Abstract
Maternal obesity and a high-fat diet (HFD) during the perinatal period have documented short- and long-term adverse outcomes for offspring. However, the mechanisms of maternal HFD effects on neonatal offspring are unclear. While the effects of maternal HFD exposure during pregnancy on the offspring are increasingly being appreciated, we do not know if maternal HFD alters the microbiota or affects neonatal susceptibility to inflammatory conditions, nor the mechanisms involved. In this study, we show that the offspring of mothers exposed to HFD develop a unique microbiota, marked by expansion of Firmicutes, and an increase in IL-17–producing type 3 innate lymphoid cells (ILC3s). The expansion of ILC3s was recapitulated through neocolonization with HFD microbiota alone. Further, the HFD offspring were susceptible to a neonatal model of inflammation that was reversible with IL-17 blockade. Collectively, these data suggest a previously unknown and unique role for ILC3s in the promotion of an early inflammatory susceptibility in the offspring of mothers exposed to HFD.
Keywords: Immunology, Inflammation
Keywords: Inflammatory bowel disease, Innate immunity, Obesity
Murine offspring exposed to high-fat diet acquire a unique microbiota, resulting in expansion of ILC3 cells.
Introduction
The average American diet, consisting of over 35% fat, has contributed to the epidemic of obesity in the United States and in other developed countries. Maternal obesity during the perinatal period has documented short- and long-term adverse outcomes for offspring (1–3). Over 50% of women of childbearing age in the United States are either overweight or obese and exposed to a high-fat diet (HFD) (4). Consumption of a HFD in adults results in alteration of the intestinal microbiome and susceptibility to colitis (5, 6). While the effects of maternal HFD exposure during pregnancy on the offspring are increasingly being appreciated (7, 8), we do not know if maternal HFD alters the microbiota or affects neonatal susceptibility to inflammatory conditions, nor the mechanisms involved. Potential mechanisms include alteration of the intestinal microbiota and differential host-commensal immune responses. We sought to begin to understand this relationship by examining how maternal HFD influences development of the microbiota in neonatal offspring and characterize the consequences of this change.
Increased abundance of intestinal Firmicutes is associated with several diseases, namely inflammatory bowel disease, obesity, and metabolic syndrome (9–12). Interestingly, members of the Firmicutes phylum, segmented filamentous bacteria (SFB), have been shown to promote the differentiation and expansion of Th17 cells in the gut and contribute to the inflammation seen in ulcerative colitis in adults (13–15). Firmicutes expansion precedes the development of necrotizing enterocolitis (NEC) (16), a devastating inflammatory disease of the intestine in preterm infants that carries substantial morbidity and mortality. Of note, mice are only susceptible to NEC prior to 2 weeks of age, which is the equivalent of less than 30 weeks human gestation (17, 18). Thus, we hypothesized that exposure to maternal HFD would alter the offspring microflora and potentially put offspring at risk for NEC.
IL-17 is a cytokine that plays a significant role in inflammatory and infectious diseases of the bowel. Type 3 innate lymphoid cells (ILC3s) are the innate counterpart to Th17 cells and can similarly produce both IL-17 and IL-22. They are found primarily in the intestinal tract and in mucosal tissues. They play a crucial role in immune homeostasis, host defense, and disease pathogenesis (19, 20). ILC3s interact with the intestinal microbiota and are present as early as the third trimester of gestation in mice. ILC3s are also found in the gut of human fetuses (21, 22). Current data suggest that commensal bacteria may have the ability to regulate functional responses of ILC3s via TLR signaling (23, 24).
Our laboratory has previously shown that neonatal mice prior to 3 weeks of age have a unique microbiota marked by an expansion of Proteobacteria (25). These mice also have an increase in baseline expression of inflammatory cytokines and are susceptible to a neonatal model of inflammation (lipopolysaccharide and platelet-activating factor [LPS/PAF] exposure). In the following experiments, we exposed breeding mice to HFD and examined the intestinal microbiota and susceptibility to inflammation in the offspring. We also utilized flow cytometric techniques and neocolonization experiments to determine the relationship between the microbiota and lamina propria (LP) immune cell populations.
Results
Maternal exposure to a HFD alters the intestinal microbiota and increases IL-17 at baseline in offspring.
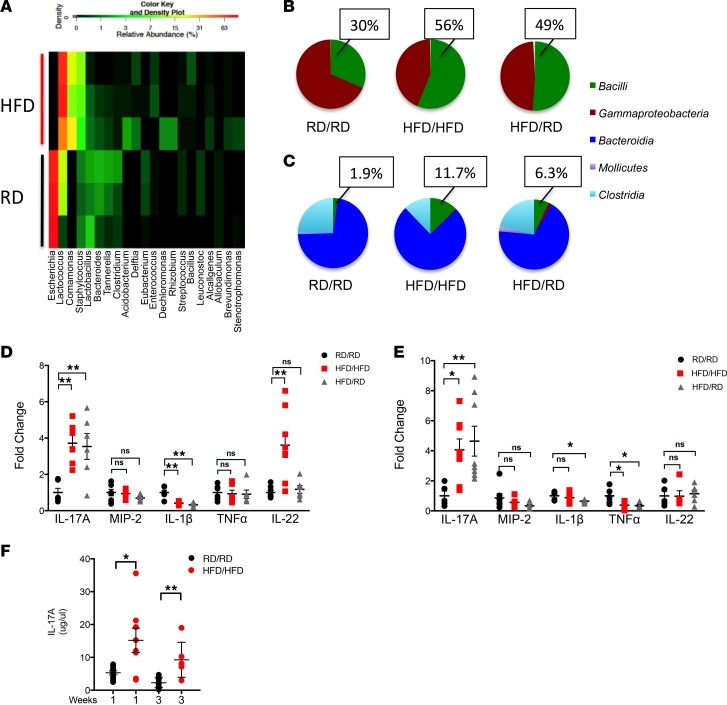
Consumption of a HFD in adults results in a unique gut microbiota marked by an increase in Firmicutes (26). This unique bacterial population is energy efficient and may be involved in the development or persistence of the obese/overweight phenotype as well as inflammation (27). In order to examine whether the maternal consumption of a HFD could change the offspring microbiota, we exposed breeding female mice to a HFD as described in the Methods. As early as 1 week of age, offsp ring of mothers exposed to a HFD not only had a unique microbiota in their mucosal fecal colonic contents (Figure 1A), but one that was different from their mothers (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99223DS1), suggesting that this was not simply acquisition of the maternal bacterial population. The unique microbiota of HFD offspring consisted most markedly of an increase in Firmicutes (primarily Lactococcus), with a decrease in Gammaproteobacteria (primarily Escherichia). The HFD offspring also showed an increase in Betaproteobacteria (primarily Comamonas). Lactococcus belong to the class Bacilli and are known to be able to adapt early to nutritionally rich environments (28) and typically occupy habitats that have high availability of sugar (29). HFD offspring may offer an intestinal habitat that benefits this phylum and competes with Proteobacteria. Since offspring are exclusively dependent on breast milk at this early time point, mice were cross-fostered to mothers on regular diet (RD) at birth to limit exposure to breast milk from HFD-fed mice. Interestingly, this early cross-fostering of HFD offspring to RD mothers (HFD/RD) also resulted in an increase in Bacilli, which belongs to the phylum Firmicutes (Figure 1B). There was an insignificant amount of SFB in both HFD and RD mice, with 40% of mice without detectable SFB by qRT-PCR (data not shown). Cross-fostering HFD offspring from birth resulted in a muted, but unique, phenotype, significant for an increase in Bacilli, which persisted to 3 weeks of age (Figure 1C).
Figure 1. HFD offspring have an altered microbiota and increased expression and production of IL-17.
(A) Heatmap of the relative abundance of the most dominant genera in the fecal colonic contents of 1-week-old mice as determined by genomic DNA analysis by next-generation sequencing. HFD offspring and RD offspring are representative of 3 mice in each group. (B) Pie chart showing the proportion of Bacilli, Gammaproteobacteria, Bacteroidia, Mollicutes, and Clostridia seen in 1-week- and (C) 3-week-old mice analyzed by next-generation sequencing in RD/RD, HFD/HFD, and HFD/RD offspring. Percentages are representative of populations of Bacilli (Firmicutes). (D) Expression of IL-17A, MIP-2, IL-1β, and TNF-α was analyzed by quantitative real-time PCR (log scale) in small intestine from 1-week- and (E) 3-week-old mice. (F) ELISA showing production of IL-17A (μg/μl) in small intestine of RD/RD and HFD/HFD mice. RD/RD represent RD offspring cross-fostered to RD mothers at birth. HFD/HFD represents HFD offspring cross-fostered to HFD mothers at birth. HFD/RD represents HFD offspring cross-fostered to RD mothers at birth. The data shown are representative of 3 experiments with 6–10 mice in each group. Data are depicted as the mean ± SEM. *P < 0.05; **P < 0.01 by 1-way ANOVA with Tukey’s post hoc test (D and E) or 2-tailed Student’s t test (F). HFD, high-fat diet; RD, regular diet; ns, not significant.
Firmicutes, and in particular SFB, promote an expansion of Th17 cells (13). To examine if the increase in Firmicutes seen in the HFD offspring was associated with an altered IL-17A response, we looked at the expression of multiple cytokines in the small intestine of 1-week- and 3-week-old offspring and found that only IL-17A was consistently and significantly increased 3- to 5-fold in the HFD offspring (Figure 1, D and E) when compared with RD control offspring. IL-17A protein production at baseline was also increased (Figure 1F). These changes persisted at 3 weeks of age and were present even when the offspring were cross-fostered to RD mothers at birth. Other cytokines were either unchanged or marginally decreased. Of note, IL-22 was transiently increased at 1 week in the HFD/HFD offspring, but this normalized at 3 weeks of age. These data suggest that HFD offspring develop a unique microbiota marked by an increased in Firmicutes, particularly Bacilli, and in the expression and production of IL-17, suggesting there might be an increase in the number of IL-17–producing cells. Therefore, we next examined the LP of these offspring to determine the cell type responsible for the elaboration of IL-17.
The ILC3 population is expanded in the LP of HFD offspring.
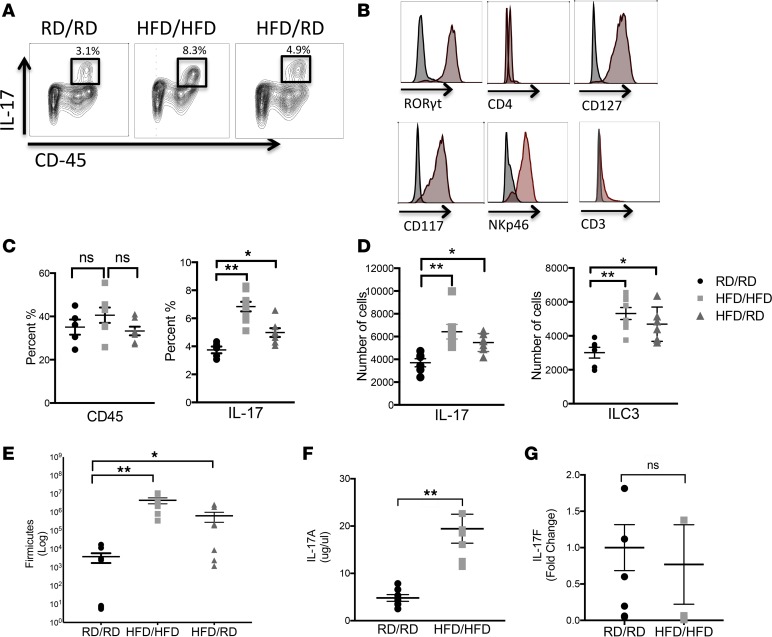
We examined immune cell populations in the LP of HFD offspring by flow cytometry at 2 weeks of age, when sufficient numbers of immune cells could be isolated for flow cytometric analysis. We found an increase in the population of IL-17–producing cells in the HFD offspring, even when cross-fostered to RD mothers (Figure 2A). Further analysis of these cells identified them as Rorγt+, CD127+, CD117+, CD3–, NKp46+, and CD4–, consistent with ILC3s (Figure 2B and Supplemental Figure 2A). Since CD45 populations were not different between these groups, these IL-17–producing cells were specifically expanded in the HFD offspring (Figure 2, C and D). To assess the contribution of Th17 cells in the increase in IL-17–producing cells, we examined the proportion of ILC3s to Th17 cells and found that the ratio decreases from 1 to 3 weeks in HFD offspring, confirming that the majority of this increase is due to an increase in the number of ILC3s and not Th17 cells (Supplemental Figure 2B). At 2 weeks of age, the HFD offspring also showed a 2–3 log increase in Firmicutes (Figure 2E) and an increase in baseline production of IL-17A (Figure 2F). To determine if the microbiota is necessary to expand the ILC3 population in offspring, we placed offspring of HFD/HFD mothers on broad-spectrum antibiotics through their dams from birth and examined the population of ILC3s in the LP at 2 weeks of age. We found that offspring exposed to antibiotics through their dams had a diminished number of ILC3s, suggesting that colonization is necessary for their expansion (Supplemental Figure 2C). To investigate the differential roles of IL-17A and IL-17F, HFD offspring were also examined for IL-17F by qRT-PCR and were found to have no difference (Figure 2G), suggesting only a preferential increase in IL-17A production. Taken together, these data suggested that the increase seen in IL-17A in the HFD offspring is due to an increase in the population of ILC3s.
Figure 2. Expansion of IL-17–producing ILC3s is seen in the lamina propria (LP) of HFD offspring.
(A) Flow cytometric analysis of LP cells from 2-week-old RD/RD, HFD/HFD, and HFD/RD offspring was performed. Live cells were stained for CD45 (x axis) and IL-17 (y axis) and representative panels are shown here. Cells positive for CD45 and IL-17 are highlighted in the bold boxes. (B) Histograms showing CD45+ and IL-17+ cells as Rorγt+, CD4–, CD3–, NKp46+, CD127+, and CD117+ when compared with isotype controls. (C) Percentage of CD45+ cells and IL-17+ cells in 2-week-old RD/RD, HFD/HFD, and HFD/RD offspring. (D) IL-17+ cells and IL-17–producing ILC3s quantified as number of cells seen in 2-week-old RD/RD, HFD/HFD, and HFD/RD offspring. (E) Quantification of Firmicutes (log scale) in the small intestine of 2-week-old mice in RD/RD, HFD/HFD, and HFD/RD offspring by qRT-PCR. (F) Quantification of IL-17A (μg/μl) in the small intestine of 2-week-old mice by ELISA in RD/RD and HFD/HFD offspring. (G) qRT-PCR of IL-17F in RD/RD and HFD/HFD offspring showing no difference. RD/RD represent RD offspring cross-fostered to RD mothers at birth. HFD/HFD represents HFD offspring cross-fostered to HFD mothers at birth. HFD/RD represents HFD offspring cross-fostered to RD mothers at birth. These data are representative of at least 3 independent experiments, each involving 6–10 mice per group. Data are depicted as the mean ± SEM. *P < 0.05; **P < 0.01 by 1-way ANOVA with Tukey’s post hoc test (C–E) or 2-tailed Student’s t test (F and G). HFD, high-fat diet; RD, regular diet; ns, not significant.
Offspring of Rag1–/– mice on HFD also have an altered intestinal microbiota and an expanded population of IL-17–producing ILC3s in the LP.
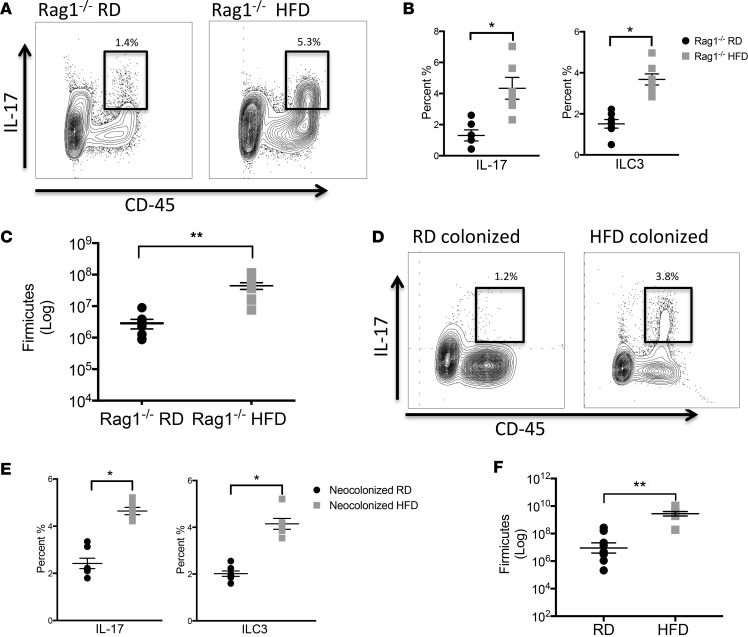
We next sought to determine if IL-17–producing cells were a result of a specific increase in ILC3s and not in Th17 cells. We utilized Rag1–/– mice that are deficient in mature T and B cells, but still have ILCs. After exposure of breeding Rag1–/– mice to either HFD or RD, the HFD offspring had an increase in IL-17–producing ILC3s (Figure 3, A and B), as well as an expansion of Firmicutes in their colonic microbiota (Figure 3C). Flow cytometric staining confirmed that these were type 3 ILCs (Rorγt+, CD127+, CD117+, NKp46+, CD4–). These data show that the HFD offspring develop a unique microbiota that is associated with a specific increase in ILC3s.
Figure 3. Rag1–/– mice on HFD have an expanded population of IL-17–producing ILC3s.
(A) Flow cytometric analysis of lamina propria (LP) cells from 2-week-old RD Rag1–/– and HFD Rag1–/– offspring was performed. Live cells were stained for CD45 (x axis) and IL-17 (y axis) and representative panels are shown here. Cells positive for CD45 and IL-17 are highlighted in the bold boxes. (B) Percentage of CD45+IL-17+ cells (left) and percentage of IL-17+Rorγt+CD4–CD127+NKp46+CD117+ ILC3s (right) in 2-week-old RD Rag1–/– and HFD Rag1–/– offspring. (C) Quantification of Firmicutes (log scale) in the small intestine of 2-week-old RD Rag1–/–and HFD Rag1–/– offspring by qRT-PCR. (D) Flow cytometric analysis of LP cells from 2-week-old neocolonized RD and neocolonized HFD offspring was performed. Cells positive for CD45 and IL-17 are highlighted in the bold boxes. (E) Percentage of IL-17+ cells and IL-17–producing ILC3s in 2-week-old neocolonized offspring. (F) Quantification of Firmicutes (log scale) in the small intestine of 2-week-old neocolonized RD and neocolonized HFD offspring by qRT-PCR. The data shown are representative of 3 experiments with 7–10 mice in each group, and are depicted as the mean ± SEM. *P < 0.05; **P < 0.01 by 2-tailed Student’s t test. HFD, high-fat diet; RD, regular diet.
Neocolonization of antibiotic-treated neonatal mice with microbiota from HFD offspring results in an expansion of IL-17–producing ILC3s.
To further evaluate the relationship between the microbiota of HFD offspring and ILC3s, we colonized neonatal mice born to mothers on broad-spectrum antibiotics. The offspring of antibiotic-exposed mothers had minimal presence of intestinal bacteria (Supplemental Figure 3). At 3 days of life, we colonized the antibiotic-exposed mice with the gut microbiota from either HFD or RD offspring. Flow cytometric analysis at 2 weeks of life showed that only mice colonized with the microbiota from HFD offspring had an expansion of IL-17–producing ILC3s (Figure 3, D and E). Additionally, the HFD neocolonized mice maintained an expansion of Firmicutes (Figure 3F). Collectively, these data suggest that the unique microbiota that develops after HFD exposure causes expansion of ILC3s.
The offspring of HFD-exposed mothers have an increased susceptibility to intestinal injury that is reversed by blocking IL-17.
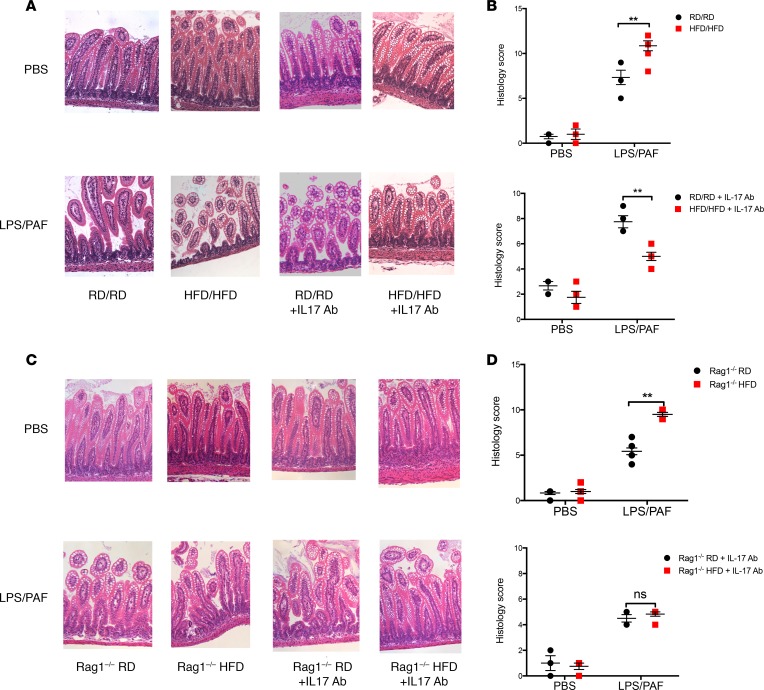
To determine if the increase in IL-17–producing ILC3s had inflammatory consequences in the offspring of HFD-exposed mothers, we exposed neonatal mice to a model of early intestinal injury consisting of LPS/PAF exposure. Notably, the offspring of HFD-exposed mothers had increased gut injury by histological scoring after LPS/PAF treatment (Figure 4, A and B). We hypothesized that this increased susceptibility to gut injury was mediated by the increase in IL-17–producing ILC3s. To test this hypothesis, mice were injected intraperitoneally with an IL-17–blocking antibody prior to LPS/PAF exposure and every other day thereafter beginning at 5 days of life. Indeed, IL-17 blockade resulted in decreased intestinal injury in susceptible mice (Figure 4, A and B). We repeated these experiments in Rag1–/– mice (Figure 4, C and D) and found similar results, with Rag1–/– mice having higher pathology scores and being rescued with IL-17 blockade. Of note, Rag1–/– mice had overall lower histology scores than wild-type mice; this is likely due to reduced injury from T cells, which are also known to play a role in intestinal inflammation in NEC. These data taken together suggest that the increased presence of IL-17–producing ILC3s is responsible for the increased susceptibility to intestinal injury seen in the offspring of HFD-exposed mothers.
Figure 4. In vivo blockade of IL-17 protects against intestinal injury induced by LPS and PAF in 2-week-old HFD offspring.
(A) Representative images of H&E staining of small intestinal sections in WT offspring exposed to LPS/PAF and PBS (above) and after IL-17 blockade (below) in RD/RD and HFD/HFD offspring. (B) Quantification of histological injury score in RD/RD and HFD/HFD offspring exposed to either LPS/PAF or vehicle control (above) and after IL-17 blockade (below). (C) Representative images of H&E staining of small intestinal sections in Rag1–/– offspring exposed to LPS/PAF and PBS (above) and after after IL-17 blockade (below) in RD/RD and HFD/HFD offspring. (D) Quantification of histological injury score in Rag1–/– mice (above) and after IL-17 blockade (below) in RD/RD and HFD/HFD offspring exposed to either LPS/PAF or vehicle control. Original magnification, ×20. The data shown are representative of 3 experiments (WT) with 4–8 mice in each group and 2 experiments (Rag1–/–) with 3–6 mice in each group. RD/RD represent RD offspring cross-fostered to RD mothers at birth. HFD/HFD represents HFD offspring cross-fostered to HFD mothers at birth. HFD/RD represents HFD offspring cross-fostered to RD mothers at birth. Data are depicted as the mean ± SEM. **P < 0.01 by 2-way ANOVA. HFD, high-fat diet; RD, regular diet; PAF, platelet-activating factor; ns, not significant.
Discussion
In this study, we have shown that exposure to a HFD during pregnancy alters the microbiota composition in offspring and increases their susceptibility to a gut inflammatory challenge. We report, for the first time to our knowledge, that offspring of mothers on HFD have a unique microbiota, with an expansion of Firmicutes. Mechanistically, these offspring have an increase in IL-17 that is due to an expansion of ILC3s. Interestingly, the microbiota of offspring of HFD-exposed mothers alone is sufficient to cause an expansion in ILC3s. Importantly, the offspring of HFD-exposed mothers have an IL-17–dependent increased susceptibility to intestinal inflammation. Taken together, these results reveal that a maternal HFD results in a microbiota-dependent expansion of ILC3s and increased susceptibility to intestinal injury in offspring mediated by IL-17.
An altered gut microbiota is associated with multiple diseases in adults. The impact of diet on changing the composition of the microbiota is now firmly established (30, 31). However, the effects of an indirect diet exposure in mothers on the establishment of the microbiota in the offspring has not been described previously. Our previous studies showed that Proteobacteria are the dominant bacterial phylum in neonatal mice, and we sought here to determine whether a maternal HFD could affect the pattern of colonization in their offspring. Indeed, we found that alteration in diet alone results in a change in the pattern of gut bacteria in the offspring. This pattern was unique and not a result of direct transmission from the mother. While our study utilizes an extreme diet (60% saturated fat), further work examining the effect of a moderately HFD (30%–40%) is also warranted. Our prior work has shown that changes in immunologic effectors, specifically IgA production, can alter the pattern of the establishment of microbiota in neonatal mice and intestinal inflammation (25). We therefore looked for an altered immune response in the offspring of HFD-exposed mothers. Surprisingly, we found a definitive expansion of IL-17–producing ILC3s in neonatal HFD offspring. Since mature T and B cells are relatively deficient in the neonatal period, the innate immune system plays a pivotal role in host immunity. While we found that IL-17A was consistently elevated in HFD offspring, there was also a transient increase in IL-22 seen only in 1-week-old HFD offspring that we speculate is for modulation of gut inflammation and epithelial repair during the initial phase of colonization, or to contain expansion of certain bacterial populations, particularly SFB (32). Notably, IL-1β can stimulate ILC3s to produce both IL-22 and IL-17 (33, 34). IL-1β was transiently decreased in HFD offspring at 1 week, when both IL-22 and IL-17 were increased. This may play an important role in shaping the microbiota in HFD offspring and warrants further investigation.
While we know that ILC3s can be influenced by the microflora, the specific mechanisms are not well characterized. Although the gut microbiota can promote ILC3s, it is unclear whether this is due to secondary mediators from intestinal epithelial cells or a direct effect (35). Our studies strongly support a role for Firmicutes in the expansion of ILC3s. The relationship between the HFD and Firmicutes expansion in offspring needs to be explored further. Bacteria from the phyla Cytophaga-Flavobacter-Bacteroidetes, along with Firmicutes, have been shown to be important in differentiation and expansion of Th17 cells (36). SFB are known to induce intestinal Th17 cells (37) and can activate ILC3s to produce IL-22 (38); however, our mice did not have a significant amount of SFB. HFD offspring had an expansion of Lactobacilli, which has been shown to utilize tryptophan to produce ligands of aryl hydrocarbon receptors (AHRs) that can promote production of IL-22 by ILCs. The expansion of ILC3s may thus be due to altered bacterial metabolite production by tryptophan-producing members of the phylum Firmicutes. Of note, short-chain fatty acids (SCFAs) can also be produced across the major phyla Actinobacteria, Bacteroidetes, and Firmicutes, with Firmicutes having the most pathways for production of SCFAs (39, 40). Further studies examining how these bacterial metabolites are sensed and the pathways by which ILC3s are promoted are under way in our laboratory.
We found a mainly NKp46+ subset of IL-17–producing ILC3s. This is consistent with work from Sawa et al., which showed that in the fetal gut, NKp46+ ILC3s are largely absent but show a marked proliferation after birth and expand to become the dominant subset in the small intestine (39, 41). Additionally, Macpherson et al. showed that immunoglobulin-bound AHR ligands can be transferred in utero and can further promote migration of NKp46+ ILC3s to the neonatal small intestine (42). AHR ligands include low-density lipoproteins, bilirubin, and arachidonic acid, which may be elevated in during exposure to HFD in mothers. The role of transferred maternal metabolites on expansion of ILC3s is worth further investigation and may provide therapeutic targets for intervention.
The observation that neonatal offspring of mothers exposed to a HFD are more susceptible to an IL-17–mediated gut inflammation may provide insight into the environmental factors that drive NEC development in preterm infants. Our study shows a substantial role for ILC3 in susceptibility to inflammation in HFD offspring; however, we cannot rule out a role for other IL-17–producing cells, including Th17 cells and IL-17–producing γδ T cells. The histology scores with IL-17 blockade in wild-type mice is lower than in Rag1–/– mice, suggesting a role for these other cell types. In adult mice, IL-17A from γδ T cells has been shown to have IL-23–independent protective effects by decreasing gut permeability in the intestine (43). Neonatal mice have significantly more γδ T cells but are relatively hyporesponsive when compared with adults (44, 45). Whether maternal HFD in human pregnancies carries an increased risk of NEC in premature infants remains to be investigated. If found to be so, our data suggest potential therapeutic approaches for NEC in preterm infants, either via modulation of the microbiota, ILC3s, or IL-17 production.
The establishment of the gut microenvironment is unique and is now shown to be influenced by diet, exposure to antibiotics, and mode of antibiotic delivery (46, 47). Our study suggests a role for maternal diet in altering the establishment of the microbiome in offspring and has significant public health consequences. The transmission of a susceptible phenotype, that is prone to inflammation and even potentially obesity or metabolic syndrome, raises the possibility for early intervention in newborns to prevent future disease. Alteration of the diet during pregnancy could also potentially result in a reduction in susceptibility to gut inflammatory responses in neonates. Future studies directed at understanding the mechanisms involved in establishing the phenotypic changes in offspring as adults as well as neonates are needed.
In summary, our study highlights the role of maternal HFD in altering the offspring microbiota with consequent microbiota-dependent expansion of IL-17–producing ILC3s and increased susceptibility to inflammation.
Methods
Mice and HFD model
C57BL/6 mice were obtained from the University of Texas Southwestern (UTSW) Medical Center Mouse Breeding Core Facility. Rag1–/– mice were originally obtained from The Jackson laboratory and had been breeding in our facility for 2 generations prior to these experiments. Female mice at 3–4 weeks of age were placed either on a HFD containing 60% fat, 20% protein, and 20% carbohydrates (Research Diets) or a control diet (chow, RD) containing 12% fat, 22% protein, and 66% carbohydrate (Envigo) ad libitum. Male mice exposed to RD were added to the cage for breeding at 6–8 weeks of age and they remained in the cages either on HFD or RD. Cross-fostering was performed shortly after birth or at birth where noted from a HFD cage to a RD cage where RD mothers would foster the pups until euthanasia. Nomenclature used for natural and adoptive mothers are as follows: (a) RD/RD, offspring born to RD mothers and maintained with them until euthanasia; (b) HFD/HFD, offspring born to HFD mothers and maintained with them until euthanasia; and (c) HFD/RD, offspring born to HFD mothers cross-fostered to RD mothers where they were maintained until euthanasia. Female Rag1–/– mice were similarly placed on RD or HFD at 3–4 weeks of age and then mated with an RD male at 6–8 weeks. HFD mice were bred for a maximum of 6 months and fasting blood glucose testing was performed monthly to ensure that breeding mice did not have impaired glucose tolerance while breeding. Pups were maintained in respective cages until euthanasia. All mice were maintained in the same animal room at the American Association of Laboratory Animal Care-accredited animal facility at the UTSW Medical Center.
qRT-PCR
Total RNA from intestinal samples was purified using TRIzol reagent and subjected to first-strand cDNA synthesis by using iScript Reverse Transcription Supermix (Bio-Rad). DNA from colonic mucosal fecal contents was extracted using ZR Fecal DNA miniprep (Zymo Research) and quantified using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific). Real-time PCR was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and the CFX Connect Real-Time system (Bio-Rad) according to the manufacturer’s instructions. For intestinal cytokines, data were analyzed by the Ct method with normalization for starting template performed using a housekeeping gene, SRP-14. For bacterial 16S rRNA analysis, samples were normalized to Eubacteria utilizing known-concentration standards. Primer sequences used are as follows: murine SRP-14 5′-AAGTGTCTGTTGAGAGCCACGGAT-3′ and 5′-CTGTCACTGTGCTGGTTTGCTCTT-3′; IL-17 5′-TCCCTCTGTGATCTGGGAA–3′ and 5′–CTCGACCCTGAAAGTGAAGG–3′; MIP-2 5′-CTCTCAAGGGCGGTCAAAAAGTT-3′ and 5′-TCAGACAGCGAGGCACATCAGGTA-3′; TNF-α 5′-CCACCACGCTCTTCTGTCTAC-3′ and 5′-TGGGCTACAGGCTTGTCACT-3′; IL-1β 5′-CCTTCCAGGATGAGGACATGA-3′ and 5′-TGAGTCACAGAGGATGG-GCTC-3′. Bacterial primers used are as follows: Eubacteria 5′-ACTCCTACGGGAGGCAGCAGT-3′ and 5′-ATTACCGCGGCTGCTGGC-3′; Enterobacteriaceae 5′-GTGCCAGCMGCCGCGGTAA-3′ and 5′-GCCTCAAGGGCACAACCTCCAAG-3′; Bacteroidetes 5′-GGTTCTGAGAGGAGGTCCC-3′ and 5′-GCTGCCTCCCGTAGGAGT-3′; Firmicutes 5′-GGAGYATGTGGTTTAATTCGAAGCA-3′ and 5′-AGCTGACGACAACCATGCAC-3′.
Next-generation sequencing
Colonic contents were extracted and genomic DNA isolated using ZR Fecal DNA miniprep (Zymo Research). Bacterial Tag-Encoded sequencing using the Ion Torrent was carried out using bar-coded primers 28F-519R for the V1–V3 region of the 16S rRNA gene by the Research and Testing Laboratory. Three thousand reads per sample were obtained. FASTA-formatted sequences were analyzed for quality and sequences that had low-quality tags, primer, ends or that failed to be at least 250 bp in length were excluded from the analysis.
Once FASTA-formatted sequences passed quality-controlled checks as described, the identity of each remaining sequence was first sorted such that the FASTA-formatted file contained reads from longest to shortest. These sequences were then clustered into OTU clusters with 96.5% identity (3.5% divergence) using USEARCH. For each cluster the seed sequence was put into a FASTA-formatted sequence file. This file was then queried against a database of high-quality sequences derived from NCBI using a distributed.NET algorithm that utilizes BLASTN+ (KrakenBLAST, www.krakenblast.com). Using a .NET and C# analysis pipeline the resulting BLASTN+ outputs were compiled and data reduction analysis performed.
Based on the sequence identity percentage derived from BLASTn+ above, the sequences were classified at the appropriate taxonomic levels based upon the following criteria. Sequences with identity scores (identity to well-characterized 16S sequences) greater than 97% (<3% divergence) were resolved at the species level, between 95% and 97% at the genus level, 90% and 95% at the family level, 85% and 90% at the order level, 80% and 85% at the class level, and 77% and 80% at the phylum level. Any match below this percentage identity was discarded. In addition, the high-scoring segment pair (HSP) was at least 75% of the query sequence or it was discarded, regardless of identity.
After resolving based on these parameters, the percentage of each organism was individually analyzed for each sample providing relative abundance information within and among the individual samples based on relative numbers of reads within each. Evaluations presented at each taxonomic level, including percentage compilations, represent all sequences resolved to their primary identification or their closest relative. Raw 16S RNA sequence data were deposited in the NCBI’s Sequence Read Archive under accession number SRP154881.
ELISA
Small intestine tissues from offspring were incubated overnight at 37°C in cell culture media (RPMI). IL-17 concentration in the supernatant was quantified using ELISA (eBioscience) according to the manufacturer’s recommendations.
Isolation of intestinal LP cells and flow cytometry
Isolation of LP cells was performed as follows. The small intestine was removed and opened longitudinally, washed of fecal contents, cut into smaller sections, and subjected to 2 sequential incubations in PBS with 0.5 mM EDTA and 0.2 mM DTT at 37°C with agitation at 220 rpm to remove epithelial cells. The solution was discarded between incubation steps and replaced. The remaining tissue was agitated in PBS and then filtered through a strainer. The tissue was pat dried and minced and placed in the incubator for 30 minutes with gentle agitation at 110 rpm in 0.4 mg/ml collagenase D and 50 mg/ml DNase I at 37°C. The samples were then washed through a strainer (100 μm) and centrifuged at 160 g at 4°C. LP cells were then washed with FACS buffer (PBS, 1% EDTA, 1% FBS) and stained with antibody cocktail for 20 minutes at 4°C. The following antibodies were used (all from eBioscience unless otherwise noted): CD45-PE-Cy7 (30-F11), c-kit-APC (180627) (R&D Systems), NKp46–PerCP (29A1.4), CD127-PE-Cy7 (A7R34), CD8a-APC (53-6.7), CD45-FITC (30-F11), and CD4-FITC (GK1.5). Biotin-conjugated CD3e (145-2C11), CD8a (53-6.7), and NKp46 (29A1.4) were obtained from Biolegend.
After extracellular staining, cells were fixed in IC fixation buffer (eBioscience), washed in permeabilization Buffer (eBioscience), and stained for intracellular antigens Rorγt-PE (AFKJS-9), IL-17-PE (eBio17B7), and anti–mIL-17-PerCP (R&D Systems) for 1.5 hours at 4°C. Samples were read on a FACSCanto (BD) and analyzed using FlowJo Software (Tree Star).
Experimental design
Colonization of microbiota-suppressed mice with HFD microbiota.
We exposed C57BL/6 breeding mice to broad-spectrum antibiotics (ampicillin, streptomycin, neomycin, metronidazole; all at 1 mg/ml and vancomycin at 0.5 mg/ml) in drinking water. Breeding, pregnant, and nursing mothers were maintained on the antibiotic water. Pups were exposed through the mothers’ breast milk and confirmed to be microbiologically suppressed through examination of fecal colonic contents by qRT-PCR (Supplemental Figure 3). For neocolonization experiments, fecal colonic contents from HFD and RD mice not exposed to antibiotics were pooled separately in PBS and homogenized for 60 seconds using a Bead Rupter 4 (Omni International). Quantification of bacterial populations was performed by serial dilution on CHROMagar plates (CHROMagar Microbiology). Microbiologically suppressed offspring were gavage fed either HFD or RD colonic contents at 1010 CFUs once at 3 days of life and sacrificed at 2 weeks. Mice were subsequently analyzed at 2 weeks of age. Two experiments were performed with a total of 4–8 mice in each group.
Neonatal LPS/PAF model of intestinal injury.
Gut mucosal injury was induced in 2-week-old mouse pups by intraperitoneal administration of LPS (1 mg/kg) and PAF (50 μg/kg). Mice were sacrificed 2 hours after LPS and PAF administration and small intestinal sections were fixed in Carnoy fixative, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Histologic preparations were reviewed on a Leica DM2000 microscope. Images were acquired at ×20 magnification using an Optronics Microfire CCD color camera and PictureFrame 2.0 acquisition software (Optronics). Histological changes were analyzed in a double-blind fashion using a 17-point scale as follows. For crypt integrity: 0, normal; 1, irregular crypts; 2, mild crypt loss; 3, severe crypt loss; 4, complete crypt loss with an intact epithelial cell layer; 5, complete loss of crypts and surface epithelium (<10 crypt width); and 6, complete loss of crypts and surface epithelium (>10 crypts). For infiltration of inflammatory cells into the mucosa: 0, normal; 1, mild; 2, modest; and 3, severe. For infiltration of the submucosa: 0, normal; 1, mild; 2, modest; and 3, severe. For infiltration of the muscle: 0, normal; 1, mild; 2, modest; and 3, severe. These scores were added, resulting in a total scoring range of 0 to 15. Three experiments were performed with 4–6 mice in each group.
In vivo blockade of IL-17.
C57BL/6 mice were treated by intraperitoneal injection with 30 μg of purified anti–mouse IL-17A (TC11-18H10.1, Biolegend) every other day starting at 5 days of life. Mice were then exposed to the LPS/PAF model of intestinal injury at 2 weeks and examined as described above. Two experiments were performed with 4–6 mice in each group.
Statistics
Data were analyzed by 1-way ANOVA with Tukey’s post hoc test, 2-way ANOVA, or unpaired 2-tailed Student’s t test, using GraphPad Prism 6. Data are expressed as mean ± SEM and significance was defined as P < 0.05.
Study approval
The Institutional Animal Care and Use Committee at the UTSW Medical Center approved all protocols involving mice.
Author contributions
STB designed and performed experiments. XN and MR performed experiments. JM designed experiments and analyzed and interpreted the data. STB and JM drafted the manuscript. LVH and RCS helped with study design and critically revised the manuscript. JM designed the study, interpreted the data, and critically revised and finalized the manuscript.
Supplementary Material
Acknowledgments
This research was supported by NIH K08 (DK100545), a Young Investigator Grant for Probiotic Research and a Children’s Medical Center Foundation Grant to JM.
Version 1. 10/04/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(19):e99223. https://doi.org/10.1172/jci.insight.99223.
Contributor Information
Sarah Thomas Babu, Email: sarahthomas056@gmail.com.
Xinying Niu, Email: xinying.niu@utsouthwestern.edu.
Julie Mirpuri, Email: julie.mirpuri@utsouthwestern.edu.
References
- 1.Bellatorre A, Scherzinger A, Stamm E, Martinez M, Ringham B, Dabelea D. Fetal overnutrition and adolescent hepatic fat fraction: the Exploring Perinatal Outcomes in Children Study. J Pediatr. 2018;192:165–170.e1. doi: 10.1016/j.jpeds.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115(3):e290–e296. doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- 3.Aune D, Saugstad OD, Henriksen T, Tonstad S. Maternal body mass index and the risk of fetal death, stillbirth, and infant death: a systematic review and meta-analysis. JAMA. 2014;311(15):1536–1546. doi: 10.1001/jama.2014.2269. [DOI] [PubMed] [Google Scholar]
- 4.Ogden CL, Yanovski SZ, Carroll MD, Flegal KM. The epidemiology of obesity. Gastroenterology. 2007;132(6):2087–2102. doi: 10.1053/j.gastro.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 5.Chu DM, Meyer KM, Prince AL, Aagaard KM. Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function. Gut Microbes. 2016;7(6):459–470. doi: 10.1080/19490976.2016.1241357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rando OJ, Simmons RA. I’m eating for two: parental dietary effects on offspring metabolism. Cell. 2015;161(1):93–105. doi: 10.1016/j.cell.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol. 2015;212(5):653.e1–653.16. doi: 10.1016/j.ajog.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasson IE, Vitins AP, Mainigi MA, Moley KH, Simmons RA. Pre-gestational vs gestational exposure to maternal obesity differentially programs the offspring in mice. Diabetologia. 2015;58(3):615–624. doi: 10.1007/s00125-014-3466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank DN, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17(1):179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 12.DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Decker GA, Rittmann BE. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83(4):460–469. doi: 10.4065/83.4.460. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov, Littman DR. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010;3(3):209–212. doi: 10.1038/mi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang XL, et al. Faecalibacterium prausnitzii supernatant ameliorates dextran sulfate sodium induced colitis by regulating Th17 cell differentiation. World J Gastroenterol. 2016;22(22):5201–5210. doi: 10.3748/wjg.v22.i22.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gálvez J. Role of Th17 cells in the pathogenesis of human IBD. ISRN Inflamm. 2014;2014:928461. doi: 10.1155/2014/928461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neu J, Pammi M. Pathogenesis of NEC: Impact of an altered intestinal microbiome. Semin Perinatol. 2017;41(1):29–35. doi: 10.1053/j.semperi.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Tanner SM, et al. Pathogenesis of necrotizing enterocolitis: modeling the innate immune response. Am J Pathol. 2015;185(1):4–16. doi: 10.1016/j.ajpath.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sodhi C, Richardson W, Gribar S, Hackam DJ. The development of animal models for the study of necrotizing enterocolitis. Dis Model Mech. 2008;1(2-3):94–98. doi: 10.1242/dmm.000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37(4):601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357(6354):912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cupedo T, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10(1):66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 22.Marquardt N, et al. Fetal CD103+ IL-17-producing group 3 innate lymphoid cells represent the dominant lymphocyte subset in human amniotic fluid. J Immunol. 2016;197(8):3069–3075. doi: 10.4049/jimmunol.1502204. [DOI] [PubMed] [Google Scholar]
- 23.Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. 2010;33(5):752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Sonnenberg GF, Fouser LA, Artis D. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol. 2010;107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 25.Mirpuri J, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5(1):28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 28.Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103(42):15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleerebezem M, Vaughan EE. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu Rev Microbiol. 2009;63:269–290. doi: 10.1146/annurev.micro.091208.073341. [DOI] [PubMed] [Google Scholar]
- 30.Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: Moving beyond associations. Cell Host Microbe. 2017;22(5):589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Statovci D, Aguilera M, MacSharry J, Melgar S. The impact of Western diet and nutrients on the microbiota and immune response at mucosal interfaces. Front Immunol. 2017;8:838. doi: 10.3389/fimmu.2017.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu J, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39(2):386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenberg GF. Regulation of intestinal health and disease by innate lymphoid cells. Int Immunol. 2014;26(9):501–507. doi: 10.1093/intimm/dxu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortez VS, Robinette ML, Colonna M. Innate lymphoid cells: new insights into function and development. Curr Opin Immunol. 2015;32:71–77. doi: 10.1016/j.coi.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buela KA, Omenetti S, Pizarro TT. Cross-talk between type 3 innate lymphoid cells and the gut microbiota in inflammatory bowel disease. Curr Opin Gastroenterol. 2015;31(6):449–455. doi: 10.1097/MOG.0000000000000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 38.Sano T, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163(2):381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Withers DR, Hepworth MR. Group 3 Innate lymphoid cells: Communications hubs of the intestinal immune system. Front Immunol. 2017;8:1298. doi: 10.3389/fimmu.2017.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reichardt N, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8(6):1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawa S, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330(6004):665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 42.Gomez de Agüero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 43.Lee JS, et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity. 2015;43(4):727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibbons DL, et al. Neonates harbour highly active gammadelta T cells with selective impairments in preterm infants. Eur J Immunol. 2009;39(7):1794–1806. doi: 10.1002/eji.200939222. [DOI] [PubMed] [Google Scholar]
- 45.Morita CT, Parker CM, Brenner MB, Band H. TCR usage and functional capabilities of human gamma delta T cells at birth. J Immunol. 1994;153(9):3979–3988. [PubMed] [Google Scholar]
- 46.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352(6285):544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.