Abstract
Alopecia areata (AA) is an autoimmune disease in which cytotoxic T cells specifically target growing hair follicles. We used high-throughput TCR sequencing in the C3H/HeJ mouse model of AA and in human AA patients to gain insight into pathogenic T cell populations and their dynamics, which revealed clonal CD8+ T cell expansions in lesional skin. In the C3H/HeJ model, we observed interindividual sharing of TCRβ chain protein sequences, which strongly supports a model of antigenic drive in AA. The overlap between the lesional TCR repertoire and a population of CD8+NKG2D+ T cells in skin-draining lymph nodes identified this subset as pathogenic effectors. In AA patients, treatment with the oral JAK inhibitor tofacitinib resulted in a decrease in clonally expanded CD8+ T cells in the scalp but also revealed that many expanded lesional T cell clones do not completely disappear from either skin or blood during treatment with tofacitinib, which may explain in part the relapse of disease after stopping treatment.
Keywords: Autoimmunity, Dermatology
Keywords: Adaptive immunity, T-cell receptor
Analysis of the T cell repertoire in the autoimmune disease alopecia areata provides insights in antigenic drive and the response of expanded T cell clones to JAK inhibitor treatment.
Introduction
Alopecia areata (AA) is one of the most common autoimmune diseases, yet the pathogenesis underlying AA is incompletely understood. T cells are considered the main pathogenic cells underlying this nonscarring form of hair loss, supported by their infiltration into the hair follicle microenvironment during the course of disease (reviewed in ref. 1). In addition, adoptive transfer studies have demonstrated the ability of T cells to precipitate AA, both in the C3H/HeJ mouse model and in SCID mice grafted with human scalp (2–4).
The normal anagen hair follicle expresses little or no MHC class Ia and no MHC class II molecules and is considered to be a site of relative local immune privilege (5, 6). We and others have shown that, in active AA lesions, the hair follicle loses its immune privilege and expresses antigen-presenting molecules, rendering it vulnerable to MHC-restricted T cells (7, 8). Further, NKG2D ligands (e.g., ULBP3 and MICA in humans; Rae-1 and H60 in mice) are upregulated in the hair follicle during active disease, where these ligands provide a stimulatory signal to NKG2D-expressing T cells and NK cells (8, 9).
Recently, using functional adoptive transfer models, we identified the predominant effector T cell population in the C3H/HeJ mouse model of AA, specifically, lymph node– and skin-infiltrating CD8+NKG2D+ T cells (8). This was consistent with our human GWAS studies, in which we identified a strong association between polymorphisms in the ULBP family of genes encoding NKG2D ligands (ULBP3/6) and AA (9, 10).
Despite these recent insights in the phenotype of pathogenic effector T cells in AA, the question remains as to whether these T cells are driven by specific antigen recognition. Unlike other genetically related autoimmune diseases, including autoimmune diabetes, thyroiditis, and rheumatoid arthritis, in which autoantigens and T cell epitopes have been described (11–14), in AA, the exact role of putative hair follicle–derived antigen recognition in the disease process is unknown. Although potential hair follicle autoantigens have been proposed in AA (15–21), it is not clear if the disease process is dependent on recognition of these (or other) antigens by infiltrating T cells.
Advances in high-throughput sequencing of T cell receptors) have transformed our ability to define the entire TCR repertoire in AA and provide a powerful technology to track T cell clones quantitatively on both a global and clonal basis. These novel methods have enabled the assessment of the T cell receptor diversity of any given T cell population (e.g., skin resident, total, or T cell subsets in blood) and have allowed the identification and quantification of unique T cell clones among heterogeneous populations. Since mature T cells only express one TCR β chain protein due to allelic exclusion, TCR β chain sequences provide a molecular fingerprint of the repertoire T cell clones (22).
Importantly for AA, TCR sequencing allows us to study the dynamics of the TCR repertoire without a priori knowledge of the antigen(s) that are recognized. The analysis of TCR diversity and dynamics of the effector T cell populations in AA invited us to ask whether AA is an antigen-driven process and to monitor pathogenic T cell populations in relation to the onset of hair loss. In this study, we used next-generation sequencing of the TCRβ repertoire in the C3H/HeJ mouse model of AA and in human AA patients to interrogate TCR dynamics during the development of AA and in response to treatment with the pan-JAK inhibitor tofacitinib. Our studies illustrate the utility of high-throughput TCR-sequencing data as an investigative and predictive tool in autoimmunity.
Results
Expanded effector T cell clones in alopecic skin overlap with the CD8+NKG2D+ T cell population.
The C3H/HeJ mouse model of AA closely resembles human AA, insofar that the disease is T cell dependent, reversible, and shows an upregulation of IFN-γ–related genes in affected skin (8, 23, 24). Due to the low frequency with which C3H/HeJ mice spontaneously develop disease (15% of females) and the protracted time course in which hair loss is achieved (9–12 months), we utilized the well-established skin graft–induced C3H/HeJ model of AA (23). In graft-induced C3H/HeJ mice, skin grafts from spontaneously affected C3H/HeJ mice are transplanted onto the backs of unaffected C3H/HeJ mice, resulting in nearly 100% of recipient mice developing disease within an accelerated time frame of approximately 6–8 weeks (23).
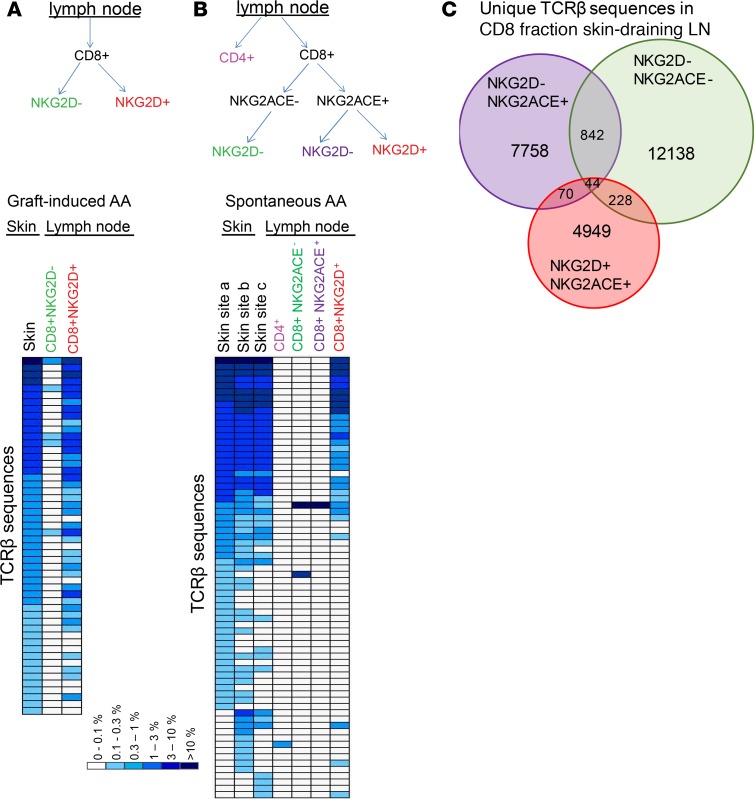
Our prior studies using adoptive T cell transfer of skin-draining lymph node cells showed that CD8+NKG2D+ T cells were both necessary and sufficient for T cell–mediated transfer of AA (8). To determine if the skin-infiltrating pathogenic T cell clones shared specificities with the CD8+NKG2D+ T cell subset, we first sorted CD8+ T cells from skin-draining lymph nodes from a mouse with graft-induced AA into NKG2D– and NKG2D+ subsets and compared the TCRβ repertoire from these subsets to the TCRβ repertoire from affected skin. Sequencing results showed that the most abundant clones in lesional skin (>0.1% of reads, 52 clones), overlapped almost completely with those in the CD8+NKG2D+ fraction, whereas only a few sequences overlapped with the CD8+NKG2D– T cell fraction (Figure 1A), supporting the notion that CD8+NKG2D+ T cells contain the pathogenic effector cells in AA.
Figure 1. Expanded clones in affected skin are predominantly NKG2D+CD8+ T cells.
Skin-draining lymph node cells from a C3H/HeJ mouse with graft-induced AA were sorted by flow cytometry into CD8+NKG2D– and CD8+NKG2D+ fractions. TCRβ chains of the sorted CD8+ T cell fractions as well as from the affected skin were sequenced by high-throughput sequencing. TCRβ sequences that represented >0.1% of the sequences in the affected skin and the frequencies of these sequences in the lymph node subsets are depicted as heatmaps (A). Skin-draining lymph node cells from a C3H/HeJ mouse with spontaneous AA were sorted into total CD4+ T cells, CD8+NKG2ACE–NKG2D–, CD8+NKG2ACE+NKG2D–, and CD8+NKG2ACE+NKG2D+. TCRβ chains of the sorted T cell fractions as well as 3 lesional skin sites (a, b, c, nonadjacent) were sequenced by high-throughput sequencing. TCRβ sequences that represented >0.1% of the sequences in the affected skin sites and the frequencies of these sequences in the lymph node subsets are depicted as heatmaps (B). Overlap between TCRβ sequences from the CD8+ T cell populations from skin-draining lymph nodes are depicted in a Venn diagram (C).
We next sorted T cells from skin-draining lymph nodes from a mouse with spontaneous AA to determine if spontaneously occurring AA would show similar features. We obtained affected skin samples from 3 nonadjacent sites and sorted skin-draining lymph node T cells into 4 subsets, 1 total CD4+ T cell population and 3 CD8+ T cell populations, based on their expression of the NKG2A, NKG2C, NKG2D, and NKG2E NK receptors (8). TCRβ sequencing showed that (a) nonadjacent lesional skin sites harbor the same expanded T cell clones and (b) these expanded clones correspond to the population of lymph node–derived CD8+NKG2D+ T cells, since 26 of 29 sequences that overlapped between skin and draining lymph nodes (>0.1% of reads) were uniquely detected in the this CD8+ subset (Figure 1B). These results complement our previous finding that CD8+NKG2D+ T cell clones represent the abundant infiltrating effector T cell population in AA skin (8) by additionally showing that these infiltrating effectors are oligoclonal and contain markedly expanded T cell clones.
TCRβ repertoire analysis of LN CD8+ fractions that were sorted based on NK cell receptors further demonstrated that there was very limited overlap between the NKG2D– and NKG2D+ CD8+ T cell populations (Figure 1C), supporting the notion that NKG2D is not merely an activation marker on CD8+ T cells in AA, but rather defines a separate CD8 T cell repertoire with distinct TCRs and specificities. Further, this analysis demonstrates that cross-referencing TCR repertoires between affected tissue and circulating T cell subsets is an effective method to identify the T cell subset(s) in which pathogenic T cells reside.
The majority of expanded T cell clones in recipient lesions are newly primed T cells.
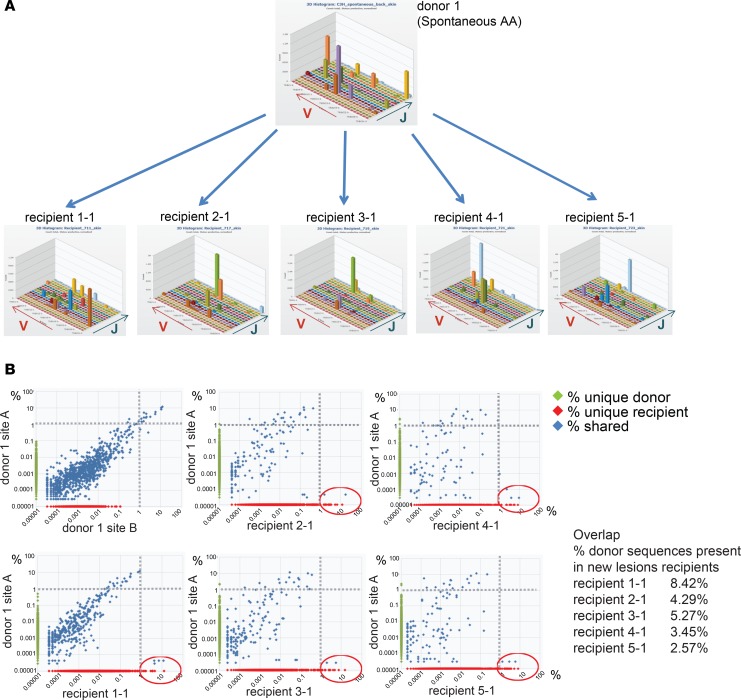
In the C3H/HeJ mouse model of AA, the hair loss initially appears on the ventral abdomen and thoracic regions, distant from the graft site on the dorsal back. The mechanism by which the graft induces hair loss at these distant skin sites (rather than spreading by extension into neighboring skin) remains unknown. There are at least two nonmutually exclusive scenarios by which this may occur, and high-throughput TCR sequencing enables the distinction between the two models. In the first scenario, pathogenic donor T cells present in the grafted skin could drain to the lymph nodes, enter the circulation, and home to unaffected skin where they induce hair loss. In the second scenario, newly primed recipient T cells are the main pathogenic effectors, which may be primed by either donor or recipient antigen-presenting cells. In this model, damaged hair follicles in the skin graft could provide a source of autoantigens and danger signals. In the first scenario, we predicted that T cell clones from the donor skin graft would expand in the new lesions in the recipient, whereas in the second scenario, we expected that the expanded T cell clones in the new lesions would be unique to the recipient (i.e., not detected in the donor skin graft). To determine which of the two scenarios occurs, we performed high-throughput TCRβ sequencing on affected skin from a donor mouse with spontaneous alopecia (2 nonadjacent sites) and on newly developing alopecic lesions from 5 recipient mice that had been grafted with the same affected donor’s skin.
Three-dimensional histograms of the TCR variable and joining region usage suggested marked clonal expansions in affected skin of both donor and recipients (Figure 2A). Plots comparing the TCR CDR3β nucleotide sequences between lesional donor skin and individual new recipient lesions revealed that a fraction of donor TCRs was detected (at lower frequencies) in recipient skin (Figure 2B). However, the high-frequency clones in the recipient lesions did not overlap significantly with those in the donor skin. Rather, the majority of the high-frequency sequences were unique to the recipient (Figure 2B). Overall, between 2.5% and 8.4% of the TCR sequences in the new lesions of recipients were also detected in the donor skin. Taken together, our TCR-sequencing data is most consistent with the second model, in which the induction of alopecia through skin grafting involves a process in which a new T cell response originates primarily from endogenous (recipient) nontransferred T cells. It is important to note, however, that a role for donor T cells transferred through the skin graft is not excluded in the initiation of this process.
Figure 2. The majority of expanded T cell clones in recipient lesions are newly primed T cells.
Skin from a donor with spontaneous AA (donor 1) was grafted onto 5 recipient mice, which subsequently developed AA. The TCRβ repertoire of donor skin and new lesions in recipients was sequenced and plotted as a 3D histogram of variable versus joining region genes versus number of reads (counts) (A). The percentages of all detected TCRβ sequences in donor and recipient skin samples were plotted, showing sequences shared between donor and recipient (percentage shared [blue]), sequences unique to donor (percentage on y axis [green]), and sequences unique to recipient (percentage on x axis [red]). The most expanded clones from the donor skin are detected at lower frequencies in recipient lesions (top left quadrant), and the most expanded clones in the recipient lesions are primarily unique to the recipient (bottom right quadrant, red circle) (B). The percentage of donor sequences present in new recipient lesions is indicated (Overlap).
T cell clonal expansion coincides with the onset of hair loss.
Although several publications have suggested an antigen-driven process in AA (15–17, 19), the role of antigen recognition in the process of hair follicle destruction by T cells has remained undefined. High-throughput TCR sequencing enabled us to investigate this question, since both an increase in clonally expanded T cells specifically coinciding with the onset of hair loss and shared TCR sequence CDR3β regions between affected mice would support the notion of an antigen-driven component of the disease.
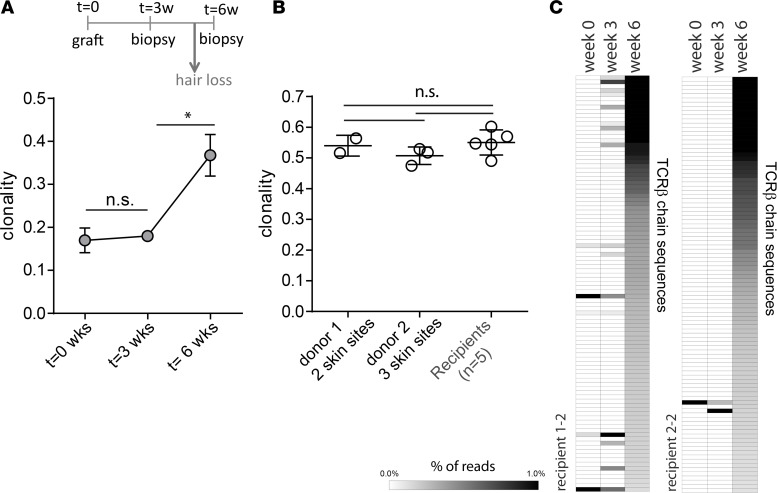
To determine the kinetics of clonal expansion, we analyzed the TCRβ repertoire of the skin of 2 recipient mice at baseline (t = 0) and 3 and 6 weeks after grafting (Figure 3A). For each sample, we determined the overall clonality, which is an inverse measure of T cell repertoire diversity, with 0 representing a diverse repertoire (lowest clonality) and 1 representing a clonal repertoire (highest clonality). The results showed that the clonality was lowest in the recipients at time points 0 and 3 weeks, when the mice do not yet display hair loss. However, at 6 weeks there was a sharp increase in clonality, coincident with the time point at which the mice begin to exhibit loss of hair. Lesional skin samples from mice with longstanding alopecia showed similar levels of clonality as those with early-stage disease (8–10 weeks) (Figure 3B), depicted in a separate set of lesional skin samples from 2 donor mice with longstanding alopecia (2 and 3 skin sites, respectively, per mouse) and 5 early-stage skin graft recipients (1 skin site each).
Figure 3. T cell clonal expansions coincide with hair loss.
Skin biopsies were taken from C3H/HeJ recipient mice at time of skin grafting t = 0 and 3 and 6 weeks after grafting, and the TCRβ chains were sequenced by high-throughput sequencing. The clonality (defined by 1 minus the normalized entropy) is plotted for recipient (n = 2) skin at the 3 different time points. *P < 0.05, 2-tailed Student’s t test (A). Clonality of affected skin samples from 2 donors with longstanding alopecia and from affected skin samples from 5 recipients with recent-onset, graft-induced alopecia. Statistical analysis was performed with 1-way ANOVA (B). The frequencies of the 100 most dominant TCRβ sequences in affected skin from 2 recipient mice at week 6 were determined at week 0 and 3. The frequencies are depicted as heatmaps (C).
The sudden increase in clonality between week 3 and 6 after grafting is likely the result of expanded pathogenic T cell clones infiltrating the skin just prior to disease onset. Analysis of the dominant TCRβ sequences in the recipients at 6 weeks after grafting showed that the majority of expanded T cell clones (top 100) in the skin at week 6 were not present at week 0 or 3, although, in recipient 1, several clones started to appear at week 3 (Figure 3C) This is consistent with the notion that expanded pathogenic T cell clones enter the skin between week 3 and 6 and that the process of hair loss coincides with an influx of expanded T cell clones that differ from the repertoire in unaffected skin. Of note, in affected animals with longstanding alopecia, the TCR repertoire was the largely similar throughout the affected skin, as evidenced by the presence of the same expanded clones in nonadjacent skin sites (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121949DS1). Overall, the appearance of expanded T cell clones at affected skin sites around the time of hair loss supports a role for an antigen-driven process in the development of disease.
Identical and near-identical TCRβ amino acid sequences in AA skin.
The CDR1, CDR2, and CDR3 regions of the TCR α and β chains interact with the composite surface of MHC-antigen complexes. The highly variable CDR3 region, which is generated through VDJ recombination, is the most important contributor to this interaction, since it primarily interacts with the peptide antigen (reviewed in ref. 25). Identical or highly similar CDR3α or CDR3β chains are occasionally observed between individuals who have been exposed to the same antigen and who share HLA-allele(s) (26). If AA is indeed an antigen-driven disease, we would expect to observe identical or similar TCRs in affected skin of individual C3H/HeJ mice.
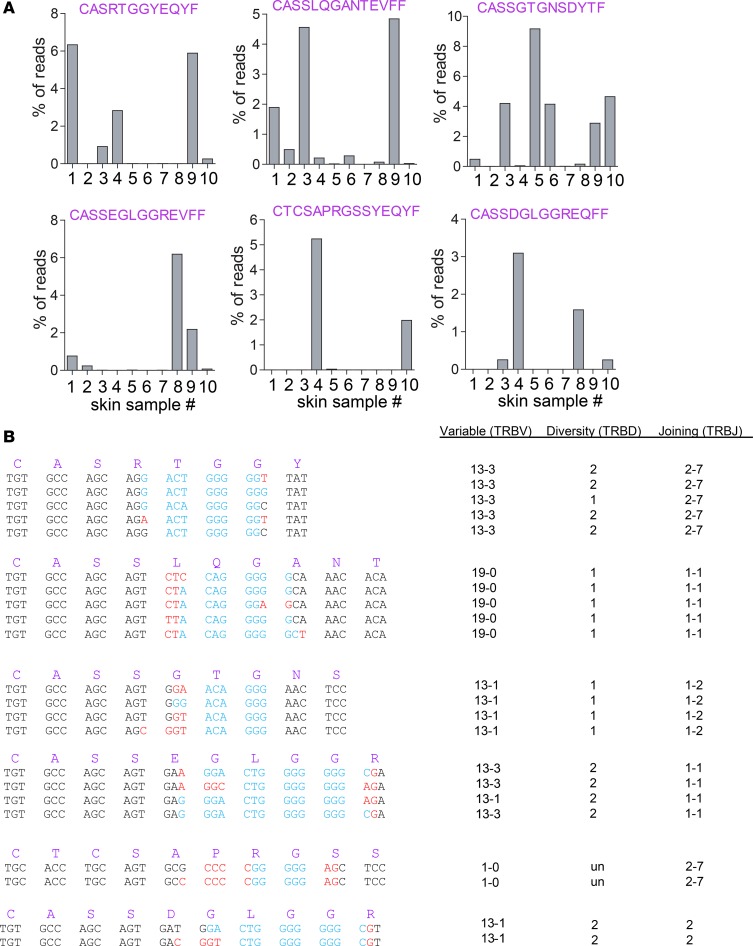
In our initial analysis of alopecic skin of C3H/HeJ mice, we found that relatively few identical nucleotide sequences were shared among individual mice. However, since distinct CDR3 nucleotide sequences (generated by distinct VDJ recombinations) can encode identical CDR3 amino acid sequences, we next determined the shared CDR3β amino acid sequences that were present among expanded clones in affected skin of individual animals (sequences that were present >0.05% in unaffected skin were excluded). Strikingly, this analysis showed that several TCRβ amino acid sequences (variable, joining, and CDR3β) were among the top 10 sequences in 2 or more individual samples (Figure 4A), suggesting that these TCR sequences play a dominant role in the recognition of specific antigens in affected skin. Despite the identical CDR3β amino acid sequences, it is important to note that these TCRs were encoded by several distinct VDJ recombinations, even within individual skin samples (Figure 4B), illustrating that the presence of these TCR chains in multiple animals is not the result of preferential formation in the thymus but rather through antigenic selection.
Figure 4. Identical CDR3β protein sequences detected in alopecic skin of individual mice.
Six different CDR3β protein sequences that were among the top 10 clones in 2 or more independent lesional skin samples (n = 10) are depicted as well as the frequencies (percentage of total reads) with which these CDR3β protein sequences are detected in the individual skin samples (A). These CDR3β protein sequences were encoded by multiple unique VDJ gene recombinations. Depicted are the different VDJ region nucleotide sequences, which encode the given CDR3β sequences (B). Black, germline encoded V- and J-regions; blue, D-region; red, N-region additions.
Among the TCR variable region usage of the most dominant shared TCRβ sequences (TRBV13-1, TRBV13-3, TRBV19-0, TRBV1-0), 3 variable regions were frequently found in lesional T cell clones (Supplemental Figure 2A), suggesting that other T cell clones harboring these variable chains may be involved in disease pathogenesis. Variable regions that were not detected in the most dominant shared lesional T cell clones did not appear to be overrepresented in lesional clones (Supplemental Figure 2A). Consistent with our finding that lesional skin T cell clones more frequently use the TRBV13-1 and TRBV13-3 variable genes, we observed that these variable regions were also more frequently used by lymph node–derived NKG2D+CD8+ T cells, which we previously identified as pathogenic effectors in the C3H/HeJ model of AA (8), than NKG2D-CD8+ T cells (data not shown).
In addition to the detection of identical CDR3β regions among the dominant T cell clones, several of these CDR3β sequences were nearly homologous to those of other dominant T cell clones (Supplemental Figure 2B). The most striking example was a highly prevalent TCRβ chain that had at least 6 variations, differing in 1 to 3 amino acids (TRB13-3 TRBJ2-7 CASSDGLGGRDYTF, CASSDGLGGREQFF, CASSDGLGGREVFF, CASSEGLGGRDYTF, CASSEGLGGREQFF, CASSEGLGGREVFF). One or more of the variations was found in all affected skin samples (both spontaneous and graft induced). It is important to note that the nongermline-encoded amino acids that differed between the variants were glutamic acid (E) and aspartic acid (D), which are the only 2 amino acids with negatively charged side chains, and their shared physicochemical properties make it likely that these 6 variants have the same or similar specificity. The detection of independently generated identical and near-identical CDR3β sequences among the most abundant clones in affected AA skin indicates convergent evolution of distinct T cell clones expressing structurally similar antigen-binding TCR structures in each individual AA mouse. Together, these findings argue strongly for the presence of dominant autoantigenic epitopes that drive disease pathogenesis and provide a lead for the identification of pathogenic T cell clones and their cognate AA autoantigens.
Human AA is characterized by an increase in clonally expanded CD8+ T cells.
In human AA, there is a wide range of clinical presentations, from a single patch of hair loss on the scalp (AA patchy [AAP]), to the loss of all scalp hair (alopecia totalis [AT]), or loss of total body hair (alopecia universalis [AU]). It is unclear if AA in humans is antigen driven, and it is not known what factors determine the course of disease. To gain insight in human AA, we performed TCRβ sequencing of scalp tissue as well as sorted CD4+ and CD8+ peripheral blood samples from a panel of AAP and AT/AU patients and healthy controls. Analysis of the scalp TCR repertoire in human disease is complicated by the fact that normal human skin contains a substantial number of T cells (27), the majority of which are αβ T cells, which will also be amplified and sequenced in the TCRβ chain sequencing. This in contrast to mouse skin, in which the majority of resident T cells are invariant γδ T cells, whose TCRs are not amplified in the high-throughput sequencing. Despite the presence of resident αβ T cells in normal scalp skin, we successfully identified disease-associated changes in the human AA scalp TCR repertoire.
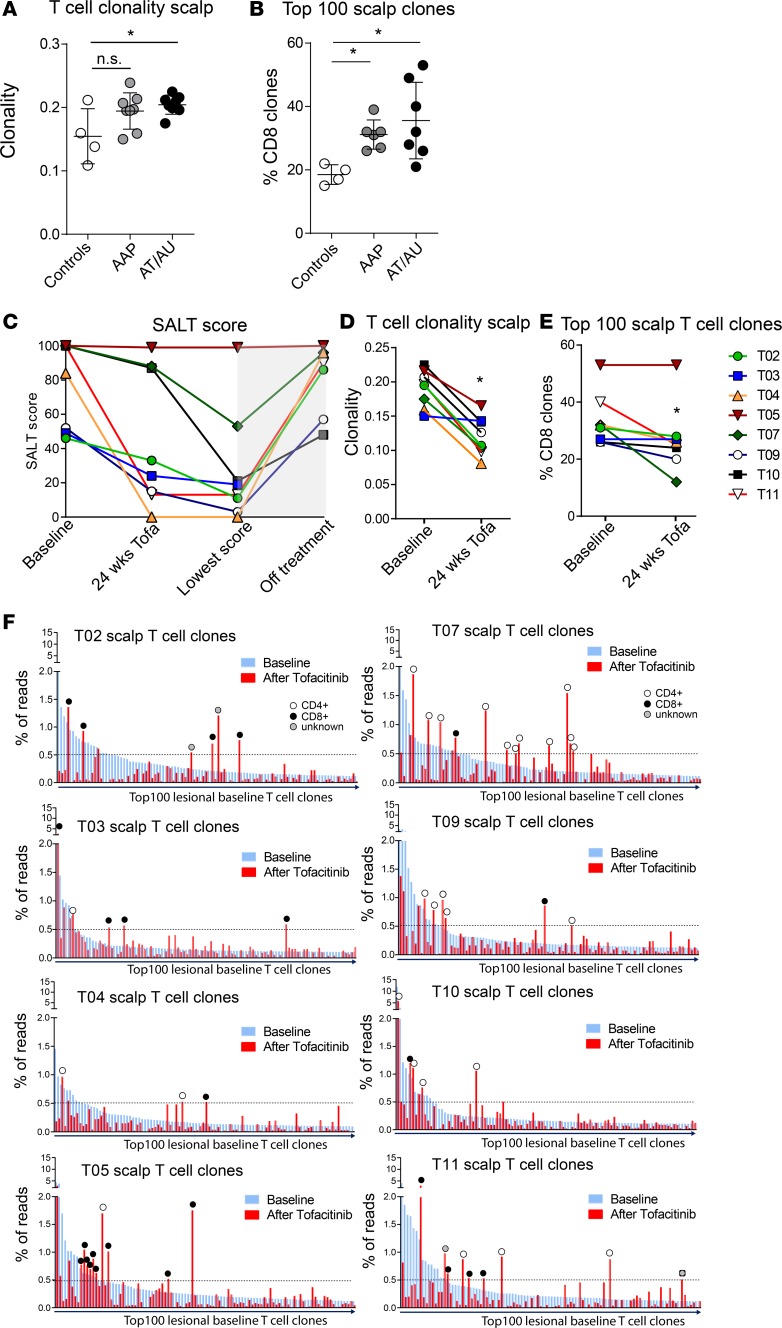
In AA lesional scalp, particularly in patients with the most severe disease (AT/AU), we observed a significantly higher T cell clonality than in the scalp of normal controls (Figure 5A), indicating increased clonal T cell expansions in lesional skin and supporting a role for antigenic drive in human AA. We next cross-referenced the scalp TCR repertoire with the peripheral blood CD4 and CD8 repertoire (both in healthy controls and in 13 of the 16 AA patients) and found that in normal scalp the majority of expanded T cell clones (top 100 clones) were actually CD4+ clones (Supplemental Figure 3). In lesional scalp of AA patients, we observed an increased number of expanded CD8+ T cell clones (Figure 5B). This is consistent with our gene expression data (8), which revealed a CTL signature in AA, with overexpression of perforin, CD8a, and granzyme B genes. However, additional information provided by TCR sequencing includes the fact that the increased CD8+ T cells observed in the lesional scalp are clonally expanded cells, pointing to an antigen-dependent process in human AA.
Figure 5. TCRβ analysis of lesional scalp and blood in AA patients, and response to treatment with oral tofacitinib.
TCRβ sequencing was performed on skin biopsies from normal scalp (n = 4), AA lesional scalp (patchy AA [AAP]) (n = 8), and alopecia totalis and alopecia universalis scalp (AT/AU) (n = 8). The clonality of the scalp T cell repertoire is indicated (A) and the number of expanded CD8+ T cell clones among the top 100 most abundant clones in the scalp is shown (B). *P < 0.05, 1-way ANOVA and Tukey’s honest significant difference. SALT scores from 8 AA patients that were treated with oral tofacitinib at baseline and at the time of last biopsy during treatment (t = 24 weeks in most patients), the lowest SALT score attained on treatment, and the highest SALT score in the 6 months following treatment cessation (C). At baseline and after 24 weeks (or more) of tofacitinib treatment the clonality of the scalp was determined (D) as well as the number of expanded CD8+ T cells among the top 100 most abundant scalp T cell clones. *P < 0.05, 2-tailed Student’s t test (E). The percentage of reads represented by the top 100 T cell clones in the lesional scalp at baseline (blue bars) and the percentage of reads represented by the same clones after treatment (red bars). The circles indicate the T cell clones that are still expanded after treatment and their coreceptor expression (white circle, CD4+; black circle, CD8+; gray circle, unknown) (F).
Effects of oral tofacitinib on scalp-resident T cell clones in AA.
Eight of the AA patients plotted in Figure 5, A and B, were enrolled in an open-label clinical trial to evaluate oral tofacitinib (pan-JAK inhibitor, 5 mg to 10 mg twice daily) in the treatment of moderate/severe AA (28) (Supplemental Table 1). In addition to the baseline samples, biopsies were obtained at 4 weeks and 24 weeks during the course of treatment (in several patients an additional scalp biopsy was obtained at a later time point). The disease severity was assessed in these patients before and during treatment using the Severity of Alopecia Tool (SALT) score, which quantifies the percentage of affected scalp surface area. The SALT scores were recorded at baseline and at the time of last biopsy; additionally, the lowest achieved SALT score while on treatment (the time point of which varied per patient) and — if available — the highest noted SALT score in the 6 months after stopping the oral tofacitinib treatment were recorded (Figure 5C). This analysis showed that all but one of the AA patients had a decrease in SALT score (increase in hair growth) in response to treatment. Upon cessation of oral tofacitinib, relapse of AA was observed within weeks to months (as indicated by the rise in SALT score, Figure 5C). TCRβ chain sequencing of the scalp biopsies showed that treatment with oral tofacitinib resulted in a significant decrease in the scalp clonality (Figure 5D), suggesting that the treatment resulted in a reduction of clonally expanded T cells in the scalp. When we specifically looked at the number of clonally expanded CD8+ T cells (among top 100 clones, which was higher in AA patients compared with healthy controls, Figure 5B), we found that treatment with tofacitinib resulted in a significant decrease in clonally expanded CD8+ T cell clones among the most expanded T cell clones in the scalp (top 100) during treatment (Figure 5E). Interestingly, the one nonresponder (T05) in this group of patients had the highest number of clonally expanded CD8+ T cells at baseline as well as after treatment. Given the small number of patients, and the fact that in this cohort there was only a single nonresponder, larger studies are needed in the future to determine if the number of expanded CD8+ T cell clones at baseline likely correlates with clinical response.
The TCR sequencing allowed us to analyze individual T cell clones before and during treatment, and we plotted the frequencies of the top 100 lesional scalp T cells clones at baseline and determined the frequencies of these same 100 T cell clones after 24 weeks of treatment (or time of last biopsy during treatment, Figure 5F). This analysis revealed that in all patients the majority of expanded T cell clones that were present at baseline were reduced in frequency in response to tofacitinib treatment. Interestingly, we found that not all clones were equally affected by tofacitinib treatment, since the frequencies of some clones were not reduced or even showed a relative increase during treatment. When we looked specifically at the baseline T cell clones that appeared resistant to tofacitinib treatment (defined as higher frequency after treatment than baseline and representing >0.5% of all reads), we found that both CD4+ and CD8+ T cells were among the resistant clones (Figure 5F).
The TCR-sequencing data revealed that tofacitinib reduced the frequencies of most expanded scalp T cell clones but also showed that the majority of these clones do not disappear completely in response to treatment. This is further underscored by the analysis of the circulating frequencies of the expanded lesional CD8+ T cell clones, which showed that (a) some of the lesional CD8+ T cell clones were detected at high frequencies in the CD8+ T cell fraction of the peripheral blood and (b) the relative frequencies of these T cell clones in the circulation were not significantly affected by tofacitinib treatment (Supplemental Figure 4). The residual lesional T cell clones observed in scalp and blood after 24 weeks or more of tofacitinib treatment may explain the relapse of AA upon cessation of treatment.
Discussion
High-throughput T cell receptor sequencing has transformed our ability to conduct a global assessment of the T cell receptor diversity in blood and in any given organ or tissue. The technology to identify and quantify unique T cell clones has provided a powerful approach to track T cells and to determine the TCR diversity in the repertoire (reviewed in ref. 29). Tracking of individual T cell clones using high-throughput sequencing has proven to be a sensitive method to monitor minimal residual disease after treatment for acute or chronic lymphoblastic leukemia and CTCL and for monitoring T cell clones among tumor infiltrating lymphocytes in solid tumors (30–37). Further, high-throughput TCR sequencing has increased our understanding of T cell repertoire diversity (38–42), T cell memory formation (43), and response to infection and aging (44–46). In studies of autoimmune diseases, several recent studies show the potential utility of this technique in ulcerative colitis, multiple sclerosis, psoriasis, and type 1 diabetes (47–52). The technique offers the potential to dynamically assess clonal frequency of pathogenic T cells in the periphery and in tissue as an immunomonitoring tool to assess disease progression and response to therapy.
Our study of the organ-specific autoimmune disease AA further demonstrates the power of this technique for determining the dynamics of skin-infiltrating T cell populations during the course of disease. This work provided insights into adoptive transfer models of autoimmunity in AA, showing that, in the skin graft model of AA, the migration of T cells from donor skin to new skin sites may occur but that, unexpectedly, priming of new T cell clones in the recipient appears to be the dominant mechanism for developing the autoimmune repertoire during disease progression. This suggests that continual recruitment of newly primed naive T cell clones may be operative in AA, suggesting that, in addition to blocking T effector memory activity, therapeutic approaches that inhibit T cell priming (e.g., CTLA4-Ig) may have a place in the treatment of chronic autoimmune states.
In AA, the role of autoantigen-specific T cells in the process of hair follicle destruction has remained enigmatic, despite the observation that autoantibodies are present in some AA patients (15–17) as well as T cells responding to candidate autoantigens (21). The shared TCR CDR3β amino acid sequences and motifs identified in CD8+NKG2D+ T cells in the C3H/HeJ mouse model strongly support the notion that the disease is driven by antigen-specific responses and that the same antigens are recognized by these TCRs. In human AA, the increased clonality of T cell repertoires in lesional skin and the presence in affected scalp of increased numbers of expanded CD8+ T cell clones are consistent with a role for antigen-specific responses as well as in murine disease. Shared TCR sequences and motifs between individual patients were less clear than in the mouse model (data not shown), which may partly be attributable to the HLA diversity in the patient population. Future studies are aimed at identifying TCRα and TCRβ chains of infiltrating CD8+ T cells in a subset of HLA-A201+ AA patients using single-cell TCR sequencing (53). This will allow us to evaluate shared dominant TCRs between patients and develop a screening tool for autoantigen discovery in human AA.
In the C3H/HeJ model, T cell infiltrates in lesional skin consist primarily of CD8+ T cells, whereas, in contrast, infiltrating T cells in human AA consist of both intrafollicular CD8+ T cells as well as perifollicular CD4+ T cells. A role for infiltrating CD8+ T cells in AA is supported by gene expression analysis, which revealed a lesional “CTL signature” (8, 24). However, there is strong evidence for the involvement for CD4+ T cells in AA, as described in a genome-wide meta-analysis of AA, which revealed the HLA-DRB1 gene as a key etiological driver (10). In addition, CD4+ Treg-associated molecules, such as CTLA4, IL-2RA, and GARP, have also been shown to be associated with AA (10). The fact that we did not identify AA-induced clonal CD4+ expansions does not exclude a role for CD4+ T cells in disease. CD4+ T cells may be involved in the licensing of CD8+ T cell effectors or functioning as regulatory T cells. In addition, the fact that normal skin already contains multiple expanded CD4+ T cell clones (presumably against skin commensals and/or pathogens) may also have obscured AA-associated CD4+ T cell expansions in this study. Although the role for CD4+ T cells in AA remains to be defined, it appears that similarities between AA and other autoimmune diseases, such as type 1 diabetes and celiac disease, involve a strong HLA–class II association/role for CD4+ T cells, combined with a CD8-mediated effector process (52, 54).
The oral pan-JAK inhibitor tofacitinib has been shown to partially and reversibly inhibit JAK-dependent cytokine signaling (55) and has been approved for the treatment of rheumatoid arthritis. Prior studies have shown an overall moderate decrease in total lymphocyte counts in the peripheral blood in response to tofacitinib (56).
In severe-to-moderate AA, we and others have shown tofacitinib treatment to be effective in the majority of patients (28, 57–60). TCR sequencing of baseline scalp and blood T cell subset samples and samples obtained after approximately 6 months of treatment provided insight into the response of lesional T cell clones to the drug treatment. Although the majority of expanded lesional scalp T cell clones were reduced in frequency after treatment, TCR sequencing further revealed that the lesional T cell clones do not entirely disappear, which may explain the recurrence of disease after stopping treatment. In addition, the data clearly indicated that not all T cells are equally sensitive to tofacitinib treatment. It is unknown whether this relates to levels of common γ chain expression and differential dependence of cytokines that signal through JAK-3. The treatment of AA using JAK inhibitors will likely be similar to the treatment of other autoimmune and inflammatory disorders, i.e., treatment is aimed at diminishing the population of pathogenic T cells and maintaining a state of remission by continuous usage.
Methods
AA patients.
For untreated samples, AA patients were recruited as part of a multicenter biomarker study. The samples from AA patients treated with tofacitinib (baseline and during treatment) were collected as part of an open-label pilot study of tofacitinib (5 mg to 10 mg orally BID) for 6 to 18 months in the treatment of moderate-to-severe AA, AT, or AU, followed by 6 months of follow-up off drug to assess for delayed response to treatment and/or incidence and timing of recurrence of disease. Further details of the pilot study are described in Jabbari et al. (28). Lesional scalp punch biopsies and peripheral blood samples were collected from AA patients. Similarly, scalp biopsies and blood samples were collected from unaffected controls. Scalp biopsies were immediately fixed in Paxgene tissue containers (PreAnalytix). Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood samples by centrifugation over Hypaque 1077 (MilliporeSigma), and CD4+ and CD8+ T cell fractions were sorted by FACS.
Transfer of AA using grafted C3H/HeJ skin.
Seven- to ten-week-old female C3H/HeJ mice (The Jackson Laboratory) were used as graft recipients and maintained under specific pathogen–free conditions. Donor mice with spontaneous alopecia were between 6 and 12 months of age. Normal-haired C3H/HeJ mice were grafted at 8 weeks of age (during the second telogen) with skin from a C3H/HeJ mouse that developed AA spontaneously, as described previously (23). In brief, mice spontaneously affected with AA were euthanized, and full-thickness skin grafts of approximately 2 cm in diameter were removed and grafted to normal-haired C3H/HeJ mice. Hair loss typically began at around 4–6 weeks after grafting.
Flow cytometric sorting.
Murine cutaneous lymph nodes were pooled; minced in RPMI; filtered through a 40-μM cell strainer; centrifuged at 400 g for 5 minutes; stained with antibodies against CD4 (GK1.5, BD), CD8α (53-6.7, BD), NKG2D (CX5, eBioscience), CD103 (2E7, eBioscience), NKG2ACE (clone 20d5, eBioscience), and CD44 (IM7, BD); and sorted on a BD Influx cell sorter.
PBMCs from AA patients were stained with antibodies against CD3 (clone OKT3, Biolegend), CD4 (clone OKT4, eBioscience), and CD8 (clone HIT8a, BD bioscience) and sorted on the FACSAria cell sorter. For several AA patients (n = 5, biomarker study), the CD4 fraction was further sorted into CD25+CD127– cells and the remaining CD4 T cells, and the CD8 fraction was further sorted into NKG2Dlo and NKG2Dhi cells.
RNA extraction, cDNA preparation, and DNA extraction.
From all samples for TCR sequencing, RNA was extracted and reverse transcribed to cDNA for TCR sequencing. Snap frozen mouse skin tissues were homogenized and RNA was extracted using the RNeasy kit (Qiagen). Fixed human scalp biopsies were homogenized and RNA was extracted using the Paxgene miRNA kit (PreAnalytix). RNA from PBMCs was isolated using the miRNeasy kit (Qiagen). RNA was quantified using NanoDrop (Thermo Fisher Scientific) and SuperScript III Reverse Transcriptase (Life Technologies) and a 2:1 mix of oligodT and random hexamers was used for first-strand cDNA synthesis.
High-throughput TCR sequencing.
TCRβ CDR3 regions were amplified and sequenced by Adaptive Biotechnologies. Briefly, the method applies a multiplex PCR system to amplify all possible rearranged TCRβ CDR3 sequences from cDNA samples, using 35 forward primers specific for all Vβ gene segments and 13 reverse primers specific for all Jβ gene segments (61). The forward and reverse primers contain at their 5′ ends the universal forward and reverse primer sequences, respectively, compatible with the Illumina HiSeq cluster station solid-phase PCR system. In this technique, a synthetic immune receptor repertoire has been used to identify amplification biases introduced by differential hybridization kinetics, and these biases are minimized and residual bias computationally removed sequencing (62). The HiSeq system generates 60 base pair reads, which cover the CDR3 lengths, sequencing from the J to the V region. The amplification and sequencing protocols have been described previously (61, 63). The raw HiSeq sequence data were preprocessed to remove errors and to compress the data. Analysis of TCRβ sequences was conducted with the Adaptive Biotechnologies TCR ImmunoSEQ assay. The TCR CDR3 region nomenclature was defined according to the International Immunogenetics collaboration (64), beginning with the second conserved cysteine encoded by the 3′ portion of the Vβ gene segment and ending with the conserved phenylalanine encoded by the 5′ portion of the Jβ gene segment. The number of nucleotides between these codons determines the length and therefore the frame of the CDR3 region. To identify which V, D, and J segments contributed to each TCR CDR3 sequence a standard algorithm was used (64).
Calculation of clonality.
Clonality was calculated from Shannon’s Entropy as previously described (65). Briefly, the entropy of each sample was normalized by dividing the entropy calculation by the log2 of the number of productive unique TCRs. The clonality is defined by 1 minus normalized entropy and is a measure of oligoclonality of the sample.
TCR-sequencing data.
All TCRβ chain–sequencing data from the C3H/HeJ model as well as from the patient studies are available at http://clients.adaptivebiotech.com/pub/deJong-2018-jciinsight
Statistics.
Analyses of the sequence data for TCRB CDR3 sequences and VDJ recombinations and their comparisons were performed using ImmunoSEQ software (provided online, Adaptive Biotechnologies). In addition, sequence frequency data were exported into Excel for further analysis, and Gene-E software (Broad Institute) was used to generate heatmaps. In the C3H/HeJ model, to determine if clonality was significantly increased around the time of hair loss, we used a 2-tailed Student’s t test, and we used a 1-way ANOVA to determine if the clonality of longstanding alopecic skin differed from that of recent-onset alopecia. To compare the clonality and percentage of expanded CD8 T cell clones among controls, AAP, and AT/AU patients, we used a 1-way ANOVA and Tukey’s honest significant difference. A P value of less than 0.05 was considered significant.
Study approval.
All procedures were performed under institutional review board–approved protocols at involved study sites (Columbia University, MD Anderson Cancer Center, University of Minnesota, University of Colorado, UCSF) and conducted under the Declaration of Helsinki principles. Informed consent was received from all subjects before inclusion in the study. Mouse experiments were performed in compliance with institutional guidelines as approved by the institutional animal care and use committee of Columbia University Medical Center.
Author contributions
ADJ, AJ, ZD, and LX were responsible in large part for performing the studies reported herein and participated in the design, execution, and interpretation of the data. JMW, MD, VP, MH, and DAN were responsible for the recruitment and classification of study subjects as well as the procurement of tissue samples. DL and MML assisted in data analysis. ADJ, RC, and AMC were responsible for conception, design, oversight, execution, and interpretation of the data for this study and for the writing, figure preparation, and editing of the final manuscript.
Supplementary Material
Acknowledgments
We are grateful for clinical research support from C. Kim, M. Furniss, B. Thompson, C. Clark, J. Bourbon, N. Nguyen, and G. Ulerio and expert technical assistance from M.J. Zhang, E.S. Chang, H.H. Lin, W.Q. Huang, and N. Fox. We thank D. Hamm (Adaptive Biotechnologies) for advice regarding the sequencing data. This work was supported in part by US Public Health Service NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants R01AR056016 (to AMC) and R21AR061881 (to AMC and RC), by Pfizer (ASPIRE grant to AMC), the Columbia University Skin Disease Research Center (P30AR069632), the Alopecia Areata Center for Research Translation (P50AR070588 to AMC), the Locks of Love Foundation, and a grant from the National Alopecia Areata Foundation (to ADJ). AJ and ADJ were recipients of Career Development Awards from the Dermatology Foundation and Irving Scholars at Columbia University. ADJ is supported by a NIAMS K01 AR068475, and AJ is supported by a NIAMS K08 AR069111. The cell sorting reported in this publication was performed in the Columbia Center for Translational Immunology Flow Cytometry Core, supported in part by a NIH-shared instrument grant (S10OD020056).
Version 1. 10/04/2018
Electronic publication
Footnotes
AJ’s present address is: Department of Dermatology and Interdisciplinary Program in Immunology, University of Iowa, Iowa City, Iowa, USA.
Conflict of interest: AMC and RC are co-inventors on US Patent no. 9,198,911 filed by Columbia University and licensed to Aclaris Therapeutics. RC is a consultant for and shareholder of Aclaris Therapeutics. AMC is a consultant for Aclaris Therapeutics and Dermira and is a shareholder of Aclaris Therapeutics. AMC has received grant support from Pfizer.
Reference information: JCI Insight. 2018;3(19):e121949. https://doi.org/10.1172/jci.insight.121949.
Contributor Information
Annemieke de Jong, Email: ad2952@cumc.columbia.edu.
Ali Jabbari, Email: ali-jabbari@uiowa.edu.
Zhenpeng Dai, Email: zd2148@columbia.edu.
Luzhou Xing, Email: lx16@columbia.edu.
Dustin Lee, Email: dleer358@gmail.com.
Mei Mei Li, Email: ml3828@cumc.columbia.edu.
Madeleine Duvic, Email: mduvic@mdanderson.org.
Maria Hordinsky, Email: hordi001@umn.edu.
Vera Price, Email: Vera.Price@ucsf.edu.
Julian Mackay-Wiggan, Email: jmackaywiggan@gmail.com.
Raphael Clynes, Email: Raphael.clynes@gmail.com.
Angela M. Christiano, Email: amc65@columbia.edu.
References
- 1.Gilhar A, Etzioni A, Paus R. Alopecia areata. N Engl J Med. 2012;366(16):1515–1525. doi: 10.1056/NEJMra1103442. [DOI] [PubMed] [Google Scholar]
- 2.Gilhar A, Shalaginov R, Assy B, Serafimovich S, Kalish RS. Alopecia areata is a T-lymphocyte mediated autoimmune disease: lesional human T-lymphocytes transfer alopecia areata to human skin grafts on SCID mice. J Investig Dermatol Symp Proc. 1999;4(3):207–210. doi: 10.1038/sj.jidsp.5640212. [DOI] [PubMed] [Google Scholar]
- 3.Gilhar A, Ullmann Y, Berkutzki T, Assy B, Kalish RS. Autoimmune hair loss (alopecia areata) transferred by T lymphocytes to human scalp explants on SCID mice. J Clin Invest. 1998;101(1):62–67. doi: 10.1172/JCI551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McElwee KJ, et al. Transfer of CD8(+) cells induces localized hair loss whereas CD4(+)/CD25(-) cells promote systemic alopecia areata and CD4(+)/CD25(+) cells blockade disease onset in the C3H/HeJ mouse model. J Invest Dermatol. 2005;124(5):947–957. doi: 10.1111/j.0022-202X.2005.23692.x. [DOI] [PubMed] [Google Scholar]
- 5.Christoph T, et al. The human hair follicle immune system: cellular composition and immune privilege. Br J Dermatol. 2000;142(5):862–873. doi: 10.1046/j.1365-2133.2000.03464.x. [DOI] [PubMed] [Google Scholar]
- 6.Paus R, Ito N, Takigawa M, Ito T. The hair follicle and immune privilege. J Investig Dermatol Symp Proc. 2003;8(2):188–194. doi: 10.1046/j.1087-0024.2003.00807.x. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Ito N, Bettermann A, Tokura Y, Takigawa M, Paus R. Collapse and restoration of MHC class-I-dependent immune privilege: exploiting the human hair follicle as a model. Am J Pathol. 2004;164(2):623–634. doi: 10.1016/S0002-9440(10)63151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing L, et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med. 2014;20(9):1043–1049. doi: 10.1038/nm.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betz RC, et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun. 2015;6:5966. doi: 10.1038/ncomms6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: Systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol. 2007;148(1):1–16. doi: 10.1111/j.1365-2249.2006.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietropaolo M, Towns R, Eisenbarth GS. Humoral autoimmunity in type 1 diabetes: prediction, significance, and detection of distinct disease subtypes. Cold Spring Harb Perspect Med. 2012;2(10):a012831. doi: 10.1101/cshperspect.a012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klareskog L, Amara K, Malmström V. Adaptive immunity in rheumatoid arthritis: anticitrulline and other antibodies in the pathogenesis of rheumatoid arthritis. Curr Opin Rheumatol. 2014;26(1):72–79. doi: 10.1097/BOR.0000000000000016. [DOI] [PubMed] [Google Scholar]
- 14.Scally SW, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med. 2013;210(12):2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung MC, Sutton CW, Fenton DA, Tobin DJ. Trichohyalin is a potential major autoantigen in human alopecia areata. J Proteome Res. 2010;9(10):5153–5163. doi: 10.1021/pr100422u. [DOI] [PubMed] [Google Scholar]
- 16.Tobin DJ. Characterization of hair follicle antigens targeted by the anti-hair follicle immune response. J Investig Dermatol Symp Proc. 2003;8(2):176–181. doi: 10.1046/j.1087-0024.2003.00805.x. [DOI] [PubMed] [Google Scholar]
- 17.Trautman S, Thompson M, Roberts J, Thompson CT. Melanocytes: a possible autoimmune target in alopecia areata. J Am Acad Dermatol. 2009;61(3):529–530. doi: 10.1016/j.jaad.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Lueking A, et al. Profiling of alopecia areata autoantigens based on protein microarray technology. Mol Cell Proteomics. 2005;4(9):1382–1390. doi: 10.1074/mcp.T500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Gilhar A, Landau M, Assy B, Shalaginov R, Serafimovich S, Kalish RS. Melanocyte-associated T cell epitopes can function as autoantigens for transfer of alopecia areata to human scalp explants on Prkdc(scid) mice. J Invest Dermatol. 2001;117(6):1357–1362. doi: 10.1046/j.0022-202x.2001.01583.x. [DOI] [PubMed] [Google Scholar]
- 20.Tobin DJ, Hann SK, Song MS, Bystryn JC. Hair follicle structures targeted by antibodies in patients with alopecia areata. Arch Dermatol. 1997;133(1):57–61. [PubMed] [Google Scholar]
- 21.Wang EH, et al. Identification of autoantigen epitopes in alopecia areata. J Invest Dermatol. 2016;136(8):1617–1626. doi: 10.1016/j.jid.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 2010;185(7):3801–3808. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McElwee KJ, Boggess D, King LE, Sundberg JP. Experimental induction of alopecia areata-like hair loss in C3H/HeJ mice using full-thickness skin grafts. J Invest Dermatol. 1998;111(5):797–803. doi: 10.1046/j.1523-1747.1998.00380.x. [DOI] [PubMed] [Google Scholar]
- 24.Jabbari A, et al. Molecular signatures define alopecia areata subtypes and transcriptional biomarkers. EBioMedicine. 2016;7:240–247. doi: 10.1016/j.ebiom.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia KC, Adams EJ. How the T cell receptor sees antigen--a structural view. Cell. 2005;122(3):333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T cell responses. Nat Rev Immunol. 2008;8(3):231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 27.Clark RA, et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J Invest Dermatol. 2006;126(5):1059–1070. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 28.Jabbari A, et al. An open-label pilot study to evaluate the efficacy of tofacitinib in moderate to severe patch-type alopecia areata, totalis, and universalis. J Invest Dermatol. 2018;138(7):1539–1545. doi: 10.1016/j.jid.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25(5):646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Emerson RO, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J Pathol. 2013;231(4):433–440. doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng WK, Armstrong R, Arai S, Desmarais C, Hoppe R, Kim YH. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci Transl Med. 2013;5(214):214ra171. doi: 10.1126/scitranslmed.3007420. [DOI] [PubMed] [Google Scholar]
- 32.Linnemann C, Mezzadra R, Schumacher TN. TCR repertoires of intratumoral T-cell subsets. Immunol Rev. 2014;257(1):72–82. doi: 10.1111/imr.12140. [DOI] [PubMed] [Google Scholar]
- 33.Schumacher JA, Duncavage EJ, Mosbruger TL, Szankasi PM, Kelley TW. A comparison of deep sequencing of TCRG rearrangements vs traditional capillary electrophoresis for assessment of clonality in T-Cell lymphoproliferative disorders. Am J Clin Pathol. 2014;141(3):348–359. doi: 10.1309/AJCP5TYGBVW4ZITR. [DOI] [PubMed] [Google Scholar]
- 34.Gros A, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124(5):2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Logan AC, et al. Immunoglobulin and T cell receptor gene high-throughput sequencing quantifies minimal residual disease in acute lymphoblastic leukemia and predicts post-transplantation relapse and survival. Biol Blood Marrow Transplant. 2014;20(9):1307–1313. doi: 10.1016/j.bbmt.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerlinger M, et al. Ultra-deep T cell receptor sequencing reveals the complexity and intratumour heterogeneity of T cell clones in renal cell carcinomas. J Pathol. 2013;231(4):424–432. doi: 10.1002/path.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107(4):1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19(10):1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estorninho M, et al. A novel approach to tracking antigen-experienced CD4 T cells into functional compartments via tandem deep and shallow TCR clonotyping. J Immunol. 2013;191(11):5430–5440. doi: 10.4049/jimmunol.1300622. [DOI] [PubMed] [Google Scholar]
- 41.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111(36):13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zvyagin, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci USA. 2014;111(16):5980–5985. doi: 10.1073/pnas.1319389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaide O, et al. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med. 2015;21(6):647–653. doi: 10.1038/nm.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remmerswaal EB, et al. Clonal evolution of CD8+ T cell responses against latent viruses: relationship among phenotype, localization, and function. J Virol. 2015;89(1):568–580. doi: 10.1128/JVI.02003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li D, et al. Profiling the T-cell receptor repertoire of patient with pleural tuberculosis by high-throughput sequencing. Immunol Lett. 2014;162(1 Pt A):170–180. doi: 10.1016/j.imlet.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Britanova OV, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol. 2014;192(6):2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 47.Lord J, Chen J, Thirlby RC, Sherwood AM, Carlson CS. T-cell receptor sequencing reveals the clonal diversity and overlap of colonic effector and FOXP3+ T cells in ulcerative colitis. Inflamm Bowel Dis. 2015;21(1):19–30. doi: 10.1097/MIB.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lossius A, et al. High-throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV-reactive CD8+ T cells. Eur J Immunol. 2014;44(11):3439–3452. doi: 10.1002/eji.201444662. [DOI] [PubMed] [Google Scholar]
- 49.Marrero I, Hamm DE, Davies JD. High-throughput sequencing of islet-infiltrating memory CD4+ T cells reveals a similar pattern of TCR Vβ usage in prediabetic and diabetic NOD mice. PLoS One. 2013;8(10):e76546. doi: 10.1371/journal.pone.0076546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toivonen R, Arstila TP, Hänninen A. Islet-associated T-cell receptor-β CDR sequence repertoire in prediabetic NOD mice reveals antigen-driven T-cell expansion and shared usage of VβJβ TCR chains. Mol Immunol. 2015;64(1):127–135. doi: 10.1016/j.molimm.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Matos TR, et al. Clinically resolved psoriatic lesions contain psoriasis-specific IL-17-producing αβ T cell clones. J Clin Invest. 2017;127(11):4031–4041. doi: 10.1172/JCI93396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seay HR, et al. Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight. 2016;1(20):e88242. doi: 10.1172/jci.insight.88242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32(7):684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han A, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc Natl Acad Sci USA. 2013;110(32):13073–13078. doi: 10.1073/pnas.1311861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghoreschi K, et al. Modulation of innate and adaptive immune responses by tofacitinib (CP-690,550) J Immunol. 2011;186(7):4234–4243. doi: 10.4049/jimmunol.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodge JA, et al. The mechanism of action of tofacitinib - an oral Janus kinase inhibitor for the treatment of rheumatoid arthritis. Clin Exp Rheumatol. 2016;34(2):318–328. [PubMed] [Google Scholar]
- 57.Kennedy Crispin M, et al. Safety and efficacy of the JAK inhibitor tofacitinib citrate in patients with alopecia areata. JCI Insight. 2016;1(15):e89776. doi: 10.1172/jci.insight.89776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jabbari A, et al. Treatment of an alopecia areata patient with tofacitinib results in regrowth of hair and changes in serum and skin biomarkers. Exp Dermatol. 2016;25(8):642–643. doi: 10.1111/exd.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu LY, Craiglow BG, Dai F, King BA. Tofacitinib for the treatment of severe alopecia areata and variants: A study of 90 patients. J Am Acad Dermatol. 2017;76(1):22–28. doi: 10.1016/j.jaad.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Ibrahim O, Bayart CB, Hogan S, Piliang M, Bergfeld WF. Treatment of alopecia areata with tofacitinib. JAMA Dermatol. 2017;153(6):600–602. doi: 10.1001/jamadermatol.2017.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlson CS, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 63.Robins H, et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods. 2012;375(1-2):14–19. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20 Suppl 1:i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- 65.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62(9):1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.