Abstract
LncRNAs have been reported to play an important role in various diseases. However, their role in the radiation‐induced intestinal injury is unknown. The goal of the present study was to analyse the potential mechanistic role of lncRNAs in the radiation‐induced intestinal injury. Mice were divided into two groups: Control (non‐irradiated) and irradiated. Irradiated mice were administered 14 Gy of abdominal irradiation (ABI) and were assessed 3.5 days after irradiation. Changes to the jejuna of ABI mice were analysed using RNA‐Seq for alterations to both lncRNA and mRNA. These results were validated using qRT‐PCR. LncRNAs targets were predicted based on analysis of lncRNAs‐miRNAs‐mRNAs interaction. 29 007 lncRNAs and 17 142 mRNAs were detected in the two groups. At 3.5 days post‐irradiation, 91 lncRNAs and 57 lncRNAs were significantly up‐ and downregulated respectively. Similarly, 752 mRNAs and 400 mRNAs were significantly up‐ and downregulated respectively. qRT‐PCR was used to verify the altered expression of four lncRNAs (ENSMUST00000173070, AK157361, AK083183, AK038898) and four mRNAs (Mboat1, Nek10, Ccl24, Cyp2c55). Gene ontology and KEGG pathway analyses indicated the predicted genes were mainly involved in the VEGF signalling pathway. This study reveals that the expression of lncRNAs was altered in the jejuna of mice post‐irradiation. Moreover, it provides a resource for the study of lncRNAs in the radiation‐induced intestinal injury.
Keywords: alternation, intestinal injury, lncRNAs, mRNAs, radiation
1. INTRODUCTION
The human genome contains relatively few protein‐coding sequences, while the majority is transcribed to produce non‐coding RNAs (ncRNAs). Long, non‐coding RNAs (lncRNAs) form a large class of ncRNAs that are more than 200 nt in length. Based on the relative chromosomal position of the gene, a lncRNA can be placed into one or more of five broad categories: (a) sense, (b) antisense, (c) bidirectional, (d) intronic or (e) intergenic.1
Although they are non‐coding, lncRNAs are able to regulate gene expression and do so through multiple mechanisms.2 These primarily include the following: (a) Chromatin modification, which includes lncRNAs HOTAIR,3 Xist4 and ANRIL,5 (b) transcriptional regulation, including Evf26 and (c) post‐transcriptional regulation, including Zeb2.7 Moreover, lncRNAs possess a variety of biological functions, such as epigenetic regulation of gene expression4 as well as the production of competing endogenous RNA. Some lncRNAs also serve as host genes for small, non‐coding RNAs that are deregulated in cancer.8 Other disease‐relevant roles (eg, aetiology and development) have been reported for lncRNAs, such as in infectious diseases,9 ophthalmological10 as well as neurodegenerative diseases,11 and various cancers.8
More than half of the cancer patients are currently treated with radiotherapy, but this approach is inexact and often damages surrounding healthy tissues. In particular, the epithelium of mammalian intestinal mucosa undergoes rapid and constant renewal throughout the life of an organism. Additionally, it acts as a physical barrier between the luminal microbiota and the rest of the body.12 As its rapid renewal and does not have much natural protection, the intestinal epithelium is sensitive to ionising radiation. As such, the intestine is one of the most sensitive organs to radiation toxicity.13 Radiotherapy of abdominal and pelvic tumours results in high radiation toxicity that can lead to the radiation‐induced intestinal injury.
Radiation‐induced tissue injury is a complex, pathophysiological process involving multiple and wide‐ranging mechanisms, which are dependent on the radiation dose and time course. These mechanisms include DNA repair, cell death, inflammation, endothelial activation, angiogenesis and matrix remodelling.14, 15 Symptomatically, the radiation‐induced intestinal injury mainly manifests as diarrhoea, dehydration, sepsis and intestinal bleeding, with eventual mortality within 10‐15 days post‐exposure.16 Due to its severity, there is a tremendous need for therapeutic measures that can prevent or treat the radiation‐induced intestinal injury.
It has been reported that some lncRNAs are involved in regulating the intestinal epithelial barrier. For instance, Su et al17 found that miR‐874 suppressed AQP3 expression, resulting in AQP3 down‐regulation. This impaired intestinal barrier integrity and may have led to intestinal barrier dysfunction via the opening of the tight junction complex.18 Interestingly, H19, a maternally expressed imprinted lncRNA,19 may function as a competing endogenous RNA (ceRNA). This would allow for finer regulation of AQP3 expression by competing for miR‐874 and may also improve intestinal barrier dysfunction. Geng et al20 found that under normal conditions, H19 is transcriptionally silent in the small intestine of adult mice. However, it becomes strongly activated after LPS treatment. H19 expression is induced by IL‐22 in intestinal epithelial cells through PKA‐ and STATE3‐associated signalling mechanisms and plays an important role in sustaining the renewal of epithelial cells during inflammation, its expression also enhances regeneration of the intestinal epithelium in colitis. Chen et al21 devised an intestinal epithelial barrier model and used dextran sodium sulphate (DSS) to induce injury. In this model, Myc‐associated zinc finger protein (MAZ) and tight junction proteins zonula occludens 1 (ZO‐1) were involved in the functioning of the intestinal epithelial barrier. However, miR‐34c targeted MAZ to inhibit its expression, while a lncRNA PlncRNA1 protected the intestinal epithelial barrier by sponging miR‐34c. Past work has also shown that fasting for 48 hours inhibits small intestinal mucosal growth in mice. Importantly, Xiao et al12 compared microarray profiles between fasted and control groups and found the lncRNA uc.173 differentially expressed between the two groups. Thus, LncRNA uc.173 can enhance intestinal mucosal growth by specifically inhibiting miRNA 195 expression.
Past work has also found lncRNAs play a role in DNA damage induced by ionising radiation. For instance, Michelini et al22 found that following DNA damage, RNA polymerase II (RNAPII) was bound to the MRE11‐RAD50‐NBS1 complex and recruited to double‐strand DNA breaks. There, it engaged in damage‐induced synthesis of long non‐coding RNAs (dilncRNAs) directed towards both DNA ends. DilncRNAs act as DNA damage response RNA (DDRNA) precursors and also recruit DDRNAs through RNA‐RNA pairing. Together, dilncRNAs and DDRNAs help focus DNA damage response (DDR) formation and its association with 53BP1. Accordingly, RNAPII inhibition prevents DDRNA recruitment, DDR activation and DNA repair. Betts et al23 found that a distal transcriptional enhancer within the 11q13 breast cancer risk region (PRE1) interacted with two oestrogen‐regulated lncRNAs, CUPID1 and CUPID2 promoter. PRE1 has also previously been shown to regulate the expression of CCND1.24 Moreover, Betts also showed that CUPID1 and CUPID2 regulated the decision to engage either the non‐homologous end joining or homologous recombination (HR) pathway.
Past work has found many lncRNAs that are involved in intestinal epithelial barrier and DNA damage induced by ionising radiation; however, there remain limited studies examining the role of lncRNAs in the radiation‐induced intestinal injury. Currently, there is no accepted approach to either prevent or treat the radiation‐induced intestinal injury. Given this, we sought to determine which lncRNAs were involved in the radiation‐induced intestinal injury and provide a better understanding of this type of injury to guide better clinical treatment of the radiation‐induced intestinal injury. In this study, we sequenced both the lncRNAs and mRNAs in murine jejuna, both at baseline and 3.5 days post‐irradiation. We selected differently expressed lncRNAs and mRNAs, as lncRNAs can act as an endogenous “sponge” that regulates the target gene of miRNAs through competition with miRNAs, we predicted lncRNAs‐targeted miRNAs and miRNAs‐targeted mRNAs, and compared these predictions with our mRNAs sequencing results. Finally, we performed GO and KEGG signalling pathway analysis and illustrated lncRNA‐miRNA‐mRNA network; this was performed in order to find out which specific lncRNAs might be involved in the radiation‐induced intestinal injury.
2. MATERIALS AND METHODS
2.1. Mouse model of the radiation‐induced intestinal injury
Male C57BL/6 mice, aged 6‐8 weeks and weighing 23‐24 g, were purchased from Huafukang Biotechnology Co. Ltd (Beijing, China). All mice were housed in a temperature‐controlled, pathogen‐free environment with a 12‐hour light/dark cycle and allowed ad libitum access to water and standard chow. Mice had been divided into two groups: Control (non‐irradiated) and irradiated. The control group included three mice that had not been irradiated, while the irradiated group contained three mice that received 14 Gy ABI. Abdominal irradiation (ABI) was performed on mice using a Cs137 γ‐ray irradiator (Atomic Energy of Canada, Chalk River, ON, Canada). Lead shielding was used to protect other body parts from irradiation. Mice were exposed to 14 Gy at 1 Gy/min at room temperature. On day 3.5 post‐irradiation, mice were killed and the jejuna were frozen in −80°C freezer. All experimental procedures and protocols were conducted according to the guidelines of our institutional animal care and use committee.
2.2. Jejuna samples and RNA isolation
RNA was isolated from the jejuna of control and irradiated mice 3.5 days post‐irradiation. RNA was isolated using TRizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA concentration from all samples was determined by assessing the OD 260/280 using a NanoDrop ND‐2000 instrument (ThermoFisher Scientific, Waltham, MA, USA). RNA integrity was assessed using denaturing agarose gel electrophoresis.
2.3. High‐throughput sequencing
High‐throughput, whole transcriptome sequencing and subsequent bioinformatics analysis were all performed with Cloud‐Seq Biotech (Shanghai, China). The following steps were used: Total RNA was used and rRNAs were removed using Ribo‐Zero rRNA Removal Kits (Illumina, San Diego, CA, USA) following the manufacturer's instructions. RNA libraries were constructed using rRNA‐depleted RNAs with TruSeq Stranded Total RNA Library Prep Kit (Illumina) according to the manufacturer's instructions. Libraries were quantified and quality‐controlled using the BioAnalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Then, 10 pM libraries were denatured to single‐stranded DNA molecules, captured on Illumina flow cells, amplified in situ as clusters and sequenced for 150 cycles using an Illumina HiSeq Sequencer according to the manufacturer's instructions.
2.4. lncRNAs sequencing analysis
After image and base recognition, the original reads were harvested from an Illumina HiSeq sequencer. 3′ adaptor‐trimming and low‐quality removal was performed with cutadapt software,25 after which the resulting high‐quality clean reads were used for lncRNA analysis. Clean reads were aligned to the mouse reference genome (UCSC MM10) using hisat2 software.26
2.5. lncRNAs identification and their differential expression
Cuffdiff software27 was used to calculate differentially expressed lncRNAs. lncRNAs that exhibited fold changes ≥2.0 with P < 0.05 and FPKM value ≥0.1 in the least in one sample from a group were classified as having a significant and differentially expressed lncRNA.
2.6. Experimental validation of lncRNAs
Quantitative real‐time polymerase chain reaction (qRT‐PCR) was used to validate lncRNA expression. Two up‐regulated, two down‐regulated lncRNAs and mRNAs were selected for validation. The housekeeping gene GAPDH was used as a reference for normalisation. All primers used are presented in Table 1. Total RNA was reverse transcribed to cDNA using PrimeScript RT Reagent Kit (Perfect Real Time; TaKaRa, Osaka, Japan) according to the manufacturer's instructions. Resulting cDNA was then subjected to qRT‐PCR analysis on a BIO‐RAD CFX Connect Real‐Time PCR System with EVA Green qPCR Mix (Abm, Vancouver, BC, Canada). The assay was performed with three independent samples, all of which were assessed in triplicate. The relative expression ratio of lncRNAs was determined using the 2−ΔΔCt method.
Table 1.
Primers used to validate lncRNA and mRNA expression
| Gene name | Primer type | Primer sequence | Product length |
|---|---|---|---|
| ENSMUST00000173070 | Forward primer | GCCGTACCCAGTAGCACAAT | 71 |
| Reverse primer | AGCAAAGCCCTTTCTTCAGGTG | ||
| AK157361 | Forward primer | TCTCAAAGCTGTTTGTGTTCTCC | 84 |
| Reverse primer | CAAATTGTAAAGCTGAGGCTAGTCT | ||
| AK083183 | Forward primer | AGTAGTAACGATGAACTGCTGGAA | 108 |
| Reverse primer | GGAATCTTGCTTTGACTTCACTACA | ||
| AK038898 | Forward primer | GGGAGGATTCTGTTGCGGTT | 109 |
| Reverse primer | TAAAGGCTTCAACGGTGGCT | ||
| Mboat1 | Forward primer | AGCCTCTCTTACCGTACCACC | 157 |
| Reverse primer | GGCTGGCTTTACCAGGATGTA | ||
| Nek10 | Forward primer | GACAGTGCCCAAAATAACATGAC | 237 |
| Reverse primer | GGATGGAGCACGATTAACCCA | ||
| Ccl24 | Forward primer | TCTTGCTGCACGTCCTTTATT | 179 |
| Reverse primer | GCATCCAGTTTTTGTATGTGCC | ||
| Cyp2c55 | Forward primer | TACATTTTGGGGCGAGTGAAAG | 120 |
| Reverse primer | AAACTCCGAATGGGGATTGTG | ||
| Gapdh | Forward primer | AGGTCGGTGTGAACGGATTTG | 95 |
| Reverse primer | GGGGTCGTTGATGGCAACA |
2.7. GO and KEGG pathway analyses for selected lncRNAs
GO and KEGG analyses were performed for the differentially expressed lncRNA‐associated genes. GO analysis included three parts: Molecular function (MF), biological process (BP) and cellular component (CC). The top 10 enriched GO terms among the two groups were presented for differential expression. KEGG pathway analysis is a process that maps molecular data sets in genomics, transcriptomics, proteomics and metabolomics onto the KEGG pathway map. This mapping allows for the interpretation of biological function of these molecules. The KEGG pathway analysis was performed with differentially expressed lncRNAs and targeted mRNAs; the analysis speculated as to the pathways involved in these lncRNAs and targeted mRNAs. P‐value <0.05 was used as our threshold for statistically significant enrichment.
2.8. lncRNA‐miRNA‐mRNA network analysis
To identify the interactions between mRNAs and lncRNAs, we constructed lncRNA‐miRNA‐mRNA networks. A prediction of the different lncRNA‐miRNA interactions was made by the popular miRNA target gene prediction software, miRNA binding sites and target mRNA prediction were performed with proprietary software based on TargetScan and miRanda. Network maps for our lncRNA‐miRNA‐mRNA analyses were illustrated using cytoscape software and were based on the lncRNA, and prediction data of miRNA and mRNA.
3. RESULTS
3.1. Differentially expressed lncRNAs and mRNAs post‐irradiation
A total of 29 007 lncRNAs and 17 142 mRNAs were detected in the control group and 3.5 days post‐irradiation. For lncRNAs, 2690 were only detected in the control group, 2510 were only detected 3.5 days post‐irradiation and 23 807 were detected in both groups (Figure 1A). Of the total 29 007 lncRNAs, 91 were significantly up‐regulated and 58 were significantly down‐regulated 3.5 days post‐irradiation when compared with controls (P < 0.05, fold change ≥2) (Figures 1B, C and 3A).
Figure 1.
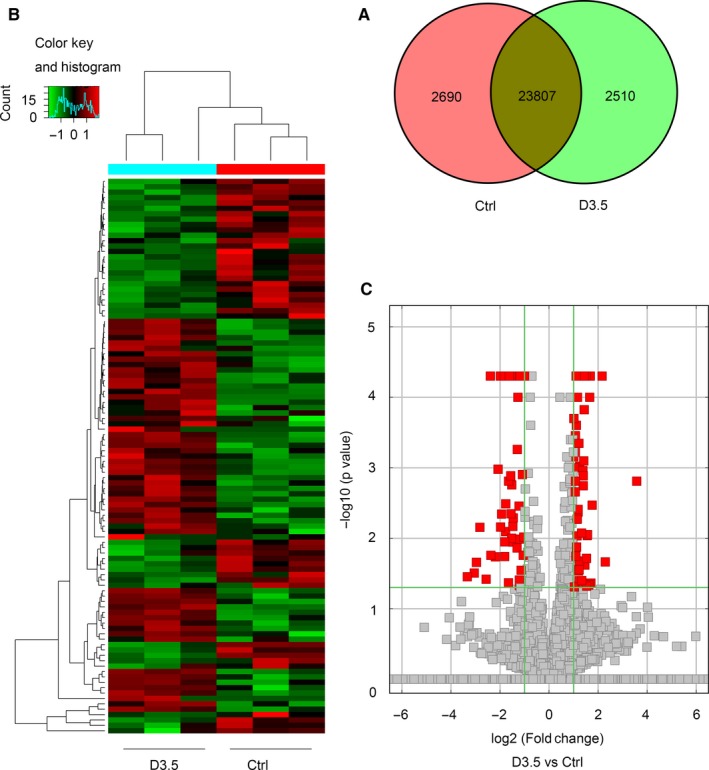
The expression profiling changes of lncRNAs in control group and irradiated group. A, Venn diagram showing the number of overlapping lncRNAs in the irradiated and control groups. B, Heat map of lncRNAs showing hierarchical clustering of altered lncRNAs in irradiated mice when compared with controls. Red represents up‐regulation and green represent down‐regulation. C, Volcano Plot indicating up‐ and downregulated lncRNAs in irradiated mice when compared with controls
For mRNAs, 282 were only detected in the control group, 346 were only detected 3.5 days post‐irradiation and 16 514 were detected in both groups (Figure 2A). Of the 17 142 total mRNAs, 752 were significantly up‐regulated and 400 were significantly down‐regulated 3.5 days post‐irradiation when compared with controls (P < 0.05, fold change ≥2) (Figures 2B, C and 3A).
Figure 2.
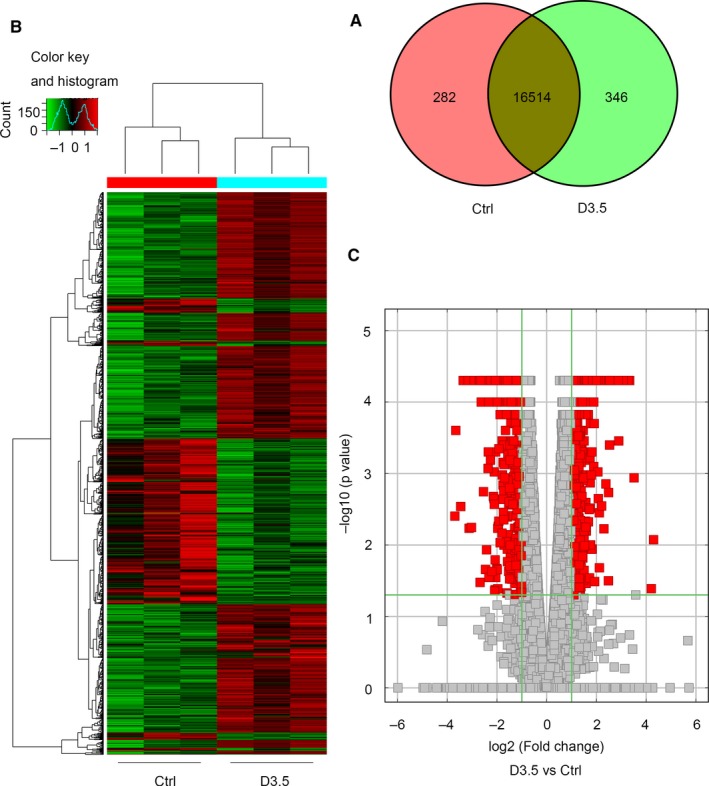
The expression profiling changes of mRNAs in control group and irradiated group. A, Venn diagram showing the number of overlapping mRNAs in irradiated and control groups. B, Heat map of mRNAs showing hierarchical clustering of altered mRNAs in irradiated mice when compared with controls. Red represents up‐regulation and green represents down‐regulation. C, Volcano Plot indicating up‐ and downregulated mRNAs in irradiated mice when compared with controls
Figure 3.
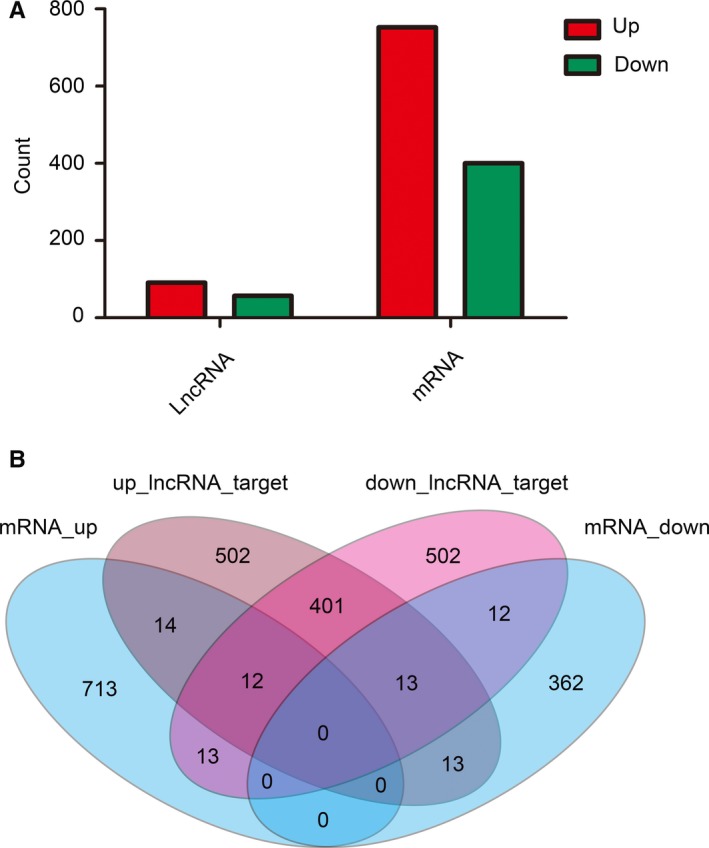
Number of relative lncRNAs and mRNAs in irradiated mice when compared with controls. A, Histogram showing the number of up‐ and downregulated lncRNAs and mRNAs in irradiated mice. B, Venn diagram showing the overlapping number of targeted mRNAs in up‐regulated lncRNAs, targeted mRNA in down‐regulated lncRNAs, up‐regulated mRNAs, and down‐regulated mRNAs
Using the differently expressed lncRNA data (top 20 for each of up‐regulated and down‐regulated lncRNAs are listed in Table 2), we next predicted their respective sponge miRNA and target mRNA. We also compared these predictions with our mRNA sequencing results. As shown in Table 3 and Figure 3B, 26 target mRNAs of up‐regulated lncRNAs were up‐regulated, while 25 target mRNAs of down‐regulated lncRNAs were down‐regulated. These were then used for our GO, KEGG pathway and lncRNA‐miRNA‐mRNA network analyses.
Table 2.
Top 20 significantly up‐ and downregulated lncRNAs
| Transcript ID | Regulation | Log2 FC | P‐value | Chr | Strand | Start | End |
|---|---|---|---|---|---|---|---|
| AK029621 | Up | 0.0001 | 9 | + | 108991901 | 109010572 | |
| ENSMUST00000182520 | Up | 649.341 | 0.04345 | 17 | |||
| TCONS_00013586 | Up | 60.8039 | 0.0395 | 17 | |||
| AK157361_1 | Up | 3.57644 | 0.00155 | 1 | |||
| ENSMUST00000173070 | Up | 2.2921 | 0.0217 | 17 | |||
| AK152734 | Up | 2.16347 | 0.00005 | 19 | |||
| AK013908 | Up | 1.7585 | 0.0034 | 13 | + | 108044473 | 108049146 |
| NR_045710 | Up | 1.70723 | 0.0434 | 3 | |||
| TCONS_00025210 | Up | 1.68216 | 0.00005 | 5 | − | 23700648 | 23712667 |
| TCONS_00006870 | Up | 1.65815 | 0.0001 | 12 | |||
| TCONS_00013542 | Up | 1.62523 | 0.0458 | 17 | + | 33909413 | 33911423 |
| uc007jjl.1 | Up | 1.57118 | 0.0089 | 11 | |||
| ENSMUST00000183030 | Up | 1.55833 | 0.0092 | 9 | |||
| AK037312 | Up | 1.53371 | 0.0479 | 5 | |||
| AK035001 | Up | 1.52899 | 0.0193 | 10 | |||
| TCONS_00025212 | Up | 1.50848 | 0.00005 | 5 | − | 23700648 | 23712667 |
| NR_045042 | Up | 1.46381 | 0.00005 | 5 | |||
| AK087220 | Up | 1.46334 | 0.0439 | 4 | − | 46379863 | 46389437 |
| ENSMUST00000172531 | Up | 1.46145 | 0.02405 | 17 | |||
| AK085607 | Up | 1.43428 | 0.00015 | 9 | |||
| TCONS_00010866 | Down | −7.37191 | 0.0053 | 15 | |||
| uc029sug.1 | Down | −3.33117 | 0.03525 | 15 | |||
| uc009bpt.1 | Down | −3.04963 | 0.03125 | 6 | + | 41059591 | 41547104 |
| AK038898 | Down | −2.95816 | 0.022 | 9 | |||
| uc009bpe.2 | Down | −2.81888 | 0.007 | 6 | + | 41227695 | 41538592 |
| TCONS_00002419 | Down | −2.56474 | 0.03815 | 10 | |||
| AK083183 | Down | −2.39063 | 0.00005 | 9 | |||
| 2_00008059 | Down | −2.36925 | 0.0175 | 13 | + | 44730837 | 44903258 |
| TCONS_00025153 | Down | −2.18865 | 0.01835 | 5 | |||
| ENSMUST00000151051 | Down | −2.07808 | 0.00105 | 2 | |||
| AK156559 | Down | −1.98272 | 0.00005 | 10 | |||
| uc029qtg.1 | Down | −1.97477 | 0.00695 | 1 | + | 131797394 | 131818115 |
| AK083360 | Down | −1.93436 | 0.00455 | 12 | |||
| AK138493 | Down | −1.87959 | 0.00005 | 2 | |||
| ENSMUST00000143673 | Down | −1.82223 | 0.00005 | 11 | |||
| AK087733 | Down | −1.80859 | 0.0182 | 18 | |||
| 2_00001168 | Down | −1.80778 | 0.01125 | 1 | + | 43934006 | 44002971 |
| TCONS_00025152 | Down | −1.76965 | 0.00805 | 5 | |||
| AK034241 | Down | −1.76488 | 0.00325 | 14 | |||
| uc007zhs.2 | Down | −1.65472 | 0.043 | 16 | + | 44765735 | 44794977 |
Table 3.
mRNAs used for the GO and KEGG signalling pathway analyses
| Gene | Regulation | Log2 FC | P‐value | Locus | Strand |
|---|---|---|---|---|---|
| Gm4724 | Up | 6.81095 | 0.00545 | chr2:175372027‐175435807 | − |
| Trmt10a | Up | 1.41578 | 0.0004 | chr3:138143447‐138159821 | + |
| Polr1e | Up | 1.22418 | 0.0023 | chr4:45018582‐45084604 | + |
| Ddah1 | Up | 3.51136 | 0.00115 | chr3:144947560‐145894277 | + |
| Ankle1 | Up | 1.13181 | 0.0008 | chr8:71406009‐71409904 | + |
| Lrp8 | Up | 1.38455 | 0.00175 | chr4:107802258‐107876840 | + |
| Ereg | Up | 1.63764 | 0.00005 | chr5:91051870‐91093649 | + |
| Pus7 | Up | 1.24149 | 0.00015 | chr5:23740647‐23783711 | − |
| Exo1 | Up | 1.42521 | 0.00005 | chr1:175880777‐175911396 | + |
| Kif24 | Up | 1.35257 | 0.00055 | chr4:41390744‐41464887 | − |
| Celsr2 | Up | 1.74194 | 0.0006 | chr3:108390850‐108415552 | − |
| Trim6 | Up | 1.28095 | 0.00015 | chr7:104218792‐104235152 | + |
| Adamts15 | Up | 1.03113 | 0.00005 | chr9:30899154‐30922452 | − |
| Pkp1 | Up | 1.38649 | 0.00005 | chr1:135871394‐135919207 | − |
| Prrg4 | Up | 1.16766 | 0.0065 | chr2:104830740‐104849876 | − |
| Fam198b | Up | 1.03435 | 0.0249 | chr3:79859292‐79946280 | + |
| Kif14 | Up | 1.38504 | 0.02075 | chr1:136181473‐137052008 | + |
| Rpusd2 | Up | 1.38904 | 0.00005 | chr2:119034789‐119039769 | + |
| Slc1a2 | Up | 2.11536 | 0.00355 | chr2:102658658‐102790784 | + |
| Orai2 | Up | 1.01472 | 0.00005 | chr5:136147463‐136170656 | − |
| Ephb2 | Up | 1.10499 | 0.00005 | chr4:136647538‐136835988 | − |
| Pgap1 | Up | 1.28283 | 0.00005 | chr1:54472999‐54557684 | − |
| Xrcc2 | Up | 1.12742 | 0.0106 | chr5:25582629‐26169460 | − |
| Zfp37 | Up | 1.03346 | 0.00145 | chr4:62189539‐62208446 | − |
| Paqr8 | Up | 1.481 | 0.00005 | chr1:20890621‐20938755 | + |
| Steap2 | Up | 1.0385 | 0.00015 | chr5:5664830‐5694578 | − |
| Klrk1 | Down | −1.62061 | 0.00905 | chr6:129610322‐129623864 | − |
| Nebl | Down | −1.49013 | 0.0299 | chr2:17343908‐17731464 | − |
| Sh2d7 | Down | −1.65355 | 0.00025 | chr9:54538983‐54560218 | + |
| Cd84 | Down | −1.0478 | 0.0017 | chr1:171839696‐171890718 | + |
| Siglech | Down | −2.62999 | 0.0001 | chr7:55768177‐55778925 | + |
| Sprr2a1 | Down | −1.46984 | 0.00005 | chr3:92215834‐92257298 | + |
| Strip2 | Down | −1.15512 | 0.0007 | chr6:29917012‐29959681 | + |
| Hpgds | Down | −1.10104 | 0.023 | chr6:65116828‐65144908 | − |
| Cd28 | Down | −1.83097 | 0.00005 | chr1:60716799‐60773359 | + |
| Inpp5d | Down | −1.15829 | 0.0106 | chr1:87620311‐87720868 | + |
| Spib | Down | −1.15289 | 0.0007 | chr7:44525994‐44532071 | − |
| Ptpn7 | Down | −1.49671 | 0.00005 | chr1:135132724‐135145326 | + |
| Cd4 | Down | −1.08809 | 0.00055 | chr6:124864691‐124888221 | − |
| Kctd14 | Down | −1.10169 | 0.0078 | chr7:97451333‐97459553 | + |
| Traf1 | Down | −1.44914 | 0.0017 | chr2:34941749‐34961772 | − |
| Srgap3 | Down | −1.44089 | 0.0004 | chr6:112717970‐112947266 | − |
| Itga4 | Down | −1.25717 | 0.00005 | chr2:79255425‐79461107 | + |
| P2rx7 | Down | −1.00178 | 0.00165 | chr5:122643910‐122691432 | + |
| Sypl2 | Down | −1.35138 | 0.0007 | chr3:108211471‐108226648 | − |
| Sh2d2a | Down | −2.02063 | 0.00005 | chr3:87846754‐87855722 | + |
| Vwa1 | Down | −1.10347 | 0.00005 | chr4:155761177‐155776161 | − |
| Ikzf2 | Down | −2.00808 | 0.00005 | chr1:69513931‐69685960 | − |
| Prkcb | Down | −1.76677 | 0.00005 | chr7:122288750‐122634402 | + |
| Ppp1r16b | Down | −1.21456 | 0.00005 | chr2:158665397‐158766334 | + |
| St6galnac3 | Down | −1.44486 | 0.009 | chr3:153205404‐153725062 | − |
3.2. Validation of lncRNAs and mRNAs expression using qRT‐PCR
We selected two up‐regulated, two down‐regulated lncRNAs and mRNAs at 3.5 days post‐irradiation for validation purposes. Our qRT‐PCR results indicated that up‐ and downregulated lncRNAs and mRNAs were consistent with our sequencing results (Figure 4).
Figure 4.
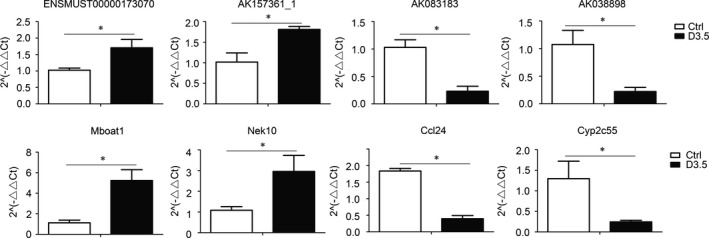
qRT‐PCR validations of two up‐regulated and two down‐regulated lncRNAs and mRNAs. ENSMUST00000173070 and AK157361 are two up‐regulated lncRNAs, AK083183 and AK038898 are two down‐regulated lncRNAs, Mboat1 and Nek10 are two up‐regulated mRNAs, Ccl24 and Cyp2c55 are two down‐regulated mRNAs. *P < 0.05
3.3. Functional analysis of miRNA target genes
We next performed a GO analysis to better understand the functional association of target genes with the differentially expressed lncRNAs (Figure 5). Our GO analysis included three parts: MF, BP and CC. As shown in Figure 5A, we determined two main functions for the first part (MF): Pseudouridine synthase activity and endodeoxyribonuclease activity. Their function with regard to biological processes (Figure 5B) was determined to be adaptive immune response and regulation of the immune response. When classified according to their respective CC classifications (Figure 5C), the cell part and cell make up the largest proportion.
Figure 5.
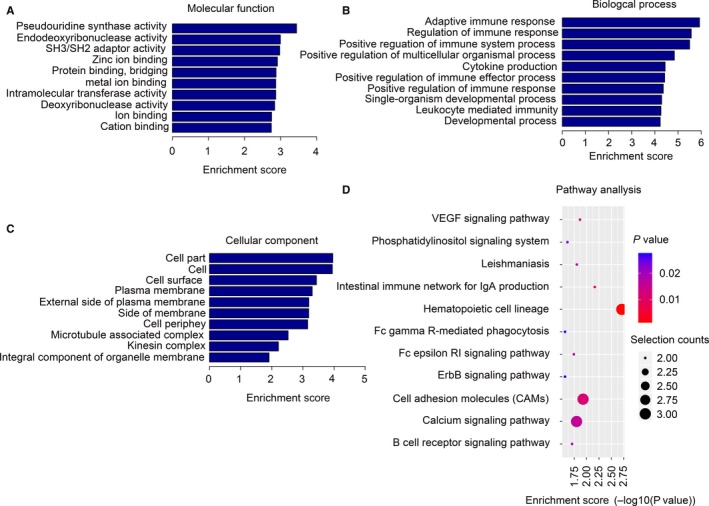
GO and KEGG signalling pathway analyses of differentially expressed lncRNAs‐targeted mRNAs. (A), Molecular function (MF), (B) Biological process (BP) and (C) Cellular component (CC) of lncRNAs‐targeted mRNAs. (D), KEGG signalling pathway analysis of lncRNAs‐targeted mRNAs
The function of the predicted target mRNAs of the identified lncRNAs was next analysed using a KEGG pathway analysis (Figure 5D). The differentially expressed lncRNAs and their target mRNAs were determined to be involved in the following biological and cellular functions: VEGF signalling pathway, phosphatidylinositol signalling system, leishmaniasis and the intestinal immune network for IgA production. With regard to the VEGF signalling pathway, two predicted mRNAs were shown to be involved: Prkcb and Sh2d2a. Prkcb is also involved in phosphatidylinositol and ErbB signalling systems. Interestingly, work using an experimental chronic colitis model induced by DSS has shown that VEGF‐C can reduce intestinal inflammation by regulating IL‐9/IL‐17 balance and improving the gut microbiota.28
3.4. Prediction of miRNA binding sites and lncRNA‐miRNA‐mRNA network analysis
We selected up‐regulated lncRNAs and their 26 targeted mRNAs as well as down‐regulated lncRNAs and their 25 targeted mRNAs to construct our lncRNAs‐miRNAs‐mRNAs network (Figure 6). As indicated, the network is complex‐ one lncRNA can associate with multiple miRNAs and one miRNA can inhibit multiple mRNAs. The miRNA mmu‐miR‐5110 combined the most lncRNAs and target mRNAs. Critically, some of these targeted mRNAs have been reported to be involved in the radiation‐induced intestinal injury or repair. Ephb2 is an intestinal stem cell marker, and it has been studied extensively in colorectal cancer.29 It has been proposed that EXO1 acts in the excision step during mismatch‐repair. After mismatch recognition in prokaryotes and eukaryotes,30 EXO1 is also involved in the 5′ to 3′ end resection at DSB (double‐strand breaks) ends to initiate HR in both yeast and mammalian systems.31 Moreover, Kobayashi et al32 found that Pgap1 was highly expressed by the follicle‐associated epithelium; given this, it may be associated with intestinal mucosal immunity. A better understanding of lncRNA function, as well as their potential intestinal target genes will need further study.
Figure 6.
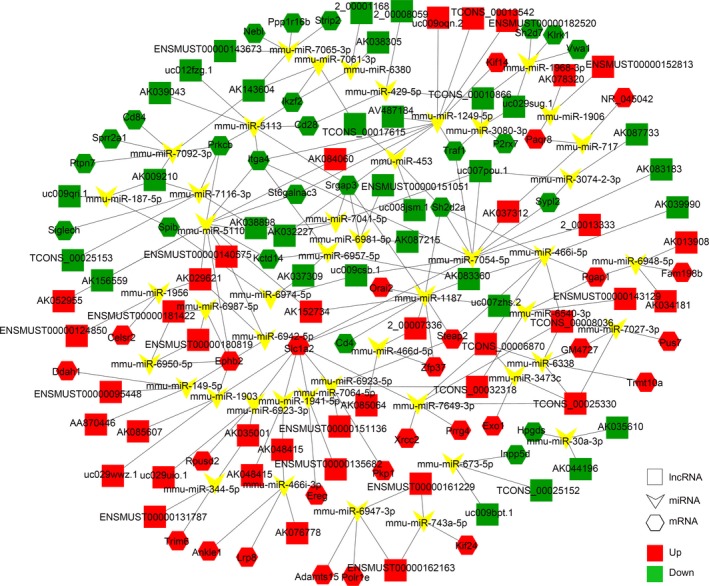
lncRNA‐miRNA‐mRNAs regulatory network analysis of lncRNAs‐targeted mRNAs. The quadrilateral represents lncRNAs, arrowheads represent miRNAs, hexagons represent mRNAs, red represents up‐regulated, and green represents down‐regulated
4. DISCUSSION
Here, we provide a basic study of lncRNAs function during the radiation‐induced intestinal injury. Using a sequencing approach, we obtained 29 007 lncRNAs from the control and irradiated groups. Of these 29 007 lncRNAs, there were 2510 lncRNAs detected only at 3.5 days post‐irradiation, while 23 807 were detected in both groups. After comparing the irradiated and control groups, we found 91 lncRNAs were significantly up‐regulated and 58 were significantly down‐regulated after irradiation.
We also obtained 17 142 mRNAs, of which 346 were only detected 3.5 days post‐irradiation and 16 514 were detected in both the groups. Comparing the two groups revealed that 752 mRNAs were significantly up‐regulated and 400 were significantly down‐regulated in the irradiated group when compared with controls.
We selected the 91 significantly up‐ and 58 down‐regulated lncRNAs, predicted their combined miRNAs and miRNA‐targeted mRNAs, and compared these with our mRNAs sequencing results, selected both lncRNAs and target mRNAs up‐regulated and down‐regulated. These lncRNAs may act as an endogenous “sponge” that regulates the target gene of miRNAs through competition with miRNAs. By comparing our predicted results with those obtained through sequencing, we found up‐regulated lncRNAs‐targeted 26 mRNAs and down‐regulated lncRNAs‐targeted 25 mRNAs. Due to consistent regulation with mRNAs, lncRNAs may function as a miRNAs sponge.
Using these data, we next performed a GO analysis and found that their biological function was relegated to an adaptive immune response and regulation of the immune response. As is well known, the intestinal mucosa harbours one of the largest, most complex immune systems in the body. Even under normal conditions, the intestinal mucosa exhibits a state of “physiological inflammation” in response to dietary and bacterial antigens that are present in the intestinal lumen.33 When exposed to radiation, immune cells are activated and inflammatory cell recruitment occurs. The result is that activated immune cells attack and destroy neighbouring cells. Given this biological function, it is likely that some genes may regulate the radiation‐induced intestinal injury through regulation of the immune response.
Our KEGG signalling pathway analysis showed that the differentially expressed lncRNAs‐targeted mRNAs were mainly involved in VEGF signalling. The VEGF signalling pathway is a well‐studied pathway that is involved in angiogenesis; researchers have also found that this pathway plays an important role in intestinal immune and inflammation. Chronic, intestinal inflammation is associated with pathological angiogenesis that further amplifies the inflammatory response. For instance, Ardelean34 showed that a VEGF monoclonal antibody could ameliorate chronic colitis induced by DSS in Endoglin heterozygous mice. Schlieve et al35 found that manipulating VEGF bioavailability led to profound effects on not only intestinal vasculature, but the epithelial stem and progenitor cells in the intestinal crypt. When taken together, these findings indicate a potential role for VEGF in the radiation‐induced intestinal injury.
The lncRNAs‐miRNAs‐mRNAs network is very complex‐ one lncRNA had multiple miRNA binding sites and could combine with multiple lncRNAs while one miRNA also combined via multiple lncRNAs. lncRNAs target different miRNAs and play different roles. Importantly, even lncRNAs from the same gene may play different roles. For example, the MBNL336 splicing factor induces lncRNA‐PXN‐AS1 exon 4 inclusion. The transcript‐lacking exon 4 binds to the PXN mRNA coding sequences, causing dissociation of translation elongation factors from PXN mRNA. This leads to inhibition of PXN mRNA translation. In contrast, the transcript containing exon 4 preferentially binds to the 3′ untranslated region of the PXN mRNA, and protecting PXN mRNA from microRNA‐24‐AGO2 complex‐induced degradation. This leads to increased PXN expression. We have performed sequencing on jejuna samples post‐irradiation and analysed the resulting lncRNAs. However, we only predicted the sponge function of lncRNAs with their target mRNAs and all will need to be verified. In addition to its miRNA sponge function, lncRNAs can also form complementary double strands with protein‐encoding transcripts, interfere with mRNA splicing and/or bind and modulate protein activity. Further work will be needed to better elucidate lncRNA function.
More than 70% of cancer patients undergo radiotherapy as intestinal mucosa undergoes rapid and constant renewal throughout the life of the organism. It is hard to protect and the intestinal epithelium is particularly sensitive to ionising radiation. Radiation‐induced intestinal toxicity results in diarrhoea, dehydration and even death. Despite this, there is currently no accepted approach to prevent or treat the radiation‐induced intestinal injury. Although our laboratory has found that mesenchymal stem cell37, 38 and 6‐shogaol39 can improve animal survival and intestinal function following irradiation injury, we still need to understand more about the effects of radiation in order to solve the problem of radiation‐induced intestinal damage from the effect itself. Therefore, our previous work has analysed the regulatory network of circRNAs40 in radiation‐induced intestinal injury, as well as some important responses of mRNAs.41 In this article, we focus on the role of lncRNAs. Researchers have found lncRNAs play an important role in ontogenesis and disease occurrence. As mentioned previously, many lncRNAs are involved in maintaining and regulating the intestinal epithelial barrier and DNA damage induced by ionising radiation. However, there has been little work carried out regarding the role of lncRNAs in the radiation‐induced intestinal injury or repair. We hope that the identification of lncRNAs that regulate the radiation‐induced intestinal injury and repair will help make a contribution to better approaches for the clinical treatment of the radiation‐induced intestinal injury.
CONFLICT OF INTEREST
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work. There is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (31670859), CAMS Innovation Fund for Medical Science (2017‐I2M‐1‐016), China Postdoctoral Science Foundation (2018M630106), Natural Science Foundation of Tianjin (18JCYBJC26800, 18JCQNJC12300), the Fundamental Research Funds for the Central Universities (10023201601602).
Lu Q, Gong W, Wang J, et al. Analysis of changes to lncRNAs and their target mRNAs in murine jejunum after radiation treatment. J Cell Mol Med. 2018;22:6357–6367. 10.1111/jcmm.13940
Contributor Information
Yan Wang, Email: wangyan@irm-cams.ac.cn.
Qiang Liu, Email: liuqiang@irm-cams.ac.cn.
References
- 1. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629‐641. [DOI] [PubMed] [Google Scholar]
- 2. Mercer TR, Dinger ME, Mattick JS. Long non‐coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155. [DOI] [PubMed] [Google Scholar]
- 3. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao J, Sun BK, Erwin JA, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng J, Bi C, Clark B, et al. The Evf‐2 noncoding RNA is transcribed from the Dlx‐5/6 ultraconserved region and functions as a Dlx‐2 transcriptional coactivator. Genes Dev. 2006;20:1470‐1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beltran M, Puig I, Peña C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1‐induced epithelial‐mesenchymal transition. Genes Dev. 2008;22:756‐769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martens‐Uzunova ES, Böttcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140‐1151. [DOI] [PubMed] [Google Scholar]
- 9. Ouyang J, Zhu X, Chen Y, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon‐stimulated gene transcription. Cell Host Microbe. 2014;16:616‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klein RH, Stephens DN, Ho H, et al. Cofactors of LIM domains associate with estrogen receptor α to regulate the expression of noncoding RNA H19 and corneal epithelial progenitor cell function. J Biol Chem. 2016;291:13271‐13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng G, Cheng C, Wang R, et al. Long noncoding RNA EBF3‐AS promotes neuron apoptosis in Alzheimer's disease. DNA Cell Biol. 2018;37:220‐226. [DOI] [PubMed] [Google Scholar]
- 12. Xiao L, Wu J, Wang JY, et al. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology 2018;154:599‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mettler FA, Voelz GL. Major radiation exposure ‐ what to expect and how to respond. N Engl J Med. 2002;346:1554‐1561. [DOI] [PubMed] [Google Scholar]
- 14. Denham JW, Hauer‐Jensen M. The radiotherapeutic injury ‐ a complex ‘wound’. Radiother Oncol. 2002;63:129‐145. [DOI] [PubMed] [Google Scholar]
- 15. Bentzen SM. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer. 2006;6:702. [DOI] [PubMed] [Google Scholar]
- 16. Leibowitz BJ, Wei L, Zhang L, et al. Ionizing irradiation induces acute haematopoietic syndrome and gastrointestinal syndrome independently in mice. Nat Commun. 2014;5:3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Su Z, Zhi X, Zhang Q, et al. LncRNA H19 functions as a competing endogenous RNA to regulate AQP3 expression by sponging miR‐874 in the intestinal barrier. FEBS Lett. 2016;590:1354‐1364. [DOI] [PubMed] [Google Scholar]
- 18. Zhang W, Xu Y, Chen Z, et al. Knockdown of aquaporin 3 is involved in intestinal barrier integrity impairment. FEBS Lett. 2011;585:3113‐3119. [DOI] [PubMed] [Google Scholar]
- 19. Juan V, Crain C, Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221‐1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geng H, Bu HF, Liu F, et al. In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology. 2018;155:144‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T, Xue H, Lin R, et al. MiR‐34c and PlncRNA1 mediated the function of intestinal epithelial barrier by regulating tight junction proteins in inflammatory bowel disease. Biochem Biophys Res Commun. 2017;486:6‐13. [DOI] [PubMed] [Google Scholar]
- 22. Michelini F, Pitchiaya S, Vitelli V, et al. Damage‐induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double‐strand breaks. Nat Cell Biol. 2017;19:1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Betts JA, Moradi Marjaneh M, Al‐Ejeh F, et al. Long noncoding RNAs CUPID1 and CUPID2 mediate breast cancer risk at 11q13 by modulating the response to DNA damage. Am J Hum Genet. 2017;101:255‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. French Juliet D, Ghoussaini M, Edwards Stacey L, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long‐range enhancers. Am J Hum Genet. 2013;92:489‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin M. Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet J. 2011;17:10‐12. [Google Scholar]
- 26. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X, Zhao J, Qin L. VEGF‐C mediated enhancement of lymphatic drainage reduces intestinal inflammation by regulating IL‐9/IL‐17 balance and improving gut microbiota in experimental chronic colitis. Am J Transl Res. 2017;9:4772‐4784. [PMC free article] [PubMed] [Google Scholar]
- 29. Merlos‐Suárez A, Barriga Francisco M, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511‐524. [DOI] [PubMed] [Google Scholar]
- 30. Burdett V, Baitinger C, Viswanathan M, et al. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl‐directed mismatch repair. Proc Natl Acad Sci USA. 2001;98:6765‐6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mimitou EP, Symington LS. DNA end resection‐unraveling the tail. DNA Repair (Amst). 2011;10:344‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi A, Donaldson DS, Kanaya T, et al. Identification of novel genes selectively expressed in the follicle‐associated epithelium from the meta‐analysis of transcriptomics data from multiple mouse cell and tissue populations. DNA Res. 2012;19:407‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brandtzaeg P, Halstensen TS, Kett K, et al. Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial lymphocytes. Gastroenterology. 1989;97:1562‐1584. [DOI] [PubMed] [Google Scholar]
- 34. Ardelean DS, Yin M, Jerkic M, et al. Anti‐VEGF therapy reduces intestinal inflammation in Endoglin heterozygous mice subjected to experimental colitis. Angiogenesis. 2014;17:641‐659. [DOI] [PubMed] [Google Scholar]
- 35. Schlieve CR, Mojica SG, Holoyda KA, et al. Vascular endothelial growth factor (VEGF) bioavailability regulates angiogenesis and intestinal stem and progenitor cell proliferation during postnatal small intestinal development. PLoS ONE. 2016;11:e0151396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan J, Liu X, Wang T, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA‐PXN‐AS1. Nat Cell Biol. 2017;19:820. [DOI] [PubMed] [Google Scholar]
- 37. Gong W, Guo M, Han Z, et al. Mesenchymal stem cells stimulate intestinal stem cells to repair radiation‐induced intestinal injury. Cell Death Dis. 2016;7:e2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo M, Gong W, Zhang HW, et al. Human mesenchymal stem cells promote survival and prevent intestinal damage in a mouse model of radiation injury. Rsc Advances. 2016;6:65105‐65111. [Google Scholar]
- 39. Wang J, Yao M, Wang Y, et al. 6‐Shogaol ameliorates injury to the intestinal mucosa and increases survival after high‐dose abdominal irradiation. J Funct Foods. 2017;36:63‐71. [Google Scholar]
- 40. Lu Q, Gong W, Wang J, et al. Identification of circular RNAs altered in mouse jejuna after radiation. Cell Physiol Biochem. 2018;47:2558‐2568. [DOI] [PubMed] [Google Scholar]
- 41. Wang J, Lu Q, Ji K, et al. The dynamic changes of gene expressions in radiation‐injured intestine based on RNA‐sequence analysis. Chin J Radiol Med Prot. 2018;38:81‐86. [Google Scholar]


