Abstract
RWD domains mediate protein–protein interactions in a variety of pathways in eukaryotes. In budding yeast, the RWD domain protein Csm1 is particularly versatile, assembling key complexes in the nucleolus and at meiotic kinetochores through multiple protein interaction surfaces. Here, we reveal a third functional context for Csm1 by identifying a new Csm1‐interacting protein, Dse3. We show that Dse3 interacts with Csm1 in a structurally equivalent manner to its known binding partners Mam1 and Ulp2, despite these three proteins’ lack of overall sequence homology. We theorize that the unique “clamp” structure of Csm1 and the loose sequence requirements for Csm1 binding have led to its incorporation into at least three different structural/signaling pathways in budding yeast.
Keywords: RWD domain, monopolin complex, S. cerevisiae, X‐ray crystallography, protein–protein interactions
Short abstract
PDB Code(s): http://firstglance.jmol.org/fg.htm?mol=6DEI
Introduction
In eukaryotes, the conserved RWD domain plays key roles in assembly and function of the kinetochore, a megadalton‐scale complex that links chromosomes to spindle microtubules in both mitosis and meiosis. This ~100 amino acid domain adopts a simple α + β sandwich structure, and usually forms homo‐ or heterodimers through a coiled‐coil domain situated N‐terminal to the RWD domain.1, 2 Despite the overall structural similarity between different RWD proteins and their conserved ability to scaffold protein assemblies by binding short peptide motifs, RWD proteins generally utilize distinct protein–protein interaction surfaces that are not shared with other family members.3, 4, 5
RWD domains are found in the inner‐kinetochore proteins Ctf19/CENP‐P and Mcm21/CENP‐O,6 the outer‐kinetochore proteins Spc24 and Spc25,7 and the kinetochore‐associated checkpoint signaling protein Mad1.1 Fungi possess another RWD protein, Csm1/Pcs1, that plays a key cross‐linking role in the kinetochore. Csm1/Pcs1 forms a “V”‐shaped complex with its binding partner Lrs4/Mde4, with two Csm1/Pcs1 dimers bridged at their coiled‐coil N‐termini by a dimer of Lrs4/Mde4.2 In Schizosaccharomyces pombe, the Pcs1:Mde4 complex binds kinetochores in mitosis and suppresses merotelic kinetochore‐microtubule attachments, likely by cross‐linking and co‐orienting the multiple microtubule‐binding elements within a single kinetochore.8, 9, 10.
Saccharomyces cerevisiae and its close relatives possess “point centromeres” whose kinetochores bind a single microtubule.11, 12, 13 In these fungi, Csm1:Lrs4 forms the structural core of the “monopolin” complex, which mediates the co‐orientation of sister kinetochores in Meiosis I to enable the bi‐orientation and segregation of homologs.2, 14, 15, 16, 17, 18 Csm1 is thought to directly cross‐link sister kinetochores, and binds the outer‐kinetochore protein Dsn1 through a conserved surface on its RWD domain.2, 18 Csm1 binds another monopolin complex subunit, Mam1, through a second conserved surface on its RWD domain.19 Mam1 in turn binds and recruits the CK1‐family kinase Hrr25, which mediates the specificity of sister kinetochore crosslinking through an unknown mechanism.16, 20
Uniquely among known RWD proteins, S. cerevisiae Csm1 and its binding partner Lrs4 function in a second context as part of a completely distinct protein network. In interphase, the Csm1:Lrs4 complex localizes to the nucleolus, where it stabilizes ribosomal DNA (rDNA) repeats against illegitimate recombination and also mediates rDNA transcriptional silencing. As in the kinetochore, Csm1 forms a protein interaction hub in the nucleolus, binding the rDNA‐associated protein Tof2 and (likely through Lrs4) the inner‐nuclear membrane proteins Nur1 and Heh1/Src1.2, 21, 22 More recently, we showed that Csm1 binds a SUMO peptidase, Ulp2, and recruits this protein to the nucleolus to de‐SUMOylate and thereby stabilize Tof2.23 Csm1 uses the same protein–protein interaction surfaces in both of its functional contexts: one surface binds both Dsn1 (kinetochore) and Tof2 (nucleolus), and a second surface binds Mam1 (kinetochore) and Ulp2 (nucleolus).19, 23
Here, we use a combined bioinformatic, biochemical, and structural approach to identify and characterize a novel Csm1‐binding protein, Dse3. We show that despite little to no sequence homology with known Csm1‐binding proteins, Dse3 binds Csm1 in a manner equivalent to Mam1 and Ulp2. Thus, our data identify a third major functional context for budding yeast Csm1 and further demonstrate its functional plasticity.
Results
Bioinformatic identification of Dse3 as a Csm1‐binding protein
Existing high‐throughput and targeted studies of protein–protein interactions (collated by the Saccharomyces Genome Database: http://yeastgenome.org) have identified 70 proteins that show a physical interaction with Csm1, either by yeast two‐hybrid binding assays, affinity‐capture mass spectrometry, in vitro reconstitution, or crystal structures. We sorted these interactors, and identified three categories of Csm1 binding partners that have been detected in at least two separate studies [Fig. 1(A)]. The first category comprises proteins that localize to the meiosis I kinetochore, which mostly function with Csm1 as part of the monopolin complex: Lrs4,2, 15, 16, 21, 22, 24, 25, 26, 27, 28, 29, 30 Mam1,2, 16, 19, 26, 27, 28, 29 Hrr25,16, 17 and Dsn1.2, 17, 18, 29 The second category comprises proteins that localize to the nucleolus, functioning with Csm1 to organize ribosomal DNA. These include Tof2,2, 21, 22, 28, 29, 31 Cdc14,22, 29, 32 Nur1,21, 22, 26, 31 Heh1(Src1),21, 22 Ulp2,23, 31, 33 and Spo12.24, 29 The third category comprises three proteins—Dse3,24, 26, 28, 29 Smc4,31, 34 and Plp226, 31—that do not localize to either the nucleolus or the meiotic kinetochore. Both Plp2 (Phosducin‐like protein 2)35 and Smc4, a member of the conserved condensin complex, possess coiled‐coil regions that may promiscuously interact with Csm1, though there is evidence of a functional link between the monopolin complex and condensin.34, 36, 37, 38, 39 Here, we focused on the uncharacterized protein Dse3, which was identified in four high‐throughput yeast two‐hybrid screens as a Csm1‐interacting protein.24, 26, 28, 29
Figure 1.
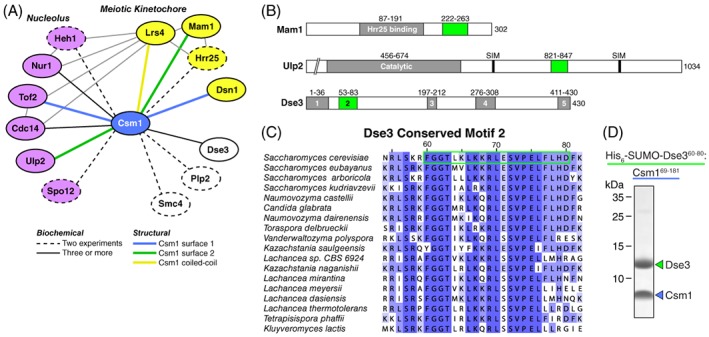
Identification of Dse3 as a Csm1‐binding protein. (A) Schematic of known Csm1‐interacting proteins from biochemical and structural information. Nucleolar proteins are shown in violet, meiotic kinetochore proteins in yellow, and others in white. Dashed and solid lines indicate that the given interaction has been detected by either two (dashed) or three or more (solid) separate studies. Colored lines indicate that the given interaction has been structurally characterized and used Csm1 Surface 1 (blue), Surface 2 (green), or N‐terminal coiled‐coil (yellow). (B) Domain schematic of Mam1 and Ulp2, which interact with Csm1 Interface 2, and the uncharacterized protein Dse3. In each case, conserved domains are shaded grey, and Csm1‐interacting regions (putative in the case of Dse3) are shaded green. (C) Sequence alignment of conserved Motif #2 in budding‐yeast Dse3 proteins. Green outline shows the region of S. cerevisiae Dse3 used for reconstitution with Csm1 (Panel D). (D) SDS‐PAGE analysis of the purified His6‐SUMO‐Dse360–80:Csm169–181 complex. The His6‐SUMO tag was removed prior to crystallization.
DSE3 (Daughter Specific Expression 3) was first identified as a gene whose expression is induced specifically in the daughter cell after S. cerevisiae budding.40 A later high‐throughput localization study showed that the Dse3 protein localizes to the bud neck, and relocalizes to the cytoplasm upon treatment with MMS or HU, two drugs that generate DNA replication stress.41 Protein sequence alignments of fungal Dse3 orthologs reveal a protein with a high degree of predicted disorder, with five short conserved regions that could constitute protein–protein interaction motifs [Fig. 1(B)]. Of these motifs, #1 and #2 are the most highly conserved [Fig. 1(C)]. To test for direct Csm1 binding, we co‐expressed His6‐SUMO‐tagged Dse3 Motif #1 (Residues 2–33) or Motif #2 (Residues 55–80 or 60–80) with an untagged Csm1 construct (Residues 69–181) missing this protein's N‐terminal coiled‐coil and C‐terminal disordered tail regions.2 Both Dse3 Motif #2 constructs robustly pulled down Csm169–181 onto Ni‐NTA affinity resin, while Motif #1 did not (not shown). We overexpressed and purified milligram quantities of a stoichiometric Dse360–80:Csm169–181 complex for biochemical and structural studies [Fig. 1(D)].
Structure of the Csm1–Dse3 complex
We identified crystallization conditions for the Dse360–80:Csm169–181 complex and determined its structure by X‐ray crystallography to 1.7 Å resolution by molecular replacement, using our prior structure of Csm12 as a search model (Table S1). The asymmetric unit contained two copies of Csm169–181, assembled as a canonical Csm1 dimer, and two copies of Dse360–80 (Fig. S1). One Dse3 protomer (chain D) is bound entirely to a single Csm1 dimer, while the second Dse3 protomer (chain C) bridges two Csm1 dimers related by crystallographic symmetry (Fig. S1). We assembled a consensus Csm1‐Dse3 model from the common regions of both Dse3 protomers [Fig. 2(A) and (B)].
Figure 2.

Structure of the Dse3–Csm1 complex. (A) Structure of Dse360–80 (green) bound to a Csm169–181 dimer (blue). The Dse3 chain shown is a composite comprising Residues 60–66 of one Dse3 protomer (chain C) and Residues 67–79 of the second Dse3 protomer (chain D). See Figure S1 for additional structural details. (B) Closeup view of the Dse3–Csm1 interaction. (C),(D) Views equivalent to (A) and (B) showing Csm1 (blue) binding Mam1 (yellow). (E) and (F) Views equivalent to (A) and (B) showing Csm1 (blue) binding Ulp2 (violet). (G) Structure‐based sequence alignment of three Csm1 Interface #2 binding regions from Ulp2, Mam1, and Dse3 (ClustalX coloring). (H) Superposition of Dse3 (green), Mam1 (yellow), and Ulp2 (violet) on Csm1, showing key Csm1‐interacting residues shared between the three proteins.
Unique among structurally characterized RWD domain proteins, Csm1 possesses two distinct protein–protein interaction surfaces (a third interaction surface on the Csm1 coiled‐coil region mediates oligomerization with Lsr4).2, 19, 23 On the Csm1 globular domain, a conserved concave hydrophobic surface (Surface 1) mediates interactions with the kinetochore protein Dsn1 and the rDNA‐associated protein Tof2.2, 23 While Surface 1 is on the “bottom” of the Csm1 dimer, directly opposed to the N‐terminal coiled‐coil domain, Surface 2 forms a “belt” around the Csm1 dimer that binds Mam1 and Ulp2.19, 23 While Mam1 and Ulp2 share little overall sequence homology, their Csm1‐binding regions share several hallmarks, including conserved hydrophobic residues and a conserved arginine that together form specific interactions with Csm1.23
When we compared the structure of Dse360–80:Csm169–181 to prior Csm1 complex structures, we found that Dse3 binds to Csm1 surface 2 [Fig. 2(A) and (B)]. The Dse3–Csm1 interface is very similar to the Mam1–Csm1 and Ulp2–Csm1 interfaces, with all three proteins sharing several hydrophobic residues and one arginine residue that makes specific hydrogen bonds with Csm1 residue D117 [Fig. 2(C–F)]. Dse3 also shares a structurally equivalent lysine residue with Ulp2 (Dse3 K67, Ulp2 K833) that in both cases hydrogen‐bonds Csm1 residue E72. Despite the overall similarity of the three interfaces, however, the three Csm1‐binding proteins show significant variability in their main‐chain conformation, with Dse3 showing the highest variation of the three [Fig. 2(G) and (H)]. Thus, while Mam1, Ulp2, and Dse3 share little overall sequence similarity, their Csm1‐binding motifs share key sequence features and the three proteins bind Csm1 Surface 2 in a structurally equivalent manner.
Discussion
The RWD domain is a conserved, yet promiscuous protein–protein interaction domain. Of known RWD domain proteins, budding‐yeast Csm1 is exceptional in its variety of known binding partners and biological functions. In interphase cells, Csm1 and its obligate binding partner Lrs4 scaffold a nucleolar protein complex with key roles in rDNA maintenance, including the suppression of rDNA recombination and rDNA silencing.2, 21, 22 Moreover, Csm1 directly recruits a SUMO peptidase, Ulp2, to this complex to stabilize it against SUMOylation‐mediated degradation.23 In the meiotic kinetochore, Csm1 scaffolds a large complex to mediate monopolar attachment of sister kinetochores.2, 14, 15, 16, 19, 20 In both of these contexts, Csm1 uses two conserved protein interaction surfaces to scaffold larger protein complexes.
Here, using bioinformatic and structural assays, we identify a new Csm1 interacting protein, Dse3, and show that Dse3 binds Csm1 in a structurally equivalent manner to the nucleolar binding partner Ulp2 and the kinetochore binding partner Mam1. While the biological function of the Csm1–Dse3 interaction is unknown, this finding indicates that Csm1 serves as a protein interaction hub in at least three different functional contexts in S. cerevisiae. Given the relatively loose sequence requirements for Csm1 binding, it seems likely that most Csm1‐interacting proteins have evolved convergently, rather than descending from a single common ancestor. Thus, the unique architecture of the Csm1:Lrs4 complex, as a “V” shaped clamp with at least 8 protein‐binding surfaces in total (four each for Surfaces 1 and 2) has been repurposed by evolution multiple times in a single organism. Future studies with other putative Csm1‐binding proteins, including Smc4 and Plp2, may reveal yet more roles for this multi‐functional complex.
Materials and Methods
Cloning and protein purification
For purification of the Dse360–80:Csm169–181 complex, coding sequences for both proteins were amplified by PCR and cloned into a polycistronic expression vector to yield Dse360–80 (sequence: FGGTLKLKKRLESVPELFLHD) tagged at its N‐terminus with a TEV protease‐cleavable His6‐SUMO tag, and Csm169–181 untagged. This vector was transformed into Escherichia coli Rosetta2 (DE3) pLysS cells (EMD Millipore), and cultures were grown at 37°C to an absorbance at 600 nm of ~0.8. The cultures were shifted to 20°C and protein expression was induced by the addition of 0.25 mM IPTG, and cells were grown ~16 h before harvesting by centrifugation.
For protein purification, cells were resuspended in protein buffer (20 mM Tris–HCl pH 7.5, 5% glycerol, 2 mM β‐mercaptoethanol) plus 300 mM NaCl/10 mM imidazole, lysed by sonication, and centrifuged 30 min at 17,000 RPM to remove cell debris. The supernatant was loaded onto a 5 mL Histrap HP column (GE Life Sciences), washed with protein buffer plus 300 mM NaCl/20 mM imidazole, then with protein buffer plus 100 mM NaCl/20 mM Imidazole. Protein was eluted with protein buffer plus 100 mM NaCl/250 mM imidazole. Protein was then loaded onto a 5 mL Hitrap Q HP column (GE Life Sciences), washed with protein buffer plus 100 mM NaCl, then eluted with a gradient to 600 mM NaCl. Peak fractions were pooled, and TEV protease42 was added to cleave His6‐SUMO tags, and the mixture was incubated 16 h at 4°C. After cleavage, the mixture was passed over Histrap HP and the flow‐through collected, concentrated by ultrafiltration (Amicon Ultra, EMD Millipore), then passed over a HiLoad Superdex 200 size exclusion column (GE Life Sciences) in protein buffer plus 300 mM NaCl (with 1 mM dithiothreitol substituting for β‐mercaptoethanol) for final purification. Protein was concentrated to ~20 mg/mL and stored at 4°C.
Crystallization and structure determination
For crystallization, purified Dse360–80:Csm169–181 at 20 mg/mL in crystallization buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM dithiothreitol) was mixed 1:1 in hanging‐drop format with crystallization buffer containing 100 mM HEPES pH 7.5, 100 mM sodium acetate, and 22% polyethylene glycol (PEG) 4000. Crystals were cryoprotected by the addition of an additional 22% PEG 400, and flash‐frozen in liquid nitrogen.
Diffraction data were collected on Beamline 9–2 at the Stanford Synchrotron Radiation Lightsource (SSRL; see support statement below). Data were processed with the SSRL autoxds script, which uses XDS43 for data indexing and reduction, AIMLESS44 for scaling, and TRUNCATE45 for conversion to structure factors (Table S1). The structure was determined by molecular replacement with PHASER,46 using a Csm1 globular‐domain dimer structure (PDB ID 3N4S, Ref. 2) as a search model. Molecular models were manually rebuilt in COOT47 and refined in phenix.refine48 using positional, individual B‐factor, and TLS refinement (Table S1). Final refined coordinates have been deposited with the Protein Data Bank with PDB ID http://firstglance.jmol.org/fg.htm?mol=6DEI.
Stanford synchrotron radiation lightsource support statement
The use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract no. DE‐AC02‐76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Supporting information
Figure S1. Structure of Dse360–80:Csm169–181.
Table S1. Data Collection and Refinement Statistics
Acknowledgment
The authors thank members of the Corbett laboratory for helpful discussions. K. D. C. acknowledges past support from the Ludwig Institute for Cancer Research and current support from the National Institutes of Health (R01 GM104141).
Current address: Synthorx Inc., 11099 North Torrey Pines Road, Suite 290, La Jolla, California 92037
References
- 1. Kim S, Sun H, Tomchick DR, Yu H, Luo X (2012) Structure of human Mad1 C‐terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci USA 109:6549–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corbett KD, Yip CK, Ee L‐S, Walz T, Amon A, Harrison SC (2010) The monopolin complex crosslinks kinetochore components to regulate chromosome‐microtubule attachments. Cell 142:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S (2012) CENP‐T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol 14:604–613. [DOI] [PubMed] [Google Scholar]
- 4. Dimitrova YN, Jenni S, Valverde R, Khin Y, Harrison SC (2016) Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell 167:1014–1027.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T (2013) CENP‐T provides a structural platform for outer kinetochore assembly. EMBO J 32:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitzberger F, Harrison SC (2012) RWD domain: a recurring module in kinetochore architecture shown by a Ctf19–Mcm21 complex structure. EMBO Rep 13:216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC (2006) Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure 14:1003–1009. [DOI] [PubMed] [Google Scholar]
- 8. Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K (2007) The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol 17:1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCollum D (2010) Prevention of merotelic chromosome attachments by the monopolin complex. Cell Cycle 9:4258–4265. [DOI] [PubMed] [Google Scholar]
- 10. Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolić‐Nørrelykke IM, Gregan J (2010) Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle 9:3997–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gordon JL, Byrne KP, Wolfe KH (2011) Mechanisms of chromosome number evolution in yeast. PLoS Genet 7:e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westermann S, Drubin DG, Barnes G (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76:563–591. [DOI] [PubMed] [Google Scholar]
- 14. Toth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K (2000) Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103:1155–1168. [DOI] [PubMed] [Google Scholar]
- 15. Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K (2003) Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4:535–548. [DOI] [PubMed] [Google Scholar]
- 16. Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K (2006) Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 126:1049–1064. [DOI] [PubMed] [Google Scholar]
- 17. Sarangapani KK, Duro E, Deng Y, Alves F de L, Ye Q, Opoku KN, Ceto S, Rappsilber J, Corbett KD, Biggins S, Marston AL, Asbury CL (2014) Sister kinetochores are mechanically fused during meiosis I in yeast. Science 346:248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarkar S, Shenoy RT, Dalgaard JZ, Newnham L, Hoffmann E, Millar JBA, Arumugam P (2013) Monopolin subunit Csm1 associates with MIND complex to establish monopolar attachment of sister kinetochores at meiosis I. PLoS Genet 9:e1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Corbett KD, Harrison SC (2012) Molecular architecture of the yeast monopolin complex. Cell Rep 1:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye Q, Ur SN, Su TY, Corbett KD (2016) Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator. EMBO J 35:2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang J, Brito IL, Villén J, Gygi SP, Amon A, Moazed D (2006) Inhibition of homologous recombination by a cohesin‐associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 20:2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mekhail K, Seebacher J, Gygi SP, Moazed D (2008) Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456:667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang J, Singh N, Carlson CR, Albuquerque CP, Corbett KD, Zhou H (2017) Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity. Genes Dev 31:802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi‐Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein‐protein interactions in Saccharomyces cerevisiae . Nature 403:623–627. [DOI] [PubMed] [Google Scholar]
- 25. Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin‐Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MHY, Butland G, Altaf‐Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae . Nature 440:637–643. [DOI] [PubMed] [Google Scholar]
- 26. Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two‐hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98:4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W (2008) Dbf4‐dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135:662–678. [DOI] [PubMed] [Google Scholar]
- 28. Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane‐Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual J‐F, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet A‐S, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi A‐L, Tavernier J, Hill DE, Vidal M (2008) High‐quality binary protein interaction map of the yeast interactome network. Science 322:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CSM, Drubin DG, Barnes G, Hazbun T (2007) A protein interaction map of the mitotic spindle. Mol Biol Cell 18:3800–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan JNY, Poon BPK, Salvi J, Olsen JB, Emili A, Mekhail K (2011) Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev Cell 20:867–879. [DOI] [PubMed] [Google Scholar]
- 31. Wysocka M, Rytka J, Kurlandzka A (2004) Saccharomyces cerevisiae CSM1 gene encoding a protein influencing chromosome segregation in meiosis I interacts with elements of the DNA replication complex. Exp Cell Res 294:592–602. [DOI] [PubMed] [Google Scholar]
- 32. Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin Z‐Y, Breitkreutz B‐J, Stark C, Liu G, Ahn J, Dewar‐Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras A‐C, Nesvizhskii AI, Tyers M (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328:1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Srikumar T, Lewicki MC, Raught B (2013) A global S. cerevisiae small ubiquitin‐related modifier (SUMO) system interactome. Mol Syst Biol 9:668–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johzuka K, Horiuchi T (2009) The cis element and factors required for condensin recruitment to chromosomes. Mol Cell 34:26–35. [DOI] [PubMed] [Google Scholar]
- 35. Flanary PL, DiBello PR, Estrada P, Dohlman HG (2000) Functional analysis of Plp1 and Plp2, two homologues of phosducin in yeast. J Biol Chem 275:18462–18469. [DOI] [PubMed] [Google Scholar]
- 36. Burrack LS, Applen Clancey SE, Chacón JM, Gardner MK, Berman J (2013) Monopolin recruits condensin to organize centromere DNA and repetitive DNA sequences. Mol Biol Cell 24:2807–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brito IL, Yu H‐G, Amon A (2010) Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics 185:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tada K, Susumu H, Sakuno T, Watanabe Y (2011) Condensin association with histone H2A shapes mitotic chromosomes. Nature 474:477–483. [DOI] [PubMed] [Google Scholar]
- 39. Dudas A, Polakova S, Gregan J (2011) Chromosome segregation: monopolin attracts condensin. Curr Biol 21:R634–R636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Colman‐Lerner A, Chin TE, Brent R (2001) Yeast Cbk1 and Mob2 activate daughter‐specific genetic programs to induce asymmetric cell fates. Cell 107:739–750. [DOI] [PubMed] [Google Scholar]
- 41. Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, Davis TN, Nislow C, Brown GW (2012) Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 14:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tropea JE, Cherry S, Waugh DS (2009) Expression and purification of soluble His(6)‐tagged TEV protease. Methods Mol Biol 498:297–307. [DOI] [PubMed] [Google Scholar]
- 43. Kabsch W (2010) XDS. Acta Cryst D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Evans PR, Murshudov GN (2013) How good are my data and what is the resolution? Acta Cryst D69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Cryst D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst, D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Afonine PV, Grosse‐Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst D68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Structure of Dse360–80:Csm169–181.
Table S1. Data Collection and Refinement Statistics


