1. INTRODUCTION AND BACKGROUND
Lysosome‐associated protein transmembrane‐4 beta (LAPTM4B) has two alleles named as LAPTM4B*1 and LAPTM4B*2 (GenBank No. AY219176 and AY219177). Allele *1 has a single copy of a 19‐bp sequence in the 5` untranslated region (5`UTR), but allele *2 contains tandem repeats of 19‐bp sequence.1 LAPTM4B gene is located on long chromosome 8 (8q22.1) and contains seven exons that encodes two isoforms of tetratransmembrane proteins, LAPTM4B‐24 and LAPTM4B‐35, with molecular weights of 25 kDa and 35 kDa respectively. The LAPTM4B‐35′s primary structure is formed by 317 amino acid residues, and LAPTM4B‐24 comprised 226 amino acids. LAPTM4B, an integral membrane protein, contains several lysosomal‐targeting motifs at the C terminus and colocalizes with late endosomal and lysosomal markers. LAPTM4B is a proto‐oncogene, which becomes up‐regulated in various cancers. Preceding studies have examined the possible link between LAPTM4B polymorphism and susceptibility to several cancers,1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 but the findings are still inconsistent. Hence, the present meta‐analysis was designed to investigate the impact of LAPTM4B polymorphism on risk of cancer.
2. METHODS
A comprehensive search in Web of Science, PubMed, Scopus, and Google Scholar databases was done for all articles describing an association between LAPTM4B polymorphism and cancer risk published up to April 2018. The search strategy was “cancer, carcinoma, tumor, neoplasms,” “LAPTM4B, Lysosome‐associated protein transmembrane‐4,” and “polymorphism, mutation, variant.” Relevant studies included the meta‐analysis if they met the following inclusion criteria: (a) Original case‐control studies that evaluated the LAPTM4B polymorphism and the risk of cancer; (b) studies provided sufficient information of the genotype frequencies of LAPTM4B polymorphism in both cases and controls. The exclusion criteria were: (a) conference abstract, case reports, reviews, duplication data; (b) insufficient genotype information provided.
Data extraction was done by two independently authors. From each study, the following data were collected: the first author's name, publication year, country, ethnicity of participants, cancer type, genotyping methods of LAPTM4B polymorphism, the sample size, and the genotype and allele frequencies of cases and controls (Table 1).
Table 1.
Characteristics of all studies included in the meta‐analysis
| Author | Year | Country | Ethnicity | Cancer type | Source of control | Genotyping method | Case/control | Cases | Controls | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *1/1 | *1/2 | *2/2 | *I | *2 | *1/1 | *1/2 | *2/2 | *1 | *2 | |||||||||
| Chen | 2016 | China | Asian | Renal cell carcinoma | PB | PCR | 180/347 | 83 | 80 | 17 | 246 | 114 | 198 | 131 | 18 | 527 | 167 | 0.538 |
| Chen | 2016 | China | Asian | Bladder cancer | PB | PCR | 91/347 | 38 | 41 | 12 | 117 | 65 | 198 | 131 | 18 | 527 | 167 | 0.538 |
| Chen | 2016 | China | Asian | B‐cell lymphoma | PB | PCR | 162/350 | 87 | 64 | 11 | 238 | 86 | 199 | 133 | 18 | 531 | 169 | 0.549 |
| Cheng | 2008 | China | Asian | Colon cancer | HB | PCR | 253/350 | 113 | 112 | 28 | 338 | 168 | 199 | 133 | 18 | 531 | 169 | 0.538 |
| Cheng | 2008 | China | Asian | Rectal cancer | HB | PCR | 237/350 | 126 | 101 | 10 | 353 | 121 | 199 | 133 | 18 | 531 | 169 | 0.539 |
| Cheng | 2008 | China | Asian | Oesophageal cancer | HB | PCR | 211/350 | 123 | 80 | 8 | 326 | 96 | 199 | 133 | 18 | 531 | 169 | 0.539 |
| Deng | 2005 | China | Asian | Lung cancer | PB | PCR | 166/134 | 54 | 91 | 21 | 199 | 133 | 67 | 59 | 8 | 193 | 75 | 0.284 |
| Ding | 2018 | China | Asian | B‐cell lymphoma | HB | PCR | 162/350 | 87 | 64 | 11 | 238 | 86 | 199 | 133 | 18 | 531 | 169 | 0.538 |
| Fan | 2012 | China | Asian | Breast cancer | HB | PCR | 732/649 | 326 | 342 | 64 | 994 | 470 | 346 | 262 | 41 | 954 | 344 | 0.355 |
| Hashemi | 2014 | Iran | Asian | Breast cancer | HB | PCR | 311/225 | 137 | 163 | 11 | 437 | 185 | 104 | 117 | 4 | 325 | 125 | 0.009 |
| Hashemi | 2016 | Iran | Asian | Prostate cancer | HB | PCR | 168/176 | 102 | 55 | 11 | 259 | 77 | 79 | 87 | 10 | 245 | 107 | 0.025 |
| Li | 2006 | China | Asian | Lung cancer | PB | PCR | 131/104 | 70 | 56 | 5 | 196 | 66 | 57 | 36 | 11 | 150 | 58 | 0.155 |
| Li | 2012 | China | Asian | Breast cancer | HB | PCR | 208/211 | 90 | 100 | 18 | 280 | 136 | 129 | 76 | 6 | 334 | 88 | 0.185 |
| Liu | 2007 | China | Asian | Gastric cancer | HB | PCR | 214/350 | 88 | 107 | 19 | 283 | 145 | 199 | 133 | 18 | 531 | 169 | 0.483 |
| Meng | 2011 | China | Asian | Cervical cancer | HB | PCR | 317/413 | 127 | 153 | 37 | 407 | 227 | 225 | 163 | 28 | 613 | 219 | 0.775 |
| Meng | 2013 | China | Asian | Endometrial cancer | HB | PCR | 283/378 | 93 | 135 | 55 | 321 | 245 | 200 | 140 | 38 | 540 | 216 | 0.072 |
| Meng | 2017 | China | Asian | Papillary thyroid carcinoma | HB | PCR | 183/697 | 90 | 73 | 20 | 253 | 113 | 397 | 264 | 36 | 1058 | 336 | 0.352 |
| Qi | 2010 | China | Asian | Liver cancer | HB | PCR | 86/78 | 27 | 51 | 8 | 105 | 67 | 36 | 34 | 7 | 106 | 48 | 0.798 |
| Shaker | 2015 | Egypt | Breast cancer | HB | PCR | 88/80 | 36 | 40 | 12 | 112 | 64 | 45 | 29 | 6 | 119 | 41 | 0.661 | |
| Sun | 2007 | China | Asian | Lymphoma | HB | PCR | 166/350 | 72 | 71 | 23 | 215 | 117 | 199 | 133 | 18 | 531 | 169 | 0.549 |
| Sun | 2008 | China | Asian | Liver cancer | PB | PCR | 190/175 | 72 | 110 | 8 | 254 | 126 | 99 | 67 | 9 | 265 | 85 | 0.586 |
| Tang | 2014 | China | Asian | NSCLC | HB | PCR | 392/437 | 158 | 171 | 63 | 487 | 297 | 226 | 176 | 35 | 628 | 246 | 0.928 |
| Wang | 2010 | China | Asian | Pancreatic cancer | HB | PCR | 58/156 | 24 | 26 | 8 | 74 | 42 | 74 | 67 | 15 | 215 | 97 | 0.976 |
| Wang | 2012 | China | Asian | Liver cancer | HB | PCR | 303/515 | 107 | 156 | 40 | 370 | 236 | 272 | 205 | 38 | 749 | 281 | 0.941 |
| Wang | 2013 | China | Asian | Nasopharyngeal carcinoma | HB | PCR | 134/327 | 74 | 48 | 12 | 196 | 72 | 163 | 145 | 19 | 471 | 183 | 0.69 |
| Wang | 2017 | China | Asian | Pancreatic cancer | HB | PCR | 233/842 | 98 | 116 | 19 | 312 | 154 | 435 | 350 | 57 | 1220 | 464 | 0.231 |
| Xu | 2012 | China | Asian | Ovarian cancer | HB | PCR | 282/365 | 122 | 115 | 45 | 359 | 205 | 231 | 108 | 26 | 570 | 160 | 0.009 |
| Yang | 2012 | China | Asian | Gallbladder cancer | HB | PCR | 91/155 | 34 | 45 | 12 | 113 | 69 | 88 | 57 | 10 | 233 | 77 | 0.850 |
| Zhai | 2012 | China | Asian | Hepatocellular carcinoma | HB | PCR | 102/135 | 37 | 52 | 13 | 126 | 78 | — | — | — | 205 | 65 | — |
| Zhang | 2014 | China | Asian | Malignant melanoma | HB | PCR | 220/617 | 101 | 102 | 17 | 304 | 136 | 336 | 246 | 35 | 918 | 316 | 0.248 |
Meta‐analysis was carried out using Revman 5.3 software (Copenhagen: The Cochrane Collaboration, 2014, The Nordic Cochrane Centre) and stata 14.1 software (Stata Corporation, College Station, TX, USA). For each study, Hardy‐Weinberg equilibrium (HWE) was determined by the chi‐squared test, in order to verify the representativeness of the study population.
The association between LAPTM4B polymorphism in relation to cancer risk was evaluated by pooled odds ratios (ORs) and their 95% confidence intervals (CIs). Pooled ORs and their 95% CIs for codominant, dominant, recessive, overdominant and the allelic comparison genetic inheritance models were calculated. The significance of the pooled OR was assessed by the Z test, and P < 0.05 was considered statistically significant. The choice of using fixed or random effects model was determined by the results of the between‐study heterogeneity test, which was measured using the Q test and I 2 statistic. If the test result was I 2 ≥ 50% or PQ < 0.1, indicating the presence of heterogeneity, the random effect model was selected; otherwise, the fixed‐effects model was chosen.
The funnel plot was used to estimate the publication bias. The degree of asymmetry was measured using Egger's test; P < 0.05 was considered significant publication bias. To measure the potential influence of each study on the overall effect size, sensitivity analysis was performed.
3. RESULTS
The characteristics and relevant data of the included studies are shown in Table 1. The results of the meta‐analysis revealed a significant association between LAPTM4B polymorphism and cancer susceptibility cancer in codominant (OR = 1.42, 95% CI = 1.27‐1.59, P < 0.00001, *1/2 vs *1/1; OR = 2.01, 95% CI = 1.69‐2.39, P < 0.00001, *2/2 vs *1/1), dominant (OR = 1.50, 95% CI = 1.34‐1.69, P < 0.00001, *1/2 + *2/2 vs *1/1), recessive (OR = 1.73, 95% CI = 1.53‐1.95, P < 0.00001, *2/2 vs *1/1 + *1/2), overdominant (OR = 1.28, 95% CI = 1.17‐1.41, P < 0.00001, *1/2 vs *1/1 + *2/2), and allele (OR = 1.40, 95% CI = 1.28‐1.53, P < 0.00001, *2 vs *1) inheritance model tested (Figure 1).
Figure 1.
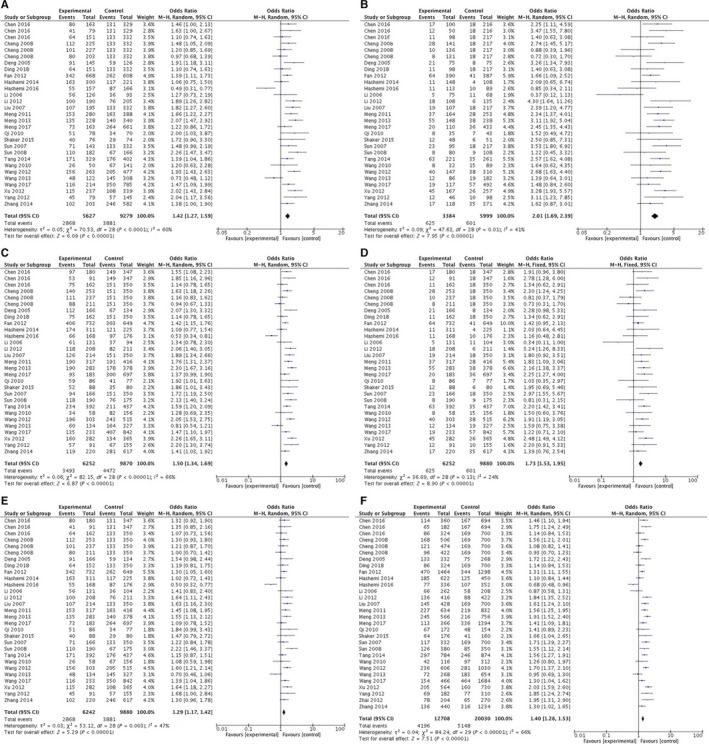
The pooled ORs and 95% CIs for the association between LAPTM4B polymorphism and cancer susceptibility. The forest plot for relationship between LAPTM4B polymorphism and cancer susceptibility for *2/2 vs *1/1 (A), *2/2 vs *1/1 (B), *1/2 + *2/2 vs *1/1 (C), *2/2 vs *1/2 + *1/1 (D), *1/2 vs *1/1 + *2/2 (E), and *2 vs *1 (F)
Stratifying according to cancer types proposed that LAPTM4B polymorphism significantly increased the risk of breast cancer, gastrointestinal cancer, gynaecological cancer, liver cancer, lung cancer, and lymphoma (data not shown).
The potential publication bias was evaluated using a Begg's funnel plot and Egger's test and the analysis suggested no publication bias for this meta‐analysis of the heterozygous codominant, dominant, recessive, overdominanat, and allele model (all P‐values for bias >0.05). We executed sensitivity analysis by neglecting a single study each time to reflect the influence of the individual data set to the pooled OR. The results indicated that the significance of pooled ORs for LAPTM4B polymorphism was not extremely influenced, suggesting the stability and reliability of the results in this meta‐analysis.
4. DISCUSSION
In the current study, we performed a meta‐analysis to find out the exact role of LAPTM4B polymorphism on risk of cancer. The results revealed that LAPTM4B polymorphism significantly increased the risk of cancer in codominant, dominant, overdominant, and allele genetic inheritance models. Stratification by cancer types suggested that LAPTM4B polymorphism is associated with the risk of breast cancer, gynaecological cancer, gastrointestinal cancer, liver cancer, lung cancer, and lymphoma. LAPTM4B is a proto‐oncogene that is overexpressed in various types of cancers. It has been proposed that overexpression of LAPTM4B‐35 promote proliferation, invasion, and migration. Its up‐regulation might be caused by gene amplification as well as transcriptional up‐regulation. LAPTM4B alleles have the same sequence except for one 19‐bp fragment for LAPTM4B *1 and two tight tandem fragments for LAPTM4B *2 in the 5′UTR of exon 1.23 The 19‐bp alteration in 5′UTR of the first exon of the LAPTM4B gene can shift the open reading frame (ORF), resulting in two alternate protein isoforms, LAPTM4B‐35 and LAPTM4B‐40. In conclusion, the finding of this meta‐analysis illustrated that LAPTM4B polymorphism may affect the risk of development of cancers.
CONFLICT OF INTEREST
The authors declare no competing of interests.
ACKNOWLEDGEMENTS
SG was supported by the operating grant from CHRIM, general operating grant from Health Science Foundation, and Research Manitoba New Investigator operating grant. MJŁ acknowledges the support from NCN grant #: 2016/21/B/NZ1/02812, by LE STUDIUM Institute for Advanced Studies (region Centre‐Val de Loire, France) through its Smart Loire Valley General Program, cofunded by the Marie Skłodowska‐Curie Actions, grant #: 665790.
Hashemi M, Bahari G, Tabasi F, et al. LAPTM4B gene polymorphism augments the risk of cancer: Evidence from an updated meta‐analysis. J Cell Mol Med. 2018;22:6396–6400. 10.1111/jcmm.13896
Contributor Information
Mohammad Hashemi, Email: mhd.hashemi@gmail.com.
Marek J. Łos, Email: mjelos@gmail.com.
REFERENCES
- 1. Hashemi M, Amininia S, Ebrahimi M, et al. Association between LAPTM4B gene polymorphism and breast cancer susceptibility in an Iranian population. Med Oncol. 2014;31:111. [DOI] [PubMed] [Google Scholar]
- 2. Xu Y, Liu Y, Zhou R, et al. LAPTM4B polymorphisms is associated with ovarian cancer susceptibility and its prognosis. Jpn J Clin Oncol. 2012;42:413‐419. [DOI] [PubMed] [Google Scholar]
- 3. Fan M, Liu Y, Zhou R, Zhang Q. Association of LAPTM4B gene polymorphism with breast cancer susceptibility. Cancer Epidemiol. 2012;36:364‐368. [DOI] [PubMed] [Google Scholar]
- 4. Li X, Kong X, Chen X, et al. LAPTM4B allele *2 is associated with breast cancer susceptibility and prognosis. PLoS ONE. 2012;7:e44916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang H, Zhai G, Ji X, Xiong F, Su J, McNutt MA. Correlation of LAPTM4B polymorphisms with gallbladder carcinoma susceptibility in Chinese patients. Med Oncol. 2012;29:2809‐2813. [DOI] [PubMed] [Google Scholar]
- 6. Liu Y, Zhang QY, Qian N, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of gastric cancer. Ann Oncol. 2007;18:311‐316. [DOI] [PubMed] [Google Scholar]
- 7. Cheng XJ, Xu W, Zhang QY, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of colorectal and esophageal cancers. Ann Oncol. 2008;19:527‐532. [DOI] [PubMed] [Google Scholar]
- 8. Wang B, Xu J, Zhou R, Zhang Q. Association of LAPTM4B gene polymorphism with nasopharyngeal carcinoma susceptibility in a Chinese population. Med Oncol. 2013;30:470. [DOI] [PubMed] [Google Scholar]
- 9. Li C, Zhou Q, Wang Y, Chen X, Yang X. Zhu D [Relationship between LAPTM4B gene polymorphism and susceptibility of lung cancer]. Zhongguo Fei Ai Za Zhi. 2006;9:109‐112. [DOI] [PubMed] [Google Scholar]
- 10. Ding H, Cheng X, Ding N, et al. Association between LAPTM4B gene polymorphism and susceptibility to and prognosis of diffuse large B‐cell lymphoma. Oncology Letters. 2018;15:264‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen D, Chang Y, Xu J, Zhang Q. Relationship between LAPTM4B gene polymorphism and susceptibility of renal cell carcinoma and bladder cancer. Int J Clin Exp Med. 2016;9:14507‐14514. [Google Scholar]
- 12. Chen D, Chang Y, Xu J, Zhang Q. Association between LAPTM4B gene polymorphism and susceptibility to and prognosis of diffuse large B‐cell lymphoma. Int J Clin Exp Med. 2016;9:14507‐14514. [Google Scholar]
- 13. Deng LJ, Zhang QY, Liu B, Zhou RL. [Relationship between LAPTM4B gene polymorphism and susceptibility of lung cancer]. Beijing Da Xue Xue Bao Yi Xue Ban. 2005;37:302‐305. [PubMed] [Google Scholar]
- 14. Meng F, Li H, Zhou R, Luo C, Hu Y, Lou G. LAPTM4B gene polymorphism and endometrial carcinoma risk and prognosis. Biomarkers. 2013;18:136‐143. [DOI] [PubMed] [Google Scholar]
- 15. Meng F, Song H, Luo C, et al. Correlation of LAPTM4B polymorphisms with cervical carcinoma. Cancer. 2011;117:2652‐2658. [DOI] [PubMed] [Google Scholar]
- 16. Meng Y, Zhou R, Xu J, Zhang Q. LAPTM4B*2 allele is associated with the development of papillary thyroid carcinoma in Chinese women. Oncol Lett. 2017;14:3421‐3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang H, Tian H, Yue W, et al. LAPTM4B polymorphism is associated with nonsmall cell lung cancer susceptibility and prognosis. Oncol Rep. 2014;31:2454‐2460. [DOI] [PubMed] [Google Scholar]
- 18. Wang B, Wang S, Liang G, Xu J, Zhou R, Zhang Q. Association of lysosomal protein transmembrane 4 beta gene polymorphism with pancreatic carcinoma susceptibility in the Chinese population. Tumour Biol. 2017;39:1010428317705518. [DOI] [PubMed] [Google Scholar]
- 19. Wang S, Zhang QY, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of primary liver cancer. Ann Oncol. 2012;23:1864‐1869. [DOI] [PubMed] [Google Scholar]
- 20. Xia LZ, Yin ZH, Ren YW, et al. The relationship between LAPTM4B polymorphisms and cancer risk in Chinese Han population: a meta‐analysis. Springerplus. 2015;4:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhai G, Yang H, Ji X, et al. Correlation of LAPTM4B polymorphisms with hepatocellular carcinoma in Chinese patients. Med Oncol. 2012;29:2744‐2749. [DOI] [PubMed] [Google Scholar]
- 22. Zhang M, Zhou R, Xu J, Zhang Q. Relationship between LAPTM4B Gene polymorphism and susceptibility of malignant melanoma in Chinese patients. Transl Oncol. 2014;7:638‐643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hashemi M, Rezaei M, Narouie B, et al. Association between LAPTM4B gene polymorphism and prostate cancer susceptibility in an Iranian population. Mol Cell Oncol. 2016;3:e1169342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun G, Li Z, Hao W, Niu J, Yin J, Yan Y. Relationship between lysosome‐associated protein transmembrane 4 beta polymorphism and susceptibility to liver cancer. World Chin J Digestology. 2008;16:908‐911. [Google Scholar]
- 25. Sun L, Zhang Q, Liu Y, Qian N. Relationship between Human Novel Gene LAPTM4B gene polymorphism and susceptibility of Lymphoma. Cancer Res Prev Treat. 2007;34:245‐248. [Google Scholar]
- 26. Qi R, Shi X. LAPTM4B gene polymorphism and liver cancer susceptibility. (postgraduate's thesis). 2010.


