Abstract
Philodendron s.l. (Araceae) has been recently focus of taxonomic and phylogenetic studies, but karyotypic data are limited to chromosome numbers and a few published genome sizes. In this work, karyotypes of 34 species of Philodendron s.l. (29 species of Philodendron and five of Thaumatophyllum), ranging from 2n = 28 to 36 chromosomes, were analyzed by fluorescence in situ hybridization (FISH) with rDNA and telomeric probes, aiming to understand the evolution of the karyotype diversity of the group. Philodendron presented a high number variation of 35S rDNA, ranging from two to 16 sites, which were mostly in the terminal region of the short arms, with nine species presenting heteromorphisms. In the case of Thaumatophyllum species, we observed a considerably lower variation, which ranged from two to four terminal sites. The distribution of the 5S rDNA clusters was more conserved, with two sites for most species, being preferably located interstitially in the long chromosome arms. For the telomeric probe, while exclusively terminal sites were observed for P. giganteum (2n = 30) chromosomes, P. callosum (2n = 28) presented an interstitial distribution associated with satellite DNA. rDNA sites of the analyzed species of Philodendron s.l. species were randomly distributed considering the phylogenetic context, probably due to rapid evolution and great diversity of these genomes. The observed heteromorphisms suggest the accumulation of repetitive DNA in the genomes of some species and the occurrence of chromosomal rearrangements along the karyotype evolution of the group.
Introduction
Araceae (ca. 3600 species) is a widely distributed monocot family with high ecological diversity, occurring mainly in tropical regions [1, 2]. Philodendron s.l. is the second largest group within the family (ca. 500 species), presenting a wide Neotropical distribution, with the Amazon region as its probable center of origin [2–4]. The genus was traditionally divided into three monophyletic subgenera: P. subg. Meconostigma (now recognized as the genus Thaumatophyllum, with 21 species), mostly with the diploid number 2n = 36; P. subg. Pteromischum (82 species), with chromosome counts only for two species (both with 2n = 32); and P. subg. Philodendron (ca. 400 species), as the most morphologically diverse group, with chromosome number ranging from 2n = 26 to 40 chromosomes, although 2n = 32 and 34 have been reported as the most common numbers [5–10]. It is suggested that x = 16 is the basic number for Philodendron s.l. (including Thaumatophyllum) and that the karyotype evolution of the group has been driven by both ascending and descending dysploidy events, since no polyploidy was reported so far [8, 9].
The ribosomal RNA (rRNA) genes have been physically mapped in the chromosome of species of several plant groups [11–13], being useful for understanding the general patterns of karyotype evolution among related species and for cytotaxonomic approaches [14–16]. In Araceae, studies involving the physical location of 5S and 35S rDNA by FISH (Fluorescent In Situ Hybridization) are available for just a few species, including 10 species of Typhonium [17] and 17 other species from different groups, such as P. hederaceum (Jacq.) Schott (as P. scandens Koch & Sello), two species of Anthurium, three species of Spathiphyllum and two species of Ulearum [18, 19]. Overall, all analyzed species presented only one pair of 5S rDNA sites, located in the subterminal or interstitial regions, whereas for 35S rDNA there was a predominance of four terminal sites, with few exceptions.
Thus, based on the chromosome number and genome size variation previously mentioned for Philodendron and Thaumatophyllum [8, 9, 20], we have addressed the following questions: (I) Is the distribution of rDNA sites in Philodendron and the sister genus Thaumatophyllum conserved? (II) Does the distribution pattern of rDNA sites agree with the available phylogenetic data? To answer these questions, molecular cytogenetic data were generated for the first time for 29 Philodendron and five Thaumatophyllum species by FISH with rDNA and telomeric probes. In addition, these data were plotted in a recently published phylogenetic tree of the group [21], aiming to better understand the patterns of karyotype evolution of both genera.
Materials and methods
Plant material and chromosomal preparations
Thirty-four species were analyzed by FISH. Sixteen species were sampled from the living collection kept at the Royal Botanic Gardens, Kew (Richmond, United Kingdom), and 18 were collected in different regions of Brazil, as licensed by the Brazilian authorities (ICMBio license numbers 14311–2 and 31038–4). The collected Brazilian plants are being cultivated in the living collection of the Laboratory of Plant Genetics and Biotechnology, Department of Genetics, UFPE (Recife, Brazil). The provenance of the plants and accession numbers are shown in Table 1.
Table 1. Species of Philodendron and Thaumatophyllum with their respective section and accession, chromosome complement size, chromosome range size, diploid number and number of rDNA sites.
| Genus | Section | Species | Provenance and accession number | 2n | Number of rDNA sites | |
|---|---|---|---|---|---|---|
| 35S | 5S | |||||
| Thaumatophyllum | T. corcovadense (Kunth) Sakur., Calazans & Mayo | Taquaritinga do Norte, Pernambuco, Brazil; Cultivated at LGBV; SV314 | 36 | 4 | 2 | |
| T. lundii (Warm.) Sakur., Calazans & Mayo | Morro do Chapéu, Bahia, Brazil; cultivated at LGBV; SV089 | 36 | 2 | 2 | ||
| T. mello-barretoanum (Burle-Marx ex G.M. Barroso) Sakur., Calazans & Mayo | Recife, Pernambuco, Brazil; Cultivated at LGBV; SV534 | 34 | 2 | 2 | ||
| T. saxicola (Krause) Sakur., Calazans & Mayo | Mucugê, Bahia, Brazil; Cultivated at LGBV; SV539 | 36 | 2 | 2 | ||
| T. spruceanum Schott | Reserva Florestal Adolpho Ducke, Amazonas, Brazil; Cultivated at LGBV; SV063 | 32 | 4 | 2 | ||
|
Philodendron subg. Philodendron |
Baursia | P. callosum K.Krause | Presidente Figueiredo, Amazonas, Brazil; Cultivated at LGBV; SV022 |
28 | - | - |
| P. glaziovii Hook.f. | Pedra Azul, Espírito Santo, Brazil; Cultivated at RBG Kew/ 1983–2011 |
34 | 6 | 2 | ||
| P. renauxii Reitz | Itapema, Santa Catarina, Brazil; Cultivated at RBG Kew/ 1983–1988 |
34 | 8 | 2 | ||
| Macrobelium | P. annulatum Croat | Cerro Jefe, Panamá; Cultivated at RBG Kew/ 1996–4421 |
32 | 2 | 2 | |
| P. barrosoanum G.S.Bunting | Reserva Florestal Adolpho Ducke, Amazonas, Brazil; Cultivated at LGBV; MC107 | 32 | 8 | 2 | ||
| P. burle-marxii G.M.Barroso | Amazonas region, Colombia; Cultivated at RGB Kew/ 1975–98 |
34 | 10 | 2 | ||
| P. eximium Schott | Taquaritinga do Norte, Pernambuco, Brazil; Cultivated at LGBV; SV293 | 32 | 6 | 2 | ||
| P. inconcinnum Schott | Cultivated at RBG Kew/ 1981–3728 | 32 | 2 | 2 | ||
| P. krugii Engl. | Trinidad and Tobago; Cultivated at RBG Kew/ 1980–1645 | 34 | 2 | 2 | ||
| P. quinquenervium Schott | Uatumã, Amazonas, Brazil; Cultivated at LGBV; SV076 | 32 | 15 | 2 | ||
| P. smithii Engl. | Tabasco, Mexico; Cultivated at RBG Kew/ 1980–1583 | 32 | 8 | 2 | ||
| P. uleanum Engl. | Napo, Ecuador; Cultivated at RGB Kew/ 1982–1568 | 34 | 12 | 2 | ||
| Philodendron | P. billietiae Croat | Cultivated at RBG Kew/ 2005–2363 | 32 | 16 | 2 | |
| P. fragrantissimum (Hook.) G.Don | Igarassu, Pernambuco, Brazil; Cultivated at LGBV; SV295 | 32 | 2 | 2 | ||
| P. giganteum Schott | Oriole trail, Montserrat; Cultivated at RBG Kew/ 2011–1735 | 30 | 10 | 2 | ||
| P. hederaceum (Jacq.) Schott | Floresta da Tijuca, Rio de Janeiro, Brazil; Cultivated at LGBV; SV248 |
32 | 2 | 2 | ||
| P. maximum K.Krause | Cultivated at RBG Kew/ 1973–381 | 34 | 12 | 2 | ||
| P. megalophyllum Schott | Uatumã, Amazonas, Brazil; Cultivated at LGBV; SV320 | 34 | 12 | 2 | ||
| P. melinonii Brongn.ex Regel | Reserva Florestal Adolpho Ducke, Amazonas, Brazil; Cultivated at LGBV; MC085 | 30 | 4 | 2 | ||
| P. schmidtiae Croat & C.E.Ceron | Napo, Ecuador; Cultivated at RBG Kew/ 1982–1573 | 32 | 14 | 2 | ||
| P. tenue K.Koch & Augustin | Costa Rica; Cultivated at RGB Kew/ 1984–612 | 34 | 6 | 2 | ||
| Polytomium | P. distantilobum K.Krause | Uatumã, Amazonas, Brazil; Cultivated at LGBV; SV318 | 32 | 2 | 2 | |
| P. lacerum (Jacq.) Schott | Guiana; Cultivated at RBG Kew/ 1979–3173 | 32 | 10 | 2 | ||
| Schizophyllum | P. bipennifolium Schott | INPA, Acre, Brazil; Cultivated at LGBV; SV307 | 32 | 8 | 3 | |
| P. nadruzianum Sakur. | Floresta da Tijuca, Rio de Janeiro, Brazil; Cultivated at LGBV; LSBC175 |
32 | 2 | 2 | ||
| P. pedatum (Hook.) Kunth | Floresta da Tijuca, Rio de Janeiro, Brazil; Cultivated at LGBV; MC081 |
32 | 4 | 2 | ||
| P. quinquelobum K.Krause | Urucu, Amazonas, Brazil; Cultivated at LGBV; MC080 | 32 | 4 | 2 | ||
| Tritomophyllum | P. angustilobum Croat & Grayum | Heredia, Costa Rica; Cultivated at RBG Kew/ 1996–4420 | 34 | 3 | 2 | |
| P. tripartitum (Jacq.) Schott | Chiapas, México; Cultivated at RBG Kew/ 1980–1524 | 34 | 6 | 2 | ||
Abbreviations: LGBV: Laboratory of Genetics and Plant Biotechnology, Federal University of Pernambuco (Recife, Brazil); RBG Kew: Royal Botanic Gardens, Kew (Richmond, United Kingdom).
Root tips were pretreated in 2 mM 8-hydroxyquinoline at 8°C for 24 h, fixed in Carnoy solution (ethanol:acetic acid, 3:1, v/v) at room temperature for 24 h, and stored at -20°C. Subsequently, the fixed root tips were washed in distilled water and digested in an enzyme solution containing 2% cellulase (w/v) (Onozuka R-10, Serva) and 20% pectinase (v/v) (Sigma-Aldrich) overnight at 37°C. Slides were prepared by squashing the meristematic tissue in 45% acetic acid. In addition, slides were stained in a solution of 2 μg mL-1 DAPI (4', 6-diamidino-2-phenylindole) and glycerol (1:1, v/v) and then analyzed. Afterwards, the best slides were de-stained and fixed in Carnoy solution for 30 min and transferred to absolute ethanol for 1 h, both at room temperature. After air drying, the slides were stored at -20°C. At least five root tips were analyzed per species.
Probes, fluorescent in situ hybridization and data analysis
The rDNA probes used for FISH were R2, a 6.5 kb fragment containing the 18S-5.8S-25S rDNA unit from Arabidopsis thaliana (L.) Heynh., and D2, a 400 bp fragment containing two 5S rDNA units from Lotus japonicus (Regel) K.Larsen [22]. Labelling was performed by nick translation with digoxigenin-11-dUTP (Roche Diagnostics) and biotin-11-dUTP (Roche Diagnostics) for 35S and 5S rDNA, respectively. The telomeric probe was amplified by PCR according to Ijdo et al. [23], with the primers (TTTAGGG)5 and (CCCTAAA)5 and labeled with Cy3-dUTP (Jena Bioscience) as described before.
The FISH procedures followed Vasconcelos et al. [24], except for the chromosome denaturing, which occurred separately from the probe in 70% formamide in 2×SSC at 80–85°C for 7 min, and then dehydrated for 5 min in an alcoholic series (ethanol 70% and 100%) at -20°C. The stringency wash was performed in 0.1×SSC at 42°C. The hybridization mix consisted of 50% (v/v) formamide, 10% (w/v) dextran sulfate, 2×SSC and 2–5 ng/μL of probe. Digoxigenin and biotin-labeled probes were detected using conjugated anti-digoxigenin rhodamine (Roche), and Alexa Fluor conjugated streptavidin (Invitrogen), respectively, in 1% (w/v) BSA. Preparations were counterstained and mounted with 2 μg/mL DAPI in Vector’s Vectashield (1:1; v/v).
Images were captured using a Leica DMLB epifluorescence microscope coupled with a Leica DFC 340FX camera, using the Leica CW 4000 software. Images were optimized for better brightness and contrast with Adobe Photoshop CS3 (Adobe Systems Incorporated). The 35S rDNA was pseudocolored in green, and the 5S rDNA was pseudocolored in red. At least eight images were analyzed per species.
Also, for a better understanding of the karyotype evolution of the genus, idiograms of the chromosomes carrying rDNA sites of each species were plotted in the phylogenetic tree previously reported for the group [21]. Thus, chromosome lengths of three metaphases for each species with similar condensation pattern were measured twice considering both chromatids, totalizing six measurements per species, using the MicroMeasure v3.3 software [25]. The Adobe Flash CS3 program (Adobe Systems Incorporated) was used for the elaboration of the idiograms.
Genomic DNA extraction, sequencing and satellite DNA analysis
Genomic DNA of P. callosum K.Krause was extracted from fresh leaves using the CTAB protocol described by Weising et al. [26]. The precipitation of contaminating polysaccharides was performed according to Michaels et al. [27]. Genomic DNA was sequenced in Illumina MiSeq (2 × 250 pb). Clusterization and characterization of the repetitive genome fraction were performed on the Galaxy/RepeatExplorer platform using the Elixir-cerit server. The TAREAN tool was used to identify satellite DNA sequences (https://repeatexplorer-elixir.cerit-sc.cz) [28–30]. Cluster sharing similarity to the telomeric motif was manually checked and the contigs were used to reconstruct the monomer.
Results
Chromosome number ranged from 2n = 28 to 36 considering the total of 34 analyzed species (Table 1): 2n = 28 (P. callosum); 2n = 30 (P. giganteum and P. melinonii), 2n = 32 (T. spruceanum and 16 Philodendron species), 2n = 34 (T. mello-barretoanum and 10 Philodendron species) and 2n = 36 (T. corcovadense, T. lundii and T. saxicola).
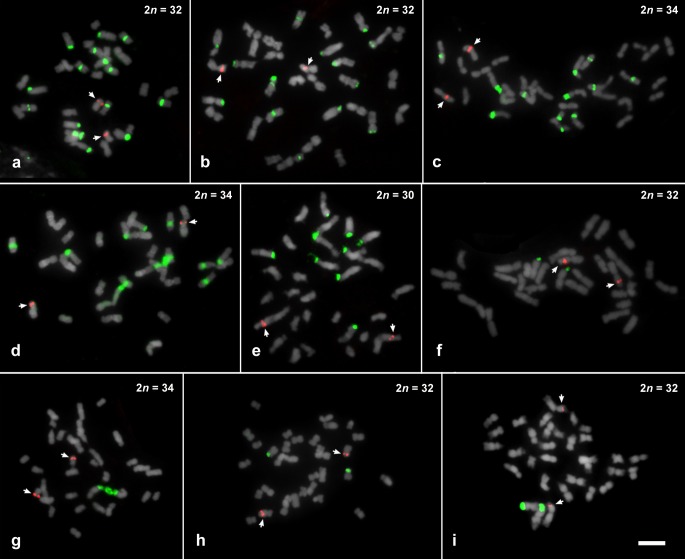
FISH using 35S rDNA probe showed a wide variation in both number and location of sites along the chromosomes (see Table 1, Figs 1–4). For Philodendron species, P. billietiae (2n = 32, Fig 1A) showed the highest number of sites (16 sites), followed by P. quinquenervium (2n = 32, Fig 1B) with 15 sites; P. schmidtiae (2n = 32, Fig 3) with 14 sites; P. maximum (2n = 34, Fig 1C), P. megalophyllum (2n = 34, Fig 3) and P. uleanum (2n = 34, Fig 1D) with 12 sites; and P. burle-marxii (2n = 34, Fig 4), P. giganteum (2n = 30, Fig 1E) and P. lacerum (2n = 32, Fig 3) with 10 sites. In the other 19 species (with chromosome numbers ranging from 2n = 30 to 34), the number of sites varied from two to eight (Table 1, Fig 3). The lowest number of 35S rDNA sites was observed for seven species: P. annulatum (Fig 3), P. distantilobum (Fig 1F), P. hederaceum (Fig 3), P. inconcinnum (Fig 4), P. krugii (Fig 1G), P. nadruzianum (Fig 1H), and P. fragrantissimum (Fig 1I), which presented only two sites. For the five Thaumatophyllum species, two presented four 35S rDNA sites (T. corcovadense and T. spruceanum, Fig 3), and the other three had two sites (T. lundii, Fig 2A; T. mello-barretoanum, Fig 2B, and T. saxicola, Fig 3).
Fig 1. Fluorescent in situ hybridization of 35S (green) and 5S rDNA (red) on mitotic chromosomes of Philodendron species, counterstained with DAPI and pseudocolored in gray.
(a) Philodendron billietiae; (b) P. quinquenervium; (c) P. maximum; (d) P. uleanum; (e) P. giganteum; (f) P. distantilobum; (g) P. krugii; (h) P. nadruzianum; (i) P. fragrantissimum. Bar in i represents 5 μm.
Fig 4. Chromosomes mapped with 35S (green) and 5S (red) rDNA probes in six species of Philodendron subgenus Philodendron not included on Vasconcelos et al. [11] phylogeny.
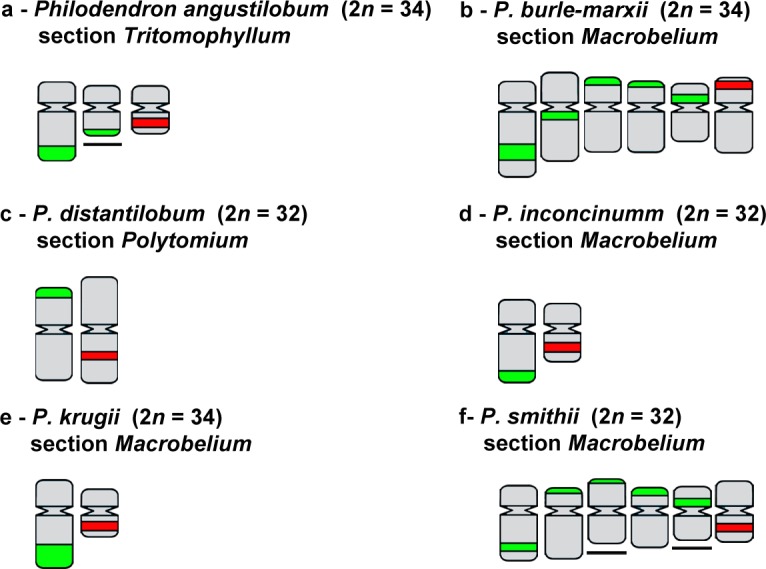
Underlined chromosomes represent single chromosomes with heteromorphism, for which it was not possible to identify their respective homologs. Non-underlined chromosomes represent the pair of homologues.
Fig 3. Chromosomal patterns for rDNA in 22 Philodendron and five Thaumatophyllum species plotted in a modified phylogeny based on Vasconcelos et al. [11].
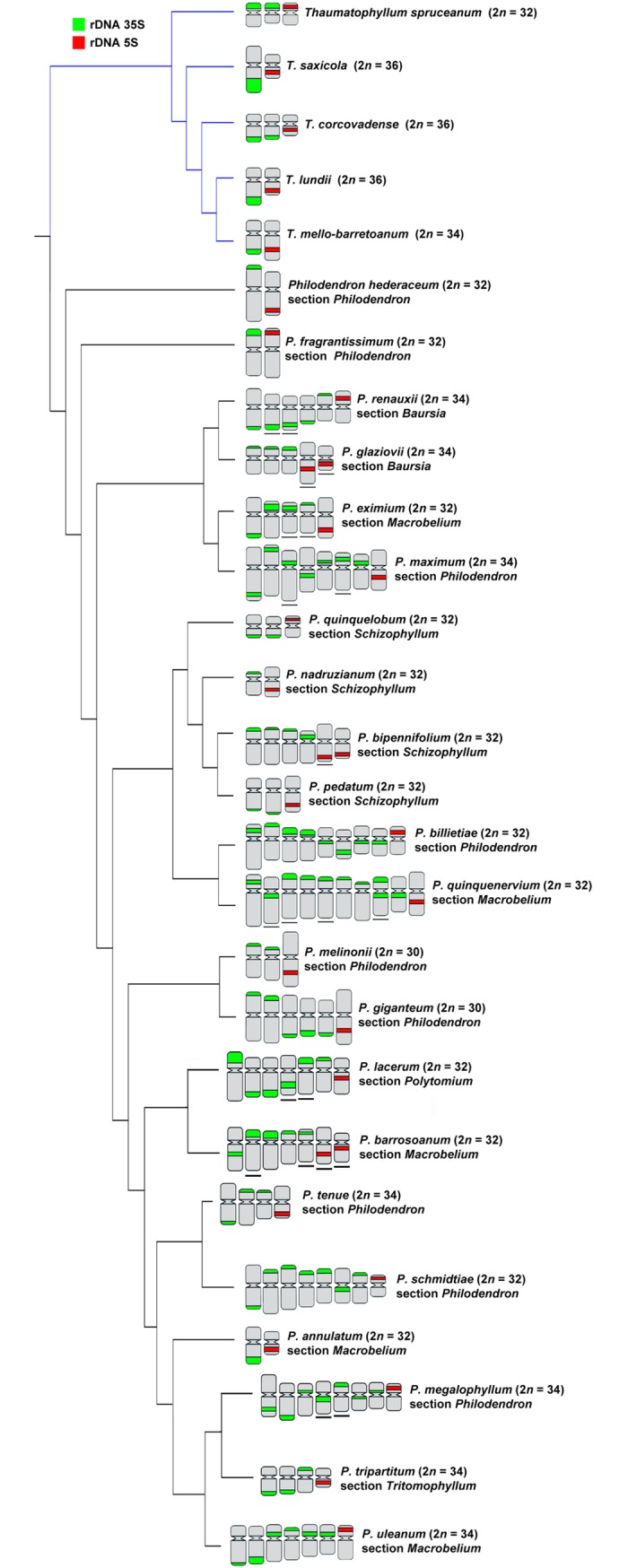
Underlined chromosomes represent single chromosomes with heteromorphism, for which it was not possible to identify their respective homologs. Each non-underlined chromosome represents a pair of homologs.
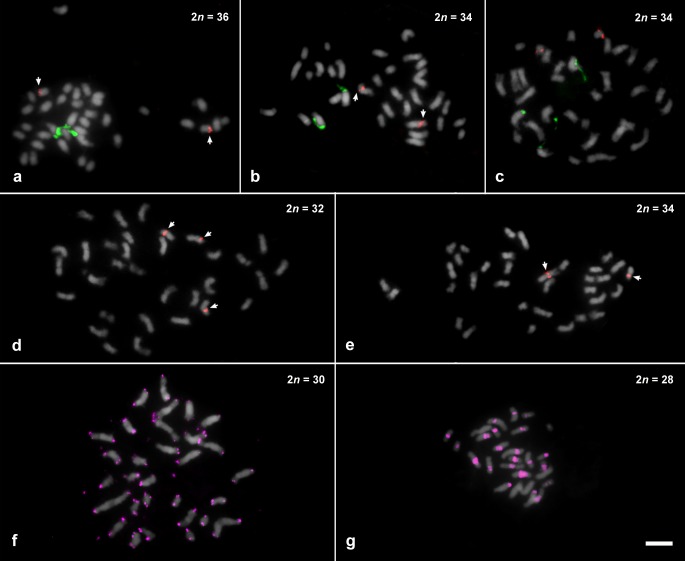
Fig 2. Distribution of repetitive sequences in species of Thaumatophyllum and Philodendron species, counterstained with DAPI and pseudocolored in gray (35S rDNA in green, 5S rDNA in red, and telomeric probe in pink).
(a) T. lundii; (b) T. mello-barretoanum; (c) P. angustilobum; (d) P. bipennifolium; (e) P. glaziovii; (f) P. giganteum; (g) P. callosum. Bar in g represents 5 μm.
In Philodendron, the 35S rDNA sites were predominantly located in the terminal region of the short arm (21 out of the 29 species, Figs 1–4), although subterminal, interstitial or proximal sites were also observed. In Thaumatophyllum, the 35S rDNA sites were located only in the terminal region, predominantly in the long arm (four out of the five species, Figs 2 and 3). In addition, we did not observe a clear pattern of distribution of numbers of rDNA sites in the phylogenetic tree of Philodendron s.l. (Fig 3).
Heteromorphisms of number and distribution of 35S rDNA sites were identified in nine of the analyzed species. Philodendron angustilobum, for example, presented an odd number of 35S rDNA (three sites), hampering the identification of the homologous chromosomes (Fig 2C). Meanwhile, other species, as P. eximium and P. smithii, showed a chromosome pair bearing a 35S rDNA site in the terminal region of the short arm of one chromosome and in the proximal region of the short arm of the other one (Figs 3 and 4); P. lacerum and P. megalophyllum, presented sites located on opposite chromosome arms in supposed homologs (Fig 3); and P. quinquenervium was the only species with two 35S rDNA sites in the same chromosome (Fig 3).
In turn, for the 5S rDNA, most species showed two sites, except for P. bipennifolium (Fig 2D), which presented three sites. For Philodendron, we observed a high variation in the position of the 5S rDNA sites, although being most frequently observed in the interstitial position of the long arm (in 14 out of 29 analyzed species, Figs 1–4). For Thaumatophyllum, the 5S rDNA sites were located in the interstitial region predominantly in the long arm (four out of 5 species, Figs 2 and 3).
Both species P. bipennifolium and P. glaziovii stood out due to 5S rDNA heteromorphisms. The first had three 5S rDNA sites, being two located in the subterminal region of the long arm and one in the proximal region of the short arm (Fig 2D). In turn, P. glaziovii, had two 5S rDNA clusters, as most of the analyzed species, but the chromosomes bearing those sites were heteromorphic in size and morphology (Figs 2E and 3).
The telomeric DNA revealed no interstitial telomeric repeat (ITR) in P. giganteum (2n = 30), which exhibited only terminal marks in both extremities of all chromosomes (Fig 2F). In turn, P. callosum (2n = 28) presented no visible terminal hybridization signals, although exhibiting large pericentromeric marks in almost all chromosomes (Fig 2G). Analysis of the repetitive fraction of this species revealed the presence of one satellite DNA related to the telomeric repeat. The manual inspection of the contigs that composed the cluster CL228 allowed the identification of Arabidopsis-like telomeric motifs within the sequence, henceforth called PcSat1 satellite DNA. In all three monomers, the telomeric repeat was represented in variable amounts (unit 1 –U1, eight times; U2, 20 times; U3, nine times), making the monomer size slightly variable (Fig 5).
Fig 5. Features of the satellite DNA PcSat1 from Philodendron callosum.
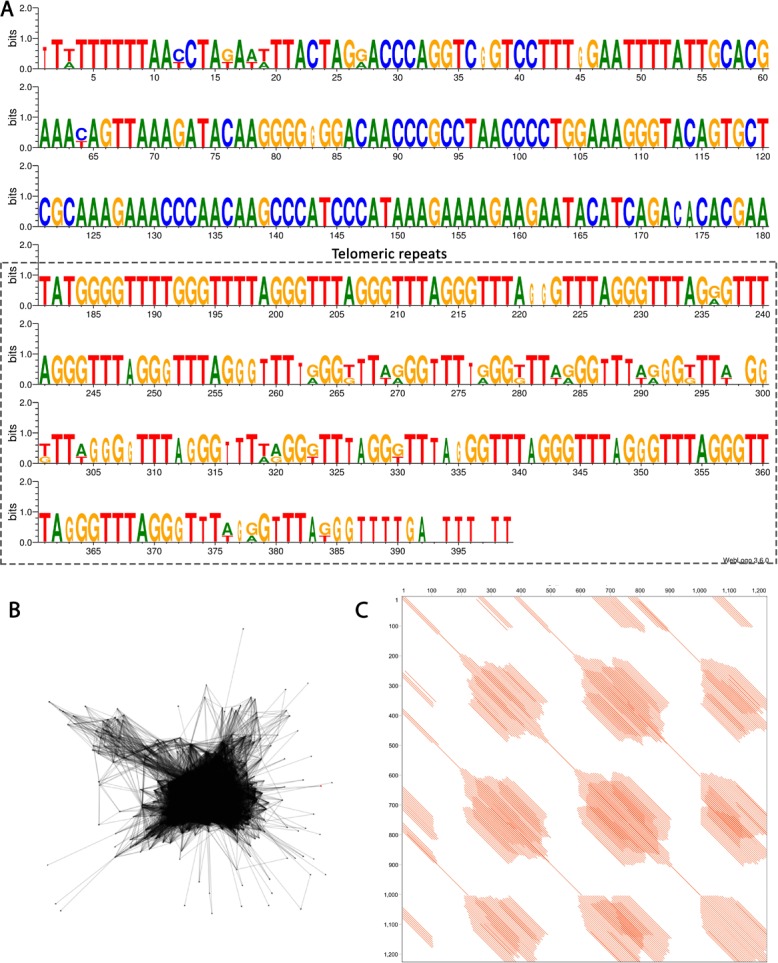
(a) Reconstructed monomer using WebLogo [63]. A cluster of conserved and degenerated plant telomeric motifs is present at the repeat unit and represents more than half of its full size (box). (b) Layout of cluster 228 obtained from Repeat Explorer output (Vasconcelos et al. unpublished data) and (c) Dotplot graph showing the internal organization of the sequence.
Discussion
The present work is the first extensive cytogenetic study analyzing the localization of rDNA sites in chromosomes of Philodendron and Thaumatophyllum species, except for P. hederaceum [18]. Chromosome numbers are all in accordance with previously published data [9]. The results revealed an enormous karyotypic variability in relation to the distribution of the 35S rDNA in Philodendron s.l., opposed to a high stability in number and position of the 5S rDNA sites. These results are in consonance with the distribution pattern of rDNA sites in angiosperms and confirm the lower dispersion capacity of 5S rDNA proposed by Roa and Guerra [11, 12]. The presence of only one chromosome pair bearing 5S rDNA as found here seems to be conserved for the Araceae family (51 out of 54 analyzed species), even though only 12 out of 144 genera were analyzed up to date [17, 19].
Considering P. subg. Philodendron (with ca. 7,25% of its species analyzed), the amplification and distribution of 35S rDNA sites seem to occur frequently and independently along the different subgroups. The number of 35S rDNA sites (ranging from two to 16) varied significantly within and among the clades of the subg. Philodendron, with no clear phylogenetic pattern, considering the relationships presented by Vasconcelos et al. [21]. Also, there was not any correspondence of our data to the traditional subdivision of the subgenus in morphological sections.
Thaumatophyllum had fewer species analyzed, but better represented, with ca. 23.8% of the species of the genus. It showed a smaller variation in the number of rDNA sites (two to four sites), indicating a higher homogeneity among karyotypes. A similar homogeneity was observed in Typhonium (four 35S rDNA sites in eight out of 10 analyzed species), the aroid genus better studied cytogenetically so far [17].
In the present study, two of the 29 Philodendron species analyzed showed an odd number of rDNA sites, whereas for other 11 species, we observed heteromorphisms in site position or in the morphology of the chromosome bearing the rDNA. Similar polymorphisms have been previously described in Araceae and in other angiosperm families [19, 31, 32]. Considering that the rDNA sites are considered fragile, recombination hotspots may occur due to the highly repetitive nature of the locus, resulting in breaks followed by chromosomal rearrangements [33–35]. This could explain the interstitial position found for the 35S rDNA in some of our analyzed species, which is relatively uncommon in plant chromosomes [12]. Several hypotheses have been proposed for such polymorphisms, including the model of amplification, dispersion and deletion, the action of transposable elements, as well as non-homologous recombination, mainly related to the preferential terminal positions of the 35S rDNA sites on the chromosomes [34, 36–40]. The expansions and contractions of the repetitive DNA have frequently been associated to changes in the chromosome morphology, also resulting in changes in the position of rDNA sites [41], without necessarily changing the gene order [42].
Additionally, the diversity in the distribution of rDNA sites and the polymorphisms observed in Philodendron could be related to natural hybridizations that may have occurred throughout the evolutionary history of the group, although none of the analyzed species has a recognized hybrid origin. In Citrus L. (Rutaceae) and related genera, heteromorphism of chromosomal types is considered to be a reliable indicator of interspecific crosses, where variation in the number and location of CMA positive bands and rDNA sites is related to the fact that most species are apomictic hybrids [16, 31, 43–45]. Rapid changes in 35S rDNA loci in response to interspecific or intergeneric hybridization, when comparing to the progenitors, have also been reported in Potamogeton L. (Potamogetonaceae) [46], Rosa L. (Rosaceae) [47] and Lolium L. × Festuca L. [48, 49] hybrids.
Aiming to test the hypothesis of the existence of descending dysploidy events in Philodendron species generating lower chromosome numbers (2n = 28; 30) associated with centric or end-to-end fusion [19, 50, 51], we applied telomeric DNA probes to verify the possible presence of ITRs. Such internal telomeres were observed in dysploid series in Nothoscordum Kunth and Ipheion Raf., both belonging to the family Amaryllidaceae [52], as well as in Typhonium [17, 19]. However, the expected ITRs were not visible in P. giganteum (2n = 30), thus suggesting that the remnants of ITRs were lost along the karyotype evolution of this species by losing the chromosome extremities in translocation events, or by elimination or dispersion after insertion, as a result of high recombination rates in these regions, as suggested for Phaseolus leptostachyus Benth. [53]. Another plausible hypothesis would be that the ITRs are present in short arrays not detectable by FISH, as reported for tomato and for the bat Carollia perspicillata L. [54, 55].
In addition, FISH with the telomeric probe in P. callosum revealed several pericentromeric blocks related to a satellite DNA sequence. Telomeric repeats within satellite DNA arrays are not uncommon, being also reported in Rumex induratus Boiss & Reuter (Polygonaceae), Jatropha curcas L. (Euphorbiaceae), and in Solanum lycopersicum L. (tomato), S. tuberosum L. (potato) and S. melongena L. (eggplant) (Solanaceae) [54, 56–58]. AT-rich degenerated telomeric repeats were identified in the repeat St49 of Solanum L. species, indicated as an ancient sat-DNA derived from a telomeric-like sequence identified in the telomeres and centromeres of tomato and potato chromosomes, respectively [54, 58].
On the other hand, the absence of signals at chromosome termini of P. callosum may be due to the presence of small standard telomere arrays (TTTAGGG)n not detected by FISH. However, the occurrence of a different telomere sequence in P. callosum chromosomes cannot be dismissed, as already described for several plant species [59–61]. This last scenario would imply in an interspecific variation within Philodendron, which was already reported for species of Genlisea A.St.-Hil. (Lentibulariaceae), a genus of carnivorous plants with a high variation level in the genome structures among species [59, 62].
Conclusions
Our data revealed a substantial variation in the number and location of the 35S rDNA sites in Philodendron indicating a rapid karyotype evolution within P. subg. Philodendron. More homogeneous karyotypes were observed in species of the sister genus Thaumatophyllum. Variation was more prominent because no clear trend regarding the 35S rDNA sites was evident, neither considering the traditional infrageneric taxonomy nor considering the most recent molecular phylogenetic data. Also, the identification of heteromorphisms in the number and position of 35S rDNA sites suggests the occurrence of expansions and/or contractions of repetitive DNA in the genomes of some species, or chromosomal rearrangements, possibly associated with natural hybridization events. Furthermore, a better understanding of the importance of the repetitive DNA during the karyotypic evolution of Philodendron s.l. will be allowed by future analyses considering the evolution of chromosome numbers and genome sizes in a phylogenetic framework, besides a characterization of the composition of the repetitive DNA among species of the group, which are currently in development.
Acknowledgments
The authors are grateful to the Royal Botanic Gardens, Kew, especially to Ilia J. Leitch, Ralf Kynast, Simon J. Mayo and Marcelo Sellaro for providing access to the sampled materials. We thank CNPq (National Council for Scientific and Technological Development), CAPES (Coordination of Higher Level Personnel), and FACEPE (Foundation for Support to Science and Technology of the State of Pernambuco) for financial support and fellowships.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was financially supported by fellowships from CNPq (National Council for Scientific and Technological Development), CAPES (Coordination of Higher Level Personnel), and FACEPE (Foundation for Support to Science and Technology of the State of Pernambuco). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mayo SJ, Bogner J, Boyce PC. The genera of Araceae. Richmond: Royal Botanic Gardens, Kew, 1997; 370 p. 10.2307/4110410 [DOI] [Google Scholar]
- 2.Boyce PC, Croat TB. The Überlist of Araceae, totals for published and estimated number of species in aroid genera. 2018. http://www.aroid.org/genera/180211uberlist.pdf 11 Feb 2018. [Google Scholar]
- 3.Calazans LSB, Sakuragui CM, Mayo SJ. From open areas to forests? The evolutionary history of Philodendron subgenus Meconostigma (Araceae) using morphological data. Flora. 2014;209: 117–121. 10.1016/j.flora.2013.12.004 [DOI] [Google Scholar]
- 4.Loss-Oliveira L, Sakuragui C, Soares ML, Schrago CG. Evolution of Philodendron (Araceae) species in Neotropical biomes. PeerJ. 2016;4: e1744 10.7717/peerj.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayo SJ. A revision of Philodendron subgenus Meconostigma (Araceae). Kew Bulletin. 1991;46: 601–681. [Google Scholar]
- 6.Grayum MH. Revision of Philodendron subgenus Pteromischum (Araceae) for Pacific and Caribbean tropical America. Syst Bot Monogr. 1996;47: 1–233. 10.2307/25027858 [DOI] [Google Scholar]
- 7.Croat TB. A revision of Philodendron subgenus Philodendron (Araceae) for Mexico and Central America. Ann Mo Bot Gard. 1997;84: 311–704. 10.2307/2992022 [DOI] [Google Scholar]
- 8.Correia-da-Silva M, Vasconcelos S, Soares MLC, Mayo SJ, Benko-Iseppon AM. Chromosomal diversity in Philodendron (Araceae): taxonomic significance and a critical review. Plant Syst Evol. 2014;300: 1111–1122. 10.1007/s00606-013-0949-9 [DOI] [Google Scholar]
- 9.Vasconcelos EV, Brasileiro-Vidal AC, Benko-Iseppon AM, Vasconcelos S. Updating the list of chromosome numbers for Philodendron (Araceae). Acta Bot Bras. 2017;31: 309–312. 10.1590/0102-33062016abb0431 [DOI] [Google Scholar]
- 10.Sakuragui CM, Calazans LSB, Loss-Oliveira L, Morais EB, Benko-Iseppon AM, Vasconcelos S, et al. Recognition of the genus Thaumatophyllum Schott − formerly Philodendron subg. Meconostigma (Araceae) − based on molecular and morphological evidence. PhytoKeys. 2018;98: 51–71. 10.3897/phytokeys.98.25044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roa F, Guerra M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol Biol. 2012;12: 225 10.1186/1471-2148-12-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roa F, Guerra M. Non-random distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet Genome Res. 2015;146: 243–249. 10.1159/000440930 [DOI] [PubMed] [Google Scholar]
- 13.Garcia S, Kovařík A, Leitch AR, Garnatje T. Cytogenetic features of rRNA genes across land plants: analysis of the Plant rDNA database. Plant J. 2017;89: 1020–1030. 10.1111/tpj.13442 [DOI] [PubMed] [Google Scholar]
- 14.Guerra M. Cytotaxonomy: the end of childhood. Plant Biosyst. 2012;146: 703–710. 10.1080/11263504.2012.717973 [DOI] [Google Scholar]
- 15.Siljak-Yakovlev S, Godelle B, Zoldos V, Vallès J, Garnatje T, Hidalgo O. Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: a case study in Reichardia genus. PLoS One 2017;12: e0182318 10.1371/journal.pone.0182318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi KU, Kim HB, Song KJ. Karyotype diversity of Korean landrace mandarins by CMA banding pattern and rDNA loci. Sci Hortic. 2018;228: 26–32. 10.1016/j.scienta.2017.10.001 [DOI] [Google Scholar]
- 17.Sousa A, Cusimano N, Renner SS. Combining FISH and model-based predictions to understand chromosome evolution in Typhonium (Araceae). Ann Bot. 2014;113: 669–680. 10.1093/aob/mct302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakshmanan PS, Laere KV, Eeckhaut T, Huylenbroeck JV, Bockstaele EV, Khrustaleva L. Karyotype analysis and visualization of 45S rRNA genes using fluorescence in situ hybridization in aroids (Araceae). Comp Cytogenet. 2015;9: 145–160. 10.3897/CompCytogen.v9i2.4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sousa A, Renner SS. Interstitial telomere-like repeats in the monocot family Araceae. Bot J Linn Soc. 2015;177: 15–26. 10.1111/boj.12231 [DOI] [Google Scholar]
- 20.Bennett MD, Leitch IJ. 2012. Plant DNA C-values database (Release 6.0, Dec. 2012). http://data.kew.org/cvalues/. 24 June 2014. [Google Scholar]
- 21.Vasconcelos S, Soares ML, Sakuragui CM, Croat TB, Oliveira G, Benko-Iseppon AM. New insights on the phylogenetic relationships among the traditional Philodendron subgenera and the other groups of the Homalomena clade (Araceae). Mol Phylogeneti Evol. 2018;127: 168–178. 10.1016/j.ympev.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 22.Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A. Chromosomal map of the model legume Lotus japonicus. Genetics. 2002;161: 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ijdo JW, Wells RA, Baldini A, Reeders ST. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991;13: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasconcelos EV, Fonsêca AFA, Pedrosa-Harand A, Bortoleti KCA, Benko-Iseppon AM, Costa AF, et al. Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosome Res. 2015;23: 253–266. 10.1007/s10577-014-9464-2 [DOI] [PubMed] [Google Scholar]
- 25.Reeves A. MicroMeasure: a new computer program for the collection and analysis of cytogenetic data. Genome. 2001;44: 439–443. 10.1139/g01-037 [DOI] [PubMed] [Google Scholar]
- 26.Weising K, Nybom H, Wolff K, Kahl G. DNA Fingerprinting in plants, principles, methods, and applications. 2nd ed Boca Raton: CRC Press, 2005; 444p. [Google Scholar]
- 27.Michaels SD, John MC, Amasino RM. Removal of polysaccharides from plant DNA by ethanol precipitation. Biotechniques. 1994;17: 274–276. [PubMed] [Google Scholar]
- 28.Novák P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. Bioinformatics. 2010;11: 378 10.1186/1471-2105-11-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bionformatics. 2013;29: 792–793. 10.1093/bioinformatics/btt054 [DOI] [PubMed] [Google Scholar]
- 30.Novák P, Ávila LR, Koblížková A, Vrbová I, Neumann P, Macas J. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017;45: e111 10.1093/nar/gkx257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barros e Silva AE, Dos Santos Soares Filho W, Guerra M. Linked 5S and 45S rDNA sites are highly conserved through the subfamily Aurantioideae (Rutaceae). Cytogenet Genome Res. 2013;140: 62–69. 10.1159/000350695 [DOI] [PubMed] [Google Scholar]
- 32.Monkheang P, Chaveerach A, Sudmoon R, Tanee T. Karyotypic features including organizations of the 5S, 45S rDNA loci and telomeres of Scadoxus multiflorus (Amaryllidaceae). Comp Cytogenet. 2016;10: 637–646. 10.3897/CompCytogen.v10i4.9958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang M, Li H, Zhang L, Gao F, Wang P, Hu Y, et al. Plant 45S rDNA clusters are fragile sites and their instability is associated with epigenetic alterations. PLoS One. 2012;7: e35139 10.1371/journal.pone.0035139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dvořáčková M, Fojtová M, Fajkus J. Chromatin dynamics of plant telomeres and ribosomal genes. Plant J. 2015;83: 18–37. 10.1111/tpj.12822 [DOI] [PubMed] [Google Scholar]
- 35.Lan H, Chen C-L, Miao Y, Yu C-X, Guo W-W, Xu Q, et al. Fragile sites of ‘Valencia’ sweet orange (Citrus sinensis) chromosomes are related with active 45S rDNA. PLoS One. 2016;11: e0151512 10.1371/journal.pone.0151512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall KJ, Parker JS. Stable chromosome fission associated with rDNA mobility. Chromosome Res. 1995;3: 417–422. 10.1007/BF00713891 [DOI] [PubMed] [Google Scholar]
- 37.Eickbush TH, Eickbush DG. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics. 2007;175: 477–485. 10.1534/genetics.107.071399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen S, Houben A, Segal D. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 2008;53: 1027–1034. 10.1111/j.1365-313X.2007.03394.x [DOI] [PubMed] [Google Scholar]
- 39.Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, et al. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci. 2008;105: 5833–5838. 10.1073/pnas.0709698105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosato M, lvarez I, Feliner GN, Rosselló JA. High and uneven levels of 45S rDNA site number variation across wild populations of a diploid plant genus (Anacyclus, Asteraceae). PLoS One. 2017;12: e0187131 10.1371/journal.pone.0187131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendes S, Moraes AP, Mirkov TE, Pedrosa-Harand A. Chromosome homeologies and high variation in heterochromatin distribution between Citrus L. and Poncirus Raf. as evidenced by comparative cytogenetic mapping. Chromosome Res. 2011;19: 521–530. 10.1007/s10577-011-9203-x [DOI] [PubMed] [Google Scholar]
- 42.Navratilova A, Neumann P, Macas P. Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. Ann Bot. 2003;91: 921–929. 10.1093/aob/mcg099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho R, Soares Filho WS, Brasileiro-Vidal AC, Guerra M. The relationship among lemons, limes and citron: a chromosomal comparison. Cytogenet Genome Res. 2005;109: 276–282. 10.1159/000082410 [DOI] [PubMed] [Google Scholar]
- 44.Brasileiro-Vidal AC, dos Santos-Serejo JA, Soares Filho WS, Guerra M. A simple chromosomal marker can reliably distinguishes Poncirus from Citrus species. Genetica. 2007;129: 273–279. 10.1007/s10709-006-0007-4 [DOI] [PubMed] [Google Scholar]
- 45.Barros e Silva AE, Marques A, Santos KGB, Guerra M. The evolution of CMA bands in Citrus and related genera. Chromosome Res. 2010;18: 503–514. 10.1007/s10577-010-9130-2 [DOI] [PubMed] [Google Scholar]
- 46.Wan T, Zhang X-L, Gregan J, Zhang Y, Guo P, Guo Y-H. A dynamic evolution of chromosome in subgenus Potamogeton revealed by physical mapping of rDNA loci detection. Plant Syst Evol. 2012;298: 1195–1210. 10.1007/s00606-012-0621-9 [DOI] [Google Scholar]
- 47.Ding XL, Xu TL, Wang J, Luo L, Yu C, Dong GM, et al. Distribution of 45S rDNA in modern rose cultivars (Rosa hybrida), Rosa rugosa, and their interspecific hybrids revealed by fluorescence in situ hybridization. Cytogenet Genome Res. 2016;149: 226–235. 10.1159/000448063 [DOI] [PubMed] [Google Scholar]
- 48.Akiyama Y, Kimura K, Yamada-Akiyama H, Kubota A, Takahara Y, Ueyama Y. Genomic characteristics of a diploid F4 Festulolium hybrid (Lolium multiflorum × Festuca arundinacea). Genome. 2012;55: 599–603. 10.1139/g2012-048 [DOI] [PubMed] [Google Scholar]
- 49.Książczyk T, Zwierzykowska E, Molik K, Taciak M, Krajewski P, Zwierzykowski Z. Genome-dependent chromosome dynamics in three successive generations of the allotetraploid Festuca pratensis × Lolium perenne hybrid. Protoplasma. 2015;252: 985–996. 10.1007/s00709-014-0734-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lysak MA, Schubert I. Mechanisms of chromosome rearrangements. Plant Genome Diversity 2013;2: 137–147. 10.1007/978-3-7091-1160-4_9 [DOI] [Google Scholar]
- 51.Bolzán AD. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat Res. 2017;773: 51–65. 10.1016/j.mrrev.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 52.Souza G, Vanzela ALL, Crosa O, Guerra M. Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae). Genetica. 2016;144: 157–166. 10.1007/s10709-016-9886-1 [DOI] [PubMed] [Google Scholar]
- 53.Fonsêca A, Ferraz ME, Pedrosa-Harand A. Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma. 2016;125: 413–421. 10.1007/s00412-015-0548-3 [DOI] [PubMed] [Google Scholar]
- 54.He L, Liu J, Torres GA, Zhang H, Jiang J, Xie C. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Res. 2013;21: 5–13. 10.1007/s10577-012-9332-x [DOI] [PubMed] [Google Scholar]
- 55.Calixto M da S, de Andrade IS, Cabral-de-Mello DC, Santos N, Martins C, Loreto V, et al. Patterns of rDNA and telomeric sequences diversification: contribution to repetitive DNA organization in Phyllostomidae bats. Genetica. 2014;142: 49–58. 10.1007/s10709-013-9753-2 [DOI] [PubMed] [Google Scholar]
- 56.Navajas-Pérez R, Schwarzacher T, Rejón MR, Garrido-Ramos MA. Characterization of RUSI, a telomere-associated satellite DNA, in the genus Rumex (Polygonaceae). Cytogenet Genome Res. 2009;124: 81–89. 10.1159/000200091 [DOI] [PubMed] [Google Scholar]
- 57.Kikuchi S, Tsujimoto H, Sassa H, Koba T. JcSat1, a novel subtelomeric repeat of Jatropha curcas L. and its use in karyotyping. Chromosome Sci. 2010;13: 11–16. 10.11352/scr.13.11 [DOI] [Google Scholar]
- 58.Gong ZY, Wu YF, Koblížková A, Torres GA, Wang K, Iovene M, et al. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell. 2012;24: 3559–3574. 10.1105/tpc.112.100511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran TD, Cao HX, Jovtchev G, Neumann P, Novák P, Fojtová M, et al. Centromere and telomere sequence alterations reflect the rapid genome evolution within the carnivorous plant genus Genlisea. Plant J 2015;84: 1087–1099. 10.1111/tpj.13058 [DOI] [PubMed] [Google Scholar]
- 60.Fajkus P, Peska V, Sitová Z, Fulnecková J, Dvorácková M, Gogela R, et al. Allium telomeres unmasked: the unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. Plant J 2016;85: 337–347. 10.1111/tpj.13115 [DOI] [PubMed] [Google Scholar]
- 61.Peška V, Sitová Z, Fajkus P, Fajkus J. BAL31-NGS approach for identification of telomeres de novo in large genomes. Methods 2017;114: 16–27. 10.1016/j.ymeth.2016.08.017 [DOI] [PubMed] [Google Scholar]
- 62.Veleba A, Bures P, Adamec L, Smarda P, Lipnerova I, Horova L. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytol 2014;203: 22–28. 10.1111/nph.12790 [DOI] [PubMed] [Google Scholar]
- 63.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.