Abstract
Our understanding of the molecular pathogenesis of childhood cancers has advanced substantially, but their fundamental causes remain poorly understood. Recently, multiple mechanisms of DNA damage and repair have been associated with mutations observed in human cancers. Here, we review the physiologic functions and oncogenic activities of transposable genetic elements. In particular, we focus on the recent studies implicating DNA transposases RAG1/2 and PGBD5 as oncogenic mutators that promote genomic rearrangements in childhood leukemias and solid tumors. We outline future studies that will be needed to define the contributions of transposons to mutational processes that become dysregulated in cancer cells. In addition, we discuss translational approaches, including synthetic lethal strategies, for identifying and developing improved clinical therapies to target oncogenic transposons and transposases.
Introduction
Childhood cancers are diverse, originating from different tissue types, cell lineages and differentiation states, and, increasingly, genetic mutations. Childhood cancer is the most common disease-related cause of death in children under the age of 15 (1). Despite recent advances in the understanding of the molecular pathogenesis of childhood cancers, their fundamental causes and how this knowledge can be used to improve clinical outcomes remain poorly understood. Environmental exposures, such as infections, have been proposed to contribute to childhood cancer development, but in most cases, these associations appear relatively weak or lacking in biologic plausibility (2). Major efforts over the years have focused on defining genetic mutations in human tumors to better understand their origin. Targeting therapy to tumor-specific mutations holds the promise of improved precision and efficacy in eradicating cancer cells, while sparing patients the acute and long-term sequelae of cytotoxic chemo- and radiotherapy. However, genome-wide studies are revealing striking differences in the prevalence of somatic mutations among tumor types; they are numerous among adult cancers, such as melanomas, and relatively infrequent among cancers of infants and young children, such as rhabdoid tumors (3, 4).
Despite the relative paucity of recurrent gene mutations, recent genomic analyses have begun to identify the essential functions of genetic rearrangements that affect noncoding elements in some pediatric tumors (5–8). In addition, recurrent chromosomal and complex genomic rearrangements, including chromothripsis, double minute chromosomes, and others, are observed in many childhood tumors (9, 10). Indeed, how genomic rearrangements observed in some pediatric tumors can occur at such a young age in otherwise genetically stable cells remains a conundrum. In contrast with adult cancers, most childhood tumors lack apparent mutations of TP53 and other canonical tumor-suppressor genes regulating genome stability (11). Whereas defects in DNA damage repair have been suggested to explain the increased incidence of some cancers in relatively young people, the causes of complex genomic rearrangements in most cancers in young children remain largely undefined.
Here, we review the evidence that endogenous nucleases related to DNA transposases contribute to the formation of genomic rearrangements in childhood tumors by acting as oncogenic mutators. In addition to discussing their functions in childhood tumorigenesis, we discuss actionable therapeutic strategies arising from synthetic lethality resulting from their nuclease activities. Finally, we propose future studies that take advantage of forward genetic screens, developmentally accurate animal models, and improved techniques for genome analysis to define other oncogenic mutators that may contribute to the development of childhood cancers. We omit the discussion of germline activity of transposable elements with respect to cancer predisposition (12–14).
Active human transposases
Transposons are mobile genetic elements that are found in nearly all living organisms and comprise about half of the human genome (12, 15, 16). Generally, these elements are classified as RNA-based transposons related to retroviruses (17) and “cut-and-paste” or “copy-and-paste” DNA transposons (12, 18). The latter are substrates for a large family of DNA-dependent nucleases, termed “DNA transposases” or “DNA recombinases.” Many DNA transposases use an RNase H-like domain that catalyzes hydrolysis to accomplish DNA excision, strand exchange, and site-specific integration (19–21). Although the majority of genes that encode transposase enzymes tend to become catalytically inactive and most transposon substrates tend to become immobile in the course of evolution, some can maintain their activities (18).
The Drosophila telomere retroelements HeT-A, TART, and TAHRE were the first active RNA transposable elements found to have cellular physiologic functions (22). Human telomerase and retrotransposon reverse transcriptase also exhibit a number of structural and mechanistic similarities and likely share a common origin (22–24). Similar to retrotransposons, human telomerase uses RNA templates to synthesize DNA at telomeres to maintain chromosomal integrity. This prevents the activation of aberrant DNA damage repair and resultant mitotic failure and genomic instability. Indeed, many tumors, including childhood tumors, rely on telomerase overexpression to maintain telomeres, frequently caused by activating somatic promoter mutations (25). It is unknown whether, in addition to maintaining telomere repeats, oncogenic telomerase activation also acts at other genetic loci. Nonetheless, telomerase is a domesticated retrotransposon that can be aberrantly activated in human cancer.
Other retrotransposons, most notably some long interspersed element-1 (LINE-1) transposons, remain active in human cells, including neural progenitor cells (26), and several human cancers (27–29). This activity can promote mobilization of LINE-1 transposons themselves as well as Alu and short interspersed element transposons. Indeed, the first discovered example of disease-associated retrotransposition was a de novo LINE-1 insertion in the F8 clotting factor gene, leading to familial hemophilia (30). In cancer, most of the observed transposition events appear genetically neutral, but at least some are clonal and can cause inactivating mutations of tumor suppressor genes (31, 32).
In addition to retrotransposons, the human genome also contains over 20 genes with recognizable similarity to DNA transposases, some of which remain active (12) (Table 1). For example, the human Transib-like DNA transposase RAG1/2 catalyzes somatic recombination of the Ig and T cell receptor genes in lymphocytes (33, 34). RAG1 endonuclease and its cofactor RAG2 are targeted to the variable, diversity, and joining [V(D)J] sites by specific binding of the recombination signal sequences (RSSs) consisting of a highly conserved heptamer (CACAGTG) and a less conserved nonamer (ACAAAAACC) motif, separated by 12- or 23-bp sequence–independent spacers (35) (Figure 1). RAG1/2 cleaves DNA at the boundary between the RSSs and the flanking coding sequence, thereby generating 2 blunt and 2 hairpin ends (35). Processing of these ends can involve the addition of nontemplated sequences at the breakpoint by terminal deoxynucleotidyl transferase in a process that results in somatic diversification of the V(D)J loci (36), which allows for the development of lymphocytes capable of recognizing a diverse range of antigens. Although DNA transposition by RAG1/2 can be observed with engineered reactions in vitro, this activity is almost completely impaired in cells (33). Thus, RAG1/2 is a domesticated DNA transposase that has evolved to carry out excision of endogenous transposon-like substrates in a coordinated manner to contribute to immune receptor diversification.
Table 1. Known human transposase-derived genes and their observed activities.
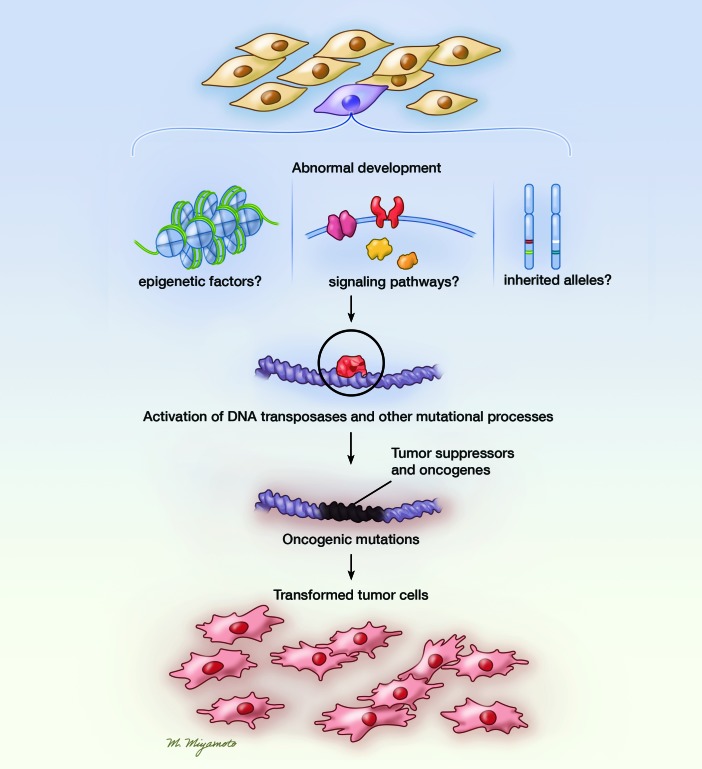
Figure 1. Schematic delineating how endogenous mutational processes, including activities of endogenous DNA transposases, can contribute to the transformation of healthy progenitor/stem cells into tumor cells.
Dysregulation of progenitor cells by abnormal developmental and epigenetic factors as well as other unknown processes (e.g., inherited alleles) leads to the activation of endogenous mutational processes, including oncogenic DNA transposase mutators, leading to mutations that initiate and/or cooperate to induce tumors. Illustrated by Mao Miyamoto.
Another example for a DNA transposase that has retained some of its original catalytic activity is SETMAR/METNASE. The Mariner-derived transposase SETMAR functions in single-stranded DNA resection during DNA repair and replication in human cells and has been found to catalyze DNA transposition in vitro but not in cells (37, 38). Thus, the human SETMAR protein preserves most, but not all, of the activities of the ancestral Hsmar1 transposase and has acquired new functions contributing to human cellular DNA repair (38). SETMAR appears to promote chromosome decatenation (38), DNA replication, and chromosomal stability. Because SETMAR is one of many cellular endonucleases that can resolve damaged DNA ends, such as ARTEMIS, which resolves RAG1/2-induced hairpins (39), its specific functions in DNA damage repair still need further definition. In addition to RAG1/2 and SETMAR, human THAP9 was also recently found to support “cut-and-paste” DNA transposition in human cells (40). THAP9 is related to the Drosophila P-element transposase and can mobilize Drosophila P-elements in both fly and human cells. The physiologic and potential pathophysiologic functions of THAP9 are currently unknown.
In addition, the human genome contains five paralogous genes, PGBD1–5, derived from piggyBac-related DNA transposases (41, 42). PGBD1 and PGBD2 appear to have invaded a common mammalian ancestor, whereas PGBD3 and PGBD4 are restricted to primates. PGBD1–4 all exist as single coding exons, fused in frame with endogenous host genes (15). One such transposase-derived fusion gene is the Cockayne syndrome B gene–PGBD3 (CSB-PGBD3) (42). CSB-PGBD3 is capable of binding human endogenous piggyBac-like transposon sequences but does not appear to have catalytic activity, though biochemical and genetic evidence indicates that it, similar to SETMAR, may participate in the DNA damage response (43, 44).
PGBD5 appears to be distinct from other human piggyBac-derived genes. First, PGBD5 was domesticated much earlier in vertebrate evolution, approximately 500 million years ago (42, 45). Furthermore, PGBD5 expression appears largely restricted to the early embryo, certain areas of the embryonic and adult brain, as well as embryonic solid tumors (45–47). Remarkably, PGBD5 encodes an enzymatically active DNA transposase, and 3 aspartic acid residues in its RNase H-like transposase domain are required for enzymatic nuclease activity similar to that of the looper moth piggyBac DNA transposase (46). PGBD5 catalyzes DNA transposition of synthetic piggyBac substrates in human cells, with a preference for TTAA sites genome wide (46). It is currently unknown whether PGBD5 can perform DNA transposition of endogenous human genomic DNA elements and whether it has physiologic functions related to its enzymatic nuclease activity in vertebrate neurons.
RAG1/2 induces oncogenic chromosomal translocations and deletions of tumor suppressor genes in lymphoblastic leukemias and lymphomas
In 1946, Barbara McClintock first suggested that transposons could cause chromosomal breakage in plants (48–51), which was subsequently supported by findings that transposons can promote chromosomal aberrations, such as translocations, inversions, deletions, duplications, and the formation of chromosomal fragments (52). The first evidence of transposase-induced chromosomal aberrations in humans was found in acute lymphoblastic leukemias (ALLs). ALL is caused by recurrent chromosomal translocations and deletions, with some occurring during fetal development (53). Recent genomic profiling studies have found copy number aberrations (CNAs) to be common in ALL (54, 55). Because functioning heptamer or nonamer sequence motifs outside the immunoglobulin receptor loci have been associated with alternative mechanisms of RAG1/2 recruitment and subsequent genomic rearrangements (56, 57), it has been proposed that aberrant RAG1/2 activity might contribute to leukemogenesis (Figure 2). Indeed, some CNAs in ALL cells have apparent V(D)J recombination sequence motifs close to the CNA breakpoints (58). This notion was further supported by the identification of RAG1/2 recognition sequences near the breakpoint junctions of CNAs and chromosomal translocations in ETV6-RUNX1 leukemias (59). Interestingly, RAG1/2 continues to be highly expressed by ETV6-RUNX1 leukemia cells, resulting in diverse and ongoing oligoclonal V(D)J rearrangements (60). It remains to be formally determined whether RAG1/2 expression is sufficient to induce CNAs and leukemic mutations. Nonetheless, it is highly probable that RAG1 and RAG2 are transposase-derived genes with oncogenic mutator functions in lymphoid cancers. In addition, RAG1/2 is required for leukemia development in genetically engineered mouse models, at least in part in response to inflammatory signaling (61). Intriguingly, this suggests that immune signaling and physiologic states may contribute to the dysregulation of RAG1/2 activity, contributing to the induction of leukemogenic mutations.
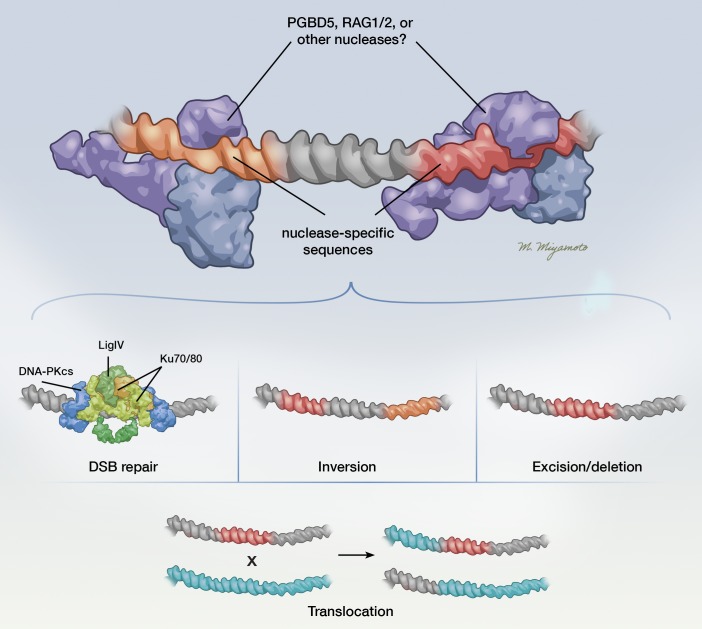
Figure 2. Possible mechanisms by which PGBD5 and RAG1/2 induce genomic rearrangements.
PGBD5 and RAG1/2 are DNA transposases that can promote oncogenic genomic rearrangements. RAG1/2 binds to RSSs, and PGBD5 binds to PSSs and through cleavage induces a variety of possible changes, such as small insertion or deletion polymorphisms at the site of a double-strand break (DSB) repair, large insertions and deletions, inversions, and translocations. Illustrated by Mao Miyamoto.
PGBD5 promotes site-specific deletions, inversions, and translocations in rhabdoid tumors
Similar to RAG1/2, which is exclusively expressed in lymphoid cells and lymphoid tumors, PGBD5 also exhibits developmentally restricted expression in neuronal tissues and in the majority of childhood solid tumors, such as medulloblastoma, neuroblastoma, ependymoma, Ewing sarcoma, and rhabdoid tumors (45–47). Remarkably, PGBD5 expression is sufficient to promote oncogenic genomic rearrangements and induce anchorage-independent cell growth in vitro and tumor formation in vivo (62). This activity requires the putative catalytic residues in the PGBD5 transposase domain and depends on cellular nonhomologous end joining (NHEJ) DNA repair. Based on the use of alignment-free algorithms to detect genome rearrangements (63, 64), PGBD5-induced cell transformation was found to be associated with the occurrence of distinct intrachromosomal deletions, inversions, and translocations, significantly enriched for noncoding regulatory elements important for embryonal tissue development (62). Some of these rearrangements involved tumor suppressor genes that were required for the survival of transformed cells (Figure 2). Importantly, some of the observed rearrangement breakpoints exhibited significant sequence homology and included distinct sequences that were termed “PGBD5-specific signal sequences” (PSSs), with similarity to the human sequence substrates of PGBD5, as identified using a forward genetic screen (62, 65). However, the genomic rearrangements did not show signs of reinsertion (transposition) of excised fragments, suggesting that PGBD5 induces mutations through its nuclease activity rather than performing DNA transposition of genomic DNA in cancer cells. Consistent with these findings, genomic loci physically associated with the PGBD5 protein complex in rhabdoid tumor cells were also significantly enriched for PSS motifs (62). Indeed, primary human rhabdoid tumors expressing PGBD5 also exhibited recurrent genomic rearrangements with PSS sequences at the breakpoints, involving known tumor suppressor genes (62). These data provided compelling support that PGBD5 promotes sequence-specific oncogenic mutations and thus acts as an oncogenic mutator in rhabdoid tumors. Presumably, similar mechanisms operate in other PGBD5-expressing human cancers, which is the subject of ongoing studies.
Active DNA transposases confer synthetic lethal dependencies as targets for improved therapy
Could tumors with active DNA transposases depend on active DNA damage repair and signaling? Individuals with constitutional defects in DNA repair have pleiotropic phenotypes, including immunodeficiency, neurodegeneration, developmental abnormalities, and cancer predisposition. For example, ataxia telangiectasia (AT) is an autosomal recessive disorder linked to loss of ATM and characterized by immunodeficiency, progressive cerebellar ataxia, oculocutaneous telangiectasias, clinical radiosensitivity, chromosome instability, and increased risk of developing lymphoid malignancies (66). Immunodeficiency resulting from defective DNA repair is understood to be a result of inefficient and genotoxic RAG1/2-mediated DNA recombination activity in developing lymphocytes. In addition, somatic retrotransposition might explain some of the neurologic defects occurring in AT patients with loss of ATM (67).
This model is consistent with the notion that DNA transposases rely on cellular DNA repair to mediate DNA rearrangements, including ligation of DNA target sites (15). In humans, this repair activity is principally carried out by NHEJ DNA repair machinery, involving the heterodimeric Ku70/Ku80 (XRCC6/XRCC5) complex and signaling factors such as the ATM and Rad3-related kinases (68–70). Based on the above considerations, we recently tested the hypothesis that cells expressing active PGBD5 would depend on ongoing DNA damage repair and signaling. We found that expression of PGBD5 but not its catalytically inactive mutant induced DNA damage and apoptosis in cells deficient in Ku80, Atm, or Atr (71). Furthermore, we found that PGBD5 expression in pediatric tumor cells was required to render them susceptible to pharmacologic inhibition of DNA damage signaling (71). In particular, PGBD5 expression conferred susceptibility of rhabdoid tumor, medulloblastoma, neuroblastoma, and Ewing sarcoma cells to the ATR-selective kinase inhibitor AZD6738 (71). AZD6738 treatment of xenograft mouse models of some but not all PGBD5-expressing childhood solid tumors led to significant reductions and occasional regressions of tumor growth in vivo (71). These experiments provided a proof-of-principle demonstration that expression of an active oncogenic mutator DNA transposase is sufficient to confer susceptibility to pharmacologic inhibitors of DNA damage repair signaling. It is conceivable that additional synthetic, functional relationships between active DNA transposases and their cellular cofactors and signaling dependencies might be similarly developed for improved cancer therapy (72).
What mutational processes and oncogenic mutators are responsible for the increased incidence of distinct cancers in children and young adults?
With the development and application of high-throughput DNA sequencing, we are beginning to be able to define comprehensive landscapes of mutational processes in human cancer cells. It is now evident that human cancers are characterized by more than 50 mutational patterns involving nucleotide substitutions and at least 17 classes of DNA deletions and rearrangements (6, 73). Some of the observed mutational processes appear to be due to impaired DNA damage repair and DNA replication, and many have unknown causes. For example, so-called signature 18 (SBS18) is exclusively observed in neuroblastomas, presumably due to as-yet-undefined oxidative processes in neuroblastoma cells. Similarly, deletion signatures ID6 and ID8 exhibit features of NHEJ recombination, with as-yet-unknown nucleases. Six additional insertion and deletion patterns of mutation have been observed, whose mechanisms are also undefined. In addition, current evidence indicates that RAG1/2 mediates chromosomal translocations and deletions in lymphoid cancers, and PGBD5 promotes deletions and genomic rearrangements in rhabdoid and presumably other PGBD5-expressing solid tumors (Figure 1). The human genome encodes numerous DNA transposase–related genes, including some such as THAP9 that remain active and many that appear to be expressed in human cancers. Thus, it is possible that additional nucleases, recombinases, or transposases may function as oncogenic mutators in specific human cancers.
Conclusions and perspectives
Despite recent studies demonstrating that transposase-derived genes can act as oncogenic mutators, the activities of most human DNA transposase– related genes remain largely undefined. The identification of oncogenic mutators involved in the generation of genomic rearrangements in childhood cancers is important because these processes and molecules that mediate them may represent effective therapeutic targets, as suggested for PGBD5 (71). We anticipate that future studies using forward genetic screens and oncogenic transformation assays will define the functions of DNA nucleases and transposases expressed in human cancer (46, 47, 65). Furthermore, high-throughput long-read DNA sequencing and improved assembly-based algorithms for its analysis could be used to comprehensively define oncogenic genomic rearrangements and mutational processes (47). Additionally, developmentally accurate animal models should help define the functions of transposase-related genes in initiating oncogenic mutations and/or contributing to tumor maintenance. Because small molecules that target specific DNA damage repair signaling pathways are being developed, a resultant more detailed and integrated understanding of transposase-induced DNA damage could help identify promising new targets. Finally, considering the distinct expression patterns of endogenous DNA transposases in healthy human tissues, further study of their physiologic functions may reveal as-yet-unknown somatic or germline developmental processes.
Acknowledgments
We thank Alejandro Gutierrez for comments on the manuscript. AGH is supported by the Deutsche Forschungsgemeinschaft (German Research Foundation 398299703), the Berliner Krebsgesellschaft e.V., and the Berlin Institute of Health. AK acknowledges the support of the St. Baldrick’s Foundation, Damon Runyon–Richard Lumsden Foundation, Josie Robertson Investigator Program, Burroughs Wellcome Fund, Matthew Larson Foundation, Cycle for Survival, Cookies for Kids’ Cancer Foundation, Alex’s Lemonade Stand Foundation, Geoffrey Beene Cancer Research Center, Starr Cancer Consortium, and the Rita Allen Foundation.
Version 1. 10/18/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Published: October 18, 2018
Reference information: JCI Insight. 2018;3(20):e123172. https://doi.org/10.1172/jci.insight.123172.
Contributor Information
Anton G. Henssen, Email: anton.henssen@charite.de.
Alex Kentsis, Email: kentsisa@mskcc.org.
References
- 1. Noone AM, et al. SEER Cancer Statistics Review, 1975–2015. National Cancer Institute. NIH website. https://seer.cancer.gov/csr/1975_2015/ Updated September 10, 2018. Accessed September 18, 2018.
- 2.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6(3):193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 3.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huether R, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346(6215):1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511(7510):428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 10.Kohl NE, et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35(2 pt 1):359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 11.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9(6):657–663. doi: 10.1016/S0959-437X(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brodeur GM, Nichols KE, Plon SE, Schiffman JD, Malkin D. Pediatric cancer predisposition and surveillance: an overview, and a tribute to Alfred G. Knudson Jr. Clin Cancer Res. 2017;23(11):e1–e5. doi: 10.1158/1078-0432.CCR-17-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- 18.Muñoz-López M, García-Pérez JL. DNA transposons: nature and applications in genomics. Curr Genomics. 2010;11(2):115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keith JH, Schaeper CA, Fraser TS, Fraser MJ. Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase. BMC Mol Biol. 2008;9:73. doi: 10.1186/1471-2199-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27(7):1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman AB, Dyda F. Mechanisms of DNA Transposition. Microbiol Spectr. 2015;3(2):MDNA3–M0034. doi: 10.1128/microbiolspec.MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardue ML, Danilevskaya ON, Traverse KL, Lowenhaupt K. Evolutionary links between telomeres and transposable elements. Genetica. 1997;100(1–3):73–84. [PubMed] [Google Scholar]
- 23.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277(5328):911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277(5328):955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 25.Peifer M, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526(7575):700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 27.Schauer SN, et al. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res. 2018;28(5):639–653. doi: 10.1101/gr.226993.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns KH. Transposable elements in cancer. Nat Rev Cancer. 2017;17(7):415–424. doi: 10.1038/nrc.2017.35. [DOI] [PubMed] [Google Scholar]
- 29.Ewing AD, et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015;25(10):1536–1545. doi: 10.1101/gr.196238.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 31.Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016;26(6):745–755. doi: 10.1101/gr.201814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee E, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94(4):463–470. doi: 10.1016/S0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 34.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 35.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 36.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261(5125):1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 37.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27(3):1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaheen M, Williamson E, Nickoloff J, Lee SH, Hromas R. Metnase/SETMAR: a domesticated primate transposase that enhances DNA repair, replication, and decatenation. Genetica. 2010;138(5):559–566. doi: 10.1007/s10709-010-9452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohapatra S, et al. Trimming of damaged 3′ overhangs of DNA double-strand breaks by the Metnase and Artemis endonucleases. DNA Repair (Amst) 2013;12(6):422–432. doi: 10.1016/j.dnarep.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majumdar S, Singh A, Rio DC. The human THAP9 gene encodes an active P-element DNA transposase. Science. 2013;339(6118):446–448. doi: 10.1126/science.1231789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A. 1996;93(4):1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar A, et al. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol Genet Genomics. 2003;270(2):173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- 43.Bailey AD, et al. The conserved Cockayne syndrome B-piggyBac fusion protein (CSB-PGBD3) affects DNA repair and induces both interferon-like and innate antiviral responses in CSB-null cells. DNA Repair (Amst) 2012;11(5):488–501. doi: 10.1016/j.dnarep.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray LT, Fong KK, Pavelitz T, Weiner AM. Tethering of the conserved piggyBac transposase fusion protein CSB-PGBD3 to chromosomal AP-1 proteins regulates expression of nearby genes in humans. PLoS Genet. 2012;8(9):e1002972. doi: 10.1371/journal.pgen.1002972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavelitz T, Gray LT, Padilla SL, Bailey AD, Weiner AM. PGBD5: a neural-specific intron-containing piggyBac transposase domesticated over 500 million years ago and conserved from cephalochordates to humans. Mob DNA. 2013;4(1):23. doi: 10.1186/1759-8753-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henssen AG, et al. Genomic DNA transposition induced by human PGBD5. Elife. 2015:4. doi: 10.7554/eLife.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henssen AG, et al. PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat Genet. 2017;49(7):1005–1014. doi: 10.1038/ng.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McClintock B. Maize genetics. Year B Carnegie Inst Wash. 1946;45:176–186. https://www.ncbi.nlm.nih.gov/pubmed/20293579 Accessed October 11, 2018.
- 49.Halpern ME. Barbara McClintock on defining the unstable genome. Genetics. 2016;204(1):3–4. doi: 10.1534/genetics.116.194092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 51.McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. Proc Natl Acad Sci U S A. 1942;28(11):458–463. doi: 10.1073/pnas.28.11.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16(10):461–468. doi: 10.1016/S0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 53.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3(9):639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mullighan CG. The genomic landscape of acute lymphoblastic leukemia in children and young adults. Hematology Am Soc Hematol Educ Program. 2014;2014(1):174–180. doi: 10.1182/asheducation-2014.1.174. [DOI] [PubMed] [Google Scholar]
- 56.Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol Cell Biol. 2005;25(14):5904–5919. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135(6):1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322(5906):1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hübner S, et al. High incidence and unique features of antigen receptor gene rearrangements in TEL-AML1-positive leukemias. Leukemia. 2004;18(1):84–91. doi: 10.1038/sj.leu.2403182. [DOI] [PubMed] [Google Scholar]
- 61.Swaminathan S, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16(7):766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henssen AG, et al. Human PGBD5 DNA transposase promotes site-specific oncogenic mutations in rhabdoid tumors. Nat Genet. 2017;49(7):1005–1014. doi: 10.1038/ng.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhuang J, Weng Z. Local sequence assembly reveals a high-resolution profile of somatic structural variations in 97 cancer genomes. Nucleic Acids Res. 2015;43(17):8146–8156. doi: 10.1093/nar/gkv831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moncunill V, et al. Comprehensive characterization of complex structural variations in cancer by directly comparing genome sequence reads. Nat Biotechnol. 2014;32(11):1106–1112. doi: 10.1038/nbt.3027. [DOI] [PubMed] [Google Scholar]
- 65.Henssen AG, et al. Forward genetic screen of human transposase genomic rearrangements. BMC Genomics. 2016;17:548. doi: 10.1186/s12864-016-2877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87(2):423–438. [PubMed] [Google Scholar]
- 67.Coufal NG, et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci U S A. 2011;108(51):20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9):a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9(8):616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henssen AG, et al. Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Sci Transl Med. 2017;9(414):eaam9078. doi: 10.1126/scitranslmed.aam9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278(5340):1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 73. Alexandrov L. The Repertoire of Mutational Signatures in Human Cancer. BioRxiv website. https://www.biorxiv.org/content/early/2018/05/15/322859 Published May 15, 2018. Accessed September 18, 2018.