Abstract
The incidence of liver disease is increasing globally. The only curative therapy for severe end-stage liver disease, liver transplantation, is limited by the shortage of organ donors. In vitro models of liver physiology have been developed and new technologies and approaches are progressing rapidly. Stem cells might be used as a source of liver tissue for development of models, therapies, and tissue-engineering applications. However, we have been unable to generate and maintain stable and mature adult liver cells ex vivo. We review factors that promote hepatocyte differentiation and maturation, including growth factors, transcription factors, microRNAs, small molecules, and the microenvironment. We discuss how the hepatic circulation, microbiome, and nutrition affect liver function, and the criteria for considering cells derived from stem cells to be fully mature hepatocytes. We explain the challenges to cell transplantation and consider future technologies for use in hepatic stem cell maturation, including 3-dimensional biofabrication and genome modification.
Keywords: Liver Development, Differentiation, Hepatocyte, Culture
The only proven treatment for patients with endstage liver disease is organ transplantation, which is hampered by the lack of donors. Therefore, mortality on the waiting list can be as high as 10%.1 Primary hepatocytes are the standard cells used for in vitro studies and are a resource for liver cell transplantation and bioartificial liver devices.2 However, the complexity of the maturation process, the instability of cultured hepatocytes, and inconsistent protocols limit their application.
Hepatocytes perform essential liver functions, including plasma protein secretion, bile production, detoxification, metabolic homeostasis, and storage of vitamins and minerals. They are the most predominant parenchymal cell type, accounting for approximately 80% of the adult liver mass, and are polygonal, approximately 20–30 μm in diameter with a volume of 3000 μm3. Like other epithelial cell types, hepatocytes are polarized, with distinct apical, lateral, and basal membranes. Hepatocytes are polyploid; approximately 20%–30% of human hepatocytes are tetraploid and octaploid, compared with 85% in rats and mice.3,4
Hepatocytes originate from definitive endoderm during embryonic development, after acquisition of hepatic competence by ventral foregut endoderm and specification of those epithelial cells into hepatic endoderm.5,6 Primordial liver cells transition into a non-polarized cellular phenotype (hepatoblasts), which subsequently generate liver buds. Hepatoblasts are bipotent and express fetal liver genes and genes associated with both hepatocyte and cholangiocyte lineages (cholangiocytes and epithelial cells that line the bile ducts and hepatocytes)7 and can differentiate into both cell types. Lineage determination relies on signaling pathways, including Notch and transforming growth factor-β, which promote biliary differentiation.8–10 Down-regulation of these pathways promotes specification of hepatoblasts toward hepatocyte fate.11–13
Unfortunately, adult hepatocytes have limited in vitro proliferation ability and quickly de-differentiate, hampering their use ex vivo. Several cell types and in vitro techniques have been proposed for generating a potential alternative to hepatocytes, which we collectively refer to as hepatocytelike cells (HLCs). We review the available strategies for HLC production and strategies for overcoming the immature hepatic phenotypes of HLCs. We propose ways to improve the generation of mature HLCs and defined criteria for HLCs.
Cell Sources for Hepatocyte Generation
Pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced PSCs (iPSCs), can differentiate into all cell types of the body, while maintaining genetic stability and are therefore a promising cell source for regenerative medicine, development of models, and drug discovery. Although the application of human ESCs is hotly debated, human iPSCs are generated by forced expression of specific pluripotency transcription factors, circumventing ethical concerns.14 Great progress has been made exploring the differentiation capacity of PSCs toward the hepatocyte lineage. Although specific culture conditions can vary, most protocols share a 3-step strategy based on ontogenetic liver development.15–17 The first is efficient induction of PSCs into a definitive endoderm fate (inducers include activin A, fibroblast growth factor [FGF] 2, WNT3A, BMP4, and LY294002). The second step is hepatic specification, which gives rise to hepatoblast-like cells (inducers include hepatocyte growth factor [HGF], FGF2, FGF4, and BMP4). The final step is to induce differentiation toward hepatocytes (oncostatin M [OSM], dexamethasone, HGF, and follistatin).
A second source of HLCs are mesenchymal stromal or stem cells (MSCs), which are present in easily accessible tissues like fat, blood, and bone marrow. They have stem cell–like characteristics, such as self-renewal and multipotent differentiation capacity, including toward HLCs. In most protocols, MSCs can be directed toward hepatocyte lineage through a hepatic commitment stage (induced by FGF and HGF) and maturation stage (induced by OSM and dexamethasone).18,19 Besides the heterogenic population of the HLCs after differentiation, they also reflect an immature phenotype.20
A third attractive cell source are hepatic progenitor/ stem cells (HPCs), which can be derived from single Lgr5+ (mouse) or EpCAM+ (human) cholangiocytes and from bile duct fragments from mouse and human livers.11,12,21 A 3-dimensional (3D) culture system based on laminin and collagen IV–enriched Matrigel has been established to support the long-term and genetically stable expansion of HPCs as organoids.11,12 Those HPCs can differentiate into the hepatocyte lineage by inhibition of Notch and Wnt signaling and activation of FGF signaling.11,12 This single-step differentiation process requires less effort than PSC-based methods and reaches high efficiency.11 However, similar to other sources, HPC-derived HLCs also partially resemble hepatocytes.12
There has been no direct comparison of the maturation level of HLCs generated from these sources. Furthermore, different standards were used for characterizing HLCs in different studies. It is therefore not clear which source is optimal.
In Vivo Liver Maturation
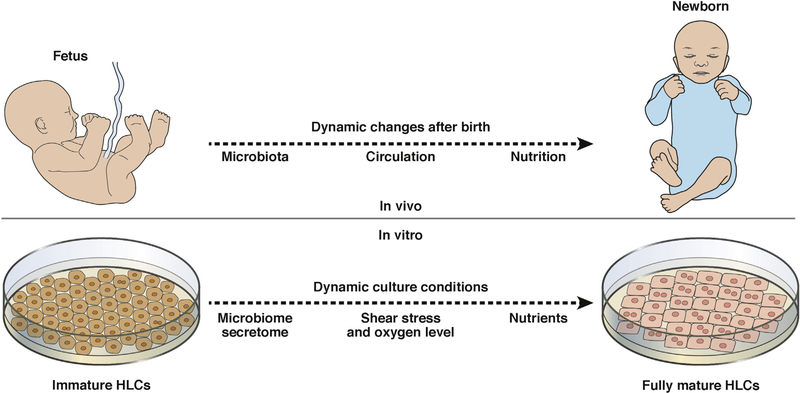
Unfortunately, stem cell–derived HLCs do not possess all functions of mature hepatocytes. Liver maturation is complex and stage-dependent; full maturation takes as long as 2 years from birth and involves expression of several signaling pathways, such as those responsible for bile acid synthesis, drug metabolism, and amino acid transport.22,23 Large modifications in homeostasis can therefore alter liver maturation. Following birth, the newborn liver is affected by changes in circulation, microbiome, and nutrition that might also promote hepatic maturation (Figure 1).
Figure 1.
Changes in liver maturation from fetus to newborn. Upon birth, the human fetal liver undergoes many changes, including changes in circulation, the microbiome, and nutrition, which are required for hepatic maturation. Increasing our understanding of these changes might provide information needed to develop more efficient in vitro liver maturation methods.
Hepatic Circulation
Nutrition, circulation, and oxygen levels change radically at birth, permanently change the environment of the newborn, and result in new metabolic and homeostatic requirements. Most features of the fetal circulation, such as the umbilical cord vessels (ductus arteriosus and ductus venosus), the foramen ovale, and the placenta, are no longer needed after birth. The change from the fetal to the newborn circulation is therefore a remarkable process of homeostatic coordination. In fetal circulation, blood from the placenta returns to the fetus through the umbilical vein, which has a lower oxygen saturation compared with adult arterial circulation (approximately 80% and 98% saturation, respectively). The liver is bypassed by the ductus venosus during fetal development. To compensate for lower oxygen conditions, fetal hemoglobin has a considerably higher affinity for oxygen than adult hemoglobin.24 This observation, combined with circulation dynamics, indicates that the functional heterogeneity of the parenchymal cells in the fetal liver depend directly on microcirculation.25
The right liver lobes are perfused with poorly oxygenated blood, mostly from portal blood; the left portion receives most of its blood from the umbilical vein, has a higher content of oxygen-dependent enzymes, and is more active in drug binding and metabolism.25,26 The hepatic artery contributes approximately 2% to fetal liver circulation and, at birth, this increases 18-fold, comprising nearly 25% of hepatic blood flow with highly oxygenated blood.27 Similarly, contribution of the portal vein increases from 20%–25% to an average of 75% and with closure of the ductus venosus, its blood distribution through the liver becomes homogenous.26,28,29
Oxygenation has been shown to increase albumin and urea synthesis in rat hepatocytes.30 In addition, shear stress, while not usually investigated, is known to maintain or even increase function of parenchymal cells, for example, through polarization, which in turn increases liver function.31,32 Oxygen levels and mechanical stimulation experienced by liver cells might therefore be applicable for hepatocyte maturation. Both of these features have now been recognized in adult hepatocytes33 and incorporated into some protocols of human ESC hepatic differentiation.15,34 Interestingly, other vascular mediators generated from liver circulation (eg, prostaglandins, nitric oxide, and catecholamines) could also be involved in hepatic growth and maturation, recently shown in a bioengineered liver.35
The Intestinal Microbiome
During the first week after birth, the immature liver undergoes a large reorganization process.36 The anatomical and functional specialization occurs among hepatic acinar units, leading to formation of 3 distinct metabolic/functional zones.22,23,37 Levels of hormones, oxygen, and nutrients in the periportal blood are important in this process,36 but the initial stimuli that generate this metabolic patterning and the microbiome’s contribution is still undetermined.
Cytochrome P450 (CYP) expression in the liver is an indication of metabolic activity, for example, the breakdown of toxic compounds. The demands of a changing environment and xenobiotic clearance are considered as some of the driving forces for CYP maturation. CYP expression is low at birth (approximately 30% of adult levels) and progressively increases, reaching adult levels by 1 year of age.23 It was recently discovered that germ-free mice have distinct CYP expression levels compared to wild-type mice.38–40 This appears to correlate with the bacterial strains that colonize the gut of newborn and adult mice, which have the ability to change drug metabolism and drug response at various stages of life.40
Studies of the link between postnatal liver maturation and bacterial colonization of the gut found that microbialderived lithocholic acid and vitamin K2 promote maturation of PSC-derived hepatocytes and fetal hepatoblasts.41 Other bacterial-derived substances might have similar effects.
Nutrition
Nutritional input received by the fetal liver changes greatly after birth. In contrast with the fully processed nutrients received from maternal blood in utero, newborn nutrition is dependent on digestion and intestinal absorption of maternal milk. Levels of lipids, carbohydrates, proteins, and other nutrients in portal circulation are periodically increased during the postprandial period, exceeding normal levels for a newborn liver. Lipids for example, are important for liver development and homeostasis, and direct binding of free fatty acids to peroxisome proliferator–activated receptors or hepatocyte nuclear factor 4a (HNF4A), or the role of sterols, steroids, and bile salts and their nuclear receptors (eg, retinoid X receptor, farnesoid X receptor, pregnane X receptor [PXR]) might have larger effects on hepatocyte maturation than reported previously.42–45 Concomitantly, many hormones and growth factors are secreted during this period and most of these hepatotrophic factors (eg, insulin, glucagon, and gastrin) directly affect hepatocyte proliferation, phenotype, and metabolism.25
Strategies to Promote Hepatocyte Differentiation and Maturation
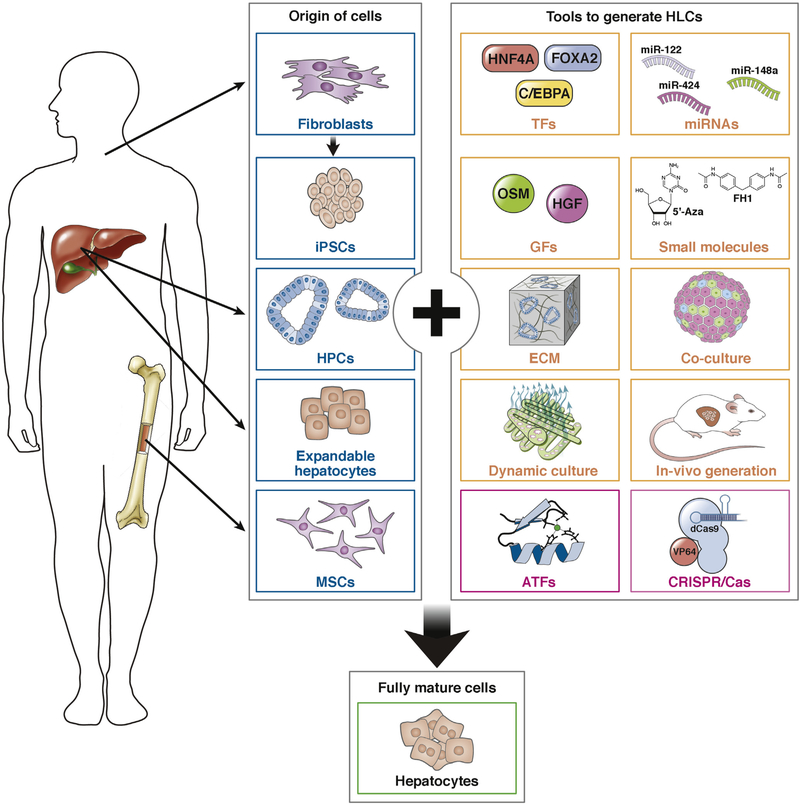
The generation of HLCs, irrespective of cell source, is a complex process. Mature hepatocytes do not proliferate often, posing challenges to the in vitro generation of these cells. Components that promote hepatocyte differentiation and maturation include growth factors, transcription factors, microRNAs, small molecules, and the microenvironment (Figure 2).
Figure 2.
Cells and methods to generate HLCs. Blue boxes show cells, and yellow and pink boxes show methods, used to generate HLCs.
Growth Factors
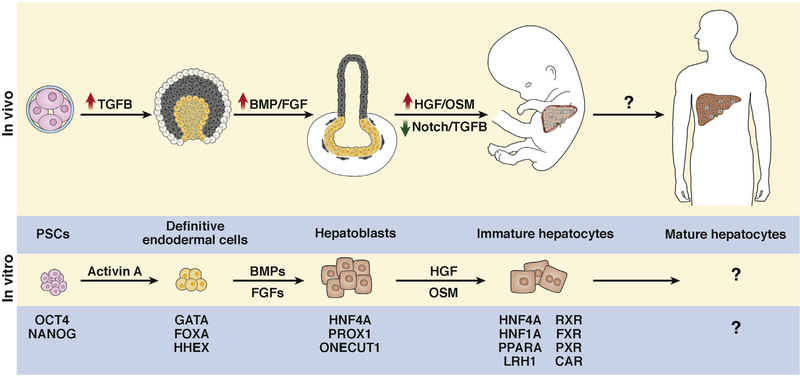
Growth factors regulate embryonic development. Culture media supplementation is used to remodel cell fate. We discuss the 3 important regulators of hepatocyte specification and maturation (Figure 3).
Figure 3.
Molecular changes during hepatocyte differentiation. Transcription factors and signaling molecules that regulate each stage of hepatocyte differentiation. Hepatocyte-generating cells are colored and supporting tissue are black and gray. Question marks indicate pathways under investigation.
The role of OSM, an interleukin-6 family cytokine in hepatocyte maturation was well defined by Kamiya et al,46 who demonstrated that OSM up-regulates the expression of albumin, glucose-6-phosphate dehydrogenase, and tyrosine aminotransferase in fetal hepatocytes isolated from the embryonic murine liver (embryonic day 14.5). Fetal hepatocytes incubated with OSM have a similar morphology to mature hepatocytes, such as tight intracellular contacts, highly condensed and granulated cytosol, and clear roundshaped nuclei. In addition, OSM induces hepatocytespecific functions, including glycogen synthesis, ammonia clearance, lipid synthesis, detoxification, and enhancement of homophilic cell adhesion.47 Interestingly, OSM promotes massive proliferation and dedifferentiation of hepatocytes, dictated by maturation stage. Progenitor cells receiving OSM do not mature. In contrast, mature hepatocytes receiving OSM dedifferentiate; when OSM was withdrawn, hepatocyte functions were rescued.48 These data indicate that OSM is important for early stages of hepatic maturation.
HGF is important throughout liver development. Knockout of HGF leads to embryonic lethality and the embryonic liver is reduced in size by loss of hepatocytes.49 In the presence of dexamethasone, HGF up-regulates expression of several mature hepatocyte markers, such as carbamoyl-phosphate synthase 1, glucose-6-phosphate dehydrogenase, and tyrosine aminotransferase in fetal murine hepatocytes.47 During in vitro PSC-derived hepatocyte generation, HGF facilitates the transition into the hepatocyte specification stage by binding to its receptor (MET), which activates the STAT3 and AKT and regulates the expression of hepatocyte markers.50
Insulin is routinely included in HLC and hepatocyte culture. Although this factor promotes survival of most cell types, insulin also preserves many hepatocyte-specific functions, including amino acid transport, protein synthesis, glycogenesis, and lipogenesis.51–53 Moreover, insulin has an important role in secretion of albumin by hepatocytes.54
These growth factors are essential for hepatic specification and/or maturation of stem cells and appear to be differentiation stage–dependent. Unfortunately, growth factors alone do not induce a hepatic phenotype in HLCs comparable to freshly isolated hepatocytes.
Transcription Factors
Liver development involves the progressive activation of transcription factors. Liver-enriched transcription factors (LETFs) regulate hepatic cell fate commitment and maintenance of a mature status. LETFs include HNF4A, constitutive androstane receptor, eosinophil-associated, ribonuclease A, peroxisome proliferator–activated receptor–α, retinoid X receptor-α, farnesoid X receptor, PXR, small heterodimer partner, and liver receptor homolog 1 (Figure 3).
One of the most studied LETFs, HNF4A, is highly expressed in hepatocytes and regulates hundreds of targets involved in hepatocyte function. Using chromatin immunoprecipitation and promoter microarrays, Odom et al55 found that 43% of actively transcribed genes in the human liver are regulated by HNF4A. Furthermore, overexpression of only HNF4A resets the hepatocyte transcription factor network in damaged hepatocytes from rats after administration of CCl4.56 These findings indicate that HNF4A is an important regulator of hepatocyte function.
Other LETFs control the expression of a range of effectors, including glucose and cholesterol homeostasis, urea cycle, synthesis of plasma proteins, coagulation factors, and most drug-processing proteins.57,58 Transcription factors regulate expression of each other.56,59–66 For example, HNF1A represses HNF4A-mediated activation of transcription, whereas HNF4A promotes HNF1A-mediated activation of transcription.65,66 In addition, HNF1A is a direct target of HNF4A64 and vice versa,67 so these proteins act in a positive-feedback loop. Transcription factor networks that sustain particular cell types might therefore be determined and designed.
Ectopic expression of tissue-specific transcription factors68 can reprogram non-hepatic lineage cells into hepatocyte-like cells.69,70 However, all non-hepatic lineage cells reprogrammed into hepatocyte-like cells have immature features compared to freshly isolated primary hepatocytes, limiting their application. This is possibly caused by a lack of transcription factor network activators that define hepatocyte identity.
Godoy et al71 compared genome-wide gene expression profiles of ESC-derived HLCs (ES-HLCs) to primary human hepatocytes freshly isolated and cultured for 14 days. They identified groups of genes whose expression correlated with mature liver functions and shared expression levels with primary hepatocytes. These included SRY-box 11, forkhead box Q1, and Y box binding protein 3, which may be new targets for further maturation of ES-HLCs.71 Other gene products that regulate hepatic maturation were not expressed at levels observed in mature hepatocytes, leaving ES-HLCs too far removed from a bona fide hepatocyte. Notably, there are differences in messenger RNA and protein levels (up to 40%72), indicating an imbalance in the ESHLC system. Therefore, transcriptome data are useful for identifying candidates, but confirmation at translational and functional levels is necessary.
Transcription factors regulate hepatocyte maturation, but measurements of their expression are not sufficient to determine hepatic maturation because most are expressed at near-adult levels in liver during mid and late gestation. Proper phenotype characterization and determination of liver-specific functions are required to accurately measure liver maturation.
MicroRNAs
MicroRNAs are a class of small non-coding RNAs that regulate cell differentiation, proliferation, and survival. Zhou et al73 generated HLCs by introducing 5 microRNAs (MIR122, MIR148a, MIR424, MIR542–5p, and MIR1246) into human umbilical cord–derived MSCs. MIR122 is specifically and abundantly expressed in the liver, and accounts for 72% of all the microRNA in liver. MIR122 is required for gene expression patterns associated with maintenance of differentiation in the liver.74 Inhibition of MIR122 reduces plasma levels of cholesterol, increases hepatic fatty acid oxidation, and decreases rates of hepatic fatty acid and cholesterol synthesis.75 In contrast, overexpression of MIR122 in mouse ES-HLCs significantly up-regulates expression of several hepatocyte markers, including albumin and CYP3A4, which reach levels of roughly 80% of normal expression in hepatocytes.76 Additionally, liver function, including CYP metabolism, increases in cells that overexpress MIR122. Interestingly, MIR122, FOXA1, and HNF4A form a positive feedback loop, in which MIR122 upregulates expression of FOXA1, whereas FOXA1 upregulates expression of HNF4A and HNF4A increases expression of MIR122.76
MIR194 is also strongly up-regulated during hepatocyte differentiation. Overexpression accelerates the differentiation toward hepatocytes in HepaRG cells (a human liver progenitor cell line) and in human ESCs. Furthermore, MIR194 reduces expression of YAP1, a factor in the Hippo signaling pathway, which regulates liver development.77 MicroRNAs regulate hepatocyte maturation and can be used as biomarkers of this process. Although their use in HLC generation is feasible, efficient and robust techniques are needed to transfer microRNAs into HLCs.
Small Molecules
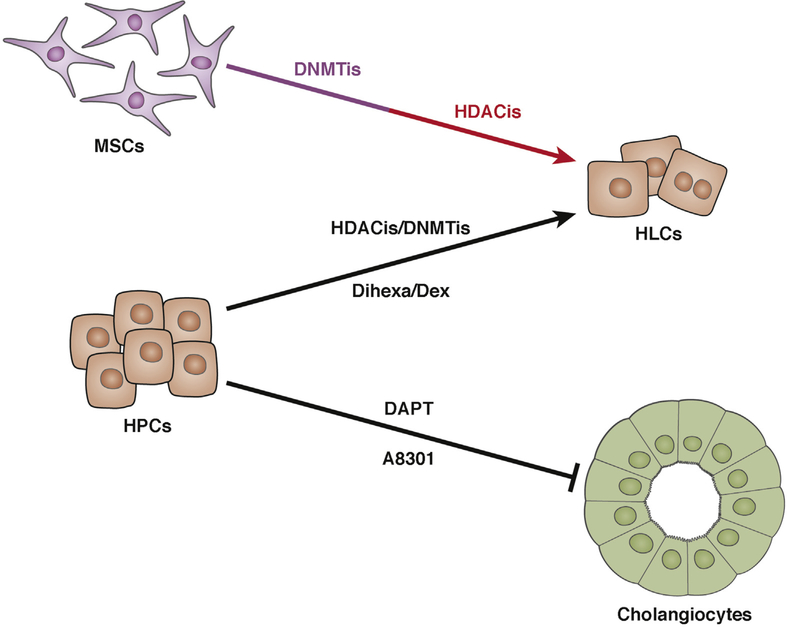
Natural and synthetic small molecules, identified by cellbased phenotypic screens, are useful chemical tools for controlling and manipulating cell fate—they are safer, more efficient, robust, and cost effective. We can separate the direct targets of small molecules during hepatocellular differentiation and maturation into 3 categories (Figure 4).
Figure 4.
Small molecules that generate HLCs or promote HLC maturation. Several small molecules can promote differentiation of HPCs and MSCs into HLCs. DNMTis prime differentiation of MSCs into HLCs. HDACis act during or after differentiation into HLCs. DAPT and A8301 block differentiation of HPCs into cholangiocytes.
Molecules related to epigenetic modification, mainly histone acetylation and DNA methylation, contribute hepatic maturation. There have been many studies of the effects of histone deacetylase inhibitors (HDACis), such as trichostatin A and dimethylsulphoxide, on hepatocyte differentiation.78–84 HDACis have a broad range of effects, including up-regulating expression of LETFs, such as HNF4A, HNF1A, CEBPA, and FOXA2,79 and increasing transcriptional activity by binding to promoter regions of CYP genes and glucose-6-phosphate dehydrogenase.80–82 Additionally, HDAC is regulate liverspecific expression of MIR122.78
HDACi-induced differentiation is always associated with proliferation arrest,85 which is an undesired phenotype of adult hepatocytes in vitro. Inhibitors of DNA methylation (DNMTis), such as 5-azacytidine and 5-aza-2’-deoxycytidine, also induce transcription of hepatocyte-specific genes.86,87 Ideally, HDACis and DNMTis can be applied together: DNMTis would be used as preconditioning agents before hepatic differentiation, whereas HDACis would be applied during or after differentiation.88
Signaling pathway-specific antagonists and agonists including Notch, HGF and its receptor c-Met, and dexamethasone are important for the specification of hepatoblasts to either hepatocytes or cholangiocytes. Notch activation increases expression of the biliary regulator HNF1B, and reduces expression of hepatocyte regulators HNF1A, HNF4A, and CEBPA.8 Studies in humans, mice, and dogs confirmed that Notch inhibition induced differentiation of Lgr5+ liver adult stem cells toward HLCs.
A8301 inhibits transforming growth factor-β, which promotes the epithelial to mesenchymal transition and dedifferentiation of hepatocytes.89 Siller et al90 generated functional HLCs with the HGF receptor agonist, N-hexanoicTyr, Ile-(6) aminohexanoic amide (dihexa). In particular, dexamethasone, a synthesized glucocorticoid, induces CYP3A, which mediates drug metabolism and synthesis of cholesterol, steroids, and other lipids in hepatocytes.91 It also promotes hepatocyte morphogenesis and formation of intercellular junctions, which are required for xenobiotic biotransformation, albumin secretion, and ammonia detoxification,92 and up-regulates expression and transcriptional activity of LETFs, such as CEBPA, HNF1A, HNF4A, and FOXA1.93 Dexamethasone also activates glucocorticoid receptor signaling; glucocorticoid receptor–dependent regulatory sequences are found within promoters of many hepatocyte-specific genes, including the albumin gene and CYP2B.94 For these reasons, dexamethasone is used in almost all protocols for end-stage differentiation of hepatocytes.
The third category consists of chemicals that are known to promote hepatocyte maturation, with unknown mechanisms of action. Shan et al95 developed a high-throughput screening platform based on albumin secretion level and proliferation of human primary hepatocytes. From a library of 12,480 small molecules, they identified 2 agents that induced proliferation and albumin secretion (FPH1 and FPH2) and 1 that induced albumin secretion alone (FH1). Further tests on iPSC-derived HLCs confirmed that FPH1 and FH1 promote hepatocyte functions.95 Both molecules contain the same N-phenyl-2-(Nphenylmethylsulfonamido)-acetamide core, but have different substitutions around the sulfonamide and amide phenyl rings, providing options for optimizing hepatocyte maturation.
Small molecules may be an alternative to transcription factor or growth factor–induced differentiation with improved reproducibility and reduced costs. Progress has been made replacing growth factors in the differentiation of PSCs toward definitive endoderm.96,97 Recently, Siller et al90 developed a stepwise growth factor–free protocol for differentiating human PSCs toward a hepatic lineage, which reached similar levels of liver function compared to growth factor–based approaches.
Microenvironment
Hepatocytes in vivo are surrounded by extracellular matrix (ECM). Natural ECM-based hydrogels, including collagen type I and Matrigel, are routinely used for coating culture dishes for primary hepatocytes and HLCs. The collagen (type I) sandwich culture can mimic the 3D microenvironment for hepatocytes and is the standard for maintaining hepatocyte features in vitro.98 Cell-derived matrices contain a complex but organized mixture of bioactive and biocompatible materials, and are therefore more effective than pure collagen sandwich cultures for functional HLCs maturation. Guo et al99 demonstrated that endothelial cell–derived matrix promotes the metabolic maturation of human adipose stem cell–derived HLCs through activating hepatocyte-enriched transcription factors, FOXA2, HNF4A, and PXR. Scaffolds generated from decellularized human and rat liver ECM contain not only collagen, laminin, or fibronectin, but also ECM-bound growth factors, improving the physiology of the in vivo microenvironment.100,101 This also seems to be a tissue ECM–specific effect, because ECM derived from other tissues does not induce differentiation to the same extent.102 Recently, we created human liver organoids that were self-assembled in vitro from liver progenitor cells seeded onto decellularized liver ECM discs. These 3D liver organoids recapitulated several aspects of hepatobiliary organogenesis and resulted in concomitant formation of progressively more differentiated hepatocytes and bile duct structures.103 Applications of the ECM-based 3D culture are limited by the mass transfer barriers formed by the top layer, batch-to-batch variation of ECM, and uncontrollable coating.
Many hepatocyte functions are regulated by substances released from neighboring nonparenchymal cells, such as Kupffer cells, sinusoidal endothelial cells, stellate cells, and hepatic MSCs.104 Cytokines secreted by nonparenchymal cells comprise acidic FGF (stellate cells),105 HGF (stellate and endothelial cells),105,106 tumor necrosis factor (Kupffer cells),107 OSM (Kupffer cells),108 interleukin-6 (Kupffer, macrophages, and endothelial cells),109,110 transforming growth factor-β (Kupffer cells),111 interleukin-1 (Kupffer cells),110 and WNT3A (macrophages).112 Of these, HGF and OSM are considered to be inducers of hepatocyte maturation.46,47 Co-culture systems might be better models at the tissue level. Takebe et al113 showed that co-cultured human MSCs, human umbilical vein endothelial cells, and human iPSCs self-organized into 3D liver buds with functional liver properties. In addition, the liver buds were vascularized and, when transplanted, the vasculatures connected to host vessels and demonstrated clear liver function.
Single-cell RNA sequencing can be used to investigate transcriptome variations among cells. This technology was used to determine how genetic factors and molecules interact to control liver organoid formation.114 Researchers used single-cell RNA sequencing analyses to determine the complex patters of communication between the 3D microenvironment and different cell types. Interactions between vascular endothelial growth factor and its receptor induce angiogenesis in the developing liver. Although this approach has been used to study primarily early developmental processes, these tools might be used in generation of healthy and viable human liver tissue from human PSCs.
Required Functions of Adult Hepatocytes
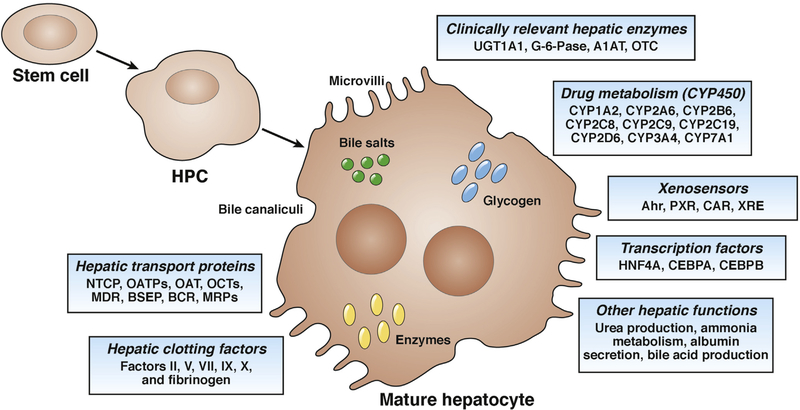
The availability of a homogeneous source of human hepatocytes is considered the most precious tool for toxicity screening. Hepatocytes also provide a renewable, cell-based source to examine other key factors of compound attrition, including metabolism of xenobiotics by CYP enzymes, drug interactions, hepatotoxicity, the activity of drug transporters, and regenerative medicine. What are the features that define hepatocytes structurally and functionally? These hepatocyte characteristics should be taken into consideration during the generation and maturation of human HLCs and freshly isolated primary human hepatocytes should always be used as the standard for comparison (Figure 5 and Table 1).
Figure 5.
Structural, metabolic, and secretory functions of mature hepatocytes. Mature hepatocytes have a typical epithelial cell structure, including polygonal shape and presence of epithelial markers, highly express genes related to drug metabolism (phase 1, 2, and 3), secrete albumin, and produce urea and bile acid.
Table 1.
Characteristics of Mature Human Hepatocytes
| Characteristic | Description |
|---|---|
| Morphology | |
| Epithelial morphology | Polygonal in shape; microvilli on cell surface |
| Polarization | Tight junctions (ZO1) in apical tip of the lateral membrane; adherens junctions (E-cadherin) at lateral membrane; adaptor proteins (α- and β-catenin) at basolateral membrane |
| Polyploidization | Presence of 2 or more nuclei in fraction of the cells |
| Gene expression | |
| Serum proteins | ALB, TF, TTR |
| Hepatocytic TFs | HNF4A, HNF1A, CEBPA/B |
| Metabolism enzymes | CYP3A4, CYP7A1, G6P |
| Absence of biliary markers | HNF1B, CK7, SOX9 |
| Absence of stem cell markers or hepatic immature markers | LGR5, OCT4, CD133, AFP, CYP3A7 |
| In vitro testing | |
| Phase 1 CYP activities | Metabolism of phenacetin (CYP1A2), bupropion (CYP2B6), tolbutamide (CYP2C9) diclofenac (CYP2G9), diclofenac (CYP3A4) |
| Phase 2 transferase activities | Metabolism of bilirubin (UGT1A1), dopamine (SULT1A1), p-aminobenzoic acid (NAT1), 1-chloro-2,4-dinitrobenzene (GSTs) |
| Phase 3 transporter activities | Transport of indocyanine green (OATP1B1), rhodamine 123 (MDR1), 5(6)-carboxy-2,070-dichlorofluorescein (MRP2) |
| Bile acid synthesis | Formation of cholic acid and chenodeoxycholic acid (CYP8A1, AKR1D1, AKR1C4) |
| Glycogen storage | Periodic acid–Schiff staining for glycogen |
| Serum protein synthesis | Secretion of albumin, α−1 antitrypsin, fibronectin, transferrin, apoprotein, transthyretin, complement factors, coagulation factors |
| Cholesterol metabolism | Formation of 7α-hydroxycholesterol (CYP7A1) |
| Lipid uptake | Transport of low-density lipoprotein |
| Urea metabolism | Synthesis of urea in cell extract |
| Coagulation factors | Factors II, V, VII, IX, X |
| Other clinically relevant enzymes | OCT, A1AT, BSEP, UGT1A1, G-6-Pase, FAH, ATP7B |
| In vivo testing | |
| Engraftment and repopulation | Human-specific markers or reporters to track transplanted HLCs in the liver (human-specific gene expression) |
| Restoration of liver function | Extended survival; presence of human proteins in the serum. Recovery of repopulating cells for genome-wide and functional metabolic studies |
| Tumorigenicity | Tumorigenesis in long-term transplantation experiments including several generations of re-transplantation |
Metabolic and Secretory Functions
The presence of hepatic enzymes with clinical implications would be useful for hepatic maturation categorization and disease modeling, for example, when studying UDPglucuronosyltransferase, glucose-6-phosphatase, or α1-antitrypsin deficiency.115 Additionally, the entire hepatic drug–metabolizing enzyme system provides an in vitro model for analyzing drug metabolism and predicting hepatotoxicity. CYPs are expressed primarily in the liver and CYP3A4 is the most abundant isoenzyme in human adult liver. Evaluation of CYPs in hepatocytes classified as phase 1 metabolism may include CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A7, and CYP7A1. The enzymes of greatest importance for drug metabolism belong to families 1–3, and are responsible for 70%–80% of all phase 1–dependent metabolism of clinically used drugs.116 Finally, urea cell cycle–related enzymes might be important when hepatic function of HLCs is evaluated; these include the ornithine transcarbamylase gene, which is located in mitochondria, along with carbamylphosphate synthetase I and argininosuccinate synthetase.117
Hepatic transport proteins and measurement of bile acids can serve as indicators of hepatic function and maturation. However, not all hepatic functions mature simultaneously and some hepatic transporters are expressed early in development and may not be exclusive for liver.118 Some important hepatic transport proteins can be classified as follows: the solute carrier SLC family, including Na+taurocholate co-transporting polypeptides, organic aniontransporting polypeptides, organic anion transporters, and organic cation transporters; the adenosine triphosphate– binding cassette transporter family, including the multidrug resistance proteins and bile salt export pump (both belong to the ABCB family), breast cancer resistance protein (belongs to the ABCG or White family); and the multi-drug resistance–associated proteins belonging to the ABCC family.
Chemical sensors like the aryl hydrocarbon receptor, PXR, and the constitutive androstane receptor are integral to the regulation and induction of the main CYPs and their analysis may provide strong evidence of the maturation state of HLCs. Similar to LETFs (eg, HNF4A and CEBPA),119 hepatic clotting factors (II, V, VII, IX, X, and fibrinogen), albumin production, urea production or ammonia metabolism, and glycogen storage may provide additional evidence of effective hepatic maturation and functional capacity in HLCs.
HLCs generated using different strategies show some degree of hepatic phenotypes, and because the features mentioned are characteristic of freshly isolated primary human hepatocytes, it is critical that they are analyzed comprehensively.
Cell Engraftment and Liver Repopulation
Patients with acute liver failure and inborn errors of hepatic metabolism are ideal candidates for cell therapy.120 Numerous studies in rodents and few clinical trials have demonstrated the long-term safety of this procedure.120–122 However, hepatocyte transplants in humans have only partially restored metabolic disorders.121,123 The efficacy of the procedure has been limited by poor cell engraftment soon after transplantation and our inability to monitor and predict rejection episodes.121 The problems of cell engraftment and repopulation of donor hepatocytes after cell transplantation might be solved by liver-directed radiation, which would inhibit host hepatocyte proliferation and induce post-mitotic hepatocyte death, allowing donor hepatocytes to proliferate and repopulate the irradiated host liver.124 Additional approaches to overcome engraftment and repopulation challenges involve expression of transgenes to provide cells with a repopulation advantage.125 Nygaard et al125 showed that hepatocytes carrying transgenes encoding coagulation factor 9 and a selection marker (a small hairpin RNA that makes cells resistant to a smallmolecule inhibitor of fumarylacetoacetate hydrolase) expressed these gene products after transplantation into mice. These experiments indicate that it is possible to genetically modify liver cells to facilitate repopulation of transplanted hepatocytes. Preclinical tests for HLCs in animal models of liver failure and/or regeneration is of great importance to demonstrate regeneration response, safety, and efficacy of HLCs after transplantation.
Immune-compromised mice with different types of liver injury have been used to study strategies to provide proliferative advantages to transplanted cells.126–130 The ability of transplanted cells to functionally repopulate livers of immune-deficient mice is the standard to establish that they are hepatocytes, rather than cells with hepatocyte-like features, which are unable to repopulate livers. To date, only limited repopulation of human HLCs has been reported.69,131–134
The ability of human HLCs to engraft, repopulate liver, and function should be evaluated in different animal models of liver injury (for review, see Rezvani et al135). Fumarylacetoacetate hydrolase–deficient mice are commonly used to evaluate the regenerative and functional capacities of human HLCs. Fah–/– mice have been crossed with Rag–/–/ Il2rg–/– mice to produce immune-compromised triplemutant mice (FRG). Livers of these mice can be repopulated up to 90% with transplanted human primary hepatocytes.127 When human HLCs were transplanted into and evaluated in FRG mice, the reported achieved liver repopulation ranged from 2% to 4% after transplantation.131
Severe combined immunodeficient mice, with liver injury caused by expression of the urokinase-type plasminogen activator, are also used in studies of liver repopulation by human HLCs. In these mice, as much as 20% of the liver can be repopulated, which was quantified by immunostaining after transplantation of 4 million HLCs.134 HLCs also repopulate livers of immune-suppressed Nagase analbuminemic rats.133 These rats were given injections of retrorsine and partial (70%) hepatectomies to induce proliferation of the transplanted cells. However, repopulation efficacy was not reported and the human serum albumin levels remained low. A similar lack of HLC repopulation was reported in Gunn rats, a model for Crigler-Najjar syndrome (deficient in bilirubin metabolism).132,136 In these experiments, researchers irradiated approximately 30% of the liver area and regeneration was induced by expression of hepatocyte growth factor (HGF) from an adenoviral vector. Transplanted HLCs repopulated 7.5% of the irradiated median liver lobe, quantified by immunostaining, and reduced levels of bilirubin by 60%.132
Future Directions
Biofabrication
Considerable effort has been dedicated to creation of hepatic tissue by 3D bioprinting.137–141 These novel techniques allow for the precise and simultaneous addition of cells and matrix molecules in a designed spatial configuration. With proper spatial control, bioprinters generate hepatic tissues with distinct cell types in discrete locations that resemble hepatic tissue architecture and cell distribution.138 These bioprinted, often called mini-livers, have some features of hepatocyte function, including albumin and urea secretion and drug metabolism to a certain level.138,140,141 However, the low resolution of available bioprinters and methods for bioprinting the vasculature (such as sacrificial hydrogel) make it impossible to print hepatic tissue with an integrated and perfusable vascular network with complex branching in a range of diameters from large vessels to capillaries.142,143 The same holds true for the periportal to pericentral zonal distrubition.142,143 The technology to transcend these challenges might be years away, but the potential of these biofabrication techniques is clear and compelling when considering whole-liver bioengineering. They allow not only the generation of organs on demand, but also a more exact and rapid 3D recreation of hepatic tissue with all of its refinements, and are only limited by our knowledge of what and where to print.
Although 3D bioprinting of whole livers lies in the future, 3D bioprinters and other biofabrication techniques might be used to generate more functional HLCs. ECM microarrays, modular assembly, rapid prototyping techniques to construct defined microarchitectures, and microchips to recreate the hepatic environment are already available.144–147 The biomechanical stimulation produced by culture medium perfusion in these platforms, associated with the small volumes used, produces highly efficient differentiation of iPSCs into several cell fates, including drugmetabolizing hepatocytes.148 These techniques might be used to accurately recreate the hepatic microenvironment and identify mechanisms of hepatic maturation, as well as to generate entirely functional HLCs.
Genetic Programming of Hepatic Maturation
To elucidate mechanisms of human liver maturation, we must learn more about the mechanisms that regulate hepatocyte differentiation and function. Induction of pluripotency and self-renewal in human iPSCs provides evidence for a powerful reprogramming approach. Our ability to control transcription of specific genes in human HLCs could lead to identification of factors that maintain a specific cell state or promote cell maturation. Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated Cas genes can be customized to precisely regulate gene expression and examine functions of gene products in cells. These approaches will facilitate high-throughput analyses of gene activation or repression in human HLCs and their differentiation intermediates.149,150
Balboa et al150 generated a chemically controllable dCas9 activator, by fusion with the dihydrofolate reductase destabilization domain. They showed that the destabilized dCas9 activator can be used to control human PSC differentiation into endodermal lineages. These CRISPR/Cas9 systems, next-generation genome sequencing, and stem cell technologies can be used in studies of liver development to identify genes that control functions of mature hepatocytes and determine how they are regulated.
Conclusions
The demand for functional liver tissue replacement options continues to grow and, despite advances in the differentiation of HLCs in vitro, generating stem cell–derived cells and tissues with primary adult hepatocyte function remains challenging. The complexity of liver tissue, together with the need for stable mature liver cells with high proliferation potential currently limit broader application of HLCs. A greater understanding of the factors involved in hepatocyte biology coupled with emerging technologies, such as biofabrication and (epi)genetic programming, may provide the tools necessary to advance the development and application of human stem cell–derived hepatocytes.
Acknowledgments
Funding
This work was supported by grants from the National Institutes of Health (DK099257) to A.S.-G.; Marie Curie Actions (H2020-MSCA-IF-2014 Project #660554), project PI15/00563 and CIBEREHD EH16PI02 from Instituto de Salud Carlos III, Spain to P.M.B.; Dutch Research Council NWO STW (15498) and Dutch Research Council NWO ZON/MW (116004121) to B.S., and China Scholarship Council (CSC201306310019) to C.C.
Abbreviations used in this paper:
- CRISPR
clustered regularly interspaced short palindromic repeats
- CYP
cytochrome P450
- 3D
3-dimensional
- DNMTi
DNA methylation inhibitor
- ECM
extracellular matrix
- ESC
embryonic stem cell
- ES-HLC
embryonic stem cell-derived hepatocyte-like cell
- FGF
fibroblast growth factor
- HDACi
histone deacetylase inhibitor
- HGF
hepatocyte growth factor
- HLC
hepatocyte-like cell
- HNF4A
hepatocyte nuclear factor 4a
- HPC
hepatic progenitor/stem cell
- iPSC
induced pluripotent stem cell
- LETF
liver-enriched transcription factor
- MSC
mesenchymal stromal or stem cell
- OSM
oncostatin M
- PSC
pluripotent stem cell
- PXR
pregnane X receptor
Footnotes
Conflicts of interest
This author discloses the following: A.S.-G. has a provisional patent application that describes hepatic differentiation of human pluripotent stem cells and liver repopulation. A.S.-G. is a co-founder and has a financial interest in Von Baer Wolff, Inc, a company focused on biofabrication of autologous human hepatocytes from stem cells technology. A.S.-G.’s interests are managed by the Conflict of Interest Office at the University of Pittsburgh in accordance with their policies. The remaining authors disclose no conflicts.
References
- 1.Kim WR, Therneau TM, Benson JT, et al. Deaths on the liver transplant waiting list: an analysis of competing risks. Hepatology 2006;43:345–351. [DOI] [PubMed] [Google Scholar]
- 2.Soldatow VY, LeCluyse EL, Griffith LG, et al. In vitro models for liver toxicity testing. Toxicol Res 2013;2: 23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidotti JE, Bregerie O, Robert A, et al. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem 2003;278:19095–19101. [DOI] [PubMed] [Google Scholar]
- 4.Gentric G, Desdouets C. Polyploidization in liver tissue. Am J Pathol 2014;184:322–331. [DOI] [PubMed] [Google Scholar]
- 5.Zaret KS. From endoderm to liver bud: paradigms of cell type specification and tissue morphogenesis. Curr Top Dev Biol 2016;117:647–669. [DOI] [PubMed] [Google Scholar]
- 6.Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet 2008;9:329–340. [DOI] [PubMed] [Google Scholar]
- 7.Schmelzer E, Zhang L, Bruce A, et al. Human hepatic stem cells from fetal and postnatal donors. J Exp Med 2007;204:1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanimizu N, Miyajima A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J Cell Sci 2004; 117:3165–3174. [DOI] [PubMed] [Google Scholar]
- 9.Decaens T, Godard C, de Reyniès A, et al. Stabilization of β-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology 2008;47:247–258. [DOI] [PubMed] [Google Scholar]
- 10.Clotman F, Jacquemin P, Plumb-Rudewiez N, et al. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev 2005;19:1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huch M, Dorrell C, Boj SF, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013;494:247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huch M, Gehart H, Van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nantasanti S, Spee B, Kruitwagen HS, et al. Disease modeling and gene therapy of copper storage disease in canine hepatic organoids. Stem Cell Rep 2015;5:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872. [DOI] [PubMed] [Google Scholar]
- 15.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010;51:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Touboul T, Hannan NRF, Corbineau S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010;51:1754–1765. [DOI] [PubMed] [Google Scholar]
- 17.Song Z, Cai J, Liu Y, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res 2009;19:1233–1242. [DOI] [PubMed] [Google Scholar]
- 18.Banas A, Teratani T, Yamamoto Y, et al. Adipose tissuederived mesenchymal stem cells as a source of human hepatocytes. Hepatology 2007;46:219–228. [DOI] [PubMed] [Google Scholar]
- 19.Lee K-D, Kuo TK-C, Whang-Peng J, et al. In vitro hepatic differentiation of human mesenchymal stem cells. Hepatology 2004;40:1275–1284. [DOI] [PubMed] [Google Scholar]
- 20.Wu X-B, Tao R. Hepatocyte differentiation of mesenchymal stem cells. Hepatobiliary Pancreat Dis Int 2012; 11:360–371. [DOI] [PubMed] [Google Scholar]
- 21.Lu W-Y, Bird TG, Boulter L, et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 2015;17:971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beath S Hepatic function and physiology in the newborn. Semin Neonatol 2003;8:337–346. [DOI] [PubMed] [Google Scholar]
- 23.Grijalva J, Vakili K. Neonatal liver physiology. Semin Pediatr Surg 2013;22:185–189. [DOI] [PubMed] [Google Scholar]
- 24.Lautt WW, ed. Hepatic Circulation Physiology and Pathophysiology. San Rafael, CA: Morgan and Claypool Life Sciences, 2009. [PubMed] [Google Scholar]
- 25.Emery JL. Functional asymmetry of the liver. Ann N Y Acad Sci 1963;111:37–44. [DOI] [PubMed] [Google Scholar]
- 26.Rudolph AM. Hepatic and ductus venosus blood flows during fetal life. Hepatology 1983;3:254–258. [DOI] [PubMed] [Google Scholar]
- 27.Lautt WW, Greenway CV. Conceptual review of the hepatic vascular bed. Hepatology 1987;7:952–963. [DOI] [PubMed] [Google Scholar]
- 28.Lind J Changes in the liver circulation at birth. Ann N Y Acad Sci 2006;111:110–120. [DOI] [PubMed] [Google Scholar]
- 29.<Zink J. The fetal and neonatal hepatic circulation In: Lautt WW, ed. Hepatic Circulation in Health and Disease. New York: Raven Press, 1981:227–248. [Google Scholar]
- 30.Tilles AW, Baskaran H, Roy P, et al. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol Bioeng 2001;73:379–389. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka Y, Yamato M, Okano T, et al. Evaluation of effects of shear stress on hepatocytes by a microchipbased system. Meas Sci Technol 2006;17:3167–3170. [Google Scholar]
- 32.Goral VN, Yuen PK. Microfluidic platforms for hepatocyte cell culture: new technologies and applications. Ann Biomed Eng 2012;40:1244–1254. [DOI] [PubMed] [Google Scholar]
- 33.Kidambi S, Yarmush RS, Novik E, et al. Oxygenmediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc Natl Acad Sci 2009;106:15714–15719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.<j/>Mallanna SK, Duncan SA. Differentiation of hepatocytes from pluripotent stem cells In: Schlaeger T, ed. Current Protocols in Stem Cell Biology. Vol 1 Hoboken, NJ: John Wiley & Sons, Inc, 2013:1G.4.1–1G.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baptista PM, Moran EC, Vyas D, et al. Fluid flow regulation of revascularization and cellular organization in a bioengineered liver platform. Tissue Eng Part C Methods 2016;22:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.<j/>Monga SPS, ed. Molecular Pathology of Liver Diseases. Boston, MA: Springer, 2011. [Google Scholar]
- 37.Katz N, Teutsch HF, Jungermann K, et al. Heterogeneous reciprocal localization of fructose-1,6-bisphosphatase and of glucokinase in microdissected periportal and perivenous rat liver tissue. FEBS Lett 1977;83:272–276. [DOI] [PubMed] [Google Scholar]
- 38.Selwyn FP, Cui JY, Klaassen CD. Special section on drug metabolism and the microbiome RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos 2015;43:1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selwyn FP, Cheng SL, Klaassen CD, et al. Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos 2016;44:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selwyn FP, Cheng SL, Bammler TK, et al. Developmental regulation of drug-processing genes in livers of germfree mice. Toxicol Sci 2015;147:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avior Y, Levy G, Zimerman M, et al. Microbial-derived lithocholic acid and vitamin K 2 drive the metabolic maturation of pluripotent stem cells-derived and fetal hepatocytes. Hepatology 2015;62:265–278. [DOI] [PubMed] [Google Scholar]
- 42.Jump DB, Ren B, Clarke S, et al. Effects of fatty acids on hepatic gene expression. Prostaglandins Leukot Essent Fatty Acids 1995;52:107–111. [DOI] [PubMed] [Google Scholar]
- 43.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med 2001;7:584–590. [DOI] [PubMed] [Google Scholar]
- 44.Laffitte BA, Chao LC, Li J, et al. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc Natl Acad Sci U S A 2003;100:5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang W, Ma K, Zhang J, et al. Nuclear receptordependent bile acid signaling is required for normal liver regeneration. Science 2006;312:233–236. [DOI] [PubMed] [Google Scholar]
- 46.Kamiya A, Kinoshita T, Ito Y, et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J 1999;18: 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamiya A, Kinoshita T, Miyajima A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett 2001;492:90–94. [DOI] [PubMed] [Google Scholar]
- 48.Levy G, Bomze D, Heinz S, et al. Long-term culture and expansion of primary human hepatocytes. Nat Biotech 2015;33:1264–1271. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt C, Bladt F, Goedecke S, et al. Scatter factor/ hepatocyte growth factor is essential for liver development. Nature 1995;373:699–702. [DOI] [PubMed] [Google Scholar]
- 50.Kitade M, Factor VM, Andersen JB, et al. Specific fate decisions in adult hepatic progenitor cells driven by MET and EGFR signaling. Genes Dev 2013;27:1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ballard FJ, Wong SSC, Knowles SE, et al. Insulin inhibition of protein degradation in cell monolayers. J Cell Physiol 1980;105:335–346. [DOI] [PubMed] [Google Scholar]
- 52.Balavoine S, Feldmann G, Lardeux B. Regulation of RNA degradation in cultured rat hepatocytes: effects of specific amino acids and insulin. J Cell Physiol 1993;156:56–62. [DOI] [PubMed] [Google Scholar]
- 53.Dahn MS, Hsu CJ, Lange MP, et al. Factors affecting secretory protein production in primary cultures of rat hepatocytes. Proc Soc Exp Biol Med 1993;203:38–44. [DOI] [PubMed] [Google Scholar]
- 54.Flaim KE, Hutson SM, Lloyd CE, et al. Direct effect of insulin on albumin gene expression in primary cultures of rat hepatocytes. Am J Physiol 1985;249:E447–E453. [DOI] [PubMed] [Google Scholar]
- 55.Odom DT, Zizlsperger N, Gordon DB, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 2004;303:1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishikawa T, Bell A, Brooks JM, et al. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest 2015;125:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev 2002;54:129–158. [DOI] [PubMed] [Google Scholar]
- 58.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev 2004;56:291–330. [DOI] [PubMed] [Google Scholar]
- 59.Sumi K, Tanaka T, Uchida A, et al. Cooperative interaction between hepatocyte nuclear factor 4 alpha and GATA transcription factors regulates ATP-binding cassette sterol transporters ABCG5 and ABCG8. Mol Cell Biol 2007;27:4248–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alder O, Cullum R, Lee S, et al. Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Rep 2014;9:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson DA, Rowader KE, Stevens K, et al. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol Cell Biol 1993;13: 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metzger S, Halaas JL, Breslow JL, et al. Orphan receptor HNF-4 and bZip protein C/EBP alpha bind to overlapping regions of the apolipoprotein B gene promoter and synergistically activate transcription. J Biol Chem 1993; 268:16831–16838. [PubMed] [Google Scholar]
- 63.Samadani U, Costa RH. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol Cell Biol 1996;16:6273–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuo CJ, Conley PB, Chen L, et al. A transcriptional hierarchy involved in mammalian cell-type specification. Nature 1992;355:457–461. [DOI] [PubMed] [Google Scholar]
- 65.Ktistaki E, Talianidis I. Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science 1997;277:109–112. [DOI] [PubMed] [Google Scholar]
- 66.Eeckhoute J, Formstecher P, Laine B. Hepatocyte nuclear factor 4α enhances the hepatocyte nuclear factor 1α-mediated activation of transcription. Nucleic Acids Res 2004;32:2586–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hatzis P, Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol Cell Biol 2001;21:7320–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 69.Huang P, Zhang L, Gao Y, He Z, et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 2014;14:370–384. [DOI] [PubMed] [Google Scholar]
- 70.Du Y, Wang J, Jia J, Song N, Xiang C, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 2014;14:394–403. [DOI] [PubMed] [Google Scholar]
- 71.Godoy P, Schmidt-Heck W, Natarajan K, et al. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J Hepatol 2015;63:934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogel C, Marcotte EM. Insights into regulation of protein abundance from proteomics and transcriptomis analyses. Nat Rev Genet 2013;13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou X, Cui L, Zhou X, et al. Induction of hepatocyte-like cells from human umbilical cord-derived mesenchymal stem cells by defined microRNAs. J Cell Mol Med 2016; 21:881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002;12:735–739. [DOI] [PubMed] [Google Scholar]
- 75.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87–98. [DOI] [PubMed] [Google Scholar]
- 76.Deng XG, Qiu RL, Wu YH, et al. Overexpression of miR122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4apositive feedback loop. Liver Int 2014;34:281–295. [DOI] [PubMed] [Google Scholar]
- 77.Jung KH, McCarthy RL, Zhou C, et al. MicroRNA regulates hepatocytic differentiation of progenitor cells by targeting YAP1. Stem Cells 2016;34:1284–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alizadeh E, Eslaminejad MB, Akbarzadeh A, et al. Upregulation of miR-122 via trichostatin A treatments in hepatocyte-like cells derived from mesenchymal stem cells. Chem Biol Drug Des 2016;87:296–305. [DOI] [PubMed] [Google Scholar]
- 79.Henkens T, Papeleu P, Elaut G, et al. Trichostatin A, a critical factor in maintaining the functional differentiation of primary cultured rat hepatocytes. Toxicol Appl Pharmacol 2007;218:64–71. [DOI] [PubMed] [Google Scholar]
- 80.Bort R, Gómez-Lechón MJ, Castell JV, et al. Role of hepatocyte nuclear factor 3 gamma in the expression of human CYP2C genes. Arch Biochem Biophys 2004; 426:63–72. [DOI] [PubMed] [Google Scholar]
- 81.Rodríguez-Antona C, Bort R, Jover R, et al. Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol Pharmacol 2003;63: 1180–1189. [DOI] [PubMed] [Google Scholar]
- 82.Massillon D, Arinze IJ, Xu C, et al. Regulation of glucose-6phosphatase gene expression in cultured hepatocytes and H4IIE cellsbyshort-chainfattyacids:roleofhepatic nuclear factor-4alpha. J Biol Chem 2003;278:40694–40701. [DOI] [PubMed] [Google Scholar]
- 83.Soto-Gutiérrez A, Kobayashi N, Rivas-Carrillo JD, et al. Reversal of mouse hepatic failure using an implanted liver-assist device containing ES cell-derived hepatocytes. Nat Biotechnol 2006;24:1412–1419. [DOI] [PubMed] [Google Scholar]
- 84.Alizadeh E, Zarghami N, Eslaminejad MB, et al. The effect of dimethyl sulfoxide on hepatic differentiation of mesenchymal stem cells. Artif Cells Nanomed Biotechnol 2014:1–8. [DOI] [PubMed] [Google Scholar]
- 85.Papeleu P, Loyer P, Vanhaecke T, et al. Trichostatin A induces differential cell cycle arrests but does not induce apoptosis in primary cultures of mitogen-stimulated rat hepatocytes. J Hepatol 2003;39:374–382. [DOI] [PubMed] [Google Scholar]
- 86.Jin B, Park DW, Nam KW, et al. CpG methylation of the mouse CYP1A2 promoter. Toxicol Lett 2004;152:11–18. [DOI] [PubMed] [Google Scholar]
- 87.Sgodda M, Aurich H, Kleist S, et al. Hepatocyte differentiation of mesenchymal stem cells from rat peritoneal adipose tissue in vitro and in vivo. Exp Cell Res 2007; 313:2875–2886. [DOI] [PubMed] [Google Scholar]
- 88.Snykers S, Henkens T, De Rop E, et al. Role of epigenetics in liver-specific gene transcription, hepatocyte differentiation and stem cell reprogrammation. J Hepatol 2009;51:187–211. [DOI] [PubMed] [Google Scholar]
- 89.Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology 2009;50:2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siller R, Greenhough S, Naumovska E, et al. Smallmolecule-driven hepatocyte differentiation of human pluripotent stem cells. Stem Cell Rep 2015;4:939–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pascussi JM, Drocourt L, Gerbal-Chaloin S, et al. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur J Biochem 2001; 268:6346–6358. [DOI] [PubMed] [Google Scholar]
- 92.Vinken M, Papeleu P, Snykers S, et al. Involvement of cell junctions in hepatocyte culture functionality. Crit Rev Toxicol 2006;36:299–318. [DOI] [PubMed] [Google Scholar]
- 93.Fraczek J, Bolleyn J, Vanhaecke T, et al. Primary hepatocyte cultures for pharmaco-toxicological studies: at the busy crossroad of various anti-dedifferentiation strategies. Arch Toxicol 2013;87:577–610. [DOI] [PubMed] [Google Scholar]
- 94.Schuetz EG, Schmid W, Schutz G, et al. The glucocorticoid receptor is essential for induction of cytochrome P-4502B by steroids but not for drug or steroid induction of CYP3A or P-450 reductase in mouse liver. Drug Metab Dispos 2000;28:268–278. [PubMed] [Google Scholar]
- 95.Shan J, Schwartz RE, Ross NT, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol 2013;9:514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Borowiak M, Maehr R, Chen S, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 2009;4: 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tahamtani Y, Azarnia M, Farrokhi A, et al. Treatment of human embryonic stem cells with different combinations of priming and inducing factors toward definitive endoderm. Stem Cells Dev 2013;22:1419–1432. [DOI] [PubMed] [Google Scholar]
- 98.j/>Herrero A, Gerbal-Chaloin S, Daujat-Chavanieu M. Bioinspired liver tissue engineering In: Jabbari E, Kim DH, Lee LP, et al. , eds. Handbook of Biomimetics and Bioinspiration. World Scientific Publishing Company, 2014:1177–1239. [Google Scholar]
- 99.Guo X, Li W, Ma M, et al. Endothelial cell-derived matrix promotes the metabolic functional maturation of hepatocyte via integrin-Src signalling. J Cell Mol Med 2017; 11:2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang B, Jakus AE, Baptista PM, et al. Functional maturation of induced pluripotent stem cell hepatocytes in extracellular matrix—a comparative analysis of bioartificial liver microenvironments. Stem Cells Transl Med 2016;5:1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Soto-Gutierrez A, Zhang L, Medberry C, et al. A wholeorgan regenerative medicine approach for liver replacement. Tissue Eng Part C Methods 2011;17:677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shamis Y, Hasson E, Soroker A, et al. Organ-specific scaffolds for in vitro expansion, differentiation, and organization of primary lung cells. Tissue Eng Part C Methods 2011;17:861–870. [DOI] [PubMed] [Google Scholar]
- 103.Vyas D, Baptista PM, Brovold M, et al. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 2018;1:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Godoy P, Hewitt NJ, Albrecht U, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013;87:1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 2008; 88:125–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bussolino F, Di Renzo MF, Ziche M, et al. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J Cell Biol 1992;119:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roberts RA, Ganey PE, Ju C, et al. Role of the Kupffer cell in mediating hepatic toxicity and carcinogenesis. Toxicol Sci 2007;96:2–15. [DOI] [PubMed] [Google Scholar]
- 108.Henkel J, Gärtner D, Dorn C, et al. Oncostatin M produced in Kupffer cells in response to PGE2: possible contributor to hepatic insulin resistance and steatosis. Lab Invest 2011;91:1107–1117. [DOI] [PubMed] [Google Scholar]
- 109.Gregory SH, Wing EJ, Danowski KL, et al. IL-6 produced by Kupffer cells induces STAT protein activation in hepatocytes early during the course of systemic listerial infections. J Immunol 1998;160:6056–6061. [PubMed] [Google Scholar]
- 110.Feder LS, Todaro JA, Laskin DL. Characterization of interleukin-1 and interleukin-6 production by hepatic endothelial cells and macrophages. J Leukoc Biol 1993; 53:126–132. [DOI] [PubMed] [Google Scholar]
- 111.De Bleser PJ, Niki T, Rogiers V, et al. Transforming growth factor-beta gene expression in normal and fibrotic rat liver. J Hepatol 1997;26:886–893. [DOI] [PubMed] [Google Scholar]
- 112.Boulter L, Govaere O, Bird TG, et al. Macrophagederived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 2012;18:572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takebe T, Zhang R-R, Koike H, et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat Protoc 2014;9:396–409. [DOI] [PubMed] [Google Scholar]
- 114.Camp JG, Sekine K, Gerber T, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017;546:533–538. [DOI] [PubMed] [Google Scholar]
- 115.Ghouse R, Chu A, Wang Y, et al. Mysteries of alpha-1antitrypsin deficiency: emerging therapeutic strategies for a challenging disease. Dis Model Mech 2014;7:411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sinz M, Wallace G, Sahi J. Current industrial practices in assessing CYP450 enzyme induction: preclinical and clinical. AAPS J 2008;10:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raghuveer TS, Garg U, Graf WD. Inborn errors of metabolism in infancy and early childhood: an update. Am Fam Physician 2006;73:1981–1990. [PubMed] [Google Scholar]
- 118.van Montfoort JE, Hagenbuch B, Groothuis GMM, et al. Drug uptake systems in liver and kidney. Curr Drug Metab 2003;4:185–211. [DOI] [PubMed] [Google Scholar]
- 119.Duncan SA, Navas MA, Dufort D, et al. Regulation of a transcription factor network required for differentiation and metabolism. Science 1998;281:692–695. [DOI] [PubMed] [Google Scholar]
- 120.Soltys KA, Soto-Gutiérrez A, Nagaya M, et al. Barriers to the successful treatment of liver disease by hepatocyte transplantation. J Hepatol 2010;53:769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soltys KA, Setoyama K, Tafaleng EN, et al. Host conditioning and rejection monitoring in hepatocyte transplantation in humans. J Hepatol 2017;66:987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Puppi J, Strom SC, Hughes RD, et al. Improving the techniques for human hepatocyte transplantation: report from a consensus meeting in London. Cell Transplant 2012;21:1–10. [DOI] [PubMed] [Google Scholar]
- 123.Fox IJ, Chowdhury JR, Kaufman SS, et al. Treatment of the Crigler-Najjar syndrome type I with hepatocyte transplantation. N Engl J Med 1998;338:1422–1426. [DOI] [PubMed] [Google Scholar]
- 124.Yamanouchi K, Zhou H, Roy-Chowdhury N, et al. Hepatic irradiation augments engraftment of donor cells following hepatocyte transplantation. Hepatology 2009; 49:258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nygaard S, Barzel A, Haft A, et al. A universal system to select gene-modified hepatocytes in vivo. Sci Transl Med 2016;8:342ra79–342ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tateno C, Yoshizane Y, Saito N, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol 2004;165:901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Azuma H, Paulk N, Ranade A, et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol 2007;25:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bissig K-D, Wieland SF, Tran P, et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest 2010;120:924–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hasegawa M, Kawai K, Mitsui T, et al. The reconstituted “humanized liver” in TK-NOG mice is mature and functional. Biochem Biophys Res Commun 2011;405: 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Strom SC, Davila J, Grompe M. Chimeric mice with humanized liver: tools for the study of drug metabolism, excretion, and toxicity. Methods Mol Biol 2010;640: 491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu S, Rezvani M, Harbell J, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature 2014;508:93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen Y, Li Y, Wang X, et al. Amelioration of hyperbilirubinemia in Gunn rats after transplantation of human induced pluripotent stem cell-derived hepatocytes. Stem Cell Reports 2015;5:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Basma H, Soto-Gutiérrez A, Yannam GR, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology 2009; 136:990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carpentier A, Tesfaye A, Chu V, et al. Engrafted human stem cell–derived hepatocytes establish an infectious HCV murine model. J Clin Invest 2014;124:4953–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rezvani M, Grimm AA, Willenbring H. Assessing the therapeutic potential of lab-made hepatocytes. Hepatology 2016;64:287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chowdhury JR, Kondapalli R, Chowdhury NR. Gunn rat: a model for inherited deficiency of bilirubin glucuronidation. Adv Vet Sci Comp Med 1993;37:149–173. [PubMed] [Google Scholar]
- 137.Faulkner-Jones A, Fyfe C, Cornelissen D-J, et al. Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D. Biofabrication 2015; 7:44102. [DOI] [PubMed] [Google Scholar]
- 138.Nguyen DG, Funk J, Robbins JB, et al. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One 2016;11:e0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ma X, Qu X, Zhu W, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci 2016;113:2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhong C, Xie H-Y, Zhou L, et al. Human hepatocytes loaded in 3D bioprinting generate mini-liver. Hepatobiliary Pancreat Dis Int 2016;15:512–518. [DOI] [PubMed] [Google Scholar]
- 141.Massa S, Sakr MA, Seo J, et al. Bioprinted 3D vascularized tissue model for drug toxicity analysis. Biomicrofluidics 2017;11:44109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ji S, Guvendiren M. Recent advances in bioink design for 3D bioprinting of tissues and organs. Front Bioeng Biotechnol 2017;5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang YS, Yue K, Aleman J, et al. 3D bioprinting for tissue and organ fabrication. Ann Biomed Eng 2017; 45:148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods 2005;2:119–125. [DOI] [PubMed] [Google Scholar]
- 145.McGuigan AP, Sefton MV. Vascularized organoid engineered by modular assembly enables blood perfusion. Proc Natl Acad Sci U S A 2006;103:11461–11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol 2008; 26:120–126. [DOI] [PubMed] [Google Scholar]
- 147.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014;32:760–772. [DOI] [PubMed] [Google Scholar]
- 148.Giobbe GG, Michielin F, Luni C, et al. Functional differentiation of human pluripotent stem cells on a chip. Nat Methods 2015;12:637–640. [DOI] [PubMed] [Google Scholar]
- 149.Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015;12:326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Balboa D, Weltner J, Eurola S, et al. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem cell reports 2015;5:448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]