ABSTRACT
Hepatocellular carcinoma (HCC), one of the most common type of cancers, is highly refractory to most systemic therapies. Understanding the genomic dysregulations, in particularly non-coding RNA (ncRNA) dysregulations, in HCC may provide novel strategies to HCC treatment. In our previous study, we demonstrated the key role of miR-200a-mediated HMGB1/RAGE signaling in HCC carcinogenesis. In the present study, we identified circular RNA (circRNA)-miRNA pair that might modulate the migration of HCC cell lines based on previously reported GEO database (GSE78520 and GSE43445) and investigated the function and molecular mechanism. circRNA-101368 was predicted by lncTar to target miR-200a, and the expression of circRNA-101368 was significantly upregulated in HCC tissue samples; the overexpression of circRNA-101368 was correlated with poorer prognosis in patients with HCC. Moreover, circRNA-101368 knockdown suppressed the migration and the protein levels of HMGB1, RAGE and NF-κB, while increased the E-Cadherin expression in HCC cell lines. As confirmed by luciferase reporter and RNA immunoprecipitation assays, circRNA-101368 directly bound to miR-200a to negatively regulate each other. The effect of circRNA-101368 knockdown on cell migration and HMGB1/RAGE signaling could be partially attenuated by miR-200a inhibition. In tissue samples, miR-200a was negatively correlated with circRNA-101368 and HMGB1, respectively, whereas circRNA-101368 and HMGB1 was positively correlated. Taken together, we demonstrated a network of circRNAs-miRNA-mRNA in HCC and provided a novel mechanism of HCC cell migration regulation.
Keywords: Hepatocellular carcinoma (HCC), HMGB1, circRNA-101368
Introduction
Hepatocellular carcinoma (HCC), one of the most common cancers of digestive system, is considered one of the leading causes of cancer-related death globally [1], partially due to its high resistance to most standard therapies. Therefore, the discovery of changes in the genome of HCC may provide new strategies for the treatment of HCC.
The high-mobility group box 1 protein (HMGB1) is a chromatin-binding factor that targets DNA and facilitates the process of transcriptional protein assembly. By acting as a damage-associated molecular pattern, HMGB1 binds with high affinity to several receptors, such as the receptor for advanced glycation end products (RAGE) and Toll-like receptor- (TLR-) 2/4/9, mediating the immune response to necrosis and immune cell invasion to trauma, pathogens, and sepsis [2]. Additionally, overexpression of HMGB1 is associated with several cancer characteristics. Binding to RAGE, HMGB1 enhances cell migration and tumor metastasis, thereby promoting cancer development [3,4]. Previously, we have demonstrated the key role of long non-coding RNA (lncRNA) TP73-AS1/miR-200a axis through its downstream HMGB1/RAGE signaling [5]. Developing in depth understanding of HMGB1/RAGE signaling-associated molecular mechanism is crucial to targeted therapy for HCC.
In the past few years, it has been recognized that abnormal expression of non-coding RNAs (ncRNAs) frequently occurs in many types of cancers, suggesting the key roles of ncRNAs in human tumorigenesis [6]. As we have mentioned, a lncRNA-miRNA pair, namely TP73-AS1-miR-200a, modulated HCC cell proliferation [5]. Besides lncRNAs and miRNAs, circular RNAs (circRNAs), a newly-described series of ncRNAs generated by ‘backsplicing’ of protein-coding mRNAs or linear ncRNAs by joining an upstream 3ʹ splice site and downstream 5ʹ splice site to form a covalently closed continuous loop [7], has also been regarded as key players in HCC tumorigenesis. By using integrated microarray analysis, Lin et al. constructed a circRNA-miRNA-mRNA regulatory network in HCC and revealed the network might be involved in the procession of carcinogenesis of HCC. In addition, 3 top-ranked circRNAs related networks were identified to be highly correlated with the pathogenesis of HCC [8].
Here, we analyzed the expression profiling of circRNAs (GSE78520) and identified a total of 13 upregulated circRNAs obtained a fold-change of > 3.0. Of them, circRNAs 101368 was predicted by lncTar to target miR-200a, which was reported by GSE43445 to be downregulated in metastasis HCC samples. Next, the function and mechanism of circRNAs 101368 and miR-200a in HCC cell lines, as well as the predicted binding between circRNAs 101368 and miR-200a were validated. In summary, we revealed a circRNAs-miRNA-mRNA network that might modulate the migration of HCC cell lines.
Results
Selection of HCC-associated circRNA and miRNA
According to previous study, a total of 13,617 circRNAs have been reported to be abundant in the human liver cancer (GSE78520), of them 366 obtained a fold-change of > 1.5 and were shown in Figure 1(a); the red line indicated that 13 circRNAs obtained a fold-change > 3.0 (Figure 1(a), Table S1). Another microRNA profiling (GSE43445) in primary and metastatic HCC cell lines indicated that a total of 142 miRNAs (227 data with significant difference) were differentially expressed (P < 0.05, Figure 1(b)). Of 205 miRNAs predicted by miRWalk to target HMGB1, 9 were in the above 142 differentially expressed miRNAs (miR-200a, let-7g, miR-193b, let-7b, let-7a, miR-224, miR-532, miR-186 and miR-34a, Figure 1(c)). Previously, we have reported that miR-200a/HMGB1/RAGE axis was involved in HCC cell proliferation [5]; here, miR-200a was selected for further experiments. By using lncTar, circRNA 101368 was predicted to target miR-200a (data not shown). In a small sample size verification in 15 paired tumor (T) and non-cancerous (N) tissue samples, the expression of circRNA 101368 was significantly upregulated in tumor samples (Figure 1(d)). Thus, circRNA 101368 was selected for further experiments.
Figure 1.
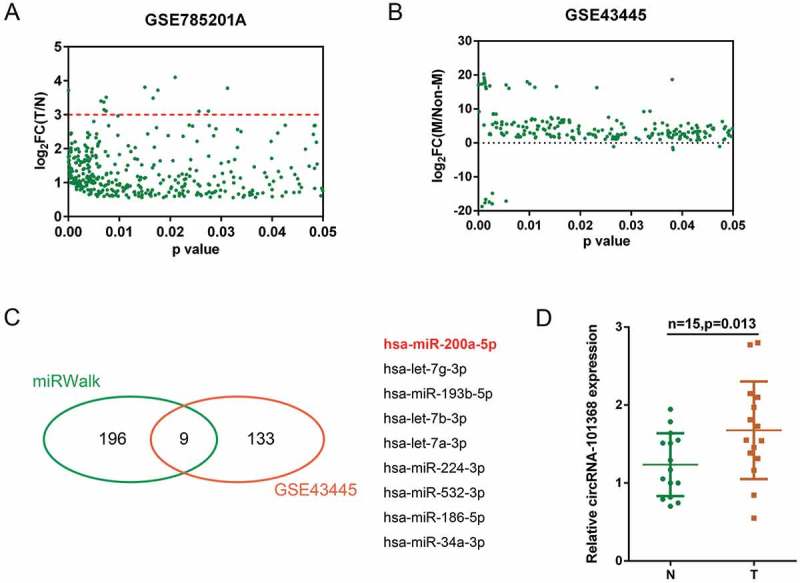
Selection of HCC-associated circRNA and miRNA (a) A total of 366 dysregulated circRNAs in HCC cell lines obtained a fold-change of > 1.5 were shown (GSE78520); the red line indicated that 13 circRNAs obtained a fold-change > 3.0. (b) A total of 142 miRNAs (227 data with significant difference) were differentially expressed in metastatic HCC cell lines compared to those in primary HCC cell lines (GSE43445). (c) Of 205 miRNAs predicted by miRWalk to target HMGB1, 9 were in the above 142 differentially expressed miRNAs (miR-200a, let-7g, miR-193b, let-7b, let-7a, miR-224, miR-532, miR-186 and miR-34a). (d) The expression of circRNA 101368 in tumor and non-cancerous tissue specimens was examined.
circRNA 101368 is overexpressed in HCC tissues and cell lines and is correlated with poorer prognosis
To verify the role of circRNA 101368 in HCC, its expression was examined in 51 paired HCC specimens and non-cancerous tissue samples. Consistent with the results in small sample size verification, circRNA 101368 expression was significantly upregulated in tumor tissues (Figure 2(a)). Moreover, circRNA 101368 expression was remarkably upregulated in specimens in advanced stages (stage III + IV, Figure 2(b)). The fold-change of circRNA 101368 expression was shown in Figure 2(c) as log2 (tumor/normal). Enrolled cases were then grouped according to circRNA 101368 expression: a high circRNA 101368 group (circRNA 101368 expression higher than the median value, n = 26) and a low circRNA 101368 group (circRNA 101368 expression lower than the median value, n = 25). Higher circRNA 101368 expression was correlated to distant metastasis (P = 0.048), advanced TNM stage (P = 0.017) and larger tumor size (P = 0.008) (Table 1). The correlation between circRNA 101368 expression and clinical features was then analyzed using COX risk proportional regression model, Kaplan-Meier analysis and log-rank test. Univariate analysis showed that TNM stage, tumor size and circRNA 101368 expression caused obvious difference in survival time; further multivariate analysis showed that circRNA 101368 expression might be a risk factor of overall survival in patients with HCC (Table 2). As shown in Figure 2(d), patients with higher circRNA 101368 expression obtained a significantly shorter overall survival compared to patients with lower circRNA 101368 expression (P = 0.0002).
Figure 2.
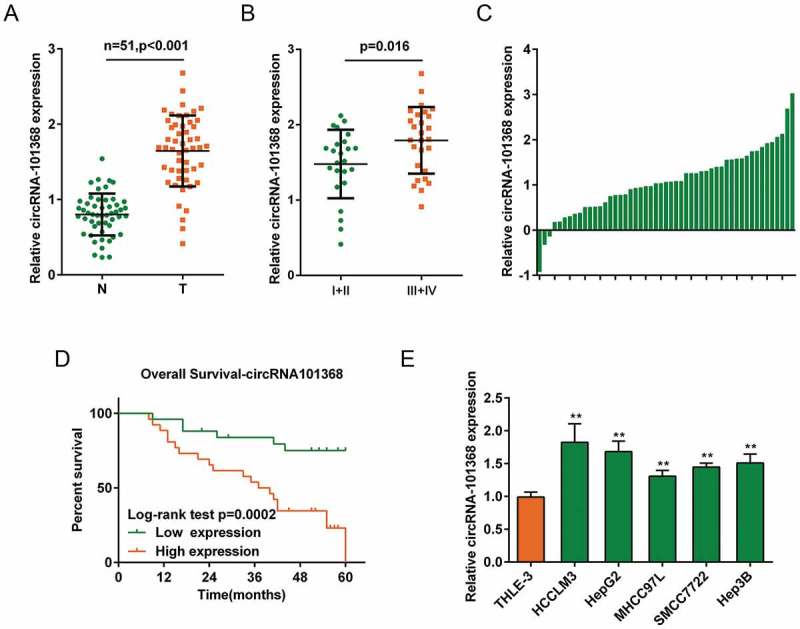
circRNA 101368 is overexpressed in HCC tissues and cell lines and is correlated with poorer prognosis (a) Expression of circRNA 101368 in 51 paired tumor and non-cancerous tissue specimens was examined using real-time PCR. (b) Expression of circRNA 101368 in 51 cases of HCC tissues was analyzed according to TNM stages. (c) The fold-change of circRNA 101368 expression in 51 cases of HCC tissues was shown as log2 (tumor/normal). (d) Kaplan-Meier overall survival curves for 51 cases grouped by circRNA 101368 expression. (e) circRNA 101368 expression in five HCC cell lines and a normal hepatocyte was examined.
Table 1.
Correlation of the expression of circRNA101368 with clinicopathologic features.
| Clinic-pathological parameters | circRNA101368 expression |
p-value | ||
|---|---|---|---|---|
| high | low | |||
| Gender | female | 7 | 7 | 0.931 |
| male | 19 | 18 | ||
| Age | <50 | 10 | 12 | 0.492 |
| ≥ 50 | 16 | 13 | ||
| Distant metastasis | Negative | 14 | 20 | 0.048 |
| Positive | 12 | 5 | ||
| TNM stage | Ⅰ+Ⅱ | 8 | 16 | 0.017 |
| Ⅲ+Ⅳ | 18 | 9 | ||
| Differentiation | Moderately – Well | 13 | 19 | 0.055 |
| Poorly | 13 | 6 | ||
| Tumor size | ≥ 5 | 19 | 9 | 0.008 |
| <5 | 7 | 16 | ||
Table 2.
Univariate and multivariate analysis for factors related to overall survival using the COX proportional hazard model.
| Univariate analysis |
Multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|
| Variable | p-value | HR | 95%CI | p-value | HR | 95%CI | |
| Gender | female vs male | 0.592 | 0.779 | 0.312–1.942 | |||
| Age(years) | ≥ 50 vs <50 | 0.286 | 0.643 | 0.286–1.445 | |||
| Distant metastasis | Negative vs Positive | 0.526 | 0.774 | 0.350–1.710 | |||
| TNM stage | Ⅰ+Ⅱ vs Ⅲ+Ⅳ | 0.079 | 0.483 | 0.214–1.088 | 0.248 | 0.596 | 0.248–1.432 |
| Differentiation | Moderately – Well vs Poorly | 0.941 | 0.969 | 0.430–2.188 | |||
| Tumor size | <5 vs ≥ 5 | 0.015 | 0.350 | 0.150–0.817 | 0.200 | 0.533 | 0.204–1.395 |
| circRNA101368 | High vs Low | 0.001 | 4.908 | 1.930–12.480 | 0.033 | 3.246 | 1.098–9.594 |
To investigate further molecular mechanism, circRNA 101368 expression in five human HCC cell lines, HCCLM3, MHCC97L, SMMC7722, Hep3B and HepG2, was examined. Consistent with results in tissue samples, circRNA 101368 expression was significantly upregulated in HCC cell lines, more upregulated in HCCLM3 and HepG2 cell lines (Figure 2(e)). Thus, HCCLM3 and HepG2 cell lines were selected as cell models for further experiments.
circRNA 101368 knockdown suppressed the cell migration and HMGB1/RAGE signaling in HCC cell lines
To examine the functions of circRNA 101368, HCCLM3 and HepG2 cell lines were transfected with si-circRNA 101368–1/2 to achieve circRNA 101368 knockdown, as confirmed using real-time PCR (Figure 3(a)). The migration of HCC cell lines could be significantly suppressed by both siRNAs, more suppressed by si-circRNA 101368–1 (Figure 3(b,c)); thus, si-circRNA 101368–1 was used as siRNA for circRNA 101368.
Figure 3.
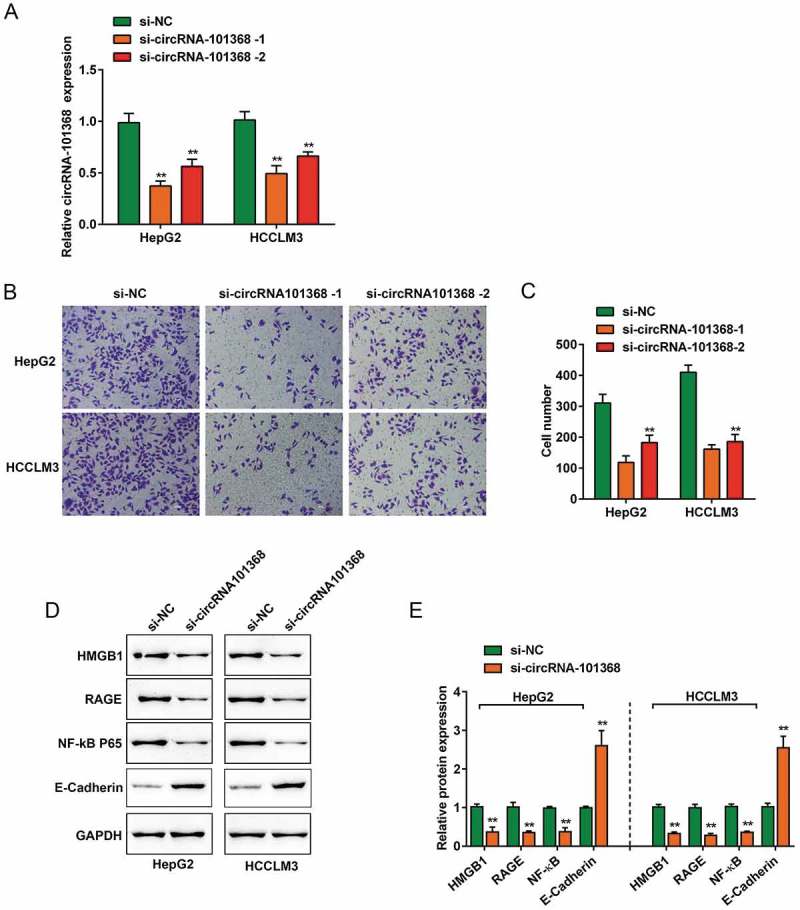
circRNA 101368 knockdown suppressed the cell migration and HMGB1/RAGE signaling in HCC cell lines (a) circRNA 101368 knockdown in HepG2 and HCCLM3 cell lines was achieved by transfection of si-circRNA 101368–1 or si-circRNA 101368–2, as confirmed using real-time PCR. (b-c) The cell migration of HepG2 and HCCLM3 cell lines transfected with si-circRNA 101368–1 or si-circRNA 101368–2 was examined using Transwell assays. (d-e) The protein levels of HMGB1, RAGE, NF-κB and E-Cadherin in HepG2 and HCCLM3 cell lines transfected with si-circRNA 101368 were examined using Immunoblotting assays.
Given the role of miR-200a/HMGB1/RAGE axis in HCC cell proliferation [5]; here, the effect of circRNA 101368 knockdown on the protein levels of HMGB1, RAGE, NF-κB (p65) and E-Cadherin was examined. In both two cell lines, circRNA 101368 knockdown increased E-Cadherin expression and reduced HMGB1, RAGE and NF-κB (p65) proteins levels. (Figure 3(d)).
miR-200a is a direct downstream target of circRNA 101368
Since lncTar predicted that circRNA 101368 may target miR-200a; here, the interaction between circRNA 101368 and miR-200a was validated. HCCLM3 and HepG2 cell lines were transfected with miR-200a mimics or miR-200a inhibitor to achieve miR-200a expression, as confirmed using real-time PCR (Figure 4(a)). In both HCC cell lines, the expression of circRNA 101368 was negatively regulated by miR-200a (Figure 4(b)). Similarly, circRNA 101368 knockdown significantly increased miR-200a expression, indicating that circRNA 101368 and miR-200a negatively regulated each other.
Figure 4.
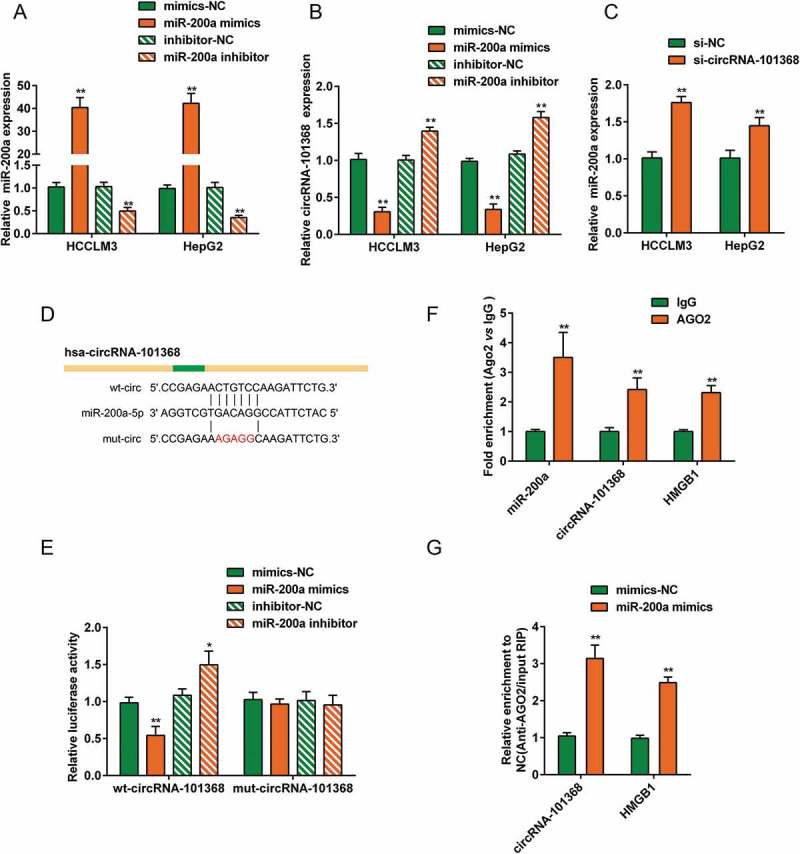
miR-200a is a direct downstream target of circRNA 101368 (a) HepG2 and HCCLM3 cell lines were transfected with miR-200a mimics or miR-200a inhibitor to achieve miR-200a expression, as confirmed using real-time PCR; (b) transfected HCC cell lines were examined for circRNA 101368 expression. (c) HepG2 and HCCLM3 cell lines were transfected with si-circRNA 101368 and examined for miR-200a expression. (d) The miR-200a binding site in circRNA 101368 predicted by lncTar was shown. A wild-type and a mutant-type circRNA 101368 luciferase reporter vector were constructed, containing wild-type or mutant-type miR-200a binding site, respectively. (e) HEK293 cells were co-transfected with the above luciferase reporter vectors and miR-200a mimics/inhibitor and examined for luciferase activity. (f-g) Association of circRNA 101368, miR-200a and HMGB1 with AGO2. Detection of AGO2 and IgG using Immunoblotting and detection of circRNA 101368, miR-200a and HMGB1 using real-time PCR.
Next, to further confirm the predicted binding between circRNA 101368 and miR-200a, luciferase reporter and RIP assays were employed. The predicted binding site was shown in Figure 4(d); a wild-type (wt-circ) and a mutant-type (mut-circ) circRNA 101368 luciferase reporter vector were constructed. After co-transfection with miR-200a mimics or miR-200a inhibitor, the luciferase activity of wt-circ was dramatically inhibited by miR-200a overexpression while enhanced by miR-200a inhibition; however, the alterations of the luciferase activity were abolished by the mutation in the predicted miR-200a binding site (Figure 4(e)). To further confirm the binding, RIP assays were performed with AGO2 antibody. RNA was extracted from the protein precipitate, and the levels of circRNA 101368, miR-200a and HMGB1 were more than 2-fold higher than IgG (Figure 4(f)). Moreover, upon miR-200a overexpression, the levels of circRNA 101368 and HMGB1 were significantly higher protein precipitate than those in mimics-NC group (Figure 4(g)). The above findings suggest that circRNA 101368 binds to miR-200a to regulate its expression in an AGO2-dependent manner.
The effect of circRNA 101368 on HCC cell migration could be partially reversed by miR-200a through HMGB1/RAGE signaling
After confirming the binding between circRNA 101368 and miR-200a, their dynamic effect on HCC cell migration and downstream HMGB1/RAGE signaling was evaluated. HCCLM3 and HepG2 cell lines were co-transfected with si-circRNA 101368 and miR-200a inhibitor, and then examined for cell migration and protein levels of HMGB1, RAGE, p65 and E-Cadherin. Consistently, circRNA 101368 knockdown significantly increased E-Cadherin expression and suppressed the cell migration, as well as the protein levels of HMGB1, RAGE and p65, whereas miR-200a inhibition exerted an opposing effect (Figure 5(a-d)); the effect of circRNA 101368 knockdown was partially attenuated by miR-200a inhibition (Figure5(a-d)), suggesting that circRNA 101368 exert its effect on HCC cell migration through miR-200a and downstream HMGB1/RAGE signaling.
Figure 5.
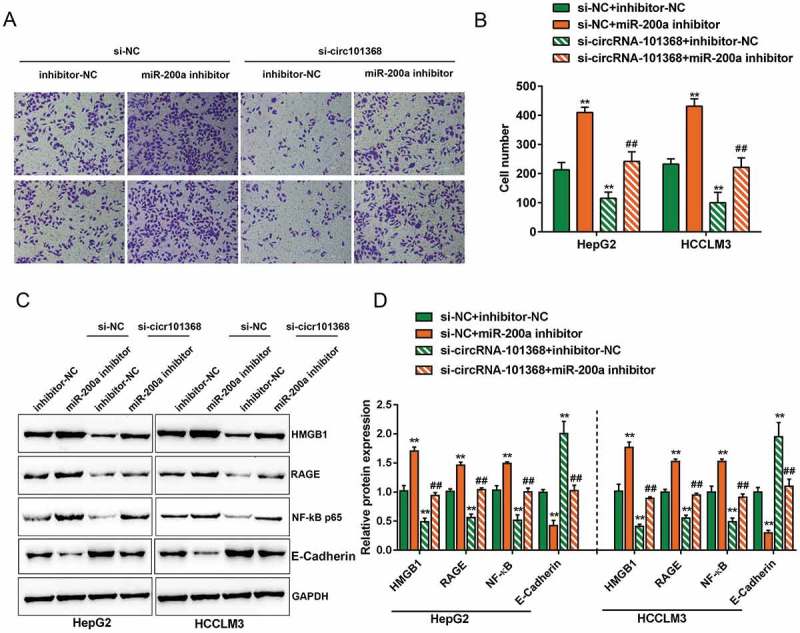
The effect of circRNA 101368 on HCC cell migration could be partially reversed by miR-200a through HMGB1/RAGE signaling HepG2 and HCCLM3 cell lines were co-transfected with si-circRNA 101368 and miR-200a inhibitor and examined for cell migration (a-b) and protein levels of HMGB1, RAGE, NF-κB (p65) and E-Cadherin (c-d) .
Expression of HMGB1 and miR-200a and their correlation in tissue specimens
As a further confirmation, the expression of HMGB1 and miR-200a in tumor and non-cancerous tissue specimens were examined. HMGB1 mRNA expression was significantly upregulated whereas miR-200a expression was downregulated in tumor tissue specimens (Figure 6(a,b)). In tissue specimens, miR-200a was negatively correlated with HMGB1 and circRNA 101368, respectively; HMGB1 was positively correlated with circRNA 101368 (Figure 6(c-e)).
Figure 6.
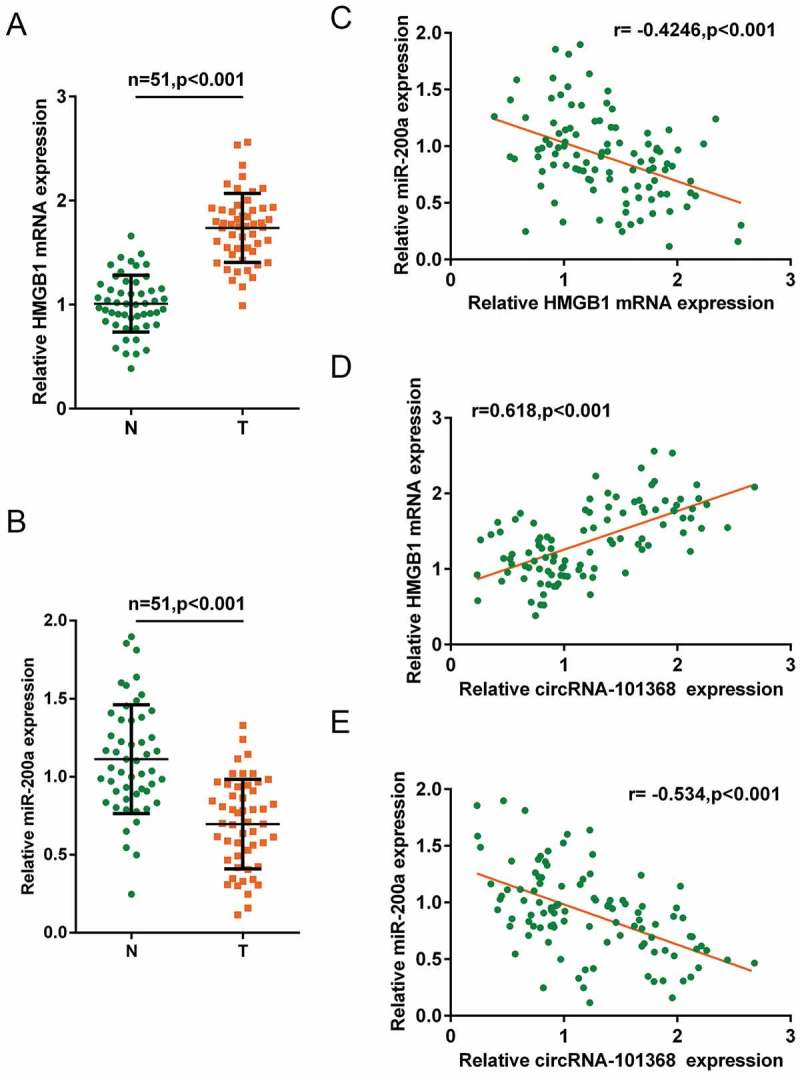
Expression of HMGB1 and miR-200a and their correlation in tissue specimens (a-b) Expression of HMGB1 and miR-200a in tissue specimens were examined using real-time PCR. (c-e) Correlation of circRNA 101368, HMGB1 and miR-200a was analyzed using Spearman’s rank correlation analysis.
Discussion
Here, we identified a circRNA-miRNA-mRNA network, namely circRNA 101368-miR-200a-HMGB1, that modulates the cell migration of HCC cell lines through HMGB1/RAGE signaling. HCC cell migration could be suppressed by circRNAs 101368 knockdown while promoted by miR-200a inhibition; the effect of circRNAs 101368 knockdown was partially attenuated by miR-200a inhibition. Moreover, miR-200a expression was negatively correlated with circRNAs 101368 or HMGB1, and circRNAs 101368 was positively correlated with HMGB1 in tissue specimens, suggesting that knocking circRNAs 101368 down may rescue miR-200a expression thus inhibiting HCC cell migration through HMGB1/RAGE signaling.
Accumulating evidence has revealed that circRNAs could act as miRNA sponges to counteract miRNA mediated repression of mRNA [9]. Lastly, the regulatory axes of circRNAs, miRNAs, and mRNAs are gradually demonstrated in different kinds of diseases [10–12], including HCC [6,8]. According to previous studies, a huge number of ncRNAs, including circRNAs and miRNAs, are dysregulated in HCC [12–14]. In the present study, we downloaded two microarray analysis results (GSE78520 and GSE43445) and found that a total of 13 circRNAs upregulated in HCC tumor tissues obtained a fold-change > 3.0, and a total of 142 miRNAs were differentially expressed in HCC tissues with metastasis. Of the 142 miRNAs, 9 were predicted to target HMGB1, a chromatin-binding factor that has been reported to contribute to HCC progression [15–17]. In our previous study, we demonstrated the key role of miR-200a/HMGB1 axis in HCC migration [5]. Since we found that miR-200a expression was downregulated in HCC tissues [5]; herein, we used lncTar to investigate the potential upstream regulatory circRNAs of miR-200a. Interestingly, cicrRNA_101368, one of the above upregulated circRNAs in HCC, was predicted to target miR-200a. Consistent with previous study, the expression of cicrRNA_101368 was significantly upregulated in HCC tissues, particularly in specimens with metastasis. Moreover, cicrRNA_101368 overexpression was correlated with poorer prognosis in patients with HCC.
Regarding the molecular mechanism, circRNAs may modulate the cell behavior of HCC cell lines through targeting different miRNAs and their downstream signaling. Through miR-4641/PCK1 pathway, circC3P1 can suppress the growth and metastasis of HCC [18]. Another circRNAs, namely circ_0067934, can promote the tumor growth and metastasis in HCC through modulating miR-1324/FZD5/Wnt/beta-catenin axis [19]. In the present study, consistent with its high expression in HCC tissue samples, cicrRNA_101368 knockdown significantly suppressed the cell migration of HCC cell lines. Moreover, as predicted by lncTar, cicrRNA_101368 could directly bind to miR-200a in an AGO2-dependent manner; therefore, cicrRNA_101368 and miR-200a negatively regulated each other. In HCC cell lines, miR-200a inhibition exerted an opposing effect on cell migration and downstream HMGB1/RAGE signaling to cicrRNA_101368 knockdown; the effect of cicrRNA_101368 knockdown could be partially attenuated by miR-200a, indicating that cicrRNA_101368 serves as a sponge of miR-200a to counteract miR-200a-mediated HMGB1 repression, therefore modulating HCC migration.
As a further confirmation of these data, miR-200a expression was significantly downregulated, whereas HMGB1 mRNA expression was upregulated in HCC tissue samples. MiR-200a was negatively correlated with cicrRNA_101368 and HMGB1, respectively; cicrRNA_101368 was positively correlated with HMGB1. Taken together, cicrRNA_101368 acts as a sponge of miR-200a to suppress miR-200a expression, thereby promoting HCC cell migration through miR-200a downstream HMGB1/RAGE signaling. We provide experimental and theoretical basis for targeted therapy for HCC from the perspective of circRNAs-miRNA-mRNA network.
Materials and methods
Cell lines, cell culture and cell transfection
Human HCC cell line HCCLM3 was obtained from cell bank of Zhongshan Hospital (Shanghai, China) and cultured in Dulbecco’s modified eagle’s medium (DMEM) (GIBCO, Life Technologies, Grand Island, NY, USA) with high glucose, containing 10% foetal calf serum (FCS), penicillin (50 IU/mL) and streptomycin (50 µg/mL). MHCC97L was obtained from The China Center for Type Culture Collection (Wuhan, China) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS; GIBCO), 1% l-glutamine, 1% penicillin-streptomycin, and 1% nonessential amino acids. SMMC7721 was purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM supplemented with 10% FBS, 1% l-glutamine, 1% penicillin-streptomycin, and 1% nonessential amino acids. Hep3B and HepG2 were purchased from American Type Culture Collection (ATCC) and cultured in Eagle’s Minimum Essential Medium (ATCC) supplemented with 10% FBS. A normal hepatocyte, THLE-3, was obtained from ATCC and cultured in DMEM supplemented with 10% FBS. Cell lines used in the present study were cultured at 37°C with 5% CO2.
Expression of miRNA was achieved by transfection of miRNA mimics or inhibitor (Genepharma Company, Shanghai, China); RNA interfering was achieved by transfection of siRNA (Genepharma Company, Shanghai, China). All transfection was performed with the help of Lipofectamine™ 2000 transfection reagent (Invitrogen, USA) following the protocols. Cells were harvested and used 48 h after transfection.
Tissue specimens
Fifty-one paired HCC specimens and matched non-cancerous tissue specimens were collected from tumor surgical resection in the Xiangya Hospital (the Central South University, Changsha, China) with the approval of the Medical Ethics Committee of Xiangya hospital at Central South University. Informed consent was obtained from all patients enrolled.
RNA extraction and real-time PCR
Total RNA was isolated from target cells or tissue samples using TRIZOL Reagent (Invitrogen, USA) following the protocols. Thereafter, RNA samples were reversely transcribed by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). The quantitative RT-PCR was performed using the Fast Start Universal SYBR Green Master (Roche, USA). The relative fold-changes were analyzed using 2−ΔΔCT method. The levels of GAPDH were used as endogenous gene for circRNA and mRNA. The levels of U6 were used as endogenous gene for miRNA.
Immunoblotting assays
Cells were lysed by RIPA buffer (Sigma-Aldrich, USA) with Complete Protease Inhibitor Cocktail (Roche, USA) and kept at −20°C before use. Proteins separation was conducted using SDS-PAGE with 5% stacking gel and 10% running gel. Proteins were then transferred onto PVDF membrane and probed with the following antibodies: anti-HMGB1 antibody (ab18256, Abcam, Cambridge, MA, USA), anti-RAGE antibody (ab3611, Abcam), anti-NF-κB antibody (ab16502, Abcam), anti-E-Cadherin antibody (ab1416, Abcam) and anti-GAPDH antibody (ab8245, Abcam). The blots were visualized using ECL Substrates (Millipore, USA). Image J software (National Institutes of Health, Bethesda, MD, USA) was employed to perform the gradation analysis.
Transwell migration
5 × 104 target cells were plant into the 24-well plate inserted with 8-μm pores (Becton Dickinson, Germany) where they can migrate towards the conditioned medium. Remove non-migratory cells with cotton swabs. Migratory cells on the lower part of the membrane were fixed with 5 % glutaraldehyde in phosphate-buffered saline (PBS) for 15 min and stained with methylene blue. Five non-overlapping fields per insert were counted manually under Olympus BX53 (Olympus Corporation, Tokyo, Japan).
Luciferase reporter assay
The fragment of circRNA 101368 was cloned to the downstream of the Renilla psiCHECK2 vector (Promega, Madison, WI, USA). With the help of the Lipofectamine™ 2000 transfection reagent, HEK293 cells were co-transfected with the above described vectors (wt-circ, mut-circ containing wild-type or mutant-type miR-200a binding site) and miRNA mimics or inhibitor. Forty-eight hours post-transfection, luciferase activity was examined using the Dual Luciferase Reporter Assay System (Promega). Renilla luciferase activity was normalized to Firefly luciferase activity for each transfected well.
RNA immunoprecipitation (RIP)
RNA immunoprecipitation was performed with AGO2 antibody using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (17–700, Millipore) following the protocols. RNA for in vitro experiments was transcribed using a T7 High YieldRNA Synthesis Kit (E2040S, NEB) following the protocols. HMGB1, circRNA 101368 and miR-200a levels in the immunoprecipitates were measured by qRT-PCR.
Statistics
Experimental results from at least three parallel experiments are presented as Mean ± SD. Differences between two groups were analyzed using two-tail Student’s T-test, and differences among multiple groups were evaluated by one-way ANOVA. Differences were considered statistically significant at P < 0.05.
Funding Statement
This work was supported by the National Natural Science Foundation of China [81108129].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Khemlina G, Ikeda S, Kurzrock R.. The biology of hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lotze MT, Tracey KJ.. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. [DOI] [PubMed] [Google Scholar]
- [3].Tang D, Kang R, Zeh HJ 3rd, et al. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ying S, Xiao X, Chen T, et al. PPAR ligands function as suppressors that target biological actions of HMGB1. PPAR Res. 2016;2016:2612743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li S, Huang Y, Huang Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137–151. [DOI] [PubMed] [Google Scholar]
- [7].Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lin X, Chen Y. Identification of potentially functional circRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. 2018;24:70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [10].Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wei X, Li H, Yang J, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8:e3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen Y, Yuan B, Wu Z, et al. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. 2017;629:35–42. [DOI] [PubMed] [Google Scholar]
- [13].Cui S, Qian Z, Chen Y, et al. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol Lett. 2018;15:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fu L, Yao T, Chen Q, et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yan W, Chang Y, Liang X, et al. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55:1863–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen M, Liu Y, Varley P, et al. High-mobility group box 1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res. 2015;75:1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li H, Huang W, Luo R. The microRNA-325 inhibits hepatocellular carcinoma progression by targeting high mobility group box 1. Diagn Pathol. 2015;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [18].Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499:1044–1049. [DOI] [PubMed] [Google Scholar]
- [19].Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497:626–632. [DOI] [PubMed] [Google Scholar]


