Abstract
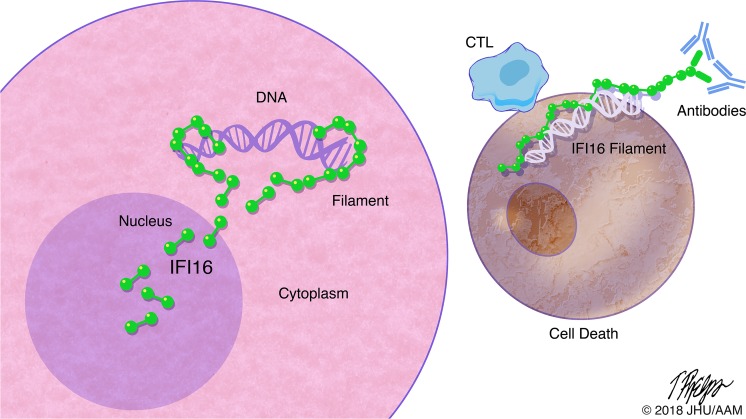
IFN-inducible protein 16 (IFI16) is an innate immune sensor that forms filamentous oligomers when activated by double-stranded DNA (dsDNA). Anti-IFI16 autoantibodies occur in patients with Sjögren’s syndrome (SS) and associate with severe phenotypic features. We undertook this study to determine whether the structural and functional properties of IFI16 play a role in its status as an SS autoantigen. IFI16 immunostaining in labial salivary glands (LSGs) yielded striking evidence of filamentous IFI16 structures in the cytoplasm of ductal epithelial cells, representing the first microscopic description of IFI16 oligomerization in human tissues, to our knowledge. Transfection of cultured epithelial cells with dsDNA triggered the formation of cytoplasmic IFI16 filaments with similar morphology to those observed in LSGs. We found that a majority of SS anti-IFI16 autoantibodies immunoprecipitate IFI16 more effectively in the oligomeric dsDNA-bound state. Epitopes in the C-terminus of IFI16 are accessible to antibodies in the DNA-bound oligomer and are preferentially targeted by SS sera. Furthermore, cytotoxic lymphocyte granule pathways (highly enriched in the SS gland) induce striking release of IFI16•dsDNA complexes from cultured cells. Our studies reveal that IFI16 is present in a filamentous state in the target tissue of SS and suggest that this property of DNA-induced filament formation contributes to its status as an autoantigen in SS. These studies highlight the role that tissue-specific modifications and immune effector pathways might play in the selection of autoantigens in rheumatic diseases.
Keywords: Autoimmunity
Keywords: Autoimmune diseases, Rheumatology
Sjögren’s syndrome autoantigen IFI16 is present in an activated, filamentous form in epithelial cells in Sjögren’s salivary glands.
Introduction
Sjögren’s syndrome (SS) is a systemic autoimmune disease characterized by lymphocytic infiltration of the lacrimal and salivary glands (1). The presence of an IFN signature in the target organs affected by this disease has been well described (2–4), suggesting that IFN system activation may contribute to disease pathogenesis (5). While the specific stimuli responsible for the sustained activation of IFN signaling in SS remain unclear, innate sensing of nucleic acids is a potent stimulus for type I IFN production (6–8). Cytoplasmic DNA sensing, in particular, has recently been identified as a pathway of interest for its potential contribution to the IFN signature in autoimmune diseases (9–11).
SS is also characterized by the presence of a well-defined repertoire of autoantibodies, including the hallmark SSA (Ro52/60) and SSB (La) specificities (12). IFN-inducible protein 16 (IFI16) is an additional autoantigen targeted by patients with SS, and anti-IFI16 antibodies are associated with more severe manifestations of this disease (13–15). IFI16 is a member of the AIM-like receptor (ALR) family of innate pattern recognition receptors, which are sensors of cytoplasmic double-stranded DNA (dsDNA) (16). The 4 human ALR proteins include IFI16, absent in melanoma 2 (AIM2), IFN-inducible protein X (IFIX), and myeloid cell nuclear differentiation antigen (MNDA) (17). These proteins share a common structure composed of N-terminal PYRIN domains that mediate oligomerization and C-terminal HIN200 domains that bind dsDNA (hematopoietic IFN inducible nuclear antigen [HIN] with 200 amino acids). Upon directly detecting cytoplasmic dsDNA, IFI16 monomers assemble into oligomeric filaments, which have been well characterized at the biophysical and suprastructural levels (18, 19).
IFI16 has been implicated as a molecule of potential importance in autoimmunity (13, 20, 21), and elevated expression of IFI16 in the tissues (14) and serum (13) of patients with SS and other inflammatory diseases (22) has been described. However, no direct observation of IFI16 activation in these diseases has been previously reported.
Using confocal microscopy, we found that IFI16 exists in filamentous structures in the cytoplasm of ductal epithelial cells, suggesting that IFI16 had been activated by cytoplasmic DNA at this site. We found that similar cytoplasmic IFI16 structures could be readily generated in the cytoplasm of cultured epithelial cells upon transfection with long dsDNA, supporting the idea that the interaction between IFI16 and dsDNA leads to cytoplasmic filament formation. Moreover, we found that dsDNA remains accessible to nuclease degradation while bound in the IFI16 oligomer in vitro and that IFI16 filaments do not dissociate after dsDNA is degraded, suggesting that, once assembled, these filaments persist regardless of the presence of dsDNA. To test whether filamentous oligomerization of IFI16 is associated with its status as an autoantigen in SS, we analyzed the binding of human anti-IFI16 antibodies to IFI16 protein with or without dsDNA. We found that the ability of anti-IFI1–positive SS sera to immunoprecipitate IFI16 was enhanced (sometimes dramatically) when it was oligomerized on dsDNA, suggesting that SS autoantibodies bind epitopes that are preferentially accessible in filamentous IFI16. Additionally, we show that epitopes in the C-terminus, but not the N-terminus of IFI16, remain accessible to antibody binding in the DNA-bound IFI16 filament, complementing a previous report that SS anti-IFI16 antibodies mainly target C-terminal epitopes (14). Cytotoxic lymphocyte (CTL) granule pathways (highly enriched in the SS gland) induce striking release of IFI16•dsDNA complexes from cultured cells, suggesting a mechanism that could promote an anti-IFI16 immune response. With these data, we propose that dsDNA-induced oligomerization of IFI16 plays a role in establishing its status as an autoantigen in SS. Our data provide further support for the hypothesis that tissue-specific antigen modifications contribute to propagation of anti–self-immune responses in rheumatic disease.
Results
Cytoplasmic IFI16 filaments are found in SS salivary epithelial cells.
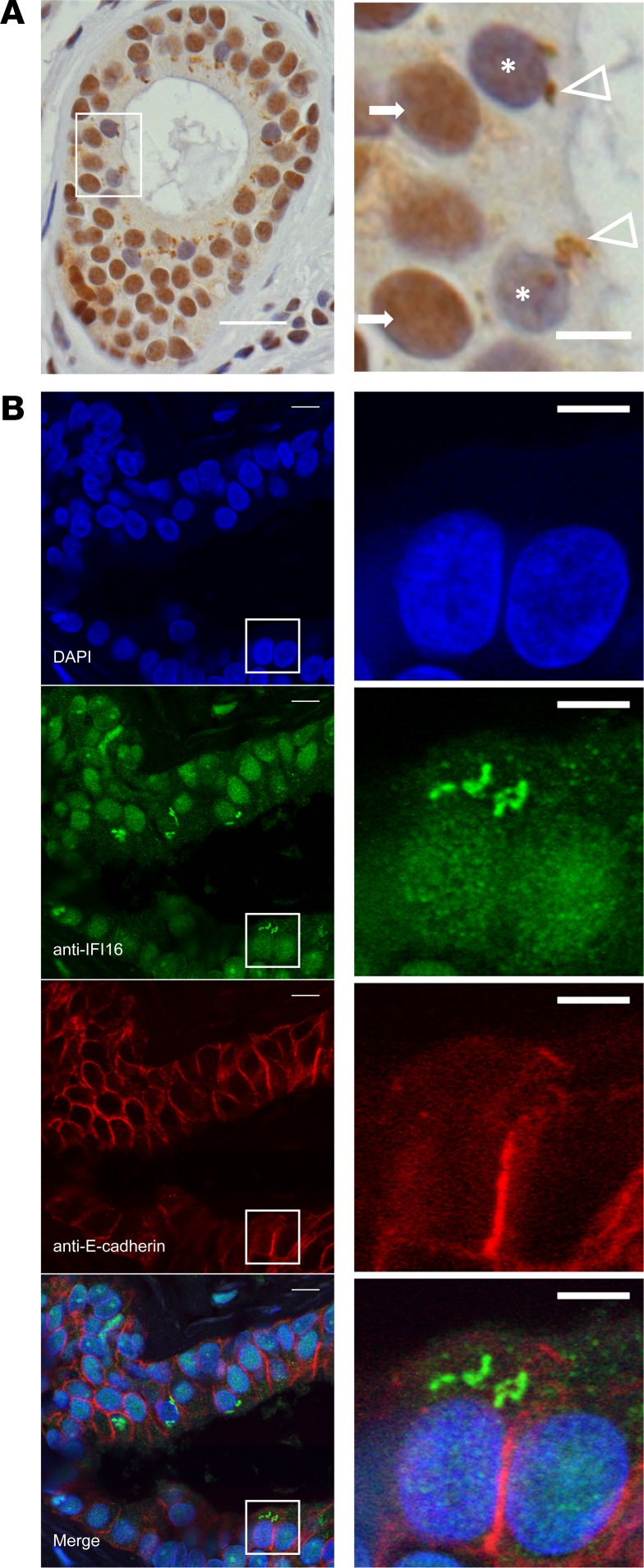
Immunostaining of IFI16 in labial salivary glands (LSGs) demonstrated a striking pattern of expression and localization: IFI16 staining was nuclear in most cells, including ductal and acinar epithelial cells and lymphocytes within inflammatory foci. In a subset of epithelial cells within the salivary ducts, IFI16 was found in irregularly shaped cytoplasmic structures (Figure 1A and Supplemental Video 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120179DS1). These cells were located in the most apical layer of the polarized epithelium within the ducts. Nuclear IFI16 staining was often absent in cells with large cytoplasmic IFI16-containing structures, consistent with a previous observation that translocation from the nucleus into the cytoplasm occurs with activation of IFI16 by dsDNA (23). Immunoflourescence costaining with an antibody against E-cadherin confirmed that the cells containing cytoplasmic IFI16 filaments were epithelial in origin (Figure 1B). No cytoplasmic filamentous structures were identified in acinar epithelial cells or cells within lymphocytic foci. Low levels of the filamentous staining were seen in rare sicca control salivary gland sections (Supplemental Table 1). No staining was visualized using a mouse isotype control or fluorescent secondary antibody alone (Supplemental Figure 1). Interestingly, many biopsies included fragments of oral mucosa, containing epithelial cells. These mucosal epithelial cells expressed nuclear IFI16, but no cytoplasmic filaments were identified in this cell layer (Supplemental Figure 1). These data provide evidence of IFI16 filament formation in vivo in salivary glands, which are the target organ of SS and are known to have a type I IFN signature in SS patients (24). Furthermore, our findings suggest that IFI16, which is an autoantigen in SS, undergoes a significant conformational change in the salivary gland, as well as intracellular redistribution to the cytoplasm in ductal epithelial cells. We found this modification to be specific to the salivary epithelial cells, as they were not visualized in neighboring oral mucosal epithelium.
Figure 1. Cytoplasmic IFI16 filament localization in ductal epithelial cells in labial salivary gland paraffin sections obtained from patients with SS.
(A) Immunostaining of IFI16 in labial salivary gland tissue demonstrates diffuse nuclear staining in cells of the basal layers of the duct (arrows) and irregularly shaped cytoplasmic structures in some cells in the apical portion of the duct (arrowheads). Nuclear IFI16 staining is frequently absent in cells with cytoplasmic IFI16 (cells denoted with asterisk). Scale bar: 20 μM in low-power field (left), 5 μM in high-power field (right). (B) Immunoflourescence microscopy of IFI16 (green) and E-cadherin (red) in labial salivary gland identifies cells containing cytoplasmic IFI16 structures as epithelial in origin. Nuclei are stained with DAPI (blue). Scale bar: 10 μM in low-power views (left) and 5 μM in high-power views (right). Features of the patients examined are reported in Supplemental Table 1.
Transfection of epithelial cells with dsDNA results in nuclear IFI16 translocation to the cytoplasm and filament formation.
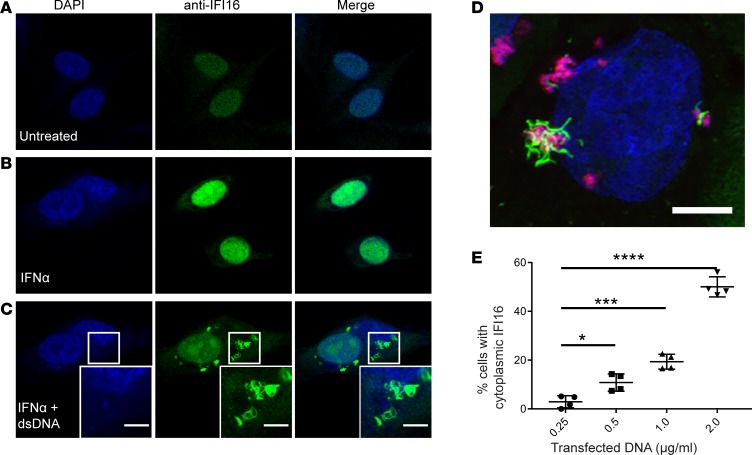
Since recombinant IFI16 assembles into filaments on dsDNA in vitro (18), our immunostaining data from human labial salivary glands supports the idea that IFI16 has been activated by sensing cytoplasmic dsDNA in epithelial cells at this site. We replicated this finding in cultured cells by transfecting dsDNA into the cytoplasm (Figure 2). Cultured human salivary gland (HSG) cells were treated with IFNα to increase basal IFI16 expression, which was confirmed by immunofluorescence (enhanced nuclear staining; Figure 2B). Transfection of the IFNα-treated cells with empty plasmid dsDNA led to the generation of cytoplasmic filaments in a dsDNA dose-dependent manner (Figure 2, C and E). Cytoplasmic IFI16 filaments were not inducible using transfected Poly(I:C), transfection reagent alone, or dsDNA without transfection reagent (Supplemental Figure 2), confirming that dsDNA was required for assembly of this structure. Both plasmid DNA and Poly(dA:dT) resulted in identical IFI16-containing structures (Figure 2D and Supplemental Video 2). To confirm that this behavior of IFI16 was not specific to the HSG cell line, primary keratinocytes were treated and analyzed, with identical results. Of note, keratinocytes express higher basal levels of IFI16 and, therefore, did not require stimulation with IFN prior to DNA transfection to generate readily visible filaments (Supplemental Figure 3). Time course experiments showed that cytoplasmic IFI16 was first visible 2 hours after transfection with dsDNA (Supplemental Figure 4); distinct cytoplasmic structures and absent nuclear staining were visible by 6 hours after transfection and remained detectable at 24 hours. Transfection with rhodamine-labeled Poly(dA:dT) and imaging by confocal microscopy confirmed that IFI16 forms filaments in direct association with DNA in the cytoplasm (Figure 2D, Supplemental Video 2). Interestingly, when using confocal microscopy to isolate individual IFI16 filaments, we found that IFI16 protein filaments without identifiable costaining DNA could be found emanating from regions of IFI16-DNA interaction (Figure 3B). We next examined IFI16 filaments in human salivary glands for evidence of colocalizing DNA, using both DAPI and an anti-DNA antibody that recognizes nuclear and mitochondrial DNA (25). These stains failed to visualize DNA in IFI16-containing filamentous structures in 8 LSG samples (Figure 3A). While these data show that interaction between dsDNA and IFI16 induces IFI16 filament formation in vivo, our observations raise a possibility that, once assembled, the protein core of the IFI16 filament may persist within cells without dsDNA.
Figure 2. Generation of IFI16 cytoplasmic filaments in response to dsDNA transfection in epithelial cells in vitro.
(A–C) Human salivary gland (HSG) cells were treated with recombinant IFNα prior to transfection with empty plasmid DNA. Cells were then fixed, permeabilized, and stained with anti-IFI16 antibody (green) and DAPI (blue). Representative confocal images at 100× magnification are shown. Camera settings were held constant between experiments. (D) Keratinocyte cultures were transfected with Rhodamine labeled Poly(dA:dT) (red) and then stained for IFI16 (green) and counterstained with DAPI (blue). A 3-dimensional rendering of a Z-stack series is shown. (E) DNA titration was performed in HSG cultures using increasing concentrations of plasmid DNA, followed by staining with anti-IFI16 (green) and DAPI (blue). Cells with cytoplasmic IFI16 were counted in 4 fields imaged at 40×. Mean values with standard deviation are indicated. *P < 0.05; ***P < 0.0005; ****P < 0.0001 as assessed by the Mann-Whitney U test. Scale bars: 5 μM. Data are representative of results of 3 experiments.
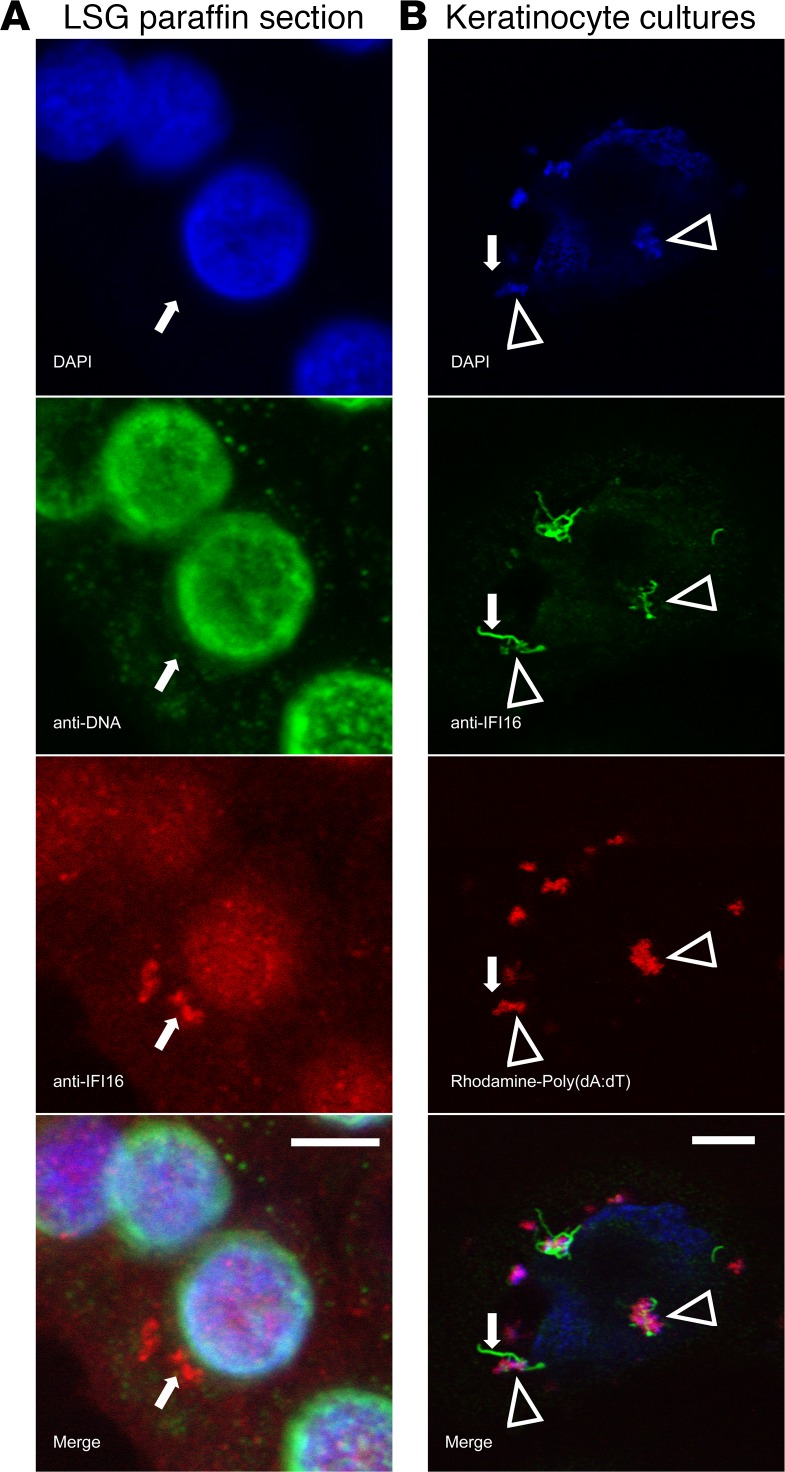
Figure 3. DNA colocalizes with IFI16 in the cytoplasm, but is not visualized in individual filaments ex vivo or in vitro.
(A) A representative ductal epithelial cell from a SS labial salivary gland (LSG) paraffin section was stained with DAPI (blue), anti-DNA monoclonal antibody (green), and anti-IFI16 monoclonal antibody (red). No DNA staining was detected in the cytoplasmic IFI16 containing structure. (B) Primary keratinocytes transfected with Rhodamine-labeled Poly(dA:dT) were stained with DAPI (blue) and anti-IFI16 (green). IFI16 was identified in association with DNA in large structures (arrowheads), but DNA was not visualized in an isolated IFI16 filament extending from a region of IFI16-DNA interaction (arrows). Scale bars: 5 μM.
DNA is susceptible to degradation by nuclease in the IFI16•dsDNA filament.
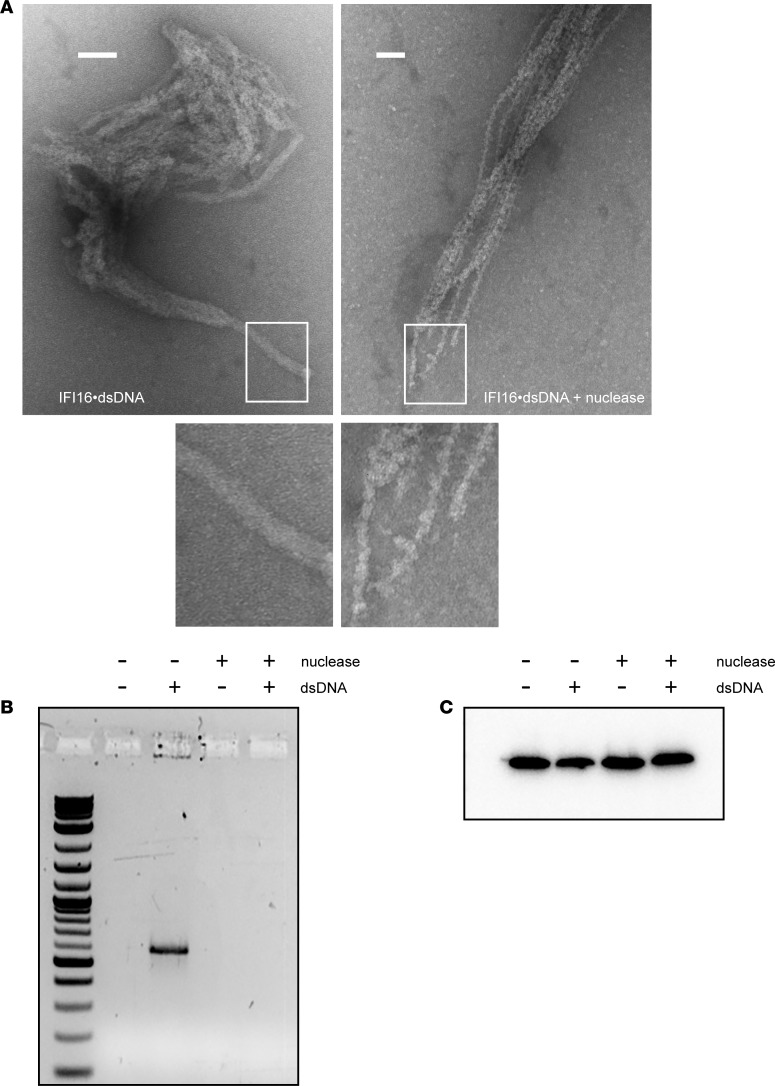
Because we did not observe IFI16 filaments in the absence of dsDNA transfection in cultured cells, and previous reports have established that dsDNA is required for filament formation in vitro (18, 26), we sought an explanation for the lack of DNA identified in IFI16 filaments in SS tissue samples. We reasoned that dsDNA might be accessible to nucleases and degraded within the nucleoprotein complex after its initial interaction with IFI16-induced filament formation. To test this idea, we generated IFI16•dsDNA filaments in vitro and visualized the samples by negative stain electron microscopy before and after treating them with a micrococcal endoexonuclease (Figure 4). The oligomerization efficiency of IFI16 is optimal at dsDNA length ≥150 (18), and we used 600 bp dsDNA in these experiments to permit imaging of larger structures by EM. Filaments were observed in both samples (Figure 4A). Strikingly, the filaments from nuclease-treated samples displayed narrower diameters than those without the treatment (8–11 nM vs. 20–25 nM). Nuclease-treated filaments demonstrated a central core fiber, with irregular protrusions emanating from this central core (Figure 4A). By contrast, the untreated samples showed smooth cylinder-like morphologies (Figure 4A). Our observations likely reflect dsDNA-free and dsDNA-bound HIN200 domains, respectively. Furthermore, agarose gel analyses of IFI16•dsDNA complexes with or without nuclease treatment confirmed dsDNA degradation (Figure 4B). Our results provide evidence that dsDNA is susceptible to degradation by nuclease in situ in the filament and that the protein component of the IFI16•dsDNA filament can persist even after the DNA template has been removed.
Figure 4. IFI16 protein filaments persist after nuclease treatment.
(A) Recombinant IFI16 was combined with dsDNA600 and then treated with micrococcal nuclease and imaged by negative stain electron microscopy. (B and C) Replicate samples were analyzed by agarose gel electrophoresis and stained with SYBR green to confirm effectiveness of nuclease treatment (B) and by SDS-PAGE and Western blotting for IFI16 (C). Scale bars: 100 nM.
Enhanced recognition of oligomeric IFI16 by SS antibodies.
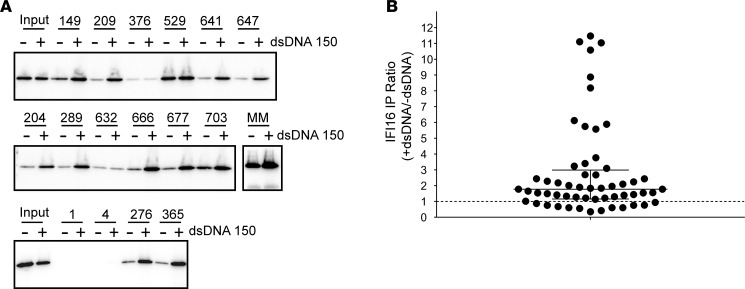
Since polyvalent molecules are particularly effective antigens, it is conceivable that the polymeric filamentous state of IFI16 that we demonstrated in minor salivary glands might be the form of IFI16 preferentially recognized by the autoantibody response in SS. To test this idea, we used anti-IFI16–positive SS sera to immunoprecipitate monomeric IFI16, as well as IFI16•dsDNA filaments (Figure 5A). The SS sera used were positive for anti-IFI16 antibodies, as determined by ELISA (14). We found that 23 of 56 (41%) sera demonstrated a 2-fold or greater enhancement in IFI16 immunoprecipitation in the presence of dsDNA (Figure 5B), while 14 of 56 (25%) had 3-fold or greater enhancement with dsDNA. The enhanced immunoprecipitation of IFI16 in the presence of DNA was not caused by anti-dsDNA antibodies, as all 56 sera were anti-DNA–negative. We also included a commercial monoclonal anti-IFI16 antibody raised against an N-terminal epitope within the pyrun domain (PYD) in this assay, and we found that its immunoprecipitation product increased only 1.4-fold with dsDNA (Figure 5A).
Figure 5. Autoantibody Immunoprecipitation of IFI16 is enhanced in the presence of dsDNA.
(A and B) Serum samples from 56 Sjögren’s Syndrome patients positive for IFI16 antibodies by ELISA were used to immunoprecipitate IFI16 in the absence or presence of 150 bp dsDNA in 10:1 IFI16/dsDNA molar ratio. Immunoprecipitation products were electrophoresed and Western Blotted with a commercial IFI16 antibody. (A) Representative data from immunoprecipitations performed with 16 patients’ sera and an N-terminal mouse monoclonal antibody (MM) are shown. Sera 1 and 4 were negative for anti-IFI16 antibodies by ELISA and were included as negative controls. Input lanes contain one-fourth protein used in each serum immunoprecipitation. (B) All 56 immunoprecipitations were quantified by densitometry. For each, the ratio of immunoprecipitated IFI16 detected in the presence versus absence of dsDNA was calculated. The dotted line indicates a ratio of 1, denoting unchanged immunoprecipitation with or without dsDNA. Median with interquartile range are indicated with solid lines.
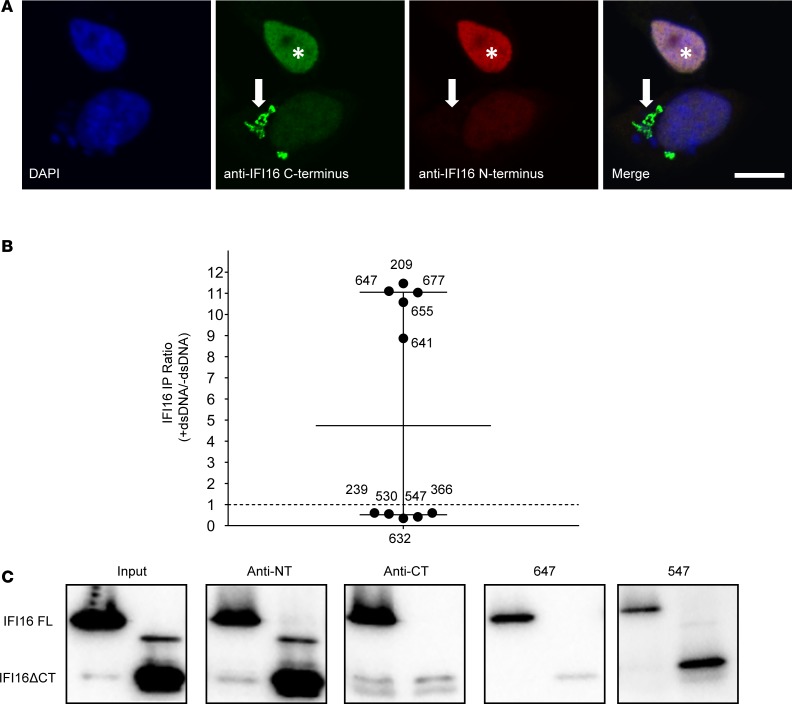
Previously published data indicate that most SS anti-IFI16 antibodies target the C-terminus of IFI16 (14). This observation prompted us to consider whether C-terminal epitopes are more accessible to antibodies when IFI16 is found in the IFI16•dsDNA filament. We therefore generated IFI16•dsDNA filaments in cultured cells and stained IFI16 with commercial antibodies directed against the N- and C-termini (Figure 6A). While both N-terminal and C-terminal antibodies stained nuclear monomeric IFI16, the N-terminal antibody was unable to bind cytoplasmic IFI16 filaments (Figure 6A), suggesting that its N-terminal epitope is masked in the filament. This result suggests that the human sera with dsDNA-induced enhancement by immunoprecipitation are likely to target C-terminal, rather than N-terminal epitopes.
Figure 6. C-terminal specificity of SS sera with property of IFI16-DNA binding.
(A) Cytoplasmic IFI16 filaments were induced in human salivary gland (HSG) cells and stained with commercial antibodies recognizing C-terminal (green) and N-terminal (red) epitopes. Both commercial antibodies recognize nuclear IFI16 (asterisk), but only the C-terminal antibody binds the cytoplasmic filament (arrow). (B) SS sera with or without the property of DNA-enhanced IFI16 immunoprecipitation were used to immunoprecipitate full-length IFI16 (FL) or an N-terminal fragment (IFI16ΔCT) lacking residues 597–729 at the C-terminus; data from Figure 5B specific to these 10 sera are shown. Commercial antibodies specific for the N-terminus (anti-NT) and C-terminus (anti-CT) were included. (C) Western blots were performed with anti-N terminal antibody. Blots were scanned by densitometry and the ratio of immunoprecipitated IFI16ΔCT/IFI16 FL in each was quantitated (Supplemental Table 2). Representative data obtained with 2 patient sera and the commercial antibodies is shown. Scale bar: 10 μM. Data are representative of results of 2 experiments.
To address this, we selected 5 SS sera with the highest DNA-induced immunoprecipitation enhancement and 5 sera without this property, and we determined their specificity for either the N- or C-terminus of the molecule (Figure 6, B and C, and Supplemental Table 2). We found that those sera with the property of dsDNA-induced immunoprecipitation enhancement target an epitope present in the C-terminus, in contrast to the sera without dsDNA-induced enhancement. Together, our results demonstrate that SS anti-IFI16 antibodies targeting the C-terminus of the molecule preferentially target the dsDNA-bound filamentous form of IFI16. Of note, this anti-C terminal specificity in SS is distinct from systemic lupus erythematosus (SLE) patients, in whom a majority of anti-IFI16 antibodies target epitopes in the N-terminus (14).
IFI16•dsDNA complexes are released from cells following CTL granule-induced cell death.
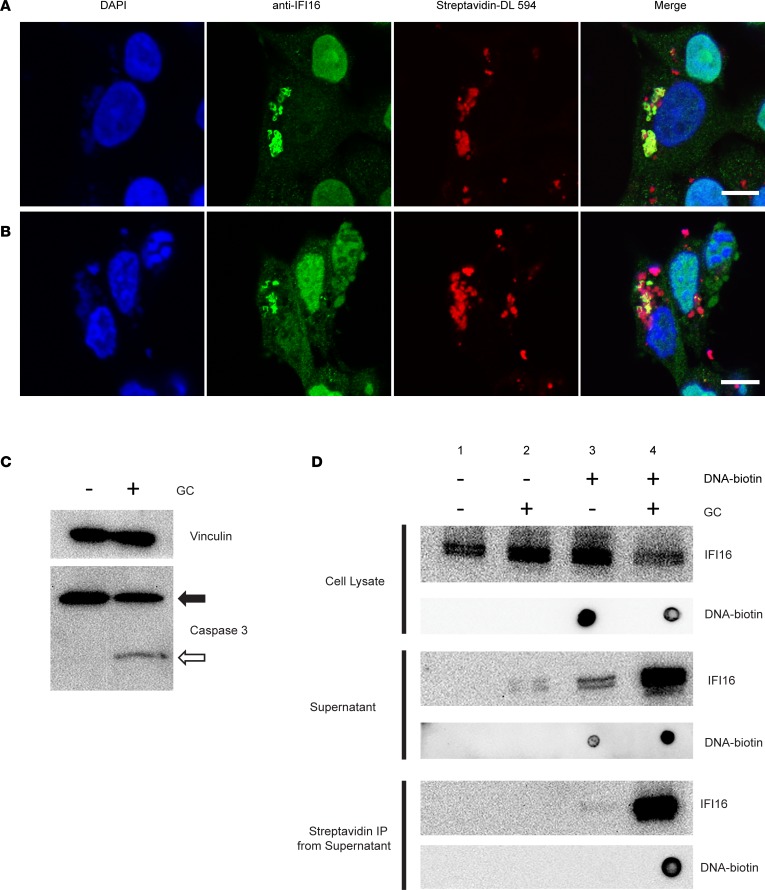
Our observation that IFI16 forms filaments in the affected salivary tissues combined with the ability of patient antibodies to bind filamentous IFI16•dsDNA oligomers prompted us to consider circumstances that might result in the release of these oligomers into the surrounding extracellular environment, where interaction with antigen presenting cells (and secondarily autoantibodies) could occur. CTLs and NK cells and their products have been implicated in the pathogenesis of SS; increased numbers of CTLs have been observed in the SS glands (27–29), along with enhanced expression of granzymes (30), supporting the notion that this immune cell subset may play an important pathogenic role in some SS patients (31). Given their membranolytic and cytotoxic functions, we hypothesized that exposure to the granule contents (GC) of CTLs might represent 1 stimulus responsible for the release of intracellular IFI16•dsDNA filaments from epithelial cells to the extracellular environment. We transfected IFN-primed HSGs with biotinylated DNA, inducing IFI16•dsDNA filaments (Figure 7A), and then treated cells with lytic amounts of YT cell GC (Figure 7B). Confocal microscopy demonstrated signs of apoptosis in treated cells, including pyknotic nuclei and disrupted cell membranes (Figure 7B). Immunoblotting for caspase 3 confirmed that GC treatment activated caspase-dependent cellular death under these conditions (Figure 7C). Cells and fragments were separated from supernatants by centrifugation, and both were analyzed by immunoblotting (Figure 7D). IFI16 was present in all cell lysates but decreased following GC treatment in dsDNA-transfected cells (lane 4, top panel, Figure 7D). Biotinylated DNA was identified only in transfected cells, and also decreased from the cellular fraction following GC treatment (lane 4, top panel, Figure 7D). Analysis of supernatants (middle panel, Figure 7D) revealed that the extracellular release of IFI16 was minimal in cells where IFI16 was exclusively nuclear (lanes 1 and 2) but increased strikingly after GC treatment in DNA-transfected cells (lane 4). In order to define whether the dsDNA and IFI16 were being released while still complexed together, we precipitated the supernatant fraction with streptavidin-coated beads to capture the transfected dsDNA, and we probed with anti-IFI16 antibody. Striking enrichment of IFI16 on the dsDNA-containing beads was seen (bottom panel, lane 4, Figure 7D), confirming that the complexes remain intact after GC-induced release. These results demonstrate that intact IFI16•dsDNA complexes, once formed intracellulary in epithelial cells, are released into the extracellular space following CTL-induced cell death. Following release from dying cells, IFI16 oligomers would be accessible for recognition by autoantibodies or susceptible to internalization by other immune cells such as DCs, B cells, or other antigen presenting cells.
Figure 7. IFI16•dsDNA filaments are released from epithelial cells following exposure to cytotoxic lymphocyte granule contents.
(A and B) IFN-treated human salivary gland (HSG) cells were transfected with biotinylated DNA to induce IFI16 filament formation and were then mock treated (A) or exposed to YT cell granule contents (GC) for 3 hours (B). Cells were fixed and stained with DAPI (blue), anti-IFI16 antibody (green), and Streptavidin-DyLight 594 (red). (C) Induction of apoptosis by GC treatment was confirmed by Western blotting for caspase 3, demonstrating intact (closed arrow) and cleaved (open arrow) forms. (D) Following GC treatment, samples of cell lysate (top panel) and supernatant (middle panel) were collected and analyzed in parallel for IFI16 and DNA content by blotting with anti-IFI16 antibody and StrepTactin-HRP. Biotinylated DNA was isolated from the supernatant of treated cells using Streptavidin DynaBeads, and the presence of IFI16•dsDNA interaction in the supernatant was confirmed by blotting for each (D, bottom panel). Scale bars: 10 μM. Data are representative of results of 3 separate experiments.
Discussion
This study provides morphologic evidence that IFI16 forms filaments in human tissues and cells. The property of filament formation has been recognized by several protein components of the innate immune system, including the ALRs IFI16 and AIM2, the inflammasome adaptor ASC, and MAVS, RIG-I, and MDA5 in the RIG-I pathway (16). Our observation of this phenomenon in SS patient samples demonstrates that filamentous assembly occurs in vivo, where it is likely tied to IFI16 function and immunogenicity. We found that IFI16 filaments were limited exclusively to epithelial cells of the ducts within the salivary glands, suggesting that this cell layer is a site of activation of the cytoplasmic DNA sensing pathway in the labial salivary gland. In contrast, mucosal epithelial cells expressed nuclear IFI16 without evidence of cytoplasmic filamentation. Salivary epithelial cells have received attention as active participants in SS pathogenesis (32), and our data indicate that IFI16 is another component of the innate immune system that appears activated in these cells.
We found that cytoplasmic IFI16 filaments assemble rapidly in cells following DNA transfection, appearing fully formed within a few hours of transfection. Once formed, the protein element of the filament persisted even after dsDNA had been removed, revealing the durability of this oligomeric structure. These data indicate that DNA serves as a template for initial filamentous assembly of IFI16, but the protein-protein interactions within the IFI16 filament dominate overall assembly. This is consistent with the previous finding that filament assembly is achieved by the non-dsDNA binding PYD (18). The morphology of the nuclease-treated IFI16 filaments was noted to be similar to the previously observed “Brussels sprout” structure of dsDNA-free AIM2 filaments, which form without a dsDNA template in a concentration-dependent manner (33). In the case of AIM2, the central core fiber is composed of PYDs, with unbound HIN domains accounting for peripheral elements of the structure. Our current data show that IFI16 generates an essentially identical conformation following nuclease treatment, consisting of a core intact fiber flanked by irregular protrusions that are presumed to also represent dsDNA-free HIN domains.
Cytoplasmic dsDNA sensing by IFI16 has been implicated in various innate immune responses, ranging from augmenting IFN pathways to inducing cell death (34–38). The observation that IFI16 filaments in SS glands lack DNA and that filaments persist in vitro after nuclease treatment suggests that IFI16 can persist in the active state even if the initial signal has been removed. This persistence of filamentous IFI16 within cells represents a memory of innate signaling events that may influence cellular function and, ultimately, cell fate. For example, it is tempting to speculate that persisting IFI16 filaments may contribute to sustained IFN signaling in the labial salivary gland in SS. Defining the mechanisms responsible for the termination of the signaling from the ALR family once oligomerization is induced, and the relevance of such pathways in diseases like SS with prominent evidence of type I IFN signaling, is an important priority.
A variety of protein modifications have been implicated in the loss of tolerance to self-antigens (39), including citrullination (40), phosphorylation (41), and protease-mediated cleavage (42). Many autoantigens targeted by antibodies in rheumatic diseases, including SS and SLE, are also nucleic acid–binding proteins (43). With these facts in mind, the discovery of IFI16 filaments in human salivary glands prompted us to consider whether IFI16 oligomerization upon sensing dsDNA was integral to its status as an autoantigen. We hypothesized that DNA-induced oligomerization exposes epitopes (likely repeated) that are targeted by anti-IFI16 antibodies in SS. Indeed, we found that 41% of SS patients exhibited ≥2-fold increased IFI16 binding when IFI16 was oligomerized in IFI16•dsDNA filaments (Figure 5B). Our results identify oligomerization as a mode of endogenous protein modification that may contribute to the antigenicity of native proteins. This property of oligomerization may be operative in the selection of autoantigens in other rheumatic diseases — such as the RNA sensor MDA5, which is targeted by antibodies in a subset of patients with dermatomyositis (44) and forms oligomeric filaments when activated by RNA (45). At the level of the B cell, oligomerization of autoantigens may generate novel epitopes unique to the oligomer (e.g., monomer-monomer interfaces) or might create an array of repeating epitopes, increasing avidity.
How might IFI16 filaments persist in salivary epithelial cells in SS? We envision 2 potential mechanisms. First, it is possible that the oligomerization of IFI16 masks ubiquitination sites that are required for proteasomal degradation. Alternatively, it has been suggested that large innate immune signaling platforms are recycled through autophagy (46–48), and it is possible that this cellular pathway is dysfunctional in SS. Data regarding the expression of autophagy proteins in SS tissues and cells are currently limited (49). Interestingly, the mitochondrial antiviral signaling protein MAVS, which forms prion-like aggregates in response to dsRNA sensing by RIG-I proteins, has been identified in high molecular weight aggregates in the peripheral blood cells of patients with SLE, where its aggregated form correlated with the expression of IFN-induced genes (50). This observation provides further evidence that impaired clearance of oligomerizing innate signaling proteins may contribute to pathogenic immune activation in rheumatic diseases. Additional study of SS tissues may determine whether the presence of filamentous IFI16 is linked to specific phenotypic features, such as the presence of a type I IFN signature.
We found that N-terminal IFI16 epitopes appear inaccessible to antibody binding in the IFI16•dsDNA filament (Figure 6A). Interestingly, our previous study of anti-IFI16 antibodies in SS and SLE revealed a difference in the predominant epitopes targeted in these 2 diseases; SS patients more frequently targeted C-terminal epitopes, while SLE sera mainly targeted N-terminal IFI16 epitopes (14). Of note, we found that SS sera with the ability to immunoprecipitate IFI16•dsDNA filaments target epitopes within the C-terminus, in contrast to SS sera without dsDNA-induced enhancement in the immunoprecipitation assay. Together, these observations support the notion that stimulus-specific structure and state of autoantigens in the target tissue play a role in shaping the autoreactive immune response in specific diseases (51). In this manner, ongoing oligomerization of IFI16 in salivary epithelial cells may yield antigenic IFI16 structures that propagate the immune response and downstream effector functions in the target tissue. Furthermore, binding to IFI16•dsDNA oligomers by anti-IFI16 antibodies provides a mechanism for coligating antigen/Fc receptors and TLRs, potentially augmenting immune responses in a manner analogous to the mechanism that has been described with other nucleic acid–binding autoantigens (52, 53). Such an amplifying role for IFI16 antibodies is consistent with the expression of more severe SS disease manifestations, including high focus score on lip biopsy, in anti-IFI16–positive patients (14).
IFI16, like most autoantigens in the rheumatic diseases (54), is expressed intracellularly, raising questions about how the filamentous form of the antigen might engage the amplification pathways noted above. CTLs and NK cells are enriched in the SS salivary gland and have been implicated in the pathogenesis of this disease (27–31). We were particularly interested in whether the granule-mediated cytotoxicity pathway (which can cause membrane lesions through the delivery of perforin) might cause the release of filamentous IFI16 from cells. When IFI16 was present as cytoplasmic filaments, exposure to GC led to the striking release of IFI16•dsDNA complexes from cells (Figure 7). These experiments suggest a pathogenic model where CTL- and NK-mediated actions on ductal epithelial cells containing IFI16 filaments leads to release of these filaments into the surrounding extracellular space, making these available for antigen presentation in disease initiation and amplifying immune responses through autoantibody binding in the propagation phase of the disease. This model suggests an interacting, positively reinforcing immune process, where cytoplasmic DNA sensing, type I and II IFNs, and immune-mediated cytotoxicity pathways focused on the glandular epithelium promote anti-IFI16 autoimmunity and tissue damage. Future study of this process of filament release will determine whether extracellular IFI16•dsDNA oligomers are also capable of propagating IFN or other inflammatory signaling pathways, similar to the observed behavior of inflammasome components (55, 56).
The identity of the dsDNA being sensed in salivary epithelial cells in SS remains unknown, but both exogenous and endogenous are possibilities (37, 57–60). Retroelements represent an alternate source of endogenous nucleic acid and have been identified as potential drivers of the IFN signature in SLE and SS (61, 62). Further, it is possible that stimuli other than nucleic acid may induce oligomerization of IFI16 without a DNA template; the closely related cytoplasmic DNA sensor AIM2 has been shown to have the capacity to auto-oligomerize without a DNA template at high concentrations (33). Additional studies are required to identify the mechanisms responsible for inducing IFI16 filament formation in SS in vivo.
Our findings provide compelling evidence that (a) the IFN-induced dsDNA sensor IFI16 undergoes oligomerization in salivary ductal epithelial cells in SS, (b) IFI16 autoantibodies in SS patient sera demonstrate enhanced binding to IFI16 oligomers (this is most striking in those directed against the C-terminus), and (c) filamentous IFI16 is released into the supernatant by epithelial cells that have been exposed to CTL granules. Together, they suggest that the combination of IFN activity, filamentation, and immune-mediated cytotoxicity may contribute to the anti-IFI16 immune response in SS. Our findings establish a link between activation of elements of the innate immune system and the anti-self humoral immune response in SS. Further study of this interplay may lead to better understanding of the pathogenesis of SS and related rheumatic diseases.
Methods
Patients and tissues.
Minor salivary gland biopsies were obtained with informed consent from patients undergoing clinical evaluation in the Johns Hopkins Jerome L. Greene Sjogren’s Center. Patients were diagnosed with SS based on the revised American-European classification criteria (63). Control subjects were referred for evaluation due to clinical signs or symptoms of SS but did not meet criteria for diagnosis and had negative salivary gland biopsies.
Cell culture.
Cells from a HSG cell line (64) (a gift from Baum, NIH/National Institute of Dental and Craniofacial Research, Bethesda, Maryland, USA), were cultured as previously described (2). Primary keratinocytes were purchased from Lonza and cultured per the vendor’s instructions. Cells were treated for 24 hours with recombinant IFNα 2a (1000 U/ml) and IFNγ (50 ng/ml) (PBL Assay Science). Transfection with empty plasmid DNA, Poly(I:C), or Poly(dA:dT) (Invivogen) was performed using Lipofectamine 2000 (Thermo Fisher Scientific) per the manufacturer’s instructions.
IHC.
Paraffin sections were processed for IHC as previously described (2) and stained with primary anti-IFI16 antibody overnight (1:75 dilution, MilliporeSigma, IFI-230). Sections were then washed in Tris buffered saline and incubated with HRP-conjugated anti-mouse secondary antibody (1:500 dilution, Dako). Staining was visualized using the diaminobenzidine substrate chromagen system (Dako). Nuclei were stained using Mayer’s hematoxylin solution (Dako).
Immunofluorescence.
Paraffin sections were deparaffinized and rehydrated as for IHC and then blocked with 5% BSA/PBS. Primary antibodies were diluted in 1% BSA/PBS and incubated overnight at 4°C. These included IFI16 (1:75 dilution, MilliporeSigma, IFI-230), E-cadherin (1:100 dilution, MilliporeSigma, EP700Y) and DNA (1:75 dilution, MilliporeSigma, AC-30-10). After washing in PBS, sections were incubated with Alexa-Fluor–labeled secondary antibodies (1:200 dilution, 1 hour at room temperature [RT]). Slides were mounted with DAPI containing ProLong Gold Antifade Mountant (Thermo Fisher Scientific).
Cultured cells were plated on coverslips and treated with IFN and DNA as described as above. Cells were fixed in 4% paraformaldehyde/PBS and were then permeabilized in ice cold acetone and blocked with 5% BSA/PBS. Primary antibody incubations were performed (1 hour, 4°C) using N-terminal IFI16 mouse monoclonal antibody (Santa Cruz Biotechnology Inc., 1G7) and C-terminal antibodies (MilliporeSigma, I1659; Santa Cruz Biotechnology Inc., C-18), both diluted 1:100 in 1% BSA/PBS. After washing, cells were incubated with Alexa-Fluor–labeled secondary antibodies (1:200 dilution, 30 minutes RT), washed, and mounted with DAPI containing ProLong Gold Antifade Mountant (Thermo Fisher Scientific).
Microscopy.
Fluorescence and light microscopy images were obtained using a Zeiss Axioskop 50 with a Zeiss AxioCam HRC camera and AxioVision 4.9.1 software. Confocal imaging was performed with Zeiss Axiovert 200 inverted microscope with LSM510-Meta confocal module. Confocal images were acquired using Zen software. Three-dimensional rendering of confocal Z stack images was performed using Imaris 7.7 software.
IFI16 immunoprecipitation and nuclease treatment.
Recombinant IFI16 (isoform 2) protein was generated as previously described, as both full-length protein and as a C-terminal truncation lacking amino acids 597-721 (18). All incubations were performed at RT. IFI16 protein was incubated with 150 bp dsDNA in a 10:1 molar ratio (20 minutes); then, 160 ng of protein was immunoprecipitated with 1 μl of human serum for 2 hours. Protein G DynaBeads (25 μl) were added and incubated for an additional hour and were then washed and boiled in gel application buffer. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting as previously described (2) using anti-IFI16 antibody (Santa Cruz Biotechnology Inc., 1G7, 1:1,000 dilution). For nuclease treatment, IFI16 and dsDNA were combined as above and were then treated with micrococcal nuclease (New England Biolabs) at a concentration of 1 × 105 units/ml (30 minutes). Nuclease activity was confirmed by agarose gel electrophoresis. Samples were analyzed by negative stain electron microscopy as previously described (18).
IFI16 ELISA.
SS sera were assayed for anti-IFI16 antibodies using an ELISA as previously described (14).
CTL granule–induced cell death and immunoprecipitation of biotinylated DNA.
YT cell GC were applied to HSG cultures as previously described (65). Briefly, HSGs grown on coverslips were treated with IFNα 2a for 24 hours and were then transfected for 6 hours with plasmid DNA labeled with PHOTOPROBE biotin (Vector Laboratories). Cells were washed in PBS and were then treated with DMEM + 1.0 mM calcium with or without GC for 3 hours. The supernatant and cell lysate fractions were then collected for analysis. For supernatant immunoprecipitation, T1 Streptavidin DynaBeads (Thermo Fisher Scientific) were incubated with the supernatant from 1 coverslip at RT for 1 hour and were then washed 3 times with PBS-T. On the final wash, the beads were divided into 2 equal aliquots for parallel analysis of DNA-protein interaction: one aliquot was eluted into a solution of 95% formamide + 10mM EDTA for analysis of isolated DNA, and the other eluted in 2× GAB for analysis of associated protein. DNA products were spotted onto nylon membranes and probed with StrepTactin-HRP (Bio-Rad). Proteins that coprecipitated with the streptavidin beads were analyzed by SDS-PAGE and immunoblotted as described above. Apoptotic effect of GC treatment was confirmed by Western blotting of cell lysates for caspase 3 using mouse monoclonal 3G2 (Cell Signaling Technologies). Biotinylated DNA was visualized in immunofluorescence assays using Streptavidin-DyLight 594 (Thermo Fisher Scientific).
Statistics.
Statistical analysis was performed using GraphPad Prism software. The Mann-Whitney U test was used to compare counts of cytoplasmic IFI16 filaments in Figure 2E, and P < 0.05 was considered significant.
Study approval.
IRB approval was obtained for the acquisition of blood and tissue from patients evaluated at the Johns Hopkins Jerome L. Greene Sjögren’s Center. Written informed consent was obtained from patients prior to their enrollment in the study.
Author contributions
BA designed research studies, conducted experiments, analyzed data, and wrote the manuscript. MM designed research studies, conducted experiments, analyzed data, and provided reagents. JS designed research studies, analyzed data, provided reagents, and wrote the manuscript. LCR designed research studies, analyzed data, wrote the manuscript, and provided mentorship of BA. AR designed research studies, analyzed data, wrote the manuscript, and provided mentorship of BA.
Supplementary Material
Acknowledgments
These studies were funded by NIH T32 AR48522, NIH RO1 DE12354 (AR), P30 AR070252, and the Jerome L. Greene Foundation. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award no. T32AR048522. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Antiochos is a Jerome L. Greene Scholar. The authors would like to thank the Johns Hopkins School of Medicine Microscope Core Facility for their invaluable assistance with the microscopy experiments performed in this study. The authors thank reviewer no. 3 for constructive review of the manuscript, which led to the experiments performed in Figure 7.
Version 1. 09/20/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(18):e120179. https://doi.org/10.1172/jci.insight.120179.
Contributor Information
Brendan Antiochos, Email: bantioc1@jhmi.edu.
Mariusz Matyszewski, Email: mmatysz1@jhu.edu.
Jungsan Sohn, Email: jsohn@jhmi.edu.
Livia Casciola-Rosen, Email: lcasciol@jhmi.edu.
Antony Rosen, Email: arosen@jhmi.edu.
References
- 1.Ramos-Casals M, Brito-Zerón P, Sisó-Almirall A, Bosch X. Primary Sjogren syndrome. BMJ. 2012;344:e3821. doi: 10.1136/bmj.e3821. [DOI] [PubMed] [Google Scholar]
- 2.Hall JC, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc Natl Acad Sci USA. 2012;109(43):17609–17614. doi: 10.1073/pnas.1209724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nezos A, et al. Type I and II interferon signatures in Sjogren’s syndrome pathogenesis: Contributions in distinct clinical phenotypes and Sjogren’s related lymphomagenesis. J Autoimmun. 2015;63:47–58. doi: 10.1016/j.jaut.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottenberg JE, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjögren’s syndrome. Proc Natl Acad Sci USA. 2006;103(8):2770–2775. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavragani CP, Crow MK. Activation of the type I interferon pathway in primary Sjogren’s syndrome. J Autoimmun. 2010;35(3):225–231. doi: 10.1016/j.jaut.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. Intracellular Nucleic Acid Detection in Autoimmunity. Annu Rev Immunol. 2017;35:313–336. doi: 10.1146/annurev-immunol-051116-052331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 8.Ori D, Murase M, Kawai T. Cytosolic nucleic acid sensors and innate immune regulation. Int Rev Immunol. 2017;36(2):74–88. doi: 10.1080/08830185.2017.1298749. [DOI] [PubMed] [Google Scholar]
- 9.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54(2):289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 10.Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46(2):269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao D, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci USA. 2015;112(42):E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fayyaz A, Kurien BT, Scofield RH. Autoantibodies in Sjögren’s Syndrome. Rheum Dis Clin North Am. 2016;42(3):419–434. doi: 10.1016/j.rdc.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alunno A, et al. Interferon gamma-inducible protein 16 in primary Sjögren’s syndrome: a novel player in disease pathogenesis? Arthritis Res Ther. 2015;17:208. doi: 10.1186/s13075-015-0722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baer AN, Petri M, Sohn J, Rosen A, Casciola-Rosen L. Association of Antibodies to Interferon-Inducible Protein-16 With Markers of More Severe Disease in Primary Sjögren’s Syndrome. Arthritis Care Res (Hoboken) 2016;68(2):254–260. doi: 10.1002/acr.22632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida K, et al. Identification of specific autoantigens in Sjögren’s syndrome by SEREX. Immunology. 2005;116(1):53–63. doi: 10.1111/j.1365-2567.2005.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sohn J, Hur S. Filament assemblies in foreign nucleic acid sensors. Curr Opin Struct Biol. 2016;37:134–144. doi: 10.1016/j.sbi.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cridland JA, et al. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci USA. 2014;111(1):E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratmann SA, Morrone SR, van Oijen AM, Sohn J. The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex. Elife. 2015;4:e11721. doi: 10.7554/eLife.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondini M, Costa S, Sponza S, Gugliesi F, Gariglio M, Landolfo S. The interferon-inducible HIN-200 gene family in apoptosis and inflammation: implication for autoimmunity. Autoimmunity. 2010;43(3):226–231. doi: 10.3109/08916930903510922. [DOI] [PubMed] [Google Scholar]
- 21.Choubey D. DNA-responsive inflammasomes and their regulators in autoimmunity. Clin Immunol. 2012;142(3):223–231. doi: 10.1016/j.clim.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanhove W, et al. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21(11):2673–2682. doi: 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci USA. 2012;109(26):10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brkic Z, Versnel MA. Type I IFN signature in primary Sjögren’s syndrome patients. Expert Rev Clin Immunol. 2014;10(4):457–467. doi: 10.1586/1744666X.2014.876364. [DOI] [PubMed] [Google Scholar]
- 25.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusakiewicz S, et al. NCR3/NKp30 contributes to pathogenesis in primary Sjogren’s syndrome. Sci Transl Med. 2013;5(195):195ra96. doi: 10.1126/scitranslmed.3005727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christodoulou MI, Kapsogeorgou EK, Moutsopoulos HM. Characteristics of the minor salivary gland infiltrates in Sjögren’s syndrome. J Autoimmun. 2010;34(4):400–407. doi: 10.1016/j.jaut.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Ciccia F, et al. Potential involvement of IL-22 and IL-22-producing cells in the inflamed salivary glands of patients with Sjogren’s syndrome. Ann Rheum Dis. 2012;71(2):295–301. doi: 10.1136/ard.2011.154013. [DOI] [PubMed] [Google Scholar]
- 30.Alpert S, Kang HI, Weissman I, Fox RI. Expression of granzyme A in salivary gland biopsies from patients with primary Sjögren’s syndrome. Arthritis Rheum. 1994;37(7):1046–1054. doi: 10.1002/art.1780370710. [DOI] [PubMed] [Google Scholar]
- 31.Seror R, et al. Low numbers of blood and salivary natural killer cells are associated with a better response to belimumab in primary Sjögren’s syndrome: results of the BELISS study. Arthritis Res Ther. 2015;17:241. doi: 10.1186/s13075-015-0750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manoussakis MN, Kapsogeorgou EK. The role of intrinsic epithelial activation in the pathogenesis of Sjögren’s syndrome. J Autoimmun. 2010;35(3):219–224. doi: 10.1016/j.jaut.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, Sohn J. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun. 2015;6:7827. doi: 10.1038/ncomms8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ansari MA, et al. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J Virol. 2013;87(15):8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orzalli MH, et al. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci USA. 2015;112(14):E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monroe KM, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343(6169):428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zavala-Cerna MG, Martínez-García EA, Torres-Bugarín O, Rubio-Jurado B, Riebeling C, Nava A. The clinical significance of posttranslational modification of autoantigens. Clin Rev Allergy Immunol. 2014;47(1):73–90. doi: 10.1007/s12016-014-8424-0. [DOI] [PubMed] [Google Scholar]
- 40.Darrah E, Andrade F. Rheumatoid arthritis and citrullination. Curr Opin Rheumatol. 2018;30(1):72–78. doi: 10.1097/BOR.0000000000000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terzoglou AG, Routsias JG, Avrameas S, Moutsopoulos HM, Tzioufas AG. Preferential recognition of the phosphorylated major linear B-cell epitope of La/SSB 349-368 aa by anti-La/SSB autoantibodies from patients with systemic autoimmune diseases. Clin Exp Immunol. 2006;144(3):432–439. doi: 10.1111/j.1365-2249.2006.03088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casciola-Rosen L, Andrade F, Ulanet D, Wong WB, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J Exp Med. 1999;190(6):815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann MH, Trembleau S, Muller S, Steiner G. Nucleic acid-associated autoantigens: pathogenic involvement and therapeutic potential. J Autoimmun. 2010;34(3):J178–J206. doi: 10.1016/j.jaut.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Nakashima R, et al. The RIG-I-like receptor IFIH1/MDA5 is a dermatomyositis-specific autoantigen identified by the anti-CADM-140 antibody. Rheumatology (Oxford) 2010;49(3):433–440. doi: 10.1093/rheumatology/kep375. [DOI] [PubMed] [Google Scholar]
- 45.Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA. 2012;109(45):18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155(3):688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris J, Lang T, Thomas JPW, Sukkar MB, Nabar NR, Kehrl JH. Autophagy and inflammasomes. Mol Immunol. 2017;86:10–15. doi: 10.1016/j.molimm.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Cheng J, et al. Autophagy regulates MAVS signaling activation in a phosphorylation-dependent manner in microglia. Cell Death Differ. 2017;24(2):276–287. doi: 10.1038/cdd.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alessandri C, et al. CD4 T lymphocyte autophagy is upregulated in the salivary glands of primary Sjögren’s syndrome patients and correlates with focus score and disease activity. Arthritis Res Ther. 2017;19(1):178. doi: 10.1186/s13075-017-1385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao WH, et al. Prion-like Aggregation of Mitochondrial Antiviral Signaling Protein in Lupus Patients Is Associated With Increased Levels of Type I Interferon. Arthritis Rheumatol. 2016;68(11):2697–2707. doi: 10.1002/art.39733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosen A, Casciola-Rosen L. Autoantigens in systemic autoimmunity: critical partner in pathogenesis. J Intern Med. 2009;265(6):625–631. doi: 10.1111/j.1365-2796.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Båve U, et al. Activation of the type I interferon system in primary Sjögren’s syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52(4):1185–1195. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 53.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6(11):823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosen A, Casciola-Rosen L. Autoantigens as Partners in Initiation and Propagation of Autoimmune Rheumatic Diseases. Annu Rev Immunol. 2016;34:395–420. doi: 10.1146/annurev-immunol-032414-112205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baroja-Mazo A, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15(8):738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 56.Franklin BS, et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat Immunol. 2014;15(8):727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen K, et al. Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33(15):1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horan KA, et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190(5):2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jakobsen MR, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110(48):E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J Immunol. 2015;194(7):3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hung T, et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350(6259):455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mavragani CP, et al. Expression of Long Interspersed Nuclear Element 1 Retroelements and Induction of Type I Interferon in Patients With Systemic Autoimmune Disease. Arthritis Rheumatol. 2016;68(11):2686–2696. doi: 10.1002/art.39795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vitali C, et al. Classification criteria for Sjögren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61(6):554–558. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu AJ, Kurrasch RH, Katz J, Fox PC, Baum BJ, Atkinson JC. Effect of tumor necrosis factor-alpha and interferon-gamma on the growth of a human salivary gland cell line. J Cell Physiol. 1994;161(2):217–226. doi: 10.1002/jcp.1041610205. [DOI] [PubMed] [Google Scholar]
- 65.Nagaraju K, Cox A, Casciola-Rosen L, Rosen A. Novel fragments of the Sjögren’s syndrome autoantigens alpha-fodrin and type 3 muscarinic acetylcholine receptor generated during cytotoxic lymphocyte granule-induced cell death. Arthritis Rheum. 2001;44(10):2376–2386. doi: 10.1002/1529-0131(200110)44:10<2376::AID-ART402>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.