Abstract
Flavonols are a flavonoid subfamily widely distributed in plants, including several ones of great importance in human and animal diet (apple, tomato, broccoli, onion, beans, tea). These polyphenolic nutraceuticals exert potent antimicrobial (membrane potential disruptors), antioxidant (free-radical scavengers), pharmacokinetic (CYP450 modulators), anti-inflammatory (lipoxygenase inhibitors), antiangiogenic (VEGF inhibitors) and antitumor (cyclin inhibitors) activities. Biotechnological production of these nutraceuticals, for example via heterologous biosynthesis in industrial actinomycetes, is favored since in plants these polyphenols appear as inactive glycosylated derivatives, in low concentrations or as part of complex mixtures with other polyphenolic compounds. In this work, we describe the de novo biosynthesis of three important flavonols, myricetin, kaempferol and quercetin, in the industrially relevant actinomycetes Streptomyces coelicolor and S. albus. De novo biosynthesis of kaempferol, myricetin and quercetin in actinomycetes has not been described before.
Introduction
Flavonoids (from Latin flavus, yellow) are a family of about 6000 nutraceuticals widely distributed in plant cells, many of them found in dietary plants [1–5]. All flavonoids have a generic chemical structure consisting of 15 carbon atoms (C6-C3-C6): two aromatic rings (rings A and B) connected by a heterocyclic pyran C which contains one oxygen (ring C, Fig 1) [6–11]. This basic skeleton can have multiple substituents, such as hydroxyl or methyl groups, as well as sugars [12]; indeed, chemical modifications in ring C lead to the formation of more than 9,000 flavonoid derivates [13].
Fig 1. Engineered flavonoid biosynthetic pathway in Streptomyces sp., including the different feeding experiments with naringenin (dashed lines).
Enzyme abbreviations: TAL, Tyrosine ammonia-lyase; 4CL, 4-coumaroyl CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; N3DOX, naringenin 3-dioxygenase; FLS1, flavonol synthase 1; F3’H, flavonoid 3’-hydroxylase; F3’5’H, flavonoid 3’,5’-hydroxylase.
Depending on the pattern of hydroxylation and the substituents on the heterocyclic ring C, flavonoids can be classified into several sub-groups, but in this paper we will focus on flavonols. They are an important subfamily, as some of their members, like myricetin, kaempferol and quercetin, represent the major intake of dietary flavonoids in most societies [2,3,9]. Actually, quercetin is the most common flavonoid in human diet, with an average intake of 13 mg/day from a total of 20 to 50 mg/day for all flavonoids [14]. The reason for this is that quercetin concentration is very high in some vegetables, like in onions (1.2 g/kg) and cabbage (0.6 mg/kg), but also in many other fruits such as blueberries, apples, tomatoes or peaches [2]. Kaempferol is also present in many plant products: flowers (1.2 mg/kg in bee pollen, 205 mg/kg in saffron), fruits (29 mg/kg in beans) and vegetables (13 mg/kg in broccoli, 22 mg/kg in cabbage and 131 mg/kg in cappers) [15–18]. Finally, myricetin can be found in diverse food sources such as tea (940 mg/kg), grapes (15 mg/kg), blackcurrants (71 mg/kg), cranberries (142 mg/kg) and blueberries (26 mg/kg) [19–21]. As other flavonoids, flavonols commonly appear in these foods as glucose or rhamnose conjugates [22,23].
As antioxidant compounds, myricetin and quercetin are able to induce glutathione-S-transferase, an important enzyme involved in oxidative stress resistance [24]. Also, these flavonols are able to act directly as free-radical scavengers preventing DNA, protein and membrane damages thanks to their aromatic hydroxyl groups [25].
As well as other flavonoids, flavonols possess anti-inflammatory activities. For example, kaempferol and quercetin can inhibit tyrosine kinases involved in activated macrophage proliferation [26], and myricetin and quercetin are able to inhibit lipoxygenases, which catalyze important steps during formation of pro-inflammatory leukotrienes and hepoxilins [27]. Linked to these anti-inflammatory activities, kaempferol and quercetin are able to protect also against diverse pro-inflammatory and pro-carcinogenic agents, as they can bind to the aryl hydrocarbon receptor AhR (the activator of CYP1A1 and CYP1A2 transcription), therefore protecting cells against more reactive metabolites (on DNA, etc.) generated by the action of the detoxifying cytochromes P450 on polycyclic aromatics and halogenated toxins [28,29]. These effects, together with other ones, associated to cyclins inhibition and p53 concentrations increase carried out by myricetin and quercetin, contribute to the antitumor activity shown by flavonol nutraceuticals [30]. Myricetin, kaempferol and quercetin are also able to block angiogenesis, by inhibiting VEGF, another activity linked to antitumor properties [31].
Apart from these bioactivities in eukaryotic cells, flavonols are also important antimicrobial agents in the producer plants, as they are able to modify membrane transport mechanisms, altering membrane potential and leading to bacterial death [32,33].
In planta, flavonoids are synthesized by complexes of various enzymes that are present on the cytosolic face of endoplasmic reticulum membranes. The first steps for flavonoid biosynthesis are included in the phenylpropanoid pathway, which converts L-Phe in 4-coumaroyl-CoA in three steps [13,34]. These first three steps are catalyzed by phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (4CH) and 4-coumaroyl CoA ligase (4CL) (Fig 1). However, in bacteria, the use of tyrosine ammonia lyase (TAL) is preferred for heterologous biosynthesis, as starting from L-Tyr, the need for the 4CH activity (a plant membrane-bound enzyme) does not longer exist, as this amino acid is already hydroxylated at the required position [35,36]. Then, the chalcone synthase (CHS) condenses a molecule of 4-coumaroyl-CoA with three molecules of malonyl-CoA, generating naringenin chalcone, the basic skeleton for more than 9,000 flavonoids [3,13,34,37]. The heterocycle C closure is catalyzed by chalcone isomerase (CHI), which generates naringenin, the flavanone common precursor for all flavonols.
In order to generate kaempferol from naringenin, the action of naringenin 3-dioxygenase (N3DOX) is required to produce dihydrokaempferol (aromadendrin) and then the flavonol synthase 1 (FLS1) transforms this intermediate in kaempferol (Fig 1). Kaempferol is then the substrate for the flavonoid 3',5’-hydroxylase (F3'5’H), giving rise to myricetin (Fig 1). On the other hand, dihydrokaempferol is also the substrate for flavonoid 3’-hydroxylase (F3'H), generating dihydroquercetin (taxifolin) (Fig 1), which finally is transformed in quercetin by the action of flavonol synthase 1 (FLS1) [12].
Myricetin, kaempferol, quercetin and their dihydro precursors show interesting nutraceutical activities, as it has been described above. This makes these bioactive compounds attractive targets for genetic and metabolic engineering experiments, like the heterologous expression of their plant biosynthetic gene pathways in suitable microbial factories such as actinomycetes. In this work, we have carried out this by using combinatorial biosynthesis, where genes from different organisms are grouped in an artificial gene cluster directing the production of the natural bioactive compound [34,38]. Previous studies have reported the biosynthesis of flavonols in microorganisms; for instance, myricetin has been already heterologously produced, after feeding with naringenin precursor, in a strain of E. coli containing an incomplete flavonol biosynthetic gene cluster [39]. Also, kaempferol has been already produced in E. coli [40] and Saccharomyces cerevisiae [41], needing in some cases feeding with coumaric acid or naringenin [39]. In the case of its heterologous production in Streptomyces venezuelae, feeding with naringenin was also necessary [42]. Finally, quercetin has been also produced in E. coli and in S. cerevisiae after feeding with coumaric acid or naringenin [39,41].
In this work, we have achieved de novo production of these three flavonols by means of biosynthetic pathways heterologously expressed in S. albus and S. coelicolor, without feeding with precursors.
Material and methods
Bacterial strains, plasmids and culture conditions
E. coli TOP10 (Invitrogen) and pUC57 (Fermentas) were used for routine sub-cloning while E. coli ET12567 [43] was required to obtain non-methylated DNA for later protoplasts transformation in Streptomyces coelicolor M1154 [44]. The high-copy number E. coli-Streptomyces shuttle vector pIAGO, a derivative of pWHM3 which contains the strong constitutive promoter for ermE* (PermE*) [45], was used as expression plasmid. The strain Streptomyces albus J1074 [46] was also used for the production of flavonols.
E. coli strains were grown in TSB liquid broth or TSB agar, supplemented with the corresponding antibiotics (ampicillin 100 μg/ml, Sigma Aldrich) for plasmid selection. S. coelicolor M1154 and S. albus J1074 were grown at 30°C in YEME 34% and 17% sucrose respectively, for protoplasts preparation. Both species were sporulated on SFM and Bennet medium respectively [47], supplemented with the corresponding antibiotics when necessary (thiostrepton 50 μg/mL).
For flavonols production, S. albus and S. coelicolor clones were grown on 3 ml of solid R5A medium [48], supplemented with the corresponding antibiotic, during 5 days at 30°C. Spores were previously quantified and an inoculum of 107 spores/mL was used for each culture.
DNA manipulation
Restriction enzymes were purchased from Takara Biochemicals, T4 DNA ligase from Thermo Scientific, and Dream Taq DNA Polymerase from Thermo Scientific. Synthetic genes for the following ORFs were generated by Genscript after codon optimization: TAL from Rhodobacter capsulatus (accession number WP_013066811), 4CL from S. coelicolor (accession number NP_628552), CHS from Glycine max (accession number L07647.1), CHI from G. max (accession number AY595413.1), N3DOX from Petroselinum crispum (accession number AY23248), FLS1 from Arabidopsis thaliana (accession number Q96330), F3’H from A. thaliana (accession number Q9SD85) and F3’5’H from Petunia x hybrida (accession number Z22544.1). Genbank accession numbers LT629805.1, LT629806.1, LT629807.1, LT629808.1, LT629809.1, MG748610, MG748611 and MG748612 for synthetic genes TAL, 4CL, CHS, CHI, F3’H, N3DOX, FLS1 and F3’5’H respectively. Compatible restriction sites were added at each gene cassette end, in order to facilitate construction of the recombinant flavonoids gene clusters, as well as ribosome binding sites at the 5’-ends.
Construction of plasmids for flavonoids production
All constructed plasmids described below were verified by restriction enzymes digestions and also by sequencing of the cloned regions. Streptomyces producing clones were confirmed by PCR. Primers used amplify the first two common genes: 5’- GTGATCGAGCTGGACATGAA-3’ as the forward primer and 5’- GGCGTCCACGAGGTGC-3’ as the reverse primer.
Construction of pKF
The plasmid pKF contains the ermE* promoter (PermE*) and the 6 genes responsible for kaempferol biosynthesis. All synthetic gene cassettes were independently cloned in pUC57 and plasmids were named pLMF1 (pUC19 containing TAL gene), pLMF2 (4CL), pLMF3 (CHS), pLMF-FLS (FLS1), pLMF5 (CHI) and pLMF-N3DOX (N3DOX) (Table 1). Additionally, TAL gene was subcloned into vector pSL1180 as HindIII-BamHI (pLMF7) to start with the cloning strategy. 4CL gene (from pLMF2) was cloned into pLMF7 as PstI-BamHI gene cassette, generating pLMF8. Next step was subcloning FLS1 gene cassette from pLMF-FLS1 into pLMF3 as an EcoRI DNA fragment, giving rise to pKF11. The correct orientation of each DNA fragment was always confirmed by restriction enzymes digestions and sequencing. The two gene cassettes from pKF11 (CHS and FLS1) were subcloned together into pLMF8 as SacI-BamHI DNA band, in order to get the first 4 genes together in a plasmid (pKF14). Finally, N3DOX gene was subcloned into pLMF5 (opened EcoRV-BamHI) as an EcoRI (blunt ended)-BamHI gen cassette and the two genes together (CHI and N3DOX) were subcloned into pKF14 as XbaI-BamHI resulting in the generation of pKF17, which contains the 6 genes required for kaempferol biosynthesis. As the expression host was Streptomyces, a further subcloning was required, and the BglII-BamHI DNA fragment carrying the 6 genes was finally subcloned into pIAGO plasmid, a derivative of the bifunctional replicative vector pWHM3, which contains the ermE* promoter, giving rise to the final plasmid pKF.
Table 1. Plasmids and strains used in this study.
| Plasmid | Description | Source |
|---|---|---|
| pIAGO | pWHM3 (replicative shuttle vector) carrying ermE* promoter | [45] |
| pSL1180 | E. coli vector | [61] |
| pUC57 | E. coli vector | Fermentas |
| pLMF1 | pUC57 carrying TAL | This study |
| pLMF2 | pUC57 carrying 4CL | This study |
| pLMF3 | pUC57 carrying CHS | This study |
| pLMF5 | pUC57 carrying CHI | This study |
| pLMF-FLS | pUC57 carrying FLS1 | This study |
| pLMF-N3DOX | pUC57 carrying N3DOX | This study |
| pLMF-F3H | pUC57 carrying F3’H | This study |
| pLMF-F35H | pUC57 carrying F3’5’H | This study |
| pLMF7 | pSL1180 carrying TAL | This study |
| pLMF8 | pSL1180 carrying TAL and 4CL | This study |
| pKF11 | pSL1180 carrying CHS and FLS1 | This study |
| pKF14 | pSL1180 carrying TAL, 4CL, CHS and FLS1 | This study |
| pKF16 | pSL1180 carrying CHI and N3DOX | This study |
| pKF17 | pSL1180 carrying TAL, 4CL, CHS, FLS1, CHI and N3DOX | This study |
| pKF | pIAGO carrying TAL, 4CL, CHS, FLS1, CHI and N3DOX | This study |
| pQR2 | pSL1180 carrying TAL, 4CL, CHS, FLS1, CHI, N3DOX and F3’H | This study |
| pQR | pIAGO carrying TAL, 4CL, CHS, FLS1, CHI, N3DOX and F3’H | This study |
| pMYR2 | pSL1180 carrying TAL, 4CL, CHS, FLS1, CHI, N3DOX and F3’5’H | This study |
| pMYR | pIAGO carrying TAL, 4CL, CHS, FLS1, CHI, N3DOX and F3’5’H | This study |
| pREC4 | pSEVA98c1 containing birA, accA2 and accBE | This study |
| Strains | Description | Source |
| E. coli TOP10 | Strain used for routine sub-cloning and transformation in S. albus | Invitrogen |
| E. coli ET12567 | Strain used for transformation in S. coelicolor | [43] |
| Streptomyces coelicolor M1154 | Strain used to create the flavonols-producing mutants | [44] |
| Streptomyces albus J1074 | Strain used to create the flavonols-producing mutants | [46] |
| S. coelicolor-pIAGO | S. coelicolor harboring pIAGO used as negative control | This study |
| S. albus-pIAGO | S. albus harboring pIAGO used as negative control | This study |
| S. coelicolor-pKF | S. coelicolor carrying pKF | This study |
| S. coelicolor-pQR | S. coelicolor carrying pQR | This study |
| S. coelicolor-pMYR | S. coelicolor carrying pMYR | This study |
| S. albus-pKF | S. albus carrying pKF | This study |
| S. albus-pQR | S. albus carrying pQR | This study |
| S. albus-pMYR | S. albus carrying pMYR | This study |
Construction of pQR
The plasmid pQR contains the ermE* promoter (PermE*) and the 7 genes required for the biosynthesis of quercetin. To obtain this plasmid, it was required to add one more gene to the previous plasmid pKF17, synthetized to produce kaempferol. This gene, F3’H, was cloned into pKF17 as DraI-BamHI gene cassette giving rise to pQR2. The 7 genes contained in this plasmid were subcloned in the vector pIAGO to be further expressed in Streptomyces. The gene cassette was cloned as BglII-BamHI DNA fragment to obtain the final plasmid pQR (Table 1).
Construction of pMYR
The plasmid pMYR contains the ermE* promoter (PermE*) and the 7 genes required for the biosynthesis of myricetin. To have the 7 genes together in the same vector, it was necessary to clone the F3’5’H gene into the previously constructed plasmid pKF17. The gene was cloned as DraI-BamHI gene cassette giving rise to pMYR2. The 7 genes contained in this plasmid were subcloned in the vector pIAGO to be further expressed in Streptomyces. The gene cassette was cloned as BglII-BamHI DNA fragment to obtain the final plasmid pMYR (Table 1).
Construction of pREC4 for malonyl-CoA metabolic engineering in S. albus
The plasmid pREC4 is a derivative of the E. coli-Streptomyces bifunctional vector pSEVA98c1 (colE1 and pIJ101 origins of replication for E. coli and Streptomyces respectively, both high copy number; apramycin resistance gene aac(3)IV). Vector pSEVA98c1 was digested with PacI-SacI in order to introduce the PermE* promoter for Streptomyces, giving rise to pREC3. Finally, pREC3 was digested with BamHI-HindIII, in order to introduce the 5.1 kb DNA fragment containing the S. coelicolor chromosomal genes birA (biotin ligase, SCO4927 gene), accA2 (alpha subunit of acetyl-CoA carboxylase, SCO4921 gene) and accBE (beta and epsilon subunits of acetyl-CoA carboxylase, SCO5535 and SCO5536 genes respectively) [49–51]. These plasmids were amplified by PCR from S. coelicolor chromosomal DNA, using the following primers, which contain sequences for restriction enzymes at their ends (marked in capital letters): birA-up (contains BglII recognition sequence): 5’-AGATCTcggcagtgcggtctttcccacac-3’, birA-rp (contains EcoRV-XbaI recognition sequence): 5’-GATATCaaaTCTAGAtcacgccaaccgcaggtgc-3’, accA2-up (contains EcoRV recognition sequence): 5’-GATATC taaactcggcttgtttcaagga-3’, accA2-rp (contains SpeI-XbaI recognition sequence): 5’-ACTAGTaaaTCTAGAgactgcttgatcagtccttga-3’, accBE-up (contains SpeI recognition sequence): 5’-ACTAGTgacggctcgcaatccttgctcg-3’, accBE-rp (contains XbaI recognition sequence): 5’-TCTAGAgttcgggtcagcgccagctg-3’.
Extraction and analysis of flavonols
S. albus J1074 and S. coelicolor clones harboring pKF, pQR, pMYR and pIAGO (negative control), or pREC4 (for malonyl-CoA metabolic engineering) were cultivated (three replicas for each strain were extracted and quantified separately). In the case of liquid culture experiments, feeding experiments were carried out adding 0.3 mM final concentration (in the case of naringenin or kaempferol) or 0.01 mM (in the case of dihydrokaempferol) of these flavonoid precursors at 48 h-old cultures of the corresponding S. albus strain in 5 mL R5 medium, and incubating the cultures for another 72 h (liquid cultures) or 161 h (solid cultures). Flavonols extraction was carried out using three volumes of ethyl acetate with 0.01% of formic acid. This mixture was incubated for 1 h in orbital shaking at room temperature. After this incubation, the organic phase was concentrated by rotary evaporation and kept at -20°C for later use.
Dry extracts were dissolved in 200 μl of methanol:DMSO (1:1), filtrated (0.2 μm PVDF) and analyzed by liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS/MS, Agilent technologies 1290 Infinity, Triple Quadrupole), which was carried out using a Zorbax Eclipse Plus C18 column (50 mm x 2.1 mm, 1.8 μm) in the negative ion mode. The analytes were eluted at a flow rate of 0.3 mL/min using a gradient of 0.1% (v/v) formic acid in water (A) and 0.1% (v/v) formic acid in acetonitrile (B) at 0–10% of B for 1 min, which was increased to 35% for 3 min and maintained at 35% for 1 min, then increased to 80% for 3 min and maintained at 80% for 2 min and finally decreased to 10% for 1 min. Flavonoids quantification was carried out in multiple reaction monitoring (MRM) mode in MS/MS. To accomplish this, the following ion sets were selected to detect the transitions of the parent ions to the product ions specific to the analytes: naringenin 272>119 Da and 272>151 Da; dihydrokaempferol 288>125 Da and 288>259 Da; dihydroquercetin 304>125 Da and 304>285 Da; kaempferol 286>93 Da and 286>117 Da; quercetin 302>151 Da and 302>179 Da; myricetin 318>151 Da and 318>179 Da; apigenin 270>117 Da and 270>150 Da; luteolin 286>131 Da and 286>151 Da. Authentic standards were purchased from Sigma Aldrich (naringenin and dihydrokaempferol), Calbiochem (dihydroquercetin) and Cayman Chemical Company (kaempferol, quercetin and myricetin).
Results
Heterologous expression of kaempferol
In microorganisms, the activity of four enzymes, TAL, 4CL, CHS and CHI, is required to produce naringenin, which is the main flavonols precursor (Fig 1). To obtain the flavonol kaempferol, the activity of other two enzymes is also needed. The first enzyme, N3DOX, hydroxylates naringenin in the position C3 to form the immediate kaempferol precursor, dihydrokaempferol. Later, FLS1 catalyzes de formation of a double bond between C2 and C3 to finally obtain kaempferol (Fig 1). In this work, all the six synthetic genes encoding for the enzymes required for the biosynthesis of kaempferol (with codon usage adapted to the transcription characteristics of Streptomyces) were cloned in a replicative high-copy number shuttle vector for E. coli-Streptomyces, under the control of PermE* (see Materials and Methods section). The final plasmid, pKF, was transformed and expressed in two different species of Streptomyces: S. albus and S. coelicolor.
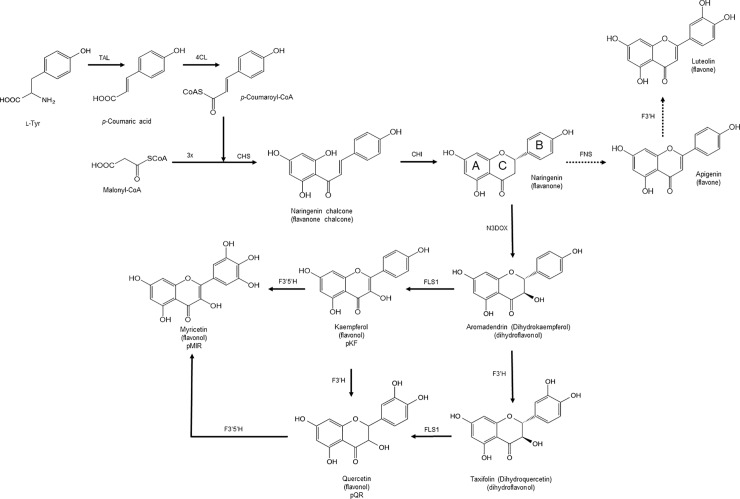
Cultures of S. albus-pKF and S. coelicolor-pKF in R5A solid medium were analyzed by HPLC-MS chromatography in multiple reaction monitoring (MRM) in MS/MS mode, in order to identify and quantify the final product, kaempferol, as well as its intermediate precursors naringenin and dihydrokaempferol. The presence of the hydroxylated form of kaempferol, quercetin, was also analyzed. In S. coelicolor-pKF no kaempferol was detected. However, low production levels (below 0.1 μM) of the precursor, dihydrokaempferol, and the flavonol quercetin were observed (Fig 2A, Table 2).
Fig 2.
A: HPLC-MS chromatogram obtained after MRM analysis of the flavonols extracted from S. coelicolor-pKF. It shows the m/z peaks corresponding to dihydrokaempferol (DHK: <0.1 μM) and quercetin (QR: <0.1 μM). B: HPLC-MS chromatogram obtained after MRM analysis of the flavonols extracted from S. albus-pKF. It shows the m/z peaks corresponding to dihydrokaempferol (DHK: 0.039 μM), quercetin (QR: <0.2 μM) and kaempferol (KF: 0.212 μM).
Table 2. Concentrations of the different flavonoids detected in the both host bacteria for heterologous biosynthesis (mean values and standard error of the mean are indicated).
| Plasmid | Host | Detected Flavonoids | Mean Concentration (μM) ± SEM | Mean Concentration (mg/L) |
|---|---|---|---|---|
| pKF | S. coelicolor | Dihydrokaempferol Kaempferol Quercetin |
Below 0.1 - Below 0.1 |
Below 0.03 - Below 0.03 |
| S. albus | Dihydrokaempferol Kaempferol Quercetin |
Below 0.1 0.212 ± 0.008 0.200 ± 0.016 |
Below 0.03 0.060 ± 0.002 0.060 ± 0.005 |
|
| pQR | S. coelicolor | Dihydrokaempferol Quercetin Kaempferol |
0.100 ± 0.010 0.100 ± 0.006 - |
0.028 ± 0.003 0.030 ± 0.002 - |
| S. albus | Dihydrokaempferol Quercetin Kaempferol |
Below 0.1 0.340 ± 0.026 0.155 ± .0006 |
Below 0.03 0.102 ± 0.008 0.044 ± 0.002 |
|
| pMYR | S. coelicolor | Dihydrokaempferol Quercetin Myricetin |
Below 0.1 Below 0.1 Below 0.1 |
Below 0.03 Below 0.03 Below 0.03 |
| S. albus | Dihydrokaempferol Kaempferol Quercetin Myricetin Apigenin (with pREC4) Luteolin (with pREC4 plus naringenin feeding) |
Below 0.1 Below 0.1 1.984 ± 0.014 0.146 ± 0.019 0.300 ± 0.021 Below 0.1 |
Below 0.03 Below 0.03 0.599 ± 0.004 0.046 ± 0.006 0.081 ± 0.006 Below 0.03 |
In S. albus-pKF, kaempferol was detected at 0.212 μM, but also dihydrokaempferol was detected below 0.1 μM and traces of quercetin (0.2 μM) (Fig 2B, Table 2). The presence of kaempferol, quercetin, myricetin and their precursors, naringenin and dihydrokaempferol was analyzed in parallel in the negative controls (S. albus-pIAGO and S. coelicolor-pIAGO). No flavonoids were detected in any case in these negative control strains.
Heterologous expression of quercetin
Quercetin is a hydroxylated form of kaempferol. For its biosynthesis, it is only necessary the activity of one extra enzyme (F3’H), able to hydroxylate kaempferol in the position B3’. After checking that our S. albus host were able to produce kaempferol, the gene encoding for the F3’H was cloned into the plasmid directing the biosynthesis of kaempferol. The new plasmid, pQR, was transformed in both S. albus and S. coelicolor protoplasts. The cultures from positive recombinant strains (in R5A solid medium) were analyzed by HPLC-MS chromatography.
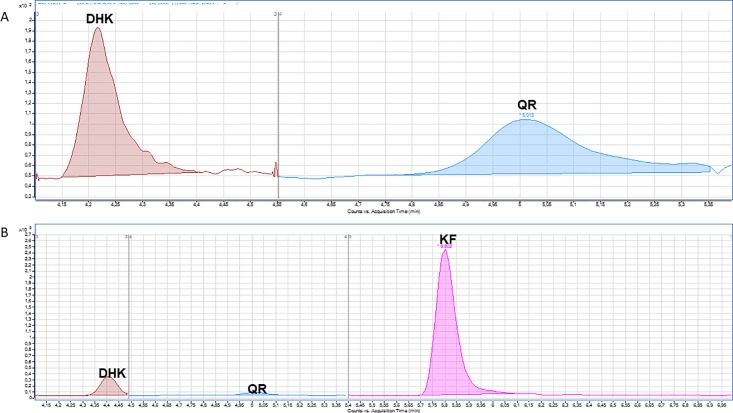
In S. coelicolor-pQR, quercetin and one of the intermediate precursor, dihydrokaempferol were observed, although at levels below 0.1 μM (Fig 3A, Table 2).
Fig 3.
A: HPLC-MS chromatogram obtained after MRM analysis of the flavonols extracted from S. coelicolor-pQR. The m/z peaks correspond to dihydrokaempferol (DHK: <0.1 μM) and to quercetin (QR: <0.1 μM). B: HPLC-MS chromatogram obtained after MRM analysis of the flavonols extracted from S. albus-pQR. The peaks correspond to dihydrokaempferol (DHK: 0.047 μM), quercetin (QR: 0.340 μM) and kaempferol (KF: 0.155 μM).
Cultures of S. albus-pQR produced quercetin (0.34 μM), and its precursors kaempferol (0.155 μM) and dihydrokaempferol (below 0.1 μM) were also detected (Fig 3B, Table 2).
Heterologous expression of myricetin
The flavonol myricetin, is also a hydroxylated form of kaempferol. In this case, there are two extra hydroxylations, while in quercetin there is only one. These two extra hydroxyl groups are in the positions B3’ and B5’, so the activity of a specific enzyme, F3’5’H, is required (Fig 1). The gene encoding this enzyme was added to the previous construction directing the biosynthesis of kaempferol and the new plasmid was further transformed in S. coelicolor and S. albus.
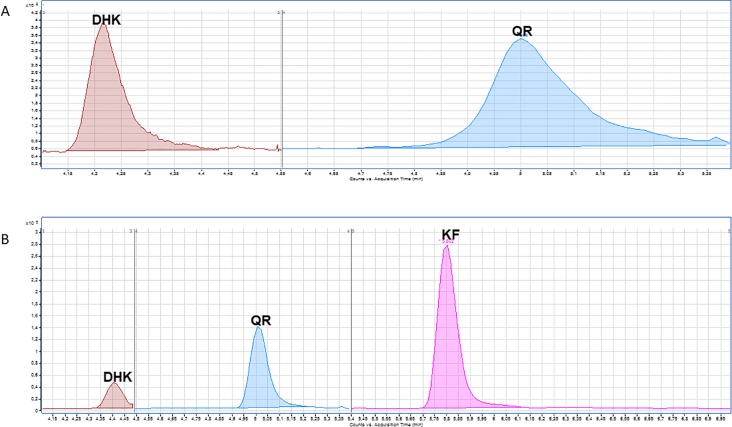
In S. coelicolor-pMYR, myricetin was detected, but also the presence of dihydrokaempferol and quercetin was observed (Fig 4A). However, the levels of production were very low (under 0.1 μM was detected in MRM analysis).
Fig 4.
A: HPLC-MS chromatogram obtained after MRM analysis of the flavonols extracted from S. coelicolor-pMYR. The peaks correspond to myricetin (MYR: <0.1 μM), dihydrokaempferol (DHK: <0.1 μM) and quercetin (QR: <0.1 μM). B: chromatogram obtained after MRM analysis of the flavonols extracted from S. albus-pMYR. The peaks correspond to myricetin (MYR: 0.146 μM), dihydrokaempferol (DHK: 0.024 μM), quercetin (QR: 1.984 μM) and kaempferol (KF: 0.034 μM). C: chromatogram obtained from extracts of S. albus-pMYR-pREC4. The peaks correspond to apigenin (0.3 μM) and luteolin (below 0.1 μM).
Nevertheless, in S. albus-pMYR, higher levels of myricetin production were observed (0.146 μM). Other precursors, kaempferol and dihydrokaempferol are also produced by S. albus-pMYR in low concentrations (below 0.1 μM). However, the most abundant flavonol produced by this recombinant strain was quercetin (1.984 μM) (Fig 4B).
S. albus-pMYR was transformed also with plasmid pREC4, which contains all the genes coding for malonyl-CoA biosynthesis enzymes (BirA and acetyl-CoA carboxylase subunits). In this strain, however, no higher production levels of myricetin were detected in liquid cultures, but 0.3 μM of the shunt product apigenin, a derivative of naringenin (the precursor of dihydrokaempferol, kaempferol and myricetin) was detected. Apigenin was not detected in S. albus-pMYR- pSEVA98c1 (control strain for metabolic engineering experiments). The same strain, S. albus-pMYR-pREC4 was cultivated in R5 liquid medium as well, but including a feeding with naringenin. In this case, no further increase in apigenin production levels was detected, but interestingly, very low amounts of luteolin (a 3’-hydroxylated derivative of apigenin) were detected.
Discussion
In this work, two different strains of Streptomyces, S. albus and S. coelicolor, have been able to biosynthesize de novo the flavonols kaempferol, quercetin (a 3’-hydroxylated kaempferol), myricetin (a double hydroxylated form of kaempferol) and the main precursor of the flavonols pathway, dihydrokaempferol.
Previous works reported the biosynthesis of kaempferol in E. coli [39,40], Saccharomyces cerevisiae [41] and S. venezuelae [42]. Two strategies were followed to produce kaempferol in E. coli. The first one consisted of adding L-Tyrosine (the first precursor of flavonoids in prokaryotes) to the culture medium, so the complete biosynthetic pathway was cloned. The levels of production obtained were up to 15 mg/L [40]. The group of Leonard et al. did not clone the gene encoding for the first enzyme of the biosynthetic pathway (tyrosine ammonia lyase, TAL), which means that feeding the cultures with precursors was needed. When p-coumaric acid (second metabolite of the biosynthetic pathway in microorganisms) was added, the yields of kaempferol reached 0.3 mg/L. The productions achieved were better (0.8 mg/L) when naringenin was used for feeding the culture [39].
Kaempferol was also produced in yeast in a similar way that it was synthetized in E. coli. The complete biosynthetic pathway was cloned in S. cerevisiae and three different precursors were added to the cultures: L-phenylalanine (first precursor of flavonoids in eukaryotes), p-coumaric acid and naringenin. The production levels achieved were 1.3 mg/L, 0.9 mg/L and 4.6 mg/L respectively [41].
Regarding the biosynthesis of kaempferol in Streptomyces, only one group succeeded to do it previously. However, the complete biosynthetic pathway was not cloned, only the genes encoding for the N3DOX and FLS1 were cloned in S. venezuelae. These enzymes are involved in the hydroxylation of naringenin to obtain dihydrokaempferol (N3DOX), and in the further formation of a double bond to form kaempferol (FLS1). So, it was required to supplement the cultures with naringenin precursor. The production levels reached 0.2 mg/L [42].
In this work we were able to produce kaempferol by cloning the complete biosynthetic pathway into S. albus without feeding the cultures with any precursor. However, the levels achieved were 0.212 μM. Also, smaller levels of its precursor dihydrokaempferol were observed, indicating that the enzyme FLS1 is not completely efficient. In S. coelicolor, no kaempferol was detected but small amounts of its precursor, dihydrokaempferol, and its hydroxylated derivative, quercetin, were detected. This may be due to the presence of an extra P450 hydroxylase naturally found in this strain that is able to use kaempferol as a substrate, thus, converting it into quercetin. This affirmation is supported by the fact that P450 systems are really well developed in Streptomyces genus, and P450 from this actinomycete has been described for regioselective hydroxylation of diverse flavonoids [52–54].
Regarding quercetin biosynthesis, it was achieved by the same groups in both E. coli and S. cerevisisae following the strategy employed to produce kaempferol [39,41]. In E. coli, the complete biosynthetic pathway, except for the first gene (encoding for TAL), was cloned, and cultures supplemented with either p-coumaric acid or naringenin. The production levels were 0.05 mg/L and 0.18 mg/L respectively [39].
Quercetin production in S. cerevisiae was higher. In this species, all the genes involved in quercetin biosynthesis were cloned. Like in the case of kaempferol production, the cultures were feed with L-Phe, p-coumaric acid and naringenin. Nonetheless, production levels were smaller. In the case of L-Phe feeding there were only traces of quercetin. When p-coumaric acid and naringenin were added, the production was up to 0.26 mg/L and 0.38 mg/L respectively [41].
In this paper, we describe the biosynthesis of quercetin in Streptomyces for the very first time. Moreover, it was de novo biosynthesis as no precursors were added to these cultures. However, only traces of quercetin were detected in the recombinant strain of S. coelicolor. In the case of S. albus, 0.1 mg/L of quercetin were observed, as well as relatively high amounts of kaempferol and dihydrokaempferol, indicating the incomplete efficiency of the P450 hydroxylases employed in this study. Although the amount of quercetin produced in this work is rather small, it is comparable to the one achieved in E. coli after being feed with naringenin [39], evidencing that our biosynthetic system is more effective.
As far as the biosynthesis of myricetin in microorganisms is concerned, this flavonol was only produced by the group of Leonard et al., in E. coli, and all the genes required to produce myricetin were introduced in E. coli and then, the cultures were supplemented with naringenin and eriodictyol. Despite this feeding, production levels only reached 0.01 mg/L [39].
In our case, we demonstrate the feasibility of de novo myricetin biosynthesis in both S. albus and S. coelicolor. Moreover, production levels (0.146 μM) in S. albus were better than those achieved in E. coli even after feeding experiments. In S. albus, not only myricetin was detected by HPLC-MS, but also high levels of its precursor, quercetin (1.984 μM). This elevated amount of precursor in comparison to the final compound may be due to a 3’-hydroxylation activity of the enzyme F3’5’H in the C3’. In fact, it is known that P. hybrida F3’5’H performs both 3’- and 3’,5’-hydroxylation reactions and can use flavonols as well as dihydroflavonols and flavanones as a substrate [39,55]. Taking into account these two considerations, the high yield of quercetin could be due to a 3’-hydroxylation of the intermediate dihydrokaempferol generating dihydroquercetin as a product which could be easily converted in quercetin by the FLS1 enzyme expressed in pMYR plasmid. Moreover, more quercetin can be produced from kaempferol due to, once again, a 3’-hydroxylation of the F3’5’H. For all these reasons, only a little quantity of kaempferol is available to be converted in myricetin by F3’5’H. Finally, it should be pointed out that P. hybrida F3’5’H has a broad substrate specificity towards dihydrokaempferol, kaempferol and quercetin but competition as well as inhibition may occur when more than one substrate is present, leading to a lower myricetin yield and quercetin accumulation [55].
In our experiments with S. coelicolor there are only traces of these compounds. These results, together with the obtained ones for the biosynthesis of kaempferol and quercetin, reveal that S. albus is a better host for flavonols biosynthesis than S. coelicolor. Also, low production levels in S. albus could be improved by metabolic engineering of the strain, facilitating the incorporation of malonyl-CoA to the flavonols biosynthetic pathway, as it is a limiting factor in flavonoid production [56]. Other authors confirmed that this strategy is useful to increase flavonoids yields [37,40,57,58]. However, the experiments with S. albus-pMYR-pREC4, where plasmid pREC4 was used to try to further increase intracellular malonyl-CoA precursor levels, and therefore to generate higher myricetin titers, were unsuccessful. In these experiments, instead of higher myricetin levels, due to the expected higher malonyl-CoA intracellular levels, a deviation from naringenin precursor towards apigenin (a shunt product in this study) was observed. A possible explanation for this result is that the enzyme N3DOX, in charge of converting naringenin intermediate towards hihydrokaempferol, shows a 79.18% identity with the enzyme FNS, which usually converts naringenin towards apigenin [59,60]. This means that genes present in pMYR can explain the generation of apigenin in S. albus-pMYR-pREC4 as a shunt product from naringenin. Once apigenin is present at those levels in S. albus-pMYR-pREC4, further feeding here with exogenous naringenin facilitates apigenin production and the low levels of the apigenin 3’-hydroxy derivative luteolin observed. This 3’-hydroxylation can be easily explained by the presence of F3’5’H hydroxylase in pMYR.
Alternatively, production levels could be also improved by facilitating hydroxylation steps carried out by F3’H and F3’5’H enzymes. These two enzymes need reducing power provided by a single redox partner (cytochrome P450 enzyme) whose presence is critical to show their optimal or maximal activities [39,54], and this reductase is not included in our plasmid constructions. In fact, there is a big hurdle to combine specific P450s with the right redox partners. Nevertheless, the partial activity of the P450 hydroxylases used in this study could be explained by a recognition of soluble endogenous redox partners since P450 systems are specially well developed in Streptomyces genus [52,54]. Further modifications of these two enzymes involving their transformation in a soluble chimera protein that fuses the P450s with suitable P450 reductases are being carried out.
Conclusions
Using a combinatorial biosynthesis approach and reconstituted plant flavonol pathways in the bacteria S. albus, this work describes the heterologous biosynthesis of the important nutraceuticals myricetin and quercetin by the first time in actinomycetes, according to public literature. Also, kaempferol biosynthesis has been achieved in this bacterium for the first time without feeding with precursors. These experiments open the way to heterologous production in actinomycetes of other flavonols and flavonoids.
Acknowledgments
Authors wish to thank the Spanish Ministry of Economy and Competitiveness (MINECO, for financial support via Grant AGL2010-20622), and also to the Government of the Principality of Asturias (program PCTI for a Technology Transfer Grant). Authors wish also to thank Sergio Cueto, PhD, from Servicios Científico-Técnicos at the University of Oviedo, for his help with HPLC chromatography and purification of compounds.
Data Availability
Data are available from the GenBank accession numbers LT629805.1, LT629806.1, LT629807.1, LT629808.1, LT629809.1, MG748610, MG748611 and MG748612 for synthetic genes TAL, 4CL, CHS, CHI, F3’H, N3DOX, FLS1 and F3’5’H respectively.
Funding Statement
The authors wish to thank the Spanish Ministry of Economy and Competitiveness (MINECO, for financial support via Grant AGL2010-20622), and also to the Government of the Principality of Asturias (program PCTI for a Technology Transfer Grant). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li A, Li S, Zhang Y, Xu X, Chen Y, Li H. Resources and Biological Activities of Natural Polyphenols. Nutrients. 2014;6: 6020–6047. 10.3390/nu6126020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manach C, Scalbert A, Morand C, Rémésy C, Jiménez L. Polyphenols: Food sources and bioavailability. American Journal of Clinical Nutrition. 2004. pp. 727–747. 10.1093/ajcn/79.5.727 [DOI] [PubMed] [Google Scholar]
- 3.Tsao R. Chemistry and biochemistry of dietary polyphenols. Nutrients. 2010;2: 1231–1246. 10.3390/nu2121231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Vallinas M, González-Castejón M, Rodríguez-Casado A, Ramírez de Molina A. Dietary phytochemicals in cancer prevention and therapy: A complementary approach with promising perspectives. Nutr Rev. 2013;71: 585–599. 10.1111/nure.12051 [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri S, Sengupta B, Taylor J, Pahari BP, Sengupta PK. Interactions of Dietary Flavonoids with Proteins: Insights from Fluorescence Spectroscopy and Other Related Biophysical Studies. Curr Drug Metab. 2013;14: 491–503. 10.2174/1389200211314040011 [DOI] [PubMed] [Google Scholar]
- 6.Fantini M, Benvenuto M, Masuelli L, Frajese G, Tresoldi I, Modesti A, et al. In Vitro and in Vivo Antitumoral Effects of Combinations of Polyphenols, or Polyphenols and Anticancer Drugs: Perspectives on Cancer Treatment. Int J Mol Sci. 2015;16: 9236–9282. 10.3390/ijms16059236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: An overview. Sci World J. 2007;73: 637–670. 10.1070/RC2004v073n07ABEH000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao B, Fang H, Xu W, Yan Y, Xu H, Liu Y, et al. Dietary fiber intake and risk of type 2 diabetes: A dose-response analysis of prospective studies. Eur J Epidemiol. 2014;29: 79–88. 10.1007/s10654-013-9876-x [DOI] [PubMed] [Google Scholar]
- 9.Liu RH. Health-Promoting Components of Fruits and Vegetables in the Diet. Adv Nutr. 2013;4: 384S–392S. 10.3945/an.112.003517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravishankar D, Rajora AK, Greco F, Osborn HMI. Flavonoids as prospective compounds for anti-cancer therapy. Int J Biochem Cell Biol. Elsevier Ltd; 2013;45: 2821–31. 10.1016/j.biocel.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 11.Verma AK, Pratap R. The biological potential of flavones. Nat Prod Rep. 2010;27: 1571–1593. 10.1039/c004698c [DOI] [PubMed] [Google Scholar]
- 12.Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26: 1001–1043. 10.1039/b802662a [DOI] [PubMed] [Google Scholar]
- 13.Chouhan S, Sharma K, Zha J, Guleria S, Koffas MAG. Recent Advances in the Recombinant Biosynthesis of Polyphenols. Front Microbiol. 2017;8 10.3389/fmicb.2017.02259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J, Zhang Y, Chen W, Zhao X. The relationship between fasting plasma concentrations of selected flavonoids and their ordinary dietary intake. Br J Nutr. 2010;103: 249–55. 10.1017/S000711450999170X [DOI] [PubMed] [Google Scholar]
- 15.Rajendran P, Rengarajan T, Nandakumar N, Palaniswami R, Nishigaki Y, Nishigaki I. Kaempferol, a potential cytostatic and cure for inflammatory disorders. Eur J Med Chem. 2014;86: 103–12. 10.1016/j.ejmech.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 16.Bhagwat S, Haytowitz DB, Holden JM. Database for the Flavonoid Content of Selected Foods Release 3 Prepared by USDA Database for the Flavonoid Content of Selected Foods Release 3. US Dep Argiculture. 2011; 1–156. [Google Scholar]
- 17.Gutiérrez-del-Río I, Villar CJ, Lombó F. Chapter 3. Therapeutic uses of kaempferol: anticancer and anti-inflammatory activity In: Garde-Cerdán T, Gonzalo-Diago A, editors. Kaempferol: Biosynthesis, food sources and therapeutic uses. Nova Science Publishers; 2016. [Google Scholar]
- 18.Calderón-Montaño JM, Burgos-Morón E, Pérez-Guerrero C, López-Lázaro M. A review on the dietary flavonoid kaempferol. Mini Rev Med Chem. 2011;11: 298–344. 10.2174/138955711795305335 [DOI] [PubMed] [Google Scholar]
- 19.Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. Metabolite profiling of grape: Flavonols and anthocyanins. J Agric Food Chem. 2006;54: 7692–702. 10.1021/jf061538c [DOI] [PubMed] [Google Scholar]
- 20.Häkkinen SH, Kärenlampi SO, Heinonen IM, Mykkänen HM, Törrönen AR. Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J Agric Food Chem. 1999;47: 2274–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/10794622 [DOI] [PubMed] [Google Scholar]
- 21.Jeganathan B, Punyasiri PAN, Kottawa-Arachchi JD, Ranatunga MAB, Abeysinghe ISB, Gunasekare MTK, et al. Genetic Variation of Flavonols Quercetin, Myricetin, and Kaempferol in the Sri Lankan Tea (Camellia sinensis L.) and Their Health-Promoting Aspects. Int J food Sci. 2016;2016: 6057434 10.1155/2016/6057434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemeth K, Piskula MK. Food content, processing, absorption and metabolism of onion flavonoids. Crit Rev Food Sci Nutr. 2007;47: 397–409. 10.1080/10408390600846291 [DOI] [PubMed] [Google Scholar]
- 23.Kahle K, Kraus M, Richling E. Polyphenol profiles of apple juices. Mol Nutr Food Res. 2005;49: 797–806. 10.1002/mnfr.200500064 [DOI] [PubMed] [Google Scholar]
- 24.Fiander H, Schneider H. Dietary ortho phenols that induce glutathione S-transferase and increase the resistance of cells to hydrogen peroxide are potential cancer chemopreventives that act by two mechanisms: the alleviation of oxidative stress and the detoxification of mutagenic. Cancer Lett. 2000;156: 117–24. Available: http://www.ncbi.nlm.nih.gov/pubmed/10880760 [DOI] [PubMed] [Google Scholar]
- 25.Limasset B, le Doucen C, Dore JC, Ojasoo T, Damon M, Crastes de Paulet A. Effects of flavonoids on the release of reactive oxygen species by stimulated human neutrophils. Multivariate analysis of structure-activity relationships (SAR). Biochem Pharmacol. 1993;46: 1257–71. Available: http://www.ncbi.nlm.nih.gov/pubmed/8216378 [DOI] [PubMed] [Google Scholar]
- 26.Comalada M, Ballester I, Bailón E, Sierra S, Xaus J, Gálvez J, et al. Inhibition of pro-inflammatory markers in primary bone marrow-derived mouse macrophages by naturally occurring flavonoids: analysis of the structure-activity relationship. Biochem Pharmacol. 2006;72: 1010–21. 10.1016/j.bcp.2006.07.016 [DOI] [PubMed] [Google Scholar]
- 27.Landolfi R, Mower RL, Steiner M. Modification of platelet function and arachidonic acid metabolism by bioflavonoids. Structure-activity relations. Biochem Pharmacol. 1984;33: 1525–30. Available: http://www.ncbi.nlm.nih.gov/pubmed/6329230 [DOI] [PubMed] [Google Scholar]
- 28.Ciolino HP, Daschner PJ, Yeh GC. Dietary flavonols quercetin and kaempferol are ligands of the aryl hydrocarbon receptor that affect CYP1A1 transcription differentially. Biochem J. 1999;340 (Pt 3: 715–22. Available: http://www.ncbi.nlm.nih.gov/pubmed/10359656 [PMC free article] [PubMed] [Google Scholar]
- 29.Lautraite S, Musonda AC, Doehmer J, Edwards GO, Chipman JK. Flavonoids inhibit genetic toxicity produced by carcinogens in cells expressing CYP1A2 and CYP1A1. Mutagenesis. 2002;17: 45–53. Available: http://www.ncbi.nlm.nih.gov/pubmed/11752233 [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Duan S, Jia H, Bai C, Zhang L, Wang Z. Flavonoids from tartary buckwheat induce G2/M cell cycle arrest and apoptosis in human hepatoma HepG2 cells. Acta Biochim Biophys Sin (Shanghai). 2014;46: 460–70. 10.1093/abbs/gmu023 [DOI] [PubMed] [Google Scholar]
- 31.Kim J-D, Liu L, Guo W, Meydani M. Chemical structure of flavonols in relation to modulation of angiogenesis and immune-endothelial cell adhesion. J Nutr Biochem. 2006;17: 165–76. 10.1016/j.jnutbio.2005.06.006 [DOI] [PubMed] [Google Scholar]
- 32.Cushnie TPT, Lamb AJ. Antimicrobial activity of flavonoids. Int J Antimicrob Agents. 2005;26: 343–56. 10.1016/j.ijantimicag.2005.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirzoeva OK, Grishanin RN, Calder PC. Antimicrobial action of propolis and some of its components: the effects on growth, membrane potential and motility of bacteria. Microbiol Res. 1997;152: 239–46. 10.1016/S0944-5013(97)80034-1 [DOI] [PubMed] [Google Scholar]
- 34.Falcone Ferreyra ML, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3: 222 10.3389/fpls.2012.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyndt JA, Meyer TE, Cusanovich MA, Van Beeumen JJ. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 2002;512: 240–4. Available: http://www.ncbi.nlm.nih.gov/pubmed/11852088 [DOI] [PubMed] [Google Scholar]
- 36.Watts KT, Lee PC, Schmidt-Dannert C. Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chembiochem. 2004;5: 500–7. 10.1002/cbic.200300783 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Chen S, Yu O. Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol. 2011;91: 949–56. 10.1007/s00253-011-3449-2 [DOI] [PubMed] [Google Scholar]
- 38.Song MC, Kim EJ, Kim E, Rathwell K, Nam S, Yoon YJ. Microbial biosynthesis of medicinally important plant secondary metabolites. Nat Prod Rep. Royal Society of Chemistry; 2014;00: 1–13. 10.1039/C4NP00057A [DOI] [PubMed] [Google Scholar]
- 39.Leonard E, Yan Y, Koffas M a G. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng. 2006;8: 172–81. 10.1016/j.ymben.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 40.Miyahisa I, Funa N, Ohnishi Y, Martens S, Moriguchi T, Horinouchi S. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechnol. 2006;71: 53–8. 10.1007/s00253-005-0116-5 [DOI] [PubMed] [Google Scholar]
- 41.Trantas E, Panopoulos N, Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng. Elsevier; 2009;11: 355–66. 10.1016/j.ymben.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 42.Park SR, Paik JH, Ahn MS, Park JW, Yoon YJ. Biosynthesis of plant-specific flavones and flavonols in Streptomyces venezuelae. J Microbiol Biotechnol. 2010;20: 1295–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/20890094 [DOI] [PubMed] [Google Scholar]
- 43.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 1992;111: 61–68. 10.1016/0378-1119(92)90603-M [DOI] [PubMed] [Google Scholar]
- 44.Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4: 207–15. 10.1111/j.1751-7915.2010.00219.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Braña AF, Méndez C, et al. Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob Agents Chemother. 2000;44: 1266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chater KF, Wilde LC. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980;116: 323–334. 10.1099/00221287-116-2-323 [DOI] [PubMed] [Google Scholar]
- 47.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. 2000. John Innes Foundation, Norwich, England. ISBN 0-7084-0623-8. [Google Scholar]
- 48.Fernández E, Weissbach U, Reillo S, Braña F, Méndez C, Rohr J, et al. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J Bacteriol. 1998;180: 4929–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arabolaza A, Shillito ME, Lin T-W, Diacovich L, Melgar M, Pham H, et al. Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor. Biochemistry. 2010;49: 7367–76. 10.1021/bi1005305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demirev A V, Lee JS, Sedai BR, Ivanov IG, Nam DH. Identification and characterization of acetyl-CoA carboxylase gene cluster in Streptomyces toxytricini. J Microbiol. 2009;47: 473–8. 10.1007/s12275-009-0135-5 [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2). Appl Environ Microbiol. 2001;67: 4166–76. Available: http://www.ncbi.nlm.nih.gov/pubmed/11526020 10.1128/AEM.67.9.4166-4176.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, et al. Cytochrome P450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem Biophys Res Commun. Academic Press; 2003;307: 610–619. 10.1016/S0006-291X(03)01231-2 [DOI] [PubMed] [Google Scholar]
- 53.Pandey BP, Roh C, Choi KY, Lee N, Kim EJ, Ko S, et al. Regioselective hydroxylation of daidzein using P450 (CYP105D7) from Streptomyces avermitilis MA4680. Biotechnol Bioeng. 2010;105: 697–704. 10.1002/bit.22582 [DOI] [PubMed] [Google Scholar]
- 54.Pandey BP, Lee N, Choi KY, Jung E, Jeong D hye, Kim BG. Screening of bacterial cytochrome P450s responsible for regiospecific hydroxylation of (iso)flavonoids. Enzyme Microb Technol. 2011;48: 386–392. 10.1016/j.enzmictec.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 55.Kaltenbach M, Schroder G, Schmelzer E, Lutz V, Schroder J. Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J. Wiley/Blackwell (10.1111); 1999;19: 183–193. 10.1046/j.1365-313X.1999.00524.x [DOI] [PubMed] [Google Scholar]
- 56.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126: 485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park SR, Ahn MS, Han AR, Park JW, Yoon YJ. Enhanced flavonoid production in Streptomyces venezuelae via metabolic engineering. J Microbiol Biotechnol. 2011;21: 1143–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/22127124 [DOI] [PubMed] [Google Scholar]
- 58.Hwang E Il, Kaneko M, Ohnishi Y, Horinouchi S. Production of Plant-Specific Flavanones by Escherichia coli Containing an Artificial Gene Cluster. Appl Environ Microbiol. 2003;69: 2699–2706. 10.1128/AEM.69.5.2699-2706.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marín L, Gutiérrez-Del-Río I, Yagüe P, Manteca Á, Villar CJ, Lombó F. De Novo Biosynthesis of Apigenin, Luteolin, and Eriodictyol in the Actinomycete Streptomyces albus and Production Improvement by Feeding and Spore Conditioning. Front Microbiol. 2017;8: 921 10.3389/fmicb.2017.00921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martens S, Mithöfer A. Flavones and flavone synthases. Phytochemistry. 2005;66: 2399–407. 10.1016/j.phytochem.2005.07.013 [DOI] [PubMed] [Google Scholar]
- 61.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8: 759–777. 10.1089/dna.1989.8.759 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the GenBank accession numbers LT629805.1, LT629806.1, LT629807.1, LT629808.1, LT629809.1, MG748610, MG748611 and MG748612 for synthetic genes TAL, 4CL, CHS, CHI, F3’H, N3DOX, FLS1 and F3’5’H respectively.