SUMMARY
Foxp3+ regulatory T cells (Treg) are essential modulators of immune responses, but the molecular mechanisms underlying their function are not fully understood. Here we show that the transcription factor Blimp-1 is a crucial regulator of the Foxp3+RORγt+ Treg subset. The intrinsic expression of Blimp-1 in these cells is required to prevent production of Th17-associated cytokines. Direct binding of Blimp-1 to the Il17 locus in Treg is associated with inhibitory histone modifications but unaltered binding of RORgt. In the absence of Blimp-1, the Il17 locus is activated, with increased occupancy of the co-activator p300 and abundant binding of the transcriptional regulator IRF4, which is required, along with RORγt, for IL-17 expression in the absence of Blimp-1. We also show that despite their sustained expression of Foxp3, Blimp-1−/− RORγt+IL-17-producing Treg lose suppressor function and can promote intestinal inflammation, indicating that repression of Th17-associated cytokines by Blimp-1 is a crucial requirement for RORγt+ Treg function.
Graphical Abstract
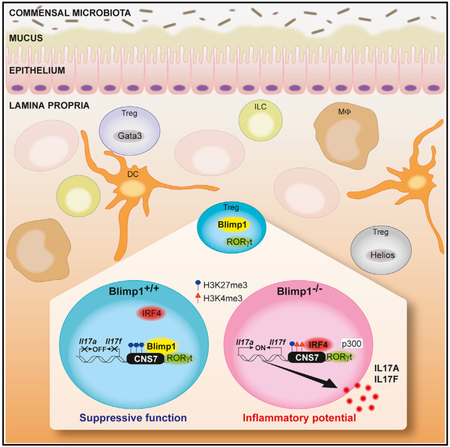
In Brief
Ogawa et al. demonstrate that the transcription factor Blimp-1 is required to prevent production of Th17-associated cytokines and inflammatory activity of microbiota-specific Foxp3+RORγt+ Treg. These findings uncover a critical role for Blimp-1 in Foxp3+Treg function and shed light on the intricate mechanisms underlying Treg phenotypic stability.
INTRODUCTION
Regulatory T cells (Treg) constitute a subset of T cells with the capability to suppress immune responses and are crucial for the maintenance of immune homeostasis. The most well-characterized Treg subpopulation is defined by the expression of the transcription factor Foxp3 (Fontenot et al., 2005). Mutations or deficiency of the Foxp3 gene in mice and humans results in severe autoimmunity (Brunkow et al., 2001) (Bennett et al., 2001). Foxp3+ Treg can develop in the thymus and in the periphery (Apostolou et al., 2002; Fontenot et al., 2005), and studies suggest that functional specification in Treg subpopulations requires the expression of transcription factors associated with the differentiation and function of effector CD4+ T cell lineages (Chaudhry and Rudensky, 2013; Levine et al., 2017; Wohlfert et al., 2011). Expression of these factors would drive the generation of effector Treg that are specifically suited to regulate immune responses mediated by their corresponding conventional effector CD4+T cell lineages. Thus, in the context of T helper type 1 (Th1)-mediated inflammation, Treg can upregulate expression of the Th1-specific transcription factor T-bet, leading to the accumulation of Foxp3+ T-bet+ Treg that were essential for control of Th1 inflammation (Levine et al., 2017). Similarly, the T helper type 2 (Th2)-associated transcription factor GATA-3 was found to play an important role for Treg function, and mice with GATA-3−/− Treg have defects in peripheral homeostasis caused by impaired Treg suppressive function (Wohlfert et al., 2011). Treg-specific deletion of the transcription factors IRF4 (Zheng et al., 2009) and STAT3 (Chaudhry et al., 2009) resulted in impaired regulation of Th2- and T helper type 17 (Th17)-dominated immune responses, respectively. Studies have identified a CD4+Foxp3+ Treg population that simultaneously expresses the transcription factor retinoic acid-related orphan receptor-γt (RORγt) (Lochner et al., 2008; Ohnmacht et al., 2015; Sefik et al., 2015), initially described as a crucial requirement for development of Th17 cells. Foxp3+RORγt+ Treg are present in both mice and humans and are enriched in the intestinal mucosa but can also be found in peripheral lymphoid organs (Ohnmacht et al., 2015; Sefik et al., 2015). Foxp3+RORγt+ Treg include microbiota-specific Treg, they can be induced in response to pathobionts (Xu et al., 2018), and their frequency is greatly diminished in germ-free mice or antibiotic-treated specific pathogen-free (SPF) mice (Ohnmacht et al., 2015; Sefik et al., 2015). This Treg subpopulation expresses the regulatory cytokine interleukin (IL) 10 and other molecules associated with Foxp3+ Treg function, such as ICOS, CTLA-4, CD39, and CD73 (Ohnmacht et al., 2015). Despite their expression of RORgt and other genes associated with the Th17 program, Foxp3+RORγt+ Treg do not produce IL-17 (Lochner et al., 2008; Sefik et al., 2015) and have a potent suppressor function, but the mechanisms underlying their phenotype are poorly understood.
B lymphocyte-induced maturation protein-1 (Blimp-1, Prdm1) is a transcription factor expressed in several hematopoietic lineages, including lymphocytes (Martins and Calame, 2008). Blimp-1 is highly expressed in CD4+ T cells and directly regulates the expression of genes associated with T cell effector and Treg function, including cytokines (IL-2, IL-10, and IL-17) (Bankoti et al., 2017; Cretney et al., 2011; Martins et al., 2006, 2008; Salehi et al., 2012; Heinemann et al., 2014; Neumann et al., 2014). In line with this, mice with T cell-specific (CD4cre or proximal LCKcre-mediated) deletion of Blimp-1 spontaneously develop chronic intestinal inflammation (Martins et al., 2006, 2008; Salehi et al., 2012). However, Foxp3+ Treg-specific deletion of Blimp-1 leads to only mild intestinal inflammation, associated with enhanced production of IL-10 by Foxp3− T effector cells (Bankoti et al., 2017), indicating differential requirements for Blimp-1 in Foxp3+ and Foxp3− T cells. This notion is supported by the observation that Blimp-1 regulates unique target genes in each cell type (Bankoti et al., 2017); however, intrinsic studies of Blimp-1’s role in Foxp3+ Treg have been hindered by the fact that only a fraction of Foxp3+ Treg express Blimp-1 under homeostatic conditions (Bankoti et al., 2017; Cretney et al., 2011).
Here, we report that Blimp-1 is preferentially expressed in the Foxp3+RORγt+ Treg subset and its expression is required and sufficient to repress Th17-associated cytokines in an IL-10-independent manner. Furthermore, we show that Blimp-1−/−IL-17-producing Foxp3+ Treg lose suppressor activity in vivo and can cause intestinal inflammation. Our findings also reveal that specific binding of Blimp-1 to the Il17a and Il17f genes in wild-type (WT) Foxp3+ Treg is associated with changes in the chromatin structure. Lack of Blimp-1 expression and the chromatin changes associated with it did not interfere with binding of RORγt but was associated with increased binding of the co-activator p300 and of the transcription factor IRF4, which was required for RORγt-mediated transcription of Il17 in the absence of Blimp-1. Thus, Foxp3+RORγt+ microbiota-specific Treg rely on Blimp-1 to suppress production of Th17-associated cytokines and maintain their regulatory function.
RESULTS
Blimp-1 Is Preferentially Expressed in Foxp3+RORγt+Helios− Treg
To characterize Blimp-1-expressing Foxp3+ Treg, we compared expression of effector T cell function-associated transcription factors in Foxp3+Blimp-1+ and Foxp3+Blimp-1− Treg. We found that the Th17-associated transcription factor RORγt was more abundantly expressed in Foxp3+Blimp-1+Tregthan in Blimp-1 −Foxp3+ Treg (Figure S1A). Foxp3+RORγt+ cells were described as peripherally induced, microbiota-specific Helios− Foxp3+ Treg that are required to control effector responses in the intestinal mucosa (Lochner et al., 2008; Ohnmacht et al., 2015; Sefik et al., 2015). Comparison of Blimp-1 mRNA expression in Foxp3+RORγt+Helios− and Foxp3+RORγt−Helios+Treg isolated from naive mice revealed consistently higher expression of Blimp-1 mRNA (as reported by yellow fluorescent protein [YFP]) in Foxp3+RORγt+Helios− cells than in Foxp3+RORγt−Helios+ cells (Figure 1A). In addition, Foxp3+RORγt+ cells sorted from the intestinal mucosa of Foxp3 and RORγt dual reporter mice had higher expression of Blimp-1 than their Foxp3+RORγt− counterparts (Figure 1B). Conversely, expression of the transcription factor Bcl6, which is directly suppressed by Blimp-1 (Cimmino et al., 2008), was less abundant in RORγt+ Treg than in RORγt− Treg (Figure S1B).
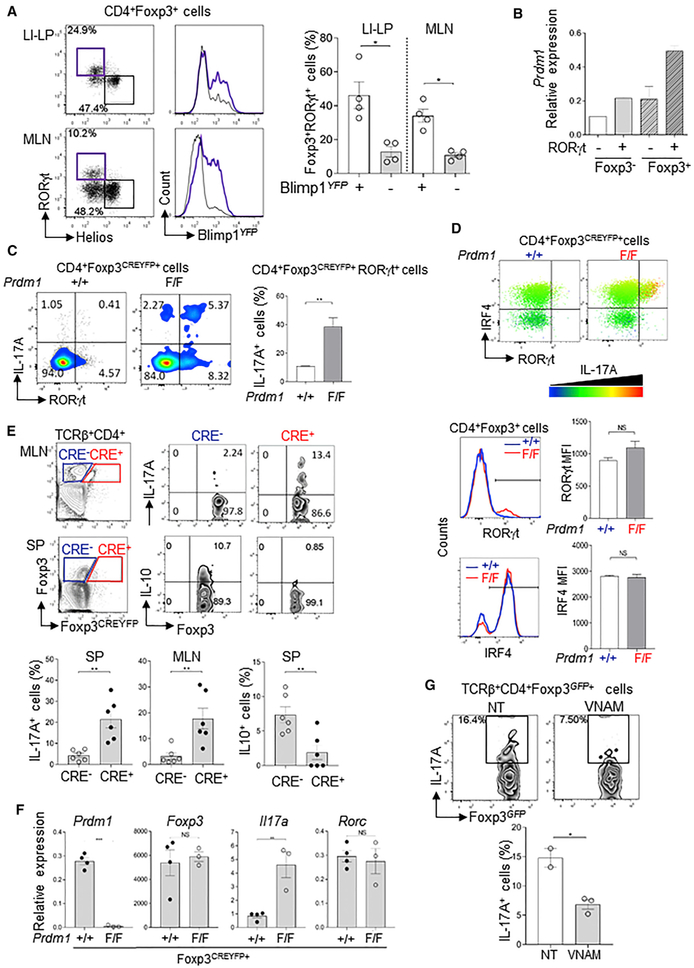
Figure 1. Blimp-1 Is Preferentially Expressed in Microbiota-Specific Foxp3+RORγt+ Treg and Is Required to Repress IL-17A Expression.
(A) Blimp-1YFP expression in RORγt+Helios− (purple) and RORγt−Helios+ (black) Foxp3+ Treg (left) and frequency of RORγt+ in Foxp3+Blimp-1YFP+ and Foxp3+Blimp-1 YFP− cells in the large intestine’s lamina propria (LI-LP) and mesenteric lymph nodes (MLNs) of Blimp-1YFP reporter mice (right).
(B) Prdm1 (Blimp-1) mRNA expression in Foxp3+RORγt+ and Foxp3+RORγt− cells sorted from the small intestine of RORγtGFPFoxp3RFP mice.
(C) IL-17A expression in splenic CD4+Foxp3+RORγt+ cells of Prdm1+/+Foxp3CREYFP+ and Prdm1F/FFoxp3 YFP−CRE+ mice.
(D) IL-17A expression in CD4+Foxp3+RORγt+IRF4+ cells (bar color indicates IL-17 expression intensity), and RORγt and IRF4 expression in Blimp-1+/+ (blue) or Blimp-1−/− (red) Foxp3+ cells.
(E) IL-17A (top) and IL-10 (middle) expression in Blimp-1+/+ (CRE−) and Blimp-1−/− (CRE+) TCRβ+CD4+Foxp3+ cells in MLNs and spleen (SP) of Prdm1F/FFoxp3YFP−CRE+ female mice. Bar graphs (bottom) show the frequency of IL17A+ (left) and IL10+ (right) cells.
(F) Expression of Prdm1, Il17a, Rorc, and Foxp3 mRNA in CD4+Foxp3+ cells sorted from Prdm1F/F and Prdm1+/+Foxp3CREYFP+ mice and re-stimulated in vitro.
(G) Expression of IL-17A in TCRβ+CD4+Foxp3+ cells from MLNs of antibiotic-treated with vancomycin, neomycin, ampycilin, and methonidazole (VNAM) or non-treated (NT) Prdm1F/FCD4cre+ mice.
Data are representative of at least two independent experiments. Error bars show average and SEM (n ≥ 3 per group); each symbol represents one mouse. *p < 0.05 and **p < 0.01, unpaired Student’s t test (A, C, D, F, and G) and paired Student’s t test (E).
Expression of Blimp-1 in Foxp3+RORγt+ Treg Is Required to Repress Th17-Associated Cytokines
To investigate Blimp-1’s role in Foxp3+RORγt+ Treg, we used Prdm1F/FFoxp3YFP−CRE+ mice, in which Blimp-1 is specifically deleted from Foxp3+ Treg (Bankoti et al., 2017). We found that the frequency of Foxp3+RORγt+ Treg was elevated in the absence of Blimp-1, whereas the frequency of RORγt−GATA− 3+Foxp3+ Treg was unaltered (Figure S1C). Analysis of cytokine expression upon in vitro stimulation revealed that Blimp-1−/−Foxp3+RORγt+ Treg produced IL-17A (Figure 1C), but not interferon-γ (IFNγ) or IL-4 (data not shown). Similar results were obtained in mice with T cell-specific deletion of Prdm1 (Prdm1F/F/CD4cre+) crossed with Foxp3RFP and IL17GFP double reporter mice, in which we detected IL-17A-expressing Fox-p3+RORγt+ cells in the mesenteric lymph nodes (MLNs) and lamina propria (LI-LP), in addition to the lungs (Figure S2A). Most IL-17F-expressing Foxp3+RORγt+ Treg also expressed IL-17A (Figure S2B), indicating that both Il17a and Il17f genes were activated in these cells. These findings were reproduced in Prdm1F/F/CD4CRE+ mice devoid of any reporter gene, in which simultaneous staining of Foxp3 and IL-17A proteins revealed significant expression of IL-17A in Foxp3+ Treg (Figure S2C).
Blimp-1 −/−Foxp3+ Treg secreted measurable amounts of IL-17A, and as expected from previous studies implicating Blimp-1 as a positive regulator of IL-10 expression (Cretney et al., 2011; Martins et al., 2006), Blimp-1 −/−Foxp3+ Treg produced reduced amounts of IL-10 (Figure S2D). All IL-17A-producing Foxp3+Treg in the Prdm1F/FFoxp3YFP−CRE+ mice expressed RORγt and coexpressed the transcription factor IRF4 (Figure 1D), which is associated with Treg function (Zheng et al., 2009) and IL-17 production in Th17 cells (Brüstle et al., 2007; Ciofani et al., 2012). However, lack of Blimp-1 did not alter the amounts of RORγt or IRF4 protein expressed on a per-cell basis (Figure 1D), indicating that Blimp-1 does not regulate the expression of Rorc or Irf4 genes in Foxp3+ Treg. This was also indicated by qRT-PCR analysis of Rorc and IRF4 mRNA in Blimp-1+/+ and Blimp-1−/−Foxp3+ Treg (Figure S2E).
Blimp-1 −/−Foxp3+ Treg from female Prdm1F/FFoxp3YFP−CRE+ also showed pronounced expression of IL-17A and decreased expression of IL-10, despite the presence of Blimp-1+/+ Treg, in which cytokine expression was not altered (Figure 1E), indicating that regulation of IL-17 expression by Blimp-1 was a cell-intrinsic effect. In addition, Blimp-1 −/− Treg expressed substantial amounts of IL-17A and IL-17F mRNA, whereas expression of Foxp3 mRNA was unaltered (Figure 1F; Figure S2F). In comparison to their IL-17-counterparts, IL-17-producing Blimp-1−/−Foxp3+RORγt+ Treg expressed higher levels of activation markers and other surface molecules associated with Treg effector function (Figure S3). This is in line with the previous observation that Blimp-1 is preferentially expressed in effector Treg (Bankoti et al., 2017; Cretney et al., 2011; Dias et al., 2017).
Addition of recombinant IL-10 to - in vitro cultured Treg did not abrogate the difference in production of IL-17A in Blimp-1−/− and Blimp-1+/+ Treg (Figures S4A and S4B), indicating that IL-17A expression by these cells is not a secondary effect of diminished IL-10 expression. In line with this, IL-10−/− Foxp3+ Treg showed only a small, non-significant increase in IL-17A expression in comparison with wild-type Foxp3+ Treg (Figure S4C). We also found that treatment of Blimp-1 conditional knock-out (Blimp-1CKO) mice with a combination of antibiotics that eliminate most intestinal microbiota led to a significant decrease in IL-17A-producing Blimp-1 −/−Foxp3+ Treg (Figure 1G). Thus, expression of Blimp-1 in Foxp3+RORγt+ Treg is intrinsically required to prevent production of Th17-associated inflammatory cytokines, and Blimp-1−/−RORγt+ IL-17-producing Treg are reduced upon disruption of the intestinal microbiota by antibiotic treatment.
Blimp-1 Binding to the Il17 Locus Results in Diminished Chromatin Accessibility
We next addressed whether Blimp-1 could function as a direct suppressor of the Il17 locus in Treg. We first determined whether enforced expression of Blimp-1 in Blimp-1 −/−Foxp3+ Treg was sufficient to abrogate IL-17 expression. We found that unlike what was observed in in vitro-differentiated Th17 cells, retroviral-driven expression of Blimp-1 led to significant repression of IL-17 in Blimp-1−/− Treg (Figure 2A). Chromatin immunopre-cipitation (ChIP) analysis of wild-type Foxp3GFP+ cells stimulated in vitro at conditions that induced Il17a and Il17f mRNA expression revealed that Blimp-1 directly binds to a consensus site located upstream of conserved non-coding sequence (CNS) 2 and strongly binds to two other consensus sites located at the Il17a promoter and at the CNS7 region (upstream of the Il17f promoter), but not to Il17a intron I, which also contained a Blimp-1 binding site (Figure 2B). Binding of Blimp-1 at these sites in Treg was significantly higher than in pathogenic (p) Th17 cells, in which Blimp-1 is expressed at very low levels (Bankoti et al., 2017) but was reported to bind to similar regions of the Il17 locus (Jain et al., 2016). Thus, Blimp-1 directly binds to conserved regulatory regions at the Il17 locus and could directly repress the expression of this locus in Foxp3+ Treg.
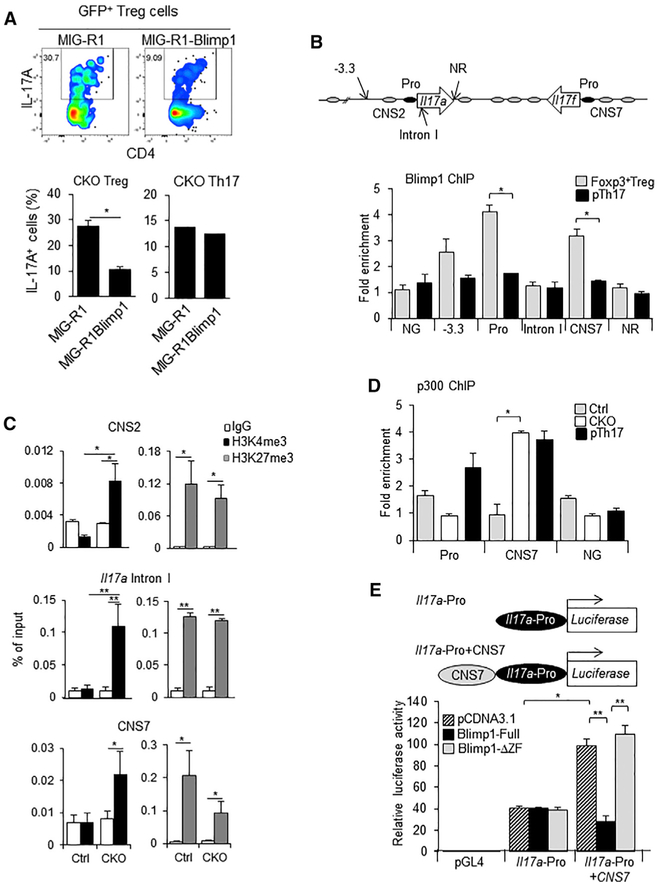
Figure 2. Binding of Blimp-1 to I117a and Il17f Genes in Foxp3+ Treg Is Associated with Decreased Locus Accessibility.
(A) IL-17A expression in Blimp-1−/− Foxp3GFP+ Treg and in wild-type Th17 cells transduced with MIG-R1 or Blimp-1-expressing MIG-R1 retrovirus (MIG-R1Blimp1).
(B) Top: representation of the Il17 locus (arrows, Il17a and Il17f genes; black ovals, promoter; gray ovals, CNS regions) indicating Blimp-1 consensus binding sites and a non-related (NR) site; bottom: ChIP of Blimp-1 in wild-type CD4+Foxp3GPP+Treg and pTh17 cells (NG, negative control: non-related gene lacking Blimp-1 binding sites).
(C) ChIP of H3K4me3 (black bars) and H3K27me3 (gray bars) at different regions of the Il17 locus in Blimp-1+/+ (Ctrl, control) or Blimp-1−/− (CKO) CD4+Foxp3GFP+ Treg (empty bars, Ctrl antibody immunoprecipitation [IP]).
(D) ChIP of p300 at the Il17a promoter and Il17 CNS7 regions in CD4+Foxp3GPP+ Treg from Ctrl and CKO mice and in wild-type pTh17 cells.
(E) Representation of promoter constructs (top) and luciferase activity assay (bottom).
Striped bars, control plasmid; black bars, full-length Blimp-1; gray bars, truncated Blimp-1 lacking the zinc finger-containing region. Data shown are from two (A) or three (B–E) independent experiments. Error bars show SEM (n ≥ 3 per group). *p < 0.05 and **p < 0.01, one-way ANOVA.
We also found that recruitment of Blimp-1 to the Il17 locus in Treg was associated with increased accumulation of histone H3 lysine 4 trimethylation (H3K4me3) in three regions of the locus, CNS2, Il17a intron I, and CNS7 (Figure 2C), and decreased deposition of histone H3 lysine 27 trimethylation (H3K27me3) specifically at CNS7, but not at CNS2 or Il17a Intron I. Thus, binding of Blimp-1 to the Il17 locus in Treg correlates with histone modifications previously associated with regulation of the activity of this locus in Th17 cells (Wei et al., 2009). Moreover, we found that binding of p300, a histone acetyltransferase that usually marks active transcription in regulatory domains, was significantly increased at the CNS7 region of the Il17 locus in Blimp-1 −/−Foxp3+Treg (Figure 2D), reaching levels similar to those the observed in wild-type pTh17 cells, in which the Il17 locus is fully active. Using a luciferase reporter construct containing the Il17 locus CNS7 region upstream of the Il17a promoter region, we found that the CNS7 region can function as an enhancer for the Il17a promoter and this activity is abrogated in the presence of a full-length, but not of a truncated, Blimp-1 construct lacking a DNA-binding domain (Figure 2E). Thus, binding of Blimp-1 at the Il17 locus CNS7 region help to prevent and/or control locus activation and therefore production of Th17-associated cytokine by Foxp3+ Treg.
Increased Binding of IRF4 at the Il17 Locus Promotes Expression of Th17-Associated Cytokines in Blimp-1−/− Treg
We next examined whether increased accessibility of the Il17 locus in Blimp-1−/− Treg could facilitate RORyt binding. ChIP of RORyt in wild-type and Blimp-1−/− Treg showed no significant increase in RORγt binding at the Il17 locus in the absence of Blimp-1 (Figure 3A). Thus, Blimp-1-mediated repression of the Il17a and Il17f genes in Treg is not achieved by impairment of RORγt binding to the locus. However, RORγt expression was required for the expression of IL-17 by Blimp-1−/− Treg, because IL-17 expression was significantly inhibited in Blimp-1 and RORγt double-knockout cells (Figure 3B).
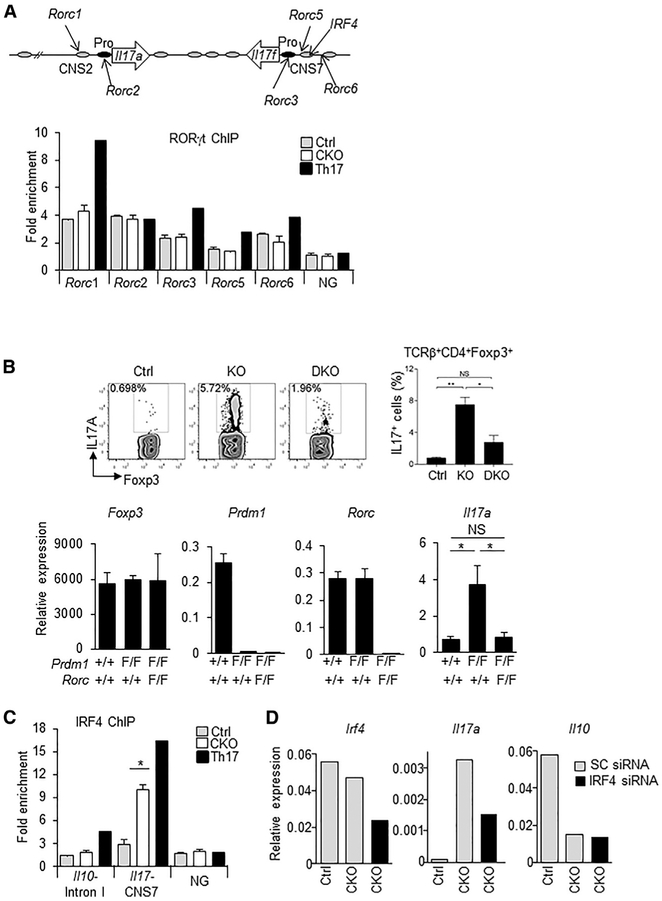
Figure 3. Increased Binding of IRF4 at the Il17 Locus and IRF4 and RORγt Requirement for IL-17A Production in Blimp-1 −/−Foxp3+ Treg.
(A) Representation of the Il17 locus showing RORγt and IRF4 consensus binding sites (top) and RORgt ChIP (bottom) in Blimp-1+/+ and Blimp-1 −/−CD4+ Foxp3GFP+ Treg and wild-type pTh17 cells.
(B) IL-17A expression in TCRβ+CD4+Foxp3+ Treg (MLNs) from Prdm1+/+Rorc+/+Foxp3CREYFP+ (Ctrl), Prdm1F/FRorc+/+Foxp3CREYFP+ (knockout [KO]), and Prdm1 F/FRorcF/FFoxp3CREYFP+ (double knockout [DKO]) mice and expression of Foxp3, Prdm1, Rorc, and Il17a mRNA (bottom graphs) in sorted and in vitro-stimulated (phorbol myristate acetate [PMA] and ionomycin for 4 hr) CD4+Foxp3CREYFP+ cells.
(C) IRF4 ChIP at the Il17 CNS7 and Il10 Intron I regions in Blimp1+/+ and Blimp1 −/−CD4+Foxp3GFP+ Treg and wild-type pTh17 cells.
(D) Irf4, Il17a, and Il10 mRNA expression following Irf4 siRNA knockdown (SC, scrambled control).
Data shown are representative of at least two independent experiments. Error bars show SEM (n ≥ 3 per group). *p < 0.05 and **p < 0.01, one-way ANOVA.
We next studied whether IRF4, which is highly expressed in Treg and one of the pioneer transcription factors required for Th17 specification (Ciofani et al., 2012), could regulate activation of the Il17 locus in Blimp-1−/− Treg. Although IRF4 expression levels were unaltered on a per-cell basis in Blimp-1−/− Treg (Figure 1D; Figure S2E), we found that Blimp-1 −/−Foxp3+ cells had a significant increase in IRF4 binding at the Il17 CNS7 region (Figure 3C). Binding of IRF4 at Il10 intron I, a region previously shown to be occupied in Treg, was not altered in Blimp-1 −/− cells. Thus, lack of Blimp-1 led to a specific increase in IRF4 binding to the Il17 locus. Knockdown of IRF4 by small interfering RNA (siRNA) in Blimp-1−/− Treg resulted in a significant decrease in IL-17A expression but unaltered IL-10 expression (Figure 3D), indicating that production of IL-17A in these cells is at least partially mediated by IRF4, which had increased binding to the Il17 locus in the absence of Blimp-1.
Blimp-1−/−IL-17-Producing Foxp3+ Treg Lose Suppressor Function and Display Inflammatory Activity In Vivo
We and others have previously shown that despite their impaired IL-10 production, peripheral Blimp-1 −/− Treg preserve their sup-pressive function (Martins et al., 2006; Kallies et al., 2006). However, these results might have been confounded by Blimp-1−/−Foxp3+ Treg comprising a heterogenous population in which only the RORγt-expressing cells produce IL-17A. To address this, we used a T cell adoptive transfer colitis induction model to test the function of IL-17A-producing Blimp-1−/−Foxp3+ Treg sorted from Foxp3RFP and IL-17AGFP double reporter Prdm1 F/F/CD4cre+ mice (Figure S5A). We found that Blimp-1−/−Foxp3RFP+ IL-17AGFP− Treg prevent colitis development, whereas Blimp-1−/−Foxp3RFP+IL-17AGFP+ Treg failed to prevent colitis induction (Figures 4A-4F). Although expression of IL-10 was reduced in Blimp-1−/−Foxp3+ Treg, expression of IL-10 in these cells did not preferentially segregate with expression of IL-17 (Figure S5B). Similarly, expression of CTLA-4, ICOS, and GITR (Figures S5C and S5D) was comparable in IL-17+ and IL-17− Blimp-1−/− Treg. In addition, although both Foxp3+IL-17A-producing and Foxp3+IL-17A-non-producing Treg partially lost Foxp3 expression, expression of Foxp3 in recovered Treg was significantly higher in mice injected with IL-17GFP+ cells (Figure 4D).
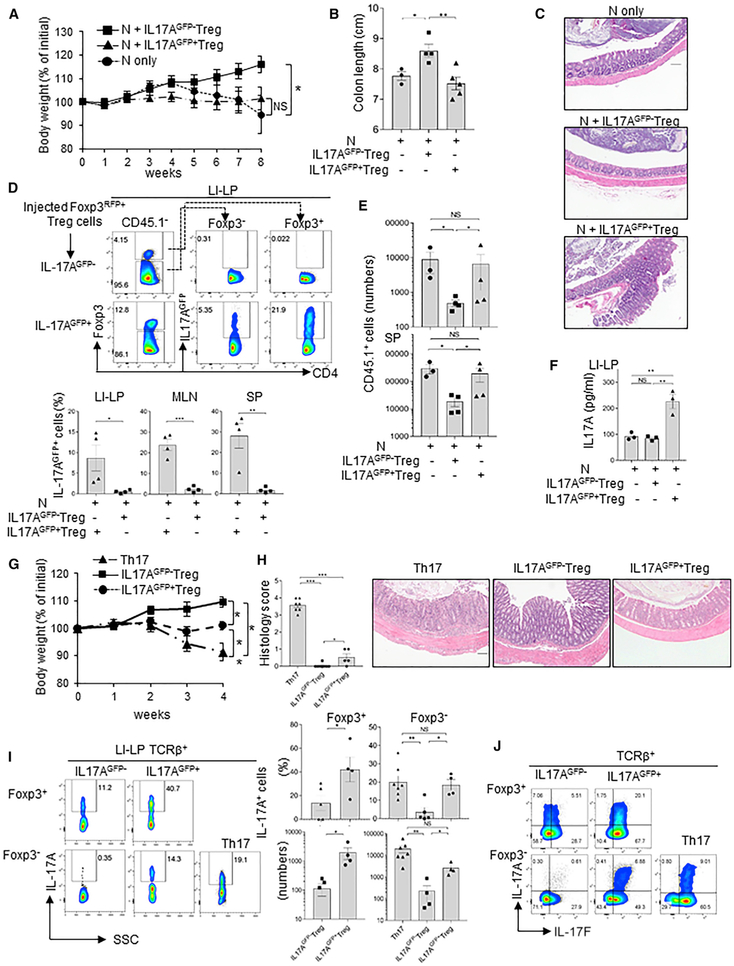
Figure 4. IL-17A-Producing Blimp1 −/− Foxp3+ Treg Have Impaired Suppressive Function and Can Cause Intestinal Inflammation.
(A) Body weight of RAG−/− mice injected with naive CD45.1+CD4+T cells alone or in combination with (CD45.2+) Blimp-1 −/−IL-17A-producing (IL17AGFP+ Treg) or non-producing (IL-17AGFP− Treg) Foxp3+ Treg from the same donor mice.
(B and C) Colon length (B) and H&E-stained histological sections (C) (10x magnification; scale bar, 100 μm) in mice shown in (A) 8 weeks after transfer.
(D) Frequency (fluorescence-activated cell sorting [FACS] plots) and absolute numbers (bar graphs) of IL-17AGFP+ cells from LI-LP of mice shown in (A)-(C).
(E) Numbers of CD45.1 + cells in the LI-LP, MLNs, and SP from mice shown in (A)–(D).
(F) IL-17A expression (ELISA) in LI-LP cells of mice shown in (A)ߝ(E).
(G) Body weight of RAG−/− mice adoptively transferred with IL-17+ Th17 cells (Th17), Blimp1 −/− IL17AGFP+, or IL17AGFP−Foxp3+ Treg.
(H) Histology scores (colon, rectum, and cecum; left), and pictures of colon histological sections (H&E stained; 10x magnification; scale bar, 100 μm) of mice shown in (G).
(I) IL-17A expression in TCRβ+ T cells from the LI-LP of mice show in (G). Foxp3+, cells that maintained Foxp3; Foxp3−, cells that had lost Foxp3 expression after transfer.
(J) Expression of IL-17A and IL-17F in TCRβ+Foxp3+ and Foxp3− cells in SP of mice shown in (G)–(I).
Data shown are from three different experiments. Error bars show SEM (n = 3–7 mice per group). *p < 0.05 and **p < 0.01, one-way ANOVA (A, B, D, and F–I) and unpaired Student’s t test (E and I).
In comparison to IL-17AGFP−Foxp3+ cells, IL-17GFP+Foxp3+ cells failed to repress expansion of the co-transferred naive cells, as indicated by the numbers of cells recovered at the experimental endpoint (Figure 4E). Foxp3+IL-17AGFP+ Treg retained IL-17A expression upon transfer (Figure 4D), which resulted in higher secretion of IL-17A in LI-LP cells of mice injected with these cells (Figure 4F). Altogether, these observations suggest that IL-17-expressing Foxp3+Treg are less efficient in suppressing effector T cell expansion in vivo.
We next tested whether Blimp-1 −/−IL-17GFP+ Treg could display inflammatory activity when adoptively transferred to RAG1−/− mice in the absence of naive CD4+ T cells. As a positive control, we transferred in vitro-differentiated IL-17GFP+ Th17 cells, which were previously shown to cause severe intestinal inflammation in this model (Gagliani etal., 2015). Blimp-1−/−IL-17GFP+Treg caused mild but noticeable intestinal inflammation, associated with impaired body weight gain (Figure 4G) and higher numbers of inflammatory cells in the LI-LP (Figure 4H; Figure S6A). This was in contrast to Blimp-1 −/− IL-17GFP~ Treg, which did not cause measurable inflammation. Mice injected with Blimp-1−/−IL17GFP+ Treg also had significantly more IL-17A- and IL-17F-producing cells (Figures 4I and4J; Figure S6B) than mice transferred with Blimp−/−IL-17GFP− Treg. Cells recovered from mice injected with IL-17GFP+ Treg retained significantly more Foxp3 expression than cells recovered from mice injected with IL-17GFP− Treg (Figure S6C). Expression of IL-10 was not significantly different between the different recipient mice groups (Figure S6D). Thus, expression of Blimp-1 in RORγt+ microbiota-specific Foxp3+ Treg is required to inhibit Th17-associated cytokines, maintain suppressive function, and prevent inflammatory activity.
DISCUSSION
Our studies described here reveal a role for the transcription factor Blimp-1 in controlling key functional aspects of a specific Foxp3+ Treg subset. We show that Blimp-1 was preferentially expressed by Foxp3+RORγt+ Treg and the intrinsic expression of Blimp-1 in these cells was required to restrain production of Th17-associated cytokines, maintain their suppressive function, and prevent inflammatory activity. Blimp-1 functioned as a direct repressor of the Il17 locus in Foxp3+ Treg, and its direct binding to the locus altered chromatin structure and restricted binding of the Th17-associated factor IRF4, which was required, along with RORγt, for IL-17 expression in Blimp-1 −/−Foxp3+ cells. These findings uncover an unappreciated aspect of Blimp1’s role in Foxp3+ Treg biology and shed light on the intricate mechanisms underlying Treg phenotypic stability.
RORγt+ microbiota-specific Foxp3+ Treg were described as an important component of the Foxp3+ Treg pool that controls homeostasis in the intestinal mucosa (Lochner et al., 2008; Sefik et al., 2015). Despite their constitutive expression of RORγt, these cells do not produce Th17-associated cytokines and have potent regulatory function under homeostatic conditions (Ohnmacht et al., 2015; Sefik et al., 2015). Here we show that Foxp3+RORγt+ Treg rely on Blimp-1 to repress expression of Th17-associated cytokines. Blimp-1 −/−Foxp3+RORγt+ Treg produce IL-17Aand IL-17F, have impaired suppressive function, and can cause intestinal inflammation when adoptively transferred to RAG1−/− mice. Moreover, we show that IL-17A production by Blimp-1 −/−Foxp3+RORγt+ Treg does not preferentially segregate with lower expression of molecules required for Treg suppression function, including IL-10, CTLA-4, GITR, and ICOS, and it was not a secondary effect of impairment of IL-10. Although we have not directly addressed IL-17-producing Blimp-1 −/− Foxp3+RORγt+Treg T cell receptor (TCR) specificity, we find that antibiotic treatment greatly reduces the number of IL-17-producing Foxp3+ Treg in Blimp-1 −/− mice, suggesting that these cells are microbiota dependent.
IL-17-producing Blimp-1 −/−RORγt+ Treg maintained expression of Foxp3 after adoptive transfer to RAG1−/− mice, suggesting that these cells were not immediately transitioning into Th17 cells, a phenotype observed in some inflammatory conditions (Komatsu et al., 2014; Kim et al., 2017). Foxp3 alone is not sufficient to repress IL-17 expression in Blimp-1 −/− Treg, although Foxp3 can repress IL-17 expression through antagonism of RORγt (Ichiyama et al., 2008; Zhou et al., 2008). This remains to be investigated but could be due to differences in the availability of Th17-inducing factors in RORγt+ Treg.
The mechanisms underlying the repression of the Il17 locus by Blimp-1 in Foxp3+ Treg do not involve repression of the expression of known Th17-promoting transcription factors such as RORγt and IRF4, because we found no significant changes in the expression of these proteins on a per-cell basis in Blimp1−/−Foxp3+ cells, although the number of Foxp3+RORγt+ Treg was elevated in Blimp-1CKO mice. These data, in addition to our finding that Blimp-1 can specifically bind to regulatory regions and suppress transcription of the Il17a and Il17f genes, points to a direct role for Blimp-1 in repressing this locus in Treg. This is also supported by the chromatin changes and p300 decreased enrichment associated with Blimp1-binding at the locus CNS7 region.
Although we cannot establish direct causality between the binding of Blimp-1 at the Il17 locus and the histone modifications discussed earlier, changes in H3K27me3 accumulation were only measurable at a locus site bound by Blimp1, suggesting that direct binding of Blimp-1 could be required for the deposition of this specific repression marker. Trimethylation on H3K27 is added by Polycomb repressive complex 2 (PRC2), containing the methyltransferase enhancer of Zeste homolog 2 (EZH2), which contains a SET domain responsible for H3K27me3. Targeting of EZH2 and PRC2 to specific loci is thought to be mediated by various sequence elements and the presence of specific transcription factor binding motifs (Yamanaka et al., 2017). Thus, Blimp-1 binding could be involved in targeting EZH2/PRC2 to the Il17a locus, but this remains to be tested.
The chromatin changes associated with Blimp-1 deficiency did not alter binding of RORγt, suggesting that the physical interaction between RORγt and Foxp3, previously reported to inhibit RORγt-mediated transcription of the Il17 locus (Ichiyama et al., 2008; Zhou et al., 2008), is preserved. Instead, we found that in the absence of Blimp1, IRF4 occupancy of the Il17 locus CNS7 region was increased and IRF4 knockdown led to significant reduction of IL-17A production, indicating the requirement of IRF4 for production of IL-17A and IL-17F in Blimp1−/− Treg. These observations, along with the finding that genetic ablation of RORγt inhibited IL-17A expression in Blimp-1 −/− Treg, support a model by which Blimp-1 modulates IRF4 function in RORγt+ Treg such that absence of Blimp-1 favors binding of IRF4 to the locus and potentially facilitates RORγt-induced transcription.
A previous study (Jain et al., 2016) reported that in vivo-differentiated pTh17 cells had a small reduction in IL-17A expression in the absence of Blimp-1, but the expression IL-17A was not evaluated in Blimp-1 −/−Foxp3+ cells. In the same study, Blimp- 1 was shown by chromatin immunoprecipitation sequencing (ChIP-seq) to be bound to different regions of the Il17 locus, including the CNS7 region in pTh17 cells. However, in our experiments, side-by-side comparison of Blimp-1 occupancy of the Il17 locus in naturally occurring Treg (n Treg) and pTh17 cells by qPCR-ChIP detected significant binding of Blimp-1 to the Il17 locus in n Treg, but not in pTh17 cells (Figure 2B). It is possible that these conflicting results could be explained by technical differences (e.g., use of different antibodies or different ChIP methodologies).
Alternatively, Blimp-1 could have opposite functions in the same locus in n Treg and pTh17 cells. Blimp-1 function can be dose dependent (Robertson et al., 2007), and the amounts of Blimp-1 protein expressed in pTh17 cells are significantly lower than those in n Treg (Bankoti et al., 2017). Blimp-1 function could also depend on the differential availability of co-factors. This could explain why enforced expression of Blimp-1 suppresses the Il17 locus in Treg, but not in Th17 cells (Figure 2; Salehi et al., 2012). These possibilities remain to be investigated.
Overall, our results support the idea that Blimp-1 is required to prevent the production of Th17-associated cytokines in RORγt+ microbiota-specific Foxp3+ Treg by a mechanism that involves direct regulation of the Il17 locus. In addition, our study provides evidence that Foxp3+Treg that acquire the capability to produce inflammatory cytokines have impaired suppressor function and gain inflammatory properties. Altogether, these findings shed light onto the mechanisms regulating Treg phenotypic stability and function.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed and will be fulfilled by the Lead Contact, Gislaine A. Martins (martinsg@csmc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6Prdm1flox/floxCD4−Cre+ and C57BL/6Prdm1flox/floxFoxp3−Cre+ were described (Bankoti et al., 2017; Salehi et al., 2012). C57BL/6 B6.129S7-Rag1tm1Mom/J (RAG−/−), B6.SJL-PtprcaPep3b/BoyJ (CD45.1+), B6.129P2-Il10tm1Cgn/J (IL10−/−), C57BL/6-Il17atm1Bcgen/J (IL17eGFP) and Foxp3RFP (C57BL/6-Foxp3tm1Flv/J) reporter mice and mice with “floxed” RORC alleles (B6(Cg)-Rorctm3Litt/J) were obtained from Jackson labs. Foxp3-IRES-GFP knock-in (Foxp3GFP) (Bettelli et al., 2008) mice were obtained from Dr. V. Kuchroo (Harvard Medical School) and crossed to IL10−/− mice to generate IL10KOFoxp3GFP. Blimp1YFP reporter mice were obtained from Dr. Eric Meffre (Yale University) and bred to Foxp3RFP mice to generated Blimp1YFPFoxp3RFP dual reporter mice. Foxp3RFP mice were also bred to ROR(γt)GFP mice or to IL17AeGFP mice to generate Foxp3RFPIL17GFP or Foxp3RFP/RORγtGFP dual reporter mice or to C57BL/6Prdm1flox/flox mice to generated Prdm1flox/floxFoxp3RFP mice. Foxp3YFP-CRE mice were bred to C57BL/ 6Prdm1flox/flox mice to generate mice with Treg-specific deletion of Blimp-1 and then bred to Rorcflox/flox mice to generate mice with Treg-specific double deficiency of Blimp-1 and Rorγt. All mice were bred and maintained in the CSMC SPF animal barrier facility and handled in accordance with the institutional guidelines.
METHOD DETAILS
Flow Cytometry
Cells were stained as previously described (Bankoti et al., 2017) with the antibodies listed in the Key Resources Table. A rabbit anti-GFP antibody was used to stain Blimp1YFP or Foxp3GFP reporter cells in some experiments. Foxp3 staining was done with eBioscience Foxp3 staining kit or BioLegend’s FOXP3 Fix/Perm Buffer Set. Samples were acquired on a LSRII analyzer and FACSymphony (BD Biosciences). FACS data was analyzed with FlowJo software.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AF700 anti-TCRβ (clone H57-597) | Biolegend | Cat#: 109224, RRID: AB_1027648 |
| APC-Cy7 anti-TCRβ (clone H57-597) | Biolegend | Cat#: 109220, RRID: AB_893624 |
| APC-Cy7 anti-CD4 (clone RM4-5) | Biolegend | Cat#: 100526, RRID: AB_312727 |
| Pacific Blue anti-CD4 (clone RM4-5) | Biolegend | Cat#: 100531, RRID: AB_493375 |
| BV510 anti-CD4 (clone GK1.5) | Biolegend | Cat#: 100449, RRID: AB_2564587 |
| APC-Cy7 anti-CD44 (clone IM7) | Biolegend | Cat#: 103028, RRID: AB_830785 |
| Pacific Blue anti-CD44 (clone IM7) | Biolegend | Cat#: 103020, RRID: AB_493683 |
| APC anti-CD44 (clone IM7) | Biolegend | Cat#: 103012, RRID: AB_312963 |
| APC-Cy7 anti-CD62L (clone MEL-14) | Biolegend | Cat#: 104427, RRID: AB_830798 |
| AF700 anti-ICOS (clone C398.4) | Biolegend | Cat#: 313527, RRID: AB_2566125 |
| APC anti-IL17A (clone TC11-18H10.1) | Biolegend | Cat#: 506916, RRID: AB_536018 |
| PE anti-IL17A (clone TC11-18H10.1) | Biolegend | Cat#: 506904, RRID: AB_315464 |
| AF488 anti-IL17A (clone TC11-18H10.1) | Biolegend | Cat#: 506909, RRID: AB_536011 |
| AF647 anti-IL17F (clone 9D3.1C8) | Biolegend | Cat#: 517004, RRID: AB_2249030 |
| APC anti-IFNg (clone XMG1.2) | Biolegend | Cat#: 505810, RRID: AB_315404 |
| APC anti-CD25 (clone PC61.5) | eBioscience | Cat#: 17-0251-82, RRID: AB_469366 |
| APC anti-Foxp3 (clone FJK-16 s) | eBioscience | Cat#: 17-5773-82, RRID: AB_469457 |
| PE anti-Foxp3 (clone FJK-16 s) | eBioscience | Cat#: 12-5773-82, RRID: AB_465936 |
| eFluor 450 anti-Helios (clone 22F6) | eBioscience | Cat#: 48-9883-41, RRID: AB_2574137 |
| PE anti-IL10 (clone JES5-16E3) | eBioscience | Cat#: 12-7101-82, RRID: AB_466176 |
| PE anti-IRF4 (clone 3E4) | eBioscience | Cat#: 12-9858-82, RRID: AB_10852721 |
| eFluor 450 anti-CD103 (clone 2E7) | eBioscience | Cat#: 48-1031-82, RRID: AB_2574033 |
| PE anti-RORγt (clone AFKJS-9) | eBioscience | Cat#: 12-6988-82, RRID: AB_1834470 |
| BV786 anti-RORγt (clone Q31-378) | BD Biosciences | Cat#: 564723, RRID: AB_2738916 |
| BV421 anti-RORγt (clone Q31-378) | BD Biosciences | Cat#: 562894, RRID: AB_2687545 |
| BUV395 anti-Gata3 (clone L50-823) | BD Biosciences | Cat#: 565448, RRID: AB_2739241 |
| BV510 anti-IL10 (clone JES5-16E3) | BD Biosciences | Cat#: 563277, RRID: AB_2738112 |
| Rabbit anti-GFP antibody | Rockland Immunochemicals | Cat#: 600-402-215, RRID: AB_828169 |
| anti-CD3ε (clone 145-2C11) | BioXcell | Cat#: BE0001-1, RRID: AB_1107634 |
| anti-CD28 (clone 37.51) | BioXcell | Cat#: BE0015-1, RRID: AB_1107624 |
| anti-IFN-γ (clone XMG1.2) | BioXcell | Cat#: BE0055, RRID: AB_1107694 |
| anti-IL4 (clone11B11) | BioXcell | Cat#: BE0045, RRID: AB_1107707 |
| anti-Blimp-1 polyclonal antibody (clone 267) | In house | N/A |
| anti-histone H3 (tri methyl K4) antibody | Abcam | Cat#: ab8580, RRID: AB_306649 |
| anti-histone H3 (tri methyl K27) antibody | Millipore | Cat#: 07-449, RRID: AB_310624 |
| anti-RORγ antibody | Santa Cruz Biotechnology | Cat#: sc-28559, RRID: AB_2285218 |
| anti-IRF-4 antibody | Santa Cruz Biotechnology | Cat#: sc-6059, RRID: AB_2127145 |
| anti-p300 antibody | Santa Cruz Biotechnology | Cat#: sc-585, RRID: AB_2231120 |
| normal rabbit IgG | Santa Cruz Biotechnology | Cat#: sc-2027, RRID: AB_737197 |
| Bacterial and Virus Strains | ||
| PGL4.10 | Promega | Cat#: E6651 |
| phRL-TK | Promega | Cat#: E6921 |
| MSCV MigR1 | Addgene | Cat#: 27490 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Vancomycin | Gold Biotechnology | Cat#: V-200 |
| Metronidazole | Sigma-Aldrich | Cat#: M1547 |
| Neomycin | Gold Biotechnology | Cat#: N-620 |
| Ampicillin | Sigma-Aldrich | Cat#: A0166 |
| Fluconazole | Sigma-Aldrich | Cat#: PHR1160 |
| Glucose | Sigma-Aldrich | Cat#: G7528 |
| PMA | Sigma-Aldrich | Cat#: P8139 |
| Ionomycin | Sigma-Aldrich | Cat#: I9657 |
| Brefeldin A | Sigma-Aldrich | Cat#: B6542 |
| polybrene | Sigma-Aldrich | Cat#: H9268 |
| rMuIL6 | Biolegend | Cat#: 575702 |
| rMuIL23 | Biolegend | Cat#: 589002 |
| rHuTGFb1 | Biolegend | Cat#: 580702 |
| rMuIL-2 | Biolegend | Cat#: 575402 |
| rMuILIβ | Biolegend | Cat#: 575102 |
| rhIL2 | Roche | Cat#: 11011456001 |
| Critical Commercial Assays | ||
| Foxp3 staining kit | eBioscience | Cat#: 42-1403 |
| FOXP3 Fix/Perm Buffer Set | BioLegend | Cat#: 421403 |
| ChIP-IT High Sensitivity kit | Active Motif | Cat#: 53040 |
| Mouse T Cell Nucleofector Medium | Lonza | Cat#: VZB-1001 |
| Neon Transfection system | Thermo Fisher Scientific | Cat#: MPK10025 |
| Dual luciferase assay system | Promega | Cat#: E1910 |
| RNeasy Mini Kit | QIAGEN | Cat#: 74106 |
| MessageBOOSTER cDNA Synthesis Kit | Epicenter | Cat#: MB060124 |
| Mouse IL-10 ELISA kit | eBioscience | Cat#: 88710586 |
| Mouse IL-17A (homodimer) ELISA kit | eBioscience | Cat#: 88737122 |
| Experimental Models: Cell Lines | ||
| Mouse: EL4 LAF cells | Tone et al., 2008 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6Prdm1flox/flox (Prdm1flox/flox) | The Jackson Laboratory | Stock#: 008100 |
| Mouse: B6Cg-Tg(Cd4-cre)1Cwi/BfluJ (CD4Cre) | The Jackson Laboratory | Stock#: 022071 |
| Mouse: Prdm1flox/floxCD4Cre | This paper | N/A |
| Mouse: Prdm1flox/floxCD4CreFoxp3GFP | This paper | N/A |
| Mouse: B6.SJL-PtprcaPep3b7BoyJ (CD45.1+) | The Jackson Laboratory | Stock#: 002014 |
| Mouse: B6.129S7-Rag1tm1Mom/J (Rag1−/− | The Jackson Laboratory | Stock#: 002216 |
| Mouse: C57BL/6-Foxp3tm4 (YFP/cre)Ay/j (Foxp3YFP-CRE) | The Jackson Laboratory | Stock#: 016959 |
| Mouse: Prdm1flox/floxFoxp3YFP-CRE | This paper | N/A |
| Mouse: C57BL/6-Il17atm1Bcgen/J (IL17AeGFP) | The Jackson Laboratory | Stock#: 018472 |
| Mouse: C57BL/6-Foxp3tm1Flv/J (Foxp3RFP) | The Jackson Laboratory | Stock#: 008374 |
| Mouse: Prdm1flox/floxCD4CreFoxp3RFP | This paper | N/A |
| Mouse: Prdm1flox/floxCD4CreFoxp3RFPIL17AeGFP | This paper | N/A |
| Mouse: B6(Cg)-Rorctm3Litt/J (Rorcflox/flox) | The Jackson Laboratory | Stock#: 008771 |
| Mouse: Prdm1flox/floxRorcflox/floxFoxp3YFP-CRE | This paper | N/A |
| Mouse: B6.129P2-Il10tm1Cgn/J (IL10−/−) | The Jackson Laboratory | Stock#: 002251 |
| Mouse: Blimp1YFP | Dr. E. Meffre lab (Yale University) | N/A |
| Mouse: Rorc(γt)Gfp | Lochner et al., 2008 | N/A |
| Mouse: Rorc(γt)GfpFoxp3RFP | This paper | N/A |
| Mouse: Foxp3-IRES-GFP knockin (Foxp3GFP) | Dr. V. Kuchroo lab (Harvard Medical School) | N/A |
| Mouse: IL10−/−Foxp3GFP | This paper | N/A |
| Oligonucleotides | ||
| See Table S1 for gene expression analysis primers | This paper | N/A |
| See Table S1 for ChIP primers | This paper | N/A |
| Il17apro-F: ACATAGCTCGAGAACAGACAGCCACATACCAAA | This paper | N/A |
| Il17apro-R: CAGTCTAAGCTTGTGGATGAAGAGTAGTGCTCCT | This paper | N/A |
| Il17CNS7-F: ACATAGGGTACCTATAGCCTGCAGCCCTGCA | This paper | N/A |
| Il17CNS7-R: CAGTCTGAGCTCGGTGTCGGACTTTACATTAGCAGAC | This paper | N/A |
| Blimp-F: ACATAGGAATTCATGAGAGAGGCTTATCTCAGATGTTG | This paper | N/A |
| Blimp-R: ACATAGCTCGAGTTAAGGATCCATCGGTTCAACTG | This paper | N/A |
| Blimp-ZFdel-R: ACATAGCTCGAGTTACTGTTTCTTCAGAGGGTAAGGAAGAG | This paper | N/A |
| Silencer Select Negative Control No. 1 siRNA | Thermo Fisher Scientific | Cat#: 4390843 |
| IRF-4 specific siRNAs | Staudt et al., 2010 | N/A |
| Recombinant DNA | ||
| pcDNA3.1(+)-Blimp-Full | This paper | N/A |
| pcDNA3.1(+)-Blimp-DZF | This paper | N/A |
| PGL4.10- Il17a promoter | This paper | N/A |
| PGL4.10- Il17a promoter+CNS7 | This paper | N/A |
| MSCV MigR1 Prdm1 | This paper | N/A |
| Software and Algorithms | ||
| FlowJo software | Tree Star, Inc. | N/A |
| rVista 2.0 software | Loots and Ovcharenko, 2004 | N/A |
| JMP software | SAS Institute | N/A |
| GraphPad Prism 7 | GraphPad Software | N/A |
| Other | ||
| LSRII analyzer | BD Biosciences | N/A |
| FACSymphony | BD Biosciences | N/A |
| BD FACS Aria II | BD Biosciences | N/A |
| BD FACS Aria III | BD Biosciences | N/A |
| ECM 830 Square Wave Electroporation System | BTX | N/A |
| 4D-Nucleofector | LONZA | N/A |
| Neon Transfection System | Thermo Fisher Scientific | N/A |
| Realplex2 Mastercycler machine | Eppendorf | N/A |
Antibiotic treatment
Mice were treated (6 wks) with a cocktail of 1mg/ml vancomycin, 0.5mg/ml metronidazole, 1mg/ml neomycin, 1mg/ml ampicillin and 0.5mg/ml Fluconazole in their drinking water.
Cytokine measurement
Cytokine production was measured in the supernatant of cell culture using eBioscience kits (IL10; IL17A). Cytokine-expressing cells were detected by intracellular staining and flow cytometry analysis. For Intracellular cytokine staining, single cell suspension from peripheral organs were stimulated (plate-bound anti-CD3, 5 μg/ml and anti-CD28, 2.5 mg/ml for 24 hr or with PMA, 2 ng/ml and Ionomycin, 0.2 ng/ml for 4 hr). Brefeldin A was added in the last 6 or 2 hr of incubation, respectively. Cells were collected, surface stained and fixed with 4% paraformaldehyde followed by staining of cytokines.
In vitro T helper 17 differentiation
Blimp1−/− Th17 cells used for retroviral transduction were stimulated as described (Salehi et al., 2012). pTh17 cells used for ChIP assays were generated as previously described (Jain et al., 2016). To generate IL17GFP+Th17 cells used in the adoptive transfer colitis experiments, naive (CD62LHighCD44LowFoxp3RFP-) cells were sorted from IL17AeGFP Foxp3RFP double report mice and cultured with mitomycin-C-treated antigen presenting cells (1:4 ratio), in the presence of soluble anti-CD3 mAb (1 μg/mL), rMuIL6 (20 ng ml), rMuIL23 (50 ng/mL) and rHuTGFβ1 (0.25 ng/mL) along with neutralizing antibodies for IFN-γ (10 μg/mL) and IL4 (10 μg ml). After 5 days of in vitro culture, pTh17 (CD4+ IL17eGFP+ Foxp3RFP−) were FACS sorted for injection into RAG1−/− mice.
Adoptive transfer-induced colitis
Lymph nodes and spleens were pooled from Blimp1−/− Foxp3RFP IL17AGFP reporter mice. CD4+Foxp3RFP+IL17AGFP− (SP) and CD4+Foxp3RFP+ IL17AGFP+ (DP) cells were sorted and co-injected (5×104, i.v.) into C57BL/6 RAG1−/− mice along with CD45RBhiCD4+T cells (1×105). In some experiments, CD4+Foxp3RFP+IL17AGFP− or IL17AGFP+ cells or in vitro differentiated wild-type IL17GFP+Th17 cells (5×104 cells/mouse, i.p.) were transferred in the absence of naive cells. Recipient mice were weighed weekly and sacrificed 4-8 weeks post-transfer. Large intestines histology samples were stained with H&E and scored as previously described (Read and Powrie, 2001), by a pathologist in a blinded fashion.
Retroviral transduction
The coding sequence of Prdm1 was cloned into the MigR1 retroviral vector. CD4+Foxp3RFP+ Treg and CD4+ Blimp1−/− Foxp3RFP−naive T cells were activated (plate-bound anti-CD3, 5 μg/ml and anti-CD28, 2.5 μg/ml) in the presence of 100 U/ml rhIL2 for Treg or with rMuIL1β (20 ng/ml), rMuIL23 (50 ng/ml), rMuIL6 (10 ng/ml), and rHuTGF-β1 (5 ng/ml) for Th17 cells. After 32h of activation, cells were resuspended in retrovirus-containing supernatants plus 8 μg/ml polybrene and rhIL2 (50 U/ml) for Treg or IL23 (50 ng/ml) for naive T cells, followed by centrifugation (7500 rpm) for 90 min at 25°C. Viral supernatant was removed and cells cultured for another 48 h with rhIL2 (200 U/ml) and rhTGF-β1 (10 pg/ml) for Treg or with media containing IL1β, IL6 and TGFβ for Th17 cells.
Chromatin Immunoprecipitation
CD4+Foxp3GFP+T cells were sort-purified from LN and SP from Control and Blimp-1CKO Foxp3GFP mice and stimulated with PMA and ionomycin before crosslinking by fixation with 1.1% paraformaldehyde for (10 min, RT). Sonicated chromatins from 1-2×106 cells were subjected to ChIP using ChIP-IT High Sensitivity kit (Active motif, Carlsbad, CA). Chromatin was incubated O/N at 4°C with 5 μL of rabbit anti-Blimp-1 polyclonal antibody (clone 267, recognizing the C-terminal of Blimpl), 5 μL of pre-immune serum, 0.8 μg of anti-histone H3 (tri methyl K4) or anti-histone H3 (tri methyl K27) antibody, 2.5 mg of anti-RORγ antibody, 2.5 μg of anti-IRF-4 antibody, 2.5 mg of anti-p300 antibody or normal rabbit IgG, after which protein G Agarose beads were added, followed by incubation at 4°C O/N. qRT-PCR was performed in DNA recovered from IP and input samples [primers sequences in Table S1 and (Salehi et al., 2012)]. Analysis of sequence homology and identification of putative Blimp1, Rorc previously confirmed binding sites, RORE consensus sites and IRF4 binding sites were performed using rVista 2.0 software. Genomic sequences were obtained from Ensembl.
Luciferase reporter assays
A 2 kb fragment containing the Il17a promoter was cloned into the PGL4.10 luciferase reporter plasmid with or without the CNS7 region. Full length wild-type Blimp-1 (Blimp-Full) or a DNA-binding-lacking construct (Blimp-ΔZF) was clone into pcDNA3.1(+) expression vector (Invitrogen, Waltham, MA). All primers used for cloning listed in the Key Resources Table. EL4 LAF T cells (5 × 106) were transfected with luciferase reporter plasmids (5 μg), Blimp1-expression plasmids (5 μg) and phRL-TK(2 μg) (Promega) as internal control plasmid. Transfections were performed with BTX ECM830 Square Wave Electroporation System (BTX, Holliston, MA). Total DNA amount was adjusted with the empty pcDNA3.1(+) vector. Transfected cells were incubated 18 h, and then stimulated, lysed and luciferase activity measured with a dual luciferase assay system.
siRNA-mediated IRF4 knock down
CD4+Foxp3GFP+ cells were sorted from Ctrl or Blimp1−/−Foxp3GFP reporter mice. A mixture of three different IRF-4 specific siRNAs (100 pmol of each siRNA) or scrambled control (SC) siRNA (300 pmol) were transfected into CD4+Foxp3GFP+ cells (2×106) by using 4D-Nucleofector System (Lonza, Basel, Switzerland) or Neon Transfection system (Thermo Fisher Scientific). Transfected cells were cultured with plate-bound anti-CD3 and anti-CD28. After 18 h post-transfection, cells are collected, and mRNA expression measured by qRTPCR. Sequences for IRF4 specific siRNAs are as previously described (Staudt et al., 2010).
Real-time quantitative PCR (qRT-PCR)
For some experiments cells were directly FACS-sorted into RLT buffer (QIAGEN) supplemented with β2-mercaptoethanol. Total mRNA was isolated using RNAeasy kits (QIAGEN) and reverse transcribed as previously described (Salehi et al., 2012). SYBR Green incorporation qRT-PCR was performed using FastStart SYBR Green Master mix (Roche) in the Realplex2 Mastercycler machine (Eppendorf). For some experiments in which cell yelled was very low, Linear amplification of mRNA was performed using the MessageBooster cDNA kit (Epicenter). Primers sequences for Il17a, Rorc, and BATF are as previously described (Salehi et al., 2012) or listed in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data for all experiments were analyzed with Prism Software V7 (GraphPad Software, La Jolla, CA) and JMP software (SAS Institute, Cary, NC). In Figures 1 and 4 “N” indicate number of mice per group. In Figures 2 and 3 “N” indicate number of experiments performed, using cells pooled form 2-3 animals in each experiment. Statistical significance was calculated by one-way ANOVA, paired or unpaired two-tailed Student’s t tests as described in the figure legends. A p ≤ 0.05 value was considered significant and are denoted in figures as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001. No statistical methods were used to predetermine sample size. No animal or sample was excluded from the analysis.
Supplementary Material
Highlights.
Blimp-1 is preferentially expressed in Foxp3+RORγt+ Treg
Blimp1−/−Foxp3+RORγt+ Treg express IL-17A and IL-17F and have inflammatory activity
Blimp-1 binding to the Il17 gene leads to chromatin changes and impairs IRF4 binding
Expression of IL-17 in Blimp1 −/− Treg requires both RORγt and IRF4
ACKNOWLEDGMENTS
We are grateful to members of the Martins laboratory for discussions; to Drs. V. Kuchroo (Harvard Medical School) and E. Meffre (Yale School of Medicine) for Foxp3GFP and Blimp1YFP single reporter mice, respectively; and to Dr. M. Fukata (CSMC) for IL17GFPFoxp3RFP dual reporter mice. We thank Drs. M. Tone, T. Araki, and T. Watanabe (CSMC) for help with luciferase assays; the CSMC Flow cytometry core (A. Lopez and Dr. J. Suda); and the Fred Hutchinson CCEH vector production core (Dr. M. Wohlfahrt). Funding for this work was provided by NIH research grant R01AI103542 and R01AI127406 (to G.A.M.) and by IBIRI-CSMC (to G.A.M.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.09.016.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Apostolou I, Sarukhan A, Klein L, and von Boehmer H (2002). Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 3,756–763. [DOI] [PubMed] [Google Scholar]
- Bankoti R, Ogawa C, Nguyen T, Emadi L, Couse M, Salehi S, Fan X, Dhall D, Wang Y, Brown J, et al. (2017). Differential regulation of effector and regulatory T cell function by Blimp1. Sci. Rep 7, 12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, White-sell L, Kelly TE, Saulsbury FT, Chance PF, and Ochs HD (2001). The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet 27, 20–21. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Korn T, Oukka M, and Kuchroo VK (2008). Induction and effector functions of T(H)17 cells. Nature 453, 1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, and Ramsdell F (2001). Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet 27, 68–73. [DOI] [PubMed] [Google Scholar]
- Brüstle A, Heink S, Huber M, Rosenplänter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, and Lohoff M (2007). The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol 8, 958–966. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, and Rudensky AY (2013). Control of inflammation by integration of environmental cues by regulatory T cells. J. Clin. Invest. 123, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, and Rudensky AY (2009). CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Martins GA, Liao J, Magnusdottir E, Grunig G, Perez RK, and Calame KL (2008). Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J. Immunol 181, 2338–2347. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, and Kallies A (2011). The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol 12, 304–311. [DOI] [PubMed] [Google Scholar]
- Dias S, D’Amico A, Cretney E, Liao Y, Tellier J, Bruggeman C, Almeida FF, Leahy J, Belz GT, Smyth GK, et al. (2017). Effector regulatory T cell differentiation and immune homeostasis depend on the transcription factor Myb. Immunity 46, 78–91. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, and Rudensky AY (2005). Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 22, 329–341. [DOI] [PubMed] [Google Scholar]
- Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limoón P, Paiva RS, Ching T, et al. (2015). Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann C, Heink S, Petermann F, Vasanthakumar A, Rothhammer V, Doorduijn E, Mitsdoerffer M, Sie C, Prazeres da Costa O, Buch T, et al. (2014). IL-27 and IL-12 oppose pro-inflammatory IL-23 in CD4+ T cells by inducing Blimp1. Nat. Commun 5, 3770. [DOI] [PubMed] [Google Scholar]
- Ichiyama K, Yoshida H, Wakabayashi Y, Chinen T, Saeki K, Nakaya M, Takaesu G, Hori S, Yoshimura A, and Kobayashi T (2008). Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J. Biol. Chem 283, 17003–17008. [DOI] [PubMed] [Google Scholar]
- Jain R, Chen Y, Kanno Y, Joyce-Shaikh B, Vahedi G, Hirahara K, Blumenschein WM, Sukumar S, Haines CJ, Sadekova S, et al. (2016). Interleukin-23-induced transcription factor Blimp-1 promotes pathogenicity of T helper 17 cells. Immunity 44, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, and Nutt SL (2006). Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol 7, 466–474. [DOI] [PubMed] [Google Scholar]
- Kim BS, Lu H, Ichiyama K, Chen X, Zhang YB, Mistry NA, Tanaka K, Lee YH, Nurieva R, Zhang L, et al. (2017). Generation of RORγt+ antigen-specific T regulatory 17 cells from Foxp3+ precursors in autoimmunity. Cell Rep 21, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, Tanaka S, Bluestone JA, and Takayanagi H (2014). Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med 20,62–68. [DOI] [PubMed] [Google Scholar]
- Levine AG, Mendoza A, Hemmers S, Moltedo B, Niec RE, Schizas M, Hoyos BE, Putintseva EV, Chaudhry A, Dikiy S, et al. (2017). Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, Riethmacher D, Si-Tahar M, Di Santo JP, and Eberl G (2008). In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med 205, 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, and Ovcharenko I (2004). rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32, W217–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G, and Calame K (2008). Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol 26, 133–169. [DOI] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, and Calame K (2006). Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol 7, 457–465. [DOI] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Liao J, Magnusdottir E, and Calame K (2008). Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J. Exp. Med 205, 1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi MF, Ahlers J, Janke M, Rudolph C, Mockel-Tenbrinck N, Kühl AA, et al. (2014). Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med 211, 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, et al. (2015). Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349, 989–993. [DOI] [PubMed] [Google Scholar]
- Read S, and Powrie F (2001). Induction of inflammatory bowel disease in immunodeficient mice by depletion of regulatory T cells. Curr. Protoc. Immunol 30, 15.13.1–15.13.10. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Charatsi I, Joyner CJ, Koonce CH, Morgan M, Islam A, Paterson C, Lejsek E, Arnold SJ, Kallies A, et al. (2007). Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development 134, 4335–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S, Bankoti R, Benevides L, Willen J, Couse M, Silva JS, Dhall D, Meffre E, Targan S, and Martins GA (2012). B lymphocyte-induced maturation protein-1 contributes to intestinal mucosa homeostasis by limiting the number of IL-17-producing CD4+ T cells. J. Immunol 189, 5682–5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, et al. (2010). Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33, 192–202. [DOI] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, and Tone M (2008). Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol 9, 194–202. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, et al. (2011). GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest 121, 4503–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Tajiri S, Fujimoto T, Matsumoto K, Fukunaga S, Kim BS, Okano HJ, and Yokoo T (2017). Generation of interspecies limited chimeric nephrons using a conditional nephron progenitor cell replacement system. Nat. Commun 8, 1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, and Rudensky AY (2009). Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 458, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. (2008). TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.