Abstract
In higher vertebrates, the circuit formed by retinal ganglion cells (RGCs) projecting ipsilaterally (iRGCs) or contralaterally (cRGCs) to the brain permits binocular vision and depth perception. iRGCs and cRGCs differ in their position within the retina and in expression of transcription, guidance and activity-related factors. To parse whether these two populations also differ in the timing of their genesis, a feature of distinct neural subtypes and associated projections, we used newer birthdating methods and cell subtype specific markers to determine birthdate and cell cycle exit more precisely than previously. In the ventrotemporal (VT) retina, i- and cRGCs intermingle and neurogenesis in this zone lags behind RGC production in the rest of the retina where only cRGCs are positioned. In addition, within the VT retina, i- and cRGC populations are born at distinct times: neurogenesis of iRGCs surges at E13, and cRGCs arise as early as E14, not later in embryogenesis as reported. Moreover, in the ventral ciliary margin zone (CMZ), which contains progenitors that give rise to some iRGCs in ventral neural retina (Marcucci et al., 2016), cell cycle exit is slower than in other retinal regions in which progenitors give rise only to cRGCs. Further, when the cell cycle regulator Cyclin D2 is missing, cell cycle length in the CMZ is further reduced, mirroring the reduction of both i- and cRGCs in the Cyclin D2 mutant. These results strengthen the view that differential regulation of cell cycle dynamics at the progenitor level is associated with specific RGC fates and laterality of axonal projection.
Keywords: ipsilateral RGCs, contralateral RGCs, neurogenesis, retina, binocular vision, RRID AB_528173, RRID AB_2315623, RRID AB_443209, RRID AB_2556549, RRID AB_2534079, RRID AB_2535812
Graphical Abstract
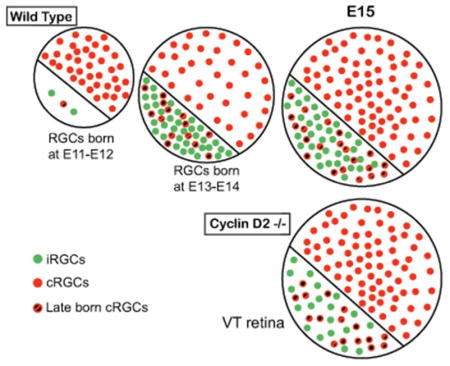
With specific markers and birthdating methods (EdU), we charted the neurogenesis of ipsi- and contralateral RGCs. Ipsi RGC neurogenesis lags behind that of contra RGCs elsewhere in the retina, but is similar to contra RGCs in ventrotemporal retina. Cyclin D2 is important for the production of RGCs from VT retina.
INTRODUCTION
The vast diversity of neurons and glia in the CNS is generated from a small, heterogeneous population of progenitors that undergo transcriptional changes during development to sequentially specify distinct cell fates (Franco & Muller, 2013; Nowakowski et al., 2017). Cell-intrinsic and -extrinsic cues regulate when and how many of each cell class and sub-class are produced to establish specific neural circuits. Previous studies in the cortex have pointed to regional differences in the timing and duration of neurogenesis, reflected in cortical layer production (Greig, Woodworth, Galazo, Padmanabhan, & Macklis, 2013; Polleux, Dehay, & Kennedy, 1997; Suter, Nowakowski, Bhide, & Caviness, 2007). In the spinal cord, extensor and flexor premotor interneurons are spatially segregated medio-laterally, a pattern that is directly related to timing of their neurogenesis (Tripodi, Stepien, & Arber, 2011). Thus, the spatiotemporal features of developmental ontogeny of neurons directly relate to the formation of functional circuits.
Retinal neurons are generated in a distinct temporal sequence from a pool of multipotent progenitor cells. Retinal ganglion cells (RGCs) are generated first, at embryonic day (E) 11, followed by cone photoreceptors, horizontal cells, and amacrine neurons (Dyer & Cepko, 2001). Bipolar neurons, rods, and Müller glia are born postnatally (Bassett & Wallace, 2012). In the retina as elsewhere in the nervous system, transcription factors and non-coding RNAs confer progenitor multipotency, and specify cell classes and subclasses (Bassett & Wallace, 2012). The competence of retinal progenitors to produce a given set of cell types changes over time as directed by specific transcription factor expression (Hafler et al., 2012; Qiu, Jiang, & Xiang, 2008).
Numerous transcription factors act as intrinsic regulators for cell fate determination of distinct retinal cell classes. The basic helix-loop-helix transcription factors Math5 and Neurod1 are required for RGC specification, whereas Islet1 and Brn3b stabilize RGC fate commitment postmitotically (L. Y. Brown, Kottmann, & Brown, 2003; N. L. Brown, Patel, Brzezinski, & Glaser, 2001; Mao, Wang, Pan, & Klein, 2008; Pan, Deng, Xie, & Gan, 2008; S. W. Wang et al., 2001). Timing is also transcriptionally regulated: In the mouse retina, the Drosophila Hunchback ortholog Ikaros regulates the shift in competence from early to later born cell classes (Elliott, Jolicoeur, Ramamurthy, & Cayouette, 2008; Kohwi, Lupton, Lai, Miller, & Doe, 2013; Li et al., 2013). In addition, ablation of Sox11 produces a delay in the birth of RGCs and cones (Usui et al., 2013).
Downstream of retinal cell class specification, retinal subclasses are defined according to their function and projection pattern (Sanes & Masland, 2015). RGCs can be divided into two subtypes with respect to their axonal laterality: contralateral RGCs (cRGCs) project to targets on the opposite side of the brain, whereas ipsilateral RGCs (iRGCs) avoid the optic chiasm midline and remain on the same side of the brain. The transcription factors Zic2 and Islet2 specify i- and cRGCs, respectively (Herrera et al., 2003; Pak, Hindges, Lim, Pfaff, & O’Leary, 2004). iRGCs can be further subdivided into early-born, transient iRGCs arising from the dorsocentral retina at E12 (Soares & Mason, 2015), and “permanent” iRGCs arising from the ventrotemporal (VT) crescent from E14 to 17. cRGCs arise from the dorsocentral retina and from dorso- and ventronasal retina up until E16, and then from the VT retina at E17 - birth (the so-called “late-born” cRGCs) (Drager & Olsen, 1980; Erskine & Herrera, 2014; Petros, Rebsam, & Mason, 2008). While specific transcription factors expressed by the early transient i- or “late-born” cRGCs are not known, SoxCs are strongly expressed by all cRGCs (Kuwajima, Soares, Sitko, Lefebvre, & Mason, 2017) and “late-born” cRGCs in VT retina are the only RGCs affected in Islet2 knockouts (Pak et al., 2004).
Despite the identification of transcription factors involved in retinal cell fate determination, the molecular mechanisms regulating the production of specific subtypes of RGCs, especially in the ventral retina where i- and cRGCs intermingle, at the appropriate time, are poorly understood. Whether i- and cRGCs derive from a common progenitor is also unknown. Recent gene profiling experiments of isolated i- and cRGCs indicate that these two populations differ not only in the expression of genes related to axon guidance but also in genes related to cell cycle regulation and cell fate determination, such as Cyclin D2 (Q. Wang, Marcucci, Cerullo, & Mason, 2016).
Cyclins Ds are a family of proteins that control cell cycle progression by regulating G1 to S phase transition via Rb phosphorylation, (Dyer & Cepko, 2001). Several lines of evidence point to Cyclin Ds as key regulators of cell cycle dynamics and cell fate determination (Tsunekawa et al., 2012), but the exact mechanisms are unclear (Tsunekawa & Osumi, 2012). Our recent work reveals that Cyclin D2 is enriched in the ventral ciliary margin zone (CMZ) (Q. Wang et al., 2016), and that in the absence of Cyclin D2, neurogenesis of i- and cRGC subtypes is diminished and the ipsi projection is decreased (Marcucci et al., 2016). Moreover, live imaging showed that during embryogenesis, cells translocate from the CMZ to the neural retina, implicating the embryonic mammalian CMZ as a source of RGCs (Marcucci et al., 2016). In other brain areas, Cyclin D2 is a cell-type specific regulator of neurogenesis (Glickstein, Monaghan, Koeller, Jones, & Ross, 2009; Petros, Bultje, Ross, Fishell, & Anderson, 2015), but its mechanism of action is not clear.
Alteration of cell cycle dynamics in the retina affects neurogenesis of specific retinal classes. For example, perturbation of Hedgehog signaling in proliferating progenitors in mouse retina leads to altered expression of different cyclins and consequently in cell cycle progression, and increased production of RGCs (Agathocleous, Locker, Harris, & Perron, 2007; Sakagami, Gan, & Yang, 2009). Specific levels of p27 (Kip1) expression are required for producing the appropriate numbers of Müller cells and bipolar neurons (Dyer & Cepko, 2001; Levine, Close, Fero, Ostrovsky, & Reh, 2000; Ohnuma, Philpott, Wang, Holt, & Harris, 1999). Hence, factors that regulate length of cell cycle or cell cycle exit determine the balance between retinal progenitor proliferation and differentiation, thereby affecting cell class fate. However, few studies have addressed whether i- and cRGC fate determination occurs through mechanisms that involve distinct timing of precursor cell cycle exit and birth.
In this study, we expanded upon previous results examining i- and cRGC neurogenesis, using EdU as a birthdating marker and RGC subtype-specific markers, to further parse neurogenic timing of i- and cRGCs. We focused specifically on the genesis of RGCs within VT retina, using dorsotemporal retina as a comparison site, at different stages of retinal development. In addition, we investigated differences in cell cycle exit between these regions as well as in the ciliary margin zone (CMZ), where Cyclin D2 is expressed, in wild type and Cyclin D2 mutants.
We found that neurogenesis in VT retina is delayed when compared to DT retina, and that within VT retina i- and cRGCs are defined by distinct times of birth, the iRGCs peaking at E13. Moreover, the so-called “late-born” VT Islet2+ cRGCs are produced earlier than anticipated. Finally, progenitors that localize to the ventral peripheral retina and ventral CMZ exit cell cycle at a slower rate than progenitors localized elsewhere in the retina, and progenitor cell cycle exit within the CMZ is dependent on Cyclin D2. Since progenitors giving rise to iRGC reside in the ventral peripheral retina and ventral CMZ, we hypothesize that the production of i- vs cRGC is determined by a differential regulation of progenitor cell cycle timing.
MATERIALS AND METHODS
Mouse breeding
Mice were housed in a timed-pregnancy breeding colony at Columbia University. Conditions and procedures were approved by the Columbia University Institutional Animal Care and Use Committee, protocol numbers AAAG8702 and AAAG9259. Females were checked for vaginal plugs at approximately noon each day. Embryonic time E0.5 corresponds to the day when the vaginal plug was detected, with the assumption that conception took place at approximately midnight. The C57BL/6 mouse line was obtained from the Jackson Laboratory (Bar Harbor, ME). Cyclin D2 deficient mice in which exons 1 and 2 have been replaced with a neomycin resistance cassette were genotyped as originally described (Sicinski et al., 1996) and were shared by the Ross lab.
Tissue preparation
Embryos younger than E16.5 were fixed by immersion with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS, pH7.4). Embryos aged E16.5 and older were intracardially perfused with 4% PFA. After fixation, tissue was washed with PBS, cryoprotected with 10% sucrose in PBS for 24 to 72 hours at 4°C, and frozen in dry ice. Consecutive coronal sections (14 μm) were cut with a cryostat (Leica Biosystems, Buffalo Grove, IL, USA) and collected on Fisherbrand Frosted microscope slides.
Immunohistochemistry (IHC)
For immunohistochemistry, slides were rinsed in PBS, and antigen retrieval was performed: slides were incubated in Sodium Citrate buffer (10mM Sodium Citrate, 0.05% Tween 20, pH 6.0) for 20 min at 96°C, followed by 20 min at room temperature. Slides were then washed in PBS for 5 min and blocked in 10% NGS, 0.2% Tween in PBS for 1 hour, and incubated in primary antibodies diluted in 0.2% Tween, 1% NGS, in PBS overnight at 4°C. After the primary antibodies incubation, the slides were washed 3 times for 10 minutes in PBS at room temperature, incubated in secondary antibodies for 2 hours at room temperature, followed by washes in PBS 3 times for 10 minutes at room temperature, and mounted with Fluor-Gel. For a list of the primary and secondary antibodies used, please refer to Table 1.
Table 1.
Antibody Table
| Antibody | Antigen | Immunogen | Source, species, clonality, cat #, RRID |
Dilution |
|---|---|---|---|---|
| Primary | Islet1 (Islet1 antibody recognizes both Islet1 and Islet2) | Rat Islet1, C-terminus, amino acids 178–349 (PEKTTRVRTVLNEKQLHTLRTCYAANPRPDALMKEQLVEMTGLSPRVIRVWFQNKRCKDKKRSIMMKQLQQQQPNDKTNIQGMTGTPMVAASPERHDGGLQANPVEVQSYQPPWKVLSDFALQSDIDQPAFQQLVNFSEGGPGSNSTGSEVASMSSQLPDTPNSMVASPIEA) | Mouse, monoclonal, Developmental Studies Hybridoma Bank, 39.4D5, RRID AB_528173 | 1:100 |
| Primary | Islet2 (Islet2 antibody recognizes Islet2 only) | Mouse Islet2, amino acids 163–176 (CGARGLHLPDAGSGR) | Rabbit, polyclonal, Gift of S. Morton and T. Jessell (Columbia University) | 1:3,000 |
| Primary | Zic2 | Human Zic2, amino acids 428–530 (SSGYESSTPPGLVSPSAEPQSSSNLSPAAAAAAAAAAAAAAAVSAVHRGGGSGSGGAGGGSGGGSGSGGGGGGAGGGGGGSSGGGSGTAGGH SGLSSNF) | Rabbit, polyclonal, RRID AB_2315623 | 1:10,000 |
| Primary | Ki67 | Human Ki67, amino acids 1,200–1,300 (LAGTLPGSKRQLQTPKEKAQALEDLAGFKELFQTPGHTEELVAAGKTTKIPCDSPQSDPVDTPTSTKQRPKRSIRKADVEGELLACRNLMPSAGKAMHTPK) | Rabbit, polyclonal, Abcam, Cat # ab15580, RRID AB_443209 | 1:1,000 |
| Secondary | Goat anti-Mouse IgG Alexa Fluor 594 | Thermo Fisher, Cat # R37121, RRID AB_2556549 | 1:500 | |
| Secondary | Goat anti-Rabbit IgG Alexa Fluor 594 | Thermo Fisher, Cat # A-11012, RRID AB_2534079 | 1:500 | |
| Secondary | Goat anti-Rabbit Alexa 647 | Thermo Fisher, Cat # A-21244, RRID AB_2535812 | 1:250 | |
Antibody characterization
Antigens for the primary antibodies used for IHC are described in Table 1.
Anti-Zic2 is a gift of Dr. Stephen Brown at University of Vermont. Zic2-specific antiserum detects Zic2 protein in CHO cells transfected with Zic2 expression plasmid but not with Zic1 or Zic3 expression plasmids (L. Y. Brown et al., 2003). In addition immunohistochemistry with anti-Zic2 labels specifically RGCs located in the VT region of the retina, and co-labels RGCs retrogradely labeled from the ipsilateral optic tract (Herrera et al., 2003). Hence Zic2-specific antiserum labels ipsilateral RGCs.
Antibodies anti-Islet1 and anti-Islet2 are a gift of Susan Morton and Dr. Thomas Jessell, Columbia University. In the mouse retina a large subset of differentiated RGCs express Islet1, and about 30–50% of differentiated RGCs express Islet2 (A. Brown et al., 2000; Herrera et al., 2003; Thor, Ericson, Brannstrom, & Edlund, 1991). The antibodies anti-Islet1 and anti-Islet2 used in this study stained the appropriate patterns of cellular morphology and distribution corresponding to both subsets of differentiated Islet1 and Islet2 RGCs (See Figures 1 and 3, and (Bhansali, Rayport, Rebsam, & Mason, 2014; Rebsam, Bhansali, & Mason, 2012)). In addition immunoreactivity for this anti-Islet1 antibody is absent from neurons located in the dorsal root ganglia in conditional mutants of Islet1 (Sun et al., 2008).
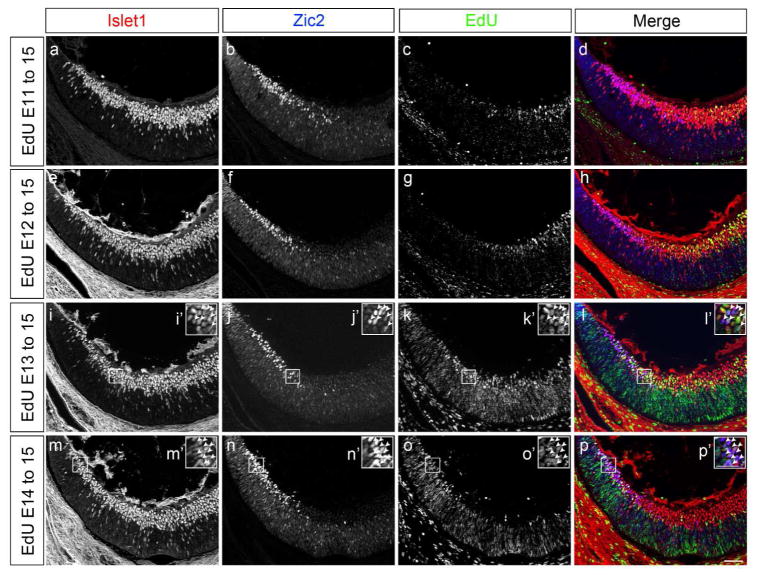
Figure 1. Neurogenesis in ventrotemporal retina increases after E13.
a to p: Frontal sections through ventrotemporal (VT) retina of C57BL/6 mice injected with EdU at E11, 12, 13 or 14, and analyzed at E15.5 with specific retinal ganglion cell (RGC) markers (see Figure 2a and c for schemes of a retinal frontal section, and EdU timeline, respectively). Frontal retinal sections were immunostained with antibodies against Islet1 (postmitotic RGCs) and Zic2 (ipsilateral RGCs), and processed for EdU detection. Panels a to d, e to h, i to l, and m to p correspond to the same frontal section.
a, e, i, m: Channel corresponding to Islet1 immunostaining. Insets i′ and m′ are magnified panels of boxed retinal areas in panels i and m respectively.
b, f, j, n: Channel corresponding to Zic2 immunostaining. Zic2+ RGCs are situated at the most peripheral zone of VT retina. Insets j′ and n′ are magnified panels of boxed retinal areas in panels j and n, respectively.
c, g, k, o: Channel corresponding to EdU detection. Note that prolonging the time interval between EdU injection and analysis results in reduced EdU signal. This is due to increased rounds of progenitor cell division and subsequent dilution of the EdU compound (see Materials and Methods section for further details on EdU detection and analysis). Insets k′ and o′ are magnified panels of boxed retinal areas in panels k and o respectively.
d, h, l, p: Merge channels. Colors are as follows: Islet1 in red, Zic2 in blue, and EdU in green. Insets l′ and p′ are magnified panels of boxed retinal areas in panels l and p, respectively. Arrows highlight Zic2+Islet1+ labeled RGCs with EdU at E13 and E14. Very few Zic2+Islet1+ RGCs are labeled with EdU when EdU is injected at E11 and E12. E, embryonic day. Scale bar in p, 50 μm, applies to panels a to p. Scale bar in p′ 25μm applies to insets i′ to p′.
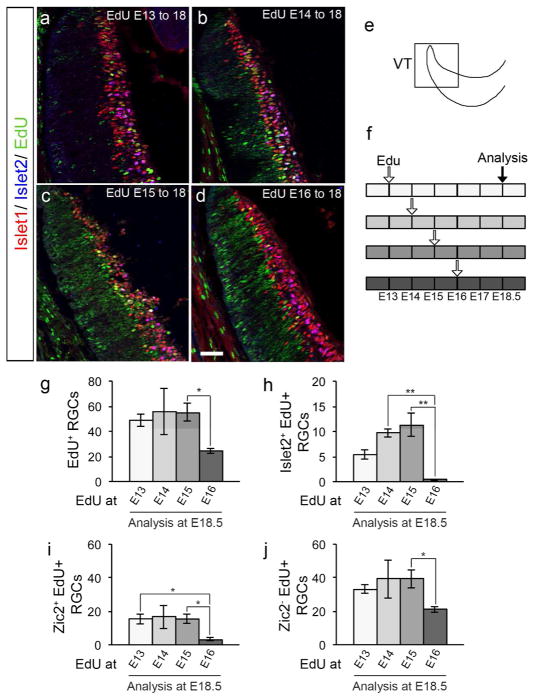
Figure 3. Contralateral Islet2+ RGCs from ventrotemporal retina are generated before E16, earlier than previously thought.
a to d: Frontal sections through ventrotemporal (VT) retina of C57BL/6 mice injected with EdU at E13 (a), 14 (b), 15 (c) or 16 (d), and analyzed at E18.5 with specific retinal ganglion cell (RGC) markers. Frontal retinal sections were immunostained with antibodies against Islet1 (postmitotic RGCs, in red) and Islet2 (contralateral RGCs, in blue), and processed for EdU detection (in green). Scale bar in d, 50 μm, applies to all panels.
e: Scheme of the VT zone of the retina in a frontal section.
f: Timeline of EdU injections at E13, 14, 15 or 16 and sacrifice at E18.5. Each box represents 12 hours, and each empty arrow represents three injections of EdU (10 am, 2 pm, 6 pm). The black arrow represents the time of analysis.
g: Bar graph comparing the number of RGCs labeled with EdU (Islet1+/EdU+ double-positive cells) in VT retina of mice injected with EdU at E13, 14, 15 or 16 and analyzed at E18.5. There is a significant decrease of RGC neurogenesis at E16, suggesting that the RGCs that comprise the VT retina at E18.5 are born before E16.
n=8 for E13, 3 for E14, 8 for E15, and 7 for E16.
h: Bar graph comparing the amount of contralateral Islet2+ RGCs labeled with EdU (Islet1+/Islet2+/EdU+ triple-positive cells) in VT retina of mice injected with EdU at E13, 14, 15 or 16 and analyzed at E18.5. There is a significant decrease of contralateral Islet2+ RGC neurogenesis at E16, suggesting that this subtype of contralateral RGCs found in VT retina after E17-E18 emerges before E16.
n=8 for E13, 5 for E14, 8 for E15, and 6 for E16.
i: Bar graph comparing the number of ipsilateral Zic2+ RGCs labeled with EdU (Islet1+/Zic2+/EdU+ triple-positive cells) in VT retina of mice injected with EdU at E13, 14, 15 or 16 and analyzed at E18.5. There is a significant reduction in ipsilateral Zic2+ RGC neurogenesis at E16, suggesting that ipsilateral RGCs are generated until E16. n=8 for E13, 3 for E14, 8 for E15, and 7 for E16.
j: Bar graph comparing the number of contralateral Zic2− RGCs labeled with EdU (Islet1+/Zic2−/EdU+ double-positive cells) in VT retina of mice injected with EdU at E13, 14, 15 or 16 and analyzed at E18.5. There is a significant decrease in contralateral Zic2− RGC neurogenesis at E16, suggesting that contralateral RGCs are generated in VT retina until E16. Of note, we expect that a population of Zic2− RGCs expresses the contralateral marker Islet2.
n=8 for E13, 3 for E14, 8 for E15, and 7 for E16.
For pairwise comparisons, * when p<0.05, and ** when p<0.01. For details on the statistical analysis, please see Table 2.
Anti-Ki67 was obtained from Abcam (Cat # ab15580). This anti-Ki67 antibody labels proliferative layers on the retina, and immunoreactivity is absent from those retinal layers containing postmitotic cells such as RGCs (Maiorano & Hindges, 2013).
Birthdating and cell cycle analysis
EdU (5-Ethynyl-2′-deoxyuridine, 2.5 mg/ml, InVitrogen) was injected into the pregnant dam intraperitoneally three times, at 10 am, 2 pm, and 6 pm, in animals at embryonic times E11, 12, 13, 14, 15 or 16. EdU, similar to BrdU and [3H]thymidine, is incorporated into DNA and labels cells undergoing S-phase (Salic & Mitchison, 2008). Embryos were sacrificed and collected at E14.5 for cell cycle exit essays or at E15.5, E16.5 or E18.5 for birthdating assays. Embryos were fixed and processed for IHC as per the procedure described below for retinal sections.
After immunostaining for Zic2 and Islet1, sections were permeabilized in 0.5% Tween in PBS for 30 minutes, washed in PBS twice for 5 minutes, and the EdU histochemistry was performed as described (Bhansali et al., 2014; Marcucci et al., 2016; Salic & Mitchison, 2008), using a fluorescent azide through a Cu(I)-catalyzed cycloaddition reaction (Click-iT EdU imaging kit, Invitrogen, Carlsbad, CA, USA), for 30 minutes at room temperature. After the EdU reaction was visible, slides were washed 3 times for 10 minutes in PBS at room temperature. Sections were then coverslipped with Fluoro-Gel. Imaging, quantification and analysis were performed as described below. The EdU signal in retinae injected at E11 or E12 and collected at E15.5, or injected at E13 and collected at E18.5, were thresholded such that only the brightest cells were counted (these are cells that became post-mitotic at E11–13), using the Meta Imaging Series software Metamorph 7.0. The EdU signal in retinae injected at E13 or E14 and collected at E15.5, or injected at E14, E15 or E16 and collected at E18.5, did not require thresholding because the time window between the age of injection and collection was too short for multiple rounds of division, and therefore had less variability in EdU signal strength amongst postmitotic RGCs.
Microscopy
All images were captured with a Zeiss AxioImager M2 microscope equipped with ApoTome, AxioCam MRm camera, and Neurolucida software (V10.40, MicroBrightField Systems, Williston, VT, USA). Brightness and contrast of photomicrographs were adjusted.
Cell number analysis of retinal sections
For quantification of Zic2, Islet1, Islet2, and EdU cell number in C57BL/6 embryos at E15.5, E16.5 or E18.5, a sector of 240 μm long (as measured in the superficial aspect of the retina) was traced starting at the most peripheral Islet1+ RGCs in dorsotemporal (DT) and/or ventrotemporal (VT) retina (Marcucci et al., 2016). Cell number was determined within the 240 μm sector. We averaged cell counts from three consecutive sections caudal to the optic nerve, and analyzed a minimum of three animals per age.
For quantification of Ki67 and EdU cell number in C57BL/6 or Cyclin D2 embryos at E14.5, cell number was determined in the ciliary marginal zone (CMZ), and two sectors from the neural retina: To delineate the region corresponding to the CMZ, a sector of 60 μm was traced ending at the most peripheral Islet1+ RGC of DT and VT retina. To delineate the regions corresponding to the neural retina, DT and VT retina were divided into 4 consecutive sectors of 60 μm in length radially (in the apical to basal axis of the retina), with the first sector traced starting at the most peripheral Islet1+ RGC (and at the border of the CMZ). The first and 4th sectors were counted and considered as neural retina and central retina, respectively. We averaged cell counts from two consecutive sections caudal to the optic nerve, and analyzed a minimum of three animals per experimental group.
Cell numbers were recorded blind to genotype within the assigned sectors using Meta Imaging Series Metamorph 7.0 (Molecular Devices, Sunnyvale, CA, USA). For EdU labeling, images were thresholded with Metamorph such that only the most strongly labeled cells were counted. For Zic2, Islet1, Islet2 and Ki67 immunolabeling, no threshold was necessary as background staining was not noticeable.
Statistical analysis
To determine statistical significance, all data were analyzed with GraphPad Instat version 3.1a (GraphPad software, La Jolla, CA) and graphs were constructed using Microsoft Excel. All error bars are expressed as the standard error of the mean (SEM). Statistical significance was determined using Krushal-Wallis test for non-parametric ANOVA with Dunn posttest if p<0.05, or on-way ANOVA for parametric ANOVA with Tukey-Kramer posttest, or using Mann-Whitney test for pairwise comparisons. For a summary of the statistical analysis performed in this study, please refer to Table 2.
Table 2.
Summary Table of Statistical Analysis
| Figure | Statistical Tests and p values |
|---|---|
| Figure 2 |
2b.
ANOVA Kruskal-Wallis test: for VT p < 0.0001 and for DT retina p=0.3493. Dunn’s multiple comparisons test for VT: E11 vs E12 p>0.05, E11 vs E13 p<0.05, E11 vs E14 p<0.001, E12 vs E13 p>0.05, E12 vs E14 p<0.01, E13 vs E14 p>0.05. Mann-Whitney for pairwise comparisons VT vs DT: E11 p=0.0022, E12 p=0.0028, E13 p=0.0152, and E14 p=0.0061 |
|
2e.
ANOVA Kruskal-Wallis test: for EdU until E15.5 p=0.0002 and for EdU until E16.5 p=0.4472. Dunn’s multiple comparisons test for EdU until E15.5: E11 vs E12 p>0.05, E11 vs E13 p<0.01, E11 vs E14 p<0.01, E12 vs E13 p>0.05, E12 vs E14 p<0.05, E14 vs E15 p>0.05. | |
|
2f.
ANOVA Kruskal-Wallis test: p=0.0011 for EdU until E15.5 and p=0.5225 for EdU until E16.5. Dunn’s multiple comparisons test for EdU until E15.5: E11 vs E12 p>0.05, E11 vs E13 p>0.05, E11 vs E14 p<0.01, E12 vs E13 p>0.05, E12 vs E14 p<0.01, E13 vs E14 p>0.05. Mann-Whitney test for pairwise comparisons: EdU E14 to E15.5 vs EdU E14 to E16.5 p>0.05, and EdU E14 to E15.5 vs EdU E15 to E16.5 p>0.05. | |
| Figure 3 |
3g. ANOVA Kruskal-Wallis test: p=0.0234. Dunn’s multiple comparisons test: E13 vs E14 p>0.05, E13 vs E15 p>0.05, E13 vs E16 p>0.05, E14 vs E15 p>0.05, E14 vs E16 p>0.05, E15 vs E16 p<0.05. |
|
3h. ANOVA Kruskal-Wallis test: p=0.0008. Dunn’s multiple comparisons test: E13 vs E14 p>0.05, E13 vs E15 p>0.05, E13 vs E16 p>0.05, E14 vs E15 p>0.05, E14 vs E16 p<0.01, E15 vs E16 p<0.01. | |
|
3i. ANOVA Kruskal-Wallis test: p=0.0079. Dunn’s multiple comparisons test: E13 vs E14 p>0.05, E13 vs E15 p>0.05, E13 vs E16 p<0.05, E14 vs E15 p>0.05, E14 vs E16 p>0.05, E15 vs E16 p<0.05. | |
|
3j. ANOVA Kruskal-Wallis test: p=0.0308. Dunn’s multiple comparisons test: E13 vs E14 p>0.05, E13 vs E15 p>0.05, E13 vs E16 p>0.05, E14 vs E15 p>0.05, E14 vs E16 p>0.05, E15 vs E16 p<0.05. | |
| Figure 4 |
4c.
Repeated measure analysis of variance: p=0.0013 for ventral retina and p=0.0344 for dorsal retina. Tukey–Kramer test for multiple comparisons: For ventral retina: CMZ vs PR p>0.05, CMZ vs CR p<0.01, PR vs CR p<0.01. For dorsal retina: CMZ vs PR p>0.05, CMZ vs CR p>0.05, PR vs CR p>0.05. |
| 4d. Mann-Whitney test: ventral CMZ Ccnd2+/+ vs Ccnd2−/− p=0.0571, dorsal CMZ Ccnd2+/+ vs Ccnd2−/−p=0.0286, | |
| 4e. Mann-Whitney test: ventral PR Ccnd2+/+ vs Ccnd2−/−p=0.4857, dorsal PR Ccnd2+/+ vs Ccnd2−/− p=0.4857. |
RESULTS
Ipsilateral and contralateral RGCs are generated at different times during retinal development
Previous analyses performed by Ursula Dräger (Drager, 1985), based on birthdating in embryos and retrograde tracing in the adult, suggested that both ipsilaterally (i) and contralaterally (c) projecting RGCs were born as early as embryonic day 11 (E11). Later tracing experiments by Collelo and Guillery showed that c-RGC axons can be found in the optic tract at E14.5, while i- RGC axons do not reach the optic chiasm until ~E16, suggesting a lag between the generation of i- and cRGCs (Colello & Guillery, 1990).
In order to revisit the birthdates of the i- and c- projecting RGCs we combined newer birthdating methods using EdU, a thymidine analog, with the subtype specific RGC markers Zic2 for iRGCs, Islet1 for all RGCs, and Islet 2 for cRGCs. We injected EdU into pregnant dams at E11, 12, 13 and 14, and collected embryos at E15.5 (Figure 1a–p, and Figure 2). We performed our analysis at E15.5 because the number of iRGCs peaks at E15–16 (Herrera et al., 2003). Accordingly, EdU injections were made from the embryonic ages when RGCs become apparent at E11, until prior to the peak of expression of Zic2, at E14.
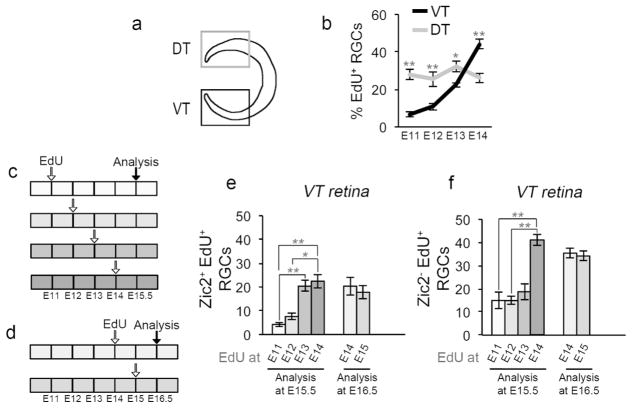
Figure 2. Ipsilateral and contralateral RGC are generated at distinct embryonic times.
a: Cartoon depicting ventrotemporal (VT) and dorsotemporal (DT) regions of retina. Zic2+ ipsilateral RGCs (iRGCs) reside in VT retina exclusively, whereas Zic2− contralateral RGCs (cRGCs) localize to the entire retina, with DT retina comprised of cRGCs only. Islet1 labels all postmitotic RGCs throughout the retina.
b: Quantification of RGC genesis from E11 to E14, in VT and DT retina. EdU was injected into pregnant dams at E11, 12, 13, or 14 as in Figure 1 (see also Figure 2c). Frontal retinal sections were immunostained with antibodies to Islet1 and processed for EdU detection at E15.5. Coincidence of Islet1 and EdU signal indicates that an RGC was born at the time of the EdU injection. RGC production was calculated as the percentage of Islet1+EdU+ double-labeled RGCs, divided by the total number of Islet1+ RGCs. Whereas the production of RGCs in DT retina is constant from E11 to E14, RGC genesis in VT retina is low at E11–12 and increases at E13.
n for VT: n=6 at E11, 9 at E12, 6 at E13, and 7 at E14. n for DT: n=6 for E11, 4 for E12, 6 for E13, and 4 for E14.
c: Timeline of 5-ethynyl-2′-deoxyuridine (EdU) injections at E11, 12, 13, or 14, and sacrifice at E15.5. Each box represents 12 hours, and each empty arrow represents three injections of EdU (10 am, 2 pm, 6 pm). The black arrow represents the time of analysis.
d: Timeline of EdU injections at E14 or 15 and sacrifice at E16.5. Each box represents 12 hours, and each empty arrow represents three injections of EdU (10 am, 2 pm, 6 pm). The black arrow represents the time of analysis.
e: Bar graph comparing the number of ipsilateral Zic2+ RGCs labeled with EdU (identified as Zic2+/Islet1+/EdU+ triple-labeled cells) in VT retina in mice injected with EdU at E11, 12, 13 or 14 and analyzed at E15.5 (left bars), or injected with EdU at E14 or 15 and analyzed at E16.5 (right bars). Ipsilateral RGC neurogenesis in VT retina increases at E13, and remains constant throughout E15. n for EdU until E15.5: n=6 at E11, 9 at E12, 6 at E13, and 7 at E14.
n for EdU until E16.5: n=6 at E14, and 4 at E15.
f: Bar graph comparing the number of Zic2− (contralateral) RGCs labeled with EdU (identified as Zic2−/Islet1+/EdU+ triple-labeled cells) in VT retina in mice injected with EdU at E11, 12, 13 or 14 and analyzed at E15.5 (left bars), or injected with EdU at E14 or 15 and analyzed at E16.5 (right bars). Contralateral RGC neurogenesis in VT retina increases at E14, one day after the surge in iRGC production, and remains constant until at least E15.5. Of note, the majority of Zic2− cRGCs labeled with EdU at E14.5 do not express Zic2 at E15.5 (left bars) or at E16.5 (right bars), suggesting that these RGCs do not upregulate the ipsilateral marker Zic2 and are in all likelihood contralateral.
n for EdU until E15.5: n=6 at E11, 9 at E12, 6 at E13, and 7 at E14. n for EdU until E16.5: n=6 at E14, and 4 at E15.
For pairwise comparisons, * when p<0.05, and ** when p<0.01. For details on the statistical analysis, please see Table 2.
First, we compared neurogenesis between two peripheral retinal zones, the dorsotemporal (DT) retina, which produces only cRGCs, and the VT retina, which produces both i- and cRGCs. While RGCs that populate the DT retina are generated at a constant rate at the ages analyzed, VT retina shows a delay in RGC production, with few cells labeled with EdU after injections at E11 or E12 and examination at E15 (Figure 1c and g). We observed that most of the RGCs that populate the VT retina at E15.5 are born between E13–14 (Figures 1k and o, 2a–b). This finding suggests a distinct temporal regulation of neurogenesis between DT and VT zones of the retina during embryogenesis.
We then specifically analyzed the time of birth of i- and cRGCs within VT retina by chronicling Zic2+ or Zic2− RGCs labeled with EdU, respectively (Figure 2c). We observed that most ipsilateral (Zic2+) RGCs found in VT retina at E15.5 are generated between E13 and E14 (Figure 2e), whereas the production of contralateral (Zic2−) RGCs significantly increases one day later at E14 (Figure 2f). To corroborate that Zic2− RGCs born at E14 and analyzed at E15.5 do not express Zic2 at a later stage, we performed EdU pulses at E14 or at E15 and analyzed Zic2+ and Zic− RGCs labeled with EdU in VT retina at E16.5 (Figure 2d, e, f). We observed a similar number of Zic2+ and Zic2− RGCs labeled with EdU from E14 or E15 to E16.5, compared to the number of RGCs labeled with EdU from E14 to E15.5. This result suggests that the majority of Zic2− RGCs produced at E14 do not express the ipsilateral marker Zic2 and are likely cRGCs. Together, these results demonstrate that within VT retina, i- and cRGCs subtypes are born in sequential and overlapping neurogenic waves, and that this process is tightly timed.
Islet2+ contralateral RGCs that reside in VT retina are generated prior to E16
Towards the end of embryogenesis, from E17 to birth, the VT retina produces RGCs that project contralaterally (Petros et al., 2008). These RGCs have been termed “late-born” cRGCs in VT retina and can be identified by the expression of Islet2, a transcription factor expressed by ~30–50% of cRGCs throughout the retina and upregulated in VT retina at E17.5 (A. Brown et al., 2000; Pak et al., 2004). We used EdU birthdating at E13, E14, E15 or E16 in combination with the cell subtype specific markers Islet2 for cRGCs, Islet1 for all differentiated RGCs, and Zic2 for iRGCs, and analyzed the retina at E18.5 to determine when late-born VT cRGCs are generated (Figure 3a–f). With increased time between EdU injection and the day of analysis, additional rounds of cell division cause dilution of the EdU label. Since EdU injections at E11 or at E12 did not produce robust and quantifiable numbers of labeled cells within VT retina at E18.5, we began EdU injections at E13.
By quantifying Islet1+EdU+ cells in VT retina at E18.5, we observed that RGC proliferation during late development is substantial until E15 and decreases thereafter (Figure 3g). When we specifically analyzed the generation of cRGCs by quantifying Islet2+EdU+ RGCs in VT retina at E18.5, we observed that Islet2+ cRGC production increases until E15 and sharply decreases at E16 (Figure 3h). Together, these experiments suggest that the so-called “late-born” Islet2+ cRGCs in VT retina are generated substantially earlier than reported (Drager, 1985; Dräger & Olsen, 1980), primarily before E15, overlapping with iRGC neurogenesis (see Figure 2). Interestingly, quantification of Islet2+EdU+ RGCs in DT retina at E18.5, suggested that proliferation of cRGCs is also nominal at E16 (data not shown).
In addition, we performed immunohistochemistry in the experimental series using antibodies to Zic2 to label Zic2+ iRGCs, and by default, Zic2− staining to represent cRGCs. We hypothesize that Islet2+ cRGCs are a subset of Zic2− cRGCs. We found that the generation of both Zic2+ iRGCs and Zic2− cRGCs is significantly reduced at E16 (Figure 3i–j). Altogether, these results suggest that the VT retina gives rise to new RGCs until E15, and that at E16 proliferation is drastically reduced, for both i- and cRGC subtypes.
Progenitors that give rise to ipsilateral RGCs exit the cell cycle at a slower rate
As cell cycle length influences time of neuronal birth (Lanctot et al., 2017), we hypothesize that progenitors that give rise to i- and c-RGCs may display differences in cell cycle parameters, in particular, differences in the timing of cell cycle exit. Although we cannot distinguish between progenitors that will generate i- or cRGCs prior to differentiation, the peripheral ventral retina houses Zic2+ iRGCs that intermingle with cRGCs. In contrast, only Zic2− cRGCs are found in the central zone of the retina surrounding the optic nerve head. Hence, we compared cell cycle exit in these two regions of the retina, peripheral and central ventral retina, to the ciliary margin zone (CMZ) at the very peripheral tip of the retina that is devoid of differentiated RGCs (Islet1+) and where progenitors that give rise to a subset of i- and cRGCs reside (Marcucci et al., 2016) (Figure 4a and b).
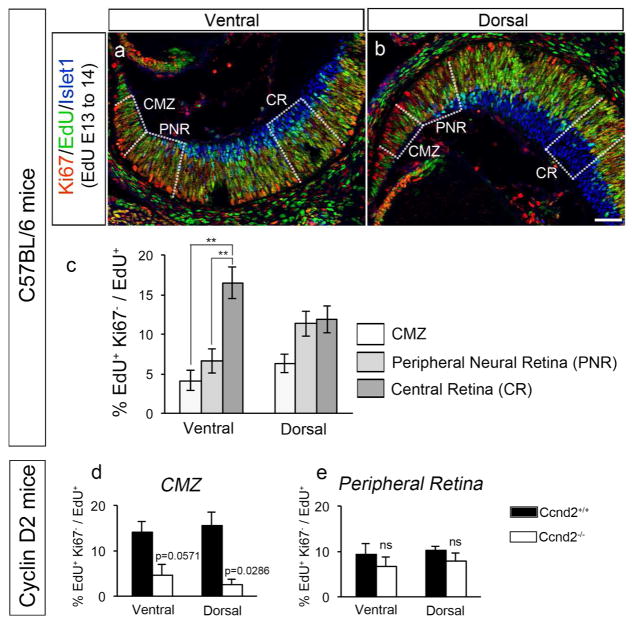
Figure 4. Fewer cells exit the cell cycle in the CMZ and peripheral ventral retina, and this process in the CMZ is Cyclin D2-dependent.
a and b: Frontal sections through ventral (panel a) and dorsal (panel b) retina of C57BL/6 mice injected with EdU at E13 and analyzed at E14.5 with specific markers. Frontal retinal sections were immunostained with antibodies against Ki67 (cells in the cell cycle, in red) and Islet1 (postmitotic RGCs, in blue) and, processed for EdU detection (in green). The three zones in which quantifications were performed for the bar graph in panel c are delineated by dashed lines: Ciliary Margin Zone (CMZ), Peripheral Neural Retina (PNR), and Central Retina (CR). Scale bar in b, 50 μm, applies to both panels.
c: Bar graph comparing the rate of cell cycle exit in the three retinal zones delineated in panels a and b for ventral and dorsal retina of C57BL/6 mice. Cell cycle exit was calculated as the ratio of the number of EdU+ cells that did not express the proliferation marker Ki67 (EdU+Ki67−) divided by total EdU+ cells (see Materials and Methods section for further details). Fewer cells exit the cell cycle in the CMZ and ventral peripheral neural retina (PNR) zone, where both i- and cRGCs reside, compared with the dorsal PNR. n= 6 for CMZ, 6 for PNR and 6 for CR.
d and e: Bar graphs comparing the rate of cell cycle exit in ventral and dorsal retina of Cyclin D2 mutant mice and wild type littermates injected with EdU at E13 and analyzed at E14.5 with specific markers. As in panels a to c, retinal sections were immunostained with antibodies against Ki67 (cells undergoing cell cycle, in red) and Islet1 (postmitotic RGCs, in blue) and processed for EdU detection (in green). Cell cycle exit was calculated as the ratio between EdU+ cells that did not express the proliferation marker Ki67 (EdU+Ki67−) divided by total EdU+ cells in the CMZ (panel d) and Peripheral Neural Retina (panel e) zones. The cell cycle regulator Cyclin D2 is expressed in both dorsal and ventral CMZ, and is particularly enriched in ventral CMZ. Nonetheless, fewer cells exit cell cycle in both ventral and dorsal CMZ in the absence of Cyclin D2. In contrast, cell cycle exit is not affected in Cyclin D2−/− peripheral neural retina, a zone of the retina that does not express Cyclin D2.
n=4 for Ccnd2+/+ and 4 for Ccnd2−/−.
For pairwise comparisons, ** when p<0.01. For details on the statistical analysis, please see Table 2.
To estimate cell cycle exit during the developmental stage when progenitors that produce i- and/or cRGCs undergo active proliferation, we injected EdU in E13 wild-type C57BL/6 pregnant dams and analyzed ventral and dorsal retina at E14.5 for Ki67, a cellular marker present during all active phases of the cell cycle (G1, S, G2 and mitosis) but absent from resting cells (cells in G0). We quantified EdU+Ki67− (cells that were proliferating at E13, but have left the cell cycle by E14) and total EdU+ cells (these comprise all proliferating progenitors at E13) in three sectors corresponding to the peripheral and central retina and the central ciliary margin zone (CMZ), an area of the retina that is devoid of differentiated RGCs (Islet1+) and where progenitors giving rise to a subset of i- and cRGCs reside (Marcucci et al., 2016) (Figure 4a–b).
Within the ventral retina, there is a significant decrease in the percentage of progenitors that exit cell cycle in the ventral peripheral neural retina and the CMZ, when compared with the ventral central neural retina (Figure 4c). This reduced rate of cell cycle exit is specific to the ventral peripheral zone as in the dorsal peripheral zone the number of cells exiting cycle in the dorsal CMZ and peripheral neural retina is comparable to that in the dorsal central zone. Since the ventral CMZ and ventral peripheral retina harbor progenitors giving rise to both i- and cRGCs, while the dorsal CMZ and dorsal peripheral retina house progenitors giving rise to cRGC only, we hypothesize that the observed reduced cell cycle exit is associated with progenitors destined to produce primarily iRGCs. Therefore, we speculate that in wild type conditions, progenitors giving rise to iRGC exit cell cycle at a slower rate.
Cyclin D2 influences cell cycle exit in the CMZ
Several lines of evidence point to Cyclin D2 as a key regulator of cell cycle progression and neurogenesis (Glickstein et al., 2009; Komada, Iguchi, Takeda, Ishibashi, & Sato, 2013; Petros et al., 2015; Tsunekawa et al., 2012; Tsunekawa & Osumi, 2012). Gene profiling revealed that Cyclin D2 is differentially expressed in the mouse retina, with more cells expressing Cyclin D2 in the ventral than in the dorsal CMZ (Q. Wang et al., 2016). In addition, in the absence of Cyclin D2, a reduced number of i- and cRGCs are found in VT retina and the ipsilateral projection is reduced, indicating that a subset of i- and cRGC progenitors reside in the ventral CMZ (Marcucci et al., 2016). Based on these previous results, we hypothesized that Cyclin D2 regulates neurogenesis of VT RGCs by influencing cell cycle progression, and therefore cell cycle exit, of progenitors localized in the CMZ.
We analyzed the fraction of cells that exit the cell cycle by injecting EdU at E13 and determining the coincidence of EdU and Ki67 in the CMZ at E14.5, comparing Cyclin D2−/− to Cyclin D2+/+ littermates. Cyclin D2 is enriched in ventral CMZ and expressed at low levels in dorsal CMZ, and is absent from the neural retina, including the sectors named as peripheral neural retina in Figure 4a–b (Marcucci et al., 2016; Q. Wang et al., 2016). Hence, we compared cell cycle exit in ventral and dorsal CMZ and in ventral and dorsal peripheral retina. We observed that cell exit is reduced in both ventral and dorsal CMZ of Cyclin D2−/− mice compared with cell cycle exit in wild type CMZ. However, cell cycle exit is similar in within the peripheral retina of wild type and Cyclin D2−/− mice. This result parallels the reduction of CMZ-derived RGCs and the reduction of cells in M phase as indicated by PH3 immunolabeling observed in our previous publication (Marcucci et al., 2016), and suggests that Cyclin D2 is necessary for normal cell cycle exit in RGC progenitors and consequent RGC generation. Altogether, these findings implicate regulation of cell cycle dynamics as a mechanism to balance RGC output and specification.
DISCUSSION
Time of birth is a key determinant of neuronal fate in the mammalian neocortex, olfactory bulb, spinal cord, and retina (Bassett & Wallace, 2012; Batista-Brito, Close, Machold, & Fishell, 2008; Molyneaux, Arlotta, Menezes, & Macklis, 2007; Tripodi et al., 2011). In the mouse, ipsilateral (i) axons arise exclusively from the ventrotemporal (VT) retina, whereas contralateral (c) axons arise from the entire retina. Together the balanced production of i- and c- axons enables binocular vision and depth perception (Erskine & Herrera, 2014; Petros et al., 2008). Recent work has shown that i- and cRGCs differ not only in their projection pattern but also in the expression of transcription, guidance, and activity-related factors (Garcia-Frigola & Herrera, 2010; Q. Wang et al., 2016).
Our results indicate that during the peak phase of retinal axon growth, neurogenesis in the VT retina lags behind neurogenesis in DT retina, the latter region giving rise to cRGCs only. Our results agree with the data published by Drager more than three decades ago indicating that VT retina and non-VT retina proliferate in distinct yet overlapping neurogenic waves (Drager, 1985). However, due to unavailable cellular markers at the time, the latter study could not readily distinguish between i- and cRGC populations as they differentiate and are positioned together within the VT retina. Further, with cell-specific markers, we show that within the VT retina, the generation of iRGCs is upregulated at E13 and cRGCs at E14, suggesting that these two RGCs subtypes projecting to opposite sides of the brain are produced in sequential and overlapping neurogenic waves. This finding complements recent work from the Huberman lab, where functionally distinct subtypes of RGCs with different axon growth strategies and projection patterns to visual nuclei in the brain also differ in their time of cell birth (Osterhout, El-Danaf, Nguyen, & Huberman, 2014).
Our results also indicate that the cRGCs that constitute the late phase of retinal axon growth from E17 until birth, are born at E15, earlier than previously suggested (A. Brown et al., 2000; Pak et al., 2004). Therefore, the RGC subtypes known to project binocularly from VT retina (ipsilateral Zic2+, and contralateral Zic2−, among which are the contralateral Islet2+ RGCs) are produced in the VT retina at more or less the same time. Interestingly, the transcription factor Islet2 is upregulated in the VT retina at E17.5 (Pak et al., 2004). This suggests that in the VT retina there is a lag between the time when Islet2 contralateral cells undergo their last round of cell division and when they begin to express Islet2. In future experiments, it will be of interest to address whether RGCs destined to express Islet2 extend axons before or after onset of Islet2 expression.
One caveat in our studies is the transient nature of Zic2 expression: Zic2 is upregulated when iRGCs become postmitotic and it is downregulated once their axons have reached the optic chiasm (Herrera et al., 2003). As the first ipsilateral axons are observed in the optic chiasm at around E16 (Colello & Guillery, 1990), we predict that our quantification at E15.5 (Figure 1 and 2) represents an accurate approximation of the number of Zic2+ RGCs in VT retina. However, our quantification of Zic2+ RGCs at E16.5 (Figure 2) and, in particular, at E18.5 (Figure 3), might underestimate the actual number of iRGCs in VT retina. To overcome such concerns, permanent labeling of iRGCs should be performed, for example, by using a Zic2-Cre dependent mouse line.
One question that remains unanswered is whether i- and cRGCs arise from a common progenitor pool or whether they originate from distinct fate-restricted progenitor pools. Recent evidence in the developing cortex indicates that upper cortical neurons originate from a set of Cux2+ fate-restricted progenitors, which challenges the classical view that all cortical neurons classes originate from a common multipotent progenitor pool that changes competence during time (Franco et al., 2012; Gil-Sanz et al., 2015). In recent work, we have shown that a subset of i- and cRGCs that reside in VT retina originate from a spatially segregated progenitor pool localized to the CMZ (Marcucci et al., 2016). Hence, the origin of different neuronal subtypes appears to be more heterogeneous than previously imagined. We also showed that in the absence of the cell cycle regulator Cyclin D2, fewer cells in the ventral CMZ of Cyclin D2−/− undergo mitosis and fewer iRGCs are born in VT retina (Marcucci et al., 2016). The finding in the present study that in the absence of Cyclin D2, cell cycle exit is slower in the CMZ could explain the deficit in i- and cRGC number seen in the Cyclin D2 mutant. Nonetheless, fate-mapping analysis will be important to reconcile whether the CMZ-derived precursors become both i- and cRGCs and whether they are distinct in their genetic profile from other VT-retinal derived RGCs.
Cell cycle length, and therefore cell cycle exit, is thought to regulate cellular birth for a balanced production of retinal and cortical cell types (Alexiades & Cepko, 1996; Suter et al., 2007). Since i- and cRGCs in retinal regions outside of the VT retina are born at distinct times, we hypothesized that cell cycle exit could also differ between these two RGCs subtypes. In line with this hypothesis, we find that fewer cells exit cell cycle in peripheral neural retina, from which the majority of iRGCs arise and where they reside. In comparison, cell cycle exit is more rapid in retinal sectors where cRGCs reside.
In addition, and supporting the hypothesis that modulation of cell cycle dynamics can act as a mechanism to fine tune neuronal production during development (Pauklin & Vallier, 2013), in the absence of the cell cycle regulator Cyclin D2, the fraction of cells that leave cell cycle in the CMZ, where Cyclin D2 is expressed, is reduced. This result mirrors previous studies where we found a decreased production of RGCs in Cyclin D2−/−, supporting the idea that a subset of RGCs arises from the CMZ in a Cyclin D2-dependent fashion, or from Cyclin D2+ progenitors (Marcucci et al., 2016). Accordingly, cell cycle exit in the adjacent peripheral neural retina, where Cyclin D2 is not expressed, is not affected. One limitation of the cell cycle exit analyses shown here in C57BL/6 and Cyclin D2 mice is that we did not address whether a longer cell cycle leads to a reduced fraction of progenitors that exit cell cycle. Future measurements of the length of different phases of cell cycle and total cell cycle length should provide an answer to this caveat.
We previously reported that the albino retina has fewer RGCs that express Zic2, a marker for iRGCs (Herrera et al., 2003), reflecting the decreased ipsilateral projection and abnormal axonal targeting in the dorsal lateral geniculate nucleus (Rebsam et al., 2012). Recent birthdating studies indicate that RGC production is delayed by about a day in the albino VT retina when compared to pigmented retina (Bhansali et al., 2014). This delay correlates with the decrease in the number of RGCs expressing Zic2 (Herrera et al., 2003), and with a later increase in RGCs expressing Islet2 within the VT retina, leading to a supplemental and segregated contralateral retinogeniculate projection (Bhansali et al., 2014). Interestingly, Cyclin D2 expression is diminished in albino CMZ compared with pigmented CMZ (Marcucci et al., 2016). Although we do not yet understand whether factors from the RPE, that may be altered in the albino due to the absence of melanin, influence VT genesis and subtype determination, these results further suggest that the time of birth of VT RGCs is a strong predictor of their cell fate determination associated with laterality of axonal projection.
In summary, regulation of cell cycle in progenitors localized to distinct retinal compartments emerges as a key mechanism to modulate neurogenic timing and therefore the origin of specific RGC subtypes.
Acknowledgments
We thank Dr. Peter Sicinski (Harvard) for sharing the Cyclin D2 mice through Elizabeth Ross’ lab (Cornell), Mika Melikyan for mouse breeding and genotyping, and Jane Dodd and members of the Mason and Herrera lab for discussion and comments on the manuscript. Supported by NIH grants R01 EY012736 and R01 EY015290 (CM), and Fight for Sight (FM).
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
References
- Agathocleous M, Locker M, Harris WA, Perron M. A general role of hedgehog in the regulation of proliferation. Cell Cycle. 2007;6(2):156–159. doi: 10.4161/cc.6.2.3745. 3745 [pii] [DOI] [PubMed] [Google Scholar]
- Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205(3):293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [pii] [DOI] [PubMed] [Google Scholar]
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35(9):565–573. doi: 10.1016/j.tins.2012.05.004. S0166-2236(12)00087-2 [pii] [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Close J, Machold R, Fishell G. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci. 2008;28(15):3966– 3975. doi: 10.1523/JNEUROSCI.5625-07.2008. 28/15/3966 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhansali P, Rayport I, Rebsam A, Mason C. Delayed neurogenesis leads to altered specification of ventrotemporal retinal ganglion cells in albino mice. Neural Dev. 2014;9:11. doi: 10.1186/1749-8104-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Yates PA, Burrola P, Ortuno D, Vaidya A, Jessell TM, … Lemke G. Topographic mapping from the retina to the midbrain is controlled by relative but not absolute levels of EphA receptor signaling. Cell. 2000;102(1):77–88. doi: 10.1016/s0092-8674(00)00012-x. [DOI] [PubMed] [Google Scholar]
- Brown LY, Kottmann AH, Brown S. Immunolocalization of Zic2 expression in the developing mouse forebrain. Gene Expr Patterns. 2003;3(3):361– 367. doi: 10.1016/s1567-133x(03)00043-7. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128(13):2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colello RJ, Guillery RW. The early development of retinal ganglion cells with uncrossed axons in the mouse: retinal position and axonal course. Development. 1990;108(3):515–523. doi: 10.1242/dev.108.3.515. [DOI] [PubMed] [Google Scholar]
- Drager UC. Birth dates of retinal ganglion cells giving rise to the crossed and uncrossed optic projections in the mouse. Proc R Soc Lond B Biol Sci. 1985;224(1234):57–77. doi: 10.1098/rspb.1985.0021. [DOI] [PubMed] [Google Scholar]
- Drager UC, Olsen JF. Origins of crossed and uncrossed retinal projections in pigmented and albino mice. J Comp Neurol. 1980;191(3):383–412. doi: 10.1002/cne.901910306. [DOI] [PubMed] [Google Scholar]
- Dräger UC, Olsen JF. Origins of crossed and uncrossed retinal projections in pigmented and albino mice. J Comp Neurol. 1980;191:383–412. doi: 10.1002/cne.901910306. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2(5):333–342. doi: 10.1038/3507255535072555. [pii] [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60(1):26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Erskine L, Herrera E. Connecting the retina to the brain. ASN Neuro. 2014;6(6) doi: 10.1177/1759091414562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Muller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337(6095):746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77(1):19–34. doi: 10.1016/j.neuron.2012.12.022. S0896- 6273(12)01161-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Frigola C, Herrera E. Zic2 regulates the expression of Sert to modulate eye-specific refinement at the visual targets. EMBO J. 2010;29(18):3170– 3183. doi: 10.1038/emboj.2010.172. emboj2010172 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Sanz C, Espinosa A, Fregoso SP, Bluske KK, Cunningham CL, Martinez- Garay I, … Muller U. Lineage Tracing Using Cux2-Cre and Cux2- CreERT2 Mice. Neuron. 2015;86(4):1091–1099. doi: 10.1016/j.neuron.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickstein SB, Monaghan JA, Koeller HB, Jones TK, Ross ME. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci. 2009;29(30):9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler BP, Surzenko N, Beier KT, Punzo C, Trimarchi JM, Kong JH, Cepko CL. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci U S A. 2012;109(20):7882–7887. doi: 10.1073/pnas.1203138109. 1203138109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, Mikoshiba K, … Mason CA. Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell. 2003;114(5):545–557. doi: 10.1016/s0092-8674(03)00684-6. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152(1–2):97–108. doi: 10.1016/j.cell.2012.11.049. S0092- 8674(12)01437-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M, Iguchi T, Takeda T, Ishibashi M, Sato M. Smoothened controls cyclin D2 expression and regulates the generation of intermediate progenitors in the developing cortex. Neurosci Lett. 2013;547:87–91. doi: 10.1016/j.neulet.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Kuwajima T, Soares CA, Sitko AA, Lefebvre V, Mason C. SoxC Transcription Factors Promote Contralateral Retinal Ganglion Cell Differentiation and Axon Guidance in the Mouse Visual System. Neuron. 2017;93(5):1110–1125. e1115. doi: 10.1016/j.neuron.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot AA, Guo Y, Le Y, Edens BM, Nowakowski RS, Feng Y. Loss of Brap Results in Premature G1/S Phase Transition and Impeded Neural Progenitor Differentiation. Cell Rep. 2017;20(5):1148–1160. doi: 10.1016/j.celrep.2017.07.018. [DOI] [PubMed] [Google Scholar]
- Levine EM, Close J, Fero M, Ostrovsky A, Reh TA. p27(Kip1) regulates cell cycle withdrawal of late multipotent progenitor cells in the mammalian retina. Dev Biol. 2000;219(2):299–314. doi: 10.1006/dbio.2000.9622. S0012-1606(00)99622-2 [pii] [DOI] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, … Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498(7455):456–462. doi: 10.1038/nature12319. nature12319 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano NA, Hindges R. Restricted perinatal retinal degeneration induces retina reshaping and correlated structural rearrangement of the retinotopic map. Nat Commun. 2013;4:1938. doi: 10.1038/ncomms2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Wang SW, Pan P, Klein WH. Rewiring the retinal ganglion cell gene regulatory network: Neurod1 promotes retinal ganglion cell fate in the absence of Math5. Development. 2008;135(20):3379–3388. doi: 10.1242/dev.024612. dev.024612 [pii] [DOI] [PubMed] [Google Scholar]
- Marcucci F, Murcia-Belmonte V, Wang Q, Coca Y, Ferreiro-Galve S, Kuwajima T, … Herrera E. The Ciliary Margin Zone of the Mammalian Retina Generates Retinal Ganglion Cells. Cell Rep. 2016;17(12):3153–3164. doi: 10.1016/j.celrep.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, … Kriegstein AR. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358(6368):1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S, Philpott A, Wang K, Holt CE, Harris WA. p27Xic1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell. 1999;99(5):499–510. doi: 10.1016/s0092-8674(00)81538-x. S0092-8674(00)81538-X [pii] [DOI] [PubMed] [Google Scholar]
- Osterhout JA, El-Danaf RN, Nguyen PL, Huberman AD. Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Rep. 2014;8(4):1006–1017. doi: 10.1016/j.celrep.2014.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak W, Hindges R, Lim YS, Pfaff SL, O’Leary DD. Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell. 2004;119(4):567–578. doi: 10.1016/j.cell.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135(11):1981– 1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155(1):135–147. doi: 10.1016/j.cell.2013.08.031. S0092-8674(13)01025-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros TJ, Bultje RS, Ross ME, Fishell G, Anderson SA. Apical versus Basal Neurogenesis Directs Cortical Interneuron Subclass Fate. Cell Rep. 2015;13(6):1090–1095. doi: 10.1016/j.celrep.2015.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros TJ, Rebsam A, Mason CA. Retinal axon growth at the optic chiasm: to cross or not to cross. Annu Rev Neurosci. 2008;31:295–315. doi: 10.1146/annurev.neuro.31.060407.125609. [DOI] [PubMed] [Google Scholar]
- Polleux F, Dehay C, Kennedy H. The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. J Comp Neurol. 1997;385(1):95–116. doi: 10.1002/(SICI)1096-9861(19970818)385:1<95::AID-CNE6>3.0.CO;2-7. [pii] [DOI] [PubMed] [Google Scholar]
- Qiu F, Jiang H, Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J Neurosci. 2008;28(13):3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. 28/13/3392 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsam A, Bhansali P, Mason CA. Eye-specific projections of retinogeniculate axons are altered in albino mice. J Neurosci. 2012;32(14):4821– 4826. doi: 10.1523/JNEUROSCI.5050-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami K, Gan L, Yang XJ. Distinct effects of Hedgehog signaling on neuronal fate specification and cell cycle progression in the embryonic mouse retina. J Neurosci. 2009;29(21):6932–6944. doi: 10.1523/JNEUROSCI.0289-09.2009. 29/21/6932 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY, … Weinberg RA. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384(6608):470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- Soares CA, Mason CA. Transient ipsilateral retinal ganglion cell projections to the brain: Extent, targeting, and disappearance. Dev Neurobiol. 2015;75(12):1385–1401. doi: 10.1002/dneu.22291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11(11):1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Nowakowski RS, Bhide PG, Caviness VS. Navigating neocortical neurogenesis and neuronal specification: a positional information system encoded by neurogenetic gradients. J Neurosci. 2007;27(40):10777–10784. doi: 10.1523/JNEUROSCI.3091-07.2007. 27/40/10777 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S, Ericson J, Brannstrom T, Edlund T. The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron. 1991;7(6):881–889. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Stepien AE, Arber S. Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature. 2011;479(7371):61–66. doi: 10.1038/nature10538. [DOI] [PubMed] [Google Scholar]
- Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan SS, Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012;31(8):1879–1892. doi: 10.1038/emboj.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa Y, Osumi N. How to keep proliferative neural stem/progenitor cells: a critical role of asymmetric inheritance of cyclin D2. Cell Cycle. 2012;11(19):3550–3554. doi: 10.4161/cc.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui A, Mochizuki Y, Iida A, Miyauchi E, Satoh S, Sock E, … Watanabe S. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development. 2013;140(4):740–750. doi: 10.1242/dev.090274. dev.090274 [pii] [DOI] [PubMed] [Google Scholar]
- Wang Q, Marcucci F, Cerullo I, Mason C. Ipsilateral and Contralateral Retinal Ganglion Cells Express Distinct Genes during Decussation at the Optic Chiasm. eNeuro. 2016;3(6) doi: 10.1523/ENEURO.0169-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, … Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15(1):24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]