Abstract
Cardiac allograft vasculopathy (CAV) accounts for about 30% of all heart-transplant (HTx) patient deaths. For patients at high risk for CAV complications after HTx, therapy must be initiated early to be effective. Therefore, new phenotyping approaches are needed to identify such HTx patients at the earliest possible time. Coronary optical coherence tomography (OCT) images were acquired from 50 HTx patients 1 and 12 months after HTx. Quantitative analysis of coronary wall morphology used LOGISMOS segmentation strategy to simultaneously identify three wall-layer surfaces for the entire pullback length in 3D: luminal, outer intimal, and outer medial surfaces. To quantify changes of coronary wall morphology between 1 and 12 months after HTx, the two pullbacks were mutually co-registered. Validation of layer thickness measurements showed high accuracy of performed layer analyses with layer thickness measures correlating well with manually-defined independent standard (=0.93, y=1.0x − 6.2μm), average intimal+medial thickness errors were 4.98±31.24 μm, comparable with inter-observer variability.
Quantitative indices of coronary wall morphology 1 month and 12 months after HTx showed significant local as well as regional changes associated with CAV progression. Some of the newly available fully-3D baseline indices (intimal layer brightness, medial layer brightness, medial thickness, intimal+medial thickness) were associated with CAV-related progression of intimal thickness showing promise of identifying patients subjected to rapid intimal thickening at 12 months after HTx from OCT-image data obtained just 1 month after HTx. Our approach allows quantification of location-specific alterations of coronary wall morphology over time and is sensitive even to very small changes of wall layer thicknesses that occur in patients following heart transplant.
Keywords: Cardiac allograft vasculopathy (CAV), optical coherence tomography (OCT), LOGISMOS, CAV progression, CAV prediction
1. Introduction
Cardiac allograft vasculopathy (CAV) represents the leading cause of late morbidity and mortality in heart transplant (HTx) recipients (Chih et al., 2016; Wever-Pinzon et al., 2014). Overall, CAV accounts for about 30% of all HTx patient deaths. For patients at high risk for CAV complications after HTx, therapy must be initiated early to be effective. Once CAV causes allograft dysfunction, the only long-term therapeutic solution is a re-transplantation. Therefore, development of a methodology for quantitative detection of early CAV progression, sufficiently sensitive to initially small changes of the intimal and medial layers and offering high measurement accuracy is of utmost importance. We report a highly automated approach to quantitative analysis of coronary optical coherence tomography (OCT) images in HTx patients, yielding accurate quantitative analysis of coronary wall layer properties.
Historically, a number of successful coronary imaging approaches reached full clinical-care acceptance, starting with selective X-ray coronary angiography that allowed visualization of coronary vessel lumen and coronary stenoses in clinical setting (Sones and Shirey, 1962). While coronary angiography can only image the coronary lumen, intravascular ultrasound (IVUS) imaging brought 3-D wall imaging to routine clinical care in the late 1980’s (Bom and Lancee, 1972). While clinical use is almost entirely limited to simultaneous visualization of the axial pullback at a selected angle together with displaying cross-sectional 2D wall locations, geometrically correct representation of IVUS data can be obtained by data fusion of 2-plane coronary angiography and IVUS (Wahle et al., 1999b,a; Slager et al., 2000; Wahle et al., 2006; Chandran et al., 2006; Stone et al., 2007; Zhang et al., 2015). Intracoronary OCT is the most recent tomographic imaging modality (Huang et al., 1991) that is transforming routine clinical care for patients with cardiovascular diseases. Intracoronary OCT is in many ways similar to IVUS while offering much better resolution (but much lower penetration depth). As a result, vascular wall layers close to the lumen can be visualized in never-before available detail, allowing, e.g., quantification of intimal or medial layer thicknesses.
Several methods have recently been reported to analyze coronary OCT images. Lumen and intima-media border were segmented using edge detection and dynamic programming with an ellipse shape fit in (Olender et al., 2017). Its performance was affected by segmentation sensitivity to large intensity variations of similar tissues in OCT. In (Zahnd et al., 2017), intima, media and adventitia layers were identified and assessed in healthy coronary wall regions by utilizing gradient-based dynamic programming, with healthy and diseased regions automatically distinguished by adaptive boosting with feature selection. Both of these methods targeted OCT image analysis in atherosclerotic cardiovascular disease, which exhibits substantially different characteristics of the wall appearance in OCT images compared to CAV. Recently, an HTx OCT study was reported that characterized morphological changes in coronary artery due to CAV development (Clemmensen et al., 2017, 2018). The analysis was performed on discrete OCT frames or using relatively short segments (not along the entire length of the available OCT pullbacks) and the clinical conclusions invariably relied in part on expert-assessed qualitative analysis or only-local quantitative measurements. Due to the diffuse and progressive character of CAV disease and its effect on coronary wall morphology, extending the analysis from isolated segments to the full available length of the coronary wall coverage is an important next step. In the process, relying on automatically generated quantitative indices of coronary wall morphology, tissue characterization, and changes thereof must be accomplished to overcome the tedious and irreproducible character of the manual and qualitative analyses. Our work reported here is addressing this very issue.
The reported method provides a diagnostic support for a notion that the earliest changes of coronary wall morphology combined with relevant biomarkers are likely indicators of clinically significant CAV development in the future (Starling et al., 2016). Presence of severe intimal thickening has been repeatedly shown indicative of cardiac events in CAV (Mehra et al., 1995; Tsutsui et al., 2001). However, limitations of the employed IVUS imaging did not allow reliable detection of small early changes of the intimal layer and the previous analysis methods relied on single-location measurements thus not supporting fully 3D analyses of longer vessel segments. Our method uses OCT imaging that offers much higher image resolution, clearly depicts even the thinnest intimal layers in undiseased or minimally diseased coronary arteries following HTx, and when combined with novel accurate quantitative 3D image analysis, allows highly sensitive assessment of coronary wall changes (Chen et al., 2017; Pazdernik et al., 2017a,b; Clemmensen et al., 2017; Woo et al., 2015).
The main novel contributions of the reported research are the development and validation of the first highly automated, inherently three-dimensional, accurate, and efficient method for multi-layer segmentation of coronary OCT in heart transplant patients including OCT–OCT image pullback registration; introduction of novel quantitative indices reflecting CAV status and CAV progression; demonstrating applicability of these quantitative indices to describe early changes of coronary wall morphology globally, regionally, and locally; and assessing their promise to predict future CAV progression in HTx patients.
2. Methods
Quantitative analysis of coronary wall morphology depicted by intravascular OCT uses our previously reported LOGISMOS segmentation strategy (Li et al., 2006; Yin et al., 2010) that was further developed for coronary OCT. In HTx patients, three wall-layer surfaces are segmented simultaneously for the entire pullback length in 3D: lumen, outer intimal (inner medial) surface, and outer medial surface. Since changes of coronary morphology between 1 and 12 months are of importance in HTx patients, an additional step of 1-month/12-month pullback registration is necessary.
2.1. LOGISMOS Segmentation
The LOGISMOS (Layered Optimal Graph-based Image Segmentation for Multiple Objects and Surfaces) segmentation approach was introduced in 2006 (Li et al., 2006) and has been continuously developed and improved ever since (Yin et al., 2010; Sun et al., 2013; Oguz and Sonka, 2014; Sonka and Abramoff, 2016; Kashyap et al., 2018). Its brief description is provided here for completeness. LOGISMOS is a general approach for optimally segmenting multiple surfaces that mutually interact within individual objects and/or between objects. The problem is modeled by a complex multi-layered graph in which solution-related costs are associated with individual graph nodes. When constructing a graph for segmentation of multiple cylindrical surfaces as is the case of coronary wall imaged by OCT, the graph construction follows the polar co-ordinate system. Columns oriented along the OCT beam direction are used with angular in-frame separation of θp, their lengths correspond to the region of interest in the OCT image. Intra-surface and inter-surface relationships are represented by context-specific graph arcs. Smoothness of the resulting surfaces can be controlled by smoothness constraint parameters between adjacent columns (Δa) and across adjacent frames (Δb). Additionally, a-priori information about maximum or minimum anatomical thicknesses of individual layers can be incorporated in the graph construction – the surface separation constraints. A single graph holding all relationships and surface cost elements is constructed, in which the segmentation of all desired surfaces is performed simultaneously in a single optimization process. The LOGISMOS method employed in this work is fundamentally 3D, surfaces are segmented utilizing inter-frame 3D context rather than being built from independently segmented 2D contours.
For each target surface, a cost function is designed to assign each node in the graph (node-spacing along the column of 10 μm) a cost value that indicates the unlikeliness of the surface passing through the voxel. Because each target surface passes through each column of nodes exactly once (along OCT beam direction in Fig. 1a), the objective of the multi-surface segmentation is to find the set of surfaces that have the globally minimal total cost and are subject to smoothness and separation constraints derived from prior knowledge. Following this automated step, the resulting optimized graph state (called residual graph) is kept in memory for the purposes of the subsequent Just-Enough Interaction (JEI) step.
Figure 1:
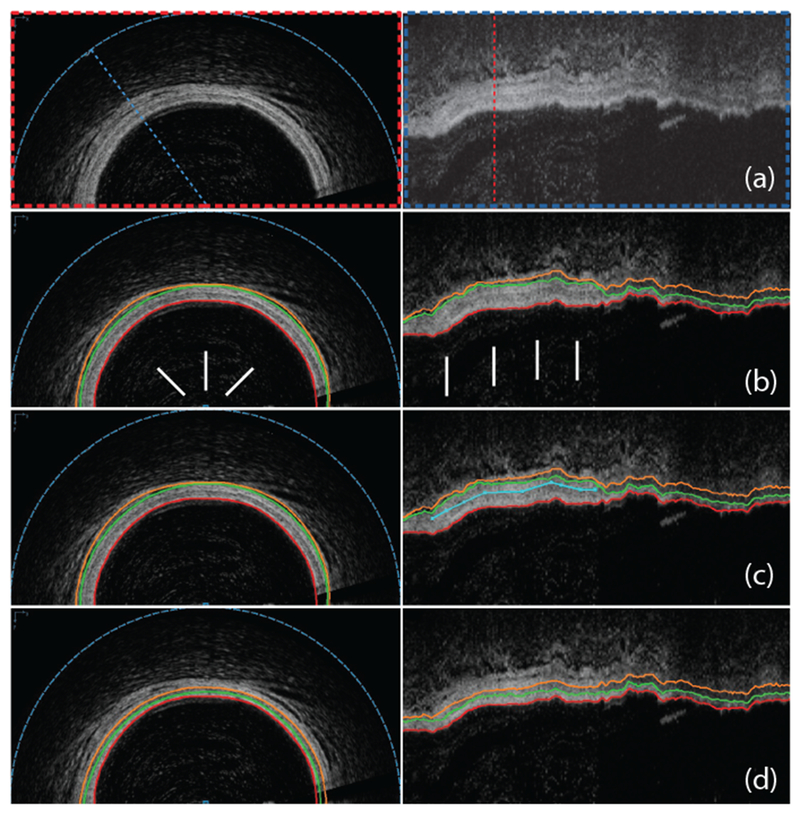
Automated OCT segmentation followed by JEI yielding clinically acceptable 3D segmentation of coronary wall layers. (a) Original cross-sectional and axial views of a 3D OCT dataset. (b) Automated 3-surface 3D LOGISMOS approach shows a regional segmentation inaccuracy (arrows) with lumen in red, outer intima in green and outer media in orange. (c) JEI interactions shown in turquoise color provide a suggested position for the outer media (orange) surface in the axial view. (d) Multi-surface 3D segmentation is re-optimized every time a set of correction points is provided – the few identified points shown completely corrected the inaccuracy in 3D. Note that all JEI modifications are optional such that the full segmentation workflow can be completed either without any interaction (fully automated) or using human expertise to guide the segmentation via JEI when needed. Important to realize is that the experts interact with the algorithm, they never directly retrace the borders in the image.
2.2. Efficiently Improving LOGISMOS Segmentation Results via Expert Guidance – Just Enough Interaction
Our JEI approach (Sun et al., 2013) starts with an initial automated LOGISMOS segmentation (Section 2.1). The user guidance for JEI functionality is achieved by expert-visual inspection of the segmentation result in 3D, identification of local or regional segmentation inaccuracies, and drawing correction line(s) by placing a set of points on the image to indicate approximate locations through which a specific segmentation surface should pass (Fig. 1b,c,d). The user feedback embedded in the correction line is incorporated into the algorithm by modifying cost values associated with graph node locations around the correction line, and then modifying capacities of corresponding terminal edges in the underlying residual graph (Kohli and Torr, 2007). The cost modification will make the expert-suggested locations more attractive to the underlying graph optimization process than was the case before. Since the suggested modifications are regional and not global, re-optimization is very fast and is completed in milliseconds. This allows real-time response to the operator and provides a good interactive use experience. Importantly, the obtained results still guarantee solution optimality with respect to the employed (JEI-modified) cost function and satisfy the desired surface smoothness and separation constraints.
2.3. Coronary OCT Segmentation
Using the LOGISMOS approach described above, layer segmentation in coronary OCT consists of the following main steps, all of which are performed in 3D.
Preprocessing: To remove OCT imaging catheter ring and decrease OCT-image noise, the OCT catheter was masked out (always the same size, always in the center of the image frame); 2D denoising median and mean filters were applied in 3×3 neighborhoods of each image-frame pixel.
Single-surface segmentation of the coronary lumen, determination of luminal centerline.
Simultaneous 3-surface segmentation yielding luminal, outer intimal, and outer medial surfaces (segmenting intimal and medial layers).
Identification of angular exclusion regions for each frame (angular wedges in which layers are not visible, Section 2.4).
Expert inspection of the automated segmentation results followed by the minimally-interactive JEI step to correct any local segmentation inaccuracies if present (Fig. 1).
As a result of these steps, layer thicknesses can be measured for each location of the coronary wall and used for calculation of wall-morphology description indices that are presented in Section 3.6. To measure morphological changes between 1-month and 12-month scans for HTx patients, the two OCT image pullbacks must be mutually registered as described in Section 2.5.
2.4. Deep-Learning-Based Exclusion Regions
Frequently, portions of the OCT-imaged wall are not analyzable due to e.g., guidewire shadow, the increased intimal layer thickness of which prevents OCT image formation due to limited OCT penetration, blood artifacts, etc. Such wall portions must be excluded from analysis. Note that only angular regions of the wall are excluded, not entire OCT image frames. Exclusion regions are automatically determined by transfer learning using ImageNet network (Sermanet et al., 2013; Sharif-Razavian et al., 2014) after feeding new datasets derived from our OCT images. For each cross-sectional frame, angular range of exhibiting or not exhibiting a layered wall appearance was determined by an expert cardiologist. After lumen segmentation, lumen-centered 60° wedge patches with 2.2 mm radius (2.0 mm tissue penetration starting at the lumen border and 0.2 mm lumen offset) were extracted from each frame and were labeled by majority vote using all local expert labels within the wedge using 1-degree partitioning. This information was used for training the deep-learning network. To achieve high learning efficiency, 36 overlapping 60° wedge patches (10° offset) were identified for each cross-sectional OCT frame. These angular patches were warped and resized to square image patches of 227 × 227 pixels required by network input. The weights of our network were initialized from an ImageNet pre-trained model using Caffe framework (Jia et al., 2014) and further fine-tuned by continuing the backpropagation with all available training-set patches. The resulting label was derived for each 1-degree partitioning of the wall on each analyzed OCT frame using a weighted-majority vote favoring central patches among all involved wedge patches.
2.5. 1-Month – 12-month Pullback Registration
Mutual registration of 1-month and 12-month post-HTx OCT image pullbacks with respect to location and orientation of corresponding OCT frames builds on expert-identification of at least 3 matching landmark pairs such as bifurcations, microvessels, etc. visible in both pullbacks (Fig. 2). The matching frame locations and orientations are computer-determined via joint axial and rotational linear interpolation of all OCT images positioned between each pair of the matching landmarks. Linear extrapolation based on two closest landmarks is used proximally or distally outside of the landmark pairs. Since this process relies on expert identification of matching landmarks, correctness of the obtained registrations were visually assessed by a second expert observer who was allowed to recompute/improve the registration by correcting existing or defining additional pairs of matching landmarks until full satisfaction.
Figure 2:
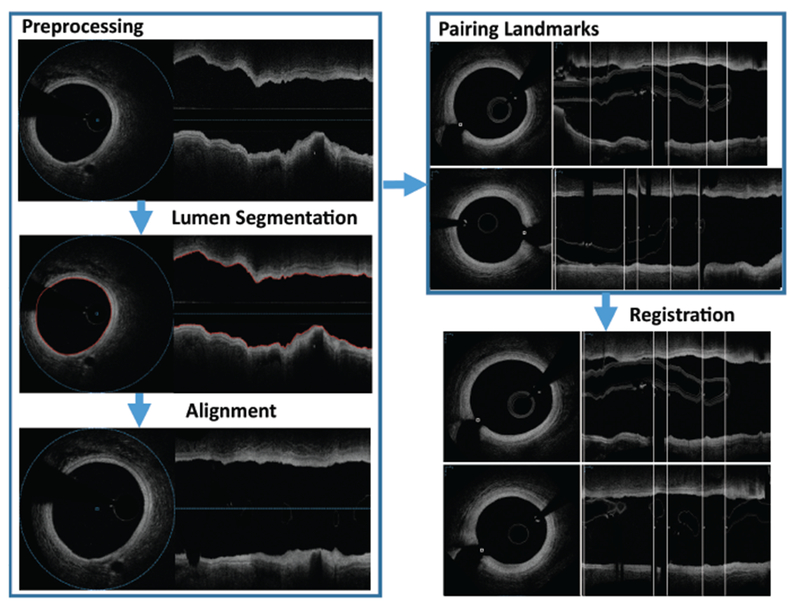
OCT–OCT registration process results in axial and orientation match between individual locations of two independent OCT pullbacks depicting the same coronary vessel segment. White vertical lines present matching landmarks. Corresponding frames in 12-month pullback are determined and properly rotated to achieve locational registration.
3. Experimental Methods
3.1. Image Data
Heart transplant subjects were recruited from the Transplant Center at IKEM, Prague, Czech Republic and the Center for Cardiovascular and Transplantation Surgery, Brno, Czech Republic, registered as clinical trial NCT02503566. Between October 2014 and December 2015, 50 subjects were enrolled. All HTx recipients ≥18 years of age were deemed eligible for inclusion in the study provided they were able and willing to give their informed consent. Exclusion criteria included renal insufficiency ≥stage IV (Glomerular filtration 30 ml/min), unfavorable post-transplant clinical conditions such as episodes of severe rejection or nosocomial sepsis with prolonged antibiotic treatment during the first month, ongoing need for circulatory support using a ventricular assist device, and acute allograft failure. Coronary arteries of these 50 heart-transplant patients were OCT-imaged 1 month and 12 months after HTx (Ilumien Optis intracoronary OCT system – Dragonfly Optis, St. Jude Medical, St. Paul, MN). Coronary imaging was preferably performed in the left anterior descending artery (LAD), if that was not possible, another coronary vessel was imaged. Overall, proximal segments of 40 LAD, 6 left circumflex artery (LCx), and 4 right coronary artery (RCA) segments were imaged. Data were exported as DICOM images with no overlays or markers included. The OCT analysis of the available 100 OCT pullbacks (540 frames/pullback, 54,000 OCT frames in total, frame-spacing of 0.1 mm, in-frame resolution of ~10 μm/pixel) focused on 3D segmentation of the intimal and medial layers and on quantitative 3D assessment of layer thicknesses. Out of these, 43,873 frames contained OCT image data with at least a portion of the coronary wall visualized (e.g., OCT frames inside of the guiding catheter were excluded). No image frames were excluded from analysis after baseline-follow-up co-registration.
All 100 pullbacks (50 1-month–12-month pairs) were used for assessment of coronary morphology and its changes. In the 50 analyzed HTx pullback pairs, the median (IQR) angular range in which layered structure was visible and thus analyzable was 250 degrees (180–351) when all 54,000 OCT frames from all registered pullbacks were evaluated.
3.2. Independent Standard – Coronary Wall Segmentation
The independent standard of coronary wall layer segmentation was obtained by averaging manual tracings provided by two expert observers (MP and ZG). In each pullback OCT dataset, about 10 randomly selected image frames were independently traced by the two observers who were allowed to edit the traced borders until full satisfaction. The intimal-medial, and outer-medial border were manually traced by each expert observer in 394 frames from 38 pullbacks. The observers were blind to each other’s tracings and also blind to the results of the automated segmentation. Each pair of tracings was averaged to form the independent standard, which was subsequently used in the validation process. As such, the independent standard was available for all 394 frames and allowed point-based and frame-based validation. The lumen surface is well-defined in OCT images, its segmentation is far less difficult, and therefore a smaller validation dataset of 135 frames from 6 pullbacks was used to validate the luminal segmentations, again averaging tracings from two experts to form the independent standard.
3.3. Parameters of the LOGISMOS Method
The OCT image segmentation method described in Section 2 used the following parameters:
Initial lumen segmentation: For each OCT frame, there were np = 120 columns at equal column intervals of θp = 3°, column length was equal to the A-scan length, intra-frame smoothness constraints was Δa = 6 nodes, inter-frame smoothness constraints Δb = 5 nodes.
Final 3-surface segmentation: The graph was constructed with the same per-frame column density. The column length was 2.2 mm (based on limited OCT tissue penetration depth of about 2.0 mm plus 0.2 mm of columns located inside of the pre-segmented lumen surface), intra- and inter-frame smoothness constraints were Δa = 2 nodes, Δb = 2 nodes, respectively. Pixel spacing (~10 μm) was used as in-column node spacing. The minimal layer thickness parameter was 0.02 mm for all layers, the maximum layer thickness for intima was 0.80 mm, for media was 0.20 mm.
Surface costs used in the 3-surface LOGISMOS segmentation were computed as directional 2D Sobel edge detector magnitudes after smoothing.
3.4. Validation, Independent Standard – Exclusion Regions
Five-fold cross-validation procedure was used to determine performance of our deep network for automated determination of wall-exclusion regions. The training/testing process was based on 43,873 frames from all available 100 pullbacks (50 1-month/12-month pairs), the regions with visible layered wall appearance (angular segments in each frame) were manually identified by expert cardiologist and served as the independent standard. The entire patient cohort was randomly divided in 5 groups. The leave-20%-out approach repeatedly utilized 4 groups for training and the remaining group for testing and performance assessment. Training-testing processes were repeated 5 times by using each of the five groups as testing data in each run. Each run determined whether or not the computer-determined exclusion regions agreed with the independent standard. The overall classification correctness was computed from a confusion matrix. Additionally, the 394 frames of 38 pullbacks, in which manual identification of exclusion regions was performed by two experts (Section 3.2), were used to derive inter-observer variability of manual identification of exclusion regions and that was compared with the performance of the deep learning classification correctness.
3.5. Validation of Layer Segmentation
Correctness of all quantitative indices of coronary wall morphology listed in Section 3.6 is directly dependent on the correctness of the layer-based segmentation. Signed and unsigned surface positioning errors of the automated and JEI segmentations are reported as mean ± standard deviation in μm. Surface positioning errors were defined as the shortest in-frame distances between computer-determined surfaces and the independent standard. Similarly, local intimal and medial thickness errors were determined as the difference between corresponding thickness measurements derived from the independent standard and from computer/JEI analyses. Inter-observer variability of each of the above-listed assessment indices was determined from the independent standard defined by two expert observers (Section 3.2) and compared with errors of the computer analysis.
3.6. Quantitative Indices of Coronary Morphology, Visualization of Results
The following indices of coronary wall morphology were measured to demonstrate wall-layer-quantification ability.
- Measured at each point of the wall of each pullback and available as local, regional, or global averages/standard deviations, minima, maxima:
- Layer-specific thickness (intimal, medial, combined intimal plus medial – I+M)
- Intimal-to-medial thickness ratio (IMratio)
- Normalized intimal thickness
- Intimal surface roughness
- Layer-specific brightness (intimal, medial)
- Histograms of intimal, medial, I+M thickness
- Measured as progression/regression describing differences between pairs of timepoints (1- and 12-months after HTx in our case), available as local, regional, or global averages/standard deviations, minima, maxima:
-
7.Intimal, medial, I+M thickening
-
8.Normalized intimal thickness change
-
9.Intimal surface roughness changes
-
10.Histograms of intimal, medial, I+M thickening
-
7.
Feature Carpets: Any of the above-listed indices assessing morphology or progression/regression can be visualized as unwrapped luminal surfaces (called feature carpets) with location-mapped color-coded values giving additional insight by providing spatial context (Fig. 3). Additional feature-carpet based measures can be derived from spatial feature analysis, e.g., inhomogeneity of local/regional indices (allowing to distinguish between diffuse and focal disease, as well as quantification of the focality extent), size distribution of specific features on the luminal wall, etc. Using the feature carpets, a variety of regional and global indices can be designed.
Figure 3:
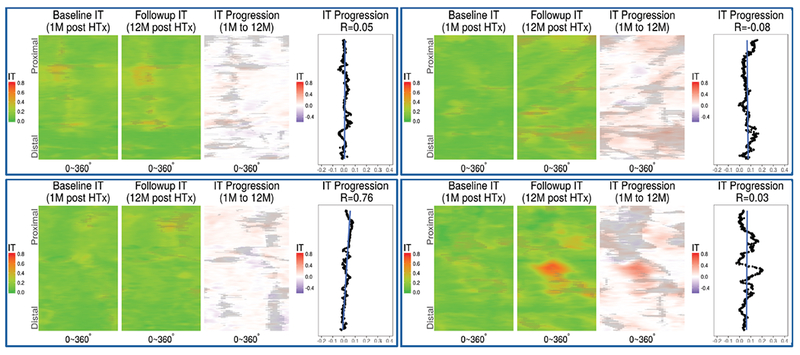
Feature carpets of intimal thickness (IT) and thickening. Mutually registered intimal thickness at 1 and 12 months (M), and their differences shown color coded on an axially unwrapped vessel wall. Shadowed areas represent non-measurable exclusion regions caused by guidewire shadow, residual blood, intimal layer thickness exceeding OCT penetration depth, etc. Rightmost chart in each panel shows frame-based IT progression, blue straight line shows a linear fit of frame-based IT progression along the co-registered vessel portion.
3.7. Assessment and Prediction of CAV Progression
Progression of CAV was assessed by comparison of the above-listed local, regional, and global quantitative indices of coronary wall morphology between 1 and 12 months. Statistical significance of measurement differences was determined to identify those indices that do and do not exhibit changes during the first year after HTx. Ability to predict CAV-related changes of intimal thickness ΔIT at 12 months solely from the 1-month vessel wall morphology indices was examined.
3.8. Analysis Time
To determine the analysis-time requirements associated with OCT segmentation, the following operations were timed and reported as mean±standard deviation in seconds: Time required for the automated segmentation step yielding complete multi-layer segmentation; time required for the JEI stage including visual review of segmentation results and placement of interaction points; and computer response time.
3.9. Statistical Analysis
Discrete variables are reported as n (%) and continuous variables as mean (±stdev) or median (and interquartile range IQR), as appropriate. Global patient-level changes of the measured morphological features were investigated using Mann-Whitney U test. Whenever multiple segments were analyzed per vessel, generalized linear mixed models with patient as random effect were employed to investigate the differences between 1-month and 12-month regional measurements. The assessment of the power of 1-month quantitative indices to predict morphological changes of intimal thickness 12 months after HTx employ correlation tests when assessed globally and mixed-effect analysis when determined regionally. Correlation coefficient R and associated slope β are reported. Since multiple pairwise tests were performed, Bonferroni correction was applied to reduce the chances of obtaining type I errors. Probability level p<0.05 determined statistical significance. All statistical tests were conducted in R (R Core Team, 2016).
4. Results
4.1. Validation of Coronary Wall Morphology
The analysis performance of the wall morphology layer segmentation was assessed in the validation sets described in Section 3.2. All validation datasets were segmented using the described fully-automated framework and also using the optional JEI refinement. Fig. 4 shows an example of the wall segmentation and allows visual comparison with the independent standard. The signed and unsigned surface positioning errors of the obtained wall layer segmentations are given in Table 1. The subpixel signed positioning errors (pixel size ~10 μm) attest to a small bias of surface detection. Statistical analysis showed that when mean errors were considered, the automated and JEI approaches provided results that were not distinguishable from those obtained by manual tracing.
Figure 4:
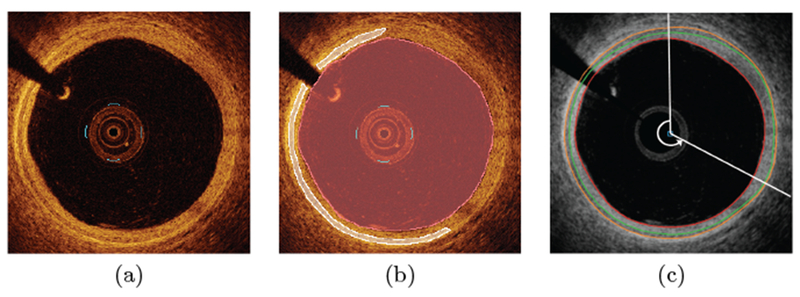
OCT segmentation. (a) Original OCT image (one frame of a 540-frame long pullback). (b) Expert-defined independent standard. (c) Result of the automated multi-layer segmentation with automatically-determined exclusion region (12 to 4 o’clock).
Table 1:
Mean signed and unsigned errors of multi-layer HTx coronary wall segmentation in comparison with the independent standard in μm.
| ERRORS (μm) | Signed Lumen | Signed Outer intima | Signed Outer media | Unsigned Lumen | Unsigned Outer intima | Unsigned Outer media |
|---|---|---|---|---|---|---|
| Automated vs. Manual | 0.57±1.82 | 1.22±30.41 | 6.45±33.88 | 2.37±1.84 | 13.61±27.22 | 16.43±30.32 |
| Automated+JEI vs. Manual | 0.51±1.73 | 1.26±31.78 | 7.26±33.43 | 2.14±1.71 | 13.75±28.68 | 15.63±30.43 |
| Manual_1 vs. Manual_2 | 1.59±1.09 | 3.92±17.63 | 7.41±18.16 | 3.15±0.84 | 13.45±12.06 | 14.74±12.94 |
When assessing the accuracy of layer thickness measurements, regression analysis showed excellent correlation between frame-based intimal+medial thicknesses derived from the independent standard, automated segmentation, and after JEI analysis (=0.93, y=1.0x − 6.2μm; =0.96, y=1.0x −6.3μm). The average intimal+medial thickness errors for the automated and JEI approaches were 4.98±31.24 μm and 5.38±28.54 μm. Inter-observer variability of manual tracing by two observers was 6.76±10.61 μm. The intimal+medial thickness errors of our automated and JEI analyses were statistically indistinguishable from the reproducibility of manual tracing.
As expected, the JEI stage showed its benefits when maximum errors of local intimal+medial thickness were considered. The JEI approach provided lower maximum errors than the automated approach at near-statistical significance (55.47±56.66 μm compared to 63.71±66.09 μm, p=0.06). In comparison, the inter-observer variability of maximum thickness errors was 58.33±28.97 μm.
4.2. Validation of Exclusion Regions
The performance of the automated definition of exclusion regions was assessed in the validation sets described in Section 3.4. The accuracy of the automated method was 81.2% which is comparable with inter-observer variability of 83.2%. when the same task was performed by two experts.
4.3. Analysis Time
Automated segmentation of a single 540-frame long OCT pullback required 91.0±19.4 seconds. The JEI step required 16 interactive edits per OCT pullback on average, which required 473±289 seconds per pullback, out of which 471±282 seconds was the human interaction time (observation+operation) and 1.5±6.3 seconds was the JEI-associated computation time. Automated definition of exclusion regions needed less than 1 minute per pullback. In addition to morphologic analyses, registration of each OCT pullback 1-month–12-month pair had 7.2 corresponding landmarks manually identified, which required about 15 minutes of expert-registration effort per pullback pair.
4.4. Assessment of CAV Progression: 1M–12M Comparisons
Comparison of quantitative indices of coronary wall morphology 1 month and 12 months after HTx is given in Table 2. The table shows significant changes of several well-established indices associated with CAV progression like decrease in luminal size and increase in intimal thickness as well as of a number of previously unavailable indices like intimal or medial surface roughness.
Table 2:
Comparison of global (patient-level) coronary wall properties between 1-month and 12-month post HTx. Advanced_IT is defined as IT>120μm, threshold identified to give the strongest quantitative differences between 1-month and 12-month measurements. Statistically significant differences shown in bold, p-values reported after Bonferroni correction.
| 1-month | 12-month | Difference | p-value | |
|---|---|---|---|---|
| Lumen Area (mm2) | 8.6±2.5 | 7.6±2.3 | −1.0±1.6 | 0.039 |
| Intimal thickness (μm) | 103.0±31.8 | 129.6±41.7 | 26.5±25.9 | 0.001 |
| Medial thickness (μm) | 81.3±19.4 | 81.5±18.6 | 0.2±11.1 | 0.966 |
| Intimal+Medial thickness (μm) | 184.3±47.4 | 211.0±56.5 | 26.7±33.2 | 0.012 |
| Normalized Intimal thickness (μm/mm2) | 13.9±7.0 | 19.6±9.8 | 5.7±7.3 | 0.001 |
| IMratio | 1.4±0.3 | 1.7±0.4 | 0.3±0.3 | <0.001 |
| Intimal roughness | 0.36±0.10 | 0.42±0.10 | 0.06±0.15 | 0.002 |
| Medial roughness | 0.37±0.10 | 0.43±0.09 | 0.06±0.13 | 0.004 |
| Intimal brightness | 40.4±13.8 | 38.1±12.7 | −2.3±17.7 | 0.392 |
| Medial brightness | 40.4±14.1 | 38.4±13.5 | −2.0±18.6 | 0.461 |
| Intimal-to-Medial brightness ratio | 1.04±0.05 | 1.04±0.05 | 0.01±0.07 | 0.587 |
| Advanced_IT (%) | 25.1%±20.1% | 43.1%±26.1% | 18.0%±17.4% | <0.001 |
| Absolute area of Advanced_IT (mm2) | 43.9±37.7 | 71.3±48.1 | 27.4±27.2 | 0.002 |
| Largest area of Advanced_IT (mm2) | 27.9±36.3 | 53.1±48.1 | 25.2±34.4 | 0.004 |
Table 3 gives absolute and relative differences of regional maximal and minimal measurements (in 3 mm long adjacent vessel segments), thus focusing on most pronounced regional changes of vessel wall morphology. Many of these indices show significant changes between 1-month and 12-month time points. Changes in both the established and novel indices have been observed and quantified.
Table 3:
Comparison of regional minima/maxima of quantitative indices of coronary wall properties between 1 month and 12 months post HTx measurements (in co-registered 3 mm long segments). Mixed effect analysis employed, statistically significant differences shown in bold.
| 1-month | 12-month | Difference | Difference(%) | 1M vs. 12M (p) | |
|---|---|---|---|---|---|
| Intimal thickness MIN | 47.8±26.2 | 55.2±35.8 | 7.4±22.8 | 17.1%±49.7% | 0.002 |
| Intimal thickness MAX | 231.5±78.0 | 282.3±104.6 | 50.9±83.8 | 26.6%±44.3% | <0.001 |
| Medial thickness MIN | 33.2±15.1 | 32.2±15.2 | −1.0±14.7 | 3.8%±44.8% | 1.000 |
| Medial thickness MAX | 150.2±30.9 | 161.3±31.0 | 11.1±31.6 | 10.2%±25.3% | <0.001 |
| Intimal+Medial thickness MIN | 109.9±51.1 | 123.0±61.3 | 13.1±40.9 | 17.1%±50.2% | 0.007 |
| Intimal+Medial thickness MAX | 323.4±88.2 | 374.6±115.1 | 51.2±88.9 | 18.2%±30.3% | <0.001 |
| Normalized Intimal thickness MIN | 9.5±5.8 | 17.4±13.7 | 7.9±12.2 | 95.2%±123.1% | <0.001 |
| Normalized Intimal thickness MAX | 20.5±21.4 | 25.2±23.3 | 4.7±15.0 | 34.4%±76.0% | 0.004 |
| IMratio MIN | 0.46±0.20 | 0.50±0.27 | 0.03±0.21 | 13.3%±57.2% | 0.338 |
| IMratio MAX | 5.57±3.08 | 7.15±4.05 | 1.58±4.16 | 50.3%±99.2% | <0.001 |
| Intimal roughness MIN | 0.11±0.10 | 0.20±0.21 | 0.10±0.16 | 116.9%±153.4% | <0.001 |
| Intimal roughness MAX | 0.89±0.57 | 0.57±0.58 | −0.31±0.68 | −25.3%±60.1% | <0.001 |
| Medial roughness MIN | 0.11±0.10 | 0.21±0.22 | 0.10±0.18 | 111.0%±153.3% | <0.001 |
| Medial roughness MAX | 0.90±0.56 | 0.59±0.62 | −0.32±0.69 | −25.5%±56.5% | <0.001 |
Changes in distribution (histograms) of local/regional (3 mm long vessel segments) intimal thickness; local/regional IMratio, and distribution of local/regional ΔIT and ΔIMratio are shown in Figs. 5, 6. They clearly show a notable increase of thicker intimal presence – globally, locally, and regionally.
Figure 5:
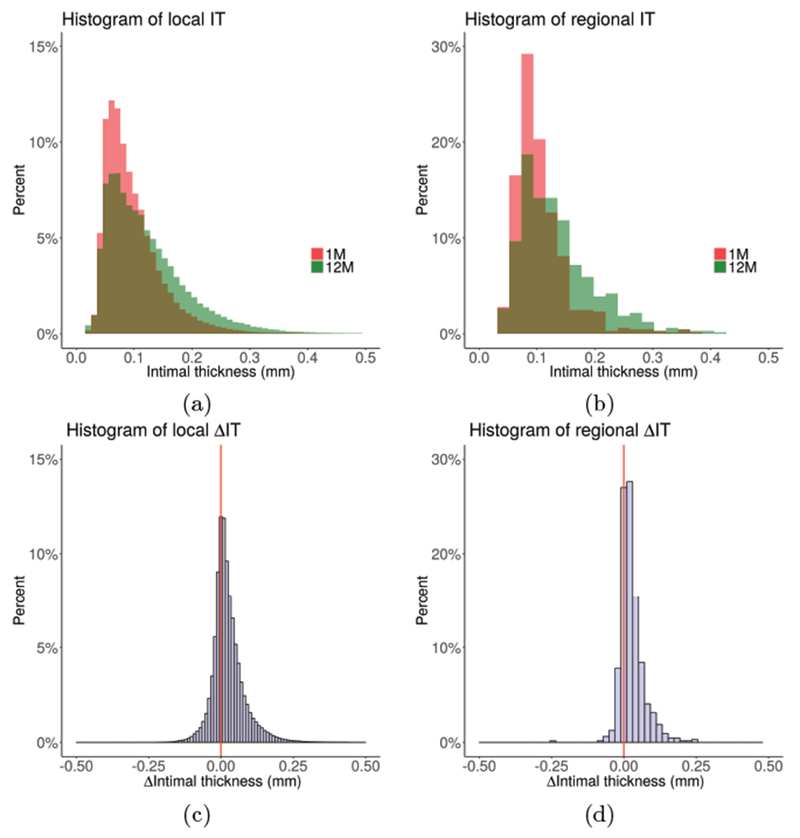
Distribution of local and regional (3 mm long vessel segments) intimal thickness across the cohort at 1-month and 12-month (80,600 local and 13 regional measurements per pullback on average; e.g., histograms in panels (a) and (c) are based on over 4 million locally co-registered measurements). (a) Histograms of local IT. (b) Histograms of regional IT. (c) Histogram of local ΔIT. (d) Histogram of regional ΔIT. Note the clear indication of intimal thickening between 1M and 12M, both locally and regionally.
Figure 6:
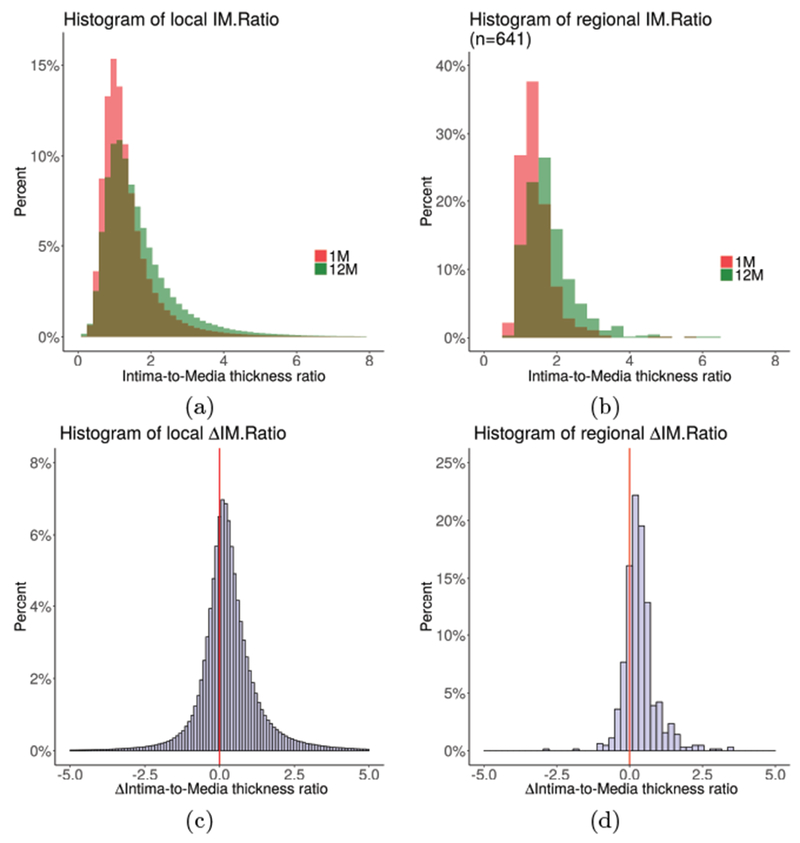
Distribution of local and regional (3 mm long vessel segments) intimal thickness across the cohort at 1-month and 12-month (80,600 local and 13 regional measurements per pullback on average). (a) Histograms of local IMratio. (b) Histograms of regional IMratio. (c) Histogram of local ΔIMratio. (d) Histogram of regional ΔIMratio. Note the clear indication of increased IMratio between 1M and 12M, both locally and regionally.
4.5. CAV Morphological Predictive Factors
To study the promise of quantitative indices of wall morphology at 1 month to serve as predictors of CAV risk at 12 months, we used data from 1 month after HTx to statistically assess their global/regional associations with CAV progression (ΔIT). Global analysis has shown that several quantitative baseline indices measured just 1 month after HTx are statistically significantly associated with indicators of CAV progression during the first 12 months after HTx. The following baseline global indices can be considered strong predictors of CAV progression during the first year: normalized OCT intimal and medial brightness (R=−0.367, p=0.009 and R=−0.368, p=0.009, respectively). Regional analysis associating ΔIT with 1-month morphological features revealed that baseline regional indices of medial thickness and intimal+medial thickness showed statistical significance (β=0.26, p=0.028; β=0.12, p=0.041).
5. Discussion
Discussion focuses on the following aspects of our work: 1) OCT multi-layer segmentation and co-registration of 1-month and 12-month OCT images, 2) segmentation sensitivity to parameter selection, 3) assessment of small wall changes over time leading to prediction of CAV development from 1-month OCT images, 4) limitations of performed research, and 5) novelty of comprehensive quantitative 3D analysis and future contribution of resulting indices to quantitative studies of CAV development.
Intracoronary OCT imaging offers unprecedented resolution clearly depicting the intimal and medial layers of the coronary wall. At the same time, its depth of tissue penetration is limited to 2 mm and the wall is not imaged in the presence of light-beam obstructing fluid in the lumen, including residual blood. Similarly, plaque-related changes of the intimal layer may not allow the intima or media to be visualized and thus layer thicknesses to be correctly measured. Our approach was to exclude such areas of the OCT-imaged wall from analysis. Alternatively, these exclusion areas may be independently analyzed, visual analysis of such wall regions was reported in (Clemmensen et al., 2017). Another aspect of our OCT image analysis approach is a point-to-point registration of coronary wall locations imaged at two different time points – 1 and 12 months after HTx in our case. This co-registration relied on visual assessment of corresponding landmarks that were subsequently used for computerized registration. While we have previously reported fully automated IVUS-IVUS pullback registration (Zhang et al., 2015), automated OCT–OCT pullback registration is substantially more difficult in HTx for at least three reasons. The first is the limited penetration of OCT that does not allow to rely on outer-wall structures to be used for OCT image appearance matching; the second is the minimal presence of focal wall disease that would be consistently present at the two OCT pullback images; and the third is the fact that OCT image pullbacks are not EKG gated. As a result, employing expert-cardiologist knowledge was needed, yielding the required accuracy of the co-registration step.
Most parameters given in Section 3.3 are dictated by the characteristics of OCT imaging and should not be considered free parameters of the image segmentation method: node spacing was directly associated with the in-frame pixel size of OCT images (~10 μm) to facilitate sub-pixel accuracy; pre-segmentation column length equaled the A-scan length in OCT images to cover the complete scanned region; final-segmentation column lengths coincided with the OCT beam penetration depth; the minimal and maximal values of wall layer thicknesses were determined from a priori knowledge of coronary wall morphology in healthy and CAV coronaries and knowledge of OCT imaging beam penetration depth; and np = 120 and θp = 3° reflected that the contour of a given layer on each frame was defined by 120 points residing at uniform 3° angle increments circumferentially thus guaranteeing highly-detailed and more-than sufficient level of layer-surface shape detail. The only parameters to which the performance of our method may be sensitive are the smoothness constraints utilized in the final segmentation. Coronary layer segmentation must allow for in-frame (Δa) and frame-to-frame (Δb) surface variability. Results reported in the paper are based on smoothness constraints of Δa = 2, Δb = 2, which were initially determined by parameter optimization in a small dataset of 5 pullbacks and subsequently applied to all 100 pullbacks analyzed in the reported work. When considering the sensitivity analysis design, smoothness constraint of 1 was not deemed appropriate as it would have resulted in surfaces that would not reflect the coronary wall layer anatomy and would substantially limit the capability of subsequent JEI editing. Smoothness values of 2, 3 and 4 were used for sensitivity testing to assess the influence of smoothness parameter selection on accuracy of automated multi-layer OCT segmentation. Compared to original Δa = 2, Δb = 2 constraints, when smoothness constraints were changed to Δa = 2, Δb = 3 or Δa = 3, Δb = 2, slightly larger signed and unsigned errors were observed while maintaining sub-pixel segmentation accuracy and overall segmentation performance statistically indistinguishable from inter-observer variability (p=NS). For larger values of the smoothness parameters, segmentation errors surpassed the pixel size of OCT images and were significantly larger than inter-observer variability (p<0.05). The segmentation errors associated with these larger values of smoothness parameters were mainly caused by outside-adventitia micro-vessels that exhibited stronger gradient costs than the intima-media layers. These errors were avoided when using the smaller smoothness constraints that better reflected frame-to-frame continuity and layer-specific context.
The outcome of our automated layer segmentation, the efficient JEI correction of any local/regional inaccuracies of the automated segmentation, the ability to achieve co-registration of coronary wall locations between 1-month and 12-month OCT pullbacks, and resulting quantitative analysis of coronary wall morphology and its location-specific changes over time yielded novel insight in coronary layer behavior after HTx. We have shown that some of the newly designed baseline indices (intimal layer brightness, medial layer brightness, medial thickness, intimal+medial thickness) were associated with CAV-related progression of intimal thickness. When indices of coronary layer morphology are further combined with patient- and donor-associated biomarkers, some of them (cellular rejection, aldosteron, creatinin, diabetes, medication), all measured at baseline only, further increase the likelihood of predicting rapid intimal thickening at 12 months. These preliminary results show promise of identifying patients subjected to rapid intimal thickening at 12 months after HTx from OCT-image and biomarker-data obtained just 1 month after HTx.
This study was not free of several limitations. Our analysis excluded areas of the wall that did not exhibit multi-layer structure. The lack of multi-layer visualization on OCT may be due to imaging artifacts or may be caused by limited OCT penetration due to focal atherosclerotic disease. While not a focus of the presented work, the currently excluded wall areas will receive attention of our future research. Another concern is the size of the analyzed cohort. A result of substantial recruiting effort, our studied group of 50 patients is already relatively large considering the available sizes of post-HTx cohorts and we have clearly demonstrated our ability to accurately quantify coronary wall layer parameters in 3D. At the same time, larger cohort is needed to increase predictive power of our work and we are actively recruiting additional patients.
To the best of our knowledge, the novelty of the presented approach – in comparison to all previously published approaches Olender et al. (2017); Zahnd et al. (2017); Clemmensen et al. (2017, 2018) – is its fully 3D character, both with respect to utilizing frame-to-frame image data context during the segmentation (rather than analyzing each 2D OCT frame independently), its simultaneous multi-surface segmentation, the ability to efficiently employ the Just-Enough Interaction strategy in 3D, its overall computational efficiency, the resulting fully 3D analysis of the complete length of the OCT pullback (rather than analyzing a small subset of available OCT frames), and the analysis of longitudinal data after axial and angular registration. Perhaps most importantly, the barrage of novel never-before available quantitative indices now allows to begin studies of the effect and consequences that local/regional/global indices of coronary morphology, biomarkers, donor information, genetic data, etc. have on CAV development. This forward-looking aspect of our work is perhaps the most exciting part of the presented research.
6. Conclusion
We have reported, validated, and demonstrated performance of – to the best of our knowledge – the first fully 3D coronary OCT wall layer analysis approach that facilitates computation of local, regional, and/or global indices of coronary wall morphology. This approach allows quantification of location-specific alterations of coronary wall morphology over time and is sensitive even to very small changes of wall layer thicknesses that occur in patients following heart transplant. As a result, studies of CAV progression in co-registered locations of the entire imaged coronary wall are now possible, which may facilitate prediction of patient-specific CAV risk and consequent administration of relevant patient-specific treatment at the earliest possible time.
Highlights.
The first fully 3D multi-layer coronary wall OCT analysis approach is reported.
Location-specific changes of coronary wall morphology over time are quantified.
Association of wall morphology indices with vasculopathy progression demonstrated.
Acknowledgment
This work was supported in part by the grants from the National Institutes of Health (R01-EB004640), Ministry of Health of the Czech Republic (16-27465A, 16-28525A, 17-28784A – all rights reserved), and IKEM (MH-CZ-DRO-IKEM-IN 00023001).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bom N, Lancee CT, 1972. Apparatus for ultrasonically examining a hollow organ. UK Patent 1402192.
- Chandran KB, Wahle A, Vigmostad SC, Olszewski ME, Rossen JD, Sonka M, 2006. Coronary arteries: Imaging, reconstruction, and fluid dynamic analysis. Critical Reviews in Biomedical Engineering 34, 23–103. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wahle A, Guo Z, Zhang H, Karmazin V, Tomasek A, Bedanova H, Lopez JJ, Kovarnik T, Pazdernik M, Sonka M, 2017. Highly automated analysis of intimal and medial thickness in heart-transplant coronary OCT facilitates longitudinal studies of CAV progression. The Journal of Heart and Lung Transplantation 36, S155. [Google Scholar]
- Chih S, Chong AY, Mielniczuk LM, Bhatt DL, Beanlands RS, 2016. Allograft Vasculopathy: The Achilles’ Heel of Heart Transplantation. J. Am. Coll. Cardiol. 68, 80–91. [DOI] [PubMed] [Google Scholar]
- Clemmensen TS, Holm NR, Eiskjær H, Jakobsen L, Berg K, Neghabat O, Løgstrup BB, Christiansen EH, Dijkstra J, Terkelsen CJ, et al. , 2018. Detection of early changes in the coronary artery microstructure after heart transplantation: A prospective optical coherence tomography study. The Journal of Heart and Lung Transplantation 37, 486–495. [DOI] [PubMed] [Google Scholar]
- Clemmensen TS, Holm NR, Eiskjær H, Løgstrup BB, Christiansen EH, Dijkstra J, Barkholt TØ, Terkelsen CJ, Maeng M, Poulsen SH, 2017. Layered Fibrotic Plaques Are the Predominant Component in Cardiac Allograft Vasculopathy: Systematic Findings and Risk Stratification by OCT. JACC Cardiovasc Imaging 10, 773–784. [DOI] [PubMed] [Google Scholar]
- Huang D, Swanson E, Lin C, Schuman J, Stinson W, Chang W, Hee M, Flotte T, Gregory K, Puliafito C, Fujimoto J, 1991. Optical Coherence Tomography. Science 254, 1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T, 2014. Caffe: Convolutional architecture for fast feature embedding. arXiv preprint arXiv:1408.5093 .
- Kashyap S, Zhang H, Rao K, Sonka M, 2018. Learning-based cost functions for 3-d and 4-d multi-surface multi-object segmentation of knee mri: Data from the osteoarthritis initiative. IEEE transactions on medical imaging 37, 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli P, Torr PH, 2007. Dynamic graph cuts for efficient inference in Markov Random Fields. IEEE Trans. Pattern Anal. 29, 2079–2088. [DOI] [PubMed] [Google Scholar]
- Li K, Wu X, Chen DZ, Sonka M, 2006. Optimal surface segmentation in volumetric images-a graph-theoretic approach. IEEE Trans. Pattern Anal. 28, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Eggermont J, Wolterbeek R, Broersen A, Busk CA, Precht H, Lelieveldt BP, Dijkstra J, 2016. Analysis and compensation for the effect of the catheter position on image intensities in intravascular optical coherence tomography. Journal of biomedical optics 21, 126005. [DOI] [PubMed] [Google Scholar]
- Mehra MR, Ventura HO, Stapleton DD, Smart FW, Collins TC, Ramee SR, 1995. Presence of severe intimal thickening by intravascular ultrasonography predicts cardiac events in cardiac allograft vasculopathy. J. Heart Lung Transplant. 14, 632–639. [PubMed] [Google Scholar]
- Oguz I, Sonka M, 2014. LOGISMOS-B: layered optimal graph image segmentation of multiple objects and surfaces for the brain. IEEE Trans Med Imaging 33, 1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender ML, Athanasiou LS, José M, Camarero TG, Cascón JD, Consuegra-Sanchez L, Edelman ER, 2017. Estimating the internal elastic membrane cross-sectional area of coronary arteries autonomously using optical coherence tomography images, in: Biomedical & Health Informatics (BHI), 2017 IEEE EMBS International Conference on, IEEE; pp. 109–112. [Google Scholar]
- Pazdernik M, Kovarnik T, Chen Z, Wahle A, Karmazin V, Melenovsky V, Kautzner J, Tomasek A, Bedanova H, Sonka M, 2017a. Increased heart rate after heart transplant is not associated with early progression of cardiac allograft vasculopathy (CAV) — a prospective study using highly automatic coronary optical coherence tomography segmentation software in 3D. The Journal of Heart and Lung Transplantation 36, S297–S298. [Google Scholar]
- Pazdernik M, Kovarnik T, Sonka M, Wahle A, Chen Z, Karmazin V, Kautzner J, Tomasek A, Melenovsky V, Bedanova H, 2017b. Should we pharmacologically modulate renin-aldosterone-angiotensin system (RAAS) to attenuate cardiac allograft vasculopathy (CAV)? A prospective study using highly automated coronary optical coherence tomography segmentation software in 3D. The Journal of Heart and Lung Transplantation 36, S292. [Google Scholar]
- R Core Team, 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: URL: https://www.R-project.org. [Google Scholar]
- Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y, 2013. Over-feat: Integrated recognition, localization and detection using convolutional networks. arXiv preprint arXiv:1312.6229 .
- Sharif-Razavian A, Azizpour H, Sullivan J, Carlsson S, 2014. CNN features off-the-shelf: An astounding baseline for recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Workshops, pp. 806–813. [Google Scholar]
- Slager CJ, Wentzel JJ, Schuurbiers JC, Oomen JA, Kloet J, Krams R, von Birgelen C, van der Giessen WJ, Serruys PW, de Feyter PJ, 2000. True 3-dimensional reconstruction of coronary arteries in patients by fusion of angiography and IVUS (ANGUS) and its quantitative validation. Circulation 102, 511–516. [DOI] [PubMed] [Google Scholar]
- Sones FM, Shirey EK, 1962. Cine coronary arteriography. Mod Concepts Cardiovasc Dis 31, 735–738. [PubMed] [Google Scholar]
- Sonka M, Abramoff MD, 2016. Quantitative analysis of retinal OCT. Med Image Anal 33, 165–169. [DOI] [PubMed] [Google Scholar]
- Starling RC, Stehlik J, Baran DA, Armstrong B, Stone JR, Ikle D, Morrison Y, Bridges ND, Putheti P, Strom TB, Bhasin M, Guleria I, Chandraker A, Sayegh M, Daly KP, Briscoe DM, Heeger PS, 2016. Multicenter Analysis of Immune Biomarkers and Heart Transplant Outcomes: Results of the Clinical Trials in Organ Transplantation-05 Study. Am. J. Transplant 16, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone PH, Coskun AU, Kinlay S, Popma JJ, Sonk a.M., Wahle A, Yeghiazarians Y, Maynard C, Kuntz RE, Feldman CL, 2007. Regions of low endothelial shear stress are the sites where coronary plaque progresses and vascular remodelling occurs in humans: an in vivo serial study. Eur Heart J 28, 705–710. [DOI] [PubMed] [Google Scholar]
- Sun S, Sonka M, Beichel RR, 2013. Graph-based IVUS segmentation with efficient computer-aided refinement. IEEE Trans Med Imaging 32, 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Ziada KM, Schoenhagen P, Iyisoy A, Magyar WA, Crowe TD, Klingensmith JD, Vince DG, Rincon G, Hobbs RE, Yamagishi M, Nissen SE, Tuzcu EM, 2001. Lumen loss in transplant coronary artery disease is a biphasic process involving early intimal thickening and late constrictive remodeling: results from a 5-year serial intravascular ultrasound study. Circulation 104, 653–657. [DOI] [PubMed] [Google Scholar]
- Wahle A, Lopez JJ, Olszewski ME, Vigmostad SC, Chandran KB, Rossen JD, Sonka M, 2006. Plaque development, vessel curvature, and wall shear stress in coronary arteries assessed by x-ray angiography and intravascular ultrasound. Medical Image Analysis 10, 615–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle A, Prause GP, von Birgelen C, Erbel R, Sonka M, 1999a. Fusion of angiography and intravascular ultrasound in vivo: establishing the absolute 3-D frame orientation. IEEE Trans Biomed Eng 46, 1176–1180. [DOI] [PubMed] [Google Scholar]
- Wahle A, Prause PM, DeJong SC, Sonka M, 1999b. Geometrically correct 3-D reconstruction of intravascular ultrasound images by fusion with biplane angiography–methods and validation. IEEE Trans Med Imaging 18, 686–699. [DOI] [PubMed] [Google Scholar]
- Wever-Pinzon O, Romero J, Kelesidis I, Wever-Pinzon J, Manrique C, Budge D, Drakos SG, Pina IL, Kfoury AG, Garcia MJ, Stehlik J, 2014. Coronary computed tomography angiography for the detection of cardiac allograft vasculopathy: A meta-analysis of prospective trials. J. Am. Coll. Cardiol. 63, 1992–2004. [DOI] [PubMed] [Google Scholar]
- Woo V, Chen Z, Hirai T, Weber JR, Kovarnik T, Wahle A, Sonka M, Lopez JJ, 2015. [TCT-355] An automated computational method for quantification of total fibrous cap volume and mean fibrous cap thickness with optical coherence tomography. Journal of the American College of Cardiology 15, B143–B144. [Google Scholar]
- Yin Y, Zhang X, Williams R, Wu X, Anderson D, Sonka M, 2010. LOGISMOS-Layered optimal graph image segmentation of multiple objects and surfaces: Cartilage segmentation in the knee joint. IEEE Trans. Med. Imaging 29, 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnd G, Hoogendoorn A, Combaret N, Karanasos A, Péry E, Sarry L, Motreff P, Niessen W, Regar E, Van Soest G, et al. , 2017. Contour segmentation of the intima, media, and adventitia layers in intracoronary oct images: application to fully automatic detection of healthy wall regions. International journal of computer assisted radiology and surgery 12, 1923–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wahle A, Chen Z, Zhang L, Downe RW, Kovarnik T, Sonka M, 2015. Simultaneous registration of location and orientation in intravascular ultrasound pullbacks pairs via 3D graph-based optimization. IEEE Transactions on Medical Imaging 34, 2550–2561. doi:10.1109/TMI.2015.2444815. [DOI] [PMC free article] [PubMed] [Google Scholar]


