Abstract
The Survival Motor Neuron (SMN) protein is essential for survival of all animal cells. SMN harbors a nucleic acid-binding domain and plays an important role in RNA metabolism. However, the RNA-binding property of SMN is poorly understood. Here we employ iterative in vitro selection and chemical structure probing to identify sequence and structural motif(s) critical for RNA–SMN interactions. Our results reveal that motifs that drive RNA–SMN interactions are diverse and suggest that tight RNA–SMN interaction requires presence of multiple contact sites on the RNA molecule. We performed UV crosslinking and immunoprecipitation coupled with high-throughput sequencing (HITS-CLIP) to identify cellular RNA targets of SMN in neuronal SH-SY5Y cells. Results of HITS-CLIP identified a wide variety of targets, including mRNAs coding for ribosome biogenesis and cytoskeleton dynamics. We show critical determinants of ANXA2 mRNA for a direct SMN interaction in vitro. Our data confirms the ability of SMN to discriminate among close RNA sequences, and represent the first validation of a direct interaction of SMN with a cellular RNA target. Our findings suggest direct RNA–SMN interaction as a novel mechanism to initiate the cascade of events leading to the execution of SMN-specific functions.
INTRODUCTION
Humans carry two nearly identical copies of Survival Motor Neuron genes: SMN1 and SMN2 (1). While SMN1 codes for full-length SMN protein, SMN2 codes for a truncated SMNΔ7 protein due to predominant skipping of SMN2 exon 7 (2–6). SMNΔ7 is less stable due to the absence of the critical C-terminal sequences (7). Hence, low levels of SMN caused by deletion or mutation of SMN1 leads to spinal muscular atrophy, a neurodegenerative disease of children and infants (8–10). Aberrant expression and/or localization of SMN have been also linked to other pathological conditions including amyotrophic lateral sclerosis (ALS), osteoarthritis, and male infertility (11–14). The most well-characterized function of SMN is the assembly of small nuclear ribonucleoproteins (snRNPs). Other less studied but nonetheless important functions of SMN include transcription, pre-mRNA splicing, stress granule formation, translation, mRNA trafficking, DNA repair, intracellular trafficking, and the assembly and regulation of numerous RNPs such as telomerase, the signal recognition particle, snoRNPs and scaRNPs (14). Most of the implicated functions of SMN involve interactions with RNA. However, very limited studies have been done to decipher the nature of sequence and structural motifs for a direct RNA–SMN interaction.
SMN encompasses several functional domains, including a nucleic acid binding region, tudor-domain, proline-rich region and a conserved YG box (14). The N-terminus of SMN, particularly the region encoded by exons 2A and 2B, displays a propensity for interaction with nucleic acids in vitro (15,16). However, SMN lacks a classical RNA-interacting motif generally present in RNA binding proteins (RBPs). In fact, other than an α-helix required for interaction with Gemin2 (17), the N-terminal region encompassing the nucleic acid-binding of SMN is predicted to be largely disordered (18). This general lack of a defined structure is a hallmark of the arginine/glycine-rich (RGG/RG) motifs present in other disordered RBPs including fused in sarcoma (FUS) and fragile X mental retardation protein (FMRP), which play roles in ALS and fragile X syndrome, respectively (19). Interestingly, the RNA binding region of SMN lacks RGG/RG motifs but encompasses lysine-rich motifs. Hence, uncovering novel determinants of RNA–SMN interactions may reveal general rules by which specificity (of RNA interactions) is attained by disordered proteins.
SMN exhibits a preference for poly(rG) oligonucleotides as demonstrated by early in vitro studies (15,16). With significance to the wide-ranging indirect interactions with RNA, SMN associates with many RBPs including Hu-antigen D (HuD), heterogenous nuclear ribonucleoprotein R (hnRNP R), FUS and TAR DNA-binding protein 43 (TDP43) (14,20). More recently, several in vivo RNA targets of SMN were identified by UV crosslinking and pulldown followed by microarray analysis (RIP-Chip) (21). These experiments employed mouse motor neuron-like NSC-34 cells carrying an inducible FLAG-tagged form of SMN. RIP-Chip of FLAG-tagged SMN revealed a large number of transcripts targeted by SMN (21). Surprisingly, the majority of SMN targets were protein-coding mRNAs, and the interaction highly correlated with altered mRNA distribution in cultured neurons upon knockdown of SMN (21). A follow up study showed ANXA2 mRNA as one of the specific targets of SMN, although additional proteins were suggested to facilitate SMN interaction with this mRNA (22). Thus far, a direct interaction between SMN and a cellular RNA target has not been demonstrated.
Iterative in vitro selection, also known as systematic evolution of ligands through exponential enrichment (SELEX), is a powerful technique to identify high-affinity nucleic acid sequences (ligands) that bind to specific proteins (23–25). A SELEX experiment where a purified protein of interest and RNA transcripts are used provides confirmation of a direct RNA–protein interaction. Hence, SELEX remains an unparalleled approach for deciphering critical sequence and structural motifs for RNA–protein interactions (26–29). One of the known disadvantages of SELEX is the selection of artificial motifs that may not exist in nature. Crosslinking and immunoprecipitation (CLIP) is an alternative approach to identify in vivo interactions through irreversible crosslinking a protein to its RNA targets (30). High-throughput sequencing (HITS) combined with CLIP (HITS-CLIP) has potential to generate transcriptome-wide mapping of RNA–protein interactions (31,32). Unlike SELEX, results of HITS-CLIP are impacted by several factors including nature and number of interacting partners of the RBP under investigation. In general, in vitro experiments and HITS-CLIP provide complementary information towards a better understanding of RNA–protein interactions.
Here, we employ SELEX, RNA structure probing, and HITS-CLIP to identify and characterize determinants of RNA–SMN interaction. Our results (of SELEX) showed an unexpected diversity of sequence and structural motifs recognized by SMN. The binding affinity of the selected sequences was demonstrably higher than the poly(rG) oligonucleotides previously reported to be the best SMN-interacting RNA target. Further analysis using site-specific mutations and RNA structure probing of a top SMN ligand as tested by binding assays implicated the role of both sequence and structural motifs as contributors of high affinity and specificity for RNA–SMN interaction. Our subsequent results of HITS-CLIP revealed diverse cellular RNA targets including messenger RNAs (mRNAs), small nucleolar RNAs (snoRNAs), long noncoding RNAs (lncRNAs), and snRNAs. We observed a complex sequence preference for RNA–SMN interaction, with enrichment of G- and A-rich sequence motifs near crosslinking sites. Among targets identified by HITS-CLIP and bearing G-rich motifs matching the results of in vitro selection was the ANXA2 mRNA. Indeed, results of our in vitro binding confirmed that ANXA2 mRNA has higher affinity for SMN than the top ligand isolated by SELEX. These results independently validate for the first time a direct interaction of SMN with a natural RNA target. Our findings support a wider role of direct RNA–SMN interactions in cellular metabolism.
MATERIALS AND METHODS
SMN protein purification
SMN protein was expressed and purified using the IMPACT protein purification kit (NEB) following the manufacturer's guidelines. The detailed purification methods used are described in Supplementary Materials and Methods.
SELEX initial pool generation and in vitro transcription
The double-stranded DNA template for the initial pool (P0) was generated by PCR using Taq polymerase (NEB) in a total volume of 1 ml containing 0.5 nmol of template oligonucleotide. Limited amplification (four cycles) was carried out to reduce bias. After amplification, the PCR product was concentrated by ethanol precipitation, separated on an 8% acrylamide gel, and purified by the crush and soak method (Supplementary Materials and Methods). The recovered DNA was resuspended in 40 μl of nuclease-free water. A 1 μl aliquot of DNA was then run on an acrylamide gel and quantified by visual comparison to a DNA ladder of known concentration. For transcription, 10.5 μg of PCR product was used as a template in a 100 μl reaction using Megashortscript T7 (Ambion) in the presence of 7.5 mM each of ATP, CTP, GTP, and UTP and 1X T7 reaction buffer. Transcription was carried out overnight at room temperature. The reaction was then run on a 8% denaturing urea–PAGE gel, and RNA products were visualized by UV shadowing and purified using the crush and soak method (Supplementary Materials and Methods). After gel purification, RNA was resuspended in 50 μl of RNase-free water and further purified using an RNase-free Micro Bio-spin column (Biorad).
In vitro selection
Variable amounts of RNA and protein (See Figure 1A) were combined together in Binding Buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5 mM DTT). The initial round of selection was carried out in four reactions of 1 ml each. Subsequent rounds of selection were carried out in a single 1 ml reaction. Prior to addition of SMN and DTT, RNA was denatured by briefly heating to 95°C, followed by refolding at 37°C for 1 hour. Binding was carried out for 20 min at room temperature. Protein–RNA complexes were then captured by passing through a Protran BA-85 nitrocellulose filter (Whatman) pre-soaked in Wash Buffer 1 (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2) and washed once with 800 μl of Wash Buffer 1, followed by two washes with 800 μl of Wash Buffer 2 (20 mM Tris–HCl pH 7.5, 450 mM NaCl, 5 mM MgCl2). Filters were then collected and placed in a 1.5 ml microfuge tube for RNA extraction.
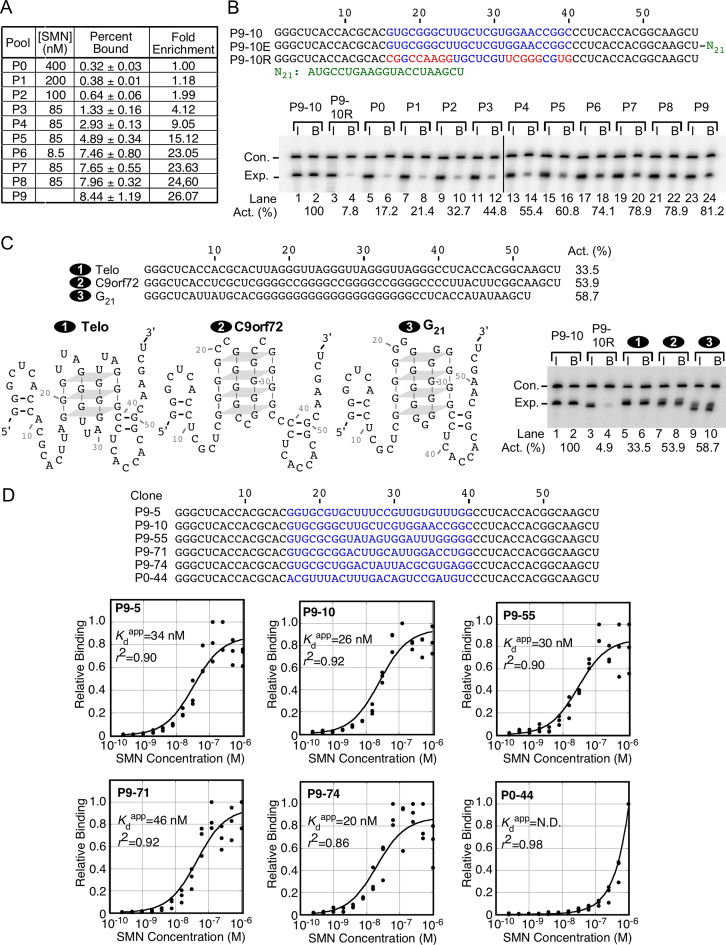
Figure 1.
In vitro selection of high-affinity RNA targets of SMN. (A) Table summarizing the selection process. The RNA concentration was kept constant at 100 nM. SMN concentration is indicated, as well as the percentage of RNA bound in a separate binding assay and the relative enrichment from the initial RNA pool. (B) Competitive binding of each RNA pool. P9-10 is a representative clone from the final selected pool and serves as a positive control. P9-10E control is a modified version of P9-10 that is extended at the 3′ end by 21 bases (N21) and is included in every sample as an internal control. P9-10R is a modified version of P9-10 in which the selected region has been flipped and serves as a negative control. Top panel: Sequences of RNA transcripts used in competitive binding assays. Black bases indicate the constant 5′ and 3′ regions, blue bases indicate the selected region. Red bases indicate bases which are altered from the original P9-10 clone, green bases indicate additions to the P9-10 sequence. Bottom panel: autoradiogram of denaturing PAGE gel depicting results of competitive binding. Experimental RNA identity is indicated at the top. Bands are labeled at the left side. Lane numbers and binding activity relative to P9-10 is indicated at the bottom. I: input RNA. B: bound and recovered RNA. (C) Comparison of P9-10 binding with other G-rich sequences. Top panel: Sequences of RNA transcripts used in competitive binding assays. Percent binding activity of each RNA is indicated at the right. Bottom left panel: Predicted structures of RNA transcripts used in competitive binding assays. Potential G quadruplexes are indicated by stacked quartets connected by grey planes which represent Hoogsteen base pairing. Grey dashes are included between sequential G residues within G quadruplexes for ease of following the sequence. Bottom right panel: Results of competitive binding assays. Labeling is the same as in (B). (D) Sequences and binding characteristics of the 5 highest affinity molecules. Base coloring is the same as in (B). Binding curves to determine the apparent Kd of the top 5 binders as well as an unselected control (P0-44) are shown below sequences. Clone designation is indicated at the top left of each graph. Y axis indicates relative binding, corrected for the highest value observed during each set of experiments. X axis indicates the SMN concentration for each reaction, on a log10 scale. Each point represents one binding reaction, lines are drawn by Kaleidagraph software by fitting the individual points to the Michaelis-Menten equation. Apparent Kd and r2 values are given.
To extract selected RNA from nitrocellulose filters, 400 μl of Tris–EDTA buffer (pH 7.5) and 600 μl of phenol:chloroform (OmniPur) was added to the filters and the samples were vortexed for three pulses of 30 s each. Tubes were then centrifuged at maximum speed for 5 min in a tabletop microcentrifuge. The aqueous phase containing RNA was transferred to a new tube, subjected to ethanol precipitation, and resuspended in 10 μl of RNase-free water. Half of the RNA was used as a template for reverse transcription using Superscript III (Invitrogen) following the manufacturer's instructions. Half of the resulting cDNA was used as template for PCR in a total volume of 1 ml. For each round, the number of PCR cycles ranged from 8 to 10 and was optimized by small-scale amplification before amplifying the entire pool. The pool was then purified and used for transcription as described above. To identify selected sequences, pools were cloned into the pUC19 vector and sequenced by Sanger sequencing.
Nitrocellulose filter paper binding assays
For direct measurement of SMN–RNA binding, 10 pmol each of T7-transcribed RNA (Supplementary Materials and Methods) and protein were combined together in 100 μl Binding Buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5 mM DTT) for a final concentration of 100 nM each. Prior to addition of SMN and DTT, RNA was denatured by briefly heating to 90°C, followed by refolding at 25°C for 10 min. Binding was carried out as described for in vitro selection. Filters were then collected and Cerenkov emissions were measured in a Packard Tri-Carb 3170TR/SL liquid scintillation counter alongside a portion of input and a negative control reaction where protein was omitted.
For determination of the dissociation constant, the same procedure as described above was carried out, except that RNA was transcribed using the high specific activity procedure and 30 000 cpm of RNA (an estimated 0.1 nM final concentration) as determined by Cerenkov counting was used as input for each assay. SMN input started at 100 pmol (1 μM final concentration) and followed a 2-fold dilution series until a minimum final concentration of 0.244 nM was reached. Final relative binding values were determined by subtracting the negative control value from each bound sample, dividing by input, then dividing again by the maximum value obtained for each set of assays.
For the competitive binding assay, RNA input was 10 pmol for both the experimental RNA and the P9-10E control. SMN input was an estimated 5 pmol (50 nM final concentration). After binding and washing, Protran BA-85 nitrocellulose filters (Whatman) were collected and placed in a 1.5 ml microfuge tube. RNA extraction from filters followed the same procedure used during selection. After precipitation, RNA pellets were resuspended in 20 μl of Gel Loading Buffer II (Ambion). Input samples were prepared by setting aside 10 μl of the reaction mixture and diluting it into 400 μl of TE buffer. Samples were then purified by phenol:chloroform extraction and ethanol precipitation in parallel to the bound RNA. 4 μl each of input and bound RNA was loaded in each well of an 8% denaturing urea–PAGE gel. After running, gels were dried and exposed to phosphor image screens. Images were scanned using a Fujifilm FLA-5100 phosphorimager. Bands were quantified using Fujifilm MultiGauge software. Relative binding was determined by dividing the experimental band in each lane by the P9-10E control, then dividing the value of each ‘bound’ lane by the value of input. Final corrections were made by dividing by the obtained numbers by value of the positive control parent clone.
Enzymatic structure probing
RNA was transcribed and gel purified as described above, except that [α-32P]UTP was omitted. After transcription and gel purification, 0.55 μg of RNA was dephosphorylated with calf intestinal phosphatase (NEB) following the manufacturer's instructions. RNA was then purified by phenol:chloroform extraction and ethanol precipitation, and labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB). Labeled RNA was then gel purified once more and further purified using P-30 RNase-free spin columns (Biorad). The 32P signal was then quantified and equal amounts taken for each reaction (∼1–2% of the total labeled RNA). Structure probing was carried out using biochemistry grade RNases T1 and V1 (Ambion) following the manufacturer's instructions. Digested products were resolved on a 16% denaturing urea–PAGE gel, dried, and exposed to phosphorimage screens. Images were scanned using a Fujifilm FLA-5100 phosphorimager. Band intensities were quantified using Fujifilm MultiGauge software. Nucleotides were considered sensitive to digestion if the signal was greater than the average signal of all bands in case of RNase T1, and if signal was greater than background (undigested) in case of RNase V1. Structures with single-stranded/double-stranded constraints were predicted using the RNA structure web server (http://rna.urmc.rochester.edu/RNAstructureWeb/).
SHAPE structure probing
RNA was transcribed and gel purified as described above, except that [α-32P]UTP was omitted. 1-methyl-7-nitroisatoic anhydride (1M7) was synthesized as previously described (33,34). 60 pmol of primers used for extension were 5′ end-labeled with [γ-32P]ATP (Perkin-Elmer) using T4 polynucleotide kinase (NEB). Labeled primers were then gel purified on a 16% denaturing urea–PAGE gel using the crush and soak method. Following gel purification, primers were resuspended in 70 μl of water and further purified using P-30 RNase-free spin columns (Biorad). Structure probing was carried out as previously described (34). For detailed methods, see Supplementary Materials and Methods.
Crosslinking and immunoprecipitation
Unless otherwise stated, all tissue culture media and reagents were purchased from Life Technologies. SH-SY5Y cells were obtained from the American Type Culture Collection (ATCC) and cultured in a 50:50 mix of minimal essential media (MEM) and F12 nutrient mixture supplemented with 10% fetal bovine serum (FBS). SH-SY5Y cells were grown (∼1.2-2 × 107) in 12–20 150 mm dishes per replicate to ∼70–80% confluency. Media was removed and plates were washed 2× with ice-cold phosphate-buffered saline (PBS). After removing the second PBS wash, plates were placed on ice and exposed to 150 mJ/cm2 of 254 nm UV light in a UV Stratalinker (Stratagene). Cells were collected by scraping in 4 ml of PBS per plate and centrifugation at 1000×g for 4 min. For negative (uncrosslinked) control samples, all steps were identical except the UV crosslinking step was omitted. The remainder of the CLIP procedure was performed as described by Ule and colleagues, with the exception of immunoprecipitation, which was carried out as described by Hafner et al. (35–36). Further details are explained in Supplementary Materials and Methods. Illumina sequencing libraries were then prepared using the TruSeq small RNA library preparation kit according to the manufacturer's instructions, with the exception that an additional gel purification step was performed between 3′ and 5′ adapter ligation to prevent adapter dimers. For each replicate, PCR amplification was optimized separately using an aliquot of cDNA. For final library preparation, 20–24 cycles of PCR amplification were carried out, depending on the replicate.
Illumina sequencing, mapping and peak calling
For sequencing of CLIP libraries, the samples for all three replicates were pooled and sequenced using an Illumina MiSeq following a 50-base single-end sequencing protocol. For sequencing of total SH-SY5Y RNA, library preparation was carried out by ribosomal depletion using Ribozero Gold kit (Epicentre) followed by library generation using the TruSeq mRNA kit (Illumina). Six to seven samples were pooled per lane of an Illumina HiSeq 2000 following a 100-base single-end sequencing protocol. After sequencing, adapter and quality trimming was carried out using Cutadapt (37). Identical reads, which may indicate PCR duplication, were removed from CLIP libraries using the Fastx toolkit. Mapping to the human genome (version hg38) was carried out using Tophat (38) using the Gencode v20 transcriptome annotation (39). All reads overlapping known repeat regions as annotated by Repeatmasker (http://repeatmasker.org) were removed. Regions of interest were determined from the CLIP sequencing data by Piranha (40). Significantly enriched GO terms and KEGG pathways were identified using the WebGESTALT web server (http://www.webgestalt.org/). HITS-CLIP data has been submitted to the Gene Expression Omnibus (Accession number GSE110411). SH-SY5Y RNA-Seq data is available at the NCBI sequence read archive (Accession number SRP132513).
RESULTS
Selection of high-affinity sequences by SELEX
We employed SELEX to isolate RNA sequences that interact with SMN with high affinity. We began by purifying recombinant SMN expressed in E. coli using the IMPACT system (Supplementary Figure S1). The IMPACT system generates a tag-free protein by utilizing a self-cleaving intein tag (41). We generated the initial RNA pool from template DNA oligonucleotides that contained a T7 promoter, 5′ and 3′ constant regions for PCR amplification, and a 25 nucleotide (nt) randomized region (Supplementary Figure S2A). We analyzed 50 clones from the initial pool (P0) and did not observe any duplicate sequences or biased base composition in the randomized region (Supplementary Table S1). We used 10 randomly selected clones from P0 to establish a baseline binding affinity of SMN for unselected sequences by the nitrocellulose filter binding assay (42) (Supplementary Figure S3A).
We performed selection using nitrocellulose filter paper to capture RNA–protein complexes [Supplementary Figure S2B, (28)]. The initial RNA input for selection was an estimated 2.4 × 1014 molecules, generated from 1.2 × 1014 molecules of DNA template. In order to capture rare sequences with high affinity for SMN, we started with an excess of protein over RNA and gradually increased the selection stringency over the first four rounds of selection by reducing the amount of the protein in the reaction (Figure 1A). After six rounds, we performed a binding assay with nine randomly selected clones and discovered a mix of sequences with low and high affinity for SMN (Supplementary Figure S3B). In order to remove the majority of low-affinity ligands, we performed a highly stringent round of selection with >10-fold less protein than RNA, followed by two more rounds with near equimolar concentrations (Figure 1A). In a nitrocellulose filter-paper binding assay using equimolar concentrations of RNA and protein, the final pool (P9) bound 8.44% of the input RNA, as compared to only 0.32% of P0, representing an enrichment of over 25-fold (Figure 1A).
In order to compare binding affinities between different sequences in a single reaction, we developed a competitive binding assay. This assay assesses the relative affinity of sequences with respect to a reference RNA of a distinct size that is common in all reactions, thus minimizing reaction-to-reaction variability and the effects of protein activity levels (28). In order to ensure that RNAs compete for binding to the protein of interest, both RNAs in each reaction are supplied in molar excess to the protein. This approach allows for an unbiased determination of the relative affinities of RNAs against a given protein (28,43). To test the competitive binding assay, we first compared the relative binding affinities of a high-affinity clone from the final selected pool (P9-10) and a negative control RNA in which the selected region of P9-10 is reversed (Figure 1B), as well as each of the nine RNA pools generated during selection. We generated the P9-10E reference RNA by extending the 3′ end of P9-10 by 21 bases (Figure 1B). As expected, SMN bound to P9-10 and P9-10E (control RNA) with similar affinity (Figure 1B, lanes 1–2). P9-10R had substantially reduced binding, confirming that SMN exhibits sequence specificity (Figure 1B, lanes 3–4). In the competitive binding assay, the relative binding steadily increased from P0 to P9, consistent with the results of binding without competition (Figure 1A and B). Of note, the final pool showed only ∼4.7-fold increase in the relative affinity in the competition-binding assay compared to >25-fold enrichment shown by the non-competition assay. This is likely due to the high background binding of P0 owing to the unavoidable interaction between P0 and the tight binder (P9-10). Despite this limitation, competitive binding assay provided an independent assessment of the enrichment of the pools against a common high-affinity target.
Previous reports have shown that SMN exhibits a preference for polyG stretches (15,16). Therefore, we set out to determine how our selected sequences compared to similarly sized sequences rich in G residues (Figure 1C). G-rich sequences are unique in that they can form a structure known as G-quadruplex, in which four repeats of three or more G residues base pair by non-Watson–Crick interactions to form a higher-order structure (44). To determine whether SMN interacts with polyG because it is a G-quadruplex binding protein, we included two RNAs (Telomere and C9orf72 repeats) derived from repeat sequences known to form G-quadruplexes in vivo (44,45). Our selected high-affinity ligand, P9-10, is not predicted to form a G-quadruplex, despite being G-rich. All three of the G-quadruplex-forming RNAs were bound by SMN well over the level of background (Figure 1C, lanes 5–10), as represented by the P9-10R RNA (Figure 1C, lanes 3–4). However, P9-10 was bound with a significantly higher affinity (Figure 1C, lanes 1–2), indicating that our SELEX experiment was able to identify a superior sequence, and G-quadruplexes are not the best RNA targets of SMN.
Characterization of selected sequences
We sequenced 99 randomly selected clones from the final pool and analyzed the binding of 78 of them (Supplementary Table S2). We observed a wide range of binding affinities for individual sequences (Supplementary Table S2). There were 10 pairs of clones that had identical sequences, suggesting that the selection was nearing completion. In order to reduce bias coming from identical sequences, we counted duplicate pairs of clones only once. The selected sequences tended to be G- and U-rich with the following distribution: 39.8% G, 31.9% U, 16.5% C and 11.9% A (Supplementary Table S2). We examined the selected sequences for enrichment of specific trinucleotides (Supplementary Table S3). Our results revealed high frequencies of GUG, UGG, UGC and UUG. Since UGG was also over-represented in the initial pool (Supplementary Table S3A), we infer that the specific enrichment of UGG did not occur during selection. We searched the selected sequences for additional enriched sequence motifs using MEME (46). We observed enrichment for a single motif, GUGCG (E-value = 4.6 × 10−9). However, many of the high-affinity clones did not contain a GUGCG motif suggesting that other sequences also contributed towards the high affinity for SMN.
We selected the top 5 highest affinity ligands (RNAs) for further characterization. First, we generated binding curves using a range of SMN protein concentrations and a sub-stoichiometric concentration (∼0.1 nM) of RNA and determined their apparent dissociation constants (Kdapp) (Figure 1D). For an estimate of nonspecific binding to RNA, we used P0-44 from the initial unselected pool. The calculated dissociation constants ranged from 20 nM in the case of P9-74 to 46 nM for P9-71 (Figure 1D). In contrast, for the unselected RNA, we were unable to determine an exact Kdapp, as binding did not reach saturation, even at the highest concentration of protein tested (Figure 1D).
In order to visualize the SMN–RNA interaction, we employed an electrophoretic mobility shift assay (EMSA) as an alternative approach. In particular, we compared the mobility of P9-10 with the negative control P9-10R with and without SMN in composite agarose–acrylamide gels (47). In the presence of increasing amounts of SMN, P9-10 underwent an upward shift suggesting formation of a complex between P9-10 and SMN (Supplementary Figure S4A, lanes 1–7). We observed two complexes, especially at higher SMN concentrations (62.5–250 nM), suggesting that the slow running complex (top band) is formed by multimerization of SMN on P9-10. In addition, at the highest SMN concentrations, RNA began to accumulate near the well, suggesting that SMN is forming higher-order complexes on the RNA molecule. However, it is not possible to speculate on the actual stoichiometry due to the limitations of this assay. In contrast to P9-10, negative control (P9-10R) remained largely unbound at all concentrations we examined (Supplementary Figure S4, lanes 8–14). These results provided an additional proof of specificity of SMN towards the selected RNA sequence compared to a random RNA sequence. In order to determine whether the entire SMN protein is required for a tight RNA interaction, we performed the same set of experiments with purified SMNΔ7 protein, which lacks C-terminal sequences and is deficient in multimerization (48,49). Consistent with the N terminus alone being the key region for RNA interaction, the RNA underwent a mobility shift at only slightly higher concentrations of SMNΔ7 than in full-length SMN (Supplementary Figure S4B, lanes 1–7). However, in contrast to full-length SMN, SMNΔ7 formed higher-order RNA–protein complexes far less efficiently. These results suggest that the reduced oligomerization capacity is a limiting factor for the stability of the large ribonucleoprotein complexes formed by SMNΔ7. Similar to full-length SMN, SMNΔ7 did not bind the negative control P9-10R except at the highest concentrations tested (Supplementary Figure S4B, lanes 8–14).
Diverse structures underlie high-affinity RNA targets of SMN
We employed selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to analyze the secondary structure of the top 5 binders isolated from the final pool of SELEX. SHAPE is a chemical structure probing method that interrogates the accessibility of every single nucleotide position in a single reaction (50). SHAPE protocol takes advantage of 1M7 reagent that preferentially adds a 2′ adduct to an unpaired nucleotide (33,50). Incorporated adduct is then detected by primer extension, as reverse transcription is blocked at the modified position. Due to the requirement for a primer-annealing site, we added an identical 21-nt sequence to the 3′-end of each of the top 5 selected sequences (Figure 2A). Of note, the 21-nt sequence is the same that we used in P9-10E. As confirmed by enzymatic probing, the structure of P9-10 is not disrupted by the presence of an additional 21-nt sequence in P9-10E (Supplementary Figure S5A,B). We also confirmed that the presence of 21-nt extension in top 5 selected sequences did not alter their high affinity for SMN (Supplementary Figure S5C). We used extended versions of the selected clones for structural analysis by SHAPE (Figure 2B).
Figure 2.
Determining the secondary structure of selected high-affinity RNAs. (A) Sequences of the 3′-extended versions of the top 5 candidate RNAs from SELEX. Coloring is the same as in Figure 1B. (B) Autoradiograms of gels depicting results of SHAPE chemical structure probing of extended versions of top 5 candidate RNAs. The first four lanes of each panel depict a sequencing ladder for band identification. Positions of A residues are indicated at the left. Treatment of RNA with DMSO (–) or 1M7 reagent (+) is indicated. Base position in 10-base increments for SHAPE lanes is indicated at the right. Of note, due to the tendency of reverse transcriptase to stop extension 1 base before the modified position, the numbering is offset by 1 compared to the sequencing ladder. (C) The predicted structures for each sequence that best fits the SHAPE reactivity profiles. Relative SHAPE reactivity is presented as purple circles next to each reactive base. Base coloring is the same as in Figure 1B.
SHAPE revealed both similarity and diversity of structures of selected sequences. In particular, P9-5E, P9-10E and P9-71E exhibited a primary stem-loop formed by base pairing between the 3′ portion of the selected region and the 3′ constant region (Figure 2C). We also observed one or two smaller stem-loops at the 5′ end of the molecules (Figure 2C). One of the hallmarks of these structures was the presence of a GGCC motif within the loop portion of the 3′ stem–loop that exhibited lower than expected reactivity (Figure 2B). It is possible that the loops themselves form some sort of structure that precludes reactivity, or that this sequence interacts with some other portion of the sequence, such as in a kissing-loop or pseudoknot. Unlike the other three sequences, P9-55E and P9-74E exhibited much higher overall reactivity (Figure 2B), with prominent single-stranded regions in the central region of the selected sequence (Figure 2C). The probed structures of these two sequences also predicted extensive base-pairing between the 21-base extensions and the original sequences (Figure 2C). However, considering that binding strength is maintained in these RNAs (Supplementary Figure S5), it is likely that the altered context is still sufficient for a tight interaction with SMN. Overall, our results did not reveal any specific structural motif common to all high-affinity targets.
Identification of critical RNA residues for SMN binding
We examined the impact of various sequence and structural motifs of P9-10 RNA on SMN binding. For the ease of interpretation, the 3 stem-loop (SL) structures formed by P9-10 are mapped onto the linear sequence and labeled as SL1, SL2 and SL3 (Figure 3A). To test which minimal region of P9-10 can be bound by SMN, we generated a series of truncated RNAs with sequences removed from either the 5′ or 3′ end of the molecule (Figure 3A). Interestingly, removal of the last 10 bases of P9-10, none of which are within the selected region, resulted in a near-complete loss of SMN binding (Figure 3A, lanes 5–6). All of the removed bases participate in formation of the stem portion of SL3, suggesting that SL3 is important for SMN binding. All shorter RNAs with 3′ bases removed were bound by SMN to a greater extent than Δ48-57, albeit much less efficiently than the full-length P9-10 (Figure 3A, lanes 7–10). In contrast, removal of bases 4 to 13 (first three G residues are important for efficient transcription) resulted in only a partial loss of binding (Figure 3A, lanes 11–12). These bases participate in the formation of SL1 and SL2, suggesting that these structures are not required for a tight SMN interaction. Further deletion of 10 or 20 more bases (Δ4-23 and Δ4-33, respectively), resulted into the complete loss of binding (Figure 3A, lanes 13–16). Since SL3 is left largely intact in the Δ4-23 transcript (Figure 3A), we can infer that additional sequences upstream are also required for a tight SMN interaction.
Figure 3.
Effects of deletions and mutations of P9-10 on SMN binding. (A) Effects of deletions and truncations of P9-10 on SMN binding. Top panel: sequences of deletion and truncation mutants. The name of each construct is indicated on the left. Color coding of the bases is the same as in Figure 1B. Red dashes indicate deleted bases. Gray boxes indicate locations of base pairs contributing to SL1, SL2, and SL3, which are labeled at the top. Percent activity determined in the middle and bottom panel are given at the right. Middle panel: Results of competitive binding assay for 5′ and 3′ truncations of P9-10. Labeling and calculations are the same as in Figure 1B. Bottom panel: Results of competitive binding assay for overlapping deletions of P9-10. Labeling and calculations are the same as in Figure 1B. Abbreviations: SL, stem-loop. (B) Effects of sequence mutations of P9-10 on SMN binding. Top panel: Sequences of mutants used in the competitive binding assay. Labeling and color coding is the same as in Figure 1B. Bottom panel: Results of competitive binding assay for sequence mutations of P9-10. Labeling and calculations are the same as in Figure 1B.
To localize specific regions critical for SMN binding, we generated a series of overlapping 5-nt deletions covering the selected region of P9-10 along with 5 adjacent bases on each side (Figure 3A). All deletions we tested lost a significant amount of binding (Figure 3A, lanes 21–42), suggesting the importance of the entire molecule for a tight SMN interaction. For Δ11–15 mutant that does not affect the sequence of the selected region at all, we observed reduction in binding by more than 70% (Figure 3A, lanes 21–22). For Δ14-18 mutant, we observed the similar effect on binding as for Δ11–15 (Figure 3A, lanes 23–24). For Δ17–21 mutant, we observed >80% loss in relative binding compared to the parent clone (Figure 3A, lanes 25–26). This could be due to an altered sequence context immediately upstream of SL3, from GUGCGG to CGCACG (Figure 3A). We observed interesting results for Δ20-24 mutant, which lacked sequences from both SL2 and SL3, yet bound with >50% of the strength of P9-10 (Figure 3A, lanes 27–28). For Δ23-27 and Δ26-30 mutants, we observed similar binding, ∼30–35% of P9-10 (Figure 3A, lanes 29–32). Both Δ35–39 and Δ38–42 exhibited substantially reduced binding (<20% of P9-10) (Figure 3A, lanes 37–40). This could be due to the loss of G residues at positions 38 (38G) and 39 (39G). In contrast, Δ41-45 mutant that retains 38G and 39G, bound to SMN relatively strongly (Figure 3A, lanes 41–42). Thus, we infer that the critical regions for binding are the sequence immediately upstream of SL3, and the loop sequence, specifically 38G and 39G residues. This may be consistent with a potential dimerization of SMN on the P9-10 RNA, with one SMN contacting the loop sequence and the other SMN contacting the base of SL3 as well as sequences upstream.
We evaluated the position-specific importance of G residues within P9-10. Mutations of the first six G residues in the selected region (G1-6A mutant) caused an almost complete loss of SMN binding, down to ∼10% of P9-10 (Figure 3B, lanes 5–6). Mutations of the last five G residues in the selected region (G7-11A mutant) also had a strong negative effect on SMN binding, but not as severe (Figure 3B, lanes 7–8). To narrow down the G-specific effect, we generated sequences with smaller numbers of G-A mutations. Mutation of the first three G residues (G1-3A mutant) produced effect similar to the one we observed for G1-6A mutant (Figure 3B, lanes 9–10), suggesting that the first three G residues in the selected region are critical for SMN binding. In contrast, mutation of the next three G residues (G4-6A mutant) had little effect on binding (Figure 3B, lanes 11–12). These results are consistent with the results observed for Δ20–24 mutant, in which deletion of bases corresponding to positions 20–24 had a minimal effect on SMN binding (Figure 3A). Mutation of the next three G residues (G7-9A mutant) resulted in a large drop in SMN binding. Of note, G4-6A and G7-9A mutants partially abrogate different regions of the same stem and yet displayed different SMN affinities (Figure 3B, lanes 11–14). It is possible that, since the closing base pairs of the loop region of SL3 are affected, that structural context of binding to the loop is changed significantly. Finally, mutation of the last two G residues (G10-11A mutant) caused a large drop in relative SMN binding (Figure 3B, lanes 15–16). These results are consistent with the results of overlapping deletion mutations and further underscore the essential role of 38G and 39G residues in SMN binding (Figure 3A).
We examined the effect of residues present in the loop of SL3 employing overlapping mutations of four consecutive residues. Of note, we deliberately limited mutations to four consecutive bases to minimize the formation of new structures. Mutation of the first four (M34–37 mutant) or the last four (M42-45 mutant) residues in the loop had almost no effect on SMN binding (Figure 3B, lanes 21–22, 29–30). In contrast, two mutants with substitutions in the middle of the loop (M36-39 and M38-41 mutants) showed near total loss of SMN binding (Figure 3B, lanes 23–26). The mutated bases in both of these mutants overlap the 38G and 39G residues (Figure 3B), further confirming that these two bases are critical for SMN binding. Interestingly, binding was reduced by ∼50% in M40–43 mutant, which retains 38G and 39G residues but substitutes the next C residue (40C) in the loop (Figure 3B, lanes 27–28). These results suggest that 40C in addition to 38G and 39G is also required for strong SMN binding.
Role of secondary structure in SMN–RNA interaction
All of the structures formed by the top binding RNAs require extensive base pairing between the constant and selected regions of each RNA molecule (Figure 2). In order to investigate the effect of RNA structure on binding of SMN, we generated a set of mutants with alterations of the constant region designed to disrupt particular structural features of the P9-10 RNA. First, we tested whether mutations in the 5′ constant region designed to disrupt SL2 had any effect on binding. The first mutant, S2M1, replaced the G at position 12 with a C (Figure 4A). As enzymatic structure probing confirmed (Figure 4B, Supplementary Figure S6A), this change in sequence results in a structural alteration in the 5′ constant region. In particular, bases 11–13 are clearly digested by RNase V1 that targets double-stranded regions. Interestingly, the base of SL3 appears to be destabilized by the S2M1 mutation, as indicated by the increased susceptibility of bases 20–22 to RNase T1 digestion that targets single stranded G residue. Binding of S2M1 is reduced by ∼20% compared to P9-10 (Figure 4C, lanes 5–6). However, whether this reduction in binding is moderated by the disruption of SL2 or the destabilization of the base of SL3 is unclear. We also generated a mutant which completely alters the 5′ arm of SL3, S2M3 (Figure 4A). Despite a more extensive mutation, the structure of S2M3 more closely resembles that of P9-10 (Figures 2B, 4B, Supplementary Figure S6B). Consistently, full binding is preserved; in fact, the S2M3 mutation results in ∼10% increase in binding (Figure 4C, lanes 7–8).
Figure 4.
Mutations targeting secondary structure of P9-10. (A) Sequences of all RNAs used for competitive binding assays in (C). Coloring and labeling is the same as in Figure 1B. (B) Predicted structures of all constant region mutants are shown, annotated with results of enzymatic structure probing (See Supplementary Figure S6). Base coloring is the same as in (A). Purple circles indicate sites digested by RNase T1, green diamonds indicate sites digested by RNase V1. Sizes of circles and diamonds indicate relative sensitivity to enzymatic cleavage. (C) Competitive binding of mutants designed to disrupt P9-10 structure. Lanes corresponding to mutants are labeled with numbers as indicated in (A). Otherwise, labeling and calculations are the same as in Figure 1B.
We next asked if strengthening the stem of SL3 by adding full complementarity could have an impact on SMN binding. To achieve this, we generated the B+1D1 mutant by adding an A residue after position 50, eliminating the bulging base at position 29 and creating an unbroken helix. To maintain a constant size of the molecule, we also deleted 1 base from the loop region (Figure 4A). Consistent with a strengthening of the stem, we observe reduced susceptibility of bases 22 to 33 to T1 digestion and additional V1 cleavage products in the same region. Moreover, due to the stronger stem, bases 41–42 in the loop are no longer sensitive to RNase V1 digestion, indicating reduced heterogeneity in the structure of the loop region (Supplementary Figure S6C). We observed reduced T1 digestion even under denaturing conditions (Supplementary Figure S6C, lane 2), indicating that the modified structure is very strong. B+1D1 mutant showed an increased SMN binding (Figure 4C, lanes 9–10), indicating that a strong stem of SL3 is favorable for a tight SMN interaction. Consistently, the next two mutants, S3M6 and S3M5, with disrupted stem of SL3, as indicated by drastic changes in T1 and V1 digestion patterns (Supplementary Figure S6D, E), showed strong reductions in SMN binding (Figure 4C, lanes 11–14). Finally, we asked whether the loop portion of SL3 is needed to be fully single-stranded in order for SMN binding. We generated the L3M4 mutant, in which the 3′ portion of the loop is altered so that it pairs with bases 34–37, reducing the size of the loop from 12 bases to 4 (Figure 4A). Hence L3M4 had even more strengthened stem than B+1D1 (Supplementary Figure S6C, E). As expected, G38 and G39 in L3M4 were extremely sensitive to RNase T1 digestion, confirming the presence of the GGCC tetra-loop (Supplementary Figure S6E, lane 4). L3M4 showed drastically reduced SMN binding (Figure 4C, lanes 15–16), indicating that the interaction between SMN and SL3 is sensitive to the length of the stem and/or the size of the loop. These results clearly underscored that SMN has strong preference for RNAs with specific secondary structures.
Identification of cellular targets of SMN through HITS-CLIP
In order to identify cellular RNA targets in direct contact with the SMN, we performed HITS-CLIP in neuronal SH-SY5Y cells (Supplementary Figure S7). Our choice of a neuronal cell line was guided by the fact that identified targets would have significance to SMA, a neurodegenerative disease. The SMN protein runs at a size of 42 kilodaltons (kDa) as shown by Western blotting (Figure 5A, left panel). Due to low efficiency of the crosslinking reaction, we did not expect to visualize crosslinked SMN by Western blotting. However, imaging by autoradiography revealed a primary band at ∼48 kDa, which was consistent with the predicted size of SMN crosslinked to RNA with a modal size of 18–20 nucleotides (Figure 5A, right panel). As expected, we did not observe this 48 kDa band without UV crosslinking.
Figure 5.
HITS-CLIP identifies cellular targets of SMN. (A) Verifying pulldown of SMN protein and RNA–SMN complexes. Protein size in kilodaltons (kDa) is given. Left panel: Western blot of input (I, 1% relative to eluted protein loaded) and eluted protein (E). Right panel: Autoradiogram of eluted protein without (–) and with (+) crosslinking of SH-SY5Y cells prior to immunoprecipitation. (B) Read mapping distribution across the transcriptome (left) and protein-coding genes (right) for RNA-Seq of SH-SY5Y cells (RNA-Seq) and reads obtained from CLIP libraries (CLIP), and distribution of statistically significant enriched regions (CLIP peaks). Color code for RNA types is given on the right of the graph. (C) Top 20 enriched KEGG pathways with genes overlapping CLIP peaks. Y axis represents the Benjamini and Hochberg (B+H) corrected P value of enrichment, X axis indicates the KEGG pathway name. (D) Top enriched motifs among CLIP peaks; from left to right, most enriched 6-mer, 7-mer, and 8-mer. (E) Overview of binding screen to identify CLIP targets that are bound tightly by SMN in vitro. Y axis represents binding strength relative to P9-10 RNA. For longer RNAs, sequences used for binding are labeled with the following: UTR: Target chosen contains entire 3′UTR of the target RNA. sca: scaRNA domain of TERC. CL: construct is designed to contain the region surrounding a crosslinked CLIP target.
We recovered RNA from the crosslinked band, performed high-throughput sequencing, and mapped the resulting reads to the human genome to identify cellular SMN-interacting RNAs. As a control for total RNA abundance, we also sequenced and mapped total ribosome-depleted RNA derived from SH-SY5Y cells.
The majority of unique mapped reads, which showed enrichment compared to total SH-SY5Y RNA, overlapped known protein-coding genes (Figure 5B). The next most abundant type of reads mapped to ribosomal RNA (Figure 5B). We assume that these reads represent nonspecific pulldown of ribosomes. A non-negligible proportion of reads (11.2%) mapped to known long noncoding RNAs (lncRNAs), although they represented a smaller proportion of total HITS-CLIP reads (Figure 5B). To capture individual candidate binding regions, we analyzed mapped reads identifying peaks with significantly increased read density compared to a random distribution. Similar to the reads derived from CLIP, the majority of enriched peaks mapped to protein-coding genes (Figure 5B). A smaller proportion of peaks overlapped snoRNAs and lncRNAs, although both were enriched compared to the proportion of raw reads mapping to these RNA types (Figure 5B). Within protein-coding genes, the majority of reads mapped to introns (Figure 5B). However, a reduced proportion was concentrated into significantly enriched peaks, indicating that few of the intron-derived reads represented high-confidence binding sites (Figure 5B). Within exons, CLIP reads and peaks followed a similar distribution to the SH-SY5Y transcriptome, with a slight enrichment of reads corresponding to the 5′UTR and coding region (Figure 5B).
To delineate families of genes or pathways disproportionately bound by SMN, we examined the genes overlapping significant CLIP peaks for enriched gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The most enriched KEGG pathway is for ribosomal proteins, followed by a number of pathways related to the cytoskeleton (Figure 5C). Many of the other targeted pathways involved RNA metabolism and interaction with the extracellular environment (Figure 5C).
In order to identify significantly enriched sequence motifs within regions potentially bound by SMN, we identified each peak that overlapped in two or more of the three replicates of CLIP and extracted the genomic sequence of the surrounding region. We then searched for significantly enriched motifs of varying sizes using MEME (46). In each condition that we tested, the highest enriched motif was purine-rich, generally with a repetitive pattern [WGA] × n (Figure 5D). Interestingly, this motif does not appear to be enriched in P9 sequences, indicating that binding requirements may be altered in vivo due to higher-order RNA structure or cooperative binding between SMN and other RBPs.
SMN directly interacts with ANXA2 mRNA
In order to determine whether an interaction in HITS-CLIP is predictive of a direct SMN interaction in vitro, we performed a limited screen of 15 candidate RNAs that contained significant HITS-CLIP peaks. We chose the candidate RNAs from mRNAs, snRNAs, and snoRNAs, including known targets of SMN such as ACTB and CPG15, components of the SMN complex such as U1, U7 and U11 snRNAs, and the 7S RNA that serves as the RNA portion of the signal recognition particle (51,52). We transcribed each candidate RNA in vitro and analyzed binding of SMN by the nitrocellulose binding assay. Most of the candidates were not bound strongly by SMN in vitro (Figure 5E), suggesting that the cellular interaction of the above-mentioned transcripts likely require additional components besides SMN. We also included ANXA2 mRNA in our screening due to a recent study implicating SMN-dependent transport of ANXA2 mRNA into axons (22). ANXA2 contains 15 known exons, with an alternatively spliced 5′UTR and first coding exon (Figure 6A). The SMN-associated localization sequence in ANXA2 mRNA contains a stem-loop with a G-rich sequence in the loop, much like the putative binding motif in P9-10, and also overlaps a number of reads in SMN HITS-CLIP (Figure 6A). The construct that we used for our binding assays (referred to as ANXA2-L) contained this localization sequence, as well as upstream sequences that include a strong HITS-CLIP peak in the previous two exons (Figure 6A). We also generated a size-matched negative control (L–) derived from a portion of the 3′UTR picked randomly, which had little coverage in HITS-CLIP. In order to determine whether other regions of the ANXA2 mRNA with strong CLIP signals are also bound directly by SMN, we generated RNAs derived from two additional upstream regions (Figure 6A). We tested each of these constructs using the competitive binding assay (Figure 6C, lanes 5–12). Of these constructs, ANXA2-L exhibited the strongest interaction with SMN (Figure 6C, lanes 5–6). The affinity of ANXA2-L for SMN was slightly better than P9-10. Of note, the size-matched control also bound with somewhat low affinity (Figure 6C, lanes 7–8), suggesting either an effect of large RNA size, or a weaker interaction site that was not detected as strongly by CLIP.
Figure 6.
Characterizing ANXA2 mRNA interaction with SMN. (A) Overview of the ANXA2 mRNA, including all reported splice isoforms. The amount of signal from CLIP is indicated at the top. Exons are shown with colored boxes. The start and stop codons are indicated with green and red circles, respectively. Regions used for binding experiments are indicated at the bottom with grey rectangles. (B) Closeup of ANXA2-L sub-sequences used in competitive binding assays. Coloring and labeling are the same as in (A). Base numbering is based on isoform A. (C) Competitive binding of ANXA2-derived sequences compared to the P9-10 selected RNA and P9-10R negative control. Labeling and calculations are the same as in Figure 1B. (D) Competitive binding of mutated forms of ANXA2 881–937. Base coloring is the same as in Figure 1B. Purple box indicates sequences derived from ANXA2 exon 13, green box indicates sequences derived from ANXA2 exon 14. Calculations are the same as in Figure 1B, except that activity is expressed relative to ANXA2 881–937, rather than P9-10. (E) Probed structure of ANXA2-L RNA. Green bases indicate additional sequences required for transcription and cloning. Blue bases indicate core SMN binding region. Purple bases indicate localization signal identified by Rihan and colleagues (Rihan et al. 2017). Bold bases mark locations with at least 3 CLIP reads mapping to them. Bases circled in red indicate mutated bases that resulted in loss of SMN binding activity. The thickness of the red circle corresponds to the relative magnitude of binding loss.
We performed a systematic analysis of ANXA2 mRNA to uncover role of specific motifs critical for SMN interaction. We began by generating two constructs roughly dividing the ANXA2-L sequence into two halves: ANXA2-5′ and ANXA2-3′ (Figure 6B). ANXA2-5′ encompassed the first 119 bases of ANXA2-L, whereas ANXA2-3′ encompassed the last 135 bases of ANXA2-L. There was a 22-base overlap between the two constructs. Interestingly, ANXA2-5′ showed increased binding activity compared to ANXA2-L, whereas ANXA2-3′ showed ∼50% reduction in binding strength (Figure 6C, lanes 13–16). These results suggested that the primary SMN binding site resides within the first 119 bases of ANXA2-L, or between positions 873 and 991 of the ANXA2 mRNA. However, ANXA2-3′ was still bound by SMN well above the background level, indicating that there is likely a weaker, sub-optimal interaction site present in this region. We further narrowed down the core SMN binding region by progressively removing sequences from each end of ANXA2-5′ (Figure 6B). Of these, the RNA corresponding to bases 881 to 937 of the ANXA2 mRNA retained most of the binding activity of the original ANXA2-L construct (Figure 6C, lanes 19–20, whereas further deletion of sequences reduced binding significantly (Figure 6C, lanes 21–22). Based on these results, we conclude that the region from bases 881 to 937 makes up the core binding site for SMN. This region overlaps the junction between exons 13 and 14 of the mature ANXA2 mRNA, suggesting that splicing may need to occur before SMN can interact with this sequence. Of note, this region also corresponds closely to the region with maximum CLIP signal, and numerous CLIP reads also cross the exon-exon junction, also suggesting an interaction with the mature mRNA rather than pre-mRNA (Figure 6B).
In order to determine whether limited mutagenesis could disrupt a tight SMN interaction with ANXA2 mRNA, we generated a number of mutations within the core binding site (Figure 6D). First, we generated an extensively mutated version with 6 base changes (881-937 M6) scattered throughout the stem-loop structure. As expected, this mutation resulted in a complete loss of SMN binding (Figure 6D, lanes 3–4). Next, we designed 3 double mutants targeting either G residues (G892AG898A and G904CG913A mutants) or structural features (C895AG901C). Consistent with important contributions of both sequence and structure to SMN binding, all three mutants lost 50% or greater binding activity. G892AG898A mutant had the strongest effect, with a >80% loss of SMN binding (Figure 6D, lanes 5–6). This mutant contains mutated bases in both exons 13 and 14, suggesting that sequences in both exons may be required for a tight interaction. To determine the contributions of individual critical G residues, we made single mutations based on G892AG898A, which showed the strongest reduction in binding. G892A almost completely abolished binding, suggesting that this G residue is either absolutely necessary for binding of SMN or for formation of the secondary structure conducive to binding. In contrast, G898A reduced binding by less than 50%, suggesting that this G residue contributes to binding but is not critical.
We performed SHAPE to probe the structure of the ANXA2-L RNA (Supplementary Figure S8). We mapped the core SMN binding region, the sites of maximum CLIP signal, and the localization signal identified in the recent study (22) onto the probed structure (Figure 6E). Consistent with RNA structure playing a critical role in SMN binding, the core-binding region forms a distinct stem-loop structure in the probed structure (Figure 6D). Interestingly, the secondary structure places the core SMN binding region in close proximity to the previously identified localization signal. Both regions have CLIP reads mapping to them, so it is likely that SMN binds to both sites in vivo, possibly assisted by the oligomerization property of SMN.
DISCUSSION
SMN belongs to a select group of proteins implicated in a major genetic disease associated with children and infants. Intensive investigations over the past two decades have revealed several functions modulated by SMN. The majority of SMN-associated functions relate to different aspects of RNA metabolism (14). Early in vitro studies revealed a distinct RNA-binding domain within SMN (15,16). However, the RNA sequence and structural determinants critical for a direct RNA–SMN interaction remain unknown. While a recent transcriptome-wide analysis identified cellular RNA targets of SMN, the experimental approach that was used did not distinguish between direct and indirect interactions of SMN with RNAs (21). We conducted this study with the sole purpose of the identification and characterization of sequence and structural motifs critical for direct RNA–SMN interactions. We first employed SELEX to isolate short RNA sequences that interact with SMN with high-affinity. The selected sequences revealed diverse sequence motifs and showed an apparent dissociation constant (Kd) between 20 and 46 nM (Figure 1D). Given the fact that this falls within the typical range Kd values for most RBPs, we propose that SMN is a bona fide RBP.
Our analysis of selected sequences yielded a high enrichment for a GUGCG motif in several but not all clones. Of note, GUGCG motifs are also present in the minor spliceosomal U11 snRNA and 7SL RNA, the RNA component of the SRP (52). Both U11 and 7SL RNAs are known targets of the SMN complex (51,53). However, several mutations outside the GUGCG motif reduced affinity for SMN interaction, ruling out the presence of a GUGCG motif as the sole determinant for RNA–SMN binding (Figures 3–4). Based on the distribution of motifs, we infer that SMN contacts RNA at two or more sites. One of these sites likely serves as the core binding motif, whereas other sites provide secondary binding contacts. Such a mode of binding has been observed for the bacterial protein CsrA (29) and for the mammalian testes-specific protein RBMY (54). In some instances, secondary interactions can enhance the overall binding strength by several orders of magnitude as observed in case of LtrA protein that associates with a group II intron (55).
As we expected, our results confirmed higher affinity of SMN for P9-10, one of the top selected sequences, than several G-rich sequences including poly(rG), telomerase RNA and GGGGCC repeats. While these findings underscore that the G-quadruplex structure is not a favored target of SMN, we still observed SMN binding to all three G-rich sequences at levels above background. Although the telomere sequence is considered to be a DNA repeat, it is transcribed to produce the TERRA RNA, which plays a role in regulating telomerase function and telomere stability (56). SMN is known to regulate telomerase biogenesis (14). Hence, an interaction between SMN and the TERRA RNA may represent another layer of control in telomere maintenance. Expansion of GGGGCC repeats in the intronic region of the C9orf72 gene leads to certain familial forms of frontotemporal dementia and ALS (57). SMN and other RBPs are mislocalized in ALS motor neurons (58,59). Based on the moderate SMN interaction with GGGGCC repeats, it is possible that a direct interaction of SMN with GGGGCC repeats might play some role in the mislocalization of SMN in these disorders.
It is common for SELEX-derived sequences to recruit portions of the constant flanking regions to form structures required for binding (29). This appeared to be the case with at least 3 top ligands we examined. Supporting this point of view, mutation and truncation of the 3′-most portion of the flanking regions of P9-10 had a significant impact on SMN binding (Figures 3 and 4). Several other mutations in the flanking sequences we tested altered SMN binding, further lending credence to the impact of the constraints exerted by the constant flanking regions on the outcome of SELEX. Of all the mutants of P9-10 we examined, S2M3 and B+1D1 were the only ones that showed improved SMN binding. The S2M3 mutant is predicted to trigger rearrangement within the 5′ end of the P9-10, breaking apart the predicted SL2 (Figure 4). In B+1D1, SL3 is strengthened by providing a base-pairing partner to the U residue at position 28, which is normally unpaired and forms a bulge within the predicted stem (Figure 4). The strong increase in SMN binding in the case of B+1D1 suggested an important role of secondary structure in the modulation of RNA–SMN interactions. These results also underscored the fact that selected sequences are not always the highest binders, likely due to restrictions in the size of the sequence space.
Increasing the strength of the stem portion of SL3 was not always linked to higher SMN binding. The L3M4 mutant, which extended the stem portion of SL3 by four bases, showed reduced SMN binding, possibly due to sequestration of loop sequences. Consistently, the results of our substitution and overlapping deletion mutations confirmed the critical role of G38 and G39 residues, which reside in the loop of SL3, in SMN interaction. Of note, GG dinucleotides located at positions analogous to G38 and G39 were selected in all of the top 5 SMN ligands we examined. However, the size of the loop in which GG dinucleotides are placed varied more widely (3–11 nucleotides) than the length of the stem (6–11 nucleotides). Consistently, deletion of the last five bases of the loop portion of SL3 did not appreciably change SMN binding (Figure 4). This would indicate that the critical feature determining binding strength is the precise length of the stem, or perhaps the distance between the GG dinucleotide and upstream motifs. Such a ‘molecular ruler’ mechanism has been proposed for the recognition of pri-miRNAs by DROSHA and DGCR8 (60).
We performed HITS-CLIP to identify in vivo RNA targets of the SMN in human neuronal SH-SY5Y cells. Similar to the results obtained by a previous study utilizing RIP-Chip performed in the mouse motor neuron-like NSC34 cells (21), we observed a preponderance of reads mapping to mRNAs. Surprisingly, despite the critical role for SMN in snRNP biogenesis, reads mapping to snRNAs were underrepresented compared to RNA-Seq of total RNA from SH-SY5Y cells (Figure 5B). This could indicate that interactions with snRNAs are transient and only occur during snRNP assembly, while interactions with other types of RNAs such as mRNAs and lncRNAs could occur throughout the life of the RNA. Although slightly under-represented compared to the total RNA-Seq, we also observed a significant portion of predicted binding sites mapped to lncRNAs, which we have recently shown to be affected by low levels of SMN in a mouse model of SMA (11). snoRNAs made up the bulk of the remainder of predicted binding sites. Given that SMN interacts with snoRNP proteins Fibrillarin and GAR1 (61), an interaction with snoRNAs is not surprising. However, our results of HITS-CLIP should be interpreted with caution because some of these captured interactions could be mediated by other factors interacting with SMN. Supporting this argument, we observed very little overlap between SELEX and CLIP data with respect to the nature of selected motifs. This disparity is not uncommon as experimental conditions in vitro and protein-protein interactions in vivo provide sufficient enough basis for variable outcomes. One example that bears a striking resemblance to our results is the FUS protein. Although in vitro experiments identified GGUG as the FUS targeting motif (62), CLIP experiments revealed a much broader range of targets (63), which were shown to be driven by higher-order assemblies rather than a 1:1 interaction (64,65). Since SMN, like FUS, has a largely disordered RNA binding domain and forms higher order assemblies (18,48,49), we propose a similar mechanism of in vivo binding, highly dependent on oligomerization and possibly recruitment by the many RBP binding partners of SMN.
We analyzed CLIP data to identify functionally related groups of RNA targets of SMN. The most enriched group of RNAs code for ribosomal proteins, consistent with the results of a previous study (21). Other SMN-associated pathways revealed by CLIP pertained to the regulation and maintenance of the actin cytoskeleton (Figure 5C). In agreement with these results, β-actin mRNA has been reported to be a target of SMN (66). Local translation of actin in the growth cone is critical for neurite outgrowth and steering (67,68). A role for SMN in the transport and localized translation of actin and other cytoskeletal components is therefore compatible with the increased sensitivity of motor neurons to low levels of SMN.
The strongest candidate from HITS-CLIP that we confirmed by in vitro binding was a region of the ANXA2 mRNA. Based on the corroborating data of the RIP-Chip experiment carried out by others (21) and a recent study suggesting a role of SMN in localization ANXA2 mRNA in motor neurons (22), we conclude that ANXA2 mRNA is a bona fide target of direct SMN interaction. ANXA2 mRNA codes for the Annexin A2 protein (ANXA2), a ubiquitously expressed member of the Annexin family, which is a highly conserved family of calcium-binding membrane-associated proteins (69). Annexins form lateral assemblies on cellular membranes in a Ca2+-dependent manner, and thus likely serve as scaffolds for organizing membrane domains and localizing activity (69). ANXA2 associates with filamentous actin, serving as an organization center for microfilaments and an attachment point for actin filaments to membranes (70). Screens for modifiers of SMA have revealed that actin regulation, neuronal Ca2+ signaling, and endocytosis all play critical roles in SMA pathogenesis (71,72). Seeing as the ANXA2 protein is involved in all three processes, it is no surprise that ANXA2 was recently also identified as a potential positive modifier of SMA and other related motor neuron diseases (73). Overexpression of ANXA2 results in the disruption of Cajal bodies (74). Multiple studies have indicated that low expression of SMN results in an increase in levels of ANXA2 mRNA (21,75).
We narrowed the core binding region of SMN to the ANXA2 mRNA to a 57-nt stretch encompassing the end of exon 13 and the start of exon 14. Interestingly, mutagenesis of the core binding region revealed that, although the majority of the binding region encompasses exon 14, at least one critical G residue required for tight binding is derived from exon 13. This indicates that SMN can only interact tightly with the mature mRNA. Therefore, we propose a model wherein SMN binds to ANXA2 mRNA after splicing, after which it multimerizes on the RNA and recruits additional protein cofactors (Figure 7). Depending on the interacting proteins, ANXA2 may be redirected towards growth cones and/or axon terminals of neuronal cells, where it is locally translated, or to stress granules, of which SMN is a critical component (76) (Figure 7). Axonal ANXA2 protein then functions as an organizing center, coordinating the cytoskeleton, mRNAs, and membrane vesicles. Low SMN causes reduction in the levels of ANXA2 in axon terminals and/or growth cones, resulting in loss of axon-specific organizing functions. At the same time, levels in the cell body rise, potentially resulting in the disruption of nuclear bodies (74). A combination of these two factors could result in a pathological state in SMA motor neurons.
Figure 7.
Proposed model of SMN binding to ANXA2 mRNA in cells. The ANXA2 pre-mRNA is shown at the top. Exons are portrayed by colored boxes, introns by black lines. After splicing, SMN binds to the high-affinity core region at the junction of exons 13 and 14, after which additional proteins are recruited and SMN forms additional contacts on the mRNA. At this point, SMN, with the assistance of other protein cofactors, targets the mRNA for localization, potentially towards stress granules, axon terminals/growth cones, or other subcellular domains.
SMN is conserved throughout the animal kingdom, and low SMN levels lead to SMA-like phenotypes in a wide range of model organisms, including C. elegans, Drosophila, and zebrafish (77). Therefore, it stands to reason that any function loss that potentially leads to SMA should be similarly conserved. Consistent with the RNA binding activity of SMN playing an important role, both zebrafish and C. elegans SMN bind RNA in vitro (16). Likewise, the core binding region of the ANXA2 mRNA is highly conserved. However, the amino acid sequences of the entire annexin family are extremely well conserved, especially the calcium-binding Annexin repeat domains (69), so whether the core binding region is conserved in order to preserve SMN binding or retention of ANXA2 protein function is unknown.
The functional significance of an RBP is best understood through the specificity of interactions at the RNA–protein interface, the complexity of which increases with the increasing size of RNA as well as additional protein-protein interactions. Findings reported in this study reveal for the first time that SMN has the inherent capability to preferentially select its targets through interactions with sequence and structural motifs. The diversity of high affinity ligands isolated by SELEX support a broad spectrum of RNA–SMN interactions with implications to novel SMN functions in RNA metabolism. Given the fact that multiple contacts within RNA are required for a tight RNA–SMN interaction, it is likely that the folding of the entire SMN protein determines the interface of RNA–SMN interaction. Several SMN variants harboring its nucleic acid-binding domain are generated by alternative splicing during normal and stress-associated conditions (18,78–80). The overall folding of SMN variants are likely to be different due to the absence/difference of C-terminal residues. It will be telling to determine whether the high affinity RNA ligands of SMN also interact with other SMN isoforms with similar affinity and specificity. While results of our CLIP and in vitro studies confirm ANXA2 mRNA as the first genuine target of direct SMN interaction, this may represent only the tip of the iceberg. To a broader significance, findings reported in this study elevate the role of SMN from facilitator to initiator of critical events due to its central role in direct interactions with specific cellular RNA targets.
DATA AVAILABILITY
HITS-CLIP data has been submitted to the Gene Expression Omnibus (accession number: GSE110411). SH-SY5Y RNA-Seq data is available at the NCBI sequence read archive (accession number: SRP132513).
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge Dr Joonbae Seo for critical reading of the manuscript and for providing valuable suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 NS055925 and R21 NS080294]; Iowa Center for Advanced Neurotoxicology (ICAN) and Salsbury Endowment (Iowa State University, Ames, IA, USA) (to R.N.S.). Funding for open access charge: NIH grant and internal funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995; 80:155–165. [DOI] [PubMed] [Google Scholar]
- 2. Singh N.N., Androphy E.J., Singh R.N.. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit. Rev. Eukaryotic Gene Expression. 2004; 14:271–285. [DOI] [PubMed] [Google Scholar]
- 3. Singh N.K., Singh N.N., Androphy E.J., Singh R.N.. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006; 26:1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh R.N. Evolving concepts on human SMN Pre-mRNA splicing. RNA Biol. 2007; 4:7–10. [DOI] [PubMed] [Google Scholar]
- 5. Singh N.N., Singh R.N.. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011; 8:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh N.N., Howell M.D., Singh R.N.. Charlotte SJ, Paushkin S, Ko C-P. Transcriptional and splicing regulation of spinal muscular atrophy genes. Spinal Muscular Atrophy: Disease Mechanisms and Therapy. 2016; Elsevier Inc. [Google Scholar]
- 7. Cho S.C., Dreyfuss G.. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010; 24:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seo J., Howell M.D., Singh N.N., Singh R.N.. Spinal muscular atrophy: an update on therapeutic progress. Biochim. Biophys. Acta: Mol. Basis Dis. 2013; 1832:2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howell M.D., Singh N.N., Singh R.N.. Advances in therapeutic development for spinal muscular atrophy. Future Med. Chem. 2014; 6:1081–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahmad S., Bhatia K., Kannan A., Gangwani L.. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J. Exp. Neurosci. 2016; 10:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ottesen E.W., Howell M.D., Singh N.N., Seo J., Whitley E.M., Singh R.N.. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Scientific Rep. 2016; 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Howell M.D., Ottesen E.W., Singh N.N., Anderson R.L., Seo J., Sivanesan S., Whitley E.M., Singh R.N.. TIA1 is a gender-specific disease modifier of a mild mouse model of spinal muscular atrophy. Scientific Rep. 2017; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Howell M.D., Ottesen E.W., Singh N.N., Anderson R.L., Singh R.N.. Gender-Specific amelioration of SMA phenotype upon disruption of a deep intronic structure by an oligonucleotide. Mol. Ther. 2017; 25:1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh R.N., Howell M.D., Ottesen E.W., Singh N.N.. Diverse role of survival motor neuron protein. Biochim. Biophys. Acta. 2017; 1860:299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lorson C.L., Androphy E.J.. The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum. Mol. Genet. 1998; 7:1269–1275. [DOI] [PubMed] [Google Scholar]
- 16. Bertrandy S., Burlet P., Clermont O., Huber C., Fondrat C., Thierry-Mieg D., Munnich A., Lefebvre S.. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum. Mol. Genet. 1999; 8:775–782. [DOI] [PubMed] [Google Scholar]
- 17. Zhang R.D., So B.R., Li P.L., Yong J., Glisovic T., Wan L.L., Dreyfuss G.. Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell. 2011; 146:384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seo J., Singh N.N., Ottesen E.W., Lee B.M., Singh R.N.. A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein. Scientific Rep. 2016; 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ozdilek B.A., Thompson V.F., Ahmed N.S., White C.I., Batey R.T., Schwartz J.C.. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017; 45:7984–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hosseinibarkooie S., Schneider S., Wirth B.. Advances in understanding the role of disease-associated proteins in spinal muscular atrophy. Expert Rev. Proteomics. 2017; 14:581–592. [DOI] [PubMed] [Google Scholar]
- 21. Rage F., Boulisfane N., Rihan K., Neel H., Gostan T., Bertrand E., Bordonne R., Soret J.. Genome-wide identification of mRNAs associated with the protein SMN whose depletion decreases their axonal localization. RNA. 2013; 19:1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rihan K., Antoine E., Maurin T., Bardoni B., Bordonne R., Soret J., Rage F.. A new cis-acting motif is required for the axonal SMN-dependent Anxa2 mRNA localization. RNA. 2017; 23:899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 24. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 25. Stoltenburg R., Reinemann C., Strehlitz B.. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007; 24:381–403. [DOI] [PubMed] [Google Scholar]
- 26. Tacke R., Manley J.L.. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA-binding specificities. EMBO J. 1995; 14:3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dember L.M., Kim N.D., Liu K.Q., Anderson P.. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996; 271:2783–2788. [DOI] [PubMed] [Google Scholar]
- 28. Singh R.N., Saldanha R.J., D'Souza L.M., Lambowitz A.M.. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J. Mol. Biol. 2002; 318:287–303. [DOI] [PubMed] [Google Scholar]
- 29. Dubey A.K., Baker C.S., Romeo T., Babitzke P.. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005; 11:1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B.. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003; 302:1212–1215. [DOI] [PubMed] [Google Scholar]
- 31. Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X.N. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008; 456:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Darnell R.B. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscipl. Rev.-RNA. 2010; 1:266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mortimer S.A., Weeks K.M.. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007; 129:4144–4145. [DOI] [PubMed] [Google Scholar]
- 34. Singh N.N., Lawler M.N., Ottesen E.W., Upreti D., Kaczynski J.R., Singh R.N.. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013; 41:8144–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.C., Munschauer M. et al. Transcriptome-wide identification of RNA-Binding protein and MicroRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ule J., Jensen K., Mele A., Darnell R.B.. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005; 37:376–386. [DOI] [PubMed] [Google Scholar]
- 37. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011; 17:10–12. [Google Scholar]
- 38. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uren P.J., Bahrami-Samani E., Burns S.C., Qiao M., Karginov F.V., Hodges E., Hannon G.J., Sanford J.R., Penalva L.O.F., Smith A.D.. Site identification in high-throughput RNA–protein interaction data. Bioinformatics. 2012; 28:3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chong S.R., Mersha F.B., Comb D.G., Scott M.E., Landry D., Vence L.M., Perler F.B., Benner J., Kucera R.B., Hirvonen C.A. et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997; 192:271–281. [DOI] [PubMed] [Google Scholar]
- 42. Wong I., Lohman T.M.. A double-filter method for nitrocellulose-filter binding - application to protein nucleic-acid interactions. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh N.N., Seo J.B., Ottesen E.W., Shishimorova M., Bhattacharya D., Singh R.N.. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol. Cell. Biol. 2011; 31:935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A.J., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J.Y., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suh M.H., Ye P., Datta A.B., Zhang M.C., Fu J.H.. An agarose-acrylamide composite native gel system suitable for separating ultra-large protein complexes. Anal. Biochem. 2005; 343:166–175. [DOI] [PubMed] [Google Scholar]
- 48. Martin R., Gupta K., Ninan N.S., Perry K., Van Duyne G.D.. The survival motor neuron protein forms soluble glycine zipper oligomers. Structure. 2012; 20:1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gupta K., Martin R., Sharp R., Sarachan K.L., Ninan N.S., Van Duyne G.D.. Oligomeric properties of survival motor neuron.Gemin2 complexes. J. Biol. Chem. 2015; 290:20185–20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wilkinson K.A., Merino E.J., Weeks K.M.. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006; 1:1610–1616. [DOI] [PubMed] [Google Scholar]
- 51. Piazzon N., Schlotter F., Lefebvre S., Dodre M., Mereau A., Soret J., Besse A., Barkats M., Bordonne R., Branlant C. et al. Implication of the SMN complex in the biogenesis and steady state level of the signal recognition particle. Nucleic Acids Res. 2013; 41:1255–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keenan R.J., Freymann D.M., Stroud R.M., Walter P.. The signal recognition particle. Annu. Rev. Biochem. 2001; 70:755–775. [DOI] [PubMed] [Google Scholar]
- 53. Lotti F., Imlach W.L., Saieva L., Beck E.S., Hao L.T., Li D.K., Jiao W., Mentis G.Z., Beattie C.E., McCabe B.D. et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012; 151:440–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Skrisovska L., Bourgeois C.F., Stefl R., Grellscheid S.N., Kister L., Wenter P., Elliott D.J., Stevenin J., Allain F.H.T.. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007; 8:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsuura M., Noah J.W., Lambowitz A.M.. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001; 20:7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cusanelli E., Chartrand P.. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015; 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L.. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016; 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamazaki T., Chen S., Yu Y., Yan B.A., Haertlein T.C., Carrasco M.A., Tapia J.C., Zhai B., Das R., Lalancette-Hebert M. et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012; 2:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun S.Y., Ling S.C., Qiu J.S., Albuquerque C.P., Zhou Y., Tokunaga S., Li H.R., Qiu H.Y., Bui A., Yeo G.W. et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015; 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Han J.J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y.J., Zhang B.T., Kim V.N.. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006; 125:887–901. [DOI] [PubMed] [Google Scholar]
- 61. Pellizzoni L., Baccon J., Charroux B., Dreyfuss G.. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001; 11:1079–1088. [DOI] [PubMed] [Google Scholar]
- 62. Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F.. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001; 276:6807–6816. [DOI] [PubMed] [Google Scholar]
- 63. Hoell J.I., Larsson E., Runge S., Nusbaum J.D., Duggimpudi S., Farazi T.A., Hafner M., Borkhardt A., Sander C., Tuschl T.. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011; 18:1428–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang X.Y., Schwartz J.C., Cech T.R.. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015; 43:7535–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schwartz J.C., Wang X.Y., Podell E.R., Cech T.R.. RNA seeds Higher-Order assembly of FUS protein. Cell Rep. 2013; 5:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M.. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003; 163:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Willis D., Li K.W., Zheng J.Q., Chang J.H., Smit A., Kelly T., Merianda T.T., Sylvester J., van Minnen J., Twiss J.L.. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci. 2005; 25:778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leung K.M., van Horck F.P.G., Lin A.C., Allison R., Standart N., Holt C.E.. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 2006; 9:1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gerke V., Creutz C.E., Moss S.E.. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005; 6:449–461. [DOI] [PubMed] [Google Scholar]
- 70. Hayes M.J., Rescher U., Gerke V., Moss S.E.. Annexin-actin interactions. Traffic. 2004; 5:571–576. [DOI] [PubMed] [Google Scholar]
- 71. Hosseinibarkooie S., Peters M., Torres-Benito L., Rastetter R.H., Hupperich K., Hoffmann A., Mendoza-Ferreira N., Kaczmarek A., Janzen E., Milbradt J. et al. The power of human protective Modifiers: PLS3 and CORO1C unravel impaired endocytosis in spinal muscular atrophy and rescue SMA phenotype. Am. J. Hum. Genet. 2016; 99:647–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Riessland M., Kaczmarek A., Schneider S., Swoboda K.J., Lohr H., Bradler C., Grysko V., Dimitriadi M., Hosseinibarkooie S., Torres-Benito L. et al. Neurocalcin delta suppression protects against spinal muscular atrophy in humans and across species by restoring impaired endocytosis. Am. J. Hum. Genet. 2017; 100:297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kline R.A., Kaifer K.A., Osman E.Y., Carella F., Tiberi A., Ross J., Pennetta G., Lorson C.L., Murray L.M.. Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases. Plos Genet. 2017; 13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kazami T., Nie H., Satoh M., Kuga T., Matsushita K., Kawasaki N., Tomonaga T., Nomura F.. Nuclear accumulation of annexin A2 contributes to chromosomal instability by coilin-mediated centromere damage. Oncogene. 2015; 34:4177–4189. [DOI] [PubMed] [Google Scholar]
- 75. Fuller H.R., Mandefro B., Shirran S.L., Gross A.R., Kaus A.S., Botting C.H., Morris G.E., Sareen D.. Spinal muscular atrophy patient iPSC-derived motor neurons have reduced expression of proteins important in neuronal development. Front. Cell Neurosci. 2015; 9:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zou T., Yang X.M., Pan D.M., Huang J., Sahin M., Zhou J.H.. SMN Deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell. Mol. Neurobiol. 2011; 31:541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Edens B.M., Ajroud-Driss S., Ma L., Ma Y.C.. Molecular mechanisms and animal models of spinal muscular atrophy. Biochim. Biophys. Acta-Mol. Basis Dis. 2015; 1852:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Setola V., Terao M., Locatelli D., Bassanini S., Garattini E., Battaglia G.. Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Singh N.N., Seo J., Rahn S.J., Singh R.N.. A Multi-Exon-Skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS One. 2012; 7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Seo J., Singh N.N., Ottesen E.W., Sivanesan S., Shishimorova M., Singh R.N.. Oxidative stress triggers Body-Wide skipping of multiple exons of the spinal muscular atrophy gene. PLoS One. 2016; 11:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HITS-CLIP data has been submitted to the Gene Expression Omnibus (accession number: GSE110411). SH-SY5Y RNA-Seq data is available at the NCBI sequence read archive (accession number: SRP132513).