Abstract
Zebrafish are a powerful tool for studying muscle function owing to their high numbers of offspring, low maintenance costs, evolutionarily conserved muscle functions, and the ability to rapidly take up small molecular compounds during early larval stages. Fukutin-related protein (FKRP) is a putative protein glycosyltransferase that functions in the Golgi apparatus to modify sugar chain molecules of newly translated proteins. Patients with mutations in the FKRP gene can have a wide spectrum of clinical symptoms with varying muscle, eye, and brain pathologies depending on the location of the mutation in the FKRP protein. Patients with a common L276I FKRP mutation have mild adult-onset muscle degeneration known as limb-girdle muscular dystrophy 2I (LGMD2I), whereas patients with more C-terminal pathogenic mutations develop the severe Walker-Warburg syndrome (WWS)/muscle-eye-brain (MEB) disease. We generated fkrp-mutant zebrafish that phenocopy WWS/MEB pathologies including severe muscle breakdowns, head malformations, and early lethality. We have also generated a milder LGMD2I-model zebrafish via overexpression of a heat shock–inducible human FKRP (L276I) transgene that shows milder muscle pathology. Screening of an FDA-approved drug compound library in the LGMD2I zebrafish revealed a strong propensity towards steroids, antibacterials, and calcium regulators in ameliorating FKRP-dependent pathologies. Together, these studies demonstrate the utility of the zebrafish to both study human-specific FKRP mutations and perform compound library screenings for corrective drug compounds to treat muscular dystrophies.
Keywords: Muscle Biology
Keywords: Drug screens
Zebrafish deficient in fukutin-related protein, a model of dystroglycanopathies, was used to screen a drug library and identified corrective compounds.
Introduction
Skeletal muscle is a dynamic organ that is capable of regeneration and repair following injury due to a resident pool of muscle progenitor cells referred to as muscle satellite cells. Skeletal muscle myofibers consist of several individual multinucleated syncytia capable of transmitting intracellular force to the extracellular matrix (ECM) via a network of filamentous actin, dystrophin, and the dystrophin-associated protein complex (DAPC) (1, 2). Pathogenic loss-of-function mutations in the DAPC can cause different forms of muscular dystrophy with varying phenotypes and clinical outcomes (3, 4). One essential component of the DAPC is dystroglycan (DAG1), a membrane-associated muscle protein that links dystrophin to the laminin subunits of the ECM (5). Dystroglycan protein consists of separate α- and β-subunits that are spliced from a single poly-mRNA (6). The translated dystroglycan protein is then modified via a network of glycosyltransferase proteins resulting in the addition of O-mannose carbohydrate moieties that are essential for the proper trafficking of the α-dystroglycan subunit to the myofiber membrane (7). Human pathogenic variants in the dystroglycan (DAG1) gene or the glycosyltransferase proteins that regulate dystroglycan glycosylation result in the failure of the dystroglycan protein to traffic to the myofiber membrane. These disorders form a subset of the muscular dystrophies referred to as the dystroglycanopathies (8, 9). A specific pathogenic variant in the FKRP gene, L276I (c.826C>A), results in the improper processing of α-dystroglycan protein and leads to skeletal myofiber breakdown that leads to limb muscle weakness during the teen years that worsens over time (10, 11). In addition, cell-based evidence shows that limb-girdle muscular dystrophy 2I (LGMD2I) patients with the L276I variant fail to properly glycosylate α-dystroglycan (α-DAG1) resulting in the protein being sequestered in the Golgi apparatus (12). Patients with more C-terminal pathogenic mutations in FKRP tend to have more severe muscle, brain, retinal, and cardiac pathologies and are diagnosed as having Walker-Warburg syndrome (WWS)/muscle-eye-brain (MEB) disease (13, 14). In addition to having severely reduced levels of glycosylated α-dystroglycan protein, these patients have decreased levels of laminin matrix proteins that may explain some of the severe retinal and brain pathologies (15). While the genetic causes of both LGMD2I and WWS/MEB have been identified, the consequential disparate clinical pathologies between the 2 disorders with the same causative gene remain poorly understood (16, 17).
Zebrafish are excellent vertebrate models for studying muscle formation and human-based muscle diseases owing to their ex vivo development, the ability to manipulate their gene expression, and strong evolutionarily conserved muscle gene function (18). Zebrafish carrying mutations in dystrophin, the DAPC, or other dystroglycanopathy-related factors are extremely useful for determining the stages of muscle disease progression and what drugs might prevent or block the onset of symptoms. Morpholino and in situ hybridization (ISH) studies of FKRP and other dystroglycanopathy genes have revealed important evolutionarily conserved glycosyltransferase functional roles for these factors in early muscle formation and development (19). FKRP zebrafish morphants were shown to have a wide range of defective muscle and brain phenotypes dependent on the degree of FKRP mRNA knockdown in each embryo (20). Additional studies have shown that FKRP morphants have significant vasculature defects and an upregulation of RNA transcripts involved in the unfolded protein response (UPR) pathway (21, 22). These findings suggest that FKRP may play additional tissue-specific roles independent of its function in the regulation of the glycosylation modifications of the α-dystroglycan protein.
Previously, our laboratory demonstrated that knockdown of zebrafish FKRP mRNA resulted in a spectrum of pathologies ranging from mild (slight tail curvature) to severe (extreme myofiber disruption, head malformation, early lethality) dependent on the efficiency of the FKRP mRNA inhibition (23). Interestingly, mRNA overexpression rescue experiments in the FKRP-morphant zebrafish revealed that overexpression of the human FKRP L276I mRNA resulted in a significantly milder phenotype, particularly in the skeletal muscles, compared with the overexpression of other more severe WWS/MEB–like human FKRP mutations (A455D and C318Y) (23). These results suggested that human FKRP mRNAs harboring patient-specific pathogenic variants when overexpressed may phenocopy the FKRP-dependent dystroglycanopathies in an FKRP-deficient zebrafish background. We decided to generate an LGMD2I model in zebrafish by first generating FKRP-deficient zebrafish using a targeted-TALEN gene editing strategy. FKRP-mutant zebrafish develop severe defects in their dorsal muscle, brain, eyes, and show pericardiac edema resulting in early lethality by 7 days post fertilization (dpf). We generated transgenic zebrafish lines under the control of a heat shock–inducible, ubiquitous promoter element that drove expression of either the human FKRP wild-type open reading frame (ORF) or a separate transgenic line that carried the FKRP (L276I) ORF. Inducing overexpression of the FKRP L276I transgene on the FKRP mutant background resulted in a mild overall pathology (LGMD2I model) compared with the complete loss of FKRP protein (e.g., severe WWS/MEB–like model). Screening of the Prestwick Chemical Library (version 2) in our LGMD2I zebrafish embryos revealed several compounds that were capable of blocking or ameliorating LGMD2I-like pathologies in our zebrafish model. By generating human patient–specific pathogenic variants on an FKRP-deficient background, we have tested a model of LGMD2I in zebrafish that can be used as a paradigm for other muscular dystrophies to identify personalized therapeutic drug compounds.
Results
Generation of FKRP-mutant zebrafish using TALENs.
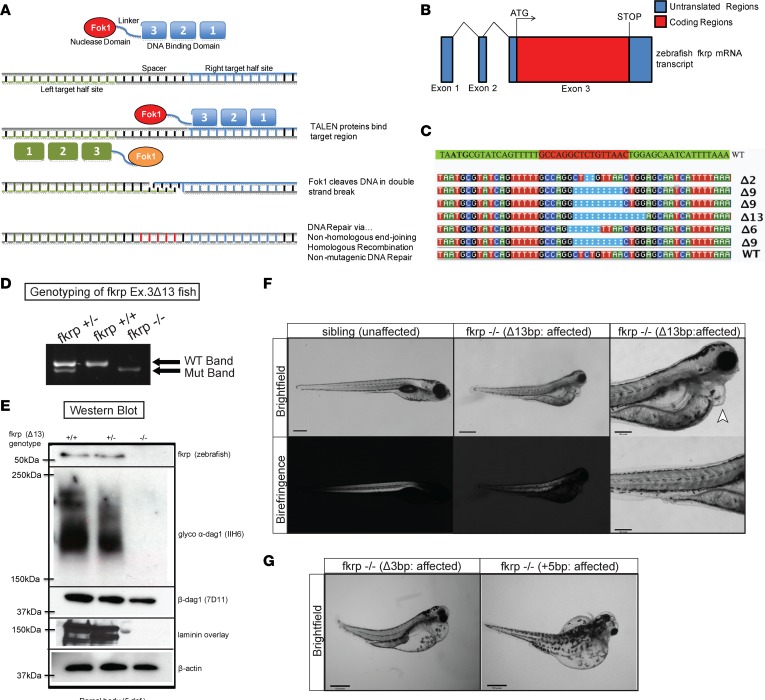
The FKRP protein is highly conserved among zebrafish, mouse, and human FKRP amino acid sequences, and they are all composed of a single coding exon (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120493DS1). To study the functional role of FKRP in zebrafish, we designed 2 TALEN constructs to target exon 3 of the zebrafish fkrp gene immediately following the ATG start codon (Figure 1, A and B). More than 300 one-cell zebrafish embryos were injected with the TALEN mRNAs and FokI nuclease and raised to adulthood as the F0 generation. We generated several different fkrp-targeted zebrafish lines that contained various deletions, insertions, and deletions plus insertions in various combinations (Figure 1C). We chose for further characterization a mutant FKRP line containing a 13-bp deletion (Δ13) with no additional DNA insertions that resulted in a frame-shift loss-of-function allele due to the ability of the mutant allele to be easily visualized via a standard PCR with agarose gel electrophoresis (Figure 1D). Sanger sequencing of the predicted top 10 homologous sequences in the zebrafish genome showed no additional mutations, indicating a high degree of specificity without off-target effects in the FKRP-mutant zebrafish (data not shown). The fkrp homozygous mutant zebrafish failed to survive past 10 dpf due to a combination of phenotypic defects likely resulting in an overall failure to thrive. We performed an immunoblot against zebrafish FKRP in the homozygous fkrp mutants, which demonstrated that no FKRP protein was produced (Figure 1E). We also noticed strongly reduced expression levels of glycosylated α-dystroglycan protein using monoclonal antibody IIH6C4 (Figure 1E). These results are consistent with previous findings in our FKRP-morphant zebrafish and in the Fkrp (P448L) severe mouse model (23, 24). Likewise, we observed significant decreases in overall laminin expression (Figure 1E), a hallmark of FKRP-dependent dystroglycanopathies, as also observed in whole-brain protein lysates of Fkrp Y307N–mutant mice (15). The fkrp Δ13 mutant line (henceforth referred to as fkrp–/– or fkrp mutant) had a complete phenocopy of the severe FKRP morphants with defects in their muscles, eyes, brain, and pericardiac edema (Figure 1F). Two additional fkrp-mutant lines (one harboring a 3-bp deletion, and one with a 5-bp insertion) were also characterized and shown to have the same pathological phenotype as the fkrp–/– line (Figure 1G). These results demonstrate an important early requirement for FKRP expression in normal muscle, eye, brain, and overall developmental growth in the zebrafish.
Figure 1. Generation of FKRP-mutant zebrafish using TALENs.
(A) Schematic showing TALEN-mediated genomic targeting of the zebrafish fkrp locus. (B) Diagram of coding and noncoding exons of the zebrafish fkrp transcribed mRNA. Exon 3 is the only coding exon of the zebrafish fkrp gene. (C) Various FKRP-mutant zebrafish lines generated using TALEN-mediated genomic engineering. (D) PCR genotyping of zebrafish tail biopsies showing the 13-bp deletion at exon 3 of the zebrafish fkrp gene locus showing fkrp WT, heterozygous, and homozygous mutant zebrafish. (E) Western blot images of protein lysates of fkrp WT, heterozygous, and homozygous mutant zebrafish at 5 dpf. Immunoblots using antibodies against zebrafish FKRP, glycosylated α-dystroglycan (α-dag1), β-dystroglycan (β-dag1), laminin, and β-actin. (F) Brightfield and birefringence images of sibling (unaffected) and fkrp–/– (Δ13bp; affected) zebrafish at 5 dpf at low (×2) and high magnification (×10). The arrowhead indicates pericardiac edema in the fkrp–/– (Δ13bp) zebrafish. (G) Two additional fkrp-mutant lines, a 3-bp deletion (Δ3bp) deletion and a 5-bp insertion (+5bp), were also generated and showed a similar phenotype to the fkrp–/– Δ13bp homozygous mutants. Scale bars: 300 μm.
FKRP-mutant zebrafish have impaired locomotion, movement, and physiological force.
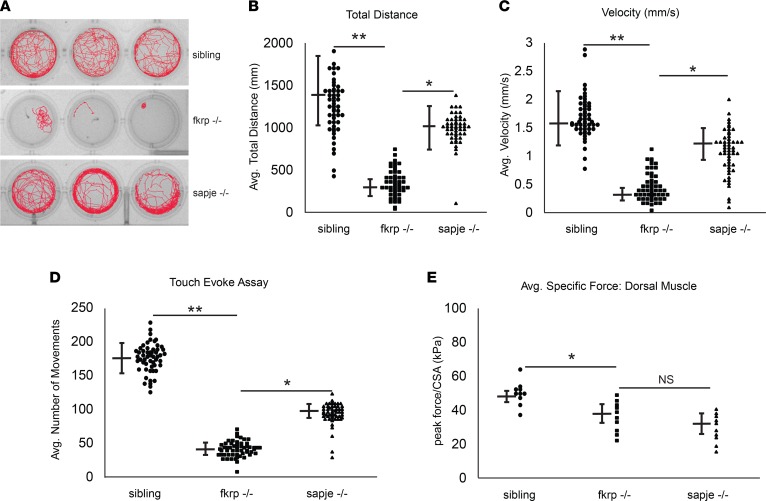
Zebrafish motion-tracking platforms and analyses of swimming behaviors are a powerful tool to determine the overall locomotive capabilities in a given time frame under nonstimulatory conditions (25). Previously, these platforms have been able to show significant decreases in overall larval motility in dystrophin-deficient (sapje) zebrafish when compared with sibling controls (26). We next tested the overall locomotion of the fkrp–/– zebrafish at 5 dpf as compared with wild-type and heterozygous sibling control fish using the DanioVision locomotive tracking platform. The fkrp–/– zebrafish larvae had overall impaired movement, remaining either motionless at the bottom of the well for the entire period of tracking time or swimming in circles in the well (Figure 2A). In comparison, the dystrophic sapje zebrafish moved significantly more than the fkrp–/– fish, although their overall distance traveled and velocity were reduced compared with control sibling zebrafish (Figure 2, B and C). The touch-evoke response assay is another highly reproducible assay to access the overall abilities of a zebrafish larva to move their dorsal muscle in response to a physical stimulus (27, 28). Touch-evoke analyses of the fkrp–/– zebrafish revealed an overall decrease in response and movement compared with sibling wild-type and heterozygous controls (Figure 2D). Next, we measured the dorsal muscle–specific force by attaching the zebrafish dorsal muscle to a force transducer using a previously established protocol (29). When stimulated to contract, the fkrp–/– zebrafish had significantly reduced dorsal muscle force per preparation cross-sectional area compared with sibling controls, but an amount of peak force reduction equivalent to that of the sapje-mutant zebrafish. (Figure 2E). Together, these studies demonstrate a requirement of FKRP expression for normal zebrafish larvae locomotion and dorsal muscle function during early developmental stages.
Figure 2. FKRP-mutant zebrafish have impaired locomotion and movement.
(A) Tracings of motion pathways of sibling (FKRP WT and heterozygotes), fkrp–/– (homozygotes), and sapje–/– (dystrophin) mutant zebrafish cohorts. (B) The average total distance traveled over the 15-minute tracking period for each individual zebrafish. (C) The average velocity (mm/s) of individual sibling, fkrp–/–, and sapje–/– zebrafish cohorts over the 15-minute tracking interval. Fifty zebrafish (n = 50) were tracked over 15 minutes in 5 separate experiments (n = 5 experimental replicates). (D) Average number of touch movements in individual sibling, fkrp–/–, and sapje–/– zebrafish cohorts 1 dpf in response to a touch stimulus. (E) Average peak force (in kilopascals [kPa]) in the dorsal skeletal muscles of individual sibling, fkrp–/–, and sapje–/– zebrafish cohorts 5 dpf normalized to cross-sectional area (CSA). Ten zebrafish (n = 10) were used for the physiological force measurements. *P < 0.05; **P < 0.005 by 2-tailed Student’s t test. NS, not significant.
FKRP-mutant zebrafish have significantly impaired vascularization and increased UPR transcripts.
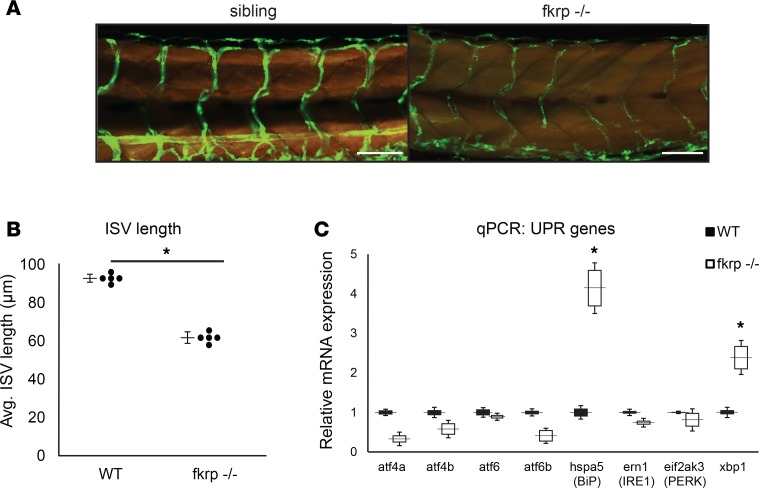
One reported finding in FKRP-morphant zebrafish was the impaired vascularization of their dorsal muscles, likely due to a dystroglycan-independent mechanism (21). We mated the fkrp–/– zebrafish onto both the Tg(actc1b:RFP) (labels skeletal myofibers red) and the flk1-GFP (labels vasculature) transgenic reporter lines to determine the overall vascularization of our fkrp–/– fish. Results indicated significantly reduced intersegmental vessel lengths and overall branching compared with sibling controls, indicating that FKRP protein may play an important role in early vascularization of the dorsal skeletal muscles of developing zebrafish (Figure 3, A and B). Another aspect of the FKRP-morphant zebrafish was an upregulation of expression of genes involved in the UPR (22). Activation of endoplasmic reticulum (ER) stress pathways and the UPR may be a general mechanism for skeletal muscle remodeling and disease processes in human myopathies (30). We detected an increase in the UPR mRNA expression levels of xbp1 and hspa5 (BiP) mRNA via quantitative real-time PCR (qPCR) analyses in the fkrp–/– zebrafish compared with wild-type controls (Figure 3C). Interestingly, we also observed a decrease in mRNA expression of several glycosyltransferases that modulate glycosylation of α-DAG1 and other target proteins (Supplemental Figure 2). Together, these findings may explain the diverse functional roles for FKRP expression in multiple tissues, and some of the phenotypic variability observed in LGMD2I patients when compared with patients having other dystroglycanopathy mutations (9, 20, 31).
Figure 3. FKRP-mutant zebrafish have significantly impaired vascularization and increased unfolded protein response (UPR) transcripts.
(A) Reduced vasculature in the fkrp–/– zebrafish compared with sibling controls. The Tg(actc1b:RFP) (labels myofibers red) and flk1-GFP (labels vasculature green) transgenic alleles were bred onto the fkrp-mutant zebrafish strain background. Scale bars: 100 μm. (B) Intersegmental vessel length (ISV) is quantified in the fkrp–/– zebrafish compared to sibling controls. n = 5 zebrafish per cohort. (C) Real-time quantitative PCR of expression levels of UPR genes (atf4a, atf4b, atf6, atf6b, hspa5/BiP, ern1/IRE1, eif2ak3/PERK, xbp1) were measured and normalized to β-actin as housekeeping control. For all experiments, 5 zebrafish (n = 5) per cohort were used. *P < 0.05 by 2-tailed Student’s t test.
Overexpression of a transgenic human FKRP L276I mutant protein induces pathogenic phenotype in FKRP-deficient zebrafish.
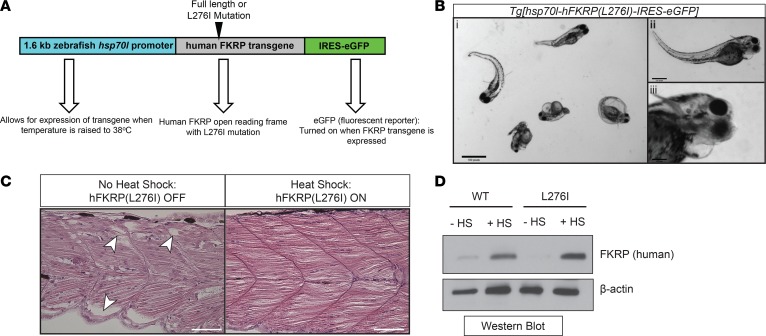
Patients with LGMD2I often contain a common L-to-I mutation at amino acid position 276 of the FKRP protein resulting in abnormal protein function (32). Zebrafish, unlike humans, do not have the leucine amino acid at position 276; however, overexpression of human FKRP mRNA containing the L276I mutation results in abnormal zebrafish pathologies, including muscle, eye, and head defects (23). Initially, we tested several noninducible constitutively overexpressing (both ubiquitous and tissue-specific) FKRP transgenic constructs; however, many of them showed toxicity (data not shown), in agreement with findings using adeno-associated viral (AAV) overexpression of FKRP in mice (33). To circumvent this problem, we generated a heat shock–inducible zebrafish that overexpresses the human FKRP (L276I mutation) transgene in a temporal manner. A control Tg[hsp70l-hFKRP(full-length/WT)-IRES-eGFP] transgenic line overexpressing full-length (WT) human FKRP protein was tested in parallel zebrafish embryo injections (Figure 4A). The Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] zebrafish were pulsed for 1 hour with heat shock at 38°C at 1 dpf, and analyzed at 2, 3, 4, and 5 dpf. Starting at 4 dpf, the Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] zebrafish developed disorganized myofibers, accompanied by eye and head malformations (Figure 4, B and C). Several Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] zebrafish developed pericardial edema, a likely secondary effect due to the disruption of the myofibers (34). The heat-shock induction of Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] showed a robust expression of human FKRP L276I protein compared with noninduced and nontransgenic controls, and overexpression similar to that of the Tg[hsp70l-hFKRP(full-length/WT)-IRES-eGFP] transgenic line (Figure 4D). Additionally, the overexpression of the FKRP L276I transgene resulted in a restoration of the overall muscle architecture, with only mild perturbations in skeletal muscle structure (Figure 4C). These results demonstrate the utility of overexpressing the human FKRP L276I pathogenic variant in the FKRP-deficient zebrafish to recapitulate the human LGMD2I phenotypes.
Figure 4. Generation of a heat shock–inducible LGMD2I-model zebrafish.
(A) Schematic of the LGMD2I-model zebrafish. A heat shock–inducible zebrafish promoter (1.6-kb zebrafish hsp70l) drives expression of either human WT FKRP (full length) or L276I open reading frame (ORF). An IRES-eGFP fluorescent reporter protein is cotranscribed upon activation of the promoter using heat shock (raising the temperature of the embryos from 28.5°C to 38°C. (B) Brightfield image of heat shock–induced Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] transgenic zebrafish showing LGMD2I pathology. These symptoms include muscle weakness resulting in tail bending (i), head and eye malformations (ii), and pericardiac edema (iii). Scale bar: 300 μm. (C) H&E histochemical staining of dorsal muscle from noninduced (no FKRP transgene expression) and induced (expression of human FKRP L276I transgene). Scale bar: 100 μm. (D) Western blot images of overexpressed (with or without heat shock [HS]) of human FKRP transgenic protein and a β-actin loading control from Tg[hsp70l-hFKRP(WT)-IRES-eGFP] and Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] transgenic zebrafish.
General muscle restorative compounds can extend the lifespan and reduce pathology in the LGMD2I zebrafish.
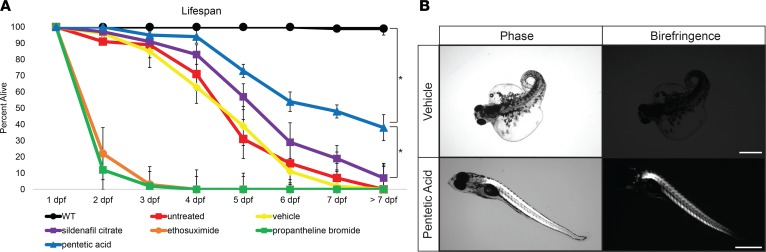
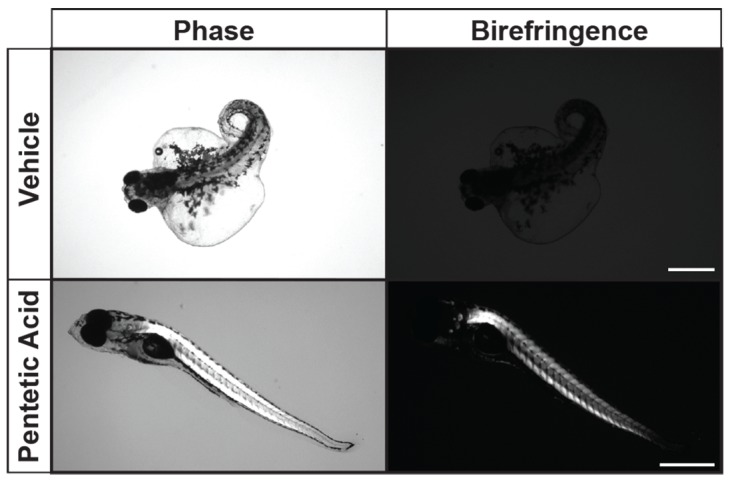
Drug library screening in zebrafish models of muscular dystrophies has proved to be a fruitful technique in identifying therapeutic compounds that can ameliorate or block dystrophic muscle pathology (18, 35, 36). Several restorative compounds have been shown to block dystrophic muscle pathology or ameliorate the muscle-wasting symptoms in both zebrafish and mouse Duchenne muscular dystrophy (DMD) models (37–39). Some of these compounds work through general mechanisms independent of the muscle mutation, such as blocking muscle ischemia, preventing inflammation, or restoring calcium homeostasis. We selectively tested in our LGMD2I zebrafish model 5 compounds (sildenafil citrate, pentetic acid, ethosuximide, aminophylline, and propantheline bromide) shown to improve dystrophic muscle pathology in sapje-mutant zebrafish and other dystrophic mutant fish through a non–mutation-specific mechanism (38). Of the compounds tested, aminophylline, propantheline bromide, and pentetic acid all showed extension of the overall lifespan of the LGMD2I-model and FKRP-mutant zebrafish (Figure 5A). However, only pentetic acid (a nonselective compound that binds to calcium and magnesium ions) showed a complete elimination of pericardiac edema pathology in both LGMD2I and FKRP-mutant zebrafish (Figure 5B). Recent studies in mice have shown that regulation of calcium genes and the influx of extracellular calcium can induce myofiber death of dystrophic muscles (40). Further research into the regulation of calcium handling in LGMD2I is warranted in uncovering the pathogenic mechanisms in FKRP-defective muscles.
Figure 5. Pentetic acid extends lifespan and ameliorates symptoms in LGMD2I zebrafish.
(A) Drug screening of select compounds in the LGMD2I-model zebrafish. Five select compounds were individually tested: sildenafil citrate (purple squares), pentetic acid (blue triangles), propantheline bromide (green squares), ethosuximide (orange circles), and aminophylline (gray diamonds) at a concentration of 2.5 μM per compound. WT (black circles) aged-matched, vehicle (0.01% DMSO in fish water; yellow diamonds), and untreated (red squares) control zebrafish cohorts were also used in comparison (n = 50 fish per treatment cohort). Zebrafish were measured for overall viability between 1 and 7 dpf. Error bars indicate SEM. (B) Ablation of LGMD2I pathology in zebrafish treated with pentetic acid (2.5 μM) compared with vehicle (0.01% DMSO in fish water) control zebrafish. Scale bar: 300 μm. *P < 0.05 by 2-way ANOVA with Tukey’s HSD test.
Drug screening of a FDA-approved drug compound library identifies potentially novel and current therapeutics for LGMD2I symptoms.
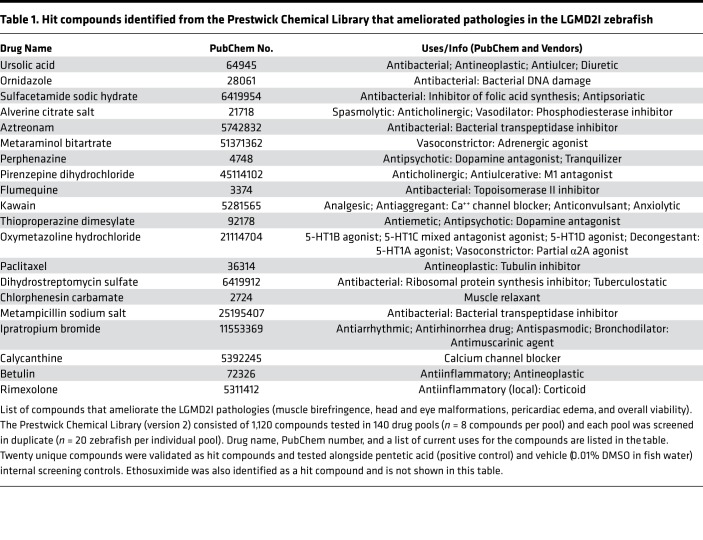
We additionally performed an unbiased, double-blinded (genotype and drug compound) drug library screen using the Prestwick library (version 2). The Prestwick library contains 1,120 compounds, including FDA-approved drugs and compounds that failed in late-stage clinical trials, and has been shown to identify corrective compounds in both DMD zebrafish models and in cell-based Utrophin-promoter reporter assays (38, 41). We performed drug screening of the Prestwick library in our LGMD2I zebrafish by incubating 20 LGMD2I heat-shocked embryos from Tg[hsp70l-hFKRP(L276I)-IRES-eGFP]:fkrp Δ13+/– heterozygote matings with an expected 50% phenotypic affected rate in all zebrafish drug pools based on previous matings (data not shown). During the initial primary screen, each pool contained 20 zebrafish embryos and 8 structurally similar compounds, and the entire library was screened in duplicate (n = 40 fish per compound pool). Each well was screened for 4 LGMD2I zebrafish phenotypic criteria: muscle birefringence, eye and head malformations, pericardiac edema, and overall viability. A pool of compounds that resulted in a reduction of affected fish in any of the 4 criteria to 10% or lower was considered a “hit.” The pool of compounds making up a hit were then tested individually in a secondary unpooled drug screening. Out of the 1,120 total compounds in the Prestwick library, 20 unique compounds (including pentetic acid as a positive control) were shown to block or ameliorate all 4 of the phenotypic criteria in the LGMD2I zebrafish (Table 1). Interestingly, most of the corrective hits identified were steroids, nonsteroidal antiinflammatory drugs (NSAIDs), and antibacterial compounds (Table 1). Interestingly, glucocorticoids have been shown to be beneficial in FKRP mouse models of LGMD2I along with being a standard treatment for other muscular dystrophies such as DMD (42, 43). Glucocorticoid treatment supplemented with alendronate (Fosamax; a bisphosphonate drug used for osteoporosis and other bone disorders) was shown to ameliorate muscle pathologies and increase myofiber size in the LGMD2I mutant mouse model (42). Further expanded preclinical testing of these 20 hit compounds along with some of their more potent derivatives in other FKRP-mutant models may prove to be effective in restoring muscle function in LGMD2I-diseased muscles and offer hope to patients dealing with this disorder.
Table 1. Hit compounds identified from the Prestwick Chemical Library that ameliorated pathologies in the LGMD2I zebrafish.
Compounds structurally similar to pentetic acid block muscle and cardiac pathologies in LGMD2I zebrafish.
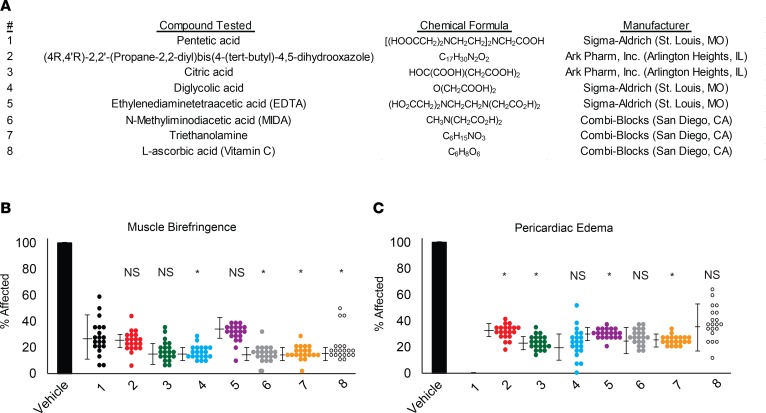
Given our findings that pentetic acid was capable of blocking overall pathologies in our LGMD2I zebrafish, we performed a chemical structure analysis for structurally similar compounds to determine a potential mechanism of action. We identified 7 chemical compounds structurally similar to pentetic acid that were readily commercially available for testing. We screened these 7 compounds in our LGMD2I zebrafish along with pentetic acid to determine if they could similarly rescue the muscle and cardiac pathologies observed in our LGMD2I model. Of these 7 compounds, 2 compounds (triethanolamine and citric acid) demonstrated the ability to reduce muscle and cardiac pathologies similarly to pentetic acid (Figure 6, A and B). The remaining 5 compounds [(4R,4′R)-2,2′-(propane-2,2-diyl)bis(4-(tert-butyl)-4,5-dihydrooxazole), diglycolic acid, ethylenediaminetetraacetic acid (EDTA), N-methyliminodiacetic acid (MIDA), and L-ascorbic acid (vitamin C)] reduced some of the LGMD2I-associated pathologies to lower than vehicle controls (Figure 6A). These findings suggest that a common mechanism of action may alleviate or block LGMD2I-related pathologies early in development. Additional chemical optimization of pentetic acid and related compounds is warranted to optimize the pharmacological abilities of these compounds in LGMD2I zebrafish and eventually human patients.
Figure 6. Compounds structurally similar to pentetic acid can ameliorate pathologies and extend lifespan in LGMD2I zebrafish.
(A) List of compounds tested and their manufacturers. (B) Dorsal muscle birefringence and (C) pericardiac edema average percentages in the LGMD2I-model drug-treated fish. Average percentages affected were normalized to vehicle-treated (100%) LGMD2I-model fish. *P < 0.05 compared with pentetic acid (compound 1; black circles) by 2-way ANOVA with Tukey’s HSD test, and the SEM is shown on the left side. NS, not significant when compared with pentetic acid (1; black circles) positive control. The following compounds were evaluated: vehicle (0.01% DMSO in fish water; black bar) was tested along with 1 (pentetic acid, black circles), 2 [(4R,4′R)-2,2′-(propane-2,2-diyl)bis(4-(tert-butyl)-4,5-dihydrooxazole); red circles], 3 (citric acid; green circles), 4 (diglycolic acid; blue circles), 5 (EDTA; purple circles), 6 (N-methyliminodiacetic acid; gray circles), 7 (triethanolamine; orange circles), and 8 (L-ascorbic acid, open circles). Twenty fish (n = 20) were tested for each compound pool.
Discussion
Our findings demonstrate the utility of genetic manipulation of zebrafish for both studying neuromuscular diseases, and for therapeutic drug compound screening to identify novel corrective drugs. We have generated what we believe is a novel zebrafish model of LGMD2I (heat shock–inducible Tg[hsp70l-hFKRP(L276I)-IRES-eGFP] transgenic line) that phenocopies human disease pathology and overall clinical symptoms. Additionally, our zebrafish LGMD2I model phenocopies the varying symptoms observed in the FKRP mouse models harboring human pathogenic variants (44). We observed muscle wasting, eye and brain malformations, pericardiac edema, and early lethality reflective of the amount of functional FKRP protein, which is similar to human patients with pathogenic FKRP variants. Questions remain as to why a large spectrum of phenotypic variation occurs as a result of the location of the FKRP pathogenic variant in the protein. In both LGMD2I and WWS/MEB, FKRP protein is expressed at relatively normal levels; however, the amount of glycosylation of α-dystroglycan is somewhat reflective of the disease severity and classification (45). There are conflicting reports as to whether or not the levels and complexity of α-dystroglycan glycosylation are faithfully representative of disease severity and classification of FKRP pathogenic variants (16). Recent work in gene therapy has shown that overexpression of full-length FKRP protein using systemic AAV delivery can block muscle wasting and cardiac symptoms associated with LGMD2I (33, 46). Additionally, AAV-mediated FKRP overexpression is capable of restoring the expression levels and complexity of α-dystroglycan glycosylation in FKRP-mutant mice (24). Nevertheless, a comprehensive functional role of FKRP in skeletal muscle along with other tissues remains to be elucidated. Our study identified steroids, NSAIDs, antibacterials, and calcium chelators as major classes of compounds that ameliorated the disease pathologies in the LGMD2I-model zebrafish. Steroid regimens have been shown to be beneficial in blocking symptoms of muscle weakness in human LGMD2I patients (47). The regulation of calcium levels appears to be essential for the progression of LGMD2I, as pentetic acid and alendronate dosing blocks symptoms in FKRP-mutant zebrafish and mice. A recent study demonstrated that tamoxifen and raloxifene (an FDA-approved osteoporosis drug that regulates calcium channels) alleviates dystrophic pathology in FKRP-mutant mice (48, 49). Another hit compound identified in our LGMD2I zebrafish screen, ursolic acid, has been shown to increase skeletal muscle mass and block muscle atrophy (50). Thus, any therapeutic treatment for LGMD2I might involve a combination therapy to not only restore FKRP expression levels, but also ameliorate the muscle weakness and pathology caused by damaged myofiber membranes.
Complete ablation of the mouse Fkrp gene is embryonic lethal; however, mice harboring human patient disease–causing mutations have been shown to mirror the varying pathologies observed in both LGMD2I and WWS/MEB (44). A recent study of the Fkrp P488Lneo mouse model revealed additional respiratory defects that were not detected in our LGMD2I zebrafish, likely due to differences in their ability to take up oxygen through their gills and skin during early larval stages (51). In contrast, our LGMD2I model zebrafish have some significant differences between their observed pathologies and the human disease progression. Notably, the vasculature and cardiac defects we observe in our zebrafish occur during early developmental stages, whereas cardiovascular issues in LGMD2I patients usually occur in their later adult years (52). Any therapeutic compound identified from our studies would have to be tested in older/adult models of FKRP-dependent dystroglycanopathies to fully validate their therapeutic effects on FKRP-defective pathologies. Another difference is the severe eye malformations we observed in the LGMD2I-model zebrafish, which are not found in LGMD2I patients but only in severe WWS/MEB patients (53). Fkrp mouse mutations carrying the common LGMD2I L276I mutation show a mild skeletal muscle weakness and cardiac pathology, with some retinal defects (44, 54, 55). In contrast, mice harboring more severe human-based FKRP pathogenic mutations (such as Y307N and P448L) have more severe muscle wasting pathology, brain defects, and cardiac defects than the FKRP L276I mice (15, 56). Detailed analyses of Fkrp-knockdown mice showed altered laminin deposits and early defects in Cajal-Retzius localization that may explain the cobblestone lissencephaly that occurs in human WWS/MEB patients (57).
Overexpression of full-length FKRP using AAV9 has been shown to rescue the skeletal muscle pathology, cardiac function, and dystroglycan glycosylation defects in FKRP-defective mice dependent on the mouse developmental stage of the rescue (46, 54, 58). A similar study using an AAV2/9 vector for FKRP overexpression demonstrated that the level of FKRP is important in preventing toxicity resulting from excessively elevated levels of FKRP protein in a mouse LGMD2I model (33). Muscle satellite cell transplantation in the FKRP L276I LGMD2I mouse model was shown to rescue muscle defects due to secretion of full-length FKRP protein in exosomes released by the transplanted satellite cells (59). Together, these studies demonstrate that overexpression of full-length FKRP protein may be an effective therapy for LGMD2I patients; however, the regulation of the amount of FKRP overexpressed is essential for the amelioration of FKRP-dependent pathologies. Intramuscular transplantation of genetically engineered muscle satellite cells can also ameliorate LGMD2I muscle pathology, although engraftment efficiency and systemic FKRP expression still remain challenging with this approach (59). It is likely that if gene therapy is approved as a clinical treatment for LGMD2I, a combinatorial treatment to ameliorate FKRP-defective tissues would be required. Our findings demonstrate that our LGMD2I-model zebrafish are a useful tool to identify corrective therapeutics through the expression of a patient-specific FKRP pathogenic mutation.
Methods
Study approval.
All zebrafish were housed in the Zebrafish Research Facility of the University of Alabama at Birmingham and the Aquatics Facility of Boston Children’s Hospital under animal protocol numbers IACUC-20320 (UAB Institutional Animal Care and Use Committee; Birmingham, Alabama) and 09-10-1534R (BCH Animal Care Resources at Boston Children’s Hospital; Boston, Massachusetts).
Zebrafish husbandry.
AB strain zebrafish were used for all injections, and served as wild-type controls when applicable. Zebrafish were housed in either 1.8- or 3-liter tanks, given shrimp feed, and maintained under 12-hour light cycles. Zebrafish were euthanized via immersion in tricaine methanesulfonate (MS222, 200 mg/l final concentration; Sigma-Aldrich), consistent with the NIH (2009) Final Report to the Office of Laboratory Animal Welfare (OLAW) on Euthanasia of Zebrafish and ethically published zebrafish euthanasia protocols (60).
Generation of FKRP-mutant fish using TALENs.
TALENs were designed using the ZiFit Targeter software (61, 62). Each TAL effector repeats array was designed to target 16-bp DNA half-sites. Fragments encoding TAL-effector repeat arrays were subcloned into wild-type FokI expression vectors (Addgene; http://www.addgene/org/talengineering/expressionvectors/). The C-terminal 0.5 TALE repeat domain varies in the different expression vectors (pJDS70, pJDS71, pJDS74, and pJDS78) from the Keith Joung lab REAL Assembly TALEN kit (Addgene; kit number 1000000017) (62, 63). TALEN DNA was linearized with PmeI for 2 hours at 37°C and purified using a Qiagen QiaQuick PCR purification kit according to the manufacturer’s instructions. Purified, linearized DNA (500 ng) was transcribed in vitro using the mMESSAGE mMACHINE T7 ULTRA kit (Ambion) according to the manufacturer’s instructions. The transcribed RNAs were polyadenylated using reagents from the same kit.
RNAs encoding each TALEN arm were combined and resuspended in nuclease-free water at 250 ng/μl concentration. Injection solution was prepared with 0.9 μl from a 10 M KCL, 10× TE mix, and 750 ng of each of the left and right half-site TALE-encoding RNA (64). One pl (125 pg RNA of each of the 2 arms, or 250 pg total) was injected into 1-cell-stage zebrafish embryos. The AB fish strain was used for all injections. Injected and uninjected (control) embryos were kept at 28.5°C overnight. On the next day, genomic DNA was extracted in pools of 12 embryos at 3 dpf. These pools were placed on ice for 5 minutes to sacrifice the fish. Next, water was completely removed and 200–300 μl of extraction buffer (1 M NaOH) was added and incubated at 95°C for 10 minutes and cooled to 4°C for 5 minutes. Ten microliters of 1 M Tris-HCL pH 8 was added to resuspend the genomic DNA. Embryos that were deformed from injection were discarded. Targeted genomic loci were amplified using primers designed approximately 100 bp upstream from the expected FokI nuclease cut site. Genomic PCR products were purified by Qiagen MinElute kit, subcloned into the TOPO vector (Invitrogen TOPO-TA kit), and transformed into XL-blue bacterial cells. Forty-eight colonies were picked and inoculated in LB-kanamycin cultures and incubated overnight at 37°C. Plasmid purification of 48 colonies was performed using a Qiagen Miniprep kit, and purified plasmid DNA was sequenced using a M13 reverse primer supplied in the TOPO-TA kit.
FKRP zebrafish mutation frequency.
The somatic mutation rate was determined by the number of clones with indel mutations/number of zebrafish embryo genomic DNA extracted. The overall somatic mutation rate of the genomic DNA was approximately 30%, which is consistent with previously published comparisons of TALEN somatic mutagenesis rates (65). Sequences that did not include both sides of the target region cut site were not included in the mutation analysis. In addition, single deletions, insertions, or substitutions were not considered mutations since they could likely result from PCR artifact. The PCR product was cloned using a TOPO-TA cloning kit (Invitrogen) according to the manufacturer’s instructions. Single colonies were sequenced to determine the mutation frequency of 1 allele.
FKRP mutant F0 (founder generation) screen.
Potential founders (injected fish) were raised to adulthood for approximately 6–8 weeks and crossed with wild-type zebrafish. At 4 dpf, progeny were lysed individually in 50 μl of lysis buffer (50 mM NaOH) to extract genomic DNA as previously described (66). The DNA solution was neutralized using 5 μl of 50 mM Tris-HCL. Samples were spun at 3,000 rpm for 5 minutes, and the supernatant of each sample was used directly for PCR amplification. Twelve embryos from each potential founder were screened for the presence of TALEN-induced mutations by PCR amplifying the region surrounding the TALEN cleavage site. PCR products were sequenced directly with forward primer located approximately 100 bp upstream of the target site. The rest of the embryos were cultured to adulthood for approximately 6–8 weeks. Zebrafish were genotyped by PCR using JumpStart DNA polymerase (Sigma-Aldrich) following the manufacturer’s protocol and primers designed to detect the zebrafish fkrp 13-bp deletion. The primers were diluted to 10 μM each in sterile water and consist of zebrafish fkrp forward 5′-TTTTCCTGAATGGGATCAGC-3′ and reverse 5′-ATCTTCAAATTCCCGAACCA-3′ resulting in an approximate 500-bp PCR amplicon.
Generation of heat shock–inducible FKRP-transgenic zebrafish.
To generate the heat shock–inducible transgenic zebrafish that overexpress human FKRP (full length or carrying the L276I mutation), we used the Tol2-based transgenesis system (67). The upstream 1.5 kb zebrafish hsp70l promoter (p5E-hsp70l; Tol2kit 222), (p3E-IRES-EGFPpA; Tol2kit 389), and middle entry clones containing either the human FKRP full-length ORF or FKRP (L276I mutation) in the pENTR/D-TOPO vector (Invitrogen) were subcloned into the pDestTol2pA2 (Tol2kit 384) using LR Clonase II plus (Invitrogen). Fifty nanograms of the plasmid along with 30 nanograms of transposase mRNA (pCS2FA-transposase; Tol2kit 396) were injected into the cells of approximately 200 AB zebrafish embryos at the 1-cell stage. Transposase mRNA was generated using the MAXIscript SP6 Transcription Kit (Life Technologies) following the manufacturer’s protocol.
Additional transgenic and mutant zebrafish lines.
The α-actin-RFP–transgenic zebrafish were a gift from H.J. Tsai (National Taiwan University; Taipei City, Taiwan) and have been previously described (68). The flk1-GFP–transgenic zebrafish were a gift from Leonard Zon (Boston Children’s Hospital) and have been previously described (69). Sapje (dystrophin) mutant zebrafish were previously described (70, 71).
Western blot analysis.
Total whole tissue protein lysates were obtained from individual zebrafish larvae after decapitation for PCR-based genotypic analyses. After decapitation, the zebrafish bodies were snap frozen in liquid nitrogen, and stored at –80°C until homogenization. Zebrafish bodies were homogenized on ice using a micro-grinder hand-held tissue homogenizer (RPI Corp) in 100 μl of M-PER buffer (ThermoFisher Scientific) supplemented with cOmplete mini-EDTA-free protease inhibitor cocktail tablets (Roche Life Sciences). Protein lysates were quantified using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific). Approximately 30 μg of whole-tissue protein lysates were diluted in NuPage LDS Sample protein buffer and electrophoretically resolved in Novex 4%–20% Tris-Glycine (Invitrogen) mini gels following the manufacturer’s guidelines. Gels were then transferred to PVDF membranes (ThermoFisher Scientific) and incubated in primary antisera overnight at 4°C. Membranes were washed for 1 hour in 1× TBS-Tween (0.1%) (Boston Bio Products), and incubated in secondary antisera for 1 hour at room temperature with gentle rocking. The membranes were then stained with the Pierce SuperSignal kit (ThermoFisher Scientific), and imaged on Blue Devil Light Film (Genesee Scientific). Films were then scanned into TIFF files and processed in Adobe Photoshop CC 2017 (Adobe Systems). Some Western blot images were directly acquired using a Typhoon Variable Mode Imager (Amersham Pharmacia).
DanioVision zebrafish motion tracking.
Five-dpf zebrafish larvae were tracked via the DanioVision (Noldus) motion tracking platform in a protocol slightly modified from a previously published protocol (28). Zebrafish were placed in 48-well plates in 300 ml of fish water and were given a 15-minute acclimation period in the DanioVision machine. After 15 minutes, the zebrafish larvae locomotion was recorded for a 15-minute period. Each experiment consisted of 10 zebrafish per tracking session and each genotype cohort was tracked during 5 independent sessions (n = 50 fish per genotype cohort; n = 5 separate experiments). Motion tracking data were analyzed using EthoVision (version XT 10; Noldus) and in both EthoVision and Excel version 2013 (Microsoft).
Zebrafish physiological force measurements.
Zebrafish dorsal muscle force measurements were performed using a protocol previously described (29). Larvae were anesthetized with tricaine and the head removed and stored for later genotyping. Physiological properties of the tail muscles were studied at 25°C in bicarbonate buffer equilibrated with 95% O2, 5% CO2. Monofilament 10-0 sutures placed at the gastrointestinal opening and anterior to the tip of the tail were used to attach the preparation horizontally between an isometric force transducer (model 403A, Aurora Scientific) and a position motor (model 308B, Aurora Scientific). Biphasic pulses (200 μs in duration) were generated by a muscle stimulator (model 701A, Aurora Scientific) and delivered to platinum electrodes that flanked the preparation. Tetanic force was evaluated with 30-ms trains of 300-Hz stimulation. The length of the preparation was carefully adjusted to optimize force. At the conclusion of data collection, the preparation was carefully rotated and its maximum width and depth measured. Preparation cross-sectional area was calculated assuming the tail was elliptical in cross section. Reported forces were normalized to cross-sectional area.
Immunofluorescent and histochemical staining and imaging.
Zebrafish images were taken on an SMZ1500 Zoom stereomicroscope (Nikon Instruments Inc.) with separate fluorescence. Some whole-mount immunofluorescence images were taken on an LSM710 confocal microscope (Carl Zeiss AG). Zebrafish H&E staining was performed using Mayer’s Hematoxylin stain (Sigma-Aldrich) following a previously published protocol (72). All images were saved as TIFF files and processed in Adobe Photoshop CC 2017.
qPCR.
All zebrafish larvae were decapitated for PCR-genotyping confirmation of their heads, while the remaining bodies of the zebrafish were snap frozen in cryovials in liquid nitrogen and stored at –80°C until confirmation by PCR genotyping. Samples were homogenized on ice using a micro-grinder hand-held tissue homogenizer, and total RNA was extracted using a Qiagen RNeasy Plus Mini Kit following the manufacturer’s protocol. One microgram of total RNA was then reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen) following the manufacturer’s guidelines. The synthesized cDNA was then mixed with SYBR Green PCR Master Mix (Applied Biosystems) following the manufacturer’s guidelines. qPCR analyses were performed in 384-well optical plates on an ABI 7900HT Real-Time PCR Machine (Applied Biosystems). Cycle times were normalized to an internal housekeeping gene as a loading control. Relative fold expression and changes were calculated using the 2–ΔΔCt method as previously described (73).
Primers.
All oligonucleotides were obtained commercially (IDT). A full list of primer sequences used in all experiments can be found in Supplemental Figure 3.
Antibodies.
A full list of antibodies (primary and secondary) used in all experiments and the dilutions they were used at can be found in Supplemental Figure 4.
Drug library screening of LGMD2I zebrafish.
The Prestwick Chemical Library (version 2) was obtained from the ICCB-Longwood Screening Facility at Harvard Medical School as a series of 96-well spotted plates with each compound diluted in a solution of 0.01% DMSO (Sigma-Aldrich) and fish water. For both primary and secondary screenings, embryo clutches were collected on day 0 and incubated at 28.5°C overnight. At 24 hours post infection (hpf), embryos were transferred into individual PCR tubes and heated on a Veriti thermal cycler (Applied Biosystems) to induce expression of the human FKRP full-length or L276I transgenic cassette. Embryos were incrementally heated by 2°C every 5 minutes until 38°C was reached, followed by incubation at 38°C for 1 hour. This protocol is a modification of a previously published protocol that was modified to avoid toxicity resulting from abnormally high levels of FKRP (full-length) or (L276I) protein (74). Fish were then returned to 28.5°C overnight. At 48 hpf, drugs were administered for screening. Primary screening was performed in compound pools in 48-well tissue-culture-grade plates (Corning Inc.) with minor modifications to a previously established protocol (38). Each pool consisted of 8 compounds per pool, and each pool was tested in duplicate. Each drug pool contained 0.5 μg/ml of each drug for a total concentration of 4 μg/ml. Each of these tested pools contained 20 embryos, cultured in each compound pool from 48 hpf through 5 dpf. At 5 dpf, fish in each drug pool were assessed for abnormal skeletal muscle development via birefringence assay, pericardiac edema, eye and head malformations, and overall viability. Primary screening pools that demonstrated 10% affected fish or less were considered hit pools. Hit pools were identified and the compounds in each pool were broken down for confirmation of individual drug efficacy using the same embryo collection, heat shock, and drug administration process as described above. Each individual drug was tested in duplicate, containing 10 embryos each (n = 20 embryos per drug). Individual drugs were tested at a total concentration of 2.4 μg/ml. As in primary screening, at 5 dpf, fish were assessed for abnormal skeletal muscle development via birefringence assay, pericardiac edema, eye and head malformations, and overall viability. Drug compounds that had shown no phenotypic malformations of affected fish were categorized as validated hit compounds. Additional compounds evaluated were obtained commercially including (4R,4′R)-2,2′-(propane-2,2-diyl)bis(4-(tert-butyl)-4,5-dihydrooxazole) and citric acid that were purchased from Ark Pharm, Inc. Diglycolic acid and EDTA were obtained from Sigma-Aldrich. MIDA, triethanolamine, and L-ascorbic acid were all purchased from Combi-Blocks.
Statistics.
All multigroup statistical tests were performed using 2-way analysis of variance (ANOVA) with Tukey’s honest significant difference (HSD) test. Direct comparisons were evaluated using a 2-tailed Student’s t test. Values of significance (P values) are stated in comparison with internal controls. P < 0.05 was considered significant. The experimenter was double-blinded to both genotype and drug compound for all library and secondary drug screening and validation steps.
Author contributions
MSA, PRS, MJF, RMH, DGP, NMV, AL, DEG, MJG, GK, and JJW performed experiments and collected data presented in the manuscript. OMC and CEAS analyzed the chemical structures of the lead drug compounds. MSA and LMK oversaw all the experiments, analyzed all of the data, and wrote and edited the manuscript. All authors have seen and approved the final version of the manuscript prior to submission.
Supplementary Material
Acknowledgments
The authors wish to thank Emanuela Gussoni, Janelle Spinazzola, and John Parant for a critical reading and helpful comments towards the manuscript. The authors wish to thank David Langenau, Keith Joung, and Finola Moore for technical assistance with the TALEN gene editing experiments. The authors wish to thank Susan Farmer, Samuel Cartner, Christian Lawrence, and the technical staff at Boston Children’s Hospital and UAB School of Medicine Aquatics facilities for general housing, care, and maintenance of zebrafish. The authors wish to thank Jennifer Smith and Caroline Shamu from the ICCB-Longwood Screening Facility for assistance with experimental design and data management. LMK is funded by the Bernard F. and Alva B. Gimbel Foundation. Research reported in this publication was supported by a combined grant from the LGMD2i Research Fund, the Samantha Brazzo Foundation, and Cure CMD. RMH is a member of the RoadMap Scholars Program at UAB (NIH grant number 5R25NS089463) and is also supported by a T32 training fellowship sponsored by the University of Alabama at Birmingham Center for Exercise Medicine. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH under award number 2T32HD071866-06. RMH is also a 2017–2018 UAB Civitan International Research Center Emerging Scholar Award winner. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. MSA is supported by a grant from the Muscular Dystrophy Association (award number 418254).
Version 1. 09/20/2018
Electronic publication
Funding Statement
R.M.H. is a member of the RoadMap Scholars Program at UAB
R.M.H. is also supported by a T32 training fellowship sponsored by the University of Alabama at Birmingham Center for Exercise Medicine. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health
L.M.K. was supported by a combined research grant from the LGMD2I Research Fund, the Samantha Brazzo Foundation, and Cure CMD
M.S.A. is supported by a grant from the Muscular Dystrophy Association.
Footnotes
Conflict of interest: LMK is a consultant for Pfizer Inc., Summit Corporation PLC, and Tarix Pharmaceuticals for muscle disease drug therapies. LMK is also a consultant for SynapDx Corporation for autism screening technologies. All other authors declare no conflicting interests.
Reference information: JCI Insight. 2018;3(18):e120493. https://doi.org/10.1172/jci.insight.120493.
Contributor Information
Daniela Garcia-Perez, Email: garc1987@ravens.benedictine.edu.
Angela Lek, Email: Angela.Lek@yale.edu.
Devin E. Gibbs, Email: degibbs@ucla.edu.
Omar Moukha-Chafiq, Email: omoukha-chafiq@southernresearch.org.
Genri Kawahara, Email: gkawahar@tokyo-med.ac.jp.
Jeffrey J. Widrick, Email: Jeffrey.Widrick@childrens.harvard.edu.
Louis M. Kunkel, Email: kunkel@enders.tch.harvard.edu.
Matthew S. Alexander, Email: malexander@peds.uab.edu.
References
- 1.Levine BA, Moir AJG, Patchell VB, Perry SV. The interaction of actin with dystrophin. FEBS Letters. 1990;263(1):159–162. doi: 10.1016/0014-5793(90)80728-2. [DOI] [PubMed] [Google Scholar]
- 2.Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338(6212):259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 3.Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345(6273):315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 4.Rahimov F, Kunkel LM. The cell biology of disease: cellular and molecular mechanisms underlying muscular dystrophy. J Cell Biol. 2013;201(4):499–510. doi: 10.1083/jcb.201212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson RA, et al. Dystroglycan is essential for early embryonic development: disruption of Reichert’s membrane in Dag1-null mice. Hum Mol Genet. 1997;6(6):831–841. doi: 10.1093/hmg/6.6.831. [DOI] [PubMed] [Google Scholar]
- 6.Endo T. Glycobiology of α-dystroglycan and muscular dystrophy. J Biochem. 2015;157(1):1–12. doi: 10.1093/jb/mvu066. [DOI] [PubMed] [Google Scholar]
- 7.Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119(Pt 2):199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 8.Godfrey C, Foley AR, Clement E, Muntoni F. Dystroglycanopathies: coming into focus. Curr Opin Genet Dev. 2011;21(3):278–285. doi: 10.1016/j.gde.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Bouchet-Séraphin C, Vuillaumier-Barrot S, Seta N. Dystroglycanopathies: About numerous genes involved in glycosylation of one single glycoprotein. J Neuromuscul Dis. 2015;2(1):27–38. [PubMed] [Google Scholar]
- 10.Bushby K. Diagnosis and management of the limb girdle muscular dystrophies. Pract Neurol. 2009;9(6):314–323. doi: 10.1136/jnnp.2009.193938. [DOI] [PubMed] [Google Scholar]
- 11.Brockington M, et al. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am J Hum Genet. 2001;69(6):1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torelli S, et al. Sub-cellular localisation of fukutin related protein in different cell lines and in the muscle of patients with MDC1C and LGMD2I. Neuromuscul Disord. 2005;15(12):836–843. doi: 10.1016/j.nmd.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 13.de Bernabé DB, et al. A homozygous nonsense mutation in the fukutin gene causes a Walker-Warburg syndrome phenotype. J Med Genet. 2003;40(11):845–848. doi: 10.1136/jmg.40.11.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beltran-Valero de Bernabé D, et al. Mutations in the FKRP gene can cause muscle-eye-brain disease and Walker-Warburg syndrome. J Med Genet. 2004;41(5):e61. doi: 10.1136/jmg.2003.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ackroyd MR, et al. Fukutin-related protein alters the deposition of laminin in the eye and brain. J Neurosci. 2011;31(36):12927–12935. doi: 10.1523/JNEUROSCI.2301-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alhamidi M, Brox V, Stensland E, Liset M, Lindal S, Nilssen Ø. Limb girdle muscular dystrophy type 2I: No correlation between clinical severity, histopathology and glycosylated α-dystroglycan levels in patients homozygous for common FKRP mutation. Neuromuscul Disord. 2017;27(7):619–626. doi: 10.1016/j.nmd.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Nigro V, Savarese M. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol. 2014;33(1):1–12. [PMC free article] [PubMed] [Google Scholar]
- 18.Berger J, Currie PD. Zebrafish models flex their muscles to shed light on muscular dystrophies. Dis Model Mech. 2012;5(6):726–732. doi: 10.1242/dmm.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore CJ, Goh HT, Hewitt JE. Genes required for functional glycosylation of dystroglycan are conserved in zebrafish. Genomics. 2008;92(3):159–167. doi: 10.1016/j.ygeno.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Thornhill P, Bassett D, Lochmüller H, Bushby K, Straub V. Developmental defects in a zebrafish model for muscular dystrophies associated with the loss of fukutin-related protein (FKRP) Brain. 2008;131(Pt 6):1551–1561. doi: 10.1093/brain/awn078. [DOI] [PubMed] [Google Scholar]
- 21.Wood AJ, et al. Abnormal vascular development in zebrafish models for fukutin and FKRP deficiency. Hum Mol Genet. 2011;20(24):4879–4890. doi: 10.1093/hmg/ddr426. [DOI] [PubMed] [Google Scholar]
- 22.Lin YY, White RJ, Torelli S, Cirak S, Muntoni F, Stemple DL. Zebrafish Fukutin family proteins link the unfolded protein response with dystroglycanopathies. Hum Mol Genet. 2011;20(9):1763–1775. doi: 10.1093/hmg/ddr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawahara G, Guyon JR, Nakamura Y, Kunkel LM. Zebrafish models for human FKRP muscular dystrophies. Hum Mol Genet. 2010;19(4):623–633. doi: 10.1093/hmg/ddp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Awano H, et al. Restoration of functional glycosylation of α-dystroglycan in FKRP mutant mice is associated with muscle regeneration. Am J Pathol. 2015;185(7):2025–2037. doi: 10.1016/j.ajpath.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y, Cattley RT, Cario CL, Bai Q, Burton EA. Quantification of larval zebrafish motor function in multiwell plates using open-source MATLAB applications. Nat Protoc. 2014;9(7):1533–1548. doi: 10.1038/nprot.2014.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipscomb L, Piggott RW, Emmerson T, Winder SJ. Dasatinib as a treatment for Duchenne muscular dystrophy. Hum Mol Genet. 2016;25(2):266–274. doi: 10.1093/hmg/ddv469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith LL, Beggs AH, Gupta VA. Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J Vis Exp. 2013;(82):50925. doi: 10.3791/50925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sztal TE, Ruparelia AA, Williams C, Bryson-Richardson RJ. Using touch-evoked response and locomotion assays to assess muscle performance and function in zebrafish. J Vis Exp. 2016;(116):54431. doi: 10.3791/54431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widrick JJ, et al. Muscle dysfunction in a zebrafish model of Duchenne muscular dystrophy. Physiol Genomics. 2016;48(11):850–860. doi: 10.1152/physiolgenomics.00088.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afroze D, Kumar A. ER stress in skeletal muscle remodeling and myopathies. FEBS J. doi: 10.1111/febs. https://doi.org/10.1111/febs.14358 [published online ahead of print December 14, 2017]. https: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poppe M, et al. The phenotype of limb-girdle muscular dystrophy type 2I. Neurology. 2003;60(8):1246–1251. doi: 10.1212/01.WNL.0000058902.88181.3D. [DOI] [PubMed] [Google Scholar]
- 32.Brockington M, et al. Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum Mol Genet. 2001;10(25):2851–2859. doi: 10.1093/hmg/10.25.2851. [DOI] [PubMed] [Google Scholar]
- 33.Gicquel E, et al. AAV-mediated transfer of FKRP shows therapeutic efficacy in a murine model but requires control of gene expression. Hum Mol Genet. 2017;26(10):1952–1965. doi: 10.1093/hmg/ddx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.North TE, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137(4):736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawahara G, Kunkel LM. Zebrafish based small molecule screens for novel DMD drugs. Drug Discov Today Technol. 2013;10(1):e91–e96. doi: 10.1016/j.ddtec.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Hromowyk KJ, Amacher SL, Currie PD. Muscular dystrophy modeling in zebrafish. Methods Cell Biol. 2017;138:347–380. doi: 10.1016/bs.mcb.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Kawahara G, et al. Dystrophic muscle improvement in zebrafish via increased heme oxygenase signaling. Hum Mol Genet. 2014;23(7):1869–1878. doi: 10.1093/hmg/ddt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawahara G, Karpf JA, Myers JA, Alexander MS, Guyon JR, Kunkel LM. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2011;108(13):5331–5336. doi: 10.1073/pnas.1102116108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waugh TA, et al. Fluoxetine prevents dystrophic changes in a zebrafish model of Duchenne muscular dystrophy. Hum Mol Genet. 2014;23(17):4651–4662. doi: 10.1093/hmg/ddu185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burr AR, Molkentin JD. Genetic evidence in the mouse solidifies the calcium hypothesis of myofiber death in muscular dystrophy. Cell Death Differ. 2015;22(9):1402–1412. doi: 10.1038/cdd.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moorwood C, Lozynska O, Suri N, Napper AD, Diamond SL, Khurana TS. Drug discovery for Duchenne muscular dystrophy via utrophin promoter activation screening. PLoS ONE. 2011;6(10):e26169. doi: 10.1371/journal.pone.0026169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu B, et al. Glucocorticoid steroid and alendronate treatment alleviates dystrophic phenotype with enhanced functional glycosylation of α-dystroglycan in mouse model of limb-girdle muscular dystrophy with FKRPP448L mutation. Am J Pathol. 2016;186(6):1635–1648. doi: 10.1016/j.ajpath.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Manzur AY, Kuntzer T, Pike M, Swan A. Glucocorticoid corticosteroids for Duchenne muscular dystrophy. Cochrane Database Syst Rev. 2008;1:CD003725. doi: 10.1002/14651858.CD003725.pub3. [DOI] [PubMed] [Google Scholar]
- 44.Blaeser A, et al. Mouse models of fukutin-related protein mutations show a wide range of disease phenotypes. Hum Genet. 2013;132(8):923–934. doi: 10.1007/s00439-013-1302-7. [DOI] [PubMed] [Google Scholar]
- 45.Brown SC, et al. Abnormalities in alpha-dystroglycan expression in MDC1C and LGMD2I muscular dystrophies. Am J Pathol. 2004;164(2):727–737. doi: 10.1016/S0002-9440(10)63160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vannoy CH, Xiao W, Lu P, Xiao X, Lu QL. Efficacy of gene therapy is dependent on disease progression in dystrophic mice with mutations in the FKRP gene. Mol Ther Methods Clin Dev. 2017;5:31–42. doi: 10.1016/j.omtm.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darin N, Kroksmark AK, Ahlander AC, Moslemi AR, Oldfors A, Tulinius M. Inflammation and response to steroid treatment in limb-girdle muscular dystrophy 2I. Eur J Paediatr Neurol. 2007;11(6):353–357. doi: 10.1016/j.ejpn.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Wu B, et al. Long-term treatment of tamoxifen and raloxifene alleviates dystrophic phenotype and enhances muscle functions of FKRP dystroglycanopathy. Am J Pathol. 2018;188(4):1069–1080. doi: 10.1016/j.ajpath.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y, Huang YL, Lai B, Zheng P, Zhu YC, Yao T. Raloxifene acutely reduces glutamate-induced intracellular calcium increase in cultured rat cortical neurons via inhibition of high-voltage-activated calcium current. Neuroscience. 2007;147(2):334–341. doi: 10.1016/j.neuroscience.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 50.Kunkel SD, et al. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 2011;13(6):627–638. doi: 10.1016/j.cmet.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Q, et al. Skeletal, cardiac, and respiratory muscle function and histopathology in the P448Lneo− mouse model of FKRP-deficient muscular dystrophy. Skeletal Muscle. 2018;8(1):13. doi: 10.1186/s13395-018-0158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahbi K, et al. Cardiac assessment of limb-girdle muscular dystrophy 2I patients: an echography, Holter ECG and magnetic resonance imaging study. Neuromuscul Disord. 2008;18(8):650–655. doi: 10.1016/j.nmd.2008.06.365. [DOI] [PubMed] [Google Scholar]
- 53.Kava M, Chitayat D, Blaser S, Ray PN, Vajsar J. Eye and brain abnormalities in congenital muscular dystrophies caused by fukutin-related protein gene (FKRP) mutations. Pediatr Neurol. 2013;49(5):374–378. doi: 10.1016/j.pediatrneurol.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 54.Qiao C, et al. Muscle and heart function restoration in a limb girdle muscular dystrophy 2I (LGMD2I) mouse model by systemic FKRP gene delivery. Mol Ther. 2014;22(11):1890–1899. doi: 10.1038/mt.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krag TO, Hauerslev S, Sveen ML, Schwartz M, Vissing J. Level of muscle regeneration in limb-girdle muscular dystrophy type 2I relates to genotype and clinical severity. Skelet Muscle. 2011;1(1):31. doi: 10.1186/2044-5040-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blaeser A, Awano H, Wu B, Lu QL. Progressive dystrophic pathology in diaphragm and impairment of cardiac function in FKRP P448L mutant mice. PLoS ONE. 2016;11(10):e0164187. doi: 10.1371/journal.pone.0164187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Booler HS, Williams JL, Hopkinson M, Brown SC. Degree of Cajal-Retzius cell mislocalization correlates with the severity of structural brain defects in mouse models of dystroglycanopathy. Brain Pathol. 2016;26(4):465–478. doi: 10.1111/bpa.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vannoy CH, Zhou H, Qiao C, Xiao X, Bang AG, Lu QL. Adeno-associated virus-mediated mini-agrin delivery is unable to rescue disease phenotype in a mouse model of limb girdle muscular dystrophy type 2I. Am J Pathol. 2017;187(2):431–440. doi: 10.1016/j.ajpath.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Frattini P, et al. Autologous intramuscular transplantation of engineered satellite cells induces exosome-mediated systemic expression of Fukutin-related protein and rescues disease phenotype in a murine model of limb-girdle muscular dystrophy type 2I. Hum Mol Genet. 2017;26(19):3682–3698. doi: 10.1093/hmg/ddx252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strykowski JL, Schech JM. Effectiveness of recommended euthanasia methods in larval zebrafish (Danio rerio) J Am Assoc Lab Anim Sci. 2015;54(1):81–84. [PMC free article] [PubMed] [Google Scholar]
- 61.Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009;4(12):1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sander JD, Yeh JRJ, Peterson RT, Joung JK. Engineering zinc finger nucleases for targeted mutagenesis of zebrafish. Methods Cell Biol. 2011;104:51–58. doi: 10.1016/B978-0-12-374814-0.00003-3. [DOI] [PubMed] [Google Scholar]
- 63.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29(8):697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 65.Moore FE, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS ONE. 2012;7(5):e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques. 2007;43(5):610. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 67.Kwan KM, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 68.Lin CY, Yung RF, Lee HC, Chen WT, Chen YH, Tsai HJ. Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev Biol. 2006;299(2):594–608. doi: 10.1016/j.ydbio.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 69.Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304(2):735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bassett D, Currie PD. Identification of a zebrafish model of muscular dystrophy. Clin Exp Pharmacol Physiol. 2004;31(8):537–540. doi: 10.1111/j.1440-1681.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 71.Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Keenan DG, Currie PD. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130(23):5851–5860. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- 72.Sullivan-Brown J, Bisher ME, Burdine RD. Embedding, serial sectioning and staining of zebrafish embryos using JB-4 resin. Nat Protoc. 2011;6(1):46–55. doi: 10.1038/nprot.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 74.Pyati UJ, Webb AE, Kimelman D. Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development. 2005;132(10):2333–2343. doi: 10.1242/dev.01806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.