Abstract
Clonal expansion of T cells harboring replication-competent virus has recently been demonstrated in patients on suppressive antiretroviral therapy (ART) regimens. However, there has not been direct evidence of this phenomenon in settings of natural control, including in posttreatment controllers who maintain control of viral replication after treatment when ART is discontinued. We present a case of an individual who has had undetectable viral loads for more than 15 years following the cessation of ART. Using near-full-genome sequence analysis, we demonstrate that 9 of 12 replication-competent isolates cultured from this subject were identical and that this identity was maintained 6 months later. A similar pattern of replication-competent virus clonality was seen in a treatment-naive HLA-B*57 elite controller. In both cases, we show that CD8+ T cells are capable of suppressing the replication of the clonally expanded viruses in vitro. Our data suggest that, while clonal expansion of replication-competent virus can present a barrier to viral eradication, these viral isolates remain susceptible to HIV-specific immune responses and can be controlled in patients with long-term suppression of viral replication.
Keywords: AIDS/HIV
Keywords: AIDS vaccine, T cells
Two subjects who control HIV without antiretrovirals have CD8+ T cells that can inhibit clonally expanded HIV-infected CD4+ T cells.
Introduction
Recent studies have demonstrated clonal expansion of plasma virus (1, 2) and CD4+ T cells containing provirus (3–6) and replication-competent (RC) virus (7–10) in patients on antiretroviral therapy (ART). This clonal expansion may represent a barrier to viral eradication. Hosmane and colleagues demonstrated that infected CD4+ T cells can divide without producing RC virus (9), potentially leading to expansion of the viral reservoir without the potential for recognition and elimination by the immune system. Posttreatment controllers (PTCs) (11–13) maintain undetectable viral loads following the discontinuation of ART, whereas elite suppressors (ESs) and viremic controllers (VCs) are treatment-naive subjects who maintain viral loads that are either undetectable (ES) or are detectable but at lower than 2,000 copies/ml (VCs) (14). While a subset of ESs and VCs have protective HLA alleles (15, 16) and robust HIV-specific CD8+ T cell responses that control viral replication (17–20), these features are not present in all HIV controllers, and additional mechanisms are likely operative in PTCs, in whom the previously defined genetic protective features are typically absent. A recent study used integration site analysis to show clonal expansion in an ES and a VC, but it is not clear whether these viruses were defective or RC clones (21). Thus, we looked for evidence of clonal expansion of RC virus in an ES to determine whether clonally expanded virus could be controlled by the immune system. We also looked for evidence of clonal expansion of RC virus in a previously described PTC who has maintained viral loads below the limits of detection for more than 15 years (22). The results presented here have implications for the development of HIV– cure strategies.
Results
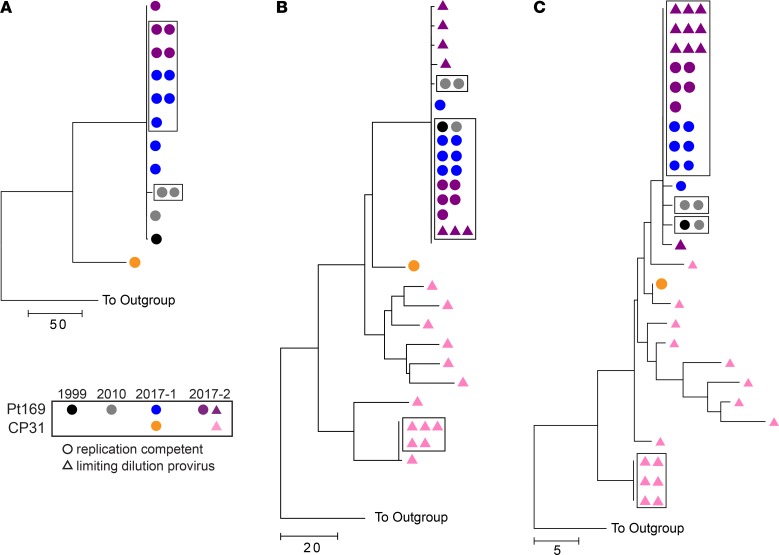
Patient 169 (Pt169) is a previously described PTC who was started on ART in primary infection when he presented with signs of viral encephalitis in 1999. He stopped ART in 2002 and has maintained undetectable viral loads since that time (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122795DS1). In 2010, 2 of the 3 RC viruses isolated from his CD4+ T cells were identical. We sought to isolate additional viral clones to look for evidence of viral evolution 7 years later. Near-full-length sequence analysis revealed that 5 of 7 isolates cultured at time point 1 in 2017 were identical (Figure 1A). Notably, the majority clone from 2017 was also 99.9% identical to one of the 2010 RC isolates (2 nucleotide changes, 1 in gag and 1 in nef; Supplemental Table 1). Five additional isolates were cultured from Pt169 at time point 2 (6 months later), four of which were identical to the majority clone seen at time point 1 (Figure 1A and Supplemental Table 1). We also analyzed a RC clone from CP31, the transmitting partner of Pt169. This virus was closely related but not identical to the expanded viral clone in Pt169 (Figure 1A). We subsequently amplified envelope (env) and nef proviral clones by limiting dilution from purified CD4+ T cells. Sequence analysis revealed that some of these proviral clones were identical to the majority RC virus clone (Figure 1, B and C). Proviral sequence analysis revealed expansion of env and nef clones in CP31, but these clones were distinct from the majority clones present in Pt169 (Figure 1, B and C).
Figure 1. Large degree of clonality in Pt169 replication-competent and proviral full-length env and nef sequences.
Phylogenetic trees of near-full-length replication-competent virus (A) and full-length env (B) and nef (C) from either proviral clones (triangles) or replication-competent virus (circles). Boxes indicate identical sequences.
To further investigate the question of clonal expansion in the context of virologic control, we investigated whether clonal expansion of RC viruses was also present in ESs, as these subjects appear to have a different mechanism of control. We have previously shown that, unlike the majority of ESs who have a very low frequency of latently infected CD4+ T cells (23), subject ES24 had a relatively large viral reservoir (IUPM 4.57; ref. 24) that was amenable to further investigation. We therefore attempted to culture multiple RC viruses from this subject. Seven RC viruses were isolated by limiting dilution, and near-full-length sequence analysis revealed that all 7 isolates were identical and differed from an isolate cultured in 2013 by only 2 nucleotides (1 env and 1 gag; Figure 2A and Supplemental Table 2). Strikingly, all proviral env and nef clones (14 and 10, respectively) that were amplified by limiting dilution from purified CD4+ T cells were also identical to the RC isolates (Figure 2, B and C).
Figure 2. Large degree of clonality in ES24 replication-competent and proviral full-length env and nef sequences.
Phylogenetic trees of near-full-length replication-competent virus (A) and full-length env (B) and nef (C) from either proviral clones (triangles) or replication-competent virus (circles). Boxes indicate identical sequences.
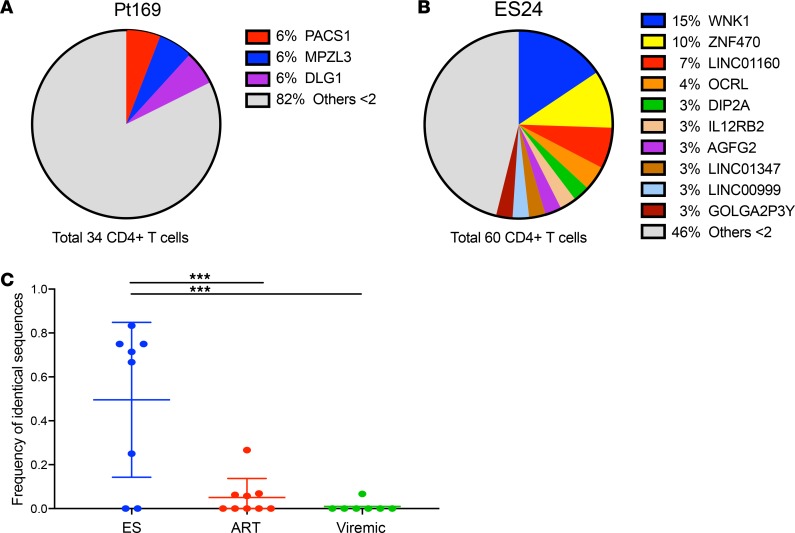
While the presence of identical sequences could theoretically be due to seeding of dominant viral clones into different CD4+ T cells during primary infection in both subjects, a recent study that analyzed full-genome sequences from patients with acute HIV-1 infection found that proviruses in this stage were extremely similar but not identical (25). Nonetheless, to obtain further evidence of clonal expansion, we performed integration site analysis on DNA from CD4+ T cells from both subjects obtained in 2017 as previously described (26–28). Integration sites from an estimated 34 and 60 infected CD4+ T cells were analyzed from Pt169 and ES24, respectively. There were 2 identical integration events each in the PACS1, MPZL3, and DLG1 genes in Pt169, providing evidence of clonal expansion in this subject (Figure 3A and Supplemental Table 3). There was even greater evidence of clonal expansion in ES24, with repeated sampling of identical integration events in each of 12 different genes (Figure 3B and Supplemental Table 4). Strikingly, 11 different cells appeared to have virus integrated at the same site in the gene WNK1, and 7 cells had virus integrated in the same site in ZNF470. To determine if there is evidence of enhanced clonal expansion in other ES individuals compared with ART-treated or viremic individuals, we sequenced proviral env clones that were amplified under limiting dilution and assessed the frequency of identical sequences. There was a statistically significant increase in the frequency of identical clones in the ES group compared with that in the ART-treated and viremic groups (Figure 3C). The high frequency of identical proviral clones seen in the ES group matches the frequency of 2 proviral clones in a prior study in which clonal expansion was confirmed by integration site analysis in HIV controllers (21).
Figure 3. Evidence of clonal expansion in Pt169 and ES24.
(A and B) Percentage of clonal integration sites for (A) Pt169 and (B) ES24. (C) Frequency of identical proviral env amplified from ESs (8 subjects, median number of clones sequenced 10.5, range 4–14), ART-treated individuals (9 subjects, median number of clones sequenced 30, range 7–44), and viremic individuals (7 subjects, median number of clones sequenced 15, range 12–19). The center line indicates the mean, and error bars show standard deviation. Statistical analysis was done using a 1-way ANOVA with Tukey’s multiple comparison test. ***P < 0.001.
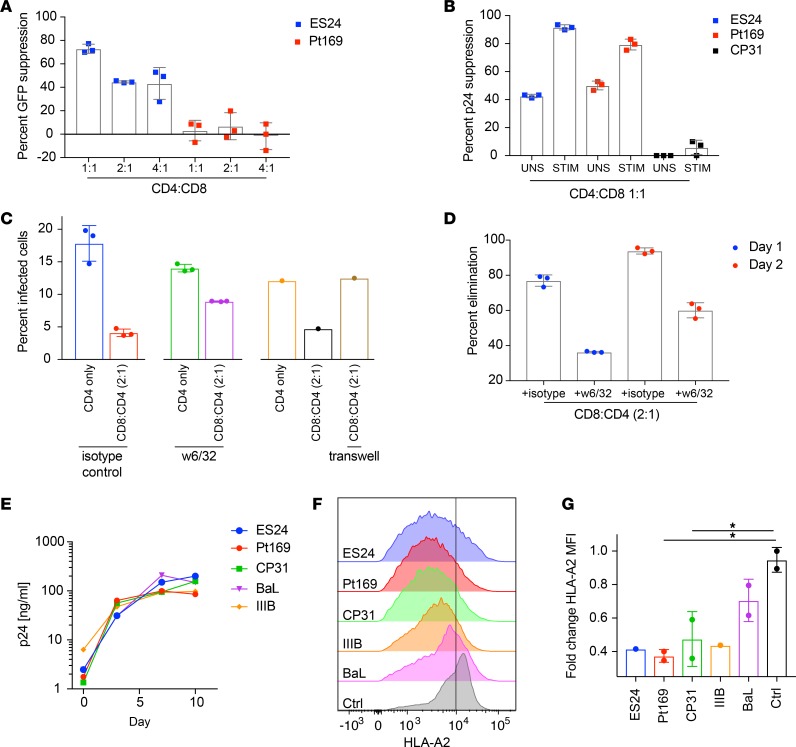
We next investigated CD8+ T cell control of these expanded viruses. We have previously shown, as well as confirmed here, that CD8+ T cells from Pt169, who does not have any protective HLA alleles, were not capable of controlling replication of a pseudotyped NL4-3 deltaENV-GFP virus in a standard viral inhibition assay (Figure 4A; ref. 22). As immune control is highly dependent on viral sequence, we tested whether his CD8+ T cells could control replication of autologous virus. We infected activated autologous CD4+ T cells from Pt169 with clonally expanded RC virus in the presence or absence of CD8+ T cells. The infected CD4+ T cells were cocultured with either autologous bulk CD8+ T cells or autologous CD8+ T cells that had been stimulated with overlapping clade B virus Gag and Nef peptides for 7 days. Stimulated CD8+ T cells effectively suppressed an autologous clonal RC virus from this subject (Figure 4B). Unstimulated CD8+ T cells also suppressed the autologous virus, but to a lesser extent. In contrast, CD8+ T cells from CP31, the transmitting partner of Pt169, did not effectively suppress replication of an autologous RC virus (Figure 4B). In separate experiments, CD8+ T cells from ES24 suppressed replication of autologous virus to a level that was similar to the level seen with CD8+ T cells from Pt169 (Figure 4B). This subject, like other ESs, was also capable of controlling the pseudotyped NL4-3 deltaENV-GFP virus in the standard suppression assay (Figure 4A). We next asked whether CD8+ T cells from Pt169 were capable of eliminating CD4+ T cells that were productively infected with autologous virus. As shown in Figure 4, C and D, his CD8+ T cells effectively reduced the percentage of infected cells. This elimination was partially blocked by MHC I–specific antibodies (Figure 4, C and D) and was dependent on contact between the effector and target cells (Figure 4C). We asked whether the ability of CD8+ T cells from Pt169 to control viral replication was due to limited fitness of the autologous clonally expanded virus. In a standard culture assay with activated primary CD4+ T cells, we saw efficient replication of the expanded isolates from Pt169 and ES24 (Figure 4E). To assess other measures of viral fitness, we tested whether the clonally expanded viruses from both subjects were capable of efficiently downregulating class I MHC molecules, a well-described mechanism of immune evasion of CD8+ T cell responses (29). Primary CD4+ T cells from an HLA-A2 HIV– subject were infected with viruses from both subjects as well as with laboratory strains, and surface HLA downregulation was measured by flow cytometry. As shown in Figure 4, F and G, clonal viruses from both subjects downregulated HLA-A2 as effectively as did IIIB and Ba-L.
Figure 4. Replication of clonal viruses can be inhibited by CD8+ T cells.
(A) Heterologous suppression of pNL4.3-deltaENV-GFP virus by unstimulated CD8+ T cells from ES24 and Pt169 at different ratios, tested in triplicate. (B) Suppression of clonal virus by unstimulated (UNS) or peptide-stimulated (STIM) CD8+ T cells from ES24, Pt169, and CP31, tested in triplicate. Pt169 CD4+ T cells were infected with clonal virus and cocultured with STIM CD8+ T cells. Coculture conditions were carried out in the presence of an isotype control, a MHC I–blocking antibody (w6/32), or in a Transwell. (C) The percentage of infected CD4+ T cells that remain after 1 day of coculture with STIM CD8+ T cells was analyzed by flow cytometry for Gag. Each symbol represents a replicate. (D) The percentage of elimination of infected cells was calculated for days 1 and 2 of coculture with STIM CD8+ T cells in the presence of isotype control or MHC I–blocking antibody, tested in triplicate. (E) Replication capacity of viral isolates from Pt169, CP31, and ES24 and laboratory strains BaL and IIIB. Representative experiment, repeated twice. (F) Downmodulation of surface HLA-A2 in primary CD4+ T cells infected by clonal isolates from Pt169, CP31, and ES24 and laboratory strains BaL and IIIB. (G) Calculated fold change in HLA-A2 MFI (n = 2). Height of bars indicates mean, and error bars show standard deviation. Statistical analysis was done using a 1-way ANOVA with Tukey’s multiple comparison test. *P < 0.05.
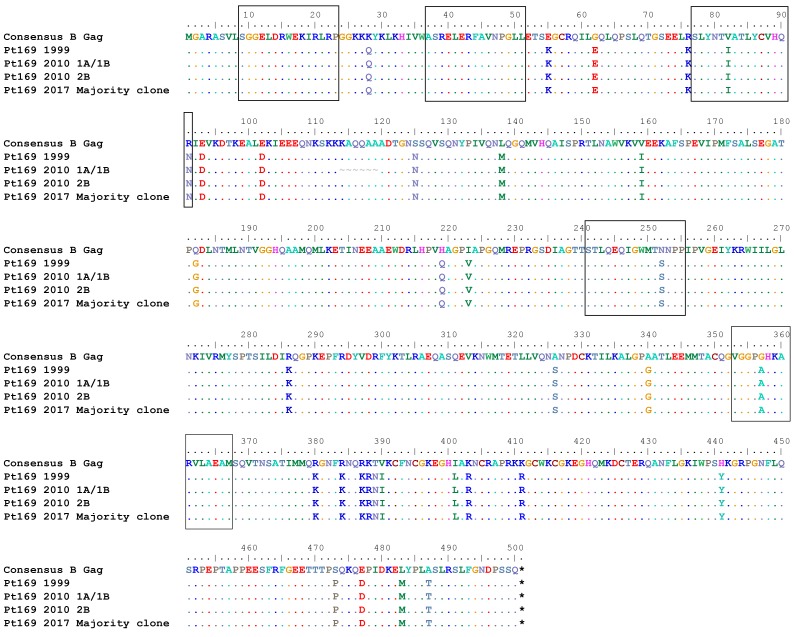
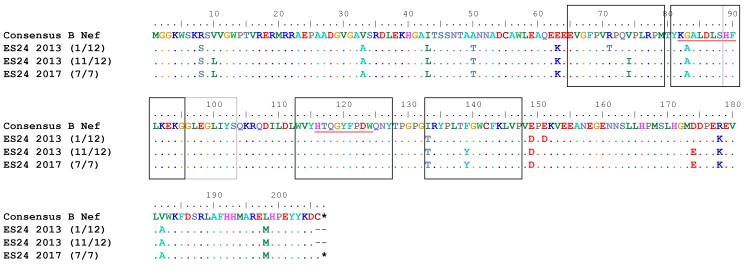
Prior studies have shown evidence of archived escape mutations in viral reservoirs in chronic progressors (CPs) that are not present in patients started on ART during primary infection (30). Pt169 was treated during primary infection but had interrupted ART for 2 weeks at 36 weeks after infection, which led to a rebound in viremia (Supplemental Figure 1). In 2010, we performed an ELISpot using overlapping Gag and Nef peptides to determine Pt169 T cell responses. Analysis of the 2017 clonally expanded virus revealed substitutions in 3 of 6 nonoverlapping Gag epitopes (Figure 5 and Supplemental Table 5) and all 3 targeted distinct Nef epitopes (Figure 6 and Supplemental Table 5). The same assay was completed on ES24 in 2017, and we found evidence of a fairly broad T cell response. Analysis of the clonally expanded virus revealed that there was a substitution in 2 of 12 nonoverlapping Gag epitopes (Figure 7 and Supplemental Table 5) and in 2 of 5 distinct Nef epitopes (Figure 8 and Supplemental Table 5). Prior studies have documented viral evolution in ESs (31, 32); therefore, we compared sequences from the 2017 clonal RC viruses from each subject to those of viruses obtained in 2010 from Pt169 and in 2013 from ES24. There was no evidence of further accumulation of mutations in Gag and Nef epitopes in either subject (Figures 5–8).
Figure 5. No evolution observed in Pt169 CD8+ T cell–targeted Gag epitopes.
Epitopes targeted in Gag by Pt169 CD8+ T cells. Black boxes indicate targeted epitopes.
Figure 6. No evolution observed in Pt169 CD8+ T cell–targeted Nef epitopes.
Epitopes targeted in Gag by Pt169 CD8+ T cells. Black boxes indicate targeted epitopes.
Figure 7. No evolution observed in ES24 CD8+ T cell–targeted Gag epitopes.
Epitopes targeted in Gag by ES24 CD8+ T cells. Black boxes indicate targeted epitopes. Red lines indicate HLA-B*57–restricted epitopes.
Figure 8. No evolution observed in ES24 CD8+ T cell–targeted Nef epitopes.
Epitopes targeted in Nef by ES24 CD8+ T cells. Black boxes indicate targeted epitopes. Red lines indicate HLA-B*B57–restricted epitopes.
Discussion
This is the first evidence to our knowledge of clonal expansion of RC virus in an ES and in a PTC. While integration site analysis showed clonal expansion in two subjects with natural virologic control (21) and identical proviral sequences have been amplified from HIV controller CD4+ T cells (2, 21, 33, 34), it has remained unclear whether or not the provirus from the clonally expanded CD4+ T cells is RC. The low frequency of HIV-infected cells has made it challenging to culture RC virus from these subjects. Very few studies have reported full-genome sequence analysis of RC isolates in HIV controllers or have reported isolation of more than a single virus from each subject (24, 35). ES24 is unusual in that he has a relatively large viral reservoir, and thus, we were able to culture and sequence 7 different isolates. Notably, his 2017 viral clone differs by just two amino acids from a full-length isolate cultured 5 years ago (24).
PTCs have likewise been reported to have a low frequency of latently infected cells (36–38), and there have been very few fully characterized RC isolates from these subjects. Pt169 has maintained a frequency of latently infected cells of approximately 1 infectious unit per million over the past 7 years, a frequency comparable to that of CPs. Here, we were able to isolate 7 viruses at one time point and 5 isolates 6 months later. Near-full-length viral sequence analysis showed a striking degree of identity within RC isolates in Pt169, similar to our results for ES24.
In addition to full-length sequence analysis of RC virus and clonal sequence analysis of proviral env and nef, we also analyzed integration sites in both subjects. While both methods provided reinforcing evidence of clonal expansion, we were not able to definitively link the expanded integration sites to the RC virus isolates. Furthermore, the frequency of identical proviral sequences was higher than the frequency of identical integration sites in Pt169 and ES24. While most studies that have determined clonal expansion by proviral sequence analysis have not simultaneously performed integration site analyses, a similar discordance was seen in a previously described CP, for whom integration site analysis was used to prove clonality of RC virus (4, 7), and in 1 of 2 PTCs in a recently published study (38). Our results can partially be explained by the fact that the different methods used analyzed different populations of proviruses. For the proviral env and nef sequence analysis, only clones of the expected size were sequenced, and hypermutated or partially deleted clones were excluded. In contrast, in the integration site analysis, all amplicons containing proviral host junctions were sequenced, regardless of whether the proviruses contained large deletions or were hypermutated.
Given the limited diversity of the viral sequences, we investigated whether these isolates were selected for because they were capable of evading the immune response. While there is strong evidence that CD8+ T cells from many HIV controllers are able to effectively suppress viral replication (17–20, 39), most of these studies have used laboratory strains of virus rather than autologous virus. This is important in light of studies that show high levels of immune escape in archived virus from CPs (30). While we and others have shown that archived virus in HLA-B*57 ESs contains few escape mutations (24, 40, 41), similar studies have not been performed in controllers who lack protective HLA alleles. Furthermore, prior data has suggested that PTCs have weak HIV-specific CD8+ T cell responses, and the mechanism of control in these patients has not been fully elucidated (36). Our data indicate that CD8+ T cells from both subjects can inhibit replication of clonal autologous viruses in autologous CD4+ T cells. Thus, the immune system is capable of controlling these isolates and may explain why both subjects have maintained undetectable viral loads for extended periods of time. It is possible that the CD4+ T cell clone harboring the virus is resistant to CD8+ T cell killing, which could explain the relative stability of the reservoir in these subjects. Alternatively, if the cell proliferates without viral antigen expression, then the immune system will not be able to recognize the cell as being infected. In the latter scenario, we would expect the accumulation of clonally expanded CD4+ T cells harboring virus in ESs, as cytotoxic T lymphocytes (CTLs) would selectively eliminate infected cells that became activated and expressed viral antigens during the process of viral replication. To test this hypothesis, we compared the frequency of identical proviral env clones in ESs, CPs on ART, and viremic subjects who were not on ART. We found a significantly higher percentage of identical env clones in ESs, suggesting that the immune response may select for these cells in these subjects.
Either mechanism of persistence, inherent resistance to killing of infected clonal cells or CD8+ T cell evasion by proliferation of infected cells without antigen expression, presents a challenge for viral eradication. However, for substantial viremia to occur, the expanded virus must be able to productively infect other CD4+ T cells. We show that CD8+ T cells from both subjects are capable of suppressing new rounds of infection. This was not seen with CD8+ T cells from CP31 and offers a potential mechanism for the long-term remission of Pt169 and ES24. This is notable, as while ES24 (HLA-A* 24/30, B* 7/57) has the protective HLA-B*57 allele, Pt169 (HLA-A* 30/68, B*42) does not. He is infected with a dual tropic virus, which may be more difficult to control. Additionally, this virus was highly pathogenic, causing severe encephalitis and a viral load of greater than 750,000 copies/ml in primary infection (22). Despite the lack of protective genetics and virulence of the virus, he has successfully controlled viral replication for more than 15 years and does not have a high level of immune activation (Supplemental Figure 2). Although this subject did not have strong CD8+ T cell responses in the classic viral inhibition assay using a pseudotyped laboratory strain, his CD8+ T cells are able to efficiently inhibit his autologous virus and eliminate autologous CD4+ T cells infected with autologous virus. We show that this CD8+ T cell control is not due to lack of viral fitness or an inability of the expanded virus to downregulate surface HLA and requires cell-to-cell contact.
In summary, we demonstrate here that long-term viral remission is possible, despite the presence of clonal expansion of RC viral isolates. Further, this control is possible in the absence of protective HLA alleles and in the context of an infectious and pathogenic virus isolate with intact immune evasion and dual tropism. Our study is limited by the fact that we have analyzed just 2 individuals, and thus our findings may not be representative of posttreatment control and/or elite control. However, elucidating the mechanisms of control of HIV-1 replication in even a single individual may advance our understanding of HIV pathogenesis. Pt169 provides an example of durable remission without obvious host advantage or defective virus. Furthermore, both individuals are unique, in that, unlike most ESs and PTCs, they have frequencies of latently infected cells that are similar to the frequency seen in CPs on ART. Clinical trials with latency-reversing agents have highlighted how challenging it is to reduce the size of the viral reservoir in vivo (42, 43). Our results suggest that clonal expansion does not preclude the possibility of achieving a functional cure in HIV-infected subjects who do not have protective HLA alleles or very small viral reservoirs.
Methods
Virus isolation and sequence analysis.
Culture of RC virus from CD4+ T cells and full-genome sequence analysis of viral isolates were performed as previously described (22, 24, 30). In brief, peripheral blood mononuclear cells (PBMCs) were isolated from patient blood and CD4+ T cells were isolated by negative selection (Miltenyi). Patient CD4+ T cells were plated at limiting dilution with 10× irradiated feeders and 0.5 μg/ml PHA for a minimum of 16 hours. The following day PHA was removed, and healthy donor activated CD4+ T cells were added. Cultures were monitored for 14 days and split when necessary. On day 14, culture supernatants were tested for p24, and positive supernatants and cells were harvested and stored at –80oC. DNA was extracted from cells using the Qiagen Gentra Purgene Cell Kit A and sequenced for HIV using previously described primer sets (35). Neighbor-joining phylogenetic analysis was performed as described previously (44).
DNA extraction and limiting dilution PCR.
DNA was extracted from patient CD4+ T cells, as described above. Limiting dilution PCRs for env and nef clones from Pt169 and ES24 obtained in 2017 was performed as previously described (45, 46). In brief, DNA was subjected to a nested limiting dilution PCR protocol using Platinum Taq HiFi (Life Technologies). The outer PCR was nearly full length at approximately 9 kB and employed a touchdown PCR protocol. The outer PCRs were diluted 1:3 with DI water, and 10 μl outer PCR DNA was used for nested amplification of both nef and env using previously published primer sets (35). Clonality was determined using Poisson statistics, and 2 positives per 12 wells amplified was considered clonal. PCR products were visualized using 1% agarose gels and isolated using the QIAquick Gel Extraction kit (Qiagen). The products were set for Sanger sequencing using previously described primer sets (35). Limiting dilution PCR for env clones from ESs, CPs, and viremic subjects (Figure 3C) obtained prior to 2017 was performed as previously described (35, 41).
Sequencing HIV integration sites.
Genomic positions for integrated proviruses were identified using the INSPIIRED protocol (27, 28). Primers for nested PCR amplification were designed to amplify the terminal portions of LTR sequences (U3 and U5) of isolated viral sequences. Genomic DNA from patient samples was randomly sheared using sonication, and linkers were ligated to the repaired DNA. DNA containing proviral host junctions was then amplified from the ligated DNA using a nested PCR, where PCR1 contained a duplex reaction (amplifying both from U3 and U5 regions) and PCR2 contained single-plex reactions for U3 or U5. Amplicons were purified from the reaction mixture, pooled, and sequenced on an Illumina MiSeq. Integration sites were determined from the sequence data using the INSPIIRED pipeline (https://github.com/BushmanLab/).
Heterologous CD8 suppression assay.
Heterologous suppression assays were performed as previously described (47). In brief, PBMCs were isolated from patient blood, CD8+ T cells were positively selected (CD8+ T cell Isolation kit, Miltenyi), and CD4+ T cells were isolated by negative selection (CD4+ T cell isolation kit, Miltenyi). CD4+ T cells were infected with 25 ng p24/100,000 cells by spinoculation at 1,200 g for 2 hours with a single round X4 tropic enveloped NL4-3 virus with GFP engineered into env. Infected CD4+ T cells were cocultured with autologous CD8+ T cells at 3 effector-to-target ratios (1:1, 2:1, 4:1) for 3 days. Flow cytometry was performed to assess the percentage of GFP+ CD4+ T cells at day 3. CD4+ T cells were determined by gating CD3+ (clone UCHT1) and CD8– (clone RPA-T8) cells.
Autologous CD8 suppression assay.
Autologous suppression assays were performed as previously described (47). In brief, PBMCs were isolated from patient blood. CD8+ T cells were positively selected (CD8+ T cell Isolation kit, Miltenyi) from one-third of the PBMCs. One-half of the isolated CD8+ T cells were cultured for 7 days with consensus B Gag and Nef pooled peptides (1 μg/ml; obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases [NIAID], catalog 12425 and 12545) the remaining CD8+ T cells were cultured without peptides. The remaining PBMCs were cultured in IL-2–rich media for 3 days and then activated with 0.5 μg/ml PHA (Remel) for 3 days. Activated CD4+ T cells were isolated by negative selection (CD4+ T cell isolation kit, Miltenyi). CD4+ T cells were infected by spinoculation at 1,200 g for 2 hours with 200 ng p24 per million CD4+ T cells of clonal autologous virus. Infected CD4+ T cells were then cocultured with autologous peptide stimulated or unstimulated CD8+ T cells at a ratio of 1:1 for 7 days. The effect of CD8+ T cells on autologous virus outgrowth was determined by measuring p24 by ELISA (Perkin Elmer) in harvested supernatants.
Autologous CD8 virus elimination assay.
Activated CD4+ T cells were infected with autologous virus as described above. On day 3 of infection, infected CD4+ T cells were cocultured with Gag- and Nef-stimulated CD8+ T cells in the presence of 10 μg/ml anti–class I MHC antibody w6/32 or an isotype control (Biolegend) at a 2:1 effector-to-target ratio. Additionally, some cells were cultured with CD8+ T cells in a 0.4-μm Transwell (Corning) at the same 2:1 effector-to-target ratio. Target cells (CD3+/CD8– cells) were stained for intracellular Gag at different time points with the Kc57 antibody (Beckman Coulter).
Viral fitness assay.
Viral fitness was analyzed as previously described (35). PBMCs from healthy donors were activated for 3 days with 0.5 μg/ml PHA. CD4+ T cells were isolated by negative selection and infected by spinoculation 1,200 g for 2 hours with equal quantities (200 ng per million CD4+ T cells) of p24 from primary patient isolates or with Ba-L or IIIB laboratory HIV-1 strains as controls. Supernatant samples were taken over the course of 7 days. Viral replication was quantified using p24 ELISA (Perkin Elmer).
HLA downregulation.
The HLA downregulation assay was performed as previously described (48). In brief, PBMCs were isolated from an HLA-A2+ healthy donor and activated for 3 days with 0.5 μg/ml PHA. CD4+ T cells were isolated by negative selection and infected by spinoculation 1,200 g for 2 hours with equal quantities (200 ng per million CD4+ T cells) of p24 from primary patient isolates or with Ba-L or IIIB laboratory HIV-1 strains as controls. Cells were harvested on day 3 and analyzed by flow cytometry for HLA-A2 (clone BB7.2) and CD4 expression (clone OKT4).
ELISpot assay.
Reactive CTL epitopes were defined by IFN-γ Elispot as previously described (49). Patient PBMCs were plated at 200,000 cells per well into precoated anti–IFN-γ ELISpot plates (MABtech). Cells were activated with overlapping peptides spanning the entire amino acid sequence of clade B consensus Gag and Nef at a concentration of 5 μg/ml (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, catalog 8117 and 5189). PBMCs were cultured overnight, and the remainder of the ELISpot was completed following the manufacturer’s protocol. Quantification of spot-forming units (SFU) was performed by the Johns Hopkins Immunology Core. Positive responses were defined as greater than 25 SFU per million PBMCs above background.
Statistics.
All statistical analyses were performed using a 1-way ANOVA with Tukey’s multiple comparison test. P values of less than 0.05 were considered significant.
Study approval.
This study was approved by the Johns Hopkins University Institutional Review Board. Informed written consent was obtained from both subjects prior to enrollment into the study.
Author contributions
RTV, RL, MS, CWP, CLN, JG, EPS, FDB, and JNB designed experiments. RTV, AKK, CG, RL, MS, CWP, CLN, and JG performed experiments. RTV, CLN, JG, FDB, JRB, and JNB analyzed the data. JNB provided materials, and RTV, EPS, and JNB wrote the manuscript.
Supplementary Material
Acknowledgments
This work was funded by the Johns Hopkins University Center for AIDS Research (P30AI094189), the Penn Center for AIDS Research (P30 AI 045008), the PennCHOP Microbiome Program, the BEAT-HIV Delaney Collaboratory (UM1AI126620), and NIAID grants AI 052845-13, AI 082020-05A1, AI 045008-15, and U19AI117950-01, (to FDB) and 2R56AI080328-05A1, 1R01AI120024-01, and AI140789-01 (to JNB).
Version 1. 09/20/2018
Electronic publication
Footnotes
MS’s present address is: IrsiCaixa AIDS Research Institute, Badalona, Spain.
CWP’s present address is: Gilead Sciences, Foster City, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(18):e122795. https://doi.org/10.1172/jci.insight.122795.
Contributor Information
Abena K. Kwaa, Email: akwaa2@jhmi.edu.
Rachel Latanich, Email: rlatani1@jhmi.edu.
Maria Salgado, Email: bersalma@hotmail.com.
Eileen P. Scully, Email: escully1@jhmi.edu.
References
- 1.Tobin NH, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol. 2005;79(15):9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Stockenstrom S, et al. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis. 2015;212(4):596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzi JC, et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci USA. 2016;113(49):E7908–E7916. doi: 10.1073/pnas.1617789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med. 2017;214(4):959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bui JK, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 2017;13(3):e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockerham LR, Hatano H, Deeks SG. Post-treatment controllers: Role in HIV “cure” research. Curr HIV/AIDS Rep. 2016;13(1):1–9. doi: 10.1007/s11904-016-0296-x. [DOI] [PubMed] [Google Scholar]
- 12.Wen Y, Bar KJ, Li JZ. Lessons learned from HIV antiretroviral treatment interruption trials. Curr Opin HIV AIDS. 2018;13(5):416–421. doi: 10.1097/COH.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 13.Rouzioux C, Hocqueloux L, Sáez-Cirión A. Posttreatment controllers: what do they tell us? Curr Opin HIV AIDS. 2015;10(1):29–34. doi: 10.1097/COH.0000000000000123. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalo-Gil E, Ikediobi U, Sutton RE. Mechanisms of virologic control and clinical characteristics of HIV+ elite/viremic controllers. Yale J Biol Med. 2017;90(2):245–259. [PMC free article] [PubMed] [Google Scholar]
- 15.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97(6):2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International HIV Controllers Study. et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3(11):1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 18.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sáez-Cirión A, et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci USA. 2007;104(16):6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migueles SA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29(6):1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boritz EA, et al. Multiple origins of virus persistence during natural control of HIV infection. Cell. 2016;166(4):1004–1015. doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salgado M, et al. Prolonged control of replication-competent dual- tropic human immunodeficiency virus-1 following cessation of highly active antiretroviral therapy. Retrovirology. 2011;8:97. doi: 10.1186/1742-4690-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blankson JN, et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81(5):2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salgado M, et al. HLA-B*57 elite suppressor and chronic progressor HIV-1 isolates replicate vigorously and cause CD4+ T cell depletion in humanized BLT mice. J Virol. 2014;88(6):3340–3352. doi: 10.1128/JVI.03380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee GQ, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. 2017;127(7):2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28(6):755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman E, et al. INSPIIRED: A pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol Ther Methods Clin Dev. 2017;4:39–49. doi: 10.1016/j.omtm.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry CC, et al. INSPIIRED: Quantification and visualization tools for analyzing integration site distributions. Mol Ther Methods Clin Dev. 2017;4:17–26. doi: 10.1016/j.omtm.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391(6665):397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 30.Deng K, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517(7534):381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol. 2010;84(14):7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mens H, et al. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol. 2010;84(24):12971–12981. doi: 10.1128/JVI.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Azevedo SSD, et al. Highly divergent patterns of genetic diversity and evolution in proviral quasispecies from HIV controllers. Retrovirology. 2017;14(1):29. doi: 10.1186/s12977-017-0354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pernas M, et al. Factors leading to the loss of natural elite control of HIV-1 infection. J Virol. 2018;92(5):e01805-17. doi: 10.1128/JVI.01805-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blankson JN, et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81(5):2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sáez-Cirión A, et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 2013;9(3):e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Persaud D, et al. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med. 2013;369(19):1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharaf R, et al. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest. 2018;128(9):4074–4085. doi: 10.1172/JCI120549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckheit RW, Siliciano RF, Blankson JN. Primary CD8+ T cells from elite suppressors effectively eliminate non-productively HIV-1 infected resting and activated CD4+ T cells. Retrovirology. 2013;10:68. doi: 10.1186/1742-4690-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migueles SA, et al. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J Virol. 2003;77(12):6889–6898. doi: 10.1128/JVI.77.12.6889-6898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203(5):1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spivak AM, Planelles V. HIV-1 eradication: Early trials (and tribulations) Trends Mol Med. 2016;22(1):10–27. doi: 10.1016/j.molmed.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen TA, Søgaard OS. Clinical interventions in HIV cure research. Adv Exp Med Biol. 2018;1075:285–318. doi: 10.1007/978-981-13-0484-2_12. [DOI] [PubMed] [Google Scholar]
- 44.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 45.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey JR, et al. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J Virol. 2008;82(15):7395–7410. doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nou E, Zhou Y, Nou DD, Blankson JN. Effective downregulation of HLA-A*2 and HLA-B*57 by primary human immunodeficiency virus type 1 isolates cultured from elite suppressors. J Virol. 2009;83(13):6941–6946. doi: 10.1128/JVI.00306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Connell KA, et al. HIV-1 evolution following transmission to an HLA-B*5801-positive patient. J Infect Dis. 2009;200(12):1820–1824. doi: 10.1086/648377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.