Abstract
Proper DNA double-strand break (DSB) repair is essential for maintaining genome integrity. Microhomology-mediated end joining (MMEJ) is an error-prone repair mechanism, which introduces mutations at break sites and contributes to chromosomal translocations and telomere fusions, thus driving carcinogenesis. Mitotic kinases PLK1, CDK1 and Aurora A are important for supporting MMEJ and are often overexpressed in various tumors. However, the functional interplay between these kinases and MMEJ has not been explored. Here, we found that MMEJ is preferentially employed to fix DSBs in cells arrested in mitosis following nocodazole treatment. We further showed that the DSB repair factor CtIP is jointly phosphorylated by CDK1/Aurora A and PLK1. CDK1/Aurora A-mediated CtIP phosphorylation at serine 327 triggers CtIP binding to the PLK1 polo-box domain, which in turn facilitates PLK1 to phosphorylate CtIP mainly at serine 723. A PLK1 phosphor-mimic CtIP mutant fails to initiate extended end resection and is thus unable to mediate homologous recombination and the G2/M checkpoint but can mediate MMEJ. These data imply that PLK1 may target CtIP to promote error-prone MMEJ and inactivate the G2/M checkpoint. These findings have helped elucidate the oncogenic roles of these factors.
INTRODUCTION
Double-strand breaks (DSBs) are repaired in mammalian cells via two main mechanisms: Ku-dependent classical non-homologous end joining (C-NHEJ) and homologous recombination (HR) (1,2). HR is initiated by processing and cutting the DSB ends to generate 3′ single-stranded DNA (ssDNA) tails, which are then bound by Rad51 recombinase to initiate homologous pairing, strand invasion and finish DSB repair by HR with the aid of a series of recombination mediator proteins and nucleases (3). Because an identical sister chromatid template is required for accurate DSB repair, HR is usually restricted to the late S/G2 phases of the cell cycle, and is considered an error-free process (2,4). Conversely, C-NHEJ can occur throughout the cell cycle to repair DSBs by direct ligation of DNA ends without extensive processing; thus, it does not require a homologous template and is associated with small alterations at junctions (1). Microhomology-mediated end joining (MMEJ) has been described as an alternative DSB repair mechanism (5,6). MMEJ is a mutagenic DSB repair process that induces a deletion or insertion around a DSB and thus contributes to the formation of chromosome rearrangements, including translocations and telomere fusion (7,8). MMEJ was originally considered as a back-up repair mechanism in Ku-deficient cells (9,10). However, recent studies have shown that it is also activated in normal, cycling cells (where both C-NHEJ and HR pathways are functional) and contributes to the survival of HR-defective tumors (11,12).
The choice between different DSB repair pathways depends on the phase of the cell cycle and the nature of the DSB. Selecting the appropriate DSB repair pathway has a critical impact on genome integrity and tumorigenesis (4,13,14). An essential determinant of DSB repair pathway choice is the 5′-3′ resection of DSB ends, which promotes HR-mediated repair and prevents Ku-dependent C-NHEJ (4,15). A two-step resection model has been established based on studies performed in several model organisms (15–18). In mammals, the Mre11-Rad50-NBS1 (MRN) complex and CtIP (CtBP-interacting protein) work together to expose short, ssDNA regions. This exposure promotes BLM-DNA2-Exo1 and RPA recruitment to these regions to generate extended 3′-ssDNA for HR-mediated repair (19). MMEJ is MRN-CtIP dependent but BLM-Exo1-RPA independent, suggesting that the limited length of ssDNA that is derived from the first step of resection is sufficient to initiate MMEJ (6,11). Inadequate 5′-3′ resection is, therefore, an important reason as to why cells unduly fix DSB damage via MMEJ, resulting in genomic instability and carcinogenesis.
CtIP function in DSB repair is tightly controlled by cell-cycle-dependent modifications. Phosphorylation of a conserved cyclin-dependent kinase (CDK) site (threonine 847; T847) at the CtIP C terminus during S/G2 phase is required for efficient end resection and resection-dependent repair via MMEJ or HR (20,21). CDK-mediated phosphorylation of CtIP at serine 327 (S327) is also critical for BRCA1 (breast cancer gene 1) binding and end resection regulation. Some debate as to the role of CtIP S327 phosphorylation, however, has been raised (22–24). In addition, phosphorylation of the five CDK sites located in the CtIP central domain permits CtIP to interact with Nbs1 (Nijmegen breakage syndrome1) via its FHA–BRCT domains, which in turn allows ATM (ataxia telangiectasia mutated) to phosphorylate CtIP and facilitate end resection upon DNA damage (21). Interestingly, a recent study found that CtIP can be sequentially phosphorylated at S327 and T847 by PLK3 (polo-like kinase 3) during G1 phase in a DNA damage-dependent manner and this phosphorylation is required for complex DSB repair to occur in G1 (25).
PLK1 (polo-like kinase 1) is a well-defined cell-cycle regulator that is expressed from early S phase to late M phase and has numerous functions during mitosis progression (26). PLK1 is activated by Bora/Aurora A during the G2/M transition and usually binds CDK-phosphorylated targets through its Polo-Box Domain (PBD) to phosphorylate them. The kinase activity of PLK1 is not essential for normal cell-cycle progression, but is indispensable for the G2/M transition in cells attempting to recover from DNA damage (27–29).
In response to DSBs, mammalian cells activate phosphatidylinositol 3-kinase-like kinases (PIKKs), including ATM and ATR (ATM and Rad3-related), to modulate checkpoint activation and DSB repair via cascades of phosphorylation (30). ATM is activated by the MRN complex and mainly responds to DSBs. ATR is activated by ATRIP (ATR-interacting protein), RPA (replication protein A), TOPBP1 (topoisomerase binding protein 1) and Claspin and responds to single-strand DNA lesions. Activated ATM and ATR phosphorylate CHK2 and CHK1, respectively, which results in downstream signal transduction and effector protein phosphorylation to initiate the S-phase and G2-phase checkpoints in mammalian cells (31–33).
In this study, we used the previously described direct repeat enhanced green fluorescent protein (DR-EGFP) reporter system (34) combined with a modified DSB induction method to study DSB repair in nocodazole-arrested mitotic cells. We found that these cells use MMEJ to fix DSB damage. DSB end resection factor CtIP is concerted phosphorylated by CDK1/Aurora A and PLK1, and PLK1-mediated phosphorylation of CtIP promotes MMEJ-mediated DSB repair. A PLK1 phosphor-mimic CtIP mutant fails to mediate extensive end resection and thus suppresses HR, promotes MMEJ and disrupts the DNA damage-induced G2/M transition checkpoint. These data suggest a new mechanism involving the mitosis kinases PLK1, CDK1 and Aurora A in DSB repair via CtIP phosphorylation. These findings further our understanding as to how the regulation of DSB repair in mammalian cells during the cell cycle can affect genomic stability and contribute to carcinogenesis.
MATERIALS AND METHODS
Cell culture, plasmid construction and recombinant protein expression
Human U2OS, HCT116 and 293T cells were cultured at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The Sf9 insect cell line was cultured in Sf-900II SFM medium (Gibco). CtIP-knockout HCT116 cells (35) and EGFP-HR and EGFP-MMEJ reporter cells (34,36) were generated and cultured as previously described (35).
CtIP, PLK1 and Aurora A cDNAs were subcloned into a pcDNA3 vector (Invitrogen). Point mutations, small hairpin RNA (shRNA) target site-resistant mutations and cDNA fragments were generated using the QuikChange Site-directed Mutagenesis Kit (Stratagene) or by polymerase chain reaction (PCR) amplification and ligation into the mammalian expression vector pcDNA3 or NBLV0051 (Novo Bio) vector containing a 3x N-terminal Flag, HA or Myc epitope. EGFP-tagged CtIP or the indicated mutants were generated using the EGFP-C1 expression vector (Clontech). Bacteria expressing His-tagged or GST-tagged PLK1, Aurora A or CtIP fragments were generated using the pET28a (Invitrogen) or pGEX6T-1 (GE Healthcare) system, respectively. Doxycycline-inducible lentiviral expression vectors containing the humanized Streptococcus pyogenes Cas9 plasmid were obtained from Addgene (#50661). The sgRNA expression vector was synthesized using two pairs of annealed oligomers (HRF5′-CACCGCTTCAAGCTTTAGGGATAAC-3′, HRR5′-AAACGTTATCCCTAAAGCTTGAAGC-3′, MMEJF5′-CACCGCGCGCCGAGTAGGGATAAC-3′, MMEJR5′-AAACGTTATCCCTACTCGGCGCGC-3′), which were inserted into the BsmBI site of the lenti Guide-puro vector (Addgene #52963). The final constructs encoded an single guide RNA (sgRNA) that targeted the I-SceI-recognizing region in the EGFP-HR or EGFP-MMEJ reporters (Supplementary Figure S1A and S1B). Stable expression of exogenous proteins in the indicated U2OS or HCT116 cells was achieved by lentiviral infection followed by relevant drug selection. Recombinant baculoviruses expressing GST-tagged wild-type (WT) CtIP or the indicated mutants were generated using the Bac-to-Bac baculovirus expression system (Invitrogen), as previously described (21,36).
shRNA interference
Silencing of endogenous CtIP and BLM was achieved by retroviral or lentiviral infection using pMKO or pLKO vectors to express corresponding shRNAs. shRNA sequences for CtIP (5′-GAGCAGACCUUUCUCAGUAUA-3′) and BLM (5′-GAGCACAUCUGUAAAUUAAUU-3′) have been previously described (11,36). The shRNA-resistant CtIP WT and indicated mutants were constructed by mutating four nucleotides in the shRNA-targeting sequence (36) by site-directed mutagenesis (Stratagene). Nocodazole (M1404) and thymidine (T1895) were purchased from Sigma; BI2536 (S1109), Roscovitine (CYC202), RO3306 (S7747) and MLN8054 (S1100) were purchased from Selleck.
Immunoblotting, immunoprecipitation and immunostaining
Whole-cell lysis, immunoblotting, immunoprecipitation and immunostaining were performed as previously described (21,34). Antibodies against CtIP, BRCA1, Chk1 and RPA were used according to previously derived conditions (21,34,36). Anti-HA (A190–208A), anti-MYC (A190–205A), anti-BLM (A300) and anti-pH3S10 (A301-844A) were purchased from Bethyl. Anti-FLAG M2 (F1804) and anti-β-actin (A5441) were purchased from Sigma. Anti-PLK1 (sc-17783) was purchased from Santa Cruz. Rabbit phospho-specific antibodies against CtIP (S327), CtIP (S723) and CtIP (S593) were generated and affinity-purified by Beijing B&M Biotech using the phospho-peptides.
Protein purification
Full-length, WT GST-CtIP was expressed in Sf9 insect cells and purified by affinity purification, as previously described (36). GST-fused CtIP fragments (GST-CtIP 200-460, GST-CtIP 460-600, GST-CtIP 600-770), full-length WT, indicated mutants and GST-BRCT proteins were expressed in BL21 cells and affinity-purified using glutathione-Sepharose4B (GE Healthcare). His-PLK1 and His-Aurora A were expressed in BL21 cells and purified using Ni-NTA agarose (Qiagen), according to the manufacturer's instructions. Protein complex purification was performed as previously described with some modifications (37). Briefly, 293T cells were incubated with nocodazole for 16 h and then lysed by sonication in lysis buffer [20 mM Tris–HCl (pH 8.0), 50 mM KCl, 0.5 mM dithiothreitol (DTT)] containing a protease inhibitor cocktail (‘PIC’, cOmplete, Roche). After centrifugation, the supernatant was loaded onto a Q sepharose FF column (GE Healthcare), equilibrated with equilibration buffer [20 mM Tris–HCl (pH 8.0), 100 mM KCl, and 0.5 mM DTT] and step-eluted in 300 mM KCl and 500 mM KCl, all in equilibration buffer. The 300 mM fraction was then added to a Superose 6 column (GE Healthcare). Fractions were collected and analyzed by western blotting using the indicated antibodies.
In vitro kinase assay
For in vitro kinase assays, bacteria-purified His-PLK1, His-Aurora A or commercially obtained CDK1-cyclinB recombinant protein (New England Biolabs) were incubated with purified protein substrates in the presence of 10 μCi γ-32P-ATP (Perkin Elmer) in kinase buffer [25 mM HEPES buffer (pH 7.4), 50 mM NaCl,1 mM Na3VO4,10 mM MgCl2, 1 mM DTT, 10 μM ATP] (21). The kinase reaction was carried out at 30°C for 30 min. Phosphorylated proteins were analyzed by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Mass spectrometry
Phosphorylation site mapping by mass spectrometry was performed as previously described (21). Briefly, 293T cells containing Flag-CtIP expression plasmids were treated with nocodazole for 16 h or not, and then the cells were harvested and lysed in NETN buffer [150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 20 mM Tris–HCl (pH 8.0), 0.5% NP-40] containing a protease inhibitor cocktail (‘PIC’, cOmplete, Roche). The cell lysate was cleared by centrifugation and the supernatant was applied to anti-Flag M2-conjugated agarose beads (Sigma) pre-equilibrated with NETN buffer containing a protease inhibitor cocktail (‘PIC’, cOmplete, Roche). After incubation at 4°C for 3 h, the beads were centrifuged and washed extensively in NETN buffer. Subsequently, Flag-CtIP was eluted with NETN buffer containing FLAG peptide and analyzed by mass spectrometry.
Laser-induced microirradiation and live-cell imaging
U2OS cells expressing the indicated EGFP-tagged or mRFP-tagged proteins were cultured in sterile glass-bottom dishes (MatTek) in DMEM supplemented with 10% FBS. DSBs were generated in live-cell nuclei by local irradiation with a 365 nm pulsed nitrogen UV laser (16 Hz pulse, 41% laser output) generated from the Micropoint System (Andor). Live-cell, time-lapse images were captured on a Nikon A1 confocal imaging system directly coupled to the Micropoint System. Raw images were imported into ImageJ (NIH of USA) for processing. The fluorescence intensity of the micro-irradiated area was determined by measuring the mean absolute intensity of the micro-irradiated areas, with the mean cellular background intensity subtracted. Each data point shown is the average of 10 independent measurements (21).
HR and MMEJ assay
U2OS cells carrying an EGFP-HR or EGFP-MMEJ reporter (34) and stably expressed doxycycline (Dox)-inducible Cas9 (38) and sgRNA were used for the HR and MMEJ assay. sgRNA directs the Cas9 endonuclease to the I-SceI recognition site to generate DSBs (Supplementary Figure S1). The cells were incubated with Dox for 24 h to induce a DSB and then recovered by trypsinization. The collected cells were analyzed using a BD Accuri C6 flow cytometer and the accompanying analysis software (BD Biosciences).
Quantitation of DNA end resection
An in vivo end resection assay at a DSB site generated via the ER-AsiSI system in U2OS cells was performed as previously described (39). Briefly, stable ER-AsiSI expression in U2OS cells was achieved by retrovirus infection followed by puromycin selection. ER-AsiSI-expressing U2OS cells (ER-AsiSI U2OS) were treated with 4-hydroxytamoxifen (4-OHT) to allow AsiSI to enter the nucleus and generate DSBs. After 4 h of 4-OHT induction, genomic DNA was isolated using a TIANamp Genomic DNA Kit (TIANGEN). RNaseH (NEB)-treated genomic DNA (3 mg) was then mock digested or digested with BsrGI or HindIII (NEB) at 37°C overnight. To quantify the extent of resection, the mock-digested and restriction enzyme-digested samples were amplified by quantitative PCR (q-PCR) using specific Taqman probes, as previously described (39).
Metaphase spread analysis
Metaphase chromosome analysis was performed as previously described (20). Briefly, cells were exposed to irradiation (2 Gy) and allowed to recover for 8 h, before 0.1 μg/ml colcemid treatment at 37°C for 1 h. Cells were collected, incubated in hypotonic solution (75 mM KCl) for 30 min, and fixed in a 3:1 methanol/acetic acid solution (three washes) and stored overnight at −20°C. The cells were then dropped onto slides, incubated for 2 h at 60°C and Giemsa-stained.
Cell-cycle distribution and G2/M transition checkpoint analysis
Cells were harvested by trypsinization, washed in phosphate-buffered saline (PBS) and fixed with ice-cold 70% ethanol for 16 h. The cells were then collected, washed twice with PBS, resuspended in propidium iodide/RNAse and the cell-cycle distribution was analyzed by flow cytometry. The G2/M transition checkpoint assay was performed as previously described with some modifications (40). Briefly, the cells were treated with 2 Gy irradiation or camptothecin (CPT) for 1 h and then cultured in fresh medium for a further 2 or 4 h. Nocodazole was added 2 h after release from irradiation or CPT. The cells were then fixed with 75% ethanol and stained with an anti-pH3S10 antibody, before incubation with a fluorescein isothiocyanate-conjugated secondary antibody. After washing, the stained cells were treated with RNase, incubated with propidium iodide and then analyzed by flow cytometry.
Quantification and statistical analysis
All statistical analyses were performed in Microsoft Excel. A two-tailed non-paired Student's t-test was used to determine significant differences between samples.
RESULTS
PLK1, CDK1 and Aurora A promote error-prone MMEJ
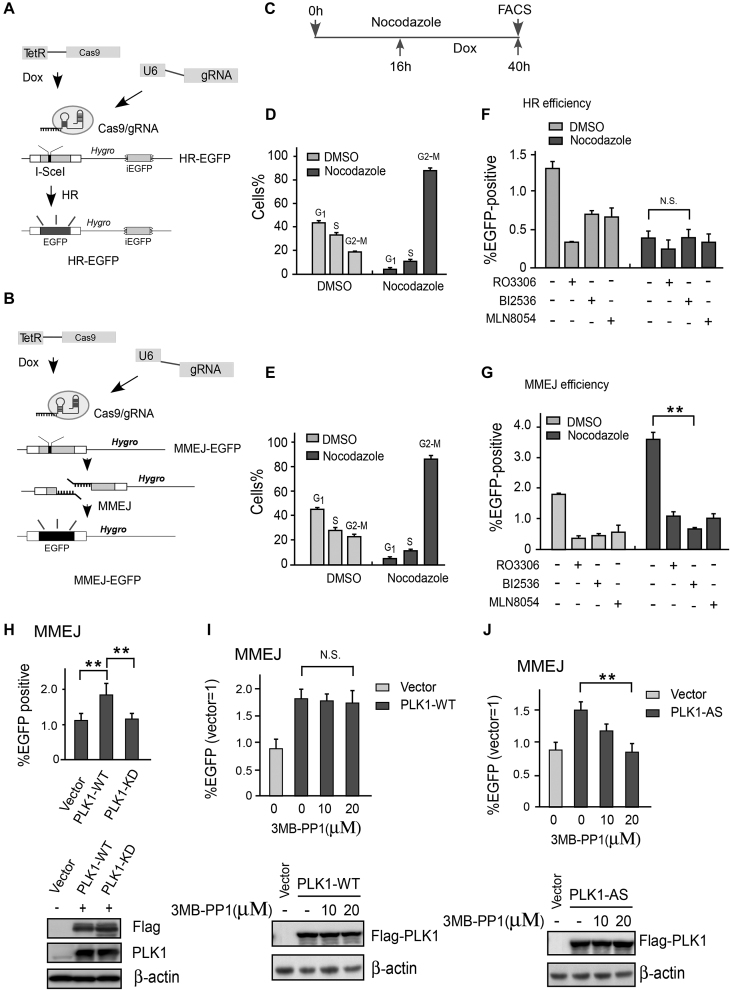
We first developed a CRISPR/Cas9-based method to generate DSBs in EGFP-HR and EGFP-MMEJ reporter cells (Figure 1A and B; Supplementary Figure S1). Following a well-designed sgRNA, Cas9 cleaved the reporter substrate at an identical region to the I-SceI endonuclease to synchronously generate a DSB on the chromosome after Dox induction. This method allowed us to measure DSB repair efficiency within a shorter time period than the classical I-SceI-mediated method (34). To study DSB repair during M phase, EGFP-HR or EGFP-MMEJ reporter cells were pre-treated with nocodazole for 16 h to arrest them in M phase, then exposed to Dox to induce DSBs and incubated in the presence of nocodazole for a further 24 h before harvesting to analyze EGFP-positive cells by fluorescence-activated cell sorting (FACS) (Figure 1C). After nocodazole treatment, >80% cells were arrested in G2/M phase (Figure 1D and E). Consistent with previous reports (41,42), reduced HR efficiency was found in nocodazole-arrested mitotic cells (Figure 1F), suggesting that the HR-mediated repair pathway was gradually shut down when cells exited S phase. Unexpectedly, using the same protocol, we observed an almost 2-fold increase in MMEJ activity in nocodazole-arrested mitotic cells (Figure 1G). CDK1, Aurora A and PLK1—major protein kinases driving cell entry into mitosis—were hyperactivated during the G2/M transition (27). We thus reasoned whether these kinases are responsible for high MMEJ activity in nocodazole-arrested mitotic cells. Indeed, after treatment with a CDK1-specific inhibitor (RO3306), Aurora A-specific inhibitor (MLN8054) or PLK1-specific inhibitor (BI25536), we found that MMEJ efficiency was drastically reduced in both control (DMSO) and nocodazole-arrested cells (Figure 1G). These data suggest that the mitosis-specific kinases CDK1, Aurora A and PLK1 may promote MMEJ-mediated DSB repair.
Figure 1.
CDK1, Aurora A and PLK1 kinase activity is required for MMEJ. (A) DR-EGFP-HR, (B) EGFP-MMEJ reporter and modified DNA DSB induction method by RNA-guided endonuclease Cas9. (C) Drug exposure and DSB repair assay. U2OS cells carrying the indicated DSB repair reporter substrate were treated with nocodazole (330 nM) or mock treated with DMSO for 16 h followed by Dox in the presence of nocodazole to induce a DSB. After 24 h Dox induction, the cells were trypsinized and FACS was performed. (D and E) After 16 h nocodazole treatment, the cell-cycle profile was determined by FACS and the mean percentage of cells in the indicated cell-cycle phases was determined. (F) HR repair efficiency in unperturbed and synchronized cells with or without the indicated drugs was determined by FACS. A CDK1 inhibitor (10 μM RO3306), PLK1 inhibitor (10 μM BI2536), Aurora A inhibitor (2 μM MLN8054) or DMSO was added to the culture before Dox induction. (G) MMEJ repair efficiency determined by the same method and strategy as the HR assay. (H) EGFP-MMEJ assay was performed in U2OS cells with overexpressed Flag-PLK1 WT or Flag-PLK1 kinase dead (K82M/D176N) mutant. Western blotting shows the expression of Flag-PLK1 variants. (I and J) U2OS cells overexpressing Flag-PLK1-WT or a PLK1-AS (analog-sensitive) mutant were treated with or without the indicated amounts of 3MB-PP1 and then analyzed by EGFP-MMEJ assay. Western blotting shows the expression of Flag-PLK1 variants. The data represent the means of three independent experiments, with error bars as SD and P values as noted: **P ≤ 0.01; n.s. not significant.
Cell-cycle analysis indicated that nocodazole treatment led to ∼80% cells with 4N DNA content and 15–35% cells with positive phospho-Histone H3 (H3pS10) staining during the whole experimental period (ranging from 16 to 40 h of nocodazole treatment, Figure 1D and E; Supplementary Figure S2A–C). These data suggest that DSB is induced and repaired mainly in the G2/M phase of the cell cycle. Co-treatment with nocodazole and CDK1 or an Aurora A inhibitor resulted in loss of H3pS10 staining in the majority of cells, but most cells still had 4N DNA content (Supplementary Figure S2A, B, D and E). This finding indicated that the cells might be forced to exit mitosis without dividing.
Inactive CDK1 or Aurora A kinases in the presence of nocodazole might result in mitotic slippage (43). We found that PLK1 inactivation using an inhibitor could only partially reduce H3S10 phosphorylation (Supplementary Figure S2D and Figure 2B), suggesting that the functions of CDK1 and Aurora A during this process are largely PLK1 independent. To show that mitotic kinase, particularly PLK1, is virtually responsible for hyperactive MMEJ in nocodazole-arrested cells, we overexpressed PLK1-WT or a PLK1-KD (kinase dead) mutant in MMEJ-reporter U2OS cells and performed MMEJ assays. As expected, forced expression of PLK1-WT but not PLK1-KD markedly promoted MMEJ activity in U2OS cells (Figure 1H). Analog-sensitive (AS) kinases are powerful tools to determine the cellular functions and phosphorylation targets of individual enzymes. A PLK1 AS mutant (C67V and L130G, hereafter referred to as PLK1-AS) but not wild-type PLK1 (PLK1-WT) was previously found to be sensitive to the purine analog 3-methylbenzyl pyrazolopyrimidine (3-MB-PP1) (44,45). The effect of PLK1-WT on MMEJ activity was not sensitive to the bulky purine analog 3-MB-PP1 (Figure 1I). However, this analog fully disrupted the effect of PLK1-AS on MMEJ (Figure 1J). These data thus confirm that PLK1 kinase activity specifically promotes MMEJ in mammalian cells.
Figure 2.
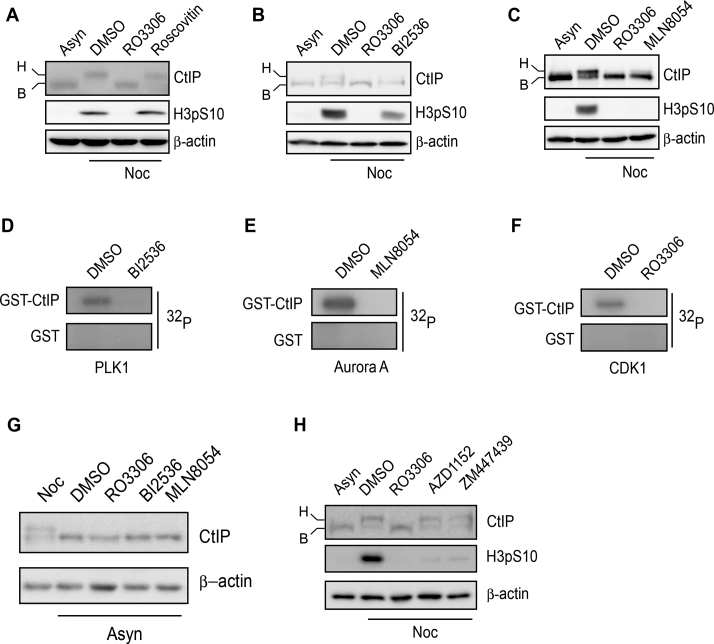
CtIP is hyperphosphorylated by mitotic kinase. (A–C) U2OS cells were incubated with nocodazole or DMSO for 16 h and then treated with the indicated drugs for a further 1 h. After drug treatment, the cells were collected, lysed and subjected to western blotting using the indicated antibodies. (D–F) Purified GST-CtIP proteins from Sf9 insect cells (Supplementary Figure S3A) were incubated with [γ-32P] ATP in the presence or absence of the indicated kinases or inhibitors for an in vitro kinase assay. The radio-labeled CtIP was visualized following SDS-PAGE. (G) U2OS cells were incubated with nocodazole or the indicated drugs for 16 h. After drug treatment, the cells were collected, lysed and subjected to western blotting using the indicated antibodies. (H) The same experiment was performed as in (A–C). Asyn, asynchronized; B, basal CtIP; H, hyperphosphorylated CtIP; Noc, nocodazole .
CtIP is phosphorylated by CDK1-Aurora A and PLK1 in nocodazole-arrested mitotic cells
CtIP is important for end resection-mediated DSB repair via both HR and MMEJ. We arrested cells in M phase with nocodazole and then performed a phosphatase-sensitive mobility shift assay for CtIP by SDS-PAGE. We found that CtIP became hyperphosphorylated after nocodazole treatment in the absence of DNA damage (Figure 2A–C). Inactivation of CDK1, Aurora A or PLK1 using specific inhibitors (RO3306, MLN8054 or BI2536, respectively) either fully abolished or reduced CtIP hyperphosphorylation (Figure 2A–C). Suppressing CDK2 activity using roscovitine had no effect on CtIP hyperphosphorylation (Figure 2A). An in vitro kinase assay also showed that purified CtIP protein (Supplementary Figure S3A) can be phosphorylated by PLK1, Aurora A and CDK1 kinases, while phosphorylation by these kinases was abolished when the cells were pre-treated with the corresponding specific inhibitors BI2536, MLN8054 and RO3306, respectively (Figure 2D–F). Inactivation of the three mitotic kinases in unperturbed cells did not lead to CtIP hyperphosphorylation (Figure 2G). Taken together, these data suggest that CtIP undergoes DNA damage-independent phosphorylation by CDK1, Aurora A and PLK1 in nocodazole-arrested cells. Nocodazole may damage microtubules and activate the spindle assembly checkpoint (SAC). Suppressing Aurora B activity, which is required for a functional SAC, using specific inhibitors AZD1152 (46) or ZM447439 (47), had no effect on CtIP hyperphosphorylation (Figure 2G). These data support that CtIP hyperphosphorylation in nocodazole-arrested mitotic cells is not due to SAC activation.
CtIP is phosphorylated by PLK1 at S723
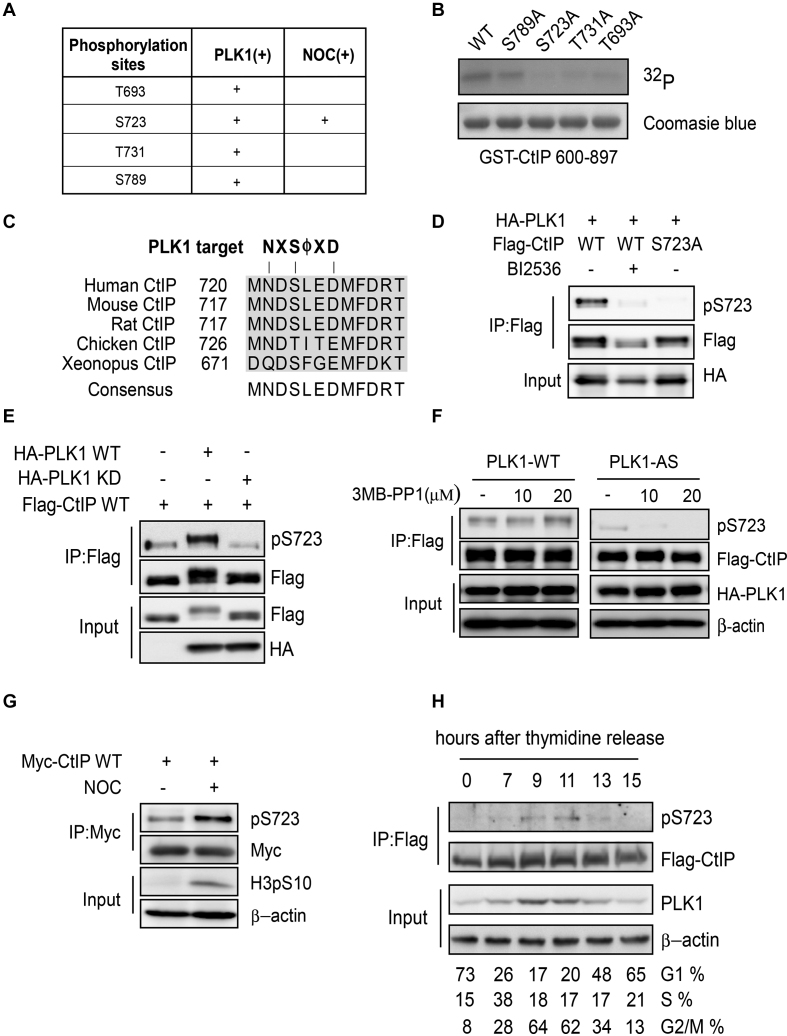
To determine the CtIP sites that are phosphorylated by PLK1, we performed an in vitro kinase reaction using recombinant CtIP purified from Escherichia coli as the substrate, and then analyzed the phosphorylated sites by mass spectrometry. We found that residues T693, S723, T731 and S789 at the CtIP C-terminus were potentially phosphorylated by PLK1 (Figure 3A). An in vitro kinase assay using GST-CtIP 600-897 fragments or indicated mutants as substrates showed that fragments carrying S723A, T731A or T693A mutations largely lost radioactive phosphorylation signals but the S789A mutant retained the phosphorylation signal (Figure 3B). These data suggest that T693, S723 and T731 are the most likely PLK1 phosphorylation sites in vivo. To test this hypothesis, we expressed Flag-tagged CtIP in U2OS cells, then arrested the cells in mitosis using nocodazole and immunopurified Flag-CtIP from the cell lysate using an anti-Flag antibody and repeated the mass spectrometry to identify the PLK1 phosphorylation sites in CtIP in vivo. Here, S723 was the only potential phosphorylation site identified (Figure 3A). This site is conserved across different species and is located in the canonical PLK1-targeted sequence (Figure 3C).
Figure 3.
CtIP is phosphorylated by PLK1. (A) CtIP phosphorylation by PLK1 was analyzed by mass spectrometry and the potential PLK1 target sites on CtIP were identified. PLK (+): purified CtIP protein was incubated with PLK1 in vitro and then subjected to mass spectrometry. NOC (+): U2OS cells stably expressing Flag-CtIP were incubated with nocodazole (NOC) for 16 h before collection. Flag-CtIP protein was immunopurified from the cell lysate using an anti-Flag antibody and subjected to mass spectrometry. (B) Purified GST-CtIP fragments (600-897) and the indicated mutants were incubated with [γ-32P] ATP in the presence of PLK1 for the in vitro kinase assay. The radio-labeled proteins were visualized following SDS-PAGE. Coomassie Blue staining indicates the protein loading. (C) Sequence alignment of the CtIP S723 surrounding region, showing a canonical PLK1 target sequence and the PLK1 target residue S723. (D) 293T cells were co-transfected with Flag-tagged CtIP and HA-tagged PLK1 expression constructs, and treated with or without 10 μM BI2536 for 1 h before harvesting. Cell extracts were subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (E) 293T cells were co-transfected with Myc-tagged CtIP and Flag-tagged PLK1 WT or PLK1-KD (kinase dead K82M/D176N) expression constructs. Cell extracts were subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (F) 293T cells were co-transfected with Flag-tagged CtIP and HA-tagged PLK1 WT or analog-sensitive PLK1 mutant (PLK1-AS) expression constructs, and treated with or without the indicated amounts of 3-MB-PP1 for 16 h. The cell extracts were subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (G) U2OS cells stably expressing Myc-tagged CtIP were treated with nocodazole for 16 h before harvesting. Cell extracts were subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (H) CtIP-knockout (KO) HCT116 cells stably expressing Flag-CtIP WT were synchronized by double thymidine-block release; CtIP S723 phosphorylation was analyzed using pS723 antibody. Cell-cycle progression was analyzed by FACS.
To further investigate CtIP phosphorylation by PLK1 in vivo, we raised a phospho-specific antibody against phosphorylated CtIP at S723 (pS723). This antibody recognized WT phosphorylated CtIP but not mutant CtIP (S723A) (Figure 3D). The PLK1 kinase-specific inhibitor BI2536 almost totally abolished the phosphorylation signal revealed by this antibody (Figure 3D). Furthermore, a catalytically inactive mutant PLK1 (K82M and D176N) failed to catalyze CtIP phosphorylation at this site (Figure 3E). These data support that the pS723 antibody specifically recognizes PLK1-mediated CtIP phosphorylation in vivo.
Both PLK1-WT and PLK1-AS were able to phosphorylate CtIP at S723 in 293T cells (Figure 3F). However, PLK1-AS was more sensitive to the purine analog than PLK1-WT. 3-MB-PP1 almost had no effect on PLK1-WT activity toward S723, but totally abolished PLK1-AS activity (Figure 3F). These data suggest that PLK1 selectively phosphorylates CtIP at the S723 site in vivo.
Furthermore, we observed a marked increase in CtIP phosphorylation levels at S723 in nocodazole-treated cells compared to asynchronous cells (Figure 3G). After double thymidine block and release, CtIP phosphorylation at S723 mainly occurred in G2/M phase, which correlates with PLK1 expression (Figure 3H). These together demonstrate that PLK1 may phosphorylate CtIP at S723 during the G2/M phase of the cell cycle.
PLK1 interacts with CtIP in a S327 phosphorylation-dependent manner
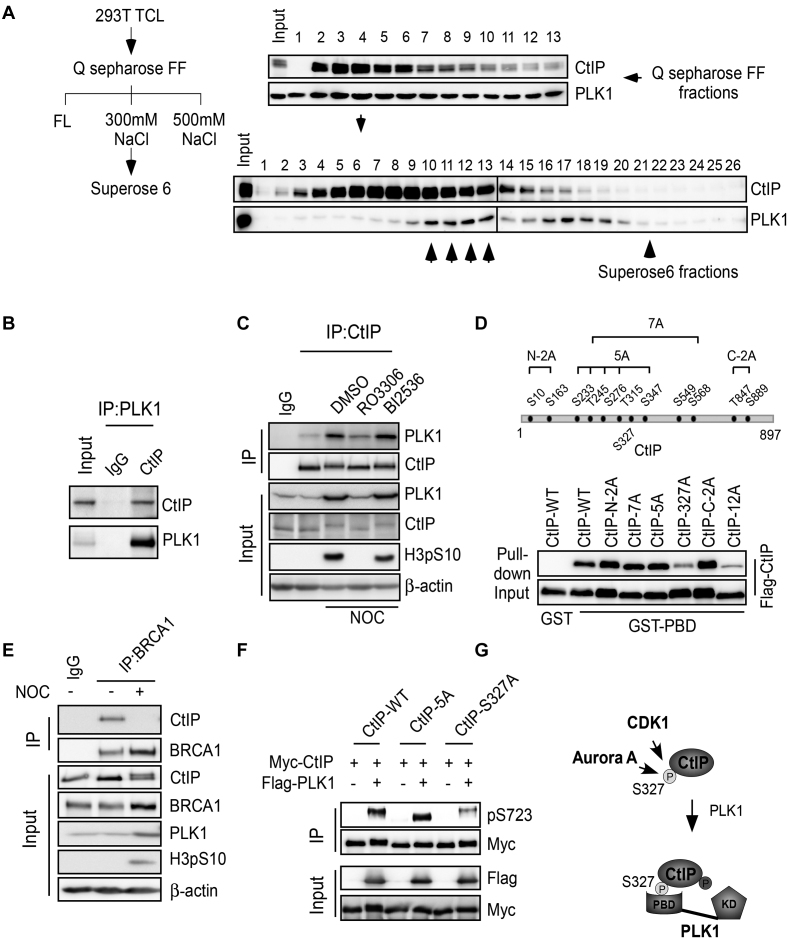
PLK1 is structurally characterized by a PBD at the C-terminus and a kinase domain at the N-terminus (Supplementary Figure S3B). PLK1 binds to protein substrates that contain phosphorylated serine or threonine residues through its PBD, and this binding is a prerequisite for the phosphorylation of the PLK1 substrates (48). Given that PLK1 phosphorylates CtIP at S723 during G2/M, we sought to determine if CtIP interacts with PLK1 during this cell-cycle stage to facilitate PLK1-mediated phosphorylation. We arrested cells in mitosis using nocodazole, collected and fractionated the cell lysate through columns and then analyzed the fractions by western blotting (Figure 4A, left). Both CtIP and PLK1 bound the Q sepharose FF column and co-fractionated at 300 mM KCl fractions after step elution. Further fractionation through a Superose 6 gel filtration column revealed that a portion of PLK1 was present in a relatively higher molecular weight fractions containing CtIP, and some PLK1 was eluted without CtIP (Figure 4A, right). These findings suggest that a portion of PLK1 associates with CtIP in nocodazole-arrested mitotic cells. Co-immunoprecipitation (co-IP) experiments were then performed to further confirm this interaction. We found that endogenous CtIP was present in the PLK1 immunocomplex (Figure 4B) and vice versa (Figure 4C). These data indicate that endogenous CtIP interacts with PLK1 in nocodazole-arrested mitotic cells.
Figure 4.
PLK1 interacts with CtIP in a S327 phosphorylation-dependent manner. (A) Left: Purification scheme. 293T cells were pre-treated with nocodazole for 16 h and lysed. Total cell lysate (TCL) was applied to a pre-equilibrated Q-sepharose FF column. After washing, the bound proteins were eluted with 300 mM NaCl, followed by fractionation on a Superose 6 gel filtration column. Right: Western blotting was performed using the indicated antibodies on alternate fractions to monitor the elution of CtIP and PLK1. Superose 6 fractions (10–13, indicated by the up-arrow) contained both CtIP and PLK1. (B) 293T cell extracts were subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (C) Unperturbed and nocodazole-arrested 293T cells, pre-treated with or without the indicated drugs, were lysed and subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (D) Top: Twelve putative CDK consensus sites (SP/TP motifs) on CtIP. Bottom: the indicated CtIP constructs were expressed in Sf9 insect cells by baculovirus infection and then used in GST pull-down assays using purified GST-PBD protein as bait. Western blotting was performed to analyze the binding between PBD and CtIP variants. (E) U2OS cells were treated with nocodazole (NOC) for 16 h and then subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (F) 293T cells were co-transfected with Myc-tagged CtIP variants and Flag-tagged PLK1 expression constructs. Cell extracts were subjected to immunoprecipitation followed by western blotting with the indicated antibodies. (G) The role of S327 phosphorylation in mediating the CtIP binding with PBD and subsequent phosphorylation of CtIP by PLK1. FL, flow through; KD, kinase domain.
We next characterized the interaction between PLK1 and CtIP. We first found that PLK1 mainly interacted with CtIP through its PBD domain (Supplementary Figure S3C), and by using purified proteins from Sf9 insect cells (GST-CtIP) and E. coli (His-PBD), we demonstrated that the interaction is direct. De-phosphorylation of CtIP by lambda phosphatase (Lamda PP) drastically reduced the interaction, and re-phosphorylation of CtIP by CDK1/CyclinB restored binding (Supplementary Figure S3D). A PLK1-WHK mutant (W414F, H538A and K540M) (Supplementary Figure S3B) carrying mutations at the critical sites for binding to the phosphor-motifs in the PLK1-PBD domain (48) failed to interact with CtIP (Supplementary Figure S3E). These findings suggest that CtIP binds PLK1 in a PBD-dependent and CDK1 phosphorylation-dependent manner. CtIP contains 12 putative CDK consensus sites (SP/TP motifs) (Figure 4D top), and the mutant CtIP-12A, in which all the 12 sites were mutated, compromised the interaction with the PLK1 PBD (Figure 4D). Other mutants, including CtIP-N2A (S10A, S163A), CtIP-7A (S233A, T245A, S276A, T315A, S347A, S549A and S568A), CtIP-5A (S233A, T245A, S276A, T315A and S347A) and CtIP-C2A (T847A, S889A), but not CtIP-S327A, could bind the PLK1 PDB at an equivalent level to WT CtIP (Figure 4D). These data support that CtIP S327 phosphorylation is important for priming the interaction between CtIP and the PLK1 PBD domain.
Concerted phosphorylation of CtIP by CDK1-Aurora A and PLK1
We used mass spectrometry to characterize CtIP phosphorylation by Aurora A. We mapped multiple Aurora A sites on CtIP, including the previously identified CDK site S327 (22) (Supplementary Figure S4A). An in vitro kinase assay indicated that S593 is the predominant site in CtIP that is targeted by Aurora A (Supplementary Figure S4B). Our phosphor-specific antibody against the phosphorylated S593 site recognized WT CtIP but not mutant CtIP (S593A) (Supplementary Figure S4C). Phosphorylation of CtIP at S593 was suppressed upon application of an Aurora A-specific inhibitor (MLN8054) or by an Aurora A kinase dead mutation (K162R) (Supplementary Figure S4C–E).
Using the phosphor-specific antibody (that specifically recognizes phosphorylated CtIP at S327), we found that the S327 site can be phosphorylated by both Aurora A and CDK1 kinase (Supplementary Figure S4F and G and data not shown). Previous reports have indicated that CtIP S327 is phosphorylated by CDKs in S/G2 phase to mediate its interaction with BRCA1 (21,22). Intriguingly, co-IP indicated that CtIP no longer associates with BRCA1 in nocodazole-arrested mitotic cells (Figure 4E). Previous reports have also indicated that the C-terminal tandem BRCT domain of BRCA1 is responsible for the phosphorylation-dependent binding between BRCA1 and CtIP (22). Consistent with this finding, we also observed an interaction between BRCT and CtIP in asynchronous cells. However, this interaction was lost upon nocodazole treatment in conjunction with an increase in S327 phosphorylation (Supplementary Figure S3F and G). This finding suggests that some proteins may compete with BRCA1 (BRCT domain) to bind CtIP in nocodazole-arrested mitotic cells. To test this hypothesis, we exposed cell lysates to different amounts of purified His-PBD and performed the same pull-down assay using GST-BRCT as bait. The binding capacity of CtIP to BRCT was suppressed when increasing amounts of His-PBD were added to the system (Supplementary Figure S3H). These data imply that overexpression of PLK1 in G2/M cells may compete with BRCA1 to interact with pCtIP-S327. CtIP may deviate from the BRCA1 complex and join the PLK1 complex at this cell-cycle stage.
We further investigated whether the S327 phosphorylation-mediated interaction is required for CtIP phosphorylation by PLK1. Compared to wild-type CtIP (CtIP-WT) or mutant CtIP-5A (S233A, T245A, S276A, T315A and S347A), which fails to bind Nbs1 FHA/BRCT domains (21), CtIP-S327A could not be fully phosphorylated by PLK1 at S723 (Figure 4F). This finding implies that the S327-mediated interaction between CtIP and the PBD is a prerequisite for PLK1 to phosphorylate CtIP. Taken together, we propose a mechanism of concerted phosphorylation of CtIP by CDK1-Aurora A and PLK1, in which CDK1-Aurora A-mediated phosphorylation of CtIP at S327 modulates the binding between CtIP and the PLK1 PBD, which in turn permits PLK1 to phosphorylate CtIP mainly at S723 (Figure 4G).
Phosphorylation of CtIP by PLK1 suppresses DSB end resection
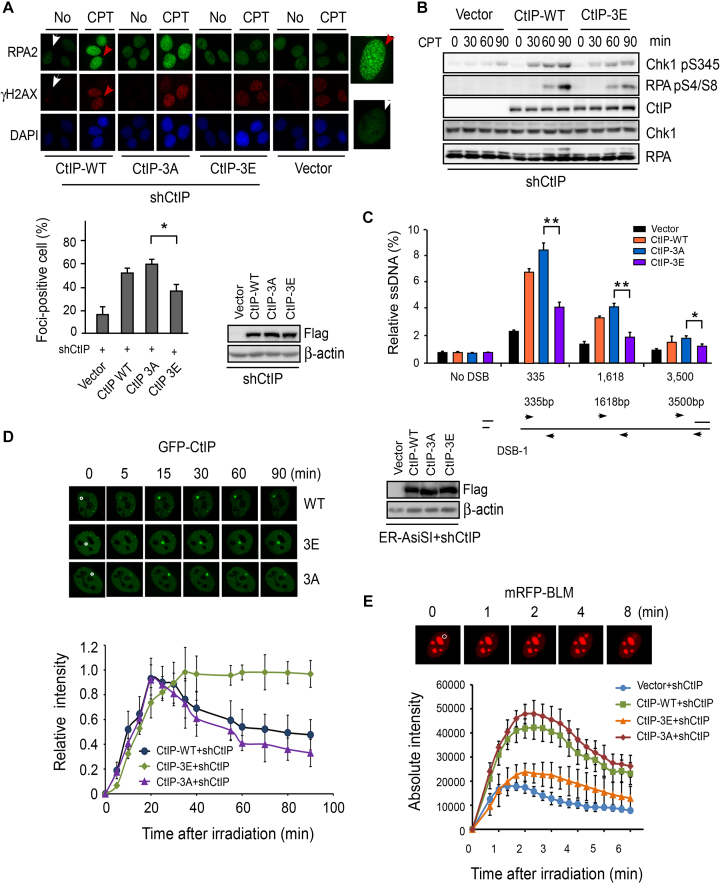
To determine whether CtIP phosphorylation by PLK1 and Aurora A affects its DSB repair function, we introduced phosphor-mimic and phosphor-block mutations into shRNA-resistant CtIP derivatives to mimic constitutive phosphorylation or to block potential phosphorylation at threonine or serine residues, respectively. Consistent with previous reports (49,50), end resection was defective in CtIP-depleted cells, as revealed by diminished RPA foci formation (Figure 5A) and RPA phosphorylation (Supplementary Figure S5A). Re-expression of CtIP-WT or CtIP-3A, in which the PLK1 phosphorylation sites T693, S723 and T731 were mutated to alanine to block potential phosphorylation, in CtIP-depleted cells could fully rescue this end-resection defect (Figure 5A and B), while re-expression of the CtIP-3E mutant, in which the PLK1 phosphorylation sites T693, S723 and T731 were mutated to negatively charged glutamic acid to mimic constitutive phosphorylation, could only partially rescue the end-resection defect with reduced RPA foci and phosphorylation (Figure 5A and B). By contrast, re-expression of the CtIP-S593D mutant did not result in any defects in RPA phosphorylation or foci formation (Supplementary Figure S5B and data not shown). These data suggest that PLK1-mediated CtIP phosphorylation but not Aurora A-mediated CtIP phosphorylation at S593 may impair DSB end resection. CtIP-3A mutant cells exhibited mild but consistently increased RPA foci (Figure 5A), suggesting that CtIP-WT but not the CtIP-3A mutant is phosphorylated by PLK1 in vivo.
Figure 5.
PLK1-mediated CtIP phosphorylation disturbs DSB end resection. (A) RPA foci formation assay was performed in U2OS cells stably expressing the indicated CtIP variants on top of endogenous CtIP silencing. Top: the indicated cells were treated with DMSO (No) or CPT (1 μM, 2 h) and fixed for immunostaining. The red and white arrows indicate representative RPA2 and H2AX foci-positive and foci-negative cells, respectively, with RPA2-immunostained cells enlarged in the right-side panels. Bottom: the percentage of RPA2 foci-positive cells among γH2AX-positive cells for each sample is shown. Data shown represent the means of three independent experiments, with error bars as SD. The western blots show the expression levels of each indicated CtIP variant. (B) U2OS cells stably expressing Flag-tagged CtIP alleles (CtIP-WT and CtIP-3E consisting of three PLK1 target sites on CtIP T693/S723/T731 mutated to E) on top of endogenous shRNA-mediated CtIP silencing were treated with CPT (2 μM) for different times followed by western blotting with the indicated antibodies. (C) Quantitation of DSB end resection at a specific DSB site. ER-AsiSI U2OS cells stably expressing the indicated CtIP variants on top of endogenous shRNA-mediated CtIP silencing were treated with 4-OH for 4 h and quantitative PCR was performed to amplify the ssDNA at various locations adjacent to a specific DSB generated by AsiSI. The bar graph shows the extent of ssDNA generated by end resection in different cell lines. The error bars indicate the SD from three independent experiments. The schematic shows the DSB site on chromosome 1 along with regions (indicated by arrow) used to amplify the ssDNA. The western blots show the expression levels of each indicated CtIP variant. (D) EGFP-CtIP WT and indicated mutants were expressed in U2OS cells on top of endogenous shRNA-mediated CtIP silencing. DSBs were induced by laser-induced micro-irradiation and recruitment of EGFP-CtIP to DSBs was monitored by live-cell imaging. Top: representative cells show the accumulation of EGFP-CtIP variants at damage regions (a disk region marked by a white circle in 0 min cell images). Bottom: accumulation curve of EGFP-CtIP variants at sites of laser-induced DSB damage. The pre-damage fluorescence level in the cell was set to 0 and the maximum level was set to 1. (E) Recruitment of mRFP-BLM to DSBs was monitored in U2OS cells stably expressing Flag-CtIP variants, in which endogenous CtIP was silenced. Representative cells show the recruitment of mRFP-BLM to micro-irradiated regions. Absolute intensities of mRFP-BLM fluorescence signals were determined. Each data point represents the average of 10 independent measurements and the error bars represent the SD.
RPA foci formation is a low-resolution technique to measure DSB end resection at random positions in the genome. To more precisely explore the extent by which end resection is impacted by PLK1-mediated CtIP phosphorylation, we used a recently developed system in which resection at specific AsiSI restriction-enzyme sites can be quantified by q-PCR (39). Here, we found that the levels of resection at a known DSB site on chromosome 1 were reduced in CtIP-3E mutant cells compared to CtIP-3A mutant cells (∼52.5, ∼50.8 and ∼32.8% reduction at position 335, 1618 and 3500 nt from DSB, respectively) and CtIP-WT cells (Figure 5C). Together these results suggest that PLK1-mediated phosphorylation indeed suppresses DSB end resection.
We next explored the cause of the end-resection defect in the CtIP-3E mutant. We performed live-cell imaging of CtIP recruitment to DSBs by monitoring fluorescently tagged CtIP. We observed that both EGFP-tagged CtIP-3A and CtIP-3E mutants accumulated in the laser-induced damage region (Figure 5D). Time-lapse imaging showed that the accumulation kinetics of the CtIP-3E mutant were indistinguishable from CtIP-WT and CtIP-3A during the acute recruitment phase (Figure 5D), suggesting that PLK1-mediated phosphorylation may not affect the initial recruitment of CtIP to DSB sites. However, when we measured retention kinetics, which may reflect the disassociation of the proteins from DSB sites, we observed significant differences between the CtIP-3A and CtIP-3E mutant. The fluorescence intensity of CtIP-3A at the damage region dropped to 3% of the maximum level in a 90-min period, whereas CtIP-3E only decreased to 90% of the maximum level. These findings support that PLK1-mediated phosphorylation may retard the disassociation of CtIP from DSB sites.
BLM protein also has important roles in end resection and HR. CtIP not only promotes BLM recruitment to DSB sites in vivo, but also enhances BLM-DNA2-mediated long-range resection in vitro (21,51). By monitoring mRFP-tagged BLM localization to DSBs in live cells, we found that BLM recruitment to damage sites was significantly reduced in CtIP-3E mutant cells compared to CtIP-3A cells (Figure 5E). Depletion of BLM by shRNA markedly reduces end resection measured by the same q-PCR-based assay as described in Figure 5C (Supplementary Figure S5C). Thus, PLK1-mediated phosphorylation of CtIP may block the transition from the initial step to the second step of end resection. As a consequence, PLK1-mediated CtIP phosphorylation affects both end resection and end resection-related repair function.
Phosphorylation of CtIP by PLK1 promotes error-prone MMEJ over error-free HR
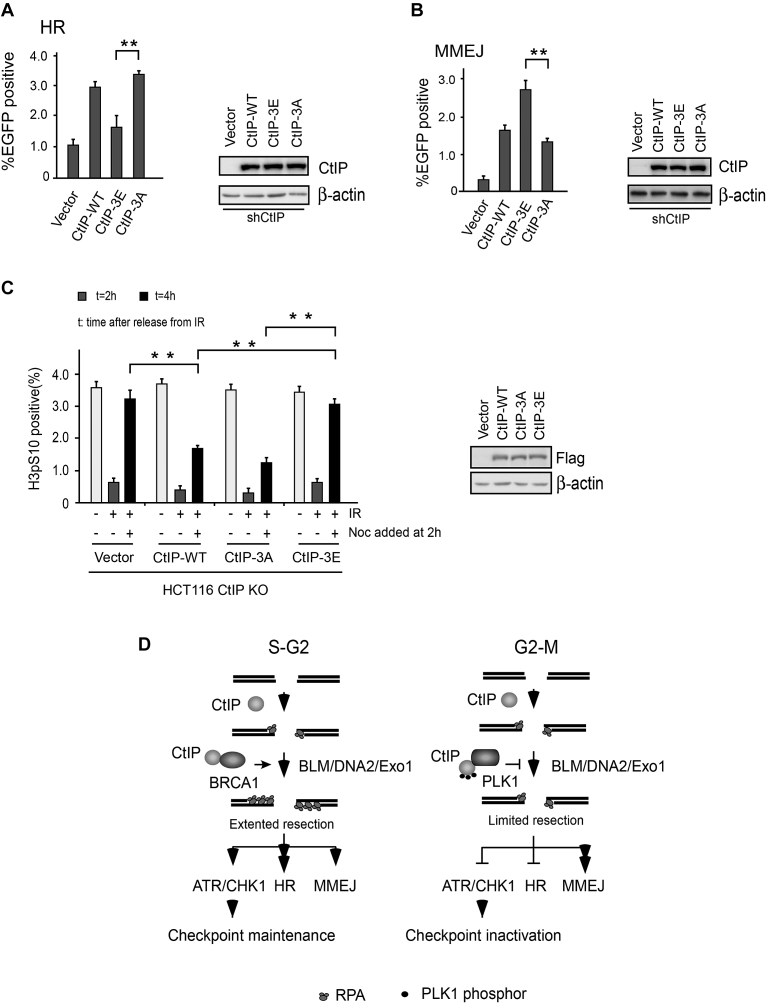
CtIP-mediated DSB end resection is required for both HR and MMEJ (34). We next considered whether PLK1-mediated CtIP phosphorylation might regulate the function of CtIP in DSB repair. To explore this, we used the U2OS cells carrying the EGFP-based HR or MMEJ substrate to perform HR and MMEJ-mediated DSB repair assays, as described in Figure 1A and B. Consistent with previous findings (34), shRNA-mediated CtIP inactivation significantly reduced HR and MMEJ activities and re-expression of CtIP-WT could fully rescue this repair defect (Supplementary Figure S5D and E). We further examined HR and MMEJ in reporter substrates containing U2OS cells expressing CtIP variants with endogenous CtIP inactivated by CtIP shRNA. In line with reduced end-resection efficiency, we found that HR was significantly impaired in CtIP-3E mutant cells but not in CtIP-WT, CtIP593D and CtIP-3A mutant cells (Figure 6A and Supplementary Figure S5D). Interestingly, we found that MMEJ was markedly increased in CtIP-3E mutant cells, but not in CtIP-WT, CtIP-3A and CtIP-S593D cells (Figure 6B and Supplementary Figure S5E). Compared with CtIP-3E cells, CtIP-3A mutant and CtIP-WT cells exhibited higher HR but lower MMEJ activities (Figure 6A and B). It is well documented that chromosomal translocations and telomere fusions are mediated by CtIP and the alt-NHEJ (MMEJ) pathway (52). Compared with cells expressing CtIP-WT or CtIP-3A mutant, CtIP-3E cells had an increased number of chromosomal fusions after 2 Gy of irradiation (Supplementary Figure S5F). These findings suggest that PLK1 may target CtIP to promote MMEJ and lead to genome instability in mammalian cells.
Figure 6.
PLK1-mediated CtIP phosphorylation disrupts the damage-inducible G2-M checkpoint and promotes MMEJ.(A) EGFP-HR or (B) EGFP-MMEJ assay was performed in U2OS cells stably expressing Flag-CtIP WT or indicated mutants, in which endogenous CtIP was silenced by shRNA. Western blotting shows the expression of Flag-CtIP variants. (C) HCT116 WT or CtIP-KO cells stably expressing Flag-CtIP WT or the indicated mutants were fixed at the indicated time points after irradiation and nocodazole (Noc) treatment. Checkpoint assays were then performed. The bar chart shows the percentage of phosphor-histone H3-positive cells measured by flow cytometry. The western blot shows the expression of Flag-CtIP variants. (D) A model to describe a role for G2/M phase-specific phosphorylation of CtIP in checkpoint regulation and DSB repair. HR, homologous recombination. The data represent the means of three independent experiments, with error bars as SD. **P < 0.01.
Phosphorylation of CtIP by PLK1 disrupted G2-M transition checkpoint
Consistent with previous findings (40), we found that depletion of CtIP by shRNA in U2OS cells can markedly reduce the level of CHK1 phosphorylation, particularly at the late time points following CPT treatment (Supplementary Figure S5A). CtIP-WT and S593D, but not the CtIP-3E mutant, could restore CHK1 activity (Figure 5B and Supplementary Figure S5B), supporting that PLK1-mediated CtIP phosphorylation may disrupt sustained ATR-CHK1 checkpoint signaling. To further address this, we introduced the CtIP expression vector into CtIP-deficient HCT116 cells (CtIP KO) (35) and generated stable cell lines expressing CtIP variants. We then performed a G2/M transition checkpoint assay. Consistent with a previous report (22), we found that CtIP depletion in HCT116 cells (CtIP KO) caused a G2/M transition checkpoint defect after irradiation and CPT treatment. The percentage of mitotic cells markedly increased 4 h after release from irradiation and CPT, despite the presence of DNA damage. Reconstitution with CtIP-WT or CtIP-3A mutant almost fully restored this G2/M checkpoint, whereas a PLK1 phospho-mimic mutant (CtIP-3E) failed to do so (Figure 6C and Supplementary Figure S6). These data suggest that PLK1-mediated phosphorylation of CtIP may disrupt the G2/M transition checkpoint. These findings are consistent with defected end resection in CtIP-3E mutant cells, as CtIP-mediated G2/M checkpoint maintenance is end-resection dependent (40).
DISCUSSION
Defective DSB repair can result in genomic instability and ultimately cancers or other relevant diseases. Cells typically employ an error-free mechanism to repair DSBs to maintain genome integrity and avoid DNA damage-induced genomic instability. In mammalian cells, DSB repair pathway choice is tightly controlled by complicated mechanisms, involving the cell cycle and DNA end resections (4,13). C-NHEJ may function throughout the cell cycle and is the predominant DSB repair pathway in G1 when resection activity is low. HR is activated only during S/G2 phase, when genomic DNA is duplicated and a sister chromatid template is available (53). Recent studies have indicated that MMEJ also exhibits substantial activity in cycling cells, and MMEJ activity increases in S phase (11). However, the function and regulation of MMEJ outside of S phase is unclear. Here, we observed a marked increase in MMEJ activity and a decrease in HR activity in nocodazole-arrested mitotic cells. This finding suggests that these cells shut down the HR pathway, but the MMEJ pathway remains available to fix DSBs in these cells. Due to a technical limitation, we were unable to detect MMEJ activity in cells enriched by mitotic shake-off. As such, it is unclear whether this DSB repair mechanism also functions in the late stages of mitosis when both C-NHEJ and HR are not utilized in order to prevent damage-induced telomere fusion (54).
We detected reduced HR activity in unperturbed cells after PLK1 inhibition, which is consistent with a previous report (55). In nocodazole-arrested cells, however, residual HR activity was no longer restricted by PLK1 activity, but application of a CDK1 inhibitor could cause a reduction in HR efficiency (Figure 1F). Different from HR, MMEJ is strongly dependent on PLK1 activity in both unperturbed cells and nocodazole-arrested cells (Figure 1G), suggesting a specific role for PLK1 activity in MMEJ regulation, particularly in nocodazole-arrested mitotic cells.
Much evidence supports that end resection has a decisive role in DSB repair pathway choice (4). Extended end resection is needed to recruit Rad51 recombinase and promote HR-mediated error-free DSB repair. However, limited resection or a resection defect encourages cells to use MMEJ or C-NHEJ as a repair mechanism, which are both error prone (4). Both damage-induced RPA phosphorylation and foci formation are classical end-resection markers (3). Differing from a previously identified CDK (T847A) mutant (20), here, end resection was just disturbed (not abolished) in PLK1 phosphor-mimic mutant (CtIP-3E) cells (Figure 5A and B). Using a q-PCR-based end-resection assay, we found that the levels of end resection (∼350 to ∼3500 bp from DSB) in CtIP-3E mutant cells are much lower than in CtIP-3A cells (Figure 5C). Depletion of the long-range resection factors Exo1 (39) or BLM (Supplementary Figure S5C) also markedly reduced end resection at the same locations adjacent to DSBs. Our data, thus, suggest that PLK1 targets CtIP to suppress long-range end resection. The MMEJ reporter allowed us to monitor DSB repair requiring <20 bp of end resection (11); this process represents the initial step of end resection. Increased MMEJ activity in CtIP 3E mutant cells (Figure 6B) thus implies that the initial step of end resection still is functional. PLK1-mediated CtIP phosphorylation may not influence this process. Furthermore, mRFP-BLM recruitment to DSB sites was drastically reduced in CtIP-3E cells (Figure 5E). A recent study indicated that CtIP may not just function as an MRN co-factor in the initial step of end resection, but also binds BLM and promotes long-range resection mediated by BLM-DNA2 (51). Our findings thus support a model whereby PLK1-mediated CtIP phosphorylation may hamper CtIP function in the second step of end resection. The CtIP-3E mutant can still work together with the MRN complex in the initial step to carry out limited end resection, which is enough to mediate MMEJ but not HR, and maintain the G2/M checkpoint (Figure 6D). Further biochemical studies are now needed to clarify the underlying mechanism.
It is well established that CtIP S327 phosphorylation induces its interaction with BRCA1, which may mediate MRN/BRCA1 complex formation and promote HR during S phase (49). Intriguingly, we found that in nocodazole-arrested mitotic cells, the BRCA1–CtIP interaction is lost, but a PLK1–CtIP interaction is established. This finding suggests that the switch between HR and MMEJ in nocodazole-arrested cells may also be caused, in part, by disassembly of the BRCA1–CtIP complex, although the role of S327 phosphorylation in HR has been questioned in the field (23,24,56,57).
CtIP is also required to maintain the G2/M transition checkpoint through end resection-dependent sustain of the ATR/CHK1 signal, but not initiation, which is mediated by factors including TopBP1 and Claspin (40). PLK1 kinase can target several components of the ATR/CHK1 pathway, including Claspin, to inactivate the DNA replication stress-induced G2/M checkpoint, or directly phosphorylate and silence CHK2 in defective ATM/CHK2 signal branches (58–60). Here, we showed that the sustained G2/M checkpoint (mediated by the ATR-CHK1 signal) is lost in CtIP-3E mutant cells. This finding suggests a new mechanism for PLK1-mediated G2/M checkpoint inactivation and supports that PLK1 has a more general role in this process than previously thought. This finding is also consistent with data obtained from budding yeast, where overexpressed CDC5 (the only polo kinase in yeast) can target Sae2 (CtIP ortholog) to suppress DSB end resection and override the DSB-induced checkpoint (61).
CtIP was originally reported as a tumor suppressor, partially due to its interaction with BRCA1 (62,63). However, a recent study suggested CtIP may facilitate mammary tumorigenesis in p53-deficient mice possibly through its function in MMEJ-mediated chromosome translocations and telomeric chromosomal fusions (64). In addition, Plk1 and Aurora A are frequently overexpressed in cancers including breast cancer (65–67). Our findings imply that excessive PLK1 or Aurora A may target CtIP to inactivate cell-cycle checkpoints and switch DSB repair to the error-prone MMEJ pathway and eventually cause genomic instability and carcinogenesis. These findings help explain the oncogenic roles of these factors and open avenues to generate novel therapeutic strategies based on targeting these factors.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Xiaohua Wu (Scripps Research Institute), Zhenghe Wang (Case Western Reserve University), Tanya T. Paull (The University of Texas at Austin), Gaëlle Legube (University of Toulouse) and Junjie Chen (Texas MD Anderson Cancer Center) for providing reagents and members of the Wang and Xu laboratories for their constructive discussions and help in the conception and interpretation of this study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China [2015CB910602 to H.W.]; National Natural Science Foundation of China [31370841 to H.W.]; Beijing Natural Science Foundation [5182003 to H.W.]; National Natural Science Foundation of China [31530016, 31761133012 to X.X.]; the National Basic Research Program of China [2015CB910601 to X.X.]; Shenzhen Science and Technology Innovation Commission [JCYJ20170412113009742 to X.X.]. Funding for open access charge: National Basic Research Program of China [2015CB910602]; National Natural Science Foundation of China [31370841].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moynahan M.E., Jasin M.. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010; 11:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 4. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 5. Wang H., Xu X.. Microhomology-mediated end joining: new players join the team. Cell Biosci. 2017; 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sfeir A., Symington L.S.. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Jasin M.. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 2011; 18:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rai R., Zheng H., He H., Luo Y., Multani A., Carpenter P.B., Chang S.. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010; 29:2598–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nussenzweig A., Nussenzweig M.C.. A backup DNA repair pathway moves to the forefront. Cell. 2007; 131:223–225. [DOI] [PubMed] [Google Scholar]
- 10. McVey M., Lee S.E.. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008; 24:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H., Razavian N., Berns M.W., Wu X.. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. PNAS. 2013; 110:7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ceccaldi R., Liu J.C., Amunugama R., Hajdu I., Primack B., Petalcorin M.I., O’Connor K.W., Konstantinopoulos P.A., Elledge S.J., Boulton S.J. et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015; 518:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat. Res. 2017; 803–805:51–55. [DOI] [PubMed] [Google Scholar]
- 14. Aparicio T., Baer R., Gautier J.. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst.). 2014; 19:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Symington L.S. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 2014; 6:a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L. et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010; 467:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C.. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010; 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mimitou E.P., Symington L.S.. Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 2009; 34:264–272. [DOI] [PubMed] [Google Scholar]
- 19. Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C.. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011; 25:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huertas P., Jackson S.P.. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009; 284:9558–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W. et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013; 9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu X., Chen J.. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004; 24:9478–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yun M.H., Hiom K.. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009; 459:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reczek C.R., Szabolcs M., Stark J.M., Ludwig T., Baer R.. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 2013; 201:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barton O., Naumann S.C., Diemer-Biehs R., Kunzel J., Steinlage M., Conrad S., Makharashvili N., Wang J., Feng L., Lopez B.S. et al. Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J. Cell Biol. 2014; 206:877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Vugt M.A., Medema R.H.. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005; 24:2844–2859. [DOI] [PubMed] [Google Scholar]
- 27. Seki A., Coppinger J.A., Jang C.Y., Yates J.R., Fang G.. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008; 320:1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Macurek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., Medema R.H.. Polo-like kinase-1 is activated by Aurora A to promote checkpoint recovery. Nature. 2008; 455:119–123. [DOI] [PubMed] [Google Scholar]
- 29. Zitouni S., Nabais C., Jana S.C., Guerrero A., Bettencourt-Dias M.. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 2014; 15:433–452. [DOI] [PubMed] [Google Scholar]
- 30. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 31. Lobrich M., Jeggo P.A.. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 2007; 7:861–869. [DOI] [PubMed] [Google Scholar]
- 32. Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S.. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004; 73:39–85. [DOI] [PubMed] [Google Scholar]
- 33. Bartek J., Lukas J.. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007; 19:238–245. [DOI] [PubMed] [Google Scholar]
- 34. Wang H., Shao Z., Shi L.Z., Hwang P.Y., Truong L.N., Berns M.W., Chen D.J., Wu X.. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 2012; 287:21471–21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu B., Cong R., Peng B., Zhu B., Dou G., Ai H., Zhang X., Wang Z., Xu X.. CtIP is required for DNA damage-dependent induction of P21. Cell Cycle. 2014; 13:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y., He J., Do J., Cho M.J., Li H., Negrete A. et al. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014; 54:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J.. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000; 14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 38. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Y., Caron P., Legube G., Paull T.T.. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014; 42:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kousholt A.N., Fugger K., Hoffmann S., Larsen B.D., Menzel T., Sartori A.A., Sorensen C.S.. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J. Cell Biol. 2012; 197:869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saleh-Gohari N., Helleday T.. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004; 32:3683–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vriend L.E., Prakash R., Chen C.C., Vanoli F., Cavallo F., Zhang Y., Jasin M., Krawczyk P.M.. Distinct genetic control of homologous recombination repair of Cas9-induced double-strand breaks, nicks and paired nicks. Nucleic Acids Res. 2016; 44:5204–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L.. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. PNAS. 2006; 103:10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burkard M.E., Randall C.L., Larochelle S., Zhang C., Shokat K.M., Fisher R.P., Jallepalli P.V.. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. PNAS. 2007; 104:4383–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D. et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000; 407:395–401. [DOI] [PubMed] [Google Scholar]
- 46. Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M.. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003; 161:281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilkinson R.W., Odedra R., Heaton S.P., Wedge S.R., Keen N.J., Crafter C., Foster J.R., Brady M.C., Bigley A., Brown E. et al. AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clin. Cancer Res. 2007; 13:3682–3688. [DOI] [PubMed] [Google Scholar]
- 48. Elia A.E., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantley L.C., Smerdon S.J., Yaffe M.B.. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003; 115:83–95. [DOI] [PubMed] [Google Scholar]
- 49. Chen L., Nievera C.J., Lee A.Y., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 50. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Daley J.M., Jimenez-Sainz J., Wang W., Miller A.S., Xue X., Nguyen K.A., Jensen R.B., Sung P.. Enhancement of BLM-DNA2-mediated long-range DNA end resection by CtIP. Cell Rep. 2017; 21:324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Badie S., Carlos A.R., Folio C., Okamoto K., Bouwman P., Jonkers J., Tarsounas M.. BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres. EMBO J. 2015; 34:828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rothkamm K., Kruger I., Thompson L.H., Lobrich M.. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003; 23:5706–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Orthwein A., Fradet-Turcotte A., Noordermeer S.M., Canny M.D., Brun C.M., Strecker J., Escribano-Diaz C., Durocher D.. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014; 344:189–193. [DOI] [PubMed] [Google Scholar]
- 55. Chabalier-Taste C., Brichese L., Racca C., Canitrot Y., Calsou P., Larminat F.. Polo-like kinase 1 mediates BRCA1 phosphorylation and recruitment at DNA double-strand breaks. Oncotarget. 2016; 7:2269–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E. et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010; 6:e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cruz-Garcia A., Lopez-Saavedra A., Huertas P.. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014; 9:451–459. [DOI] [PubMed] [Google Scholar]
- 58. van Vugt M.A., Bras A., Medema R.H.. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell. 2004; 15:799–811. [DOI] [PubMed] [Google Scholar]
- 59. van Vugt M.A., Gardino A.K., Linding R., Ostheimer G.J., Reinhardt H.C., Ong S.E., Tan C.S., Miao H., Keezer S.M., Li J. et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010; 8:e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peschiaroli A., Dorrello N.V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N.E., Pagano M.. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 2006; 23:319–329. [DOI] [PubMed] [Google Scholar]
- 61. Donnianni R.A., Ferrari M., Lazzaro F., Clerici M., Tamilselvan Nachimuthu B., Plevani P., Muzi-Falconi M., Pellicioli A.. Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway. PLoS Genet. 2010; 6:e1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chinnadurai G. CtIP, a candidate tumor susceptibility gene is a team player with luminaries. Biochim. Biophys. Acta. 2006; 1765:67–73. [DOI] [PubMed] [Google Scholar]
- 63. Chen P.L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E.Y., Lee W.H.. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 2005; 25:3535–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reczek C.R., Shakya R., Miteva Y., Szabolcs M., Ludwig T., Baer R.. The DNA resection protein CtIP promotes mammary tumorigenesis. Oncotarget. 2016; 7:32172–32183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ke Y.W., Dou Z., Zhang J., Yao X.B.. Function and regulation of Aurora/Ipl1p kinase family in cell division. Cell Res. 2003; 13:69–81. [DOI] [PubMed] [Google Scholar]
- 66. Strebhardt K., Ullrich A.. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer. 2006; 6:321–330. [DOI] [PubMed] [Google Scholar]
- 67. Wang H., Zhang X., Teng L., Legerski R.J.. DNA damage checkpoint recovery and cancer development. Exp. Cell Res. 2015; 334:350–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.