Abstract
DEAD-Box proteins (DBPs) constitute a prominent class of RNA remodeling factors that play a role in virtually all aspects of RNA metabolism. To better define their cellular functions, deletions in the genes encoding each of the Escherichia coli DBPs were combined with mutations in genes encoding different Ribonucleases (RNases). Significantly, double-deletion strains lacking Ribonuclease R (RNase R) and either the DeaD or SrmB DBP were found to display growth defects and an enhanced accumulation of ribosomal RNA (rRNA) fragments. As RNase R is known to play a key role in removing rRNA degradation products, these observations initially suggested that these two DBPs could be directly involved in the same process. However, additional investigations indicated that DeaD and SrmB-dependent rRNA breakdown is caused by delays in ribosome assembly that increase the exposure of nascent RNAs to endonucleolytic cleavage. Consistent with this notion, mutations in factors known to be important for ribosome assembly also resulted in enhanced rRNA breakdown. Additionally, significant levels of rRNA breakdown products could be visualized in growing cells even in the absence of assembly defects. These findings reveal a hitherto unappreciated mechanism of rRNA degradation under conditions of both normal and abnormal ribosome assembly.
INTRODUCTION
DEAD-Box proteins (DBPs) represent a large class of RNA-remodeling factors that are found in almost all organisms. These proteins are commonly associated with adenosine triphosphate (ATP)-dependent RNA helicase activity and have been implicated in numerous cellular processes (1–3). The characteristic features of DBPs are a set of >13 short conserved motifs, including the eponymous DEAD motif, that localize within two RecA-like domains. These domains jointly bind to RNA and ATP and mediate ATP-dependent RNA-duplex unwinding activity (4). The structures of several DBPs in complex with RNA have been determined, yielding important insights into the mechanism of RNA recognition and RNA-duplex unwinding. Nonetheless, the physiological functions of DBPs are difficult to determine based on sequence or structure alone. For many of these proteins, their biological functions remain poorly understood or even completely unknown.
My laboratory is interested in elucidating the cellular functions of DBPs from Escherichia coli. This organism encodes five DBPs: DbpA, DeaD, RhlB, RhlE and SrmB, which have been characterized to varying degrees. For example, the main property associated with DbpA is its binding to 23S rRNA, which is mediated by the C-terminal region of the protein downstream of its helicase domain (5,6). RhlB is an integral component of the RNA degradosome, and is believed to unwind RNA structures to facilitate RNA degradation (7). DeaD and SrmB are implicated in ribosome assembly at low temperatures and an absence of either protein confers a cold-sensitive growth defect (8,9). These two proteins are also implicated in gene regulation (10,11). Little is known about RhlE apart from the fact that it associates with ribosomes and functionally influences the action of DeaD and SrmB in ribosome assembly at low temperatures (12). As studies on these DBPs have been limited, it is likely that they participate in additional, yet unknown, cellular processes. We have also shown that four of these DBPs perform ATP-independent reactions in vitro, including stimulating the annealing of complementary RNA strands, promoting strand exchange and RNA chaperone activity (13). These observations suggest that these ATP-independent activities could confer additional physiological functions to these proteins inside the cell.
One approach used to define cellular function is to evaluate genetic interactions between factors of interest with those of known functions. The identification of any interactions, either positive or negative, can implicate factors of unknown or poorly characterized function into known pathways. This approach is used here to investigate E. coli DBP function, yielding new insights into the mechanistic basis of rRNA degradation. Specifically, it is shown that an absence of either DeaD and SrmB results in slower ribosome assembly, one consequence of which is enhanced rRNA breakdown initiated via endonucleolytic cleavage. The rRNA degradation fragments are rapidly removed by RNase R but can be visualized in strains that lack this enzyme. A similar breakdown of rRNA is also observed when previously characterized ribosome assembly factors (RAFs) are absent or under conditions that inhibit assembly indirectly. Overall, these studies demonstrate that the efficiency of ribosomal assembly is a critical parameter that governs rRNA degradation. Moreover, it is shown that a significant residual degree of rRNA degradation takes place inside cells even when ribosome assembly is unperturbed.
MATERIALS AND METHODS
Strains and plasmids
The wild-type (W.T.) strain used for this study is MG1655*, a derivative of MG1655 that contains a point mutation in rph that restores the reading frame of its prematurely terminated gene product (14). Deletion mutations were derived from the Keio strains collection (15) and were transferred to the MG1655* strain background by P1 transduction. A temperature sensitive pnp (pnpts) allele was derived from strain SK6639 (16) and transferred to the MG1655* background using a linked chloramphenicol-resistant marker. A Polynucleotide phosphorylase (PNPase) or RNase R-expression strain was constructed by transforming the isopropyl-β-D-thiogalactoside (IPTG)-regulated PNPase or RNase R expression plasmids (pCA24N-pnp or pCA24N-rnr, respectively) (17) into an MG1655* derivative that contains an unmarked Δpnp allele, followed by chromosomal rnr deletion by using P1 transduction in each case. The plasmid pKAK7, which expresses PNPase from its endogenous promoter, has been described (16).
Northern blot analysis
Total RNA was isolated using the hot-phenol method. RNAs were fractionated on 2% agarose gels in MOPS buffer containing 0.66 M formaldehyde. After electrophoresis, the RNA was transferred to positively charged membrane (Nytran), cross-linked using 254 nM ultraviolet light, and hybridized overnight with radiolabeled probes for 16S rRNA (5′-CGTTCAATCTGAGCATGATC-3′) or 23S rRNA (5′- TAGCTGGCGGTCTGGGTTG -3′) at 37°C.
Primer extension
Primer extension analysis was performed as described (18), using a gel-purified radiolabeled oligonucleotide complementary to 16S rRNA (5′-ACCGCTTGTGCGGGCCCCCG-3′). The reaction products were fractionated on a denaturing 10% polyacrylamide gel.
3′ RACE
Total cellular RNA was isolated from a ΔrnrΔrimM strain and ligated to a pre-adenylated universal miRNA cloning linker (NEB cat #S1315S) using T4 RNA ligase 2 truncated KQ (NEB cat #S0373S). The reaction products were annealed with primer CJ1341 (5′ CCGTGATTGATGGTGCCTACAG 3′), which is complementary to the cloning linker, and reverse-transcribed. The cDNA products were amplified with CJ1341 and a DNA primer that corresponds to the 16S rRNA nts 537–558 (5′ GGAGGGTGCAAGCGTTAATCGG 3′). The products were resolved on an agarose gel, eluted and sequenced to determine the 3′ ends of the different rRNA fragments.
RNase E digestion assays
30S or 70S fractions were collected after ultracentrifugation of cell extracts from a WT strain and concentrated using a Microcon 100 filter (Amicon). A plasmid clone expressing His-tagged RNase E protein that contains the catalytic domain (amino-acids 1–529) was obtained from Kenneth McDowall (University of Leeds) and used to prepare purified RNase E, as described (19). Digestion assays were performed with 100 ng of 30S particles or 300 ng of 70S particles in RNase E digestion buffer (25 mM Tris, pH 8, 150 mM NaCl, 10 mM MgCl2 and 0.1% Triton X-100) in 10 μl volume for 1 h at 37°C. The reactions were sequentially extracted with phenol and chloroform and analyzed by primer-extension.
Western blot analysis
Western blot analysis was performed on cells lysates prepared by heating cell pellets resuspended in sample buffer at 100°C, followed by electrophoresis and transfer to nitrocellulose membrane. The membranes were sequentially incubated with antibody to PNPase (a kind gift of A. J. Carpousis, University of Toulouse, France) and horseradish peroxidase-conjugated (HRP) anti-rabbit antibody. The blots were developed using Clarity Max Western ECL substrate (BioRad).
Ribosome pulse-labeling
Cells were grown in 1 ml LB+10 mM MgSO4 medium at 37°C and 5 μCi of 32P inorganic phosphate was added at mid-log phase. After 1 min, the cells were pelleted by centrifuging in a microfuge for 20 s, resuspended in 1 ml of pre-warmed growth medium and transferred to a shaking water bath at 37°C. 0.45 ml aliquots were removed at pre-determined times and transferred to microfuge tubes containing 1 ml of chilled saline to halt ribosome assembly. The cells were lysed by sonication and combined with unlabeled cell lysates derived from the corresponding strain. The combined lysates were layered on a 14–32% sucrose gradient and ultracentrifuged, as described (12). Sixteen fractions of ∼1.7 ml were collected after centrifugation and 0.5 ml of each was filtered through nitrocellulose membrane using a dot-blot vacuum manifold (Schleicher and Schuell).
RESULTS
Synthetic growth defects in strains containing mutations in DBPs and RNase R
Many RNAs are unstable in vivo with typical half-lives of 1–5 min, their instability determined by the presence of RNases in the cell, the sequence of the RNAs and the structures that the RNAs adopt in vivo (20,21). Specifically, most of the RNases display a preference for either single-stranded or double-stranded RNA sequences. Because of their ability to remodel RNA, one of the outcomes of DBP function could be to alter RNA degradation rates, a role that has already been attributed to RhlB (22).
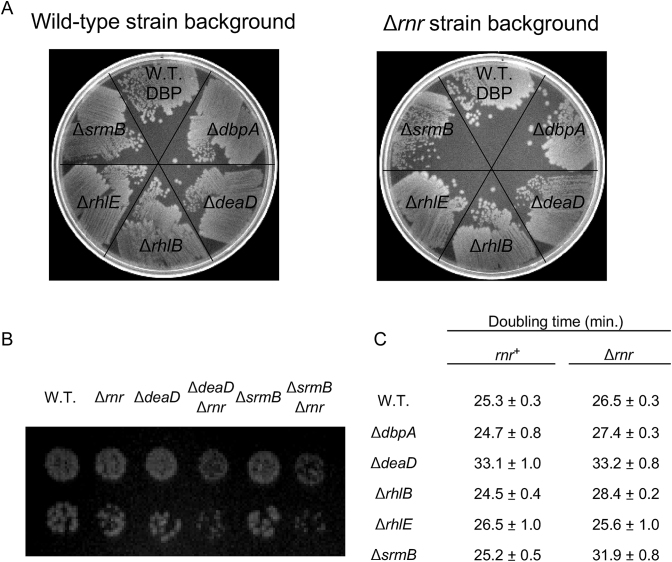
To investigate whether the E. coli DBPs influence RNA degradation through additional mechanisms, genetic interactions between DBPs and different E. coli RNases were investigated. For these purposes, mutations in genes encoding several ribonucleases; i.e. RNase II, RNase PH, RNase R, RNase T, RNase III, RNase P, RNase G, RNase E or PNPase, were combined with deletions in genes encoding each of the five E. coli DBPs, and the phenotypes of the double mutant strains were assessed by screening colonies on LB-agar plates. Among the different double mutants tested, two combinations, containing a deletion in the gene encoding RNase R (rnr) and either a DeaD or a SrmB deletion, gave rise to strains that grew more poorly on agar plates at 37°C as compared to strains containing the individual mutations (Figure 1A). Such a phenotype was not observed for combinations of the rnr deletion with an absence of the other three DBPs. To confirm the slow growth phenotype of the ΔdeaDΔrnr and ΔsrmBΔrnr strains, these strains, along with appropriate controls, were spotted at limiting dilution on LB-agar plates. Reduced growth of individual colonies was observed for the double-mutant derivatives relative to their WT or singly mutated strain counterparts (Figure 1B). Additionally, the doubling time of each of the DBP mutant strains, as well as their Δrnr derivatives, was measured in LB medium. The ΔdeaD, ΔdeaDΔrnr and the ΔsrmBΔrnr strains were found to exhibit 25–35% longer doubling times compared to the WT strain (Figure 1C). An increased doubling time for the ΔdeaD strain has been noted previously (23).
Figure 1.
Growth defects in strains lacking RNase R and DBPs. (A) Growth of strains on LB-agar plates. Deletion alleles of each of the five Escherichia coli DBPs were transferred into WT or Δrnr strain backgrounds. The derivative strains were streaked on an LB-agar plate and grown overnight at 37°C. (B) Dilutions of saturated cultures necessary to yield single colonies were spotted on LB-agar plates and grown overnight at 37°C. (C) Doubling times were determined in LB medium at 37°C. Mean values and standard errors are derived from triplicate cultures for each strain.
Accumulation of ribosomal RNA (rRNA) fragments in ΔdeaDΔrnr and ΔsrmBΔrnr strains
RNase R is one of eight E. coli 3′ to 5′ exonucleases, which, uniquely among the exonucleases, can digest through RNA containing high levels of secondary structure (24). This property is attributed to a recently identified helicase domain in this enzyme, which opens secondary structures to enable RNA digestion (25). Given the evidence of genetic interactions between rnr and either deaD or srmB, it suggested a possibility that the two DBPs could be participating in a process that operates in parallel to the RNase R-degradation pathway. Consequently, the inactivation of both RNase R and DeaD/SrmB would result in growth defects.
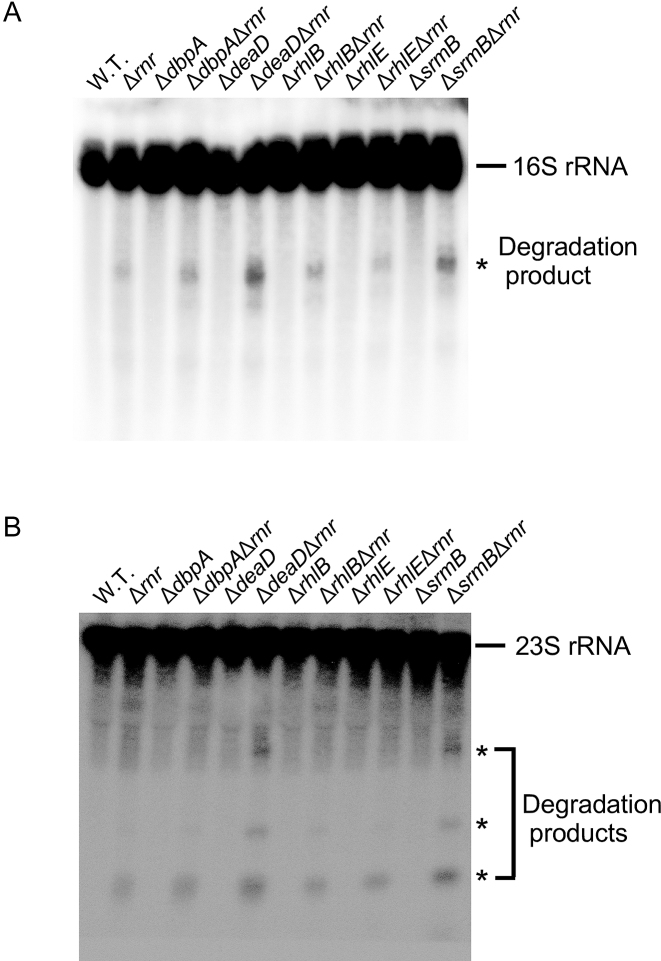
Among the substrates for RNase R inside the cell are degradation products that arise from rRNA breakdown. These RNAs are particularly important substrates, not only because rRNAs comprise a large fraction of total cellular RNA, but also because their high degree of secondary structure makes them resistant to digestion by other RNases. Thus, the growth defects observed for the ΔdeaDΔrnr and ΔsrmBΔrnr strains could be related to an accumulation of rRNA degradation products. To determine whether this was the case, total RNA was isolated from strains containing different combinations of WT, Δrnr and/or DBP deletion alleles and analyzed by northern blotting using probes for 16S or 23S rRNA.
When a probe corresponding to the 5′ region of 16S rRNA was used, primarily full-length 16S rRNA was visualized for each sample (Figure 2A). Additionally, low levels of degradation products could be seen in the WT strain, the single-deletion strains or the ΔdbpAΔrnr, ΔrhlBΔrnr or ΔrhlEΔrnr strains. However, for the samples derived from the ΔdeaDΔrnr and ΔsrmBΔrnr strains, an increased accumulation of an rRNA degradation product of ∼0.9 kb was observed. Thus, an accumulation of rRNA degradation products correlates with the growth defects of ΔdeaDΔrnr and ΔsrmBΔrnr strains (Figure 1). A similar result was also obtained using a probe for 23S rRNA. Apart from the full-length rRNA, degradation products of ∼0,35, 0.6 and 1.9 kB could be observed for each of the Δrnr strain derivatives, with increased levels of these products in the ΔdeaDΔrnr and ΔsrmBΔrnr strains (Figure 2B). These observations suggested that an absence of RNase R and either DBP leads to an enhanced accumulation of rRNA breakdown products inside the cell.
Figure 2.
Accumulation of rRNA fragments in ΔdeaDΔrnr and ΔsrmBΔrnr strains. RNA was isolated from WT or mutant strains grown in LB medium at 37°C, as indicated, and analyzed by northern blotting using probes to 16S rRNA (A) or 23S rRNA (B). The full length rRNAs and rRNA degradation products are indicated.
The rRNA fragments observed in ΔdeaDΔrnr and ΔsrmBΔrnr strains are similar to those observed in pnptsΔrnr strains
It has been previously shown that rRNA fragments can be visualized in strains that lack RNase R and contain mutations in PNPase (24). These fragments are generated by endonuclease cleavage and accumulate when both exonucleases are inactivated. In the previous studies, strains containing deletion alleles of pnp and rnr could not be used because these two genes are collectively essential. Therefore, a combination of a Δrnr and a pnpts allele was used to demonstrate an accumulation of rRNA degradation products under non-permissive conditions, i.e. a temperature-shift to 42°C.
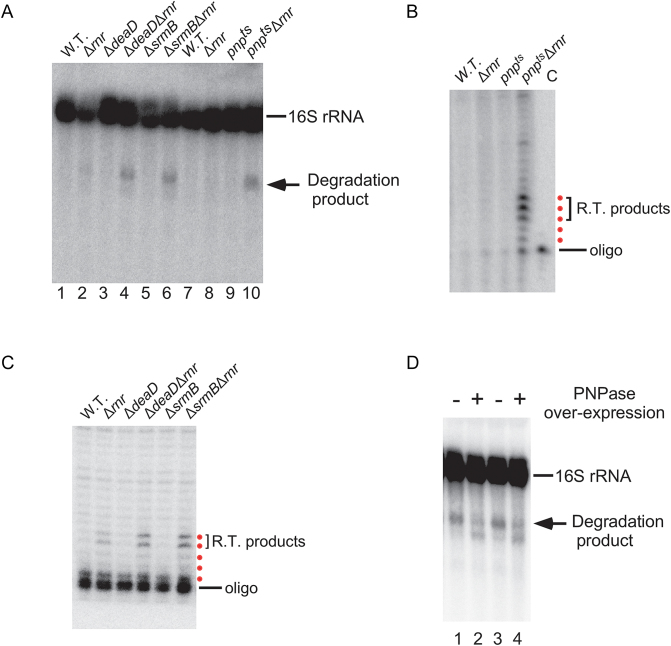
To determine whether the rRNA fragments observed in ΔdeaDΔrnr or ΔsrmBΔrnr strains could be related to those previously observed in the pnptsΔrnr strain, total RNA was extracted from a pnptsΔrnr strain after a shift to the non-permissive temperature, as well from strains containing the single mutations. These RNAs were analyzed by northern blotting alongside a subset of the RNAs analyzed in Figure 2. Low levels of 16S rRNA fragments were seen for strains containing either the individual pnpts or Δrnr mutations, but, as expected, increased levels of these products could be observed in the pnptsΔrnr strain (Figure 3A, lanes 7–10). Significantly, the size of the main fragment was similar to the one observed in the ΔdeaDΔrnr or ΔsrmBΔrnr strains (Figure 3A, lanes 1–6).
Figure 3.
The rRNA degradation products in the ΔdeaDΔrnr and srmBΔrnr strains are the same as in the pnptsΔrnr strain. (A) RNA was isolated from WT, pnpts, Δrnr and pnptsΔrnr strains after a temperature shift to 42°C for 1 h following growth at 30°C to early-log phase. The RNAs were analyzed by northern blotting using a probe for 16S rRNA (lanes 7–10). Total RNA from ΔdeaDΔrnr, ΔsrmBΔrnr or derivative strains grown at 37°C were also analyzed in parallel (lanes 1–6). (B and C) Primer extension analysis. (B) RNA was isolated from WT, pnpts, Δrnr and pnptsΔrnr strains after a temperature shift to 42°C and analyzed by primer extension using an oligonucleotide complementary to 16S rRNA. The major reverse-transcription (RT) products are denoted by a bracket. A control lane (C) containing only the oligonucleotide is also included. The products that correspond to the first five reverse-transcribed nucleotides are indicated by red dots. (C) Primer extension was performed on RNA isolated from ΔdeaDΔrnr, ΔsrmBΔrnr or derivative strains grown at 37°C, as described in (B). (D) Suppression of rRNA fragment accumulation by PNPase over-expression. A control plasmid (pBR322) or a PNPase-expression plasmid (pKAK7) were transformed into ΔdeaDΔrnr (lanes 1 and 2) or ΔsrmBΔrnr strains (lanes 3 and 4). Total RNA was isolated from the resulting strains and analyzed by northern blotting using a 16S rRNA probe. The positions of the full-length RNAs and of the main rRNA degradation product are indicated.
To further characterize the products, primer extension was performed on total RNA samples derived from each of these strains. It has been earlier proposed that the major 16S rRNA degradation product observed in the pnptsΔrnr strain is generated by endonucleolytic cleavage after nucleotide (nt) 919 (26). To confirm these findings, RNA was isolated from the pnptsΔrnr strain, as well from strains containing the individual mutations, and analyzed by primer extension using an oligonucleotide complementary to 16S rRNA nts 924–940. Significantly, an accumulation of products corresponding to nearby cleavages was observed with the pnptsΔrnr strain (Figure 3B). Interestingly, cleavages were observed not only after nt 919, but also at adjacent positions, particularly after nts 918 and 920. Next, primer extension was performed on RNA derived from ΔdeaDΔrnr or ΔsrmBΔrnr strains, or derivatives thereof. For the former set of RNAs, enhanced cleavages after positions 918 and 919 were observed (Figure 3C). Given the similar cleavage patterns observed for the pnptsΔrnr , ΔdeaDΔrnr and ΔsrmBΔrnr strains, these results strongly suggested that the rRNA fragments in the latter set of strains are generated by the same mechanism as in the pnptsΔrnr strain.
The similarities between the fragments observed in the pnptsΔrnr strain and in the ΔdeaDΔrnr/ΔsrmBΔrnr strains (Figure 3A) also suggested a possibility that an involvement of SrmB and DeaD in this process could be due to their ability to stimulate of PNPase activity. Thus, a lack of PNPase or either of these DBPs would be expected to yield a similar pattern of rRNA fragments. If that is the case, one prediction was that the inefficient degradation of rRNA fragments in the ΔdeaDΔrnr or ΔsrmBΔrnr strains might be overcome by PNPase over-expression. To test this possibility, the ΔdeaDΔrnr and ΔsrmBΔrnr strains were transformed with either a control or a multicopy PNPase plasmid, and total RNA isolated from these strains was analyzed by northern blotting. Notably, reduced levels of the main 16S rRNA degradation fragment were observed upon PNPase over-expression in each case (Figure 3D). Interestingly, a new, shorter product was observed, suggesting that it could be derived from the longer fragment, but that in the absence of DeaD or SrmB, increased PNPase activity only allows partial digestion of the main degradation product.
Two models for rRNA degradation in ΔsrmBΔrnr and ΔdeaDΔrnr strains
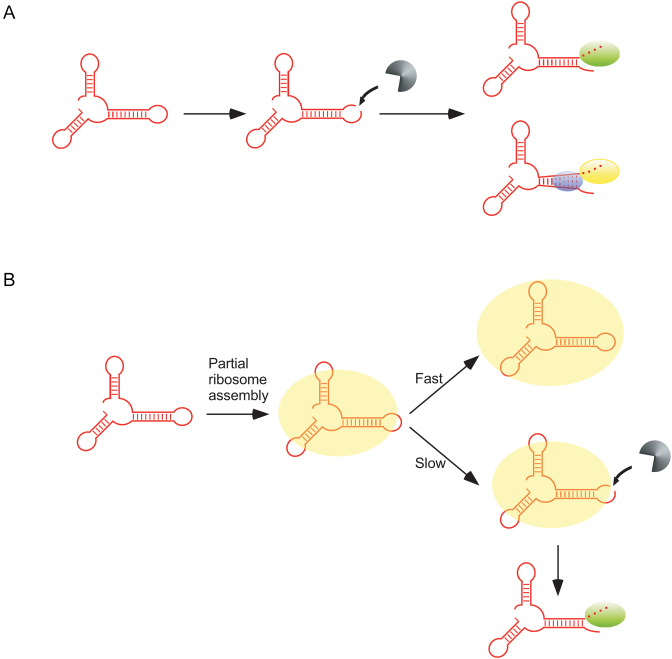
Based on the results obtained, an initial model for the accumulation of rRNA degradation products was developed (Figure 4A). In this model, rRNA degradation products arise through endonuclease cleavage and are removed by either RNase R or PNPase. SrmB and DeaD are proposed to facilitate PNPase function, possibly by acting as RNA helicases to resolve RNA duplexes and allow PNPase to digest these otherwise structured RNAs. Lacking either DBP and RNase R, rRNA fragment clearance by PNPase occurs with reduced efficiency, resulting in the observed accumulation of degradation products in the ΔdeaDΔrnr or ΔsrmBΔrnr strains (Figure 2).
Figure 4.
Two models for the accumulation of rRNA fragments in ΔdeaDΔrnr and ΔsrmBΔrnr strains. (A) A fraction of newly synthesized rRNAs (shown in red) is proposed to be cleaved by an endonuclease, depicted in gray. The resulting products are digested either by RNase R (green oval) or PNPase (yellow oval). PNPase-mediated digestion of structured rRNAs is facilitated via unwinding of base-pairing by SrmB or DeaD (blue ovals). (B) Newly synthesized rRNA is incorporated into a nascent ribosomal particle (show as a yellow oval). In the absence of delays, ribosomal subunit assembly rapidly proceeds to completion, resulting in the protection of the rRNAs from RNase action (top). When ribosome assembly is delayed, endonucleolytic cleavage occurs within exposed rRNA regions (bottom). The resulting fragments are primarily digested by RNase R (green oval) and therefore accumulate in its absence.
Although the model proposed in Figure 4A was consistent with the experimental results, some aspects of the model could not be easily explained. For example, in this model PNPase is proposed to work synergistically with SrmB and DeaD. However, since fragments were observed both in the ΔdeaDΔrnr or ΔsrmBΔrnr strains, it suggested that each factor is needed to remove the rRNA fragments. As it has been shown that DeaD and SrmB function independently as RNA helicases (27), why both factors should influence this process was not clear. Second, there is no evidence to suggest that either of these DBPs associate with PNPase to stimulate its RNase activity. Third, the one DBP known to bind to PNPase and to stimulate PNPase function is RhlB. Specifically, RhlB and PNPase form a stoichiometric complex in vitro and in vivo, and are also key constituents of the RNA degradosome (28,29). Moreover, efficient digestion of structured RNAs by PNPase in vitro has been shown to require RhlB (7). Surprisingly, the data presented in Figure 2 indicated no role for RhlB in rRNA degradation.
Based on these arguments, some doubts on the validity of the model proposed in Figure 4A were created and an alternative model based on the known role of DeaD and SrmB in ribosome assembly at low temperatures was developed (Figure 4B). In this model, it is proposed that as newly transcribed rRNAs are incorporated into ribosomal particles, the completion of ribosomal assembly confers protection from cellular RNases (30,31). However, when ribosome assembly is delayed, enhanced rRNA cleavages occur and the resulting products are primarily digested by RNase R. Consequently, when ribosome assembly is delayed and RNase R is absent, high levels of rRNA degradation fragments accumulate. Thus, in this model, the accumulation of rRNA fragments in strains that lack DeaD or SrmB is proposed to be linked to their effects on the kinetics of ribosome assembly rather than through an unwinding of structured rRNA fragments.
ΔsrmB and ΔdeaD mutants display a kinetic ribosome assembly defect
As mentioned, both DeaD and SrmB have been implicated in ribosome assembly at low temperatures and strains lacking either of these factors exhibit reduced growth at 22°C (8,9). However, neither strain was previously found to display a significant ribosomal defect at 37°C, the temperature at which the above described analyses were performed (23). Based on the available data, therefore, it was unclear whether the ΔdeaD and ΔsrmB strains undergo slower ribosome assembly at 37°C, and therefore, whether the accumulation of rRNA fragments in ΔdeaDΔrnr and ΔsrmBΔrnr strains (Figure 2) can be explained by the model described in Figure 4B.
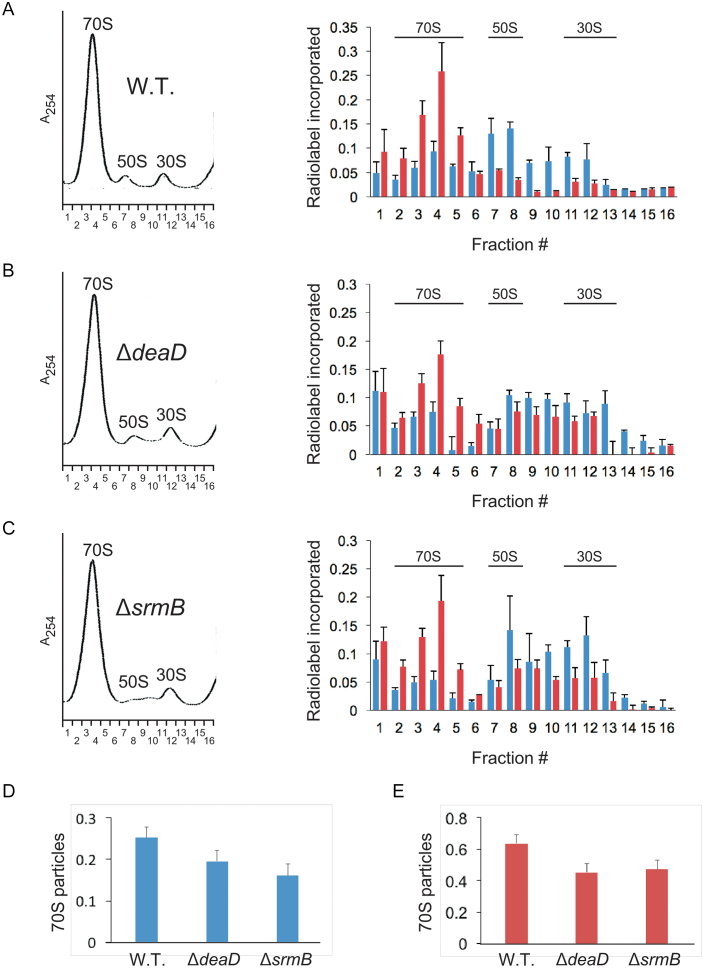
To determine whether the ΔdeaD and ΔsrmB mutations affect ribosome assembly, a pulse-labeling approach was used. Thus, WT, ΔdeaD or ΔsrmB strains were grown at 37°C and pulse-labeled with radiolabeled inorganic phosphate for 1 min. Thereafter, cell aliquots were collected 5 and 15 min after the labeling period. The cells were combined with an excess of unlabeled cells from the same strain grown at 37°C, lysed and ultracentrifuged on a sucrose-density gradient. The ultracentrifuged material was analyzed at 254 nM, and 16 fractions were collected in each case. These fractions were filtered through nitrocellulose membrane to determine the extent of radiolabeled rRNA incorporation into the different ribosomal particles (Figure 5A–C).
Figure 5.
Ribosome assembly kinetics. (A–C) Cultures of WT (A), ΔdeaD (B) or ΔsrmB (C) strains were pulse-labeled with 32P inorganic phosphate for 1 min. Aliquots of the cultures were removed thereafter 5 or 15 min post-labeling and combined with unlabeled cells. Lysates derived from these cells were individually layered on 14–32% sucrose density gradients and ultracentrifuged. (Left) Ribosome profiles were determined at 254 nM. The positions of the peaks corresponding to the 30S and 50S ribosomal subunits, and to 70S ribosomes, are indicated. Sixteen ribosomal fractions were collected in each case. A254, absorbance at 254 nM; (right) The ribosomal fractions were filtered through nitrocellulose membrane to capture labeled rRNAs present in ribosomal particles. The amount of radioactivity in each fraction was quantified by phosphorimaging of the dried membranes. Blue bars, relative amount of radiolabel in each fraction for cultures harvested 5 min after labeling; red bars, radiolabel incorporation for cultures harvested after 15 min of labeling; n = 3. The main fractions that correspond to the 70S, 50S and 30S particles are indicated. (D and E) The amount of radiolabel in 70S particles (fractions 2–5), relative to the total amount of radiolabel present in all fractions, was quantified for WT, ΔdeaD and ΔsrmB strains for cultures harvested 5 min (D) or 15 min (E) after labeling.
For the 5-min time point, a significant amount of radiolabel was found to be present in the fractions corresponding to the 30S and 50S ribosomal subunits as well in the 70S ribosome fraction in each case. However, the relative amount of label in fractions 2–5, which includes most of the 70S particles, was observed to be higher in the WT strain, suggesting reduced rates of 70S ribosome formation in the mutant strains (Figure 5D). By the 15-min time point, the amount of radiolabel in the 70S fractions was increased for each strain, indicating the progress of ribosomal assembly (Figure 5A–C). Again, however, radiolabel incorporation in the 70S fractions was found to be greater for the WT strain relative to either the ΔdeaD or ΔsrmB strain (Figure 5E). From these experiments, it can be concluded that both the ΔdeaD and ΔsrmB strains exhibit reduced rates of ribosome assembly, consistent with the model shown in Figure 4B, which invokes enhanced degradation of rRNA as a consequence of ribosome assembly delays. Furthermore, these data demonstrate that even though DeaD and SrmB have been thought to be important for ribosome assembly at low temperatures, their effects on this process at physiological growth temperatures is still significant.
rRNA degradation products are observed in strains lacking ribosome assembly factors
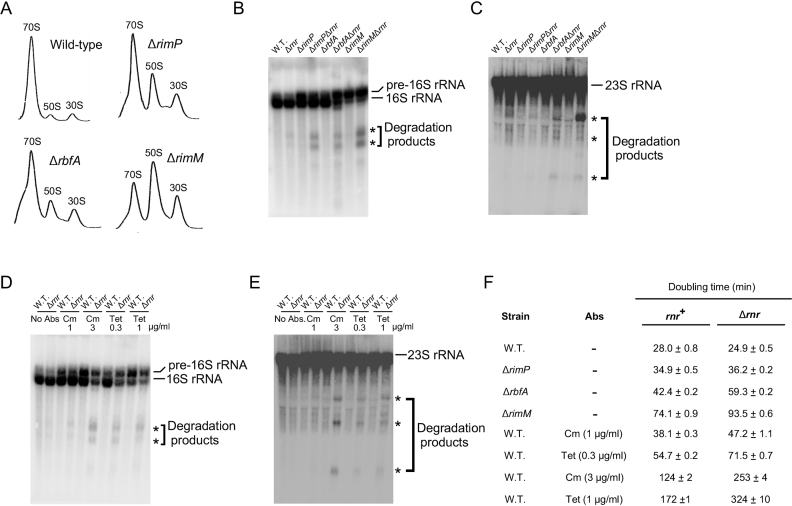
A corollary of the results obtained above is that defects in ribosome assembly that are independent of DeaD or SrmB should also result in rRNA fragmentation. To investigate this hypothesis, a series of strains containing mutations in established RAFs were analyzed. It is known that many RAFs are important for ribosome biogenesis, and the absence of such RAFs causes ribosome assembly defects (30,32). To test their effects on rRNA degradation, deletion alleles of the RimP, RbfA or the RimM RAFs were transferred to a WT or Δrnr background. Prior to using the strains for RNA analysis, ribosomal profiles were generated to verify the ribosome assembly defects expected for the mutant strains. As expected, elevated levels of subunit particles were observed for each strain, which presumably reflects the accumulation of incompletely formed subunits (Figure 6A). Next, total RNA was isolated and analyzed by northern blotting. Significantly, when a 16S-rRNA specific probe was used, increased levels of 16S rRNA degradation products could be observed with each of the strains containing RAF deletions in the Δrnr background (Figure 6B). Apart from the expected 0.9 kb fragment, the appearance of a second, smaller fragment, similar to one observed in Figure 3D, was seen. This product was analyzed by 3′ RACE and was found to contain a 3′-end that corresponds to 16S rRNA nt 764, which presumably reflects exonucleolytic digestion following cleavage ∼150 nts downstream. Additionally, in some cases, a band migrating more slowly than the mature 16S rRNA could be detected. This band likely corresponds to the previously described 17S precursor of the 16S rRNA that contains 115 unprocessed nucleotides at the 5′ end and 33 nts at the 3′ end, as it is known that mutations in RAFs can interfere with the processing of this RNA (33). The RNA samples were also analyzed by using a probe for 23S rRNA, which revealed multiple degradation fragments in the strains that contained RAF deletions in the Δrnr background (Figure 6C).
Figure 6.
Generation of rRNA fragments due to impaired ribosome assembly. (A) Ribosomal profiles of strains containing mutations in established RAFs. Cell lysates from WT, ΔrimP, ΔrbfA or ΔrimM strains were analyzed by ultracentrifugation, as described for Figure 5. (B and C) RNA was isolated from WT or Δrnr strains containing ΔrimP, ΔrbfA or ΔrimM mutations and analyzed by northern blotting as described in Figure 2 using probes for 16S rRNA (B) or 23S rRNA (C). (D and E) WT or Δrnr strains were grown in the absence or presence of the indicated amounts of antibiotics (Abs) and RNA derived from these strains was analyzed by northern blotting, as described in Figure 2 using probes for 16S rRNA (D) or 23S rRNA (E). (F) Doubling times were determined in LB medium at 37°C. Mean values and standard errors are based on triplicate cultures for each strain.
To determine whether there are any additional conditions that can cause rRNA degradation, WT or Δrnr strains were grown in the presence of sub-lethal concentrations of the antibiotics chloramphenicol (Cm) or tetracycline (Tet). Each of these antibiotics interferes with translation, and it is known that general defects in translation, by impeding ribosomal protein production, reduce the ability of cells to form functional ribosomes (34). As was observed with the RAF mutants, significant levels of 16S and 23S rRNA fragments were observed in the Δrnr strain background when cells were grown in the presence of antibiotics (Figure 6D and E). Thus, conditions that interfere with ribosome assembly indirectly also increased rRNA degradation. Overall, the results from Figure 6A–E indicate that rRNA fragments are generated when ribosome assembly is impaired, and these fragments are stabilized in a Δrnr background, supporting the notion that the rRNA fragments observed in ΔdeaDΔrnr and ΔsrmBΔrnr strains are also caused by ribosome assembly defects.
Lastly, the doubling times of the strains analyzed in Figure 6B–E were determined (Figure 6F). Each set of conditions associated with defective ribosomal assembly was found to display an increased doubling time in the Δrnr strain background relative to the rnr+ background (Figure 6F), analogous to the findings with the ΔdeaD and ΔsrmB mutations (Figure 1). Thus, with the exception of the ΔrimP mutant, for each strain or condition that reduced growth by a modest degree in the rnr+ background, (the ΔrbfA mutant, the ΔrimM mutant, Cm at 1 μg/ml or Tet at 0.3 μg/ml), doubling times in the Δrnr strain background were further increased by ∼30%. For conditions that affected growth more severely in the rnr+ background (Cm at 3 μg/ml or Tet at 1 μg/ml), doubling times increased further by ∼100% in the Δrnr background, suggesting that the effect of RNase R removal has a more profound effect on growth when ribosome assembly is severely impaired. These results indicate that there is a general deleterious effect of impaired ribosomal assembly on growth when RNase R is absent.
rRNA fragments are observed in a wild-type strain under steady-state growth conditions
Given the evidence for rRNA degradation under conditions of impaired ribosome assembly, a question of interest was whether this process also occurs through a similar mechanism under conditions of normal assembly. In this regard, significant levels of rRNA fragments have been previously observed in pnptsΔrnr strains (24). One caveat to those experiments, however, is that the fragments were observed under conditions that involved both a temperature shift to inactivate PNPase and a growth arrest after the temperature shift. It is known that the rRNAs in functional ribosomes start getting degraded when cells encounter non-growing conditions, which likely reflects a mechanism to recycle nucleotides under unfavorable growth conditions (35). These considerations suggested that the rRNA degradation products observed in the pnptsΔrnr strain could have been artificially induced by the conditions used to inactivate PNPase.
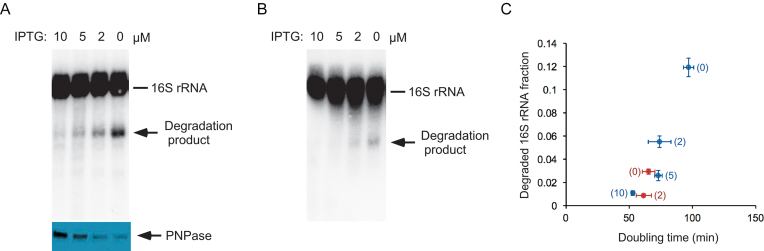
Nonetheless, a careful evaluation of RNA products in the Δrnr strain (Figures 2A, B, 3A and 6B; lane 2 each) indicated a modest but distinct accumulation of rRNA fragments. This suggested that a significant extent of rRNA breakdown could be happening even under conditions of normal growth, but the extent of degradation might be under-estimated by the residual activity of PNPase in these strains. To re-examine the issue of rRNA degradation under conditions of both balanced growth and normal ribosome assembly, first, a doubly mutated ΔpnpΔrnr strain containing an IPTG-regulated PNPase-expression plasmid was constructed. This strain was grown in LB medium containing different levels of IPTG, and the extent of 16S rRNA fragment accumulation was quantified in each case. Significantly, increased levels of rRNA fragment were observed when this strain was grown under reduced IPTG concentrations (Figure 7A). Specifically, when no IPTG was present, 12% of the 16S rRNA was found to be present as a degradation product. The IPTG-dependent synthesis of PNPase was confirmed by western-blot analysis using anti-PNPase antibodies. Similarly, when a ΔpnpΔrnr strain containing an IPTG-regulated RNase R-expression plasmid was used, degradation products were also observed (Figure 7B). In this case, lower levels of the 16S rRNA fragment were found to accumulate under reduced IPTG conditions, possibly because the basal expression or activity of RNase R is greater than that of PNPase.
Figure 7.
Visualization of rRNA fragments in the absence of ribosome assembly defects. A ΔpnpΔrnr strain containing an IPTG-regulated PNPase or RNase R-expression plasmid was grown at 37°C in LB media supplemented with different concentrations of IPTG, and aliquots of these cultures were used to prepare total RNA or total protein. (A) RNA and protein analysis of the IPTG-regulated PNPase expression strain. Top, RNAs were analyzed by northern blotting, as described in Figure 2A; bottom, total cellular protein was analyzed by western blotting using anti-PNPase antibodies. (B) RNA analysis of the IPTG-regulated RNase R expression strain. (C) Comparison of 16S rRNA fragment levels and growth rates. Doubling times for the strains analyzed in (A) and (B) were measured and plotted against the relative fraction of rRNA degradation products observed. Blue, IPTG-regulated PNPase expression strain; red, IPTG-regulated RNase R expression strain. Numbers in brackets for each data point correspond to the concentration of IPTG, in micromolar, that was present in the growth medium; n = 3.
Because PNPase and RNase R are collectively essential, it was possible that the increased accumulation of rRNA fragments under conditions of lowered PNPase or RNase R expression could be indirectly caused by reduced growth rates. This is significant because previous studies have shown that the extent of rRNA degradation is ∼10% when cells divide every 2 h or less, but degradation increases to 70% in cells that divide every 10 h (36). To determine whether the changes in the levels of the rRNA fragments observed could be attributed to growth rate differences, cell doubling times were determined and plotted against the extent of 16S rRNA fragment levels observed in Figure 7A and B (Figure 7C). For cells expressing inducible PNPase, a growth rate reduction was observed when IPTG concentration was reduced, suggesting that limiting PNPase does impair growth when RNase R is absent. However, the difference in doubling times over the range of IPTG concentrations used was <2-fold, whereas the extent of variation in fragment levels spanned more than 10-fold, strongly suggesting that the observed changes in rRNA fragment levels stem primarily from differences in PNPase expression rather than growth rate. Similarly, a 3-fold difference in rRNA fragment levels was observed in cells expressing inducible RNase R at 0 versus 2 μM IPTG concentration with negligible differences in growth rate. The levels of rRNA fragments in this strain could not be reliably quantified at higher IPTG concentrations due to their low levels. It can be concluded that even in the absence of ribosome assembly defects, a significant degree of rRNA fragmentation takes place inside the cell, which is revealed by limiting the expression of exonucleases.
In vitro digestion of ribosomal particles with RNase E
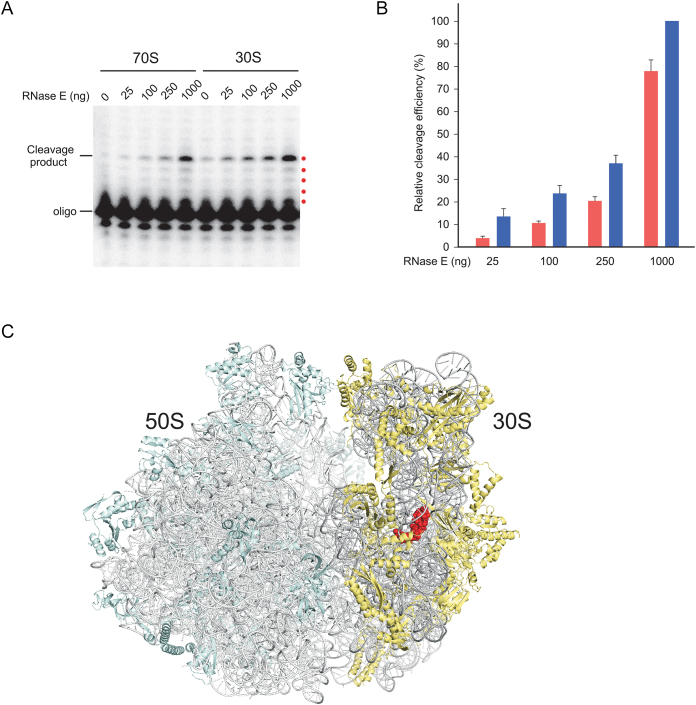
Based on the results presented in Figure 7, the next question was whether rRNA degradation can be recapitulated in vitro on ribosomal particles derived from a WT strain. As it has been earlier proposed that the cleavage of 16S rRNA in pnptsΔrnr strains is RNase E dependent (37), this enzyme was purified and its ability to cleave 16S rRNA in ribosomal particles was tested in vitro. For these purposes, cell extracts derived from a WT strain were ultracentrifuged and 70S ribosome as well as 30S ribosomal subunit fractions were collected. In these preparations, the 30S fraction is expected to contain a mixture of mature small ribosomal subunits as well as assembly intermediates en route to assembly completion. Each fraction was concentrated and digested with purified RNase E. After digestion, the rRNA was extracted and RNase E-dependent cleavages were assessed by primer extension, as in Figure 3B and C.
Treatment of each fraction with increasing amounts of RNase E resulting in a dose-dependent cleavage of 16S rRNA between nts 918 and 919 (Figure 8A), similar to those seen in the ΔdeaDΔrnr, ΔsrmBΔrnr and pnptsΔrnr strains (Figure 3B and C) and confirming that RNase E can cleave 16S rRNA within the context of ribosomal particles at this position. A quantitation of cleavage efficiency indicated that when low levels of RNase E were present, 16S rRNA in 30S particles was cleaved three to four times more efficiently than in the 70S particles, although the differences were reduced when RNase E dosage was increased (Figure 8B). These results suggest that the 30S particles are more susceptible to RNase E, especially at low concentrations, consistent with the notion that ribosomal particles become resistant to endonucleolytic attack as they mature to form complete ribosomes, as proposed in Figure 4.
Figure 8.
Digestion of ribosomal particles by RNase E. (A) 30S or 70S ribosomal fractions were concentrated and treated with different amounts of purified RNase E for 1 h at 37°C. RNA was extracted from these reactions and analyzed by primer extension, as described in the Figure 3B legend. The products that correspond to the first five reverse-transcribed nucleotides are indicated by red dots. (B) Quantitation of RNase E cleavage efficiency. The amount of cleavage products in (A) was quantified by phosphorimaging, reduced by any signal observed without RNase E addition, and normalized with respect to the amount of product generated by using 1 μg of RNase E on 30S particles. Red bars, 70S particles; blue bars, 30S particles. Mean values and standard errors are based on four independent preparations of 30S and 70S particles. (C) Visualization of a RNase E cleavage site in the high-resolution Escherichia coli 70S ribosome structure (PDB ID: 4YBB, molecule A) shown via ribbon representation. The RNA and proteins components of the ribosomal large subunit are colored light gray and light cyan, respectively, whereas the corresponding residues on the small subunit are colored dark gray and light yellow. The residues abutting the RNase E cleavage site (16S rRNA nts 916–921) are shown as red spheres. The view shown is from the tRNA E site.
To determine the structural basis for its relative resistance to endonucleolytic cleavage, the residues abutting the 16S rRNA cleavage site were mapped to the 70S ribosome (Figure 8C). Significantly, this region was found to be buried within the interior of the 30S small subunit. Hence its resistance to endonucleolytic cleavage in the mature ribosome can be attributed to an inaccessibility of RNase E to the cleavage site. Because immature 30S particles were found to be more sensitive to cleavage compared to 70S ribosomes, it may be inferred that this region is more accessible in the former form, but additional support for this hypothesis will require further investigation.
DISCUSSION
DBPs represent an important class of remodeling factors that are involved in many aspects of RNA function, but for most of the members of this family their physiological functions are not well understood. To investigate whether DBPs from E. coli could have a yet unknown role in RNA turnover, deletion mutations in the genes encoding individual E. coli DBPs were combined with a panel of mutations in different RNases. Two strains, containing either the ΔdeaDΔrnr or the ΔsrmBΔrnr mutations, were found to exhibit reduced growth on LB-agar plates, which was not observed for the individual deletion strains, suggesting a functional relationship between RNase R with DeaD and SrmB (Figure 1). Given the previously ascribed role of RNase R in rRNA metabolism, an analysis of rRNA was performed to elucidate the molecular basis of the growth defects. Both the ΔdeaDΔrnr and ΔsrmBΔrnr strains were found to exhibit enhanced levels of 16S rRNA and 23S rRNA fragments (Figure 2). Primer extension analysis of the major 16S rRNA degradation product in the ΔdeaDΔrnr and ΔsrmBΔrnr strains revealed that they are cleaved at essentially the same sites as those from a pnptsΔrnr strain (Figure 3).
These observations initially suggested a mechanism in which the RNA fragments are generated by endonuclease cleavage, and the digestion of these fragments is facilitated by DeaD and SrmB, possibly by unwinding RNA duplexes to promote PNPase digestion (Figure 4A). However, an alternative mechanism was suggested by additional observations. First, it was unclear why a loss of either DeaD or SrmB should influence PNPase function, considering that these two DBPs are thought to act independently. Second, neither factor has been found to associate with PNPase directly. Third, an absence of RhlB, the DBP that does so, showed no effect on rRNA fragment accumulation. Finally, strains lacking either DeaD or SrmB were found to display reduced kinetics of 70S ribosome formation at 37°C (Figure 5), suggesting a possibility that the rRNA fragments observed could instead have been caused by delays in ribosomal assembly. Based on these considerations, a revised model for the generation of the rRNA degradation products was developed (Figure 4B). By this model, nascent rRNA is proposed to be susceptible to ribonuclease cleavage during the ribosome assembly process. The cleaved rRNAs, incapable of undergoing ribosome assembly completion, are then removed through a salvage pathway mediated by RNase R and PNPase. Any delays in ribosome assembly completion, as occurs in ΔdeaD or ΔsrmB strains, are expected to increase the extent of rRNA cleavage. Thus, the underlying basis of rRNA fragmentation is the sensitivity of rRNA to endonucleolytic cleavage prior to incorporation into ribosomes. Further support for this model was obtained from the observation that strains containing mutations in known RAFs also exhibited enhanced rRNA degradation (Figure 6).
Apart from conditions of delayed ribosome assembly, 16S rRNA fragments could also be visualized in the absence of assembly defects when exonucleases were depleted (Figure 7). High levels of rRNA degradation fragments have been observed before in pnptsΔrnr strains (24), but those studies left open the possibility that they could have arisen due to the treatment applied to inactivate PNPase, a condition that leads to growth cessation. Similarly, rRNA fragment accumulation has also been observed in strains that lack PNPase and contain mutations in RNase R that inactivate its helicase activity, but these strains exhibit a four-fold increase in cell doubling time (38). In contrast, another study found undetectable levels of rRNA degradation over several hours of cell growth (31). However, rRNA degradation during the early stages of ribosome assembly was not measured in that study. These differences can be reconciled because, by the model presented in Figure 4B, the observed degradation occurs relatively soon after rRNA synthesis. Once ribosome assembly has been completed, the rRNA becomes highly stable, and remains so unless cells experience a stress situation. This hypothesis was supported by biochemical assays which showed that the 16S rRNA present in 30S particles, which contain a mixture of assembly intermediates and completed small subunits, is more susceptible to cleavage by RNase E than 70S ribosomes (Figure 8). However, given that cleavage was also observed in the latter, the in vitro experiments do not accurately mimic the in vivo situation where an extremely high degree of long-term resistance to rRNA degradation has been observed (31). An examination of the 70S ribosome structure revealed that the residues near the primary 16S rRNA cleavage site are not readily accessible, which provides a rationalization for why degradation occurs during the assembly phase but not in the fully assembled ribosome (39). Extrapolating these observations, it is likely that rRNA degradation during ribosome assembly, and a relative protection upon the completion of ribosome assembly, occurs in other organisms as well. Future studies using appropriate RNase-deficient strains and assembly-defective mutants will be useful to test this hypothesis.
Some aspects of this work may be related to the rRNA degradation that is observed when cells undergo starvation, a stress situation where degradation of pre-existing ribosomes can provide raw materials for further RNA synthesis. For starvation-induced rRNA degradation, it has been proposed that the trigger is the separation of 30S and 50S subunits, allowing RNases to access sensitive sites on rRNAs (40). The RNase-sensitivity of isolated subunits might explain why even though SrmB and DeaD are implicated only in 50S subunit assembly, fragments derived from 16S rRNA were also observed in the ΔdeaDΔrnr and ΔsrmBΔrnr strains (Figure 2A). The degradation of 16S rRNA, in this event, could be caused by a deficit of mature 50S particles, that results in an excess of unassociated, and thereby, endonuclease-sensitive 30S particles. Similarly, 23S rRNA degradation fragments were observed in the ΔrimPΔrnr, ΔrbfAΔrnr and ΔrimMΔrnr strain backgrounds (Figure 6C), even though each of the absent RAFs is only implicated in 30S subunit formation (30). In this case, a deficit of mature 30S particles would lead to an excess of unassociated 50S particles. An alternative hypothesis for the mutual sensitivity of both subunits, even when assembly of only one is impaired, is that final maturation only occurs when both subunits join together to initiate translation (41), and until that happens, both subunits remains endonuclease-sensitive.
The finding presented here are also relevant to studies on the ribosome assembly process itself. As an example, a question that often arises is whether the loss of a specific assembly factor will allow ribosome formation to take place, albeit at a slower pace, or whether it will lead to the formation of off-pathway products that are eventually removed through a salvage pathway (42). Based on the results presented here, even in those instances when ribosomes formation eventually occurs, slowing down the assembly process is expected to result in enhanced rRNA degradation, with a corresponding impact on ribosome yield. Ribosome assembly and rRNA degradation may therefore be considered to represent stochastic processes whereby rRNAs are either packaged into a relatively RNase-resistant form via the assembly process or else they undergo degradation, with the balance increasingly shifted toward the latter process as delays in assembly are experienced. A corollary of this model is that assays to assess rRNA degradation could be useful to identify new factors involved in ribosome assembly.
Growing cells invest a significant fraction of their material and energetic resources for ribosome synthesis. In particular, up to 20% of the cellular proteins synthesized are ribosomal proteins and as much as 70% of cellular transcription is specifically used for rRNA synthesis (43). It is also known that the ability to synthesize ribosomes is a limiting factor in determining how fast cells can grow. The economics of ribosome synthesis would suggest that cells should have evolved mechanisms to minimize rRNA degradation. It is, therefore, surprising that rRNA degradation occurs even in growing cells. However, this situation may be unavoidable because all organisms contain a milieu of RNases that perform key cellular functions and the completion of ribosome assembly takes several minutes to complete even in simple organisms, at which time the rRNAs are susceptible to RNases. Thus, it is possible that the balance between the expression of RNases and rRNA degradation has been optimized through evolution, but the degradation of a fraction of the transcribed rRNAs is unavoidable. A second, more intriguing possibility, is that rRNA cleavage reflects a mechanism to enable the rapid clearance of rRNAs, which due to their high degree of complexity, are prone to entering a non-productive folding or assembly pathway. In this scenario, endonuclease-sensitive rRNAs sites have been evolutionarily conserved so that faulty rRNAs can be removed efficiently. Indeed, a lack of rRNA fragments in any of the rnr+ strains, despite severe assembly defects (Figure 6), suggests that the clearance of such fragments is highly efficient. The two possible explanations are not mutually exclusive, and further work on this topic will be needed to determine if either reason, or any other, is responsible for the apparently costly degradation of transcribed rRNAs during steady-state growth.
Finally, is worth reiterating that the ΔdeaD and ΔsrmB mutant strains grew more slowly in the context of a Δrnr strain background compared to the rnr+ background (Figure 1). Similarly, mutations in RAFs or the addition of antibiotics conferred a greater increase in doubling time in the Δrnr background than in the rnr+ background (Figure 6F). As higher levels of fragments were observed only in the Δrnr strain background (Figures 2 and 6), the likely explanation for the growth defects observed is that an accumulation of these fragments is toxic to the cell. Exactly why rRNA fragment accumulation should reduce growth, however, remains unclear. One possibility that has been previously suggested is that these fragments compete with the pool of available ribosomal proteins, thereby interfering with ribosome assembly itself (24). Similarly, the presence of these fragments could also result in a titration of cellular RNases, with growth defects arising as a consequence of reduced availability of the RNases for their physiological substrates. Alternatively, an accumulation of rRNA degradation products might reduce the extent of nucleotides recycling, thereby impeding new transcription. Researching the basis for impaired growth due to rRNA fragment accumulation will be an interesting topic for future investigations.
ACKNOWLEDGEMENTS
I thank Dr Isabelle Iost for providing critical comments on the manuscript and Dr Arun Malhotra for assistance with rendering the ribosome structure.
FUNDING
National Institutes of Health (NIH) [GM114540]. Funding for open access charge: NIH [RO1 GM114540].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M.. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989; 17:4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cordin O., Banroques J., Tanner N.K., Linder P.. The DEAD-box protein family of RNA helicases. Gene. 2006; 367:17–37. [DOI] [PubMed] [Google Scholar]
- 3. Linder P., Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 4. Mallam A.L., Sidote D.J., Lambowitz A.M.. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife. 2014; 3:e04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsu C.A., Kossen K., Uhlenbeck O.C.. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001; 7:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diges C.M., Uhlenbeck O.C.. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001; 20:5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liou G.G., Chang H.Y., Lin C.S., Lin-Chao S.. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J. Biol. Chem. 2002; 277:41157–41162. [DOI] [PubMed] [Google Scholar]
- 8. Charollais J., Dreyfus M., Iost I.. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004; 32:2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charollais J., Pflieger D., Vinh J., Dreyfus M., Iost I.. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003; 48:1253–1265. [DOI] [PubMed] [Google Scholar]
- 10. Resch A., Vecerek B., Palavra K., Blasi U.. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol. 2010; 7:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vakulskas C.A., Pannuri A., Cortes-Selva D., Zere T.R., Ahmer B.M., Babitzke P., Romeo T.. Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol. Microbiol. 2014; 92:945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008; 14:381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao X., Jain C.. DEAD-box proteins from Escherichia coli exhibit multiple ATP-independent activities. J. Bacteriol. 2011; 193:2236–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gutgsell N.S., Jain C.. Gateway role for rRNA precursors in ribosome assembly. J. Bacteriol. 2012; 194:6875–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:doi:10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yancey S.D., Kushner S.R.. Isolation and characterization of a new temperature-sensitive polynucleotide phosphorylase mutation in Escherichia coli K-12. Biochimie. 1990; 72:835–843. [DOI] [PubMed] [Google Scholar]
- 17. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H.. Complete set of ORF clones of Escherichia coli ASKA library (A complete set of E. coli K-12 ORF archive): Unique Resources for Biological Research. DNA Res. 2005; 12:291–299. [DOI] [PubMed] [Google Scholar]
- 18. Diwa A., Bricker A.L., Jain C., Belasco J.G.. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 2000; 14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- 19. Jain C. Overexpression and purification of tagged Escherichia coli proteins using a chromosomal knock-in strategy. Protein Expr. Purif. 2006; 46:294–298. [DOI] [PubMed] [Google Scholar]
- 20. Hui M.P., Foley P.L., Belasco J.G.. Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 2014; 48:537–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohanty B.K., Kushner S.R.. Regulation of mRNA Decay in Bacteria. Annu. Rev. Microbiol. 2016; 70:25–44. [DOI] [PubMed] [Google Scholar]
- 22. Khemici V., Poljak L., Toesca I., Carpousis A.J.. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:6913–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jagessar K.L., Jain C.. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA. 2010; 16:1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng Z.F., Deutscher M.P.. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hossain S.T., Malhotra A., Deutscher M.P.. How RNase R degrades structured RNA: Role of the helicase activity and the S1 Domain. J.Biol. Chem. 2016; 291:7877–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basturea G.N., Zundel M.A., Deutscher M.P.. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011; 17:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bizebard T., Ferlenghi I., Iost I., Dreyfus M.. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004; 43:7857–7866. [DOI] [PubMed] [Google Scholar]
- 28. Lin P.H., Lin-Chao S.. RhlB helicase rather than enolase is the beta-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonucleolytic complex. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16590–16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Py B., Higgins C.F., Krisch H.M., Carpousis A.J.. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996; 381:169–172. [DOI] [PubMed] [Google Scholar]
- 30. Shajani Z., Sykes M.T., Williamson J.R.. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011; 80:501–526. [DOI] [PubMed] [Google Scholar]
- 31. Piir K., Paier A., Liiv A., Tenson T., Maivali U.. Ribosome degradation in growing bacteria. EMBO Rep. 2011; 12:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilson D.N., Nierhaus K.H.. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007; 42:187–219. [DOI] [PubMed] [Google Scholar]
- 33. Bylund G.O., Wipemo L.C., Lundberg L.A., Wikstrom P.M.. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 1998; 180:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maguire B.A. Inhibition of bacterial ribosome assembly: a suitable drug target. Microbiol. Mol. Biol. Rev. 2009; 73:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deutscher M.P. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol, Transl. Sci. 2009; 85:369–391. [DOI] [PubMed] [Google Scholar]
- 36. Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J. Mol. Biol. 1977; 115:335–354. [DOI] [PubMed] [Google Scholar]
- 37. Sulthana S., Basturea G.N., Deutscher M.P.. Elucidation of pathways of ribosomal RNA degradation: an essential role for RNase E. RNA. 2016; 22:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hossain S.T., Deutscher M.P.. Helicase activity plays a crucial role for RNase R function in vivo and for RNA metabolism. J. Biol. Chem. 2016; 291:9438–9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zundel M.A., Basturea G.N., Deutscher M.P.. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009; 15:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shetty S., Varshney U.. An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6126–E6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thurlow B., Davis J.H., Leong V., T F.M., Williamson J.R., Ortega J.. Binding properties of YjeQ (RsgA), RbfA, RimM and Era to assembly intermediates of the 30S subunit. Nucleic Acids Res. 2016; 44:9918–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ehrenberg M., Bremer H., Dennis P.P.. Medium-dependent control of the bacterial growth rate. Biochimie. 2013; 95:643–658. [DOI] [PubMed] [Google Scholar]