Abstract
Mtu1(Trmu) is a highly conserved tRNA modifying enzyme responsible for the biosynthesis of τm5s2U at the wobble position of tRNAGln, tRNAGlu and tRNALys. Our previous investigations showed that MTU1 mutation modulated the phenotypic manifestation of deafness-associated mitochondrial 12S rRNA mutation. However, the pathophysiology of MTU1 deficiency remains poorly understood. Using the mtu1 knock-out zebrafish generated by CRISPR/Cas9 system, we demonstrated the abolished 2-thiouridine modification of U34 of mitochondrial tRNALys, tRNAGlu and tRNAGln in the mtu1 knock-out zebrafish. The elimination of this post-transcriptional modification mediated mitochondrial tRNA metabolisms, causing the global decreases in the levels of mitochondrial tRNAs. The aberrant mitochondrial tRNA metabolisms led to the impairment of mitochondrial translation, respiratory deficiencies and reductions of mitochondrial ATP production. These mitochondria dysfunctions caused the defects in hearing organs. Strikingly, mtu1−/– mutant zebrafish displayed the abnormal startle response and swimming behaviors, significant decreases in the sizes of saccular otolith and numbers of hair cells in the auditory and vestibular organs. Furthermore, mtu1−/– mutant zebrafish exhibited the significant reductions in the hair bundle densities in utricle, saccule and lagena. Therefore, our findings may provide new insights into the pathophysiology of deafness, which was manifested by the deficient modifications at wobble position of mitochondrial tRNAs.
INTRODUCTION
Deficient nucleotide modifications of mitochondrial tRNAs have been linked to human diseases, including cancer, diabetes, neurological disorders, hypertension and deafness (1–5). The systematic mapping of all modifications among 22 mammalians mitochondrial tRNAs revealed 15 species of modifications in 118 positions (6). These nucleotide modifications regulate the biogenesis, structure and function of the corresponding RNAs and RNA–protein complexes (7–11). In particular, the core modifications including pseudouridinylation at position 55 at the TΨC loop primarily contributed to structural stability of tRNAs (3,7). The loss of pseudouridinylation at position 55 at the TΨC loop of tRNAGlu caused the maternally inherited deafness and diabetes (4). The nucleotide modifications at positions 34 and 37 of anticodon loop regulate the stabilization of anticodon structure, fidelity and efficiency of mitochondrial translation (3,10,12–15). Mutations in the nucleotides at position 37 including tRNAIle 4295A>G, tRNAAsp 7551A>G and tRNAMet 4435A>G mutations have been associated with hypertension and deafness (5,16–18). The defective i6A37 or m1G37 modification caused by mutations in tRNA modifying enzymes TRIT1 or TRMT5 were responsible for mitochondrial dysfunction leading to clinical phenotypes (19,20).
The nucleotides at position 34 of mitochondrial tRNAs are more prone to be modified than those at other positions of tRNAs (1,3,6). Of these, 5-formylcytidine (f5C), 5-taurinomethyluridine (τm5U) and 5-taurinomethyl-2-thiouridine (τm5s2U) modifications are present at the wobble position of human mitochondrial tRNAs (6,21–24). These post-transcriptional modifications have a pivotal role in the structure and function of tRNAs, including structural stabilization, aminoacylation, and codon recognition at the decoding site of small rRNA (6,10,25). In human mitochondria, MTO1, GTPBP3 and MTU1(TRMU) are the tRNA modifying enzymes responsible for the biosynthesis of the τm5s2U of tRNALys, tRNAGlu and tRNAGln (7,24,26,27). Indeed, MTO1 and GTPBP3 are involved in the formation of the 5-taurinomethyl group, while MTU1 is responsible for 2-thio group of τm5s2U in mitochondrial tRNALys, tRNAGlu and tRNAGln but not involved in 2-thiolation of cytoplasmic tRNAs (7,24,26–28). In contrast, Urm1/Uba4/NCS2/NCS6/TUM1 is responsible for 2-thiolation of cytoplasmic tRNAs (6–8). Our previous investigations showed that MTO1, GTPBP3, or MTU1 genes were the potential modifier genes for the phenotypic expression of deafness-associated 12S rRNA 1555A>G mutation (29–32). In particular, MTU1 is a highly conserved tRNA modifying enzyme responsible for the 2-thiolation of tRNALys, tRNAGlu, and tRNAGln with mnm5s2U34 in bacteria, mcm5s2U34 in yeast and τm5s2U in human mitochondria (27,31,33–35). Mutations in MTU1 gene were identified in patients of reversible infantile liver failure (36,37). However, Mtu1−/− knockout mice died at a developmental stage as early as E7.5-8 (38). Interestingly, Mtu1+/- knockout mice exhibited deficient 2-thiolation in mt-tRNAs, impairment of mitochondrial translation, severe disruption of mitochondrial membrane integrity and a broad decrease in respiratory complex activities in the hepatocytes (38). In the previous investigations, we identified a nuclear modifier allele (G28T and A10S) in the MTU1 gene, which interacts with the mitochondrial 12S rRNA 1555A→G mutation to cause deafness (26,32). To investigate whether defects in MTU1 cause the dysfunction of hearing organs in vivo, we generated a mtu1ko zebrafish model produced by genome editing using the CRISPER/Cas9 system. First, the mtu1ko zebrafish was assessed for the in vivo effects of defective mtu1 on mitochondrial tRNA metabolism, mitochondrial translation, and enzymatic activities of electron transport chain complexes and ATP production. The mtu1ko zebrafish was further evaluated for the effect on hearing function including the startle response and swimming behaviors, the sizes of saccular otolith and numbers of hair cells of auditory and vestibular organs as well as hair bundle densities.
MATERIALS AND METHODS
Experimental fish and maintenance
All experiments were conducted using the AB wild-type strain of zebrafish (Danio rerio) of either sex. The animal protocols used in this investigation were approved by the Zhejiang University Institutional Animal Care and Use Committee. All fish were kept in recirculating water at 26°C and fed with commercial pellets at a daily ration of 0.7% of their body weight. Embryos were reared at 28.5°C on a 14-h light/10-h dark cycle according to standard protocols (39). Embryos were staged by hours post fertilization (hpf) and days post fertilization (dpf).
Sequence retrieval and analysis
Mtu1 homologue in zebrafish was obtained from databases of NCBI using the human MTU1 amino acid sequences as queries (40). The BLAST homology analyses were performed with the available web-based programs of NCBI. Protein sequence alignments were carried out using the BioEdit program. The phylogenies of protein sequences were estimated with MEGA 6.0 using the neighbor joining method.
In situ hybridization
Probes were synthesized with digoxigenin (DIG)-labeled antisense RNA probes specific to zebrafish mtu1 (forward primer: GAGGGAGAACACCTAATC; reverse primer: TATCCACAACAAACCAGG). Whole mount in situ hybridization (WISH) was performed as detailed elsewhere (41–43). Zebrafish embryos from various age of post-fertilization were dechlorinated in 2 mg/ml pronase in E3 medium and fixed at 4°C in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight, then transferred to 100% methanol for storage at –20°C for at least 20 min before undergoing hybridization. After the hybridization procedure, embryos were washed extensively in PBS with 0.1% Tween 20, re-fixed in 4% paraformaldehyde, and then transferred to 70% glycerol. Stained embryos were visualized using stereoscopic Microscopes (SMZ18, Nikon). Zebrafish larval at 120 hpf were fixed in 10% neutral buffer formalin overnight at 4°C and then embedded in optimal cutting temperature (OCT) for frozen section using a cryostat (Leica CM 3050S). Ten μm transverse sections were obtained and used for ISH with DIG-labeled antisense RNA probes specific to zebrafish mtu1.
Cas9-based genome editing in zebrafish
The zCas9 expression plasmid pSP6-2sNLS-spCas9 was linearized by Xba I used as a templated for Cas9 mRNA in vitro synthesis with mMESSAGE mMACHINE mRNA transcription synthesis kits (Ambion). The sequence of sgRNAs was designed according to criteria as described previously (44). The gRNA transcription plasmid was pT7-gRNA. We used the CRISPR/Cas9 design tool (http://zifit.partners.org) to select specific targets to minimize off-target effects. Cas9-encoding mRNA (300 ng/μl) and gRNA (20 ng/μl) were co-injected into one-cell-stage wild-type embryos. Injected embryos were incubated at 28.5°C, and collected for making genomic DNA for genotyping at 50 hpf. Genome poof of the 50 hpf injected embryo was used as template to amplify mtu1 gene by using the following two primers: F1: ACAAATGACTTCACCATT; R1: CAACCTAATAAATGAACG. The fragments were cloned by using the TA Cloning Kit (TAKARA) and then were sequenced.
Western blot analysis
Fish were sacrificed after anesthesia, and homogenized in RIPA reagent (Invitrogen) using a homogenizer. Twenty μg of total cellular proteins were electrophoresed through 10% bis-Tris SDS-polyacrylamide gels and then transferred to a polyvinyl difluoride (PVDF) membrane. After transfer, the membrane was incubated in 1 × PBST (1 × PBS, 1% Tween 20) and 5% nonfat dry milk for 1 h. After blocking, the membrane was incubated with primary antibody overnight at 4°C following with secondary antibody. The hybridized membrane was then exposed to chemiluminescence reagent for 1 min and developed by ChemiScope 3300 mini (CLiNX, Shanghai). The antibodies were obtained from different companies including Mtu1 from Hangzhou HuaAn Biotecnology Co (HuaBio), Gapdh (SAB2701826) from Sigma-Aldrich, Nd1 (ab74257), Nd6 (ab81212), Atp5a (ab188107), Sdhb (ab151684) and Yars2 (ab127542) from Abcam, Cytb (A9762) from ABclonal, Co2 (55070-1-AP), Kars (14951-1-AP), Lars2 (17097-1-AP), Tufm (26730-1-AP), Tfb2m (24411-1-AP) and Tom20 (1802-1-AP) from Proteintech. Peroxidase Affini Pure goat anti-mouse IgG and goat anti-rabbit IgG (Jackson) were used as a secondary antibody and protein signals were detected using the ECL system (CWBIO). Quantification of density in each band was performed as detailed previously (30,42).
Mitochondrial tRNA analysis
Total RNAs were isolated from fish using Totally RNA™ Kit (Ambion, Inc). Five μg of total cellular RNAs were electrophoresed through a 10% polyacrylamide/7 M urea gel in Tris–borate–ethylenediaminetetraacetic acid (EDTA) buffer (TBE) (after heating the sample at 65°C for 10 min), and then electroblotted onto a positively charged nylon membrane (Roche) for the hybridization analysis with oligodeoxynucleotide probes. Oligodeoxynucleosides used for DIG-labeled probes (Supplementary Table S1) were zebrafish mitochondrial tRNALys, tRNAGlu, tRNAGln, tRNALeu(UUR), tRNATrp, tRNAHis, tRNAMet (GeneBank accession no. NC_002333.2) (46) and cytoplasmic tRNAGlu, tRNAAla, tRNAGly and 5S rRNA. The oligodeoxynucleotides were generated by using DIG-oligonucleotide Tailing kit (Roche). The hybridization was carried out as detailed elsewhere (16,45).
The presence of thiouridine modification in the tRNAs was verified by the retardation of electrophoretic mobility in a polyacrylamide gel that contains 0.05 mg/ml (N-acryloylaminophenyl) mercuric chloride (APM) (21,26,28,47). Total RNAs were separated by polyacrylamide gel electrophoresis and blotted onto positively charged membrane (Roche Applied Science). Each tRNA was detected with the specific DIG-oligodeoxynucleoside probe at the 3′ termini as detailed elsewhere (47). Oligonucleotide probes for mitochondrial tRNALys, tRNAGlu, tRNAGln, tRNALeu(UUR) and cytoplasmic tRNAGlu, tRNAAla were described as above. DIG-labeled oligodeoxynucleosides were generated by using the DIG Oligonucleoside Tailing Kit (Roche). APM gel electrophoresis and quantification of 2-thiouridine modification in tRNAs were conducted as detailed (21,26).
Assays of activities of respiratory complexes
The enzymatic activities of Complex I, II, III, IV and V were assayed as detailed elsewhere (48,49). Briefly, the activity of Complex I was determined through the oxidation of NADH with ubiquinone as the electron acceptor. Complex II was examined through the artificial electron acceptor DCPIP. The activity of Complex III was measured through the reduction of cytochrome c (III) by using d-ubiquinol-2 as the electron donor. The Complex IV was monitored through the oxidation of cytochrome c (II). The activity of complex V was explored through the NADH oxidation via conversion of phosphoenolpyruvate to lactate by two step reactions.
Enzyme histochemistry (EHC) staining for SDH and COX in the frozen-sections were measured as detailed elsewhere (50). In brief, 10 μm serial transverse frozen sections were obtained from mtu1−/−, mtu1+/– and wild type zebrafish at 5 dpf. For SDH staining, samples were incubated in 0.1 M phosphate buffer, pH 7.6, containing 5 mM EDTA, 1 mM potassium cyanide, 0.2 mM phenazine methosulfate, 50 mM succinic acid, 1.5 mM nitro blue tetrazolium at 37°C for 25 min. For COX staining, samples were incubated in 5 mM phosphate buffer, pH 7.6, containing 5 mM EDTA, 1 mM potassium cyanide, 0.2 mM phenazine methosulfate, 50 mM succinic acid, 1.5 mM nitro blue tetrazolium at 37°C for 60 min.
Measurement of ATP generation
ATP generation was measured using an ATP analysis kit (Beyotime) and a microplate reader (Syneregy H1, Bio-Tek) according to the manufacturer's instructions with minor modifications (40). Briefly, 20 embryos (per genotypes) were treated with the ATP synthase inhibitor oligomycin to determine cytosolic ATP generation, and same number of embryos (from same batch) were incubated in E3 medium for total cellular ATP levels measurement. After treatment, all embryos were homogenized and centrifuged at 12000g for 5 minutes at 4°C. Protein concentrations of the supernatant were determined using the bicinchoninic acid assay, and 20 μg of total protein lysate was used for ATP analysis. Mitochondrial ATP levels = (total cellular ATP levels) – (oligomycin-resistant ATP levels, cytosolic ATP).
Histological studies
For hematoxylin and eosin (H&E) staining, Zebrafish at 5 dpf were anesthetized and fixed in 4% paraformaldehyde for 24 h at room temperature. They were then dehydrated, infiltrated, embedded in paraffin, sliced into 3 μm thick by pathologic microtome (RM2016, Leica). Tissue sections were then stained by hematoxylin and eosin (H&E) to observe tissue architecture.
For phalloidin staining, adult zebrafish were anesthetized in 0.02% MS-222 and the inner ear were dissected (51,52). Then, the tissues were fixed with 4% PFA and permeabilized with 0.01% Triton X-100. The hair cells from utricle, saccule, and lagena were labeled with β-actin-staining TM Fluorescent-Phalloidin (1: 200 in PBS, 14 μM stock solution Cytoskeleton) for 20 mins at room temperature. All nuclei were stained with DAPI (1: 1000 in PBS, 10 μM stock solution, Sigma). Fluorescence was visualized with a confocal microscope (OLYMPUS, FV1000).
Hair cell vital staining
FM1-43FX dye (Life Technologies, NY) labeling of hair cells in the neuromasts was performed on live zebrafish larvae (53). Zebrafish at 5 dpf were immersed in 3 μM FM1-43FX dye in 1X larval water for 45 s and quickly rinsed off. The fish were washed three times with larval water and anesthetized with 0.02% MS-222 (Sigma-Aldrich) for imaging. The numbers of neuromasts of anterior and posterior lateral line were counted. To further evaluate the survival of hair cells in the neuromasts, 5 dpf zebrafish were stained with both FM1-43FX and phalloidin. Briefly, phalloidin staining was performed following FM1-43FX dye treatment and fixation (4% PFA). All nuclei were stained with DAPI. Fluorescence was detected with a confocal microscope (OLYMUS 1000).
DASPEI (2-[4-(dimethylamino)styryl]-N-ethylpyridinium iodide) staining of the neuromast hair cells was performed on live fish. Zebrafish larvae were incubated for 10 min at 28°C in 10 μM DASPEI. The fish were then quickly washed three times with larval water and then transferred into the 2% methyl cellulose for imaging with an inverted fluorescence microscope (DM400B, Leica). The numbers of DASPEI-labeled neuromasts of posterior lateral line were counted.
Transmission electron microscopy analysis
The mtu1−/−, mtu1+/− and mtu1+/+ zebrafish larvae at 5 dpf were fixed with 2.5% glutaraldehyde and transferred to 1% osmium tetroxide and dehydrated using a graded ethanol series followed by treatment with gradient infiltration (acetone and Spurr resin), and finally embedded in Epon-812 resin. Ultra-thin sections were obtained and stained with 3% uranyl acetate and 1% lead citrate and imaged under a Hitachi Model H-7650 transmission electron microscopy.
Statistical analysis
Statistical analysis was carried out using the unpaired, two-tailed Student's t-test contained in the Microsoft-Excel program or Macintosh (version 2007). Differences were considered significant at a P < 0.05.
RESULTS
Conservation of sequence and expression patters of zebrafish Mtu1
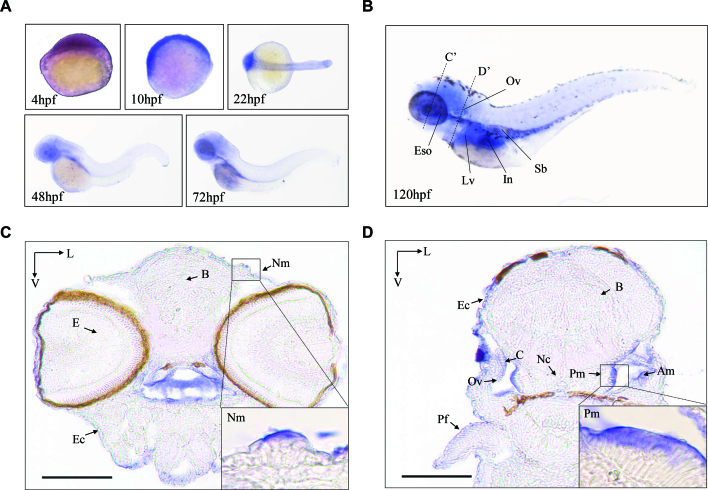
Zebrafish Mtu1 comprises 416 amino acids containing a 40-amino acid mitochondrial target signal. The protein alignments of Zebrafish Mtu1 with it homologs of other organisms, including Homo sapiens, Bos Taurus, Canis lupus, Mus musculus, Rattus norvegicus, Xenopus tropicalis, Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae revealed an extensive conservation of protein sequence (Supplementary Figure S1). In particular, zebrafish Mtu1 is 66% and 65% identical to the amino acid sequence of mouse and human MTU1, respectively. The spatial patterns of mtu1 expression in zebrafish larvae were analyzed by whole amount in situ hybridization (WISH) using DIG-labeled antisense probes. As shown in Figure 1A, the hybridization signal of mtu1 was detectable at 4 hpf stage, indicating that mtu1 is maternally expressed. Transcripts of mtu1 were also detected in the otic placodes of 22 hpf larvae. The otic placode eventually forms sensory patches and develops as the zebrafish inner ear. There was also weak expression of mtu1 in primordial cells that form neuromasts of the zebrafish lateral line organ system. At 120 hpf, the mtu1 expression was pronounced in neuromasts, inner ear region and swimming bladder. As shown in Figure 1, the staining analysis of transverse sections at 120 hpf larva, including brain, eye, otic vesicle, pectoral fin, epidermal cells, posterior macula hair cells, cristae, neuromast, notochord and anterior macula hair cells, revealed the abundant expression of mtu1 in the inner ear and neuromast but the week expression of mtu1 in the brain, eye, pectoral fin and epidermal cells. These data led us to test hypothesis that the loss of mtu1 would disrupt hair bundle morphology and hair cell function. To test this hypothesis, we targeted mtu1 to generate null mutations and characterized the phenotype of mutant Zebrafish.
Figure 1.
The expression patterns of Zebrafish Mtu1 in the sensory organs. (A) Whole-mount in situ hybridization on wild type larval zebrafish at various ages (4–72 hpf). (B) Lateral views with anterior to the left show mtu1 expression in otic vesicle (Ov), liver (Lv), esophagus (Eso), swimming bladder (Sb) and intestine (In) at 120 hpf. (C, D) Transverse frozen sections through dashed line C’ and D’ in B were used for the assessment of mtu1 expression in neuromast (Nm), posterior macula (Pm) and anterior macula (Am) hair cells, respectively. Insets show higher magnifications of Nm and Pm. Scale bars: 100 μm. V, ventral; L, lateral; B: brain; E: eye; Pf: pectoral fin; Ec: epidermal cells; C: cristae; Nc: notochord.
Generation of mtu1 knock-out zebrafish using CRISPR/Cas9 system
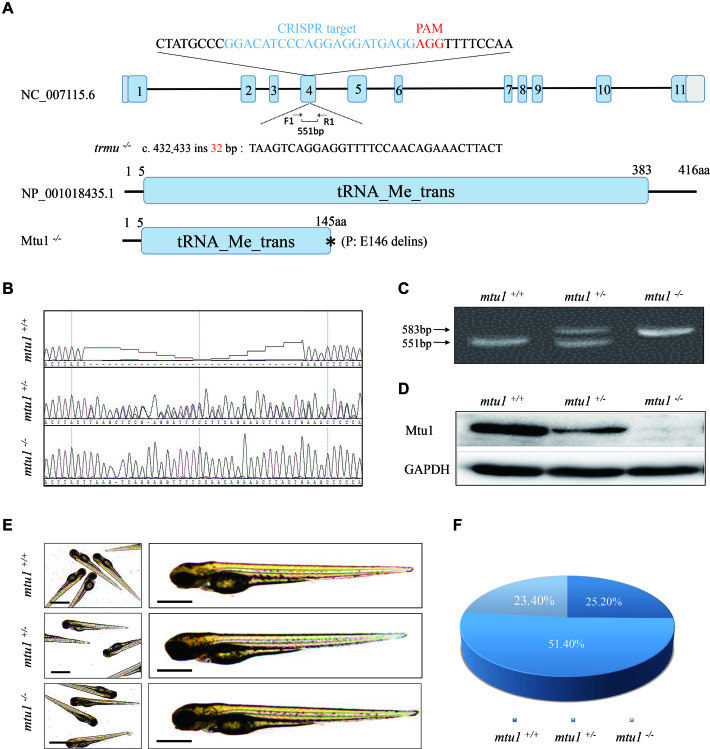
With the goal of defining the physiological roles of Mtu1, we have taken advantage of the CRISPR/Cas9 technology to generate zebrafish mutant lines where the Mtu1 ortholog was disrupted. As shown in Figure 2A, a single guide RNA (sgRNA) targeting exon 4 of mtu1 was injected into wild-type one-cell stage embryos together with Cas9 mRNA. As a result, an allele, mtu1ins32bp was generated by introducing a 32 bp insertion in the exon 4 of mtu1 (heterozygous and homozygous zebrafish described as mtu1+/−, mtu1−/− and wild type as WT). In fact, this insertion caused a frame-shift and a stop codon 145 amino acid of Mtu1 after the insertion. This allele was subsequently propagated after confirmation of the mutation by Sanger sequencing, DNA-PAGE and Western blot analyses (Figure 2B–D). Both mtu1+/− and mtu1−/− mutants were adult-viable. Furthermore, there was no notable difference in gross morphology between mutant and wild type fishes. Larvae exhibiting the mutant phenotype (described later) carried the two mutant alleles of mtu1. The ratios of genotypes/phenotype of offsprings (F2) in clutches from different F1mtu1 heterozygous crosses mirrored the Mendelian ratio and was consistent with a recessive inheritance pattern (Figure 2E and F).
Figure 2.
Generation of mtu1 knock-out zebrafish using CRISPR/Cas9 system. (A) Schematic representation of CRISPR/Cas9 target site at exon 4 as used in this study. An allele, mtu1ins32bp was produced by a 32 bp insertion in exon 4 and a truncated 145 amino acid non-functional protein. (B–D) Genotyping of mtu1ins32bp by Sanger sequence, the PAGE RFLP and Western blot analyses. (E) The morphology of mtu1−/−, mtu1+/− and mtu1+/+ zebrafish at 3 dpf. (F) The ratios of genotypes/phenotype of offsprings (F2) in clutches from different F1 mtu1 heterozygous crosses at 10 dpf (n = 350).
Deficient thiolation of mitochondrial tRNALys, tRNAGln, and tRNAGlu in mtu1 mutants
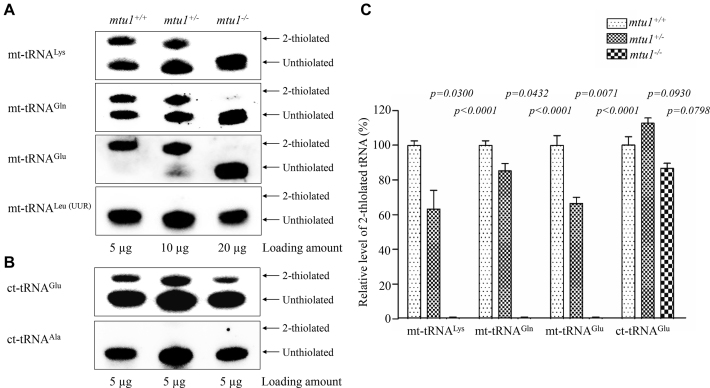
To investigate whether the loss of Mtu1 affected the 2-thiouridine modification at position 34 in tRNAs in zebrafish, the 2-thiouridylation levels of tRNAs were determined by isolating total RNAs from mtu1+/− and mtu1−/− mutants and wild type fishes, purifying tRNAs, qualifying the 2-thiouridine modification by the retardation of electrophoresis mobility in APM polyacrylamide gel (26,27,47), and hybridizing DIG-labeled probes for mitochondrial tRNAGln, tRNAGlu, and tRNALys and tRNALeu(UUR) and cytoplasmic tRNAGlu, tRNAAla as internal controls. In this system, the mercuric compound can specifically interact with the tRNAs containing the thiocarbonyl group–such as tRNAGln, tRNAGlu, and tRNALys, thereby retarding tRNA migration. As shown in Figure 3A and B, 2-thiouridylation levels of mitochondrial tRNALys, tRNAGlu, and tRNAGlu in wild type zebrafish were 54%, 94% and 51%, respectively, while the 2-thiouridylation was absent in the mitochondrial tRNALeu(UUR). However, the mtu1−/− mutant zebrafish exhibited the complete loss of 2-thiouridylation of mitochondrial tRNAGln, tRNAGlu, and tRNALys. Furthermore, the 2-thiouridylation levels of mitochondrial tRNALys, tRNAGlu and tRNAGln in mtu1+/− mutant fish were 63%, 66% and 85%, relative to those of wild type fishes, respectively. Moreover, there were no difference of 2-thiouridylation level of cytoplasmic tRNAGlu between mutant and wild type fishes, while the 2-thiouridylation was absent in the cytoplasmic tRNAAla. These results demonstrated that mtu1 in zebrafish is excursively involved in the 2-thiouridylation of mitochondrial tRNALys, tRNAGlu and tRNAGln.
Figure 3.
APM gel electrophoresis combined with Northern blotting of mitochondrial tRNAs. (A) Five μg (mtu1+/+), 10 μg (mtu1+/−) and 20 μg (mtu1−/−) of zebrafish total RNAs were separated by polyacrylamide gel electrophoresis that contains 0.05 mg/ml APM, electroblotted onto a positively charged membrane, and hybridized with a DIG-labeled oligonucleotide probe specific for the mt-tRNALys. The blots were then stripped and the membrane was re-hybridized with DIG-labeled probes for mt-tRNAGlu, mt-tRNAGln, and mt-tRNALeu(UUR), respectively. The retarded bands of 2-thiolated tRNAs and non-retarded bands of tRNA without thiolation are marked by arrows. (B) Five micrograms of total RNA from various genotype Zebrafish for APM gel to determine 2-thiolation levels of cytosolic tRNAGlu and tRNAAla. (C) Proportion in vivo of the 2-thiolated tRNA levels. The proportion values for the mutant zebrafish are expressed as percentages of the average values for the wild type zebrafish. The calculations were based on three independent determinations of each tRNA in each fish. The error bars indicate standard errors; P indicates the significance, according to Student's t test, of the difference between mutant and wild type for each tRNA.
Decreases in the steady-state levels of mitochondrial tRNAs
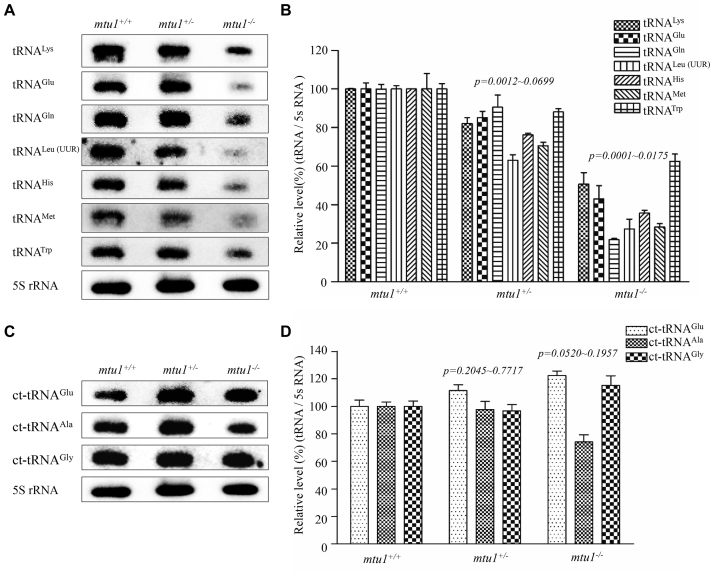
To assess if the mtu1 mutation ablated the metabolism of tRNA, we subjected total RNAs from mutant and control zebrafish to Northern blots and hybridized them with DIG-labeled oligodeoxynucleotide probes for mitochondrial tRNALys, tRNAMet, tRNATrp, tRNAHis, tRNALeu(UUR), tRNAGln and tRNAGlu and cytoplasmic tRNAGlu, tRNAAla and tRNAGly. The cytosine and guanine were present at nucleotides at positions 34 of mitochondrial tRNAMet and tRNAHis, respectively, while U34 was present in the remaining mitochondrial and cytoplasmic tRNAs as described above. As illustrated in Figure 4A and C, the loss of mtu1 caused the decreases in the steady-state levels of all seven mitochondrial tRNAs but not those of three cytoplasmic tRNAs. For comparison, the levels of each tRNA were normalized to the reference 5S RNA. As shown in Figure 4B, the average steady-state levels of tRNALys, tRNAGln and tRNAGlu, tRNALeu(UUR), tRNATrp, tRNAMet, and tRNAHis were 21%, 50%, 43%, 27%, 62%, 28% and 35% in the mtu1−/− zebrafish lines, relative to the mean values of controls (P < 0.0001 to 0.0175). Furthermore, the average steady-state levels of tRNALys, tRNAGln and tRNAGlu, tRNALeu(UUR), tRNATrp, tRNAMet, and tRNAHis were 90%, 82%, 85%, 63%, 88%, 70% and 76%, in the mtu1+/− zebrafish lines, relative to average values of controls (P < 0.0012 to 0.0699). As shown in Figure 4D, the steady-state levels of cytosolic tRNAGlu, tRNAAla and tRNAGly in these mtu1 mutant lines were comparable with those in control lines. These data indicated that the mtu1ins32bp mutation caused the global defects in mitochondrial tRNA metabolism.
Figure 4.
Northern blot analysis of mitochondrial tRNAs. (A, C). Five μg of total cellular RNA samples from mutant and wild type zebrafish were electrophoresed through a denaturing polyacrylamide gel, electroblotted and hybridized with DIG-labeled oligonucleotide probes specific for mitochondrial tRNALys, tRNAGlu, tRNAGln, tRNAMet, tRNATrp, tRNAHis, tRNALeu(UUR), cytoplasmic tRNAGlu, tRNAAla and tRNAGly as well as 5S rRNA probe, respectively. (B, D) Quantification of the levels of tRNAs. Average relative levels tRNA content were normalized to the average content in the mutant and wild type 5S rRNA, respectively. The values for the mtu1+/− and mtu1−/− zebrafish are expressed as percentages of the average values for the mtu1+/+ zebrafish. The calculations were based on three independent determinations. Graph details and symbols are explained in the legend to the Figure 3.
Reductions in the levels of mitochondrial proteins
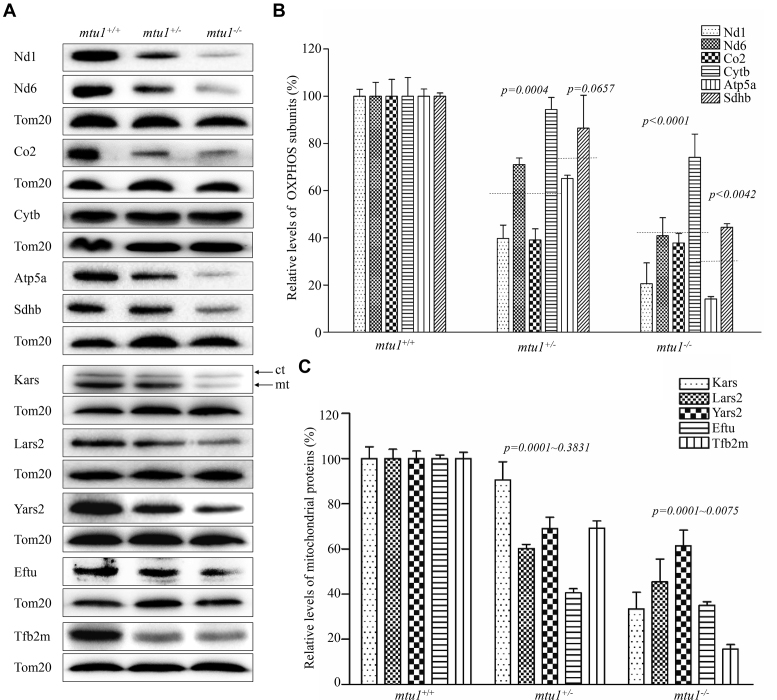
To investigate whether the aberrant mitochondrial tRNA metabolism alters mitochondrial translation, the Western blot analysis was carried out to examine the levels of subunits of OXPHOS including four mtDNA encoding polypeptides (Nd1, Nd6, Cytb and Co2) and seven nuclear encoding mitochondrial proteins (Atp5a, Sdhb, Kars, Lars2, Yars2, Tufm and Tfb2m) in mutant and control zebrafish lines with TOM20 as a loading control. As shown in Figure 5, the levels of Nd1, Nd6 (subunit 1 and 6 of NADH dehydrogenase), Cytb (apocytochrome b), Co2 (subunit II of cytochrome c oxidase), Sdhb (subunit of succinate dehydrogenase), Atp5a (subunit of H+-ATPase), Kars (lysyl-tRNA synthetase), Lars2 (leucyl-tRNA synthetase 2), Yars2 (tyrosyl-tRNA synthetase 2), Tufm (Tu translation elongation factor, mitochondrial) and Tfb2m (transcription factor B2, mitochondrial) were decreased in mutant fishes, as compared with those in control fishes. As showed in Figure 5B, the levels of Nd1, Nd6, Cytb and Co2 in mtu1−/− mutant fish lines were 21%, 41%, 74% and 38%, with the average of 43%, relative to the mean value measured in the control fish lines (P = 0.0001). Furthermore, the average levels of these mtDNA-encoding proteins in mtu1+/− lines were 61%, relative to those in control lines. Notably, the levels of Sdhb and Atp5a in the mtu1−/− mutant fishes were 45% and 14% of the average values measured in the wild type fishes. As showed in Figure 5C, the levels of Kars, Lars2, Yars2, Tufm and Tfb2m in mtu1−/− mutant fished lines were 16%, 33%, 45%, 61% and 35%, relative to the mean value measured in the control fish lines (P < 0.0001).
Figure 5.
Western blot analysis of mitochondrial proteins. (A) Twenty μg of total proteins from mutant and wild type zebrafish were electrophoresed through a denaturing polyacrylamide gel, electroblotted and hybridized with antibodies for 6 subunits of OXPHOS (four encoded by mtDNA and two encoded by nuclear genes), Kars, Lars2, Yars2, Eftu, Tfb2m as well as Tom20 as a loading control. Quantification of levels of OXPHOS subunits (B) and other mitochondrial proteins (C). Average content of Nd1, Nd6, Cytb, Co2 and Atp5a, Sdhb, Kars, Lars2, Yars2, Eftu and Tfb2m was normalized to the average content of Tom20 in mutant and wild type zebrafish. The values for the mutant zebrafish are expressed as percentages of the average values for the wild type zebrafish. The horizontal dashed lines represent the average values of four proteins encoded by mtDNA or two proteins encoded by nuclear genes for each group. The calculations were based on three independent determinations. Graph details and symbols are explained in the legend to the Figure 3.
Deficient oxidative phosphorylation
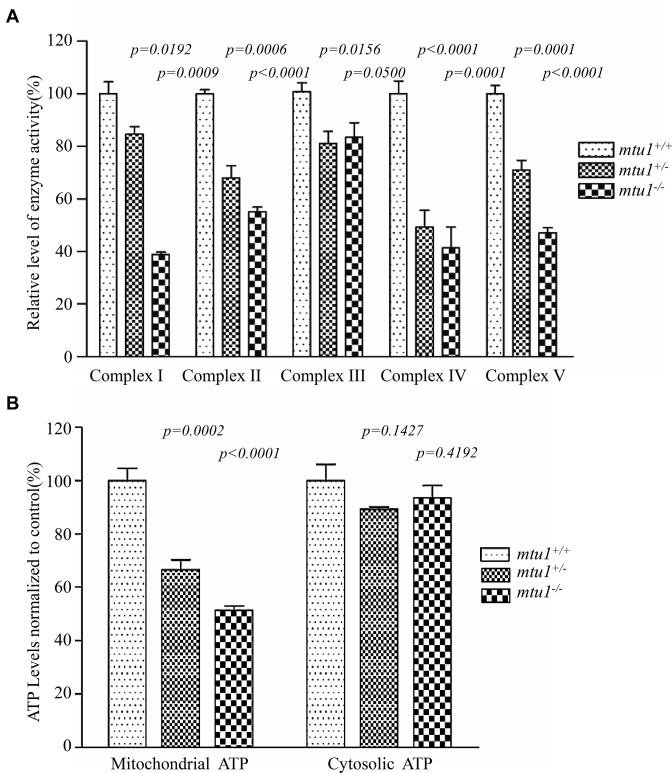
To evaluate the effect of the mtu1 mutation on the oxidative phosphorylation, we measured the activities of respiratory complexes by isolating mitochondria from mtu1+/−, mtu1−/− and wild type zebrafish. Complex I activity was determined by following the oxidation of NADH with ubiquinone as the electron acceptor (48,49). The activity of complex II (succinate ubiquinone oxidoreductase) exclusively encoded by the nuclear DNA was examined by the artificial electron acceptor DCPIP. Complex III (ubiquinone cytochrome c oxidoreductase) activity was measured as the reduction of cytochrome c (III) using D-ubiquinol-2 as the electron donor. Complex IV (cytochrome c oxidase) activity was monitored by following the oxidation of cytochrome c (II). As shown in Figure 6A, the activities of Complex I, II, III, IV and V in mtu1−/− mutant fish lines were 38.9% (P = 0.0009), 55.1% (P < 0.0001), 83.4% (P = 0.050), 41.4% (P = 0.0001), and 47.1% (P < 0.0001), relative to the mean values measured in the control fish lines, respectively. Furthermore, the activities of Complex I, II, III, IV and V in mtu1+/− mutant fish lines were 84.7% (P = 0.0192), 68.0% (P = 0.0006), 81.1% (P = 0.0156), 49.4% (P < 0.0001), and 71.0% (P = 0.0001) of the average values measured in control fish lines, respectively.
Figure 6.
Enzymatic activities of respiratory chain complexes and measurement of ATP levels. (A) The activities of respiratory complexes were investigated by enzymatic assays on complexes I, II, III, IV and V in mitochondria isolated from mtu1 mutant and wild type zebrafish. The calculations were based on three independent determinations. (B) Measurement of mitochondrial and cytosolic ATP levels. Average cytosolic ATP level (presence of oligomycin for inhibition of the mitochondrial ATP synthesis) and mitochondrial ATP level (subtraction of cytosolic ATP level from total cellular ATP levels) are shown. Three independent experiments were made for each genotype of zebrafish. Graph details and symbols are explained in the legend to Figure 3.
The effect of mtu1 mutation on the capacity of oxidative phosphorylation was further evaluated by measuring the cellular and mitochondrial ATP levels. Twenty μg of total cellular proteins derived from mutant and wild type embryos were assayed for cytosolic ATP levels (presence of oligomycin for inhibition of the mitochondrial ATP synthesis) and total cellular ATP levels. Mitochondrial ATP levels in mutant and wild type fishes were determined by subtraction of cytosolic ATP level from total cellular ATP levels. As shown in Figure 6B, the levels of mitochondrial ATP in mtu1+/− and mtu1−/− mutant fishes were 66.7% (P = 0.0002) and 51.3% (P < 0.0001), relative to the mean value measured in the wild type fish lines, respectively.
Loss of mtu1 caused the defects in the hearing organ of zebrafish at 5 dpf
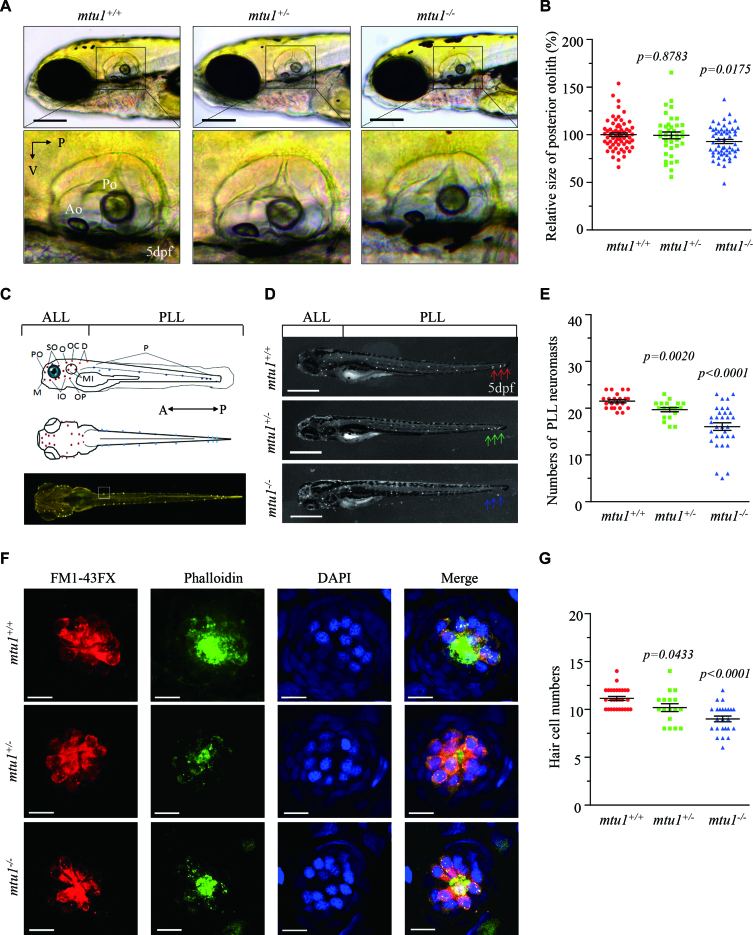
The hair cells in zebrafish are present in auditory and vestibular organs including saccule, utricle and lagena, as well as the lateral line system containing the neuromasts. To investigate if the loss of mtu1 altered the hearing and balance, we evaluated the startle response and swimming behaviors at 5 dpf. About 42% and 23% of larvae carrying the mtu1−/− or mtu1+/- mutation exhibited an abnormal swimming behavior and weak or non-responders to the startle stimulus, respectively. Larvae with weak or non-responders to the startle stimulus frequently sank to the bottom of the dishes in the head down position and slowly settled their lateral side (Supplementary Figure S2).
Zebrafish only have saccule and utricle within 5 days of development. By contrast, the lagena forms later during the juvenile stage of fish. Then, we measured the size of saccule otolith using mtu1−/− (n = 98), mtu1+/− (n = 95) and mtu1+/+ (n = 99) larvae at 5 dpf. As shown in Figure 7A and B, the sizes of saccular otolith in mtu1−/− mutant larvae were significantly reduced with average of 90%, relative to the mean sizes of wild type larvae (P = 0.0175), while there was no significant difference of saccule sizes between mtu1+/− mutant and wild type larvae. As shown in Figure 7C, the mtu1−/− (n = 32), mtu1+/− (n = 20) and mtu1+/+ (n = 21) larvae were then labeled with FM1-43FX to assess the effects on the neuromast numbers of anterior lateral line (ALL) and posterior lateral line (PLL). The numbers of neuromasts ALL in mtu1 mutant larvae were comparable with those in wild type larvae (Supplementary Figure S3). As illustrated in Figure 7D, E, numbers of PLL macula neuromasts were significantly reduced in mtu1−/− and mtu1+/− mutant larvae. Specifically, the wild type larvae have 22–23 lateral neuromasts (21.5 ± 0.3) in the posterior lateral line, whereas the lateral neuromasts in mtu1+/− and mtu1−/− mutant larvae were only 19.6 ± 0.4 and 16.1 ± 0.8, respectively.
Figure 7.
Defects in hearing organs in zebrafish at 5 dpf. (A) Otolith morphologies of mtu1−/−, mtu1+/− and mtu1+/+ zebrafish were illustrated under a Leica microscope with an objective magnification of 20 X. Lateral views of the otic vesicle were shown in the low arrows. V, ventral; P, posterior. (B) Quantification of sizes of posterior otolith in the mtu1−/−(n = 98), mtu1+/− (n = 95) and mtu1+/+ (n = 99) zebrafish. (C) Lateral and dorsal views of zebrafish larvae show the distribution of neuromasts along the body. A, anterior; P, posterior. (D) Larval of mtu1−/−, mtu1+/− and mtu1+/+ zebrafish were fluorescently labeled with FM1-43FX and observed under a stereoscopic microscope with an objective magnification of 20 X. Neuromast numbers of anterior lateral line (ALL) and posterior lateral line (PLL) in mutant and wild type fishes were counted, respectively. The arrows indicated the positions where the number of neuromasts were significantly reduced in the mutant fishes. (E) Quantification of neuromasts numbers of posterior lateral line (PLL) from the mtu1−/−, mtu1+/− mutant and wild type zebrafish. The calculations were based on the numbers of mtu1−/− (n = 32), mtu1+/− (n = 20) and mtu1+/+ (n = 21) larvae. (F) The hair cells, stereocilia and nuclei of neuromas from mutant and wild type zebrafish were stained with FM1-43FX (red), phalloidin (green) and DAPI (blue), respectively. Scale bars = 10 μm. (G) Quantification of numbers of hair cells for the mtu1−/−(n = 25), mtu1+/− (n = 17) and mtu1+/+ (n = 28) zebrafish. Graph details and symbols are explained in the legend to Figure 3.
Each neuromast of lateral line contains a cluster of hair cells surrounded by the nonsensory support cells. To examine if the mtu1 lesion changed hair cell numbers in neuromasts, mtu1−/− (n = 25), mtu1+/− (n = 17) and mtu1+/+ (n = 28) larvae were stained with FM1-43FX for hair cells, phalloidin for stereocilia and DAPI for nuclei, respectively. As shown in figure 7F, G, the numbers of hair cells in mtu1+/− and mtu1−/− neuromasts were significantly decreased, relative to the average numbers in mtu1+/+ neuromasts, by 8% and 20% (P = 0.0433, P < 0.0001), respectively. Moreover, the mtu1 mutant and wild type larvae were also stained with DASPI to assess the integrity of hair cell in lateral line neuromast and the results were consistent with the FM1-43FX staining experiments (Supplementary Figure S4).
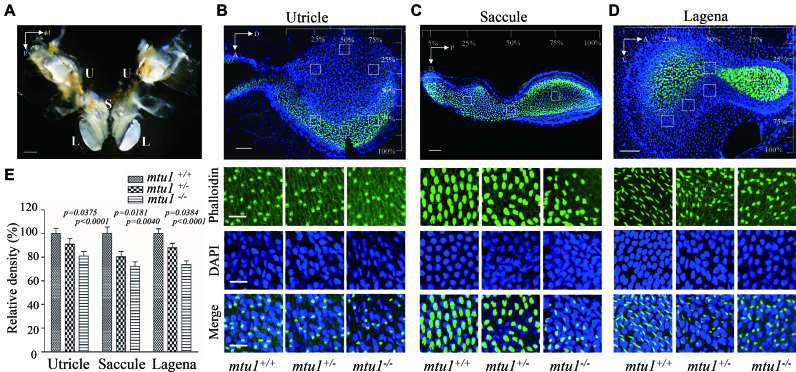
mtu1 mutants exhibited less hair bundles in zebrafish
To further investigate the effect of mtu1 mutation on hearing and balance, saccule, utricle and lagena from mutant and wild type zebrafish inner ear at the age of 10 months were labeled with phalloidin and DAPI for quantifying the hair cell bundle densities. Four discrete regions of saccule and six regions each from utricle and lagena from mtu1+/+, mtu1+/− and mtu1−/− zebrafish were selected for this analysis. As shown in Figure 8, the hair bundle densities in utricle, saccule and lagena of mtu1 mutant zebrafishes were significantly decreased, as compared with those in wild type fishes. The densities of hair bundles in the utricle, saccule and lagena of mtu1−/− zebrafish were reduced, relative to those of the average values in wild type zebrafish, by 20%, 28% and 27% (P < 0.0001, P = 0.0004, P < 0.0001), respectively. Furthermore, the densities of hair bundles in the utricle, saccule and lagena of mtu1+/− zebrafish were decreased, relative to those of the average values in wild type zebrafish, by 10%, 20% and 13% (P = 0.0375, 0.0180, 0.038), respectively.
Figure 8.
Reduced density of hair cell bundles in inner ear labyrinth. (A) Anatomy of adult wild type zebrafish inner ear labyrinth, showing two lagenas (L), two saccule (S), and two utricle (U). (B–D) Hair bundle density in lagenas, saccule and utricle. The hair cells, nuclei of from mutant and wild type zebrafish were stained with phalloidin (green) and DAPI (blue), respectively. Hair cell counts were sampled at six locations of lagenas, four locations of saccule and six locations of utricle, respectively. A 1600 μm2 box was placed at each sampling area and labeled hair cell bundles were counted within each box to determine hair cell density were selected for counting. A, anterior; D, dorsal; M, medial; P, posterior; V, ventral. (E). Quantification of density of hair cell bundles for the mtu1−/−, mtu1+/− and mtu1+/+ zebrafish. Graph details and symbols are explained in the legend to Figure 3.
Mitochondrial defects in hair cells
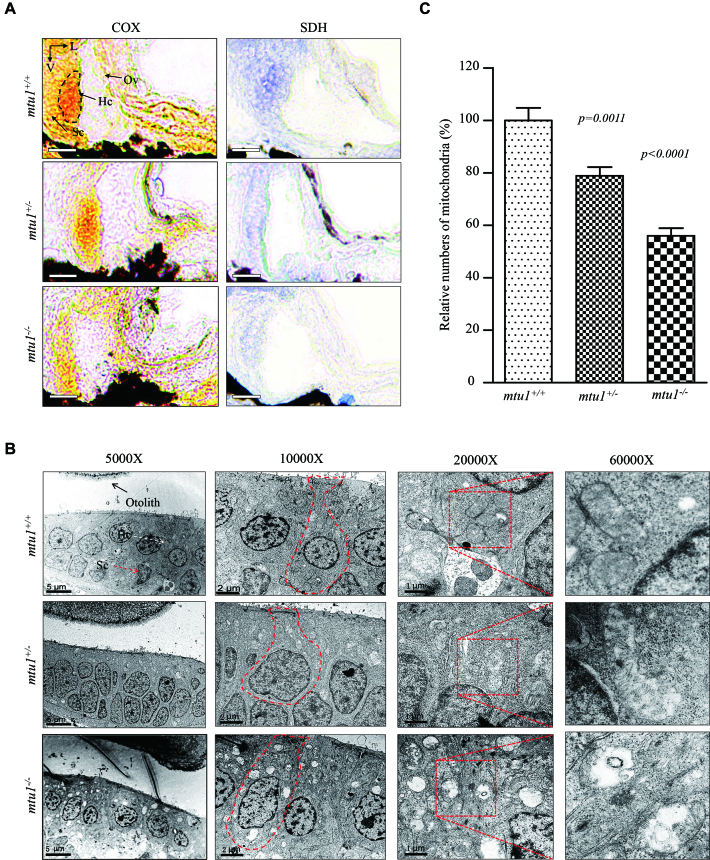
Mitochondrial dysfunction in hair cells of inner ear was assessed by enzyme histochemistry (EHC) staining for SDH and COX in the frozen-sections of posterior macula of the mtu1/−, mtu1+/− and mtu1+/+ larvae at 5 dpf. As shown in Figure 9A, the reduced activities of SDH and COX were observed in the anoterior and posterior macula of mtu1−/− and mtu1+/− mutant larvae, as compared to the wild type larvae. Mitochondrial defects in hair cells of mtu1−/− and mtu1+/− mutant larvae at 5 dpf were further evaluated by using transmission electron microscope. As shown in Figure 9B, hair cells of mtu1 mutant zebrafish exhibited abnormal mitochondrial morphology including vacuolated, fragmented mitochondria and the loss of cristae, as compared to those of wild type zebrafish. As shown in Figure 9C, mitochondrial numbers in mtu1−/− and mtu1+/− mutant cells were 78% and 56%, related to average values of control cells.
Figure 9.
Mitochondrial defects in hair cells. (A) Assessment of mitochondrial function in hair cells by enzyme histochemistry (EHC) staining for SDH and COX in the frozen-sections of posterior macula of mtu1−/−, mtu1+/− and mtu1+/+ zebrafish at 5dpf. Loss of EHC signal is indicated by arrows (magnification X400). V, ventral; L, lateral; Ov, otic vesicle; Hc, hair cell; Sc, supporting cell. (B) Mitochondrial networks from hair cells of inner electron microscopy. Ultrathin sections were visualized with 5000×, 10 000×, 20 000× and 60 000× magnifications. (C) Quantification of mitochondrial numbers of hair cells from the mtu1−/−, mtu1+/− mutant and mtu1+/+ zebrafish. The calculations were based on 50 different hair cells of mtu1−/−, mtu1+/− mutant and mtu1+/+ zebrafish, respectively. Graph details and symbols are explained in the legend to Figure 3.
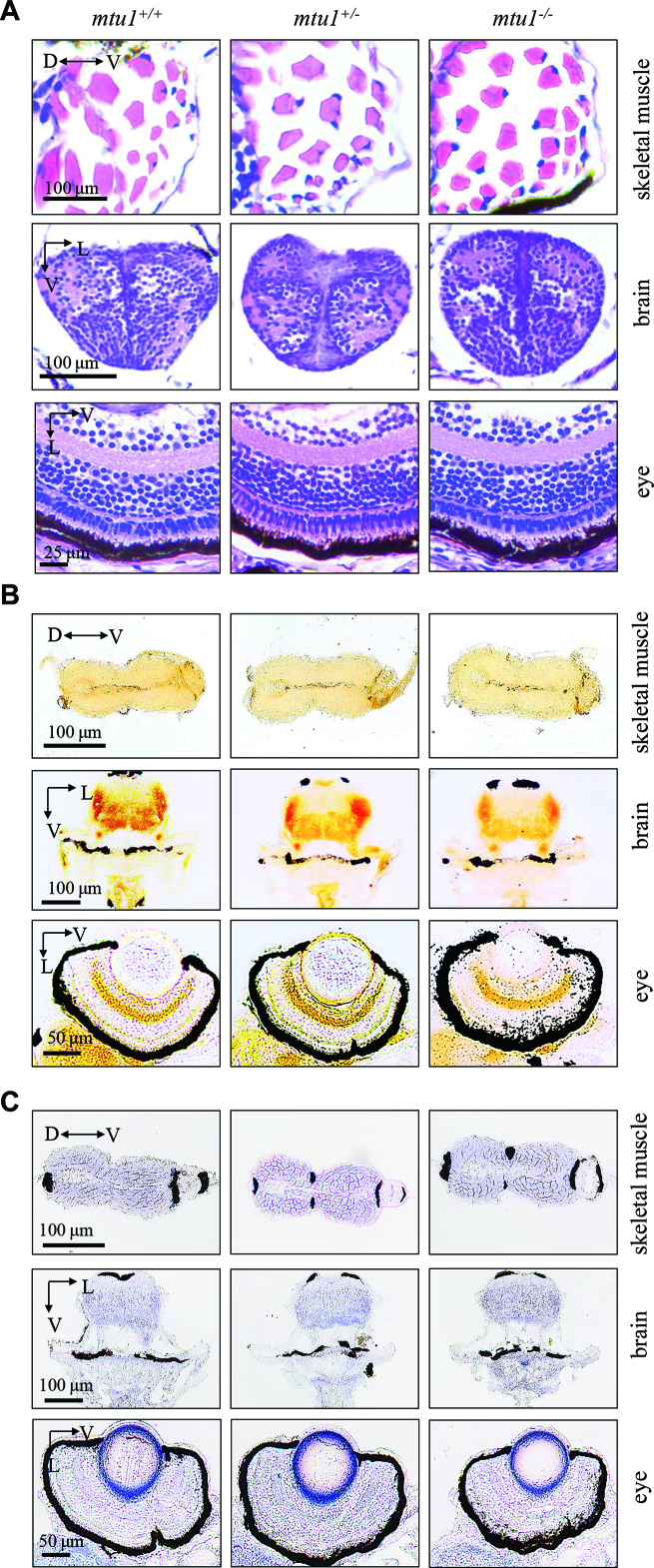
mtu1 −/− zebrafish at 5 dpf did not show the defects in brain, eyes and muscles
We then investigated the pathological effects of mtu1-knockout on other organs/tissues using the sections of skeletal muscles, brain and eyes from zebrafish at 5 dpf. As shown in Figure 10A, skeletal muscle, brain and retina sections of mtu1−/−, mtu1+/− and mtu1+/+ zebrafish at 5 dpf stained with hematoxylin/eosin (H&E) did not show obvious morphological differences. Mitochondrial dysfunctions were further examined by enzyme histochemistry (EHC) staining for SDH and COX in the frozen-sections of skeletal muscles, brain and retina of the mtu1−/−, mtu1+/− and mtu1+/+ larvae at 5 dpf. As shown in Figure 10B and C, there were no significant differences of SDH and COX activities in sections of skeletal muscles, brain, eye among mtu1−/−, mtu1+/− and mtu1+/+ zebrafish. These data suggested that the mtu1 knock-out did not affect the other organs/structures at 5 dpf.
Figure 10.
mtu1 −/− zebrafish at 5 dpf did not show the defects in muscle, brain and eyes. (A) Hematoxylin and eosin (HE) staining of skeletal muscles, brain and eye in the wild type (mtu1+/+), mtu1+/− and mtu1−/− zebrafish at 5 dpf. (B) Succinate dehydrogenase (SDH) and (C) cytochrome c oxidase (COX) staining of skeletal muscles, brain and eye in wild type (mtu1+/+), mtu1+/− and mtu1−/− zebrafish at 5 dpf. D, dorsal; P, posterior; V, ventral; L, lateral.
DISCUSSION
In the present study, we investigated the roles of MTU1 in mitochondrial biogenesis and hearing function by characterizing mtu1 knock-out zebrafish generated using CRISPR/Cas9 system. Mtu1 is a highly conserved tRNA modifying enzyme responsible for the biosynthesis of tτm5s2U at the wobble position of mitochondrial tRNAGln, tRNAGlu and tRNALys (26,33–35). Therefore, the primary defect in the mtu1 deletion was the deficient 2-thiouridine modification of U34 of tRNALys, tRNAGlu and tRNAGln. It was anticipated that the failure in tRNA metabolism caused by mtu1 deletion perturbed the mitochondrial protein synthesis and oxidative phosphorylation. In the present study, we demonstrated that the inactivation of mtu1 abolished the 2-thiouridylation in tRNALys, tRNAGlu and tRNAGln but did not affect the 2-thiouridylation in cytosolic tRNAGlu in the mtu1−/− mutant zebrafish. These data were consistent with the fact that the small interfering RNA down regulation of or other mutations of MTU1 led to the defects in 2-thiouridylation in mitochondrial tRNALys, tRNAGlu, and tRNAGln (26,27,54–56). Those unmodified tRNAs caused by the deletion of mtu1 likely leave the tRNAs more exposed to degradation, thereby lowering the steady-state level of those tRNAs (26,32,57). In this study, marked decreases in the steady-state levels of mitochondrial tRNALys, tRNAGlu and tRNAGln were significantly correlated with the deficient 2-thiouridylation of those tRNAs in mtu1−/−, and mtu1+/− zebrafish. Furthermore, the steady-state levels of mitochondrial tRNALeu(UUR), tRNATrp, tRNAMet and tRNAHis were also decreased significantly in in mtu1−/−, and mtu1+/− zebrafish. In fact, modifications other than the 2-thio modification occur in the wobble position of those tRNAs, such as 5-taurinomethyluridine in the tRNALeu(UUR) and 5-formylcytidine in the tRNAMet (6,23,24). Thus, the deletion of mtu1 likely leads to transcriptional/translational defects and thereby reduces the steady-state levels of those tRNAs, as in the case of human cells carrying the MTU1 A10S mutation (32).
It was anticipated that the deficient modifications at wobble position of tRNAs caused by the mtu1 deletion affect the fidelity and efficiency of mitochondrial translation. In fact, the τm5s2U modification is necessary for the decoding UUG codons as it serves to stabilize U:G wobble base-pairing by increasing stacking interactions (58,59). Thus, the deficient modifications at wobble position of tRNAs affected the decoding accuracy through altered codon-anticodon interactions and then led to mistranslation (60). Alternatively, these reduced levels of those tRNAs caused by the mtu1 deletion perturbed the efficiency of mitochondrial translation. In the present study, 57% and 39% reductions in the average levels of 4 mitochondrial proteins were observed in the mtu1−/− and mtu1+/− mutant zebrafish, respectively. Notably, a marked decrease was observed in the expression of Nd6, which is expressed from an mRNA rich in UUG codons, in mtu1−/− mutant zebrafish (58). These data were comparable with the reduced rates of mitochondrial translation observed in human cell lines carrying the MTU1 A10S mutation (26,32). Strikingly, the mtu1 deletion resulted in the lower levels of Atp5a of H+-ATPase and Sdhb of succinate ubiquinone oxidoreductase, Kars, Lars2, Yars2, Tufm and Tfb2m, encoded by nuclear genes. These data strongly indicated that the mtu1 deletion down-regulated the mitochondrial protein synthesis. The impairment of mitochondrial translations resulted in the respiratory deficiency, deficient ATP synthesis, and subsequent failure of cellular energetic process (61,62). In this study, the low expressions of subunits of OXPHOS, including mtDNA encoding polypeptides, Sdhb and Atp5a were apparently responsible for the defective activities of complex I, II, III, IV and V. The respiratory deficiency then gave rise to significant decreases in mitochondrial ATP production and subsequent failure of cellular energetic process in the mtu1−/− and mtu1+/− mutant zebrafish (53,54). These mitochondrial dysfunctions were further evidenced by the reduced activities of SDH and COX observed in the anoterior and posterior macula, abnormal mitochondrial morphology and reduced numbers of mitochondria in the hair cells of mtu1−/− mutant larvae. These data demonstrated that the mtu1 knock-out zebrafish recapitulated the biochemical phenotypes in human cell lines carrying the mutations and knockdown of MTU1 genes (26,27,32,54,55).
The function of auditory organs depends on a very high rate of ATP production (63–65). Therefore, it was hypothesized that the mitochondrial dysfunction caused by MTU1 deficiency particularly ablated the function of hair cells in zebrafish. The hair cells in zebrafish are present within neuromasts of both auditory and vestibular organs including saccule, utricle and lagena, as well as the lateral line system (66). Each neuromast contain develop 15–20 hair cells surrounded by the nonsensory support cells (67). In this study, the mtu1−/− mutant larvae at 5 dpf displayed the abnormal swimming behavior, weak response to the startle stimulus and the reduced sizes of saccular otolith, indicating that the loss of mtu1 altered the function of hearing and balance. However, mtu1−/− zebrafish at 5 dpf did not show the defects in brain, eyes and muscles. These defects in hearing organs were further evidenced by the fact that the mtu1−/− mutant zebrafish exhibited the significant decreases in the numbers of neuromast in the posterior lateral line and the loss of hair cells in the each neuromast of lateral lines. Furthermore, mtu1−/− mutant zebrafishes displayed the significant decreases in the hair bundle densities in utricle, saccule and lagena. These data demonstrated that the mitochondrial dysfunction caused by the deletion of mtu1 caused hearing impairment. Thus, our findings may provide new insights into the pathophysiology of deafness, which was manifested by the deficient modifications at wobble position of mitochondrial tRNAGln, tRNAGlu and tRNALys.
In summary, our findings convincingly demonstrate the molecular mechanism underlying MTU1 deficiency using the mtu1 knock-out zebrafish generated by CRISPR/Cas9 system. We demonstrated the abolished 2-thiouridine modification of U34 of mitochondrial tRNALys, tRNAGlu and tRNAGln in the mtu1 knock-out zebrafish. The elimination of τm5s2U modification mediated mitochondrial tRNA metabolisms, causing the global decreases in the levels of mitochondrial tRNAs. The aberrant mitochondrial tRNA metabolisms resulted in the impairment of mitochondrial translation, respiratory phenotypes and reductions of mitochondrial ATP production. These mitochondria dysfunctions caused the defects in hearing organs. Strikingly, the mtu1−/− mutant zebrafish displayed the abnormal startle response and swimming behaviors, significant decreases in the sizes of saccular otolith and numbers of hair cells in the auditory organs. The mtu1−/− mutant zebrafish exhibited the significant decreases in the hair bundle densities in utricle, saccule and lagena. Thus, our findings demonstrated the essential role of tRNA modification in mitochondrial biogenesis and deafness.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Priorities Program of China [2014CB541704 to M.X.G., 2014CB541702 to Y.C.]; National Natural Science Foundation of China [81330024 to M.X.G; 81470685 to Y.C.]. Funding for open access charge: National Basic Research Priorities Program of China [2014CB541704 to M.X.G., 2014CB541702 to Y.C.]; National Natural Science Foundation of China [81330024 to M.X.G; 81470685 to Y.C.].
Conflict of interest statement. None declared.
REFERENCE
- 1. Jonkhout N., Tran J., Smith M.A., Schonrock N., Mattick J.S., Novoa E.M.. The RNA modification landscape in human disease. RNA. 2017; 23:1754–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang X., Cozen A.E., Liu Y., Chen Q., Lowe T.M.. Small RNA Modifications: Integral to function and disease. Trends Mol. Med. 2016; 22:1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011; 45:299–329. [DOI] [PubMed] [Google Scholar]
- 4. Wang M., Liu H., Zheng J., Chen B., Zhou M., Fan W., Wang H., Liang X., Zhou X., Eriani G. et al. A deafness- and diabetes-associated tRNA mutation causes deficient pseudouridinylation at position 55 in tRNAGlu and mitochondrial dysfunction. J. Biol. Chem. 2016; 291:21029–21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou M., Xue L., Chen Y., Li H., He Q., Wang B., Meng F., Wang M., Guan M.X.. A hypertension-associated mitochondrial DNA mutation introduces an m1G37 modification into tRNAMet, altering its structure and function. J. Biol. Chem. 2018; 293:1425–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki T., Suzuki T.. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014; 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El Yacoubi B., Bailly M., de Crécy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 8. Duechler M., Leszczynska G., Sochacka E., Nawrot B.. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol. Life Sci. 2016; 73:3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gingold H., Pilpel Y.. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011; 7:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allner O., Nilsson L.. Nucleotide modifications and tRNA anticodon-mRNA codon interactions on the ribosome. RNA. 2011; 17:2177–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klassen R., Ciftci A., Funk J., Bruch A., Butter F., Schaffrath R.. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016; 44:10946–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drummond D.A., Wilke C.O.. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008; 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johansson M.J., Esberg A., Huang B., Bjork G.R., Bystrom A.S.. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008; 28:3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haag S., Sloan K.E., Ranjan N., Warda A.S., Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016; 35:2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chatterjee K., Nostramo R.T., Wan Y., Hopper A.K.. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim. Biophys. Acta. 2018; 1861:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang M., Peng Y., Zheng J., Zheng B., Jin X., Liu H., Wang Y., Tang X., Huang T., Jiang P. et al. A deafness-associated tRNAAsp mutation alters the m1G37 modification, aminoacylation and stability of tRNAAsp and mitochondrial function. Nucleic Acids Res. 2016; 44:10974–10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merante F., Myint T., Tein I., Benson L., Robinson B.H.. An additional mitochondrial tRNAIle point mutation (A-to-G at nucleotide 4295) causing hypertrophic cardiomyopathy. Hum. Mutat. 1986; 8:216–222. [DOI] [PubMed] [Google Scholar]
- 18. Liu Y., Li R., Li Z., Wang X.J., Yang L., Wang S., Guan M.X.. Mitochondrial transfer RNAMet 4435A>G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension. 2009; 53:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yarham J.W., Lamichhane T.N., Pyle A., Mattijssen S., Baruffini E., Bruni F., Donnini C., Vassilev A., He L., Blakely E.L. et al. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 2014; 10:e1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Powell C.A., Kopajtich R., D'Souza A.R., Rorbach J., Kremer L.S., Husain R.A., Dallabona C., Donnini C., Alston C.L., Griffin H. et al. TRMT5 mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am. J. Hum. Genet. 2015; 97:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K.. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002; 21:6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Asano K., Suzuki T., Saito A., Wei F.Y., Ikeuchi Y., Numata T., Tanaka R., Yamane Y., Yamamoto T., Goto T. et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 2018; 46:1565–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakano S., Suzuki T., Kawarada L., Iwata H., Asano K., Suzuki T.. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat. Chem. Biol. 2016; 12:546–551. [DOI] [PubMed] [Google Scholar]
- 24. Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A., Rorbach J., Lantaff R., Blanco S., Sauer S. et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016; 7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 26. Meng F., Cang X., Peng Y., Li R., Zhang Z., Li F., Fan Q., Guan A.S., Fischel-Ghosian N., Zhao X. et al. Biochemical evidence for a nuclear modifier allele (A10S) in TRMU (methylaminomethyl-2-thiouridylate-methyltransferase) related to mitochondrial trna modification in the phenotypic manifestation of deafness-associated 12S rRNA mutation. J. Biol. Chem. 2017; 292:2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T.. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005; 280:1613–1624. [DOI] [PubMed] [Google Scholar]
- 28. Wang X., Yan Q., Guan M.X.. Combination of the loss of cmnm5U34 with the lack of s2U34 modifications of tRNALys, tRNAGlu, and tRNAGln altered mitochondrial biogenesis and respiration. J. Mol. Biol. 2010; 395:1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X., Li R., Lin X., Guan M.X.. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12S rRNA A1555G mutation. J. Biol. Chem. 2002; 277:27256–27264. [DOI] [PubMed] [Google Scholar]
- 30. Li X., Guan M.X.. A human mitochondrial GTP binding protein related to tRNA modification may modulate the phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell. Biol. 2002; 22:7701–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan Q., Li X., Faye G., Guan M.X.. Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15S rRNA. J. Biol. Chem. 2005; 280:29151–29157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guan M.X., Yan Q., Li X., Bykhovskaya Y., Gallo-Teran J., Hajek P., Umeda N., Zhao H., Garrido G., Mengesha E. et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am. J. Hum. Genet. 2006; 79:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kambampati R., Lauhon C.T.. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry. 2003; 42:1109–1117. [DOI] [PubMed] [Google Scholar]
- 34. Yan Q., Guan M.X.. Identification and characterization of mouse TRMU gene encoding the mitochondrial 5-methylaminomethyl-2-thiouridylate-methyltransferase. Biochim. Biophys. Acta. 2004; 1676:119–126. [DOI] [PubMed] [Google Scholar]
- 35. Armengod M.E., Meseguer S., Villarroya M., Prado S., Moukadiri I., Ruiz-Partida R., Garzón M.J., Navarro-González C., Martínez-Zamora A.. Modification of the wobble uridine in bacterial and mitochondrial tRNAs reading NNA/NNG triplets of 2-codon boxes. RNA Biol. 2014; 11:1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeharia A., Shaag A., Pappo O., Mager-Heckel A.M., Saada A., Beinat M., Karicheva O., Mandel H., Ofek N., Segel R. et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009; 85:401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L., Smertenko T., Alston C.L., Neeve V.C., Best A. et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA. 2014; 312:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y., Wei F.Y., Kawarada L., Suzuki T., Araki K., Komohara Y., Fujimura A., Kaitsuka T., Takeya M., Oike Y. et al. Mtu1-Mediated thiouridine formation of mitochondrial tRNAs is required for mitochondrial translation and is involved in reversible infantile liver injury. PLoS Genet. 2016; 12:e1006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 2000; Eugene: University of Oregon Press. [Google Scholar]
- 40. Chen D., Li F., Yang Q., Tian M., Zhang Z., Zhang Q., Chen Y., Guan M.X.. The defective expression of gtpbp3 related to tRNA modification alters the mitochondrial function and development of zebrafish. Int. J. Biochem. Cell. Biol. 2016; 77:1–9. [DOI] [PubMed] [Google Scholar]
- 41. Moens C. Whole mount RNA in situ hybridization on zebrafish embryos: mounting. CSH Protoc. 2008; doi:10.1101/pdb.prot5038. [DOI] [PubMed] [Google Scholar]
- 42. Moens C. Whole mount RNA in situ hybridization on zebrafish embryos: hybridization. CSH Protoc. 2008; doi:10.1101/pdb.prot5037. [DOI] [PubMed] [Google Scholar]
- 43. Moens C. Whole mount RNA in situ hybridization on zebrafish embryos: probe synthesis. CSH Protoc. 2008; doi:10.1101/pdb.prot5036. [DOI] [PubMed] [Google Scholar]
- 44. Zou B., Mittal R., Grati M., Lu Z., Shu Y., Tao Y., Feng Y., Xie D., Kong W., Yang S. et al. The application of genome editing in studying hearing loss. Hear Res. 2015; 327:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gong S., Peng Y., Jiang P., Wang M., Fan M., Wang X., Zhou H., Li H., Yan Q., Huang T. et al. A deafness-associated tRNAHis mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res. 2014; 42:8039–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Broughton R.E., Milam J.E., Roe B.A.. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001; 11:1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shigi N., Suzuki T., Tamakoshi M., Oshima T., Watanabe K.. Conserved bases in the T Psi C loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J. Biol. Chem. 2002; 277:39128–39135. [DOI] [PubMed] [Google Scholar]
- 48. Birch-Machin M.A., Turnbull D.M.. Methods in Cell Biology. 2001; 65:Academic Press; 97–117. [DOI] [PubMed] [Google Scholar]
- 49. Zhang J., Jiang P., Jin X., Liu X., Zhang M., Xie S., Gao M., Zhang S., Sun Y.-H., Zhu J. et al. Leber's hereditary optic neuropathy caused by the homoplasmic ND1 m.3635G>A mutation in nine Han Chinese families. Mitochondrion. 2014; 18:18–26. [DOI] [PubMed] [Google Scholar]
- 50. Phillips M.J., Webb-Wood S., Faulkner A.E., Jabbar S.B., Biousse V., Newman N.J., Do V.T., Boatright J.H., Wallace D.C., Pardue M.T.. Retinal function and structure in Ant1-deficient mice. Invest. Ophthalmol. Vis. Sci. 2010; 51:6744–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bang P.I., Sewell W.F., Malicki J.J.. Morphology and cell type heterogeneities of the inner ear epithelia in adult and juvenile zebrafish (Daniorerio). J. Comp. Neurol. 2001; 438:173–190. [DOI] [PubMed] [Google Scholar]
- 52. Haden M., Einarsson R., Yazejian B.. Patch clamp recordings of hair cells isolated from zebrafish auditory and vestibular end organs. Neuroscience. 2013; 248:79–87. [DOI] [PubMed] [Google Scholar]
- 53. Gale J.E., Marcotti W., Kennedy H.J., Kros C.J., Richardson G.P.. FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J. Neurosci. 2001; 21:7013–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sasarman F., Antonicka H., Horvath R., Shoubridge E.A.. The 2-thiouridylase function of the human MTU1 (TRMU) enzyme is dispensable for mitochondrial translation. Hum. Mol. Genet. 2011; 20:4634–4643. [DOI] [PubMed] [Google Scholar]
- 55. Boczonadi V., Smith P.M., Pyle A., Gomez-Duran A., Schara U., Tulinius M., Chinnery P.F., Horvath R.. Altered 2-thiouridylation impairs mitochondrial translation in reversible infantile respiratory chain deficiency. Hum. Mol. Genet. 2013; 22:4602–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Navarro-Gonzalez C., Moukadiri I., Villarroya M., Lopez-Pascual E., Tuck S., Armengod M.E.. Mutations in the Caenorhabditis elegans orthologs of human genes required for mitochondrial tRNA modification cause similar electron transport chain defects but different nuclear responses. PLoS Genet. 2017; 13:e1006921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fakruddin M., Wei F.Y., Suzuki T., Asano K., Kaieda T., Omori A., Izumi R., Fujimura A., Kaitsuka T., Miyata K. et al. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 2018; 22:482–496. [DOI] [PubMed] [Google Scholar]
- 58. Kirino Y., Goto Y., Campos Y., Arenas J., Suzuki T.. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:7127–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kurata S., Weixlbaumer A., Ohtsuki T., Shimazaki T., Wada T., Kirino Y., Takai K., Watanabe K., Ramakrishnan V., Suzuki T.. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008; 283:18801–18811. [DOI] [PubMed] [Google Scholar]
- 60. Reverendo M., Soares A.R., Pereira P.M., Carreto L., Ferreira V., Gatti E., Pierre P., Moura G.R., Santos M.A.. tRNA mutations that affect decoding fidelity deregulate development and the proteostasis network in zebrafish. RNA Biol. 2014; 11:1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005; 39:359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Szczepanowska J., Malinska D., Wieckowski M.R., Duszynski J.. Effect of mtDNA point mutations on cellular bioenergetics. Biochim. Biophys. Acta. 2012; 1817:1740–1746. [DOI] [PubMed] [Google Scholar]
- 63. Guan M.X. Molecular pathogenetic mechanism of maternally inherited deafness. Ann. N.Y. Acad. Sci. 2004; 1011:259–271. [DOI] [PubMed] [Google Scholar]
- 64. Schapira A.H. Mitochondrial diseases. Lancet. 2012; 327:102–108. [DOI] [PubMed] [Google Scholar]
- 65. Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum. Mutat. 1999; 13:261–270. [DOI] [PubMed] [Google Scholar]
- 66. Nicolson T. The genetics of hearing and balance in zebrafish. Annu. Rev. Genet. 2005; 39:9–22. [DOI] [PubMed] [Google Scholar]
- 67. Raible D.W., Kruse G.J.. Organization of the lateral line system in embryonic zebrafish. J. Comp. Neurol. 2000; 421:189–198. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.