1 [Col1a1], Col3a1, and fibronectin [FiN]) in lungs from WT and Tg mice treated with control scrambled siRNA or Lama1 siRNA. (C) Representative fluorescence double-label IHC evaluation of α–smooth muscle actin (α-SMA, red), CC10 (green), and nuclei (DAPI, blue) in lungs from WT and TGF-β1 Tg mice treated with scrambled control siRNA or Lama1 siRNA (left panels). Double-label IHC evaluation of α-SMA (green), myosin (red), and nuclei (DAPI, blue) in lungs from TGF-β1 Tg mice treated with scrambled siRNA control or Lama1 siRNA (right panels). (D) α-SMA IHC staining of the lungs of TGF-β1 Tg mice with scrambled siRNA or Lama1 siRNA silencing. Arrows indicate positively stained cells in the interstitial area in the lungs of C57 TGF-β1 Tg mice. A (left) and C are representative of evaluations in a minimum of 3 mice. The values in A and B represent mean ± SEM of evaluations in a minimum of 5 mice each group. *P < 0.05, **P < 0.01. Scale bars: 100 μm (A); 25 μm (C and D).
Role of Lama1 in alternative and fibrotic (reparative) macrophage activation.
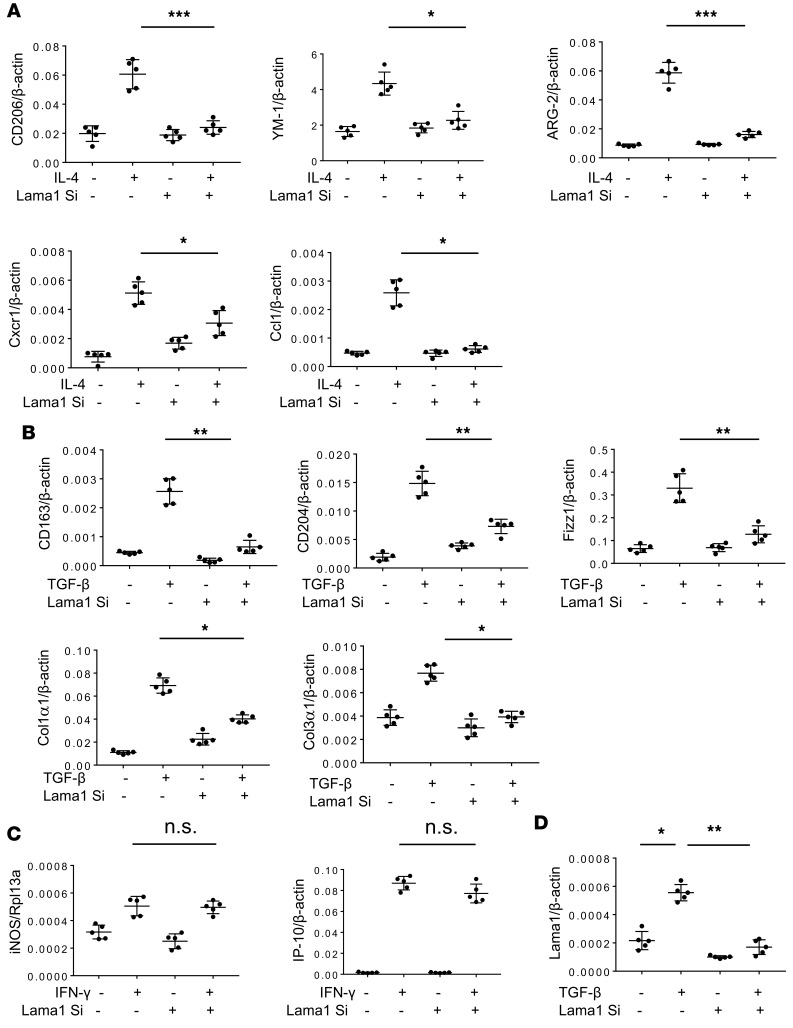
Since Lama1 is highly induced in pulmonary macrophages in C57 TGF-β1 Tg mice, we characterized the role(s) of Lama1 in macrophage activation. Lama1 induction in alveolar macrophages was significant at later fibrotic phases in the lung of TGF-β1 Tg mice (Supplemental Figure 5), supporting an important role of macrophage Lama1 in the fibrotic phase of pulmonary fibrosis. The responses of alveolar macrophages obtained from C57 mice to ligands that are known to induce specific macrophage activation paradigms were evaluated in cells that had been treated with Lama1-specific siRNA or scrambled controls in vitro. As expected, in vitro treatment of cells that had received the scrambled control with recombinant IL-4 (rIL-4) stimulated the expression of known markers of alternative (M2) activation, including CD206, YM-1, Arg-2, Cxcr1, and Ccl1 (Figure 4A). In contrast, these rIL-4–induced responses were significantly reduced in macrophages in which Lama1 had been effectively silenced (Figure 4A). TGF-β1 is also known to induce alternative and fibrotic (reparative) macrophage activation (36, 37). This includes the stimulation of a multiple markers including CD163, CD204, Fizz1, collagen type I (Col1α1), and collagen type III (Col3α1). These rTGF-β1–stimulated responses were readily apparent in cells treated with the scrambled control and significantly ameliorated in macrophages in which Lama1 was silenced (Figure 4B). Importantly, Lama1 silencing did not alter the ability of recombinant IFN-γ to induce expression levels of M1 macrophage activation (iNOS [NOS2] and IP-10) (Figure 4C). The in vitro Lama1 silencing was highly effective, as illustrated in Figure 4D. Alveolar macrophages isolated from the TGF-β Tg mice receiving Lama1 siRNA demonstrated similar decreases in fibrotic macrophage activation (Supplemental Figure 6), supporting a specific role for Lama1 in the macrophage activation process. To further evaluate the in vivo relevance of these findings, we also studied macrophages isolated from C57BL/6J-Lama1nmf223 mice with a functional missense mutation at the N-terminal end of Lama1 (38). In accordance with our findings, macrophages from mice with a functional missense mutation of Lama1 also manifested blunted alternative and fibrotic macrophage activation and unchanged classical macrophage activation responses when stimulated with IL-4, TGF-β1, or IFN-γ, respectively (Supplemental Figure 7). These Lama1 mutant mice also demonstrated reduced lung fibrosis with bleomycin challenge compared with normal controls, further supporting an in vivo functional role of Lama1 in pulmonary fibrosis (Supplemental Figure 8). These studies demonstrate that Lama1 plays a critical role in alternative and fibrotic macrophage activation.
Figure 4. Lama1 plays a critical role in alternative and fibrotic/reparative macrophage activation.
Alveolar macrophages were isolated from the C57 mice and stimulated with recombinant IL-4 (rIL-4), rTGF-β, and rIFN-γ with (+) and without (–) in vitro siRNA silencing of Lama1 (Lama1 Si). (A) Expression of macrophage alternative activation markers was evaluated by qRT-PCR after 24-hour stimulation with rIL-4 (20 ng/ml). (B) Expression of alveolar macrophage fibrotic/reparative markers evaluated by qRT-PCR after 24-hour stimulation with rTGF-β1 (20 ng/ml). (C) Expression of macrophage classical activation markers evaluated by qRT-PCR after 24-hour stimulation with rIFN-γ (100 ng/ml). (D) Lama1 siRNA silencing efficacy on TGF-β stimulated Lama1 expression in alveolar macrophages evaluated by qRT-PCR. The values represent mean ± SEM of evaluations, with a minimum of 5 mice in each group. *P < 0.05, **P < 0.01, ***P < 0.001.
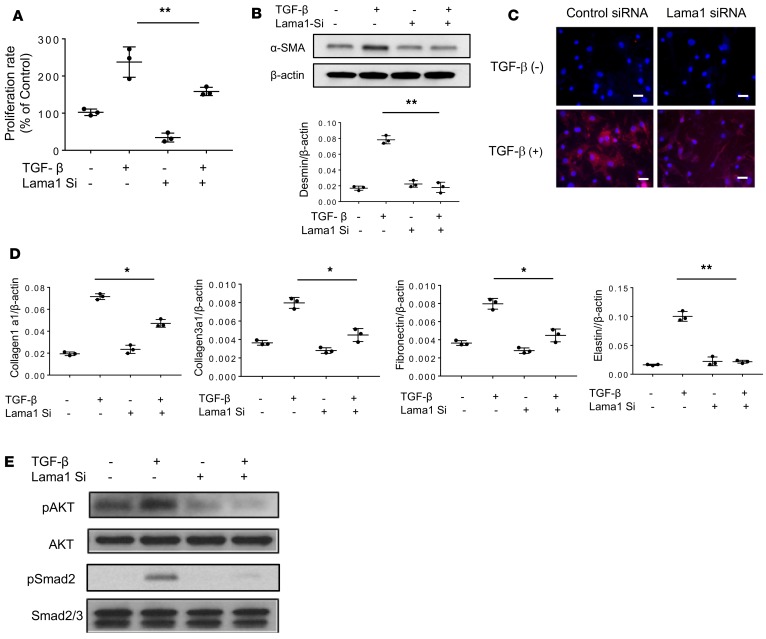
Role of Lama1 in fibroblast proliferation, myofibroblast transformation, and TGF-β1 signaling. Fibroblasts are one of the major effector cell types responsible for tissue fibrotic responses. Because TGF-β1 is a potent stimulator of fibroblast Lama1 expression in C57 but not in BALB/c mice (Figure 2C), studies were undertaken to define the roles of Lama1 in TGF-β1–induced fibroblast responses. In these experiments, we compared the responses of TGF-β1–stimulated normal human lung fibroblasts (NHLFs) when treated with Lama1 siRNA or scrambled controls. The Lama1 silencing in vitro was also highly effective in these fibroblasts, as illustrated in Figure 4D. TGF-β1 was a potent stimulator of the proliferation of cells treated with the control siRNA (Figure 5A). As noted above, TGF-β1 was also a potent stimulator of fibroblast-to-myofibroblast differentiation, which was characterized by enhanced expression of the myofibroblasts markers α-SMA and desmin (Figure 5, B and C). TGF-β1 also stimulated extracellular matrix gene expression and activated Smad2 and Akt fibroblast signaling (Figure 5, D and E). In all cases, these responses were at least partially Lama1 dependent, as treatment with Lama1 siRNA significantly decreased TGF-β1–stimulated fibroblast proliferation, myofibroblast differentiation, matrix gene expression, and fibroblast signaling. When viewed in combination, these studies demonstrate that Lama1 is a critical regulator of TGF-β1–stimulated fibroblast proliferation, myofibroblasts transformation, matrix gene expression, and the activation of its fibroblast signaling pathways.
Figure 5. Lama1 plays a critical role in TGF-β–stimulated fibroblast proliferation, myofibroblast transformation, extracellular matrix protein expression, and activation of Akt and Smad2 signaling.
Normal human lung fibroblasts (NHLFs) were treated with Lama1 siRNA or the scrambled control and stimulated for 24 hours with rTGF-β1 (20 ng/ml) or vehicle control. (A) Cellular proliferation was assessed using WST-1 evaluations. (B) Western blot evaluation of α–smooth muscle actin (α-SMA) and qRT-PCR evaluation of desmin expression. (C) IHC evaluation of α-SMA expression. Scale bars: 25 μm. (D) qRT-PCR evaluations of the expression of extracellular matrix proteins. (E) Western blot evaluation of Akt and Smad2 activation in fibroblasts treated with TGF-β1 or vehicle control with (+) and without (–) Lama1 siRNA silencing (Si). The values in A, B, and D represent mean ± SEM of triplicate evaluations in 3 separate experiments. *P < 0.05, **P < 0.01. Western blot and IHC evaluations in B, C, and E are representative of 3 separate experiments.
Increased expression of Lama1 in the lungs of IPF patients.
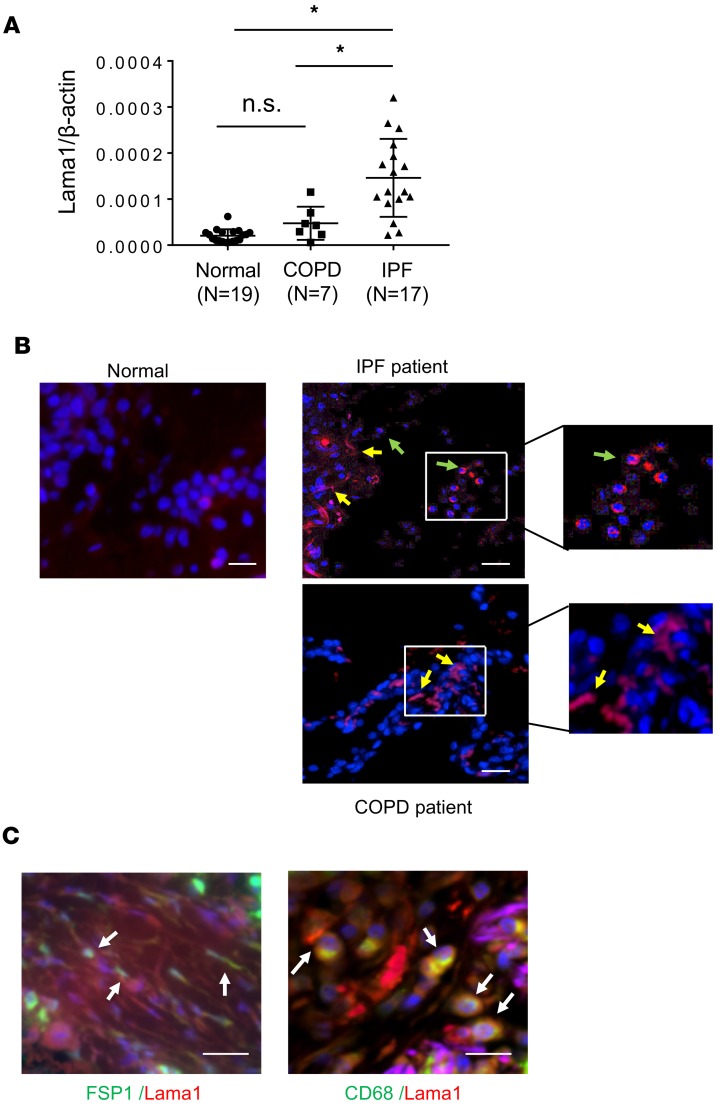
The studies noted above demonstrate that Lama1 plays a critical role in the generation of TGF-β1–induced fibroproliferative responses. This allows for the exciting hypothesis that exaggerated Lama1 expression is seen in human fibrotic disorders. To test this hypothesis, we evaluated the expression of Lama1 in lung tissues from IPF patients and compared these findings to expression in lung tissues from normal control patients and patients with COPD. As shown in Figure 6, Lama1 expression was significantly increased in the lung tissues from IPF patients compared with normal and disease controls (Figure 6A). In keeping with our findings in TGF-β1 Tg mice, Lama1 IHC also demonstrated an increase in the number of macrophage-like and interstitial fibroblast–like cells that contained Lama1 in lungs from IPF patients compared with the control cohort or COPD patients (Figure 6B). Double IHC with markers specific for fibroblasts (FSP-1) and macrophages (CD68) further confirmed that these cells were the major Lama1-positive cells in IPF patients (Figure 6C). Thus, in keeping with our murine findings, these studies demonstrate that enhanced Lama1 expression is present in lung tissues from patients with IPF.
Figure 6. Increased expression of Lama1 in lungs from IPF patients.
(A) Levels of mRNA encoding Lama1 were evaluated, using qRT-PCR, in lung tissues from normal controls, patients with COPD, and patients with IPF. (B) Representative IHC staining of Lama1 in the lungs from controls and patients with IPF and COPD. Arrows indicate Lama1+ cells. Yellow arrows, fibroblasts-like cells; green arrows, macrophage-like cells. (C) Representative double IHC staining using macrophage- (CD68) and fibroblast-specific markers with Lama1 in lungs from IPF patients. *P < 0.05. Scale bars in B and C: 50 μm.
Discussion
To further understand the pathogenesis of a fibrotic disorders, we used what we believe to be a novel Tg approach and identified a genetic modifier of TGF-β1–induced pulmonary fibrosis. Analysis of a panel of 10 inbred strains, each of which had a similar level of transgene-induced TGF-β1 overexpression, revealed that TGF-β1 was remarkably fibrogenic in some strains but minimally fibrogenic in others. By combining HBCGM and mRNA profiling, we identified Lama1 as a genetic modifier of these different TGF-β1–induced pulmonary fibrotic responses. Moreover, we demonstrate that there are strain-specific differences in Lama1 mRNA expression in macrophages and fibroblasts and that Lama1 has unique roles in alternative and fibrogenic/reparative macrophage differentiation, fibroblast proliferation and signaling, and myofibroblast differentiation. Analysis of lung tissue obtained from patients with IPF and COPD indicates that these findings may translate to human pulmonary fibrotic diseases. This is the first study to our knowledge to identify Lama1 as a genetic modifier of pulmonary fibrosis. The discovery that Lama1 plays an important role in TGF-β1–induced pulmonary fibrosis has identified unique mechanisms that underlie the effects of this genetic modifier. It also demonstrates that Lama1 could be an important new biomarker and/or therapeutic target for IPF and potentially other fibrotic disorders.
Strain-dependent susceptibility to developing tissue fibrotic responses have been demonstrated in animal models of tissue fibrosis in the liver, kidney, intestine, skin, and lung (reviewed in ref. 39). Inbred strains are susceptible or resistant to the development of pulmonary fibrosis after bleomycin challenge or adenoviral transfer of active TGF-β1, and it has been shown that bleomycin induced pulmonary fibrosis strongly in C57 strains but weakly in BALB/c mice (39–42). However, the underlying genetic factors and mechanisms that contribute to these phenotype differences have not been fully defined. A prior study demonstrated that the state of cellular activation and inducibility of lysyl oxidase modulate the effects of TGF-β1 and strain-specific injury and repair responses to bleomycin (43). Our studies add to knowledge in this area by demonstrating that Lama1 is also an important modifier of TGF-β1 tissue responses. Additional investigation will be required to define the roles of these genetic regulators, their relationships to each other, and the mechanisms they use to control tissue injury and fibroproliferative repair.
Our current genetic studies have also led to an understanding of the dichotomous role of TGF-β1 in the pathogenesis of fibrosis and emphysema. When we evaluated the degree of alveolar destructive changes by measuring chord length of all 10 strains of mice, we also observed different levels of emphysema depending on the strain (Supplemental Figure 9). Interestingly, we did not observe inverse relationships between fibrosis and emphysema phenotypes in these strains of mice. These studies led us to speculate that independent fibrogenic and emphysematous genetic modifiers determine the final outcome of the tissue phenotypes in the lungs of TGF-β Tg mice. Thus, identification of potential emphysematous genetic modifiers warrants further investigation.
In this study, we have demonstrated that haplotype analysis combined with expression profiling on a Tg model of pulmonary fibrosis could be an effective tool to identify candidate gene(s) regulating pulmonary fibrosis. Since haplotype evaluation generates multiple candidate genes based on strain-dependent tissue phenotypes, it is important to identify reasonable candidate genes that can be validated with additional screening methods. Combining array evaluation on multiple strains of mice with extreme phenotypes as we employed in this study, such as fibrosis-prone versus fibrosis-resistant strains of mice, and haplotype analysis significantly increased the selectivity of the candidate genes. However, our mRNA array evaluation as a method of candidate selection has intrinsic limitations that include the following: (i) the criteria of 2-fold-up- and -downregulated genes in array analysis exclude a number of genes with borderline changes that may have substantial expression alterations; (ii) the method excludes genes other than those involved in transcriptional regulation. In this regard, it is interesting to note that more than half of the top 20 listed genes identified by haplotype analysis (listed in Supplemental Table 1) have been reported to be associated with tissue fibrosis or extracellular matrix remodeling in various organs, including lung, liver, and heart. Among these, integrin α7 (Itga7) was reported as a high-affinity receptor for laminin (laminin111) of which Lama1 is a major component (44). This finding led us to the exciting hypothesis on the Itga7, as a signaling receptor for laminin, mediates enhanced Lama1 regulation of lung fibrosis. Thus, the top-listed genes generated by haplotype analysis warrant investigation in future studies with additional selection methods.
Laminins are heterotrimeric proteins that contain α, β, and γ subunits. As a component of the extracellular matrix, they are known to play an important role in maintaining tissue integrity and cellular differentiation, migration, and adhesion (45). Currently about 16 laminin heterotrimeric complexes are known to be expressed in a tissue-specific manner, and basement membranes are the major site of laminin expression in the adult lung (45–47). In the adult lung, alveolar epithelial cells express mainly laminin α3 and α5, and lung fibroblasts express mostly α3 chains (45–47). Recent studies also reported aberrant expression of α3 subunits in pulmonary fibrosis (48). Lama1 is a major component of the laminin 111 heterotrimeric complex, which is known to be expressed in the basement membranes of various tissues (49). However, Lama1 expression in the lung is mostly restricted to the period of lung morphogenesis and is not expressed in an obvious manner in adult lungs (47). It is believed to play a critical role in lung development, because constitutive null mutations of Lama1 are embryonically lethal (50). However, the expression and contributions of Lama1 to the pathogenesis of pulmonary fibrosis have not been investigated. Our studies add to understanding of the biology of Lama1. They are the first to our knowledge to demonstrate that Lama1 is a modifier of TGF-β1 responses and plays a critical role in TGF-β1–induced pulmonary fibrosis. They also demonstrate that Lama1 is significantly induced in the lungs of TGF-β1 Tg mice with pulmonary fibrosis and lungs from patients with IPF. Interestingly, in both settings, macrophages were the most prominent site of Lama1 expression, and Lama1 was shown to be uniquely able to regulate alternative and fibrotic macrophage activation. Both are known to be critical events in the pathogenesis of fibrotic tissue responses (36, 51–53). Lama1 was also induced by TGF-β1 in fibroblasts, where it played a critical role in fibroblast proliferation and signaling and myofibroblast differentiation. This further clarifies the mechanisms that Lama1 uses in fibrotic tissue responses. Interestingly, the siRNA silencing of Lama1 and a functional missense mutation (Y265C) that blocks the optimal interaction between Lama1 and other molecules (38) caused similar changes in macrophage activation. These data further support the contention that Lama1 plays a critical role in alternative and fibrotic macrophage activation and ultimately fibrotic tissue responses in the lung.
After tissue injury, monocytes and macrophages undergo marked phenotypic and functional changes to play critical roles in the initiation, maintenance, and resolution phases of tissue repair (36). There is also accumulating evidence that different monocyte and macrophage populations play distinct roles in tissue repair, fibrosis, and regeneration and that disturbances in macrophage function can lead to aberrant states that can contribute to a state of persistent injury that can lead to pathological fibrosis (36). A variety of pathways of macrophage differentiation have been described, including classic, alternative, and reparative phenotypes (reviewed in ref. 36). Our studies add to our understanding of macrophage biology by demonstrating that TGF-β1 is a potent stimulator of macrophage Lama1 and that Lama1 does not contribute to classic but is critical for the alternative and fibrotic/reparative macrophage activation observed at sites of injury and repair. The mechanism(s) by which Lama1 mediates these responses will require additional investigation.
In summary, our studies have established a methodology that utilizes Tg, in silico haplotype, and microarray approaches to define genetic modifiers of mediator function and used this approach to demonstrate that Lama1 is a genetic modifier of TGF-β1–induced pulmonary fibrosis. These studies also highlighted important new mechanisms by which the effects of Lama1 are mediated, by demonstrating that Lama1 plays a critical role in alternative and fibrotic macrophage activation, fibroblast proliferation and signaling, and myofibroblast transformation. Importantly, they also demonstrated that Lama1 is expressed in an exaggerated fashion in the lungs and lung macrophages of patients with IPF. When viewed in combination, these studies suggest that Lama1 is a genetic modifier of TGF-β1 responses and could be a biomarker and therapeutic target in pulmonary fibrosis. Additional studies of TGF-β1 genetic modifiers and the biology of Lama1 in fibrosis are warranted.
Methods
Mice.
WT C57, BALB/cJ, C3H/HeJ, CBA/J, 129Si/J, FVB/J, NZW/J, DBA/J, AKR/J, and A/J mice were purchased from the Jackson Laboratory and were housed at Brown University animal facilities. TGF-β1 Tg mice were generated, characterized, and maintained in our laboratory as previously described (34). TGF-β1 Tg mice on 10 different background strains were generated by continuously backcrossing C57 TGF-β1 Tg mice with other WT strains of mice for more than 10 generations. For transgene induction in TGF-β1 Tg mice, 6- to 8-week-old transgene+ and transgene– littermate controls were randomized to normal water or water containing 0.5 mg/ml doxycycline as described previously (34). Chemically induced, viable Lama1 mutant mice on a C57 background (C57BL/6J-Lama1nmf223) (38) were purchased from the Jackson Laboratory and used for macrophage preparation.
Histologic analysis.
The lungs were removed en bloc, inflated at 25-cm pressure with neutral buffered 10% formalin, fixed in 10% formalin, embedded in paraffin, sectioned, and stained. H&E and Mallory trichrome staining were performed in the Molecular Pathology Core at Brown University.
Sircol collagen assay.
Animals were anesthetized, a median sternotomy was performed, and right heart perfusion was completed with calcium- and magnesium-free PBS. The heart and lungs were then removed. Right lungs were frozen in liquid nitrogen and stored at −80°C until they were used. Collagen content was determined by quantifying total soluble collagen using the Sircol collagen assay kit (Biocolor, Accurate Chemical & Scientific Corp.) according to the instructions of the manufacturer.
HBCGM analysis.
Genetic factors were identified using the HBCGM methods previously described (35, 54). In brief, HBCGM utilizes biallelic SNPs that are polymorphic among the analyzed strains. These SNPs usually display a limited degree of variation locally among the strains. Haplotype blocks were then constructed using methods described previously (54). The pattern of genetic variation within each block was correlated with the distribution of trait values among the strains analyzed by using ANOVA-based statistical modeling. P values from the ANOVA model and the corresponding genetic effect size were calculated for each block as previously described (55). The blocks were then ranked by their P values, and those below an input threshold were used as candidate predictions.
mRNA profiling with array analysis.
The mRNA expression profiling on the fibrosis-prone strains (C57, AKR/J, A/J strains) versus fibrosis-resistant strains of mice (C3H/HeJ, 129Si/J, BALB/cJ) was done using an Affymetrix Mouse 1.0 ST array according to the manufacturer’s instructions. Three TGF-β1 Tg mice on each strain together with WT controls were subjected to Affymetrix evaluation, and the data have been filed in the NCBI’s Gene Expression Omnibus database (GEO GSE119662). Normalization was carried out using the robust multiarray average (RMA) method (56) to generate one expression measure for each probe set on each array. For each strain, the fold change in the expression level of each gene between Tg and control mice was evaluated. The average fold change for the 3 fibrosis-prone strains and that for the 3 fibrosis-resistant strains was evaluated. 127 genes that met the specified criteria (average fold change >2 in fibrosis-prone strains and <2 in fibrosis-resistant strains) were identified and intersected with the candidate gene list identified by HBCGM analysis. In this analysis, the statistical significance of the expression difference between Tg mice and controls was not assessed. We acknowledge that this practice could increase the rate of false-positive findings, but we argue that the false-negative rate will be greatly increased by enforcing a stringent statistical significance threshold, which could mistakenly filter out true genetic factors, which would not be recovered in the subsequent analysis. On the other hand, we controlled the false-positive rate by intersecting with the orthogonal information (57), which was the HBCGM analysis result in this case. We further rigorously validated the biological function of the Lama1 gene experimentally. Therefore, although we applied loose candidate selection criteria in one step of the whole analysis, we applied an additional candidate selection process and completed a functional test to ensure the validity of our findings.
Real-time quantitative RT-PCR.
mRNA levels were assessed using real-time quantitative RT-PCR (qRT-PCR) assays. In these assays, total cellular RNA from lungs or other mouse tissues were obtained using TRIzol reagent (Gibco) and RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions.
IHC evaluation.
To localize the expression of Lama1, single- or double-label IHC was undertaken with a modification of procedures described previously by our laboratory (58). First, slides were deparaffinized in xylene, rehydrated in an ethanol gradient, and washed in PBS. To unmask antigens, slides were placed for 20 minutes at high temperature under high pressure in citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6). Tissue sections were then blocked with a non-serum protein-blocking reagent (DakoCytomation Inc.) for 1 hour at room temperature (RT) and incubated with primary antibodies (anti-Lama1, 1:50 dilution, AF-3559, R&D Systems; and anti-CD68, 1:50 dilution, 12829S, Cell Signaling Technology) for 60 minutes at RT in a humid chamber. Substitution of the primary antibody with PBS served as a negative control. The slides were then washed in PBST (0.01 % Tween 20) and incubated with secondary antibodies conjugated with horseradish peroxidase (Cell Signaling Technology). Reaction products were developed using diaminobenzidine (DAB) containing 0.3% hydrogen peroxide up to 5 minutes. Cell nuclei were stained with VECTASHIELD Antifade Mounting Medium with DAPI (50 μl, H-1200, VECTOR Laboratories). Fluorescence was detected by immunofluorescence microscopy.
Immunoblot assay.
50 μg BAL fluids and/or lung lysates were subjected to immunoblot analysis using α-SMA, phosphorylated ERK (pERK), total ERK (TERK), pAkt, and total AKT (TAKT). These samples were gel fractionated, transferred to membranes, and evaluated as described previously by our laboratory (34).
Establish primary lung fibroblasts from C57 and BALB/c mice.
According to the reported protocol (59), ex vivo lung was first chopped into small pieces, then incubated in Liberase Blendzyme solution (grade III, 0.14 Wünsch U/ml, Roche) for 30 minutes under a tube rotator in a 37°C incubator. The suspension was filtered through a 70-μm nylon mesh and was centrifuged at 400 g for 3 minutes. The cell pellet was resuspended into DMEM/F12 media with 15% FBS, 1× antibiotic/antimycotic and plated in a 10-cm2 tissue culture dish at 37°C/5% CO2. Fourteen days after the beginning of the cell isolation, it was changed into EMEM with 15% FBS and 1× penicillin/streptomycin.
Isolation of alveolar macrophages from the lungs of mice.
Mice were sacrificed using intraperitoneal ketamine/xylazine injection, and the trachea was cannulated and perfused with two 0.9-ml aliquots of cold saline. The cellular contents were resuspended into RPMI1640 media with 10% FBS and 1× penicillin/streptomycin and were plated in 10-cm2 tissue flasks at 37°C/5% CO2. After 1 day, floating cells were discarded and media was changed. We identified macrophage population to be greater than 95% through the flow cytometry.
In vivo Lama1 siRNA silencing.
In vivo siRNA silencing targeting Lama1 in the lung was done according to procedures previously described by our laboratory (60, 61). In brief, scrambled siRNA and Lama1-targeted siRNAs were selected and synthesized by Bioneer Inc. and delivered to mice intranasally at a dose of 3 nmol/mouse/d, once a day for 14 days. Six-to 8-week-old TGF-β1 Tg mice and control littermates were randomized to receive either Lama1 siRNA or scrambled control siRNA. Starting on the second day of treatment, both WT and TGF-β1 Tg mice were given water containing 0.5 mg/ml doxycycline for transgene induction, as described previously (34).
In vitro Lama1 siRNA silencing.
Specific siRNA silencing was done according to previously described procedures (61). In brief, NHLFs (Lonza, CC-2512) (0.5 × 106 cells/250 μl) were seeded in each well of a 48-well plate. According to the manufacturer’s protocol for siRNA transfection, 12.5 μl OptiMEM containing 5 μM siRNA was added to 12.5 μl OptiMEM containing 2 μl Lipofectamine 2000 (Invitrogen) after a 5-minute incubation at RT. After gentle mixing, the siRNA-Lipofectamine mixture was incubated at RT for 10 minutes and added to the cells.
Profiling of IPF, COPD, and control lung tissues.
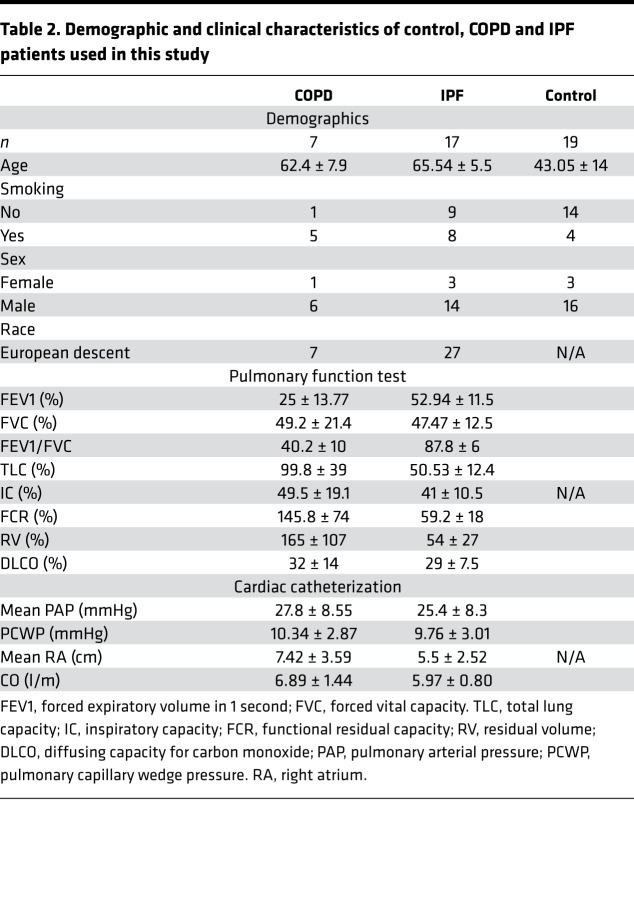
Fresh human lung tissue was obtained from subjects with end-stage lung disease undergoing transplant or donor lungs that were not implanted at the time of transplant. The demographics and clinical characteristics of the groups are shown in Table 2. All protocols were approved by the Partners HealthCare Institutional Review Board. For each subject, small pieces (<5 mm) of whole lung tissue were treated with RNAlater (QIAGEN) and snap frozen in liquid nitrogen prior to storage at –80°C.
Table 2. Demographic and clinical characteristics of control, COPD and IPF patients used in this study.
Information on the primers and antibodies used in this study.
The primer sequences used for qRT-PCR and antibodies used for immunoblot or IHC evaluation are shown in Supplemental Tables 3 and 4, respectively.
Statistics.
Normally distributed data are expressed as mean ± SEM and were assessed for significance by Student’s t test or ANOVA, as appropriate. Data that are not normally distributed were assessed for significance using a Kruskal-Wallis test, followed by a Dunn’s post hoc test for multiple comparisons or Mann-Whitney U test for a 2-group comparison.
Study approval.
Mice were housed and all experimental procedures performed in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited laboratory animal facility at Brown University. This study was approved by the Brown Institutional Animal Care and Use Committee and adhered to the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). All protocols for obtaining and use of human lung tissues were approved by the Partners HealthCare Institutional Review Board, and written informed consent was obtained from research subjects or authorized health care proxies before or at the time of tissue procurement.
Author contributions
CML, CGL, and JAE conceived and designed the study. CML, SJC, WKC, JWP, JHP, IOR, and MZ collected data. CML, AC, IOR, CGL, GP, and JAE performed analysis and interpreted data. GP, CGL, and JAE drafted the manuscript for important intellectual content.
Supplementary Material
Acknowledgments
This work was supported by NIH grants P01 HL114501, UH2 HL123876 (JAE), and R01-HL115813 (CGL), U01 DA044399 (GP).
Version 1. 09/20/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(18):e99574. https://doi.org/10.1172/jci.insight.99574.
Contributor Information
Chang-Min Lee, Email: chang-min_lee@brown.edu.
Soo Jung Cho, Email: sjc9006@med.cornell.edu.
Won-Kyung Cho, Email: wonkyungcho@amc.seoul.kr.
Jin Wook Park, Email: jinwook_park@brown.edu.
Jae-Hyun Lee, Email: jhleemd@yuhs.ac.
Ivan O. Rosas, Email: irosas@rics.bwh.harvard.edu.
Ming Zheng, Email: mzheng@stanford.edu.
Gary Peltz, Email: gpeltz@stanford.edu.
Chun Geun Lee, Email: chun_lee@brown.edu.
Jack A. Elias, Email: jack_elias@brown.edu.
References
- 1.Kaur A, Mathai SK, Schwartz DA. Genetics in idiopathic pulmonary fibrosis pathogenesis, prognosis, and treatment. Front Med (Lausanne) 2017;4:154. doi: 10.3389/fmed.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathai SK, Schwartz DA, Warg LA. Genetic susceptibility and pulmonary fibrosis. Curr Opin Pulm Med. 2014;20(5):429–435. doi: 10.1097/MCP.0000000000000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pezzolesi MG, Krolewski AS. The genetic risk of kidney disease in type 2 diabetes. Med Clin North Am. 2013;97(1):91–107. doi: 10.1016/j.mcna.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raghu G. Interstitial lung disease: a clinical overview and general approach. In: Fishman AP, Elias JA, Fishman JA, Grippi MA, R. KL, Senior RM, eds. Fishman’s Pulmonary Diseases and Disorders. New York, New York, USA: McGraw Hill Inc.; 1998:1037–1053. [Google Scholar]
- 5.Krein PM, Winston BW. Roles for insulin-like growth factor I and transforming growth factor-beta in fibrotic lung disease. Chest. 2002;122(6 suppl):289S–293S. doi: 10.1378/chest.122.6_suppl.289s. [DOI] [PubMed] [Google Scholar]
- 6.Selman M, King TE, Pardo A, American Thoracic Society, European Respiratory Society, American College of Chest Physicians Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med. 2001;134(2):136–151. doi: 10.7326/0003-4819-134-2-200101160-00015. [DOI] [PubMed] [Google Scholar]
- 7.Ling E, Robinson DS. Transforming growth factor-beta1: its anti-inflammatory and pro-fibrotic effects. Clin Exp Allergy. 2002;32(2):175–178. doi: 10.1046/j.1365-2222.2002.01287.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee CG, et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1) J Exp Med. 2001;194(6):809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M, Lefaix J, Delanian S. TGF-beta1 and radiation fibrosis: a master switch and a specific therapeutic target? Int J Radiat Oncol Biol Phys. 2000;47(2):277–290. doi: 10.1016/S0360-3016(00)00435-1. [DOI] [PubMed] [Google Scholar]
- 10.Nakao A, et al. Transient gene transfer and expression of Smad7 prevents bleomycin-induced lung fibrosis in mice. J Clin Invest. 1999;104(1):5–11. doi: 10.1172/JCI6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yehualaeshet T, O’Connor R, Begleiter A, Murphy-Ullrich JE, Silverstein R, Khalil N. A CD36 synthetic peptide inhibits bleomycin-induced pulmonary inflammation and connective tissue synthesis in the rat. Am J Respir Cell Mol Biol. 2000;23(2):204–212. doi: 10.1165/ajrcmb.23.2.4089. [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R, Sukhatme VP. Fibrosis and angiogenesis. Curr Opin Nephrol Hypertens. 2000;9(4):413–418. doi: 10.1097/00041552-200007000-00013. [DOI] [PubMed] [Google Scholar]
- 13.Khalil N, O’Connor RN, Flanders KC, Unruh H. TGF-beta 1, but not TGF-beta 2 or TGF-beta 3, is differentially present in epithelial cells of advanced pulmonary fibrosis: an immunohistochemical study. Am J Respir Cell Mol Biol. 1996;14(2):131–138. doi: 10.1165/ajrcmb.14.2.8630262. [DOI] [PubMed] [Google Scholar]
- 14.Khalil N, et al. Regulation of the effects of TGF-beta 1 by activation of latent TGF-beta 1 and differential expression of TGF-beta receptors (T beta R-I and T beta R-II) in idiopathic pulmonary fibrosis. Thorax. 2001;56(12):907–915. doi: 10.1136/thorax.56.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu YD, Hua J, Mui A, O’Connor R, Grotendorst G, Khalil N. Release of biologically active TGF-beta1 by alveolar epithelial cells results in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2003;285(3):L527–L39. doi: 10.1152/ajplung.00298.2002. [DOI] [PubMed] [Google Scholar]
- 16.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest. 1997;100(4):768–776. doi: 10.1172/JCI119590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly M, Kolb M, Bonniaud P, Gauldie J. Re-evaluation of fibrogenic cytokines in lung fibrosis. Curr Pharm Des. 2003;9(1):39–49. doi: 10.2174/1381612033392341. [DOI] [PubMed] [Google Scholar]
- 18.Kuwano K, et al. Increased circulating levels of soluble Fas ligand are correlated with disease activity in patients with fibrosing lung diseases. Respirology. 2002;7(1):15–21. doi: 10.1046/j.1440-1843.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- 19.Kuwano K, et al. The involvement of Fas-Fas ligand pathway in fibrosing lung diseases. Am J Respir Cell Mol Biol. 1999;20(1):53–60. doi: 10.1165/ajrcmb.20.1.2941. [DOI] [PubMed] [Google Scholar]
- 20.Kuwano K, et al. P21Waf1/Cip1/Sdi1 and p53 expression in association with DNA strand breaks in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1996;154(2 Pt 1):477–483. doi: 10.1164/ajrccm.154.2.8756825. [DOI] [PubMed] [Google Scholar]
- 21.Kuwano K, et al. Essential roles of the Fas-Fas ligand pathway in the development of pulmonary fibrosis. J Clin Invest. 1999;104(1):13–19. doi: 10.1172/JCI5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke DL, Murray LA, Crestani B, Sleeman MA. Is personalised medicine the key to heterogeneity in idiopathic pulmonary fibrosis? Pharmacol Ther. 2017;169:35–46. doi: 10.1016/j.pharmthera.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Flanders KC, et al. Mice lacking Smad3 are protected against cutaneous injury induced by ionizing radiation. Am J Pathol. 2002;160(3):1057–1068. doi: 10.1016/S0002-9440(10)64926-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akhurst RJ, Derynck R. TGF-beta signaling in cancer — a double-edged sword. Trends Cell Biol. 2001;11(11):S44–S51. doi: 10.1016/S0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 25.Roberts AB, Piek E, Böttinger EP, Ashcroft G, Mitchell JB, Flanders KC. Is Smad3 a major player in signal transduction pathways leading to fibrogenesis? Chest. 2001;120(1 Suppl):43S–47S. doi: 10.1378/chest.120.1_suppl.s43-a. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda H, et al. Negative effect of transforming growth factor-beta-1 on intestinal anastomotic tissue regeneration. Eur Surg Res. 2001;33(5-6):388–394. doi: 10.1159/000049735. [DOI] [PubMed] [Google Scholar]
- 27.Chan T, et al. Development, characterization, and wound healing of the keratin 14 promoted transforming growth factor-beta1 transgenic mouse. Wound Repair Regen. 2002;10(3):177–187. doi: 10.1046/j.1524-475X.2002.11101.x. [DOI] [PubMed] [Google Scholar]
- 28.Amendt C, Mann A, Schirmacher P, Blessing M. Resistance of keratinocytes to TGFbeta-mediated growth restriction and apoptosis induction accelerates re-epithelialization in skin wounds. J Cell Sci. 2002;115(Pt 10):2189–2198. doi: 10.1242/jcs.115.10.2189. [DOI] [PubMed] [Google Scholar]
- 29.Fargeas C, Wu CY, Nakajima T, Cox D, Nutman T, Delespesse G. Differential effect of transforming growth factor beta on the synthesis of Th1- and Th2-like lymphokines by human T lymphocytes. Eur J Immunol. 1992;22(8):2173–2176. doi: 10.1002/eji.1830220833. [DOI] [PubMed] [Google Scholar]
- 30.Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annu Rev Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Giangreco L, Broome HE, Dargan CM, Swain SL. Control of CD4 effector fate: transforming growth factor beta 1 and interleukin 2 synergize to prevent apoptosis and promote effector expansion. J Exp Med. 1995;182(3):699–709. doi: 10.1084/jem.182.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, et al. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax. 2004;59(2):126–129. doi: 10.1136/thorax.2003.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak JC, et al. Elevated plasma TGF-beta1 levels in patients with chronic obstructive pulmonary disease. Respir Med. 2009;103(7):1083–1089. doi: 10.1016/j.rmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Lee CG, et al. Early growth response gene 1-mediated apoptosis is essential for transforming growth factor beta1-induced pulmonary fibrosis. J Exp Med. 2004;200(3):377–389. doi: 10.1084/jem.20040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng M, et al. The role of Abcb5 alleles in susceptibility to haloperidol-induced toxicity in mice and humans. PLoS Med. 2015;12(2):e1001782. doi: 10.1371/journal.pmed.1001782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44(3):450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong D, Shi W, Yi SJ, Chen H, Groffen J, Heisterkamp N. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 2012;13:31. doi: 10.1186/1471-2172-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards MM, et al. Mutations in Lama1 disrupt retinal vascular development and inner limiting membrane formation. J Biol Chem. 2010;285(10):7697–7711. doi: 10.1074/jbc.M109.069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walkin L, et al. The role of mouse strain differences in the susceptibility to fibrosis: a systematic review. Fibrogenesis Tissue Repair. 2013;6(1):18. doi: 10.1186/1755-1536-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barth RK, Hanchett LA, Baecher-Allan CM. Mapping susceptibility genes for the induction of pulmonary fibrosis in mice. Chest. 2002;121(3 suppl):21S. doi: 10.1378/chest.121.3_suppl.21s. [DOI] [PubMed] [Google Scholar]
- 41.Haston CK, Amos CI, King TM, Travis EL. Inheritance of susceptibility to bleomycin-induced pulmonary fibrosis in the mouse. Cancer Res. 1996;56(11):2596–2601. [PubMed] [Google Scholar]
- 42.Kolb M, et al. Differences in the fibrogenic response after transfer of active transforming growth factor-beta1 gene to lungs of “fibrosis-prone” and “fibrosis-resistant” mouse strains. Am J Respir Cell Mol Biol. 2002;27(2):141–150. doi: 10.1165/ajrcmb.27.2.4674. [DOI] [PubMed] [Google Scholar]
- 43.Cheng T, et al. Lysyl oxidase promotes bleomycin-induced lung fibrosis through modulating inflammation. J Mol Cell Biol. 2014;6(6):506–515. doi: 10.1093/jmcb/mju039. [DOI] [PubMed] [Google Scholar]
- 44.Nishiuchi R, et al. Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin-integrin interactions using recombinant alpha3beta1, alpha6beta1, alpha7beta1 and alpha6beta4 integrins. Matrix Biol. 2006;25(3):189–197. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Miner JH. Laminins and their roles in mammals. Microsc Res Tech. 2008;71(5):349–356. doi: 10.1002/jemt.20563. [DOI] [PubMed] [Google Scholar]
- 46.Pierce RA, et al. Expression of laminin alpha3, alpha4, and alpha5 chains by alveolar epithelial cells and fibroblasts. Am J Respir Cell Mol Biol. 1998;19(2):237–244. doi: 10.1165/ajrcmb.19.2.3087. [DOI] [PubMed] [Google Scholar]
- 47.Pierce RA, Griffin GL, Miner JH, Senior RM. Expression patterns of laminin alpha1 and alpha5 in human lung during development. Am J Respir Cell Mol Biol. 2000;23(6):742–747. doi: 10.1165/ajrcmb.23.6.4202. [DOI] [PubMed] [Google Scholar]
- 48.Morales-Nebreda LI, et al. Lung-specific loss of α3 laminin worsens bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol. 2015;52(4):503–512. doi: 10.1165/rcmb.2014-0057OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen NM, Senior RM. Laminin isoforms and lung development: all isoforms are not equal. Dev Biol. 2006;294(2):271–279. doi: 10.1016/j.ydbio.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 50.Alpy F, et al. Generation of a conditionally null allele of the laminin alpha1 gene. Genesis. 2005;43(2):59–70. doi: 10.1002/gene.20154. [DOI] [PubMed] [Google Scholar]
- 51.Wermuth PJ, Jimenez SA. The significance of macrophage polarization subtypes for animal models of tissue fibrosis and human fibrotic diseases. Clin Transl Med. 2015;4:2. doi: 10.1186/s40169-015-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schupp JC, et al. Macrophage activation in acute exacerbation of idiopathic pulmonary fibrosis. PLoS ONE. 2015;10(1):e0116775. doi: 10.1371/journal.pone.0116775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rőszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peltz G, et al. Next-generation computational genetic analysis: multiple complement alleles control survival after Candida albicans infection. Infect Immun. 2011;79(11):4472–4479. doi: 10.1128/IAI.05666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao G, et al. In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science. 2004;306(5696):690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- 56.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng M, Shafer S, Liao G, Liu HH, Peltz G. Computational genetic mapping in mice: the ship has sailed. Sci Transl Med. 2009;1(3):3ps4. doi: 10.1126/scitranslmed.3000377. [DOI] [PubMed] [Google Scholar]
- 58.Lee CG, et al. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced tissue responses and apoptosis. J Exp Med. 2009;206(5):1149–1166. doi: 10.1084/jem.20081271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seluanov A, Vaidya A, Gorbunova V. Establishing primary adult fibroblast cultures from rodents. J Vis Exp. 2010;(44):2033. doi: 10.3791/2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, et al. Amphiregulin, an epidermal growth factor receptor ligand, plays an essential role in the pathogenesis of transforming growth factor-β-induced pulmonary fibrosis. J Biol Chem. 2012;287(50):41991–42000. doi: 10.1074/jbc.M112.356824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon PO, et al. Self-assembled micelle interfering rna for effective and safe targeting of dysregulated genes in pulmonary fibrosis. J Biol Chem. 2016;291(12):6433–6446. doi: 10.1074/jbc.M115.693671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.