Abstract
Gut microbiota–derived metabolites play important roles in health and disease. D–amino acids and their L-forms are metabolites of gut microbiota with distinct functions. In this study, we show the pathophysiologic role of D–amino acids in association with gut microbiota in humans and mice with acute kidney injury (AKI). In a mouse kidney ischemia/reperfusion model, the gut microbiota protected against tubular injury. AKI-induced gut dysbiosis contributed to the altered metabolism of D–amino acids. Among the D–amino acids, only D-serine was detectable in the kidney. In injured kidneys, the activity of D–amino acid oxidase was decreased. Conversely, the activity of serine racemase was increased. The oral administration of D-serine mitigated the kidney injury in B6 mice and D-serine–depleted mice. D-serine suppressed hypoxia-induced tubular damage and promoted posthypoxic tubular cell proliferation. Finally, the D-serine levels in circulation were significantly correlated with the decrease in kidney function in AKI patients. These results demonstrate the renoprotective effects of gut-derived D-serine in AKI, shed light on the interactions between the gut microbiota and the kidney in both health and AKI, and highlight D-serine as a potential new therapeutic target and biomarker for AKI.
Keywords: Microbiology, Nephrology
Keywords: Amino acid metabolism
Under acute kidney injury conditions, D-serine was detectable in the kidney and its levels in the circulation were correlated with the decline in kidney function.
Introduction
Recent studies revealed that the dysbiosis of the gut microbiota was closely associated with several diseases, such as inflammatory bowel disease (1), diabetes (2), cardiovascular disease (3), and kidney disease (4). To date, clinical trials on fecal microbial transplantation were shown to be effective in clinical conditions, such as Clostridium difficile infection (5), pouchitis (6), and metabolic syndrome (7). The gut microbiota might play a therapeutic role in these disorders, but the precise roles and underlying mechanisms remain unclear.
Data have revealed the important roles of bacterial metabolites in both health and disease. The short-chain fatty acids (SCFAs) produced by the gut microbiota demonstrated antiinflammatory effects, such as the promotion of regulatory T cell proliferation (8). In a murine model of acute kidney injury (AKI), SCFAs protected against tubular injury and reduced dendritic cell accumulation in the kidney (9). By contrast, the gut microbiota produces uremic toxins in chronic kidney disease (10). These findings suggest that the gut microbiota might regulate host homeostasis via their metabolites.
Recent advances have shed light on the chirality of amino acids. Free D–amino acids and their L-forms were quantified by 2D HPLC (11). Furthermore, studies have reported that D–amino acids in the murine gut were the primary microbial products (12) and that they played physiologic roles in the central nervous system (13), heart (14), and skin (15). However, the precise roles of the gut microbiota–associated D–amino acids in kidney diseases have yet to be revealed. Thus, we explored the pathophysiologic role of D–amino acids in association with the gut microbiota in humans with AKI and in a mouse model of AKI.
Results
AKI induces gut dysbiosis in a mouse infusion/reperfusion model.
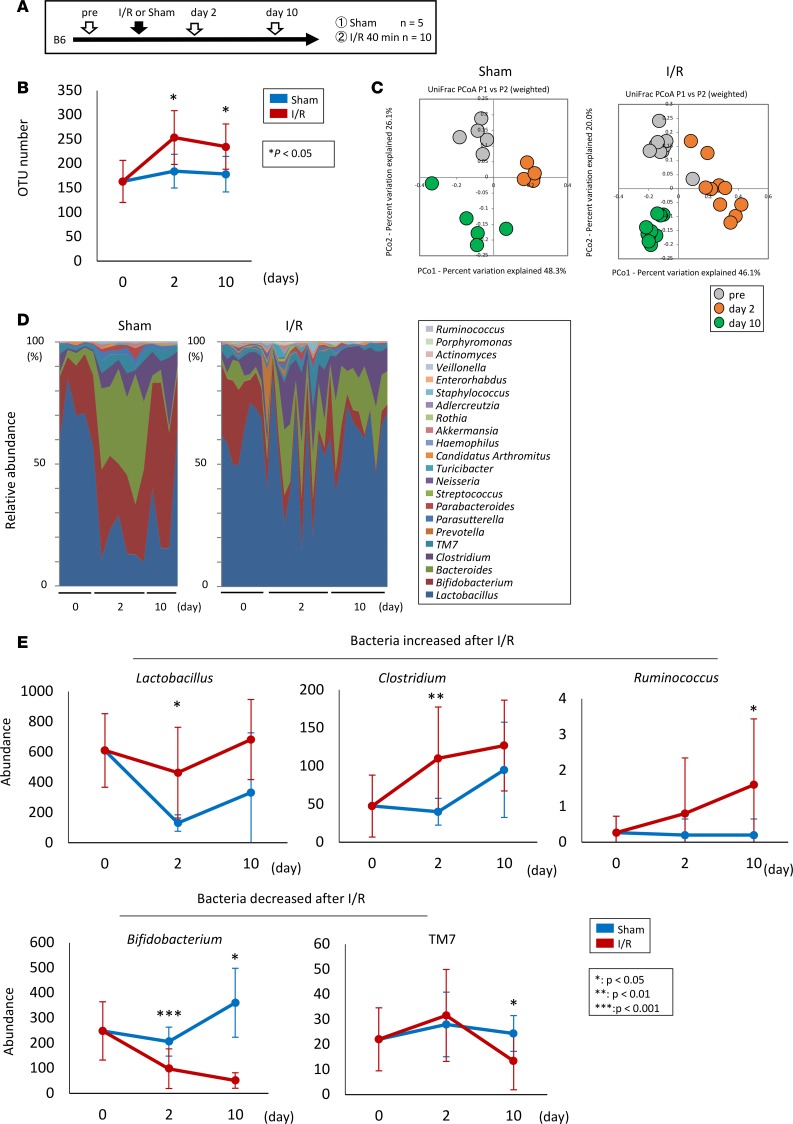
To assess the effect of AKI on gut microbiota, we performed 16S rRNA gene-sequencing analysis on mouse feces on days 0, 2, and 10 after ischemia/reperfusion (I/R) injury to an AKI model (Figure 1A). The clustering analysis of 16S reads revealed that the microbiota in the AKI group had significantly higher species richness than that in the sham-operated mice on days 2 and 10 (Figure 1B). A comparison of the overall microbiota structure of the groups showed clearly different patterns in the weighted UniFrac principal coordinate analysis (Figure 1C). Moreover, the genus-level taxonomic assignment showed a decreased abundance of Bifidobacterium and TM7 and an increased abundance of Lactobacillus, Clostridium, and Ruminococcus after I/R injury (Figure 1, D and E).
Figure 1. AKI-induced gut dysbiosis in the mouse I/R model.
(A) The protocol for I/R injury in mouse kidney. (B) Comparison of species richness between the gut microbiota in the I/R-injured and the sham-operated mice. (C) Principal coordinate analyses (PCoAs) based on the weighted UniFrac analysis of the bacterial community structures before the induction of I/R (pre) and on days 2 and 10. (D and E) Comparison of the bacterial composition (D) and the abundance of several species (E) between the I/R-injured and the sham-operated mice before I/R and days 2 and 10. Data are presented as mean ± SEM. Statistical analysis was performed using Student’s t test (B and E). *P < 0.05, **P < 0.01, ***P < 0.001. OTU, operational taxonomic unit.
Gut microbiota protects against tubular injury in mouse AKI.
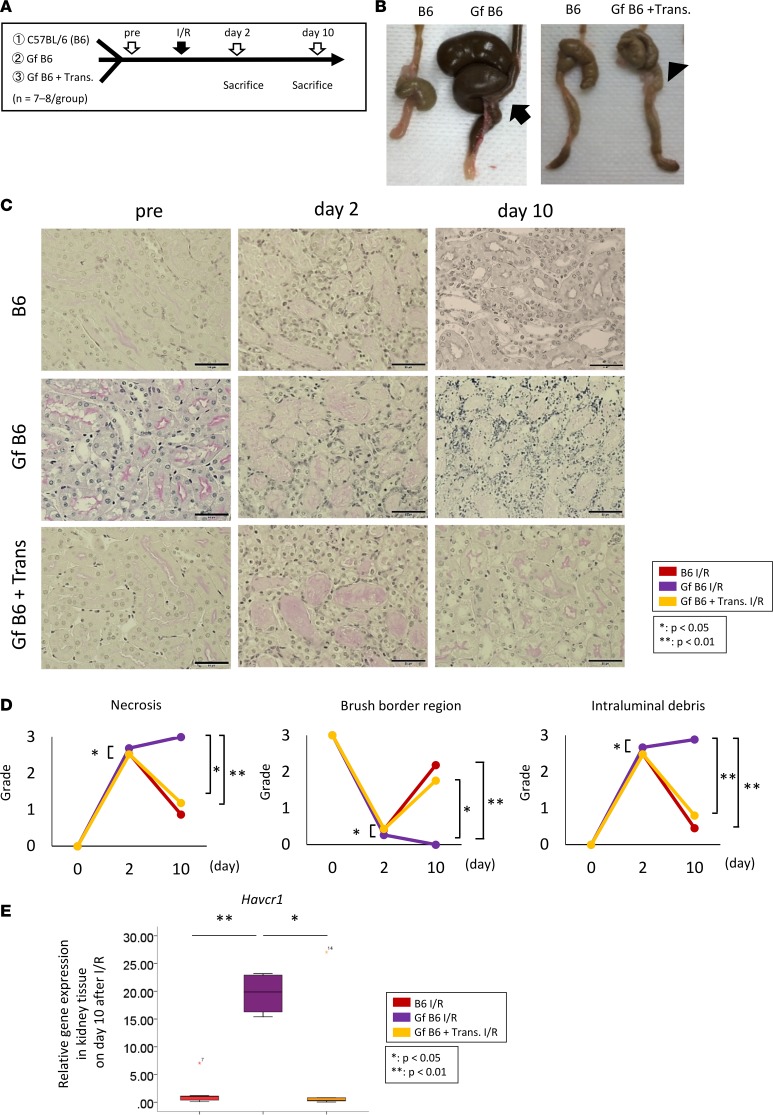
We explored the contribution of the gut microbiota to the pathogenesis of AKI. Kidney I/R injury was induced in germ-free C57BL/6 (Gf B6) mice with or without fecal transplantation from normal C57BL/6 (B6) mice (Figure 2A). The Gf B6 mice had enlarged ceca, which reverted to a normal size after fecal transplantation from normal B6 mice (Figure 2B). On day 2 after I/R, tubular injury was worse in the Gf B6 mice than in the normal B6 mice. Although the tubular injury was partially improved on day 10 in the normal B6 mice, the damage progressed to tubular necrosis in the Gf B6 mice (Figure 2, C and D). Interestingly, fecal transplantation from normal mice attenuated the renal pathology in the Gf B6 mice (Figure 2, C and D). The increased expression of hepatitis A virus cellular receptor 1 (Havcr1) mRNA in the kidneys of Gf B6 mice after I/R was also decreased by the fecal transplantation from the normal B6 mice (Figure 2E). We also evaluated the contribution of the gut microbiota to AKI in gut microbiota–depleted mice by the administration of antibiotics. In the B6 mice, antibiotic administration (Abx) reduced the gut microbiota and induced the enlargement of the cecum, similar to that observed in Gf B6 mice (Supplemental Figure 1C; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97957DS1). The acute tubular necrosis score, which is used to determine tubular injury, was higher in antibiotic-treated mice than in antibiotic-untreated mice on day 10 after I/R injury (Supplemental Figure 1, D and E).
Figure 2. Gut microbiota protects against tubular injury in a mouse model of AKI.
(A) Gf B6 mice were used to evaluate the pathophysiologic role of the gut microbiota in AKI. (B) Gf B6 mice exhibit an enlarged cecum (arrow), which reverted to the normal size in feces-transplanted Gf B6 mice (Gf + Trans, arrowhead). (C and D) The Gf B6 mice exhibited severe tubular injury on day 2 and impaired repair on day 10; these changes are attenuated by fecal transplantation from normal C57BL/6 (B6) mice. Scale bar: 50 μm. (E) The Gf B6 mice exhibit higher expression levels of hepatitis A virus cellular receptor 1 (Havcr1) mRNA in the kidney tissue 10 days after the I/R compared with the B6 mice. Box-and-whisker plots represent the median values. Boxes represent the 25th and 75th percentiles, and whiskers depict the minimum and maximum values. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple comparison test (D and E). *P < 0.05, **P < 0.01.
AKI-induced dysbiosis alters the balance of D/L–amino acids.
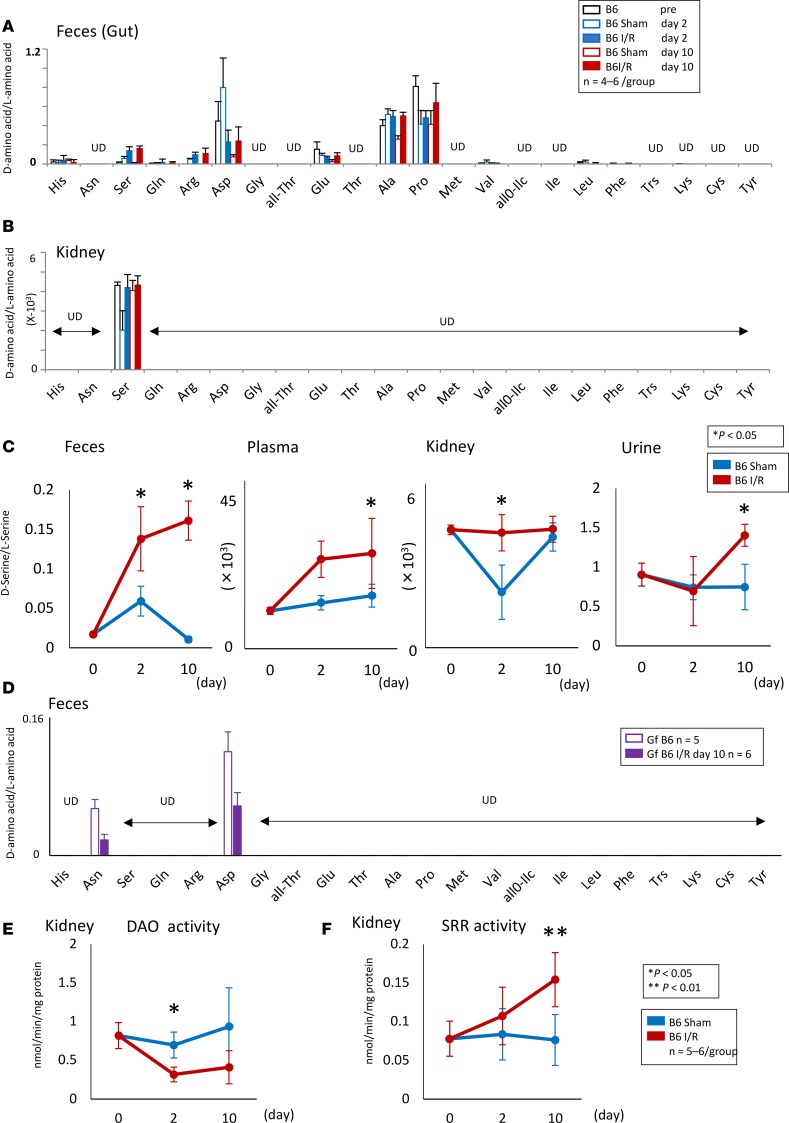
Recent studies revealed that the D–amino acids detected by 2D HPLC are produced by the gut microbiota and are involved in intestinal mucosal defenses (12). We explored whether AKI-induced dysbiosis altered the balance of D/L–amino acid profiles in the feces, plasma, kidneys, and urine of mice with I/R injury. The 2D HPLC analysis revealed that the D-serine/L-serine, D-arginine/L-arginine, and D-alanine/L-alanine ratios were increased in feces after I/R injury (Figure 3A). Surprisingly, only D-serine was detected in the kidneys by 2D HPLC (Figure 3B). The D-serine/L-serine ratio was increased in the feces, plasma, kidneys, and urine of I/R mice compared with those in sham-operated mice (Figure 3C). By contrast, no D–amino acids, except D-asparagine and D-aspartic acid, were detectable in the feces of Gf B6 mice, thus suggesting that the gut microbiota was the main source of D–amino acids, particularly D-serine (Figure 3D). The overall levels of D–amino acids and their L-forms for all animal groups are presented in Supplemental Figures 2–6.
Figure 3. Dysbiosis induced by AKI alters the balance of D/L–amino acids.
(A and B) Although some free D–amino acids were detected in the feces of B6 mice with or without I/R, only D-serine can be detected in the kidney. (C) Free D-serine is increased in the feces, plasma, kidney, and urine after the I/R. (D) No free D–amino acids, except D-asparagine and D-aspartic acid, are detected in the feces of Gf B6 mice before and after I/R. (E) DAO activity is decreased in the kidney after the I/R injury. (F) SRR activity is increased in the injured kidney. Data are shown as mean ± SEM. Statistical analysis was performed using Student’s t test (C, E, and F). *P < 0.05, **P < 0.01. UD, undetectable.
On the basis of these results, we focused on the metabolism of D-serine in the injured kidney. In addition to the gut microbiota, peripheral organs, including the kidneys, are host sites for D-serine metabolism by D–amino acid oxidase (DAO) and serine racemase (SRR). The activity of DAO, which degrades D–amino acids, including D-serine (16), was decreased in the I/R-injured kidneys (Figure 3E). On the contrary, the activity of SRR, which generates D-serine from L-serine (17), was increased in the I/R-injured kidneys (Figure 3F).
D-serine reduced tubular injury after I/R injury.
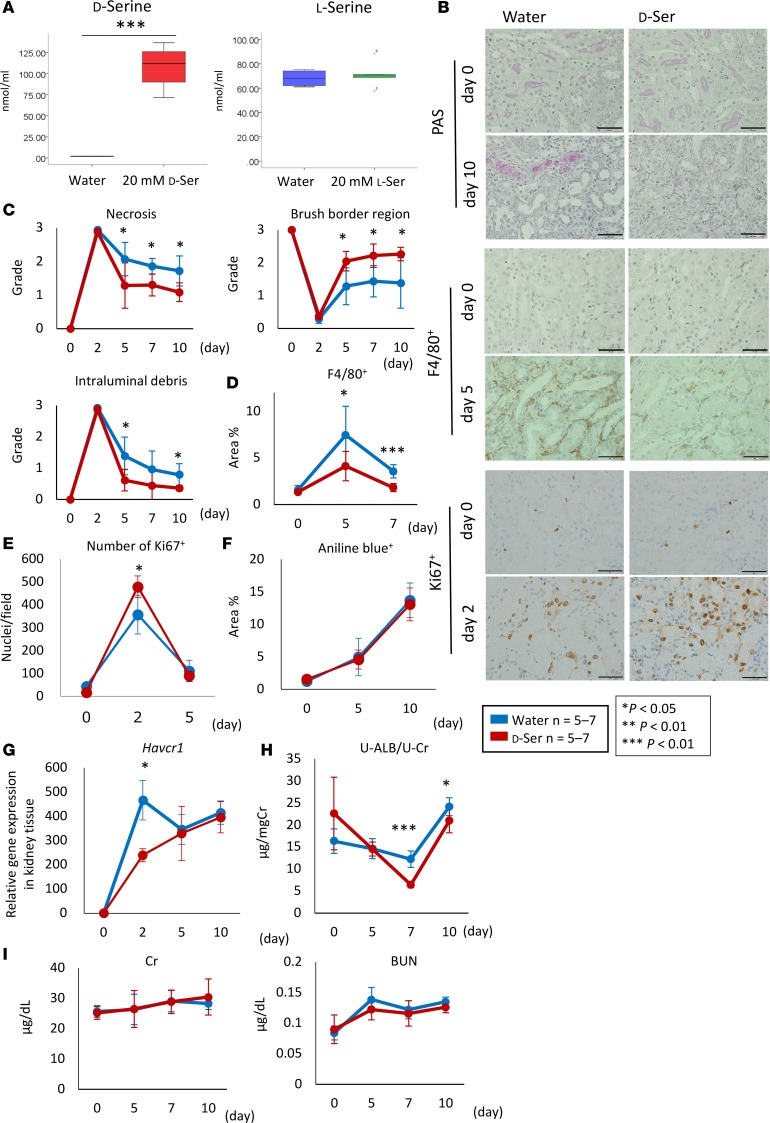
We explored the pathophysiologic function of D-serine in AKI. We administered 20 mM D- or L-serine to mice via drinking water. Oral L-serine administration did not change the plasma concentration of L-serine, whereas the oral administration of 20 mM D-serine increased the plasma levels of D-serine to 100 nM (Figure 4A), thus suggesting that D-serine was absorbed through the gastrointestinal tract and led to higher plasma D-serine levels than those in the I/R-injured mice (Supplemental Figure 4). Interestingly, the administration of 20 mM D-serine attenuated tubular injury on days 5, 7, and 10 in the I/R-injured mice (Figure 4, B and C). Furthermore, the accumulation of F4/80+ cells in the kidneys was reduced on days 5 and 7 after I/R injury following the administration of 20 mM D-serine (Figure 4, B and D). Consistent with this finding, an increased number of Ki67+ tubules and decreased levels of Havcr1 mRNA expression were detected in the D-serine–treated mice 2 days after I/R injury (Figure 4, B, E, and G). Moreover, the administration of D-serine led to a reduction in urine albumin excretion on days 7 and 10 after I/R injury (Figure 4H). The extent of interstitial fibrosis, which was evaluated by Azan staining and presented as the percentage area stained with aniline blue, and the levels of creatinine and blood urea nitrogen were similar between the groups (Figure 4, F and I). There was no significant difference in the volume of water intake between the untreated and D-serine–treated mice, thus suggesting that water intake alone was not associated with the observed renoprotection (Supplemental Figure 7).
Figure 4. D-serine reduces tubular injury after I/R.
(A) Although oral L-serine administration does not change the plasma concentration of L-serine, 20 mM D-serine increased the plasma levels of D-serine to 100 nM/ml. (B) Representative images of tissue samples stained with PAS, F4/80+, and Ki67+ are shown. Scale bar: 50 μm. (C) The grades of necrosis and intraluminal debris are decreased and that of the brush border region is increased after the administration of D-serine at 5, 7, and 10 days after the I/R injury. (D) The administration of D-serine reduces the percentage area positive for F4/80+ at 5 and 7 days after the I/R injury. (E) The number of Ki67+ nuclear/field is increased by the administration of D-serine at 2 days after the I/R injury. (F) There is no difference in the percentage area stained with aniline blue between groups. (G) Havcr1 gene expression in the kidney tissue is decreased in the D-serine treatment group at 2 days after the I/R injury. (H) Urinary albumin excretion is protected in the D-serine treatment group. (I) The levels of serum creatinine (Cr) and serum blood urea nitrogen (BUN) are similar between the groups after the administration of D-serine. Data are shown as mean ± SEM. Statistical analysis was performed using Student’s t test (A and C–I). *P < 0.05, **P < 0.01, ***P < 0.001.
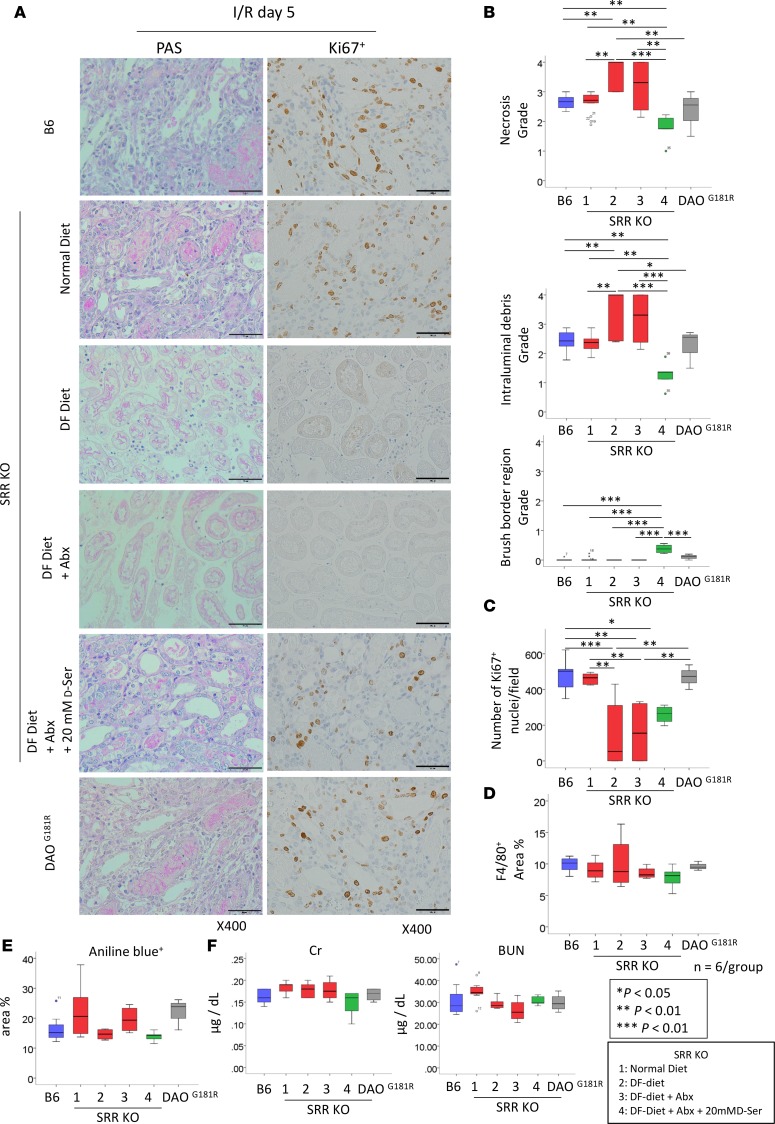
D-serine levels in the kidney can be regulated by DAO and SRR (16, 17). To elucidate the effect of intrarenal D-serine metabolism, we induced kidney I/R injury in SRR-knockout mice, which exhibited low levels of D-serine in the kidney (17). Additionally, we induced kidney I/R injury in mice with a natural variant (DAOG181R) that inactivates DAO, thereby leading to elevated levels of D-serine in the kidney (16). The extent of tubular injury and interstitial fibrosis were comparable among the DAOG181R, SRR-knockout, and control mice at 5 days after I/R injury (Figure 5). We then induced kidney I/R injury in SRR-knockout mice that were fed with a D–amino acid–free (DF) diet with or without Abx. Tubular injury was worse and the number of Ki67+ tubules was lower in SRR-knockout mice that were fed with a DF diet with or without Abx compared with those in B6 and SRR-knockout mice (Figure 5, A–C). Interestingly, the administration of 20 mM D-serine mended the kidney injury on day 5 after the I/R injury. We found that the D-serine administration did not change the percentage of F4/80+ cells, the number of Ki67+ tubules, the interstitial fibrosis, or the renal function induced by the I/R injury (Figure 5, D–F).
Figure 5. D-serine administration protects from AKI after I/R.
(A) Representative images of tissue samples stained with PAS and Ki67+ are shown. Scale bar: 50 μm. (B) The grades of necrosis and intraluminal debris are decreased and that of the brush border region is increased by the administration of D-serine at 5 days after I/R in the SRR-knockout mice compared with those in mice fed with DF diet with or without Abx. (C) The number of Ki67+ tubules are decreased in the SRR-knockout mice and those fed with DF diet with or without Abx. There are no differences in the percentage area positive for F4/80+ (D), percentage area positive for aniline blue (E), or serum creatinine (Cr) or serum blood urea nitrogen (BUN) (F) between the groups. The box-and-whisker plots represent the median values. Boxes represent the 25th and 75th percentiles, and whiskers depict the minimum and maximum values. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple comparison test (B–F). *P < 0.05, **P < 0.01, ***P < 0.001.
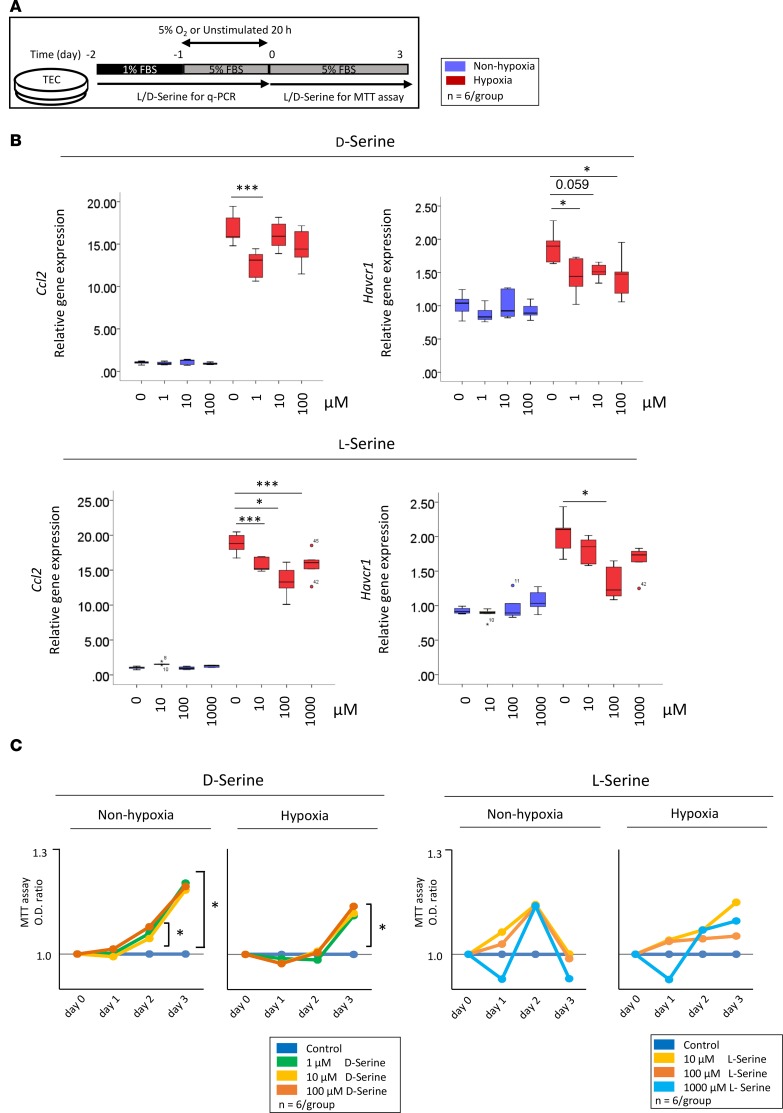
D-serine suppresses damage and promotes the hypoxia-mediated proliferation of tubular epithelial cells.
To elucidate the underlying mechanisms of D-serine–mediated renoprotection, we used tubular epithelial cells (TECs) stimulated with D-serine (Figure 6A). The mRNA expression levels of C-C motif chemokine ligand 2 (Ccl2) and Havcr1 were inhibited in hypoxia-exposed TECs at concentrations of 1–100 μM D-serine or 10–1000 μM L-serine (Figure 6B). The proliferation of posthypoxic TECs was accelerated by D-serine but not by L-serine (Figure 6C).
Figure 6. D-serine suppresses TEC damage and promotes proliferation in response to hypoxia.
(A) TECs are used to evaluate D-serine–mediated renoprotection. (B) The mRNA expression levels of Ccl2 and Havcr1 are attenuated in TECs treated with D-serine at concentrations of 1–100 μM. (C) D-serine accelerates the proliferation of posthypoxic TECs. The box-and-whisker plots represent the median values. Boxes represent the 25th and 75th percentiles, and whiskers depict the minimum and maximum values. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple comparison test (B and C). *P < 0.05, **P < 0.01, ***P < 0.001.
Despite the observed renoprotective effect of D-serine, studies reported that D-serine is associated with nephrotoxicity (18–20). Therefore, we explored the potential toxic effects of D-serine. The oral administration of D-serine at a dose of 80 mM led to the increasing of urine albumin excretion at 10 days after I/R injury in our murine model of AKI (Supplemental Figure 8A). Posthypoxic tubular proliferation was inhibited by D-serine at a dose of 100 mM (Supplemental Figure 8B).
Serum D-serine levels reflect kidney function in patients with AKI.
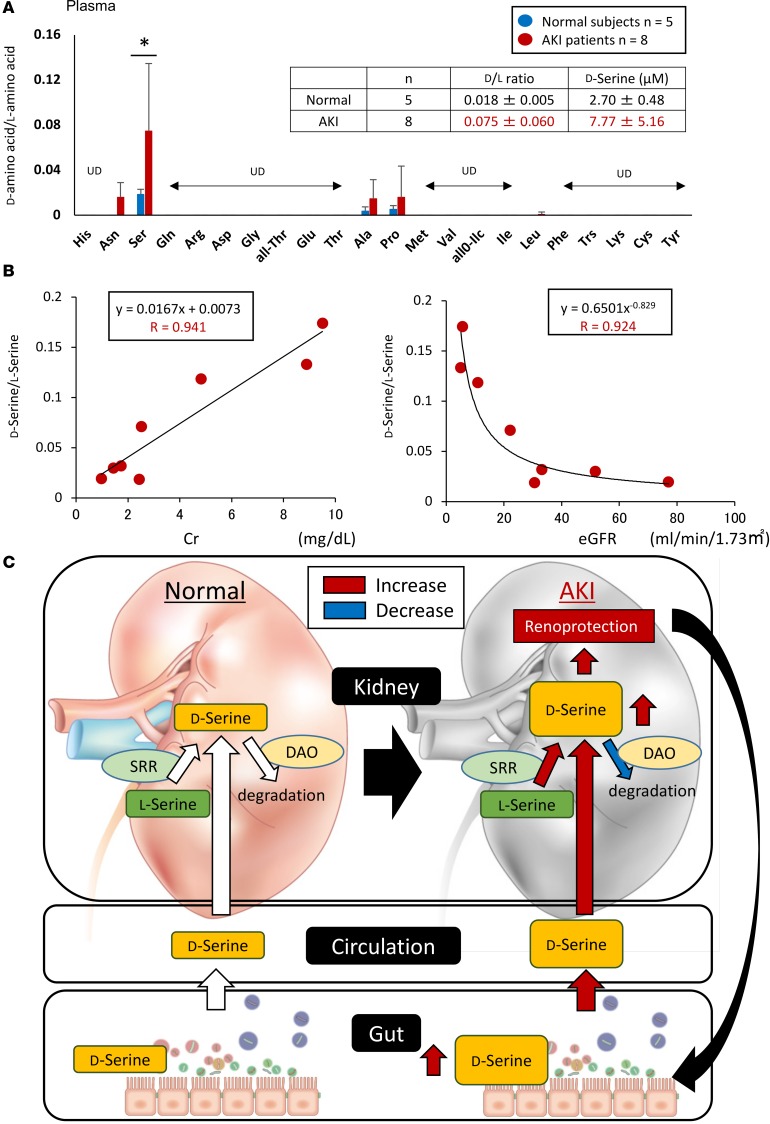
We assessed the association between D-serine and renal function in patients with AKI. D-serine, D-asparagine, D-alanine, D-proline, and D-leucine were detected in the plasma of AKI patients. The D-serine/L-serine ratio was significantly higher in the plasma of AKI patients than in the plasma of healthy subjects (Figure 7A). Moreover, the serum D-serine level was significantly correlated with an increase in creatinine and a decrease in estimated glomerular filtration rate (Figure 7B).
Figure 7. Serum levels of D-serine reflect kidney function in patients with AKI.
(A) D-serine, D-asparagine, D-alanine, D-proline, and D-leucine are detected in the plasma of AKI patients. Among these, the D-serine/L-serine ratio is increased in the plasma of AKI patients compared with that in healthy subjects. Data are shown as mean ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05. Normal subjects (n = 5) have a D-serine/L-serine ratio of 0.018 ± 0.005 and D-serine values of 2.70 ± 0.48 μm; AKI patients have a D-serine/L-serine ratio of 0.075 ± 0.060 and D-serine values of 7.77 ± 5.16 μm. (B) Serum D-serine levels are correlated with an increase in creatinine (Cr) and a decrease in estimated glomerular filtration rate (eGFR). (C) Proposed model of the relationship between gut-derived D-serine and the kidney based on the results of this study.
Discussion
This study revealed that the gut microbiota provided protection against tubular injury in the mouse kidney I/R injury model. AKI-induced gut dysbiosis altered the metabolism of D–amino acids in mice. Among the D–amino acids, only D-serine was detectable in the kidney. Furthermore, DAO activity was decreased and SRR activity was increased in the injured kidney after the I/R injury. Oral D-serine administration was protective in this mouse model of AKI. Furthermore, the DF diet with and without Abx led to the worsening of the kidney injury in the SRR-knockout mice. However, the administration of 20 mM D-serine mended the kidney injury on day 5 after the I/R. D-serine also suppressed hypoxia-induced tubular damage during the early phase of injury and promoted posthypoxic tubular cell proliferation. Finally, D-serine was correlated with the decrease in kidney function in AKI patients.
Accumulated data have demonstrated the significance of the gut microbiota in both health and disease (1–6). Our findings show that the gut microbiota also plays a central role in a mouse model of AKI. In support of our findings, the Gf B6 mice have been reported as suffering from severe kidney injury after I/R (21), whereas the depletion of gut microbiota was reported to protect the kidney from I/R injury (22). Although the precise underlying mechanisms are unclear, recent studies reported that microbial metabolites contributed to a healthy host. Among the bacterial metabolites, SCFA was reported to have protective effects in some disease models (8, 23), including AKI (9). In addition to SCFA, 2D HPLC has been used to reveal that D–amino acids were metabolized by the gut microbiota and were involved in intestinal mucosal defenses (12). The current study shows increased numbers of Lactobacillus, Clostridium, and Ruminococcus and decreased numbers of Bifidobacterium and TM7 after the I/R injury. Although several reports showed dysbiosis in chronic kidney disease, the changes in the gut bacteria during AKI remain unknown (4, 10). Moreover, the association between the activated pathways of D–amino acids, particularly D-serine, has not yet been investigated.
Although we detected D–amino acids in the feces of the normal mice with or without AKI, only D-asparagine and D-aspartic were found in the feces of the Gf B6 mice. These findings indicated that D–amino acids were metabolized by the gut microbiota, regardless of the kidney injury. Moreover, only D-serine was detected in the kidney, and the D-serine/L-serine ratio was increased in the feces, plasma, kidneys, and urine of the I/R-injured mice. Consistent with our results, Sasabe et al. (24) reported an increased D-serine/L-serine ratio in the sera of mice after I/R injury in the kidney.
D-serine administration was protective against AKI after the I/R injury. Our findings indicated that D-serine inhibited tubular damage and promoted tubular proliferation, thus leading to attenuated kidney injury after the I/R. Moreover, we demonstrated low DAO and high SRR activities in the injured kidney. Reduced DAO activity in the mouse models of AKI has been reported previously (24). However, the intrarenal metabolism of D-serine, along with the DAO deficiency, does not contribute to I/R-mediated kidney injury. Although alterations in DAO activity maintain D-serine levels in the injured kidney, this level of DAO activity might not be sufficient to protect against tubular injury. Therefore, the increased release of D-serine from the gut to the circulation might compensate for decreased D-serine levels in the injured kidney, thereby providing renoprotection. By contrast, the DF diet with or without Abx augmented the kidney injury, which was accompanied by a reduction in Ki67+ TECs in the SRR-knockout mice. The administration of 20 mM D-serine led to a recovery of kidney injury on day 5 after the I/R. However, the precise mechanisms of D-serine metabolism in AKI have not yet been investigated.
Several studies have reported the nephrotoxicity of D-serine (19). Although a peritoneal injection of 400–800 mg/kg D-serine induced tubular damage in rats (20, 22), Okada et al. (25) reported that 20 mM D-serine activated general control nonderepressible 2 kinase, thus leading to cellular senescence in human tubular cells. In agreement with those studies, we observed D-serine cytotoxicity in the current study. Although 80 mM D-serine augmented the kidney injury, 1–100 μM D-serine induced TEC proliferation in our model. These results strongly suggest that the dose of D-serine is associated with specific responses in TECs. Given that 2D HPLC provides more accurate information than conventional methods, further investigation is necessary to reveal the association between D-serine concentration and in vivo and in vitro tissue responses.
The D-serine/L-serine ratio was also increased in patients with AKI. Moreover, the ratio was inversely correlated with renal function. Although plasma D-serine levels were previously evaluated in human chronic kidney disease (26), the levels of D–amino acids were not evaluated in human AKI. Our results suggest that D–amino acids, particularly D-serine, can be potential biomarkers for AKI. The possibility remains that D-serine levels were affected by renal function because the excretion rate of D-serine was not evaluated in the current study. However, the D-serine/L-serine ratio was increased in the feces, plasma, kidneys, and urine of the I/R-injured mice compared with that in the sham-operated mice (Figure 3C). Following the administration of 20 mM D- or L-serine through drinking water, the plasma concentration of L-serine did not change, whereas the plasma D-serine levels increased to 100 nM (Figure 4A). This finding suggested that D-serine was absorbed through the gastrointestinal tract and elevated the plasma D-serine levels compared with the I/R-injured mice that did not receive D-serine. Furthermore, these results indicated that the gut microbiota might have contributed, at least in part, to the high levels of serum D-serine after I/R injury. Further studies are needed to clarify the microbial species that metabolize D-serine.
This study demonstrated the renoprotective effect of gut microbiota–derived D-serine in AKI (Figure 7C). Given the biological roles of D–amino acids produced by the gut microbiota, the present data shed light on the mechanisms that can explain the interaction between the gut microbiota and the kidney and illustrate that D–amino acids are potential therapeutic targets and biomarkers for AKI.
Methods
Mice.
B6 mice were purchased from CLEA Japan. The mice were housed and bred at Kanazawa University. The Gf B6 mice were obtained from CLEA Japan. All procedures for the Gf B6 mice were performed in a Gf isolator at CLEA Japan. To confirm the Gf B6 status, the 16S rDNA PCR of feces was performed at CLEA Japan before and after the procedures (27) (Supplemental Figure 9). As described previously, DAOG181R and SRR-knockout mouse lines were generated and maintained on a B6 background (28, 29).
Fecal microbiota transplantation was performed 12 weeks before the I/R injury. Donor B6 mice were euthanized, and the small intestines, ceca, and colons were removed under aseptic conditions. The content was diluted in 1 ml PBS, and 200 μl of the content was administered via the anus into the rectum. The transplanted Gf B6 mice were then cohoused with normal B6 mice for 12 weeks.
To model gut microbiota depletion with antibiotics, B6 mice were treated with 1 g/l ampicillin sodium salt (catalog A0166; MilliporeSigma), 1 g/l neomycin sulfate (catalog 146-08871; Wako Pure Chemical Industries), 1 g/l metronidazole (catalog M3761; MilliporeSigma), 0.5 g/l vancomycin hydrochloride (catalog 222-01303; Wako), and 0.5 g/l gentamicin sulfate (catalog G0383; TCI) in drinking water for 12 weeks prior to the I/R injury.
I/R injury.
Ischemia was induced by clamping the renal pedicle with a nontraumatic clip (Natsume Seisakusho) after the induction of anesthesia (30). After 40 minutes, the clip was removed. Body temperature was controlled at 37.0°C throughout the procedure.
Renal histopathology.
The kidneys were fixed in 10% neutral buffered formalin and embedded in paraffin; the paraffin sections were stained with periodic acid–Schiff (PAS) and Azan reagent. The PAS-stained debris at the corticomedullary junction or the brush border (i.e., corticomedullary junction and cortical region) was quantified from at least 10 different kidney sections. The acute tubular necrosis score was graded from 0 to 4 (0, none; 1, mild; 2, moderate; 3, severe; 4, necrosis) on the basis of proximal tubule dilation, brush border damage, the presence of proteinaceous casts, interstitial widening, and necrosis. The specimens were evaluated by evaluators who were blinded to the mouse strains and treatment conditions. Staining for Ki67 and F4/80 was performed as described previously (30, 31).
Murine TEC line.
The murine TEC line mProx24 was a gift by Takeshi Sugaya (St. Marianna University School of Medicine, Tokyo, Japan). The cells were cultured in DMEM supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin.
RNA isolation and quantitative real-time PCR.
Total RNA was isolated from the cultured cells and the kidneys by using the High Pure RNA isolation kit (Roche Diagnostics) and the ISOSPIN cell and tissue RNA (NIPPON Gene), respectively. Quantitative real-time PCR with iQ SYBR Green Supermix (catalog 170-8885; Bio-Rad) was performed using the Villa 7 Real-Time PCR System (Thermo Fisher Scientific). The following primers were used: Ccl2, 5′-CTTCCTCCACCACCATGCA-3′ and 5′-CCAGCCGGCAACTGTGA-3′; Havcr1, 5′-AGGAAGACCCACGGCTATTT-3′ and 5′-TGTCACAGTGCCATTCCAGT-3′ (catalog Mm00506686; Applied Biosystems TaqMan Gene Expression Assays). Data were analyzed using the ΔΔCt method (32).
Cell proliferation assay.
TEC proliferation was determined with the Cell Counting Kit-8 (catalog CK04; Dojindo) according to the manufacturer’s instructions (33).
Bacterial 16S rRNA amplicon sequencing and analysis.
We performed 16S rRNA gene-sequencing analysis of the mouse and human gut microbiota (34, 35). Bacterial genomic DNA was isolated from the feces by using the enzymatic lysis method. The isolated DNA (40 ng) was used for PCR for the V1–V2 hypervariable regions of the 16S rRNA gene by using the universal primers 27Fmod (5′-AGRGTTTGATYMTGGCTCAG-3′) and 338R (5′-TGCTGCCTCCCGTAGGAGT-3′) with 10-bp barcode tags (34). Thermal cycling was performed with a 9700 PCR system (Life Technologies Japan) by using Ex Taq polymerase (Takara Bio) with the following cycling conditions: initial denaturation at 96°C for 2 minutes; 25 cycles of denaturation at 96°C for 30 seconds, annealing at 55°C for 45 seconds, and extension at 72°C for 1 minute; and final extension at 72°C. All amplicons were purified using AMPure XP magnetic purification beads (Beckman Coulter), quantified using the Quant-iT PicoGreen dsDNA Assay kit (Life Technologies Japan), and sequenced on a 454 Genome Sequencer FLX Titanium platform (Roche Diagnostics). Reads with average quality values of <25 and inexact matches to the universal primer sequences were filtered; 3,000 reads that passed the quality filter were randomly selected from each sample and subjected to downstream analyses. The selected reads were rearranged in descending order according to the quality value (36) and clustered into operational taxonomic units with a 96% pairwise identity cutoff by using the UCLUST program (37) version 5.2.32 (http://www.drive5.com/). The taxonomic assignment of each operational taxonomic unit was determined by similarity searching against the Ribosomal Database Project and the NCBI genome database by using the GLSEARCH program. The 16S rRNA gene V1–V2 region sequences analyzed in the current study were deposited in DDBJ/GenBank/EMBL with accession DRA006069.
Determination of chiral amino acid by 2D HPLC.
The concentrations of D- and L–amino acids were evaluated with the NANOSPACE SI-2 2D HPLC system (Shiseido) (11, 38). The NBD–amino acids were isolated using a KSAARP column (1.0 mm i.d. × 500 mm, an ODS column designed by collaboration with Kyushu University and Shiseido) and an online fraction collecting system in the first dimension. The isolated fractions were automatically transferred to the second dimension, which consisted of a narrow-bore enantio-selective KSAACSP-001S column (250 mm × 1.5 mm i.d., 25 C; prepared in collaboration with Shiseido), to determine the D- and L-enantiomers. The mobile phases for the second dimension were mixed solutions of MeOH and MeCN that contained formic acid. The fluorescence detection of the NBD–amino acids was performed at 530 nm with an excitation at 470 nm.
Chemicals.
D-serine, L-serine, D-alanine, flavin adenine dinucleotide (FAD) disodium salt n-hydrate, and pyridoxal phosphate monohydrate were purchased from Wako. L-(-)-threo-3-hydroxyaspartic acid (L-THA) was purchased from Tocris Bioscience. All other chemicals were of the highest grade available and were purchased from reputable commercial sources: boric acid (Wako), potassium chloride (Wako), 2-mercaptoethanol (Wako), Glycerol (Wako), trichloroacetic acid (Wako), 2,4-dinitro phenylhydrazine (Wako), dithiothreitol (Nacalai Tesque), ATP disodium salt n-hydrate (Wako), and MgCl2 hexahydrate (Wako).
Preparation of mouse kidney homogenates.
The kidneys were homogenized in a Potter-Elvehjem homogenizer in 5 volumes of cold 10 mM borate buffer (pH 8.5) containing protease inhibitors (Nacalai Tesque). The homogenates were dialyzed for 1 day at 4°C against 1 l of 10 mM borate buffer (pH 8.5) containing 5 mM 2-mercaptoethanol and 10% (v/v) glycerol. The buffer was changed once during dialysis. The dialyzed homogenates were recovered and centrifuged at 20,000 g for 10 minutes at 4°C to pellet any denatured proteins during the dialysis. The supernatants were recovered and stored at −80°C until use.
Enzyme activity assays.
The protein concentration of kidney homogenates was determined with the Bio-Rad protein assay kit by using bovine serum albumin (MilliporeSigma) as the standard. The catalytic activity of DAO (EC 1.4.3.3) was determined using a colorimetric method that measured 2-oxo acid production (39). Appropriate amounts (0.42–1.3 mg) of the kidney homogenates were added to a reaction mixture containing air-saturated 50 mM borate buffer (pH 8.5), 60 μM FAD, 33 ng/ μl catalase from Aspergillus niger (MilliporeSigma), and 20 mM D-alanine to a final volume of 150 μl. The reaction mixture was incubated at 37°C for 30 minutes prior to the addition of 10 μl 100% (w/v) trichloroacetic acid to stop the reaction. The 2-oxo acid products were reacted with 2,4-dinitrophenylhydrazine and quantified by measuring the absorbance at 445 nm against a blank mixture that lacked amino acids.
The activity of SRR in the kidney homogenates was determined by a colorimetric method that measured 2-oxo acid production from L-THA (40) because the production of D-serine from L-serine (or vice versa) was under the detection limit by HPLC. SRR is a multifunctional enzyme that exhibits racemase activity and dehydratase activity, which is higher than the racemase activity and is highest with L-THA (41). Appropriate amounts (0.42–1.3 mg) of the kidney homogenates were added to a reaction mixture containing air-saturated 50 mM borate buffer (pH 8.5), 50 μM pyridoxal phosphate, 1 mM dithiothreitol, 1 mM ATP, 1 mM MgCl2, and 20 mM L-THA to a final volume of 150 μl. The reaction mixture was incubated at 37°C for 30 minutes or 1 hour prior to the addition of 10 μl 100% (w/v) trichloroacetic acid to stop the reaction. The 2-oxo acid products were reacted with 2, 4-dinitrophenylhydrazine and were quantified by measuring the absorbance at 445 nm against a blank mixture that lacked amino acids.
Human samples.
We obtained blood samples from patients with AKI. Supplemental Table 1 summarizes the clinical characteristics of AKI patients. Patients who were treated with immunosuppressive drugs and antibiotics were excluded. All samples were collected from 2013 to 2017 at Kanazawa University Hospital.
Statistics.
The data are presented as mean ± SEM, as determined by SPSS Statistics (ver. 23, IBM). Statistical analysis was performed by using a 2-tailed unpaired Student’s t test to compare 2 groups and by using 1-way ANOVA with Tukey’s multiple comparison test to compare more than 2 groups. P values of less than 0.05 were considered significant.
Study approval.
This study was approved by the Ethics Committee of Kanazawa University Hospital (IRB approval 1291) and was conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent and were informed about their right to withdraw from the study at any time. All animals were maintained and used in accordance with the guidelines of Kanazawa University, and the experiments were conducted under study approval no. AP-153653 and AP-163726 from Kanazawa University.
Author contributions
YN, YI, and TW designed and performed the experiments. KF, T. Miyake, NS, S. Kitajima, TT, YS, AS, T. Miyagawa, AH, MS, YK, KS, MO, SYN, and YY collected the samples and analyzed the clinical data. MM, KH, and RK performed HPLC quantify the chiral animo acids and gifted KO mice (DAO mice and SRR mice) to our laboratory. MM and KH performed HPLC to quantify the chiral amino acids. TM, MK, and HH evaluated the activities of DOA and SRR. HM, MH, and WS performed the metagenome analysis of the gut microbiota. S. Kaneko provided advice on critical revision. YN, YI, and TW wrote the manuscript. TW supervised the work.
Supplementary Material
Acknowledgments
We would like to thank Takeshi Sugaya for providing the mProx24 cell line. We also thank Maiko Nakane and Yurika Miyoshi for their assistance. This work was supported by Japan Society for the Promotion of Science KAKENHI grant 16K15326.
Version 1. 10/18/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: JCI Insight. 2018;3(17):e97957. https://doi.org/10.1172/jci.insight.97957.
Contributor Information
Yusuke Nakade, Email: nakade-yusuke@umin.ac.jp.
Yasunori Iwata, Email: yiwata7191@gmail.com.
Kengo Furuichi, Email: kfuruichi@m-kanazawa.jp.
Masashi Mita, Email: masashi.mita@to.shiseido.co.jp.
Kenji Hamase, Email: hamase@phar.kyushu-u.ac.jp.
Ryuichi Konno, Email: konno-rhcc@cc9.ne.jp.
Taito Miyake, Email: taito06090713@gmail.com.
Norihiko Sakai, Email: norin0826@yahoo.co.jp.
Shinji Kitajima, Email: kitajimajima@ybb.ne.jp.
Tadashi Toyama, Email: toyama@mbj.nifty.com.
Yasuyuki Shinozaki, Email: shinopostbox@gmail.com.
Akihiro Sagara, Email: akihiroskidney@yahoo.co.jp.
Taro Miyagawa, Email: ns50f1982@gmail.com.
Akinori Hara, Email: ahara@m-kanazawa.jp.
Miho Shimizu, Email: mshimizu@staff.kanazawa-u.ac.jp.
Yasutaka Kamikawa, Email: yasutakamikawa@gmail.com.
Kouichi Sato, Email: ksksks590307@yahoo.co.jp.
Megumi Oshima, Email: mgm_oshima@yahoo.co.jp.
Shiori Yoneda-Nakagawa, Email: yonedayone@gmail.com.
Yuta Yamamura, Email: yuta.yamamura@gmail.com.
Shuichi Kaneko, Email: skaneko@m-kanazawa.jp.
Tetsuya Miyamoto, Email: miyamotot@pharm.kitasato-u.ac.jp.
Masumi Katane, Email: katanem@pharm.kitasato-u.ac.jp.
Hiroshi Homma, Email: hommah@pharm.kitasato-u.ac.jp.
Hidetoshi Morita, Email: hidetoshi-morita@okayama-u.ac.jp.
Wataru Suda, Email: wataru_suda@cb.k.u-tokyo.ac.jp.
Masahira Hattori, Email: m-hattori@aoni.waseda.jp.
Takashi Wada, Email: twada@m-kanazawa.jp.
References
- 1.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53(1):1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 3.Jonsson AL, Bäckhed F. Role of gut microbiota in atherosclerosis. Nat Rev Cardiol. 2017;14(2):79–87. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 4.Evenepoel P, Poesen R, Meijers B. The gut-kidney axis. Pediatr Nephrol. 2017;32(11):2005–2014. doi: 10.1007/s00467-016-3527-x. [DOI] [PubMed] [Google Scholar]
- 5.Kelly CR, et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent clostridium difficile infection: a randomized trial. Ann Intern Med. 2016;165(9):609–616. doi: 10.7326/M16-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landy J, et al. Variable alterations of the microbiota, without metabolic or immunological change, following faecal microbiota transplantation in patients with chronic pouchitis. Sci Rep. 2015;5:12955. doi: 10.1038/srep12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vrieze A, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–6.e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 8.Papatriantafyllou M. Regulatory T cells: distilling regulatory T cell inducers. Nat Rev Immunol. 2013;13(8):546. doi: 10.1038/nri3506. [DOI] [PubMed] [Google Scholar]
- 9.Andrade-Oliveira V, et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol. 2015;26(8):1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briskey D, Tucker P, Johnson DW, Coombes JS. The role of the gastrointestinal tract and microbiota on uremic toxins and chronic kidney disease development. Clin Exp Nephrol. 2017;21(1):7–15. doi: 10.1007/s10157-016-1255-y. [DOI] [PubMed] [Google Scholar]
- 11.Koga R, et al. Enantioselective two-dimensional high-performance liquid chromatographic determination of N-methyl-D-aspartic acid and its analogues in mammals and bivalves. J Chromatogr A. 2012;1269:255–261. doi: 10.1016/j.chroma.2012.08.075. [DOI] [PubMed] [Google Scholar]
- 12.Sasabe J, et al. Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat Microbiol. 2016;1(10):16125. doi: 10.1038/nmicrobiol.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiriyama Y, Nochi H. D-Amino acids in the nervous and endocrine systems. Scientifica (Cairo) 2016;2016:6494621. doi: 10.1155/2016/6494621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariyoshi M, et al. D-Glutamate is metabolized in the heart mitochondria. Sci Rep. 2017;7:43911. doi: 10.1038/srep43911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue R, et al. Localization of serine racemase and its role in the skin. J Invest Dermatol. 2014;134(6):1618–1626. doi: 10.1038/jid.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi Y, et al. Simultaneous two-dimensional HPLC determination of free D-serine and D-alanine in the brain and periphery of mutant rats lacking D-amino-acid oxidase. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(29):3184–3189. doi: 10.1016/j.jchromb.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Horio M, et al. Levels of D-serine in the brain and peripheral organs of serine racemase (Srr) knock-out mice. Neurochem Int. 2011;59(6):853–859. doi: 10.1016/j.neuint.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Montesinos Guevara C, Mani AR. The role of D-serine in peripheral tissues. Eur J Pharmacol. 2016;780:216–223. doi: 10.1016/j.ejphar.2016.03.054. [DOI] [PubMed] [Google Scholar]
- 19.Maekawa M, Okamura T, Kasai N, Hori Y, Summer KH, Konno R. D-amino-acid oxidase is involved in D-serine-induced nephrotoxicity. Chem Res Toxicol. 2005;18(11):1678–1682. doi: 10.1021/tx0500326. [DOI] [PubMed] [Google Scholar]
- 20.Orozco-Ibarra M, et al. Evaluation of oxidative stress in D-serine induced nephrotoxicity. Toxicology. 2007;229(1-2):123–135. doi: 10.1016/j.tox.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Jang HR, Gandolfo MT, Ko GJ, Satpute S, Racusen L, Rabb H. Early exposure to germs modifies kidney damage and inflammation after experimental ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2009;297(5):F1457–F1465. doi: 10.1152/ajprenal.90769.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emal D, et al. Depletion of gut microbiota protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2017;28(5):1450–1461. doi: 10.1681/ASN.2016030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques FZ, et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017;135(10):964–977. doi: 10.1161/CIRCULATIONAHA.116.024545. [DOI] [PubMed] [Google Scholar]
- 24.Sasabe J, et al. Ischemic acute kidney injury perturbs homeostasis of serine enantiomers in the body fluid in mice: early detection of renal dysfunction using the ratio of serine enantiomers. PLoS One. 2014;9(1):e86504. doi: 10.1371/journal.pone.0086504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada A, et al. D-serine, a novel uremic toxin, induces senescence in human renal tubular cells via GCN2 activation. Sci Rep. 2017;7(1):11168. doi: 10.1038/s41598-017-11049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura T, et al. Chiral amino acid metabolomics for novel biomarker screening in the prognosis of chronic kidney disease. Sci Rep. 2016;6:26137. doi: 10.1038/srep26137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto K, Kuwayama E, Nozu R, Ueno M, Hayashimoto N. Effect of hypochlorous acid solution on the eradication and prevention of Pseudomonas aeruginosa infection, serum biochemical variables, and cecum microbiota in rats. Exp Anim. 2015;64(2):191–197. doi: 10.1538/expanim.14-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyoshi Y, et al. Alteration of intrinsic amounts of D-serine in the mice lacking serine racemase and D-amino acid oxidase. Amino Acids. 2012;43(5):1919–1931. doi: 10.1007/s00726-012-1398-4. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi Y, Hamase K, Tojo Y, Mita M, Konno R, Zaitsu K. Determination of D-serine and D-alanine in the tissues and physiological fluids of mice with various D-amino-acid oxidase activities using two-dimensional high-performance liquid chromatography with fluorescence detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(24):2506–2512. doi: 10.1016/j.jchromb.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 30.Iwata Y, et al. Aberrant macrophages mediate defective kidney repair that triggers nephritis in lupus-susceptible mice. J Immunol. 2012;188(9):4568–4580. doi: 10.4049/jimmunol.1102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menke J, et al. CSF-1 signals directly to renal tubular epithelial cells to mediate repair in mice. J Clin Invest. 2009;119(8):2330–2342. doi: 10.1172/JCI39087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwata Y, et al. Down-regulation of the two-component system and cell-wall biosynthesis-related genes was associated with the reversion to daptomycin susceptibility in daptomycin non-susceptible methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2017;36(10):1839–1845. doi: 10.1007/s10096-017-2999-3. [DOI] [PubMed] [Google Scholar]
- 33.Iwata Y, et al. Involvement of CD11b+ GR-1 low cells in autoimmune disorder in MRL-Fas lpr mouse. Clin Exp Nephrol. 2010;14(5):411–417. doi: 10.1007/s10157-010-0309-9. [DOI] [PubMed] [Google Scholar]
- 34.Kim SW, et al. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013;20(3):241–253. doi: 10.1093/dnares/dst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ueno, M. Kikuchi M, Oshima K, Kim SW, Morita H, Hattori M. Assessment and improvement of methods for microbial DNA preparation from faecal samples. In: de Bruijn FJ, ed. Handbook of Molecular Microbial Ecology II: Metagenomics in Different Habitats. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2010:191–198. [Google Scholar]
- 36.Said HS, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014;21(1):15–25. doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 38.Hamase K, et al. Simultaneous determination of hydrophilic amino acid enantiomers in mammalian tissues and physiological fluids applying a fully automated micro-two-dimensional high-performance liquid chromatographic concept. J Chromatogr A. 2010;1217(7):1056–1062. doi: 10.1016/j.chroma.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Katane M, Seida Y, Sekine M, Furuchi T, Homma H. Caenorhabditis elegans has two genes encoding functional d-aspartate oxidases. FEBS J. 2007;274(1):137–149. doi: 10.1111/j.1742-4658.2006.05571.x. [DOI] [PubMed] [Google Scholar]
- 40.Katane M, et al. Characterization of a homologue of mammalian serine racemase from Caenorhabditis elegans: the enzyme is not critical for the metabolism of serine in vivo. Genes Cells. 2016;21(9):966–977. doi: 10.1111/gtc.12398. [DOI] [PubMed] [Google Scholar]
- 41.Strísovský K, Jirásková J, Mikulová A, Rulísek L, Konvalinka J. Dual substrate and reaction specificity in mouse serine racemase: identification of high-affinity dicarboxylate substrate and inhibitors and analysis of the beta-eliminase activity. Biochemistry. 2005;44(39):13091–13100. doi: 10.1021/bi051201o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.