Abstract
PURPOSE:
Limited sampling strategy (LSS) is a validated method to estimate pharmacokinetic (PK) parameters from a reduced number of samples. Omeprazole is used to phenotype in vivo cytochrome P450 (CYP) 2C19 activity. This study examined a LSS using two estimation methods to determine apparent oral clearance (CL/F) and thus CYP2C19 activity.
METHODS:
Data from 7 previously published studies included healthy subjects receiving a single, oral dose of omeprazole with intensive PK sampling. CL/F was estimated using non-compartmental analysis (NCA) and population PK modeling. LSS was simulated by selecting the 1, 2, 4, and/or 6-hour post dose time points. Linear regression was performed to assess whether CL/F estimated from limited sampling could accurately predict CL/F from the full PK profile.
RESULTS:
Median CL/F was 23.7 L/h by NCA and 19.3 L/h by population PK modeling. In comparing the LSS NCA estimated versus observed CL/F, all evaluated linear regression models had unacceptable coefficients of determination (r2, range: 0.14 – 0.81). With the population PK approach, 737 plasma concentrations (n=71) and CYP2C19 genotype data were described with a one-compartment structural model with mixed zero and first order absorption and lag time. In comparing the population PK LSS estimated versus observed CL/F, all evaluated linear regression models had unacceptable r2 (range: 0.02 – 0.74). Post-hoc comparison of CYP2C19 poor metabolizers (PMs) versus CYP2C19 extensive metabolizers (EMs) resulted in significantly lower CL/F in PMs versus EMs.
CONCLUSIONS:
Omeprazole LSS performed poorly in estimating CL/F utilizing two separate estimation approaches and does not appear to be a suitable method for determining CYP2C19 activity.
Keywords: cytochrome P450, omeprazole, limited sampling strategy, CYP2C19
Introduction
Phenotyping is the preferred method to assess the clinical significance of a drug-drug interaction and to quantify real-time, in vivo cytochrome P450 (CYP) activity. Omeprazole is used as a probe drug to phenotype CYP2C19 and CYP3A activity1. Omeprazole apparent oral clearance (CL/F), metabolic ratios of omeprazole to 5-hydroxyomeprazole plasma concentrations, or area under the concentration time curve (AUC) ratios are phenotyping parameters used to evaluate CYP2C19 activity 2. Intensive sampling of omeprazole and metabolite concentrations is performed to measure these phenotyping parameters and to calculate PK parameters by non-compartmental analysis (NCA). Intensive sampling can be cumbersome, as 10 to 16 blood samples are collected up to 24-hours post dose3.
Limited sampling strategy (LSS) is a validated method to estimate PK parameters from a reduced number of blood samples. Omeprazole LSS utilizing plasma concentrations and AUC ratios have been previously published4,5. However, study design limitations (i.e., small sample sizes) existed for previously reported studies (n=18 and 37) and complicate the ability to recommend such LSS models for general use. Beyond the NCA LSS approach, a LSS with a population PK approach has been used for a phenotyping cocktail study and a phenotyping study to quantify constitutive CYP3A activity6,7. With data from seven previously published studies in healthy adult subjects, the first objective was to develop novel omeprazole limited sampling models to estimate CL/F utilizing a NCA LSS approach. The second objective was to determine the feasibility of an omeprazole LSS utilizing a population PK approach for parameter estimations. Based on the known CYP2C19 gene-dose effect 8, omeprazole CL/F estimates were examined to determine if such estimates readily discriminate between CYP2C19 extensive metabolizers (EMs) and CYP2C19 poor metabolizers (PMs).
Materials and Methods
STUDY SUBJECTS AND SAMPLING
Institutional Review Board exemption was obtained for this study. Omeprazole plasma concentration data were obtained from seven studies in healthy adult subjects who received a single, oral 20 or 40 mg dose with intensive PK sampling 0 to 24 hours post dose (Table 1, Supplemental Digital Content) 3,8–13. Omeprazole and metabolite plasma concentration detection and genotyping methods for CYP2C19*1, *2, *3, and *17 alleles are described elsewhere 3,8–13.
NON-COMPARTMENTAL ANALYSIS FOR LIMITED SAMPLING STRATEGY
Omeprazole CL/F was calculated using NCA with Phoenix® WinNonlin 7.0 (Cetara USA, Inc., Princeton, NJ, USA). Full concentration time profiles for all subjects were used to characterize omeprazole AUCinf and CL/F (Figure 1, Supplemental Digital Content). Traditional LSS entailed randomizing subject data into training (n=56) and validation (n=114) sets. The selected time points were 0, 1, 2, 4, and/or 6-hours post dose since these were observed across all studies (Table 1, Supplemental Digital Content) and were evaluated in a previous study 5. Linear regression equations to estimate CL/F as a function of partial AUCs 14,15 were derived from the training set. The estimated, log-transformed CL/F from LSS were compared with the observed CL/F calculated from the full concentration and time profiles using R version 3.2.2 (R Foundation, Vienna, Austria). Preset criterion for model selection was a coefficient of determination (r2) ≥ 0.9 16. The resulting equations were used to calculate individual CL/F estimates from the validation set. If the model equations fulfilled r2 ≥ 0.9, bias and precision were then determined by relative percent mean prediction error (%MPE,−5% to 5%) and relative percent mean absolute error (%MAE, ≤10%) 16.
LIMITED SAMPLING STRATEGY WITH A POPULATION PK APPROACH
The full concentration time data of four studies 3,8,10,12 (Table 1, Supplemental Digital Content) with available CYP2C19 genotype data (n=71) were modeled using NONMEM (version 7.3.0, GloboMax, Hanover, MD, USA) with a first-order conditional estimation method with interaction. An exponential error model was applied to describe inter-subject variability, and a mixed additive and proportional error model applied to describe the residual variability. Individual subject random effects and PK parameter estimates were generated by post hoc Bayesian estimation. Potential covariates were added univariately to the model on CL/F and V as a linear function. Covariates that were significant (change in the objective function of > 3.84, p<~0.05) and improved the model fit were retained. Covariates were retained in the final model after multivariate screening with forward selection (change in objective function by > 7.88, p<~0.005). To test whether estimated CL/F from limited time points could be predictive of CYP2C19 activity, a final model that did not evaluate CYP2C19 activity as a potential covariate was selected, in order to simulate lack of prior knowledge of CYP2C19 activity. The bootstrap procedure (n=1000) was performed using WINGS for NONMEM (UC San Francisco, San Francisco, CA, USA). Limited sampling datasets were derived using PK time points at 0, 1, 2, 4, and/or 6-hours. Estimated CL/F were then compared post hoc by linear regression to the CL/F estimated from the full profile. The same preset criterion of r2 ≥ 0.9 and acceptable limits of %MPE and %MAE were used.
EVALUATION OF OMEPRAZOLE GENE DOSE EFFECT
A gene-dose effect in omeprazole PK has been previously reported 8. To confirm that a gene-dose effect was observed in the current analyses, omeprazole CL/F was evaluated by the Wilcoxon Rank Sum test between CYP2C19 PMs versus homozygous and heterozygous CYP2C19 EMs.
Results
LIMITED SAMPLING STRATEGY WITH NON-COMPARTMENTAL ANALYSES
Data from 170 subjects with 2272 omeprazole plasma concentrations was included (Figure 1, Supplemental Digital Content). Using the full concentration time profile, median observed CL/F was 23.7 L/h and V was 35.6L (Table 2, Supplemental Digital Content). Utilizing the training set (n=56), eight linear regression models were developed and all models had unacceptable r2 (range: 0.14 – 0.81, Table 1). A representative plot of the best model with an r2 of 0.81 is illustrated in Figure 1a.
Table 1.
Linear regression model equations and r2 values for predicting omeprazole clearance from limited sampling strategies
| Model | PK post dose times (h) |
Limited Sampling linear regression equation log(CL) = |
r2 | Estimated CL/F (L/h) from limited sampling by CYP2C19 activity† Median CL/F (2.5th and 97.5th percentiles) |
|||
|---|---|---|---|---|---|---|---|
| All (n=71) |
CYP2C19 EMs (n=62) |
CYP2C19 PMs (n=9) |
p-value | ||||
| 1-NCA | 0,1 | 0.84 + 0.22 × log(CL0–1) | 0.14 | 350.2 (43.5–9455.0) | 348.7 (50.8–9596.9) | 414.9 (43.4–4047.3) | 0.872 |
| 2-NCA | 0,1,2 | 0.64 + 0.40 × log(CL0–2) | 0.29 | 70.7 (16.9–1326.4) | 86.9 (18.8–1661.5) | 32.0 (14.3–40.6) | 5.10E-4 |
| 3-NCA | 0,1,2,4 | −0.028 + 0.93 × log(CL0–4) | 0.74 | 33.9 (7.0–301.9) | 38.2 (7.3–316.9) | 8.3 (6.7–13.4) | 1.98E-5 |
| 4-NCA | 0,1,2,4,6 | 0.023 + 0.94 × log(CL0–6) | 0.81 | 29.8 (5.2–141.6) | 32.4 (5.4–155.7) | 5.8 (4.6–10.6) | 1.86E-5 |
| 5-NCA | 1,2 | 0.44 + 0.47 × log(CL1–2) | 0.35 | 93.2 (22.9–1746.0) | 109.6 (24.8–2200.9) | 32.0 (20.8–42.8) | 2.85E-5 |
| 6-NCA | 1,2,4 | −0.14 + 0.95 × log(CL1–4) | 0.78 | 43.1 (7.3–344.7) | 51.9 (8.0–391.7) | 8.6 (6.9–14.0) | 1.22E-5 |
| 7-NCA | 2,4 | −0.11 + 0.81 × log(CL2–4) | 0.73 | 83.8 (9.9–597.9) | 88.6 (12.1–719.6) | 11.1 (9.3–22.0) | 1.56E-5 |
| 8-NCA | 2,4,6 | 0.11 + 0.73 × log(CL2–6) | 0.73 | 56.6 (6.5–291.6) | 60.0 (7.6–296.4) | 7.1 (5.6–16.1) | 1.59E-5 |
| 9-Pop PK | 1 | 0.42 + 0.59 × log(CL1) | 0.02 | 27.1 (17.6–30.3) | 27.1 (17.2–29.9) | 27.2 (19.3–30.3) | 0.809 |
|
10-Pop PK |
1,2 | −0.11+ 0.97 × log(CL1,2) | 0.43 | 29.4 (8.1–44.6) | 30.2 (9.2–44.7) | 12.1 (8.7–18.6) | 5.65E-5 |
|
11-Pop PK |
1,2,4 | 0.057 + 0.91 × log(CL1,2,4) | 0.74 | 23.1 (6.7–58.1) | 23.8 (6.8–59.9) | 7.4 (6.0–15.1) | 4.53E-5 |
|
12-Pop PK |
1,2,4,6 | 0.082 + 0.97 × log(CL1,2,4,6) | 0.74 | 17.5 (5.5–39.9) | 19.7 (6.3–40.5) | 6.4 (4.3–10.4) | 4.36E-5 |
|
13-Pop PK |
2,4 | −0.036 + 0.99 × log(CL2,4) | 0.73 | 21.2 (6.5–53.5) | 22.4 (6.8–53.8) | 7.3 (5.9–15.6) | 4.20E-5 |
|
14-Pop PK |
4,6 | 0.1 + 0.97 × log(CL4,6) | 0.69 | 16.4 (5.1–40.5) | 19.3 (5.9–40.6) | 6.9 (4.9–11.4) | 1.01E-4 |
|
15-Pop PK |
2,4,6 | 0.1 + 0.96 × log(CL2,4,6) | 0.70 | 16.5 (5.2–39.0) | 19.2 (5.9–39.4) | 6.4 (4.3–11.1) | 6.79E-5 |
NCA values were derived using the training set (n=56). Population PK utilized all subjects with CYP2C19 genotype data (n=71). Median (2.5th and 97.5th percentile) estimated CL/F from limited sampling reported for all subjects with CYP2C19 genotype data. P-value by Wilcoxon Rank Sum test comparing CYP2C19 PMs to CYP2C19 EMs.
Abbreviations: CL/F=oral clearance; EMs=extensive metabolizers; PMs=poor metabolizers; NCA=non-compartmental analysis; Pop PK=population pharmacokinetics.
Figure 1.
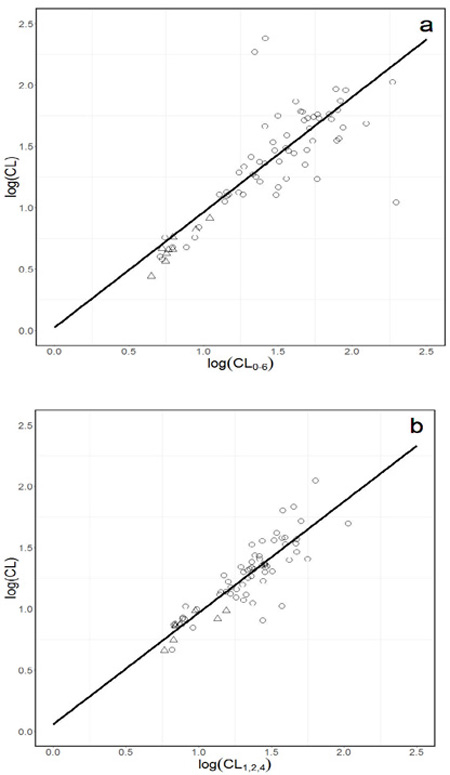
Representative full concentration-time log(CL) versus limited sampling log(CL) plots for a. NCA model of log(CL) = 0.023 + 0.94 × log(CL0–6) and b. Population PK model of log(CL) = 0.057 + 0.91 × log(CL1,2,4). Open circles represent 1 or 2 active CYP2C19 alleles and open triangles represent 0 active CYP2C19 alleles.
LIMITED SAMPLING STRATEGY WITH A POPULATION PK APPROACH
Omeprazole plasma concentrations (n=737) from 71 subjects with CYP2C19 genotype data were described with a one-compartment structural model with mixed zero and first order absorption and lag time. The median observed CL/F was 19.3 L/h and V was 23.7 L (Table 2, Supplemental Digital Content). Female sex and age were independent predictors of V. The final model described the data without significant bias (Figure 2, Supplemental Digital Content). Seven linear regression models were developed and all had unacceptable r2 (range: 0.02 – 0.74, Table 1). A representative plot of the best model, with r2 of 0.74, is illustrated in Figure 1b.
OMEPRAZOLE CLEARANCE AND CYP2C19 ACTIVITY
There were 62 CYP2C19 EMs and 9 CYP2C19 PMs. In 13 of 15 linear regression models, the LSS estimated CL/F in CYP2C19 PMs was significantly lower than CYP2C19 EMs (Table 1). The two regression models that failed to predict lower CL/F for CYP2C19 PMs used a single, 1-hour time point. Using a later single or a combination of later time point(s) allowed for better estimation of CL/F. Ad hoc evaluation of the CYP2C19 phenotype as a covariate of CL/F in population PK modeling estimated 48.7% lower CL/F in PMs than EMs. CYP2C19 activity covariate on CL/F resulted in a significant objective function value drop of 26.0 (p<0.05) from the base model.
Discussion
This study was unable to develop an acceptable omeprazole LSS model to estimate CL/F with NCA. These results add to the conflicting literature that support 5,17,18 and refute 19 the suitability of an omeprazole LSS approach. Differences in sample size, selection of omeprazole phenotyping parameter, and method of model validation exist among studies that may explain these conflicting findings. The selection of CL/F as a function of a partial AUC was done since omeprazole AUC currently lacks adequate validation, and using the AUC ratio of omeprazole to 5-hydroxyomeprazole has limited generalizability. Niioka reported three limited sampling models of plasma concentrations to estimate omeprazole AUC 5. In a separate analysis of external data sets but using the same limited sampling models 5, each model had bias and lacked precision 19. In another study, a three-time point limited sampling model accurately estimated the AUC ratio of omeprazole to 5-hydroxyomeprazole 4, but was only applicable to Caucasian CYP2C19*1/*1 subjects. To the best of our knowledge, this is the first LSS study to utilize an omeprazole CL/F as a function of a partial AUC to estimate CYP2C19 activity.
Limited sampling model selection and model validation approaches differed between the current study and previous studies. In the current study, model selection was determined by an r2 ≥ 0.916. In other studies, model selection and model validation were primarily determined from the highest r2 or correlation coefficient (r) 5,17. Selecting a limited sampling model based on the highest r2 or r may not be correct 4 since neither the r2 norr r are measures of predictive performance 20.
Although the structural population PK model was similar to another published model 21, the current model was unable to accurately characterize the absorption phase. Several structural models were tested and a lag time was incorporated in the one-compartment first-order absorption and elimination model. Due to the variability in individual observed Tmax, predicted peak concentrations did not match observed peak concentrations in the final model. None of the limited sampling models developed with the population PK approach met the preset r2 criterion. This was unexpected given that the same approach was successful in a study that used midazolam limited sampling to accurately quantify constitutive CYP3A activity 22. With the high variability in omeprazole absorption, the wide observed range of Tmax overlapped with the selected limited sampling time points. These early time points may not have accurately represented the elimination phase, and thus omeprazole CL/F estimates from the LSS population PK approach may be unreliable.
By NCA and population PK modeling, omeprazole CL/F differed significantly between CYP2C19 PMs and CYP2C19 EMs. The current study observed a 2.8–6.4 fold lower CL/F in CYP2C19 PMs versus EMs; a magnitude of the gene-dose effect consistent with previously published studies 8,23. Only nine subjects were CYP2C19 PMs, with only the variant CYP2C19*2 and *3 alleles tested. The contribution of additional CYP2C19 variant alleles on omeprazole PK is unknown, but is likely not clinically meaningful due to a low population frequency of <1% 24.
Conclusions
Omeprazole is a probe drug used for determining CYP2C19 activity. NCA or population PK LSS for omeprazole do not appear to be predictive of CL/F estimated from full concentration-time profiles.
Supplementary Material
Acknowledgements
Presented in part at the 46th Annual Meeting of the American College of Clinical Pharmacology, San Diego, CA, USA, September 2017. Dr. Lin was a post-doctoral fellow with funding supported by Pfizer Global Research and Development and the UCSD, Skaggs School of Pharmacy and Pharmaceutical Sciences, in La Jolla, CA.
Footnotes
Conflicts of Interest and Source of Funding
All authors declare no conflict of interest. Funding support was provided by a Research in Pediatric and Developmental Pharmacology NIH grant (1U54HD090259-01 to E.V.C.)
References
- 1.Gonzalez HM, Romero EM, Chavez Tde J, et al. Phenotype of CYP2C19 and CYP3A4 by determination of omeprazole and its two main metabolites in plasma using liquid chromatography with liquid-liquid extraction. J Chromatogr B Analyt Technol Biomed Life Sci 2002;780(2):459–465. [DOI] [PubMed] [Google Scholar]
- 2.Streetman DS, Bertino JS, Jr., Nafziger AN. Phenotyping of drug-metabolizing enzymes in adults: a review of in-vivo cytochrome P450 phenotyping probes. Pharmacogenetics 2000;10(3):187–216. [DOI] [PubMed] [Google Scholar]
- 3.Cho JY, Yu KS, Jang IJ, et al. Omeprazole hydroxylation is inhibited by a single dose of moclobemide in homozygotic EM genotype for CYP2C19. Br J Clin Pharmacol 2002;53(4):393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawson EB, Wu JC, Baldwin RM, et al. Omeprazole limited sampling strategies to predict area under the concentration-time curve ratios: implications for cytochrome P450 2C19 and 3A phenotyping. Eur J Clin Pharmacol 2012;68(4):407–413. [DOI] [PubMed] [Google Scholar]
- 5.Niioka T Clinical usefulness of limited sampling strategies for estimating AUC of proton pump inhibitors. Yakugaku Zasshi 2011;131(3):407–413. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen TT, Benech H, Pruvost A, et al. A limited sampling strategy based on maximum a posteriori Bayesian estimation for a five-probe phenotyping cocktail. Eur J Clin Pharmacol 2016;72(1):39–51. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Patel M, Nikanjam M, et al. Midazolam single time point concentrations to estimate exposure and cytochrome P450 (CYP) 3A constitutive activity utilizing limited sampling strategy with a population pharmacokinetic approach. J Clin Pharmacol In press. [DOI] [PubMed]
- 8.Yin OQ, Tomlinson B, Chow AH, et al. Omeprazole as a CYP2C19 marker in Chinese subjects: assessment of its gene-dose effect and intrasubject variability. J Clin Pharmacol 2004;44(6):582–589. [DOI] [PubMed] [Google Scholar]
- 9.Allegrini A, Nuzzo L, Scaringi AT, et al. Bioequivalence study of two capsule formulations of omeprazole in healthy volunteers. Arzneimittelforschung 2008;58(8):385–388. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin RM, Ohlsson S, Pedersen RS, et al. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol 2008;65(5):767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussein RF, Lockyer M, Hammami MM. Bioequivalence assessment of two capsule formulations of omeprazole in healthy volunteers. Arzneimittelforschung 2007;57(2):101–105. [DOI] [PubMed] [Google Scholar]
- 12.Johnson MD, Hamilton CD, Drew RH, et al. A randomized comparative study to determine the effect of omeprazole on the peak serum concentration of itraconazole oral solution. J Antimicrob Chemother 2003;51(2):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhim SY, Park JH, Park YS, et al. Pharmacokinetics and bioequivalence of 20 mg omeprazole capsule in 24 healthy Korean male volunteers. Int J Clin Pharmacol Ther 2009;47(1):23–29. [DOI] [PubMed] [Google Scholar]
- 14.Masters JC, Harano DM, Greenberg HE, et al. Limited sampling strategy of partial area under the concentration-time curves to estimate midazolam systemic clearance for cytochrome P450 3A phenotyping. Ther Drug Monit 2015;37(1):84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai W, Gong SL, Tsunoda SM, et al. Evaluation of partial area under the concentration time curve to estimate midazolam apparent oral clearance for cytochrome P450 3A phenotyping. Drug Metabol Drug Interact 2013;28(4):217–223. [DOI] [PubMed] [Google Scholar]
- 16.Al-Khatib M, Shapiro RJ, Partovi N, et al. Limited sampling strategies for predicting area under the concentration-time curve of mycophenolic acid in islet transplant recipients. Ann Pharmacother 2010;44(1):19–27. [DOI] [PubMed] [Google Scholar]
- 17.Bottiger Y Use of omeprazole sulfone in a single plasma sample as a probe for CYP3A4. Eur J Clin Pharmacol 2006;62(8):621–625. [DOI] [PubMed] [Google Scholar]
- 18.Niioka T, Uno T, Sugimoto K, et al. Estimation of CYP2C19 activity by the omeprazole hydroxylation index at a single point in time after intravenous and oral administration. Eur J Clin Pharmacol 2007;63(11):1031–1038. [DOI] [PubMed] [Google Scholar]
- 19.Lam LH, Allegrini A, Hammami MM, et al. Evaluation of omeprazole limited sampling strategy to estimate cytochrome P450 (CYP) 2C19 activity in healthy adults. Clinical Pharmacology and Drug Development 2015;4(S1):26. [Google Scholar]
- 20.Sheiner LB, Beal SL. Some suggestions for measuring predictive performance. J Pharmacokinet Biopharm 1981;9:503–512. [DOI] [PubMed] [Google Scholar]
- 21.Marier JF, Dubuc MC, Drouin E, Alvarez F, Ducharme MP, Brazier JL. Pharmacokinetics of omeprazole in healthy adults and in children with gastroesophageal reflux disease. Ther Drug Monit 2004;26(1):3–8. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Patel M, Nikanjam M, et al. Midazolam single timepoint concentrations to estimate exposure and cytochrome P450 (CYP) 3A constitutive activity utilizing limited sampling strategy with a population pharmacokinetic approach. Journal of Clinical Pharmacology In press 2018. [DOI] [PubMed]
- 23.Sakai T, Aoyama N, Kita T, et al. CYP2C19 genotype and pharmacokinetics of three proton pump inhibitors in healthy subjects. Pharm Res 2001;18(6):721–727. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 2017;102(4):688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


