Abstract
Transcriptional regulation enables cells to respond to environmental changes. Of the estimated 304 candidate transcription factors (TFs) in Escherichia coli K-12 MG1655, 185 have been experimentally identified, but ChIP methods have been used to fully characterize only a few dozen. Identifying these remaining TFs is key to improving our knowledge of the E. coli transcriptional regulatory network (TRN). Here, we developed an integrated workflow for the computational prediction and comprehensive experimental validation of TFs using a suite of genome-wide experiments. We applied this workflow to (i) identify 16 candidate TFs from over a hundred uncharacterized genes; (ii) capture a total of 255 DNA binding peaks for ten candidate TFs resulting in six high-confidence binding motifs; (iii) reconstruct the regulons of these ten TFs by determining gene expression changes upon deletion of each TF and (iv) identify the regulatory roles of three TFs (YiaJ, YdcI, and YeiE) as regulators of l-ascorbate utilization, proton transfer and acetate metabolism, and iron homeostasis under iron-limited conditions, respectively. Together, these results demonstrate how this workflow can be used to discover, characterize, and elucidate regulatory functions of uncharacterized TFs in parallel.
INTRODUCTION
Transcription factors (TFs) modulate gene expression in response to environmental perturbations by interacting with a combination of sigma factors, RNA polymerase (RNAP), activating metabolites, and inorganic compounds. These signals collectively lead TFs to bind to specific DNA sequences referred to as binding sequence motifs (1). Microorganisms, therefore, can quickly adapt to diverse and extreme environmental conditions. In transcriptional regulation, genes are indirectly or directly regulated by one or more TFs. A set of genes directly controlled by the same TF are considered to belong to a regulon (2), with the complete set of regulons forming the transcriptional regulatory network (TRN).
Databases such as EcoCyc (3,4), RegulonDB (5), and TEC (6) maintain large amounts of information about TFs. However, a complete TRN for individual organisms still does not exist due to challenges outlined below.
Identifying all TFs
The genome-scale annotation of genes is required for the identification of the complete set of TFs. The emergence of high-throughput DNA sequencing has created a large number of candidate protein-encoding DNA sequences, leading to an increased demand for the discovery and annotation of protein functions. However, assigning a physiological function to the sequenced but uncharacterized genes is still a substantial challenge (7,8). For example, although Escherichia coli K-12 MG1655 has one of the most widely-studied genomes, a functional annotation is still missing for approximately 30% of its genes (9). This lack of annotation includes an estimated 50–80 uncharacterized TFs in E. coli K-12 MG1655 (6). The percentage of uncharacterized genes in other strains is even higher. Thus, a new workflow is needed to predict and validate a complete set of TFs in prokaryotes.
Characterizing transcription factor binding sites (TFBS)
Genome-wide characterization of TFBS is essential for the reconstruction of a global TRN. Despite a significant amount of knowledge about microbial TFs in databases and the literature, the binding activities of many TFs remain to be discovered. Traditionally, TFBS are identified through approaches such as DNase I footprinting and electromobility shift assays, which are limited to the interactions between TFs and single targets (10). With advances in genome-wide research technologies, many TFs have been experimentally investigated using the systematic evolution of ligands by exponential enrichment (SELEX) and chromatin immunoprecipitation with microarray (ChIP-chip) or by sequencing (ChIP-seq) (6,11–16). Recently, the ChIP-seq protocol has been combined with an exonuclease treatment (ChIP-exo) to reflect in vivo regulatory interactions between TFs and target genes at a single-base-pair resolution (17). Moreover, ChIP-exo can be easily applied to investigate differential binding patterns of the same TF under different environmental conditions (18–20). Thus, ChIP-exo provides us with a robust approach to characterize TFBS at the genome-scale.
Reconstructing TRNs
Several computational approaches have been developed for the reconstruction of the TRN, including the use of gene expression data (21,22), regulon-based associations (23), and integrated analysis with metabolic models (24). The expression data-driven approach for TRN reconstruction was widely used to predict transcription factor activities in E. coli K-12 MG1655. Recently, we have supplemented ChIP-exo with transcription profiling to describe the regulons of major TFs, including Cra, ArgR, Fur, OxyR, SoxRS, OmpR, and GadEWX (20,25–29). Therefore, this well-described approach is successfully applied to TRN reconstruction.
Here, we address these three challenges through the development of an integrated computational and experimental workflow to discover uncharacterized TFs in prokaryotes. Using E. coli K-12 MG1655 as an example, we combined a previously published computational approach with biological knowledge to identify candidate TFs for experimental validation. Given the resulting list of candidate TFs, we then examined their DNA-binding domains, predicted their active conditions, and performed an in vivo experimental validation of predicted DNA-binding capabilities. This workflow resulted in the elucidation of the biological functions of three uncharacterized TFs (YiaJ, YdcI, and YeiE) through an in-depth analysis of mutant phenotypes. Together, these results demonstrate the utility of our systematic identification workflow and provide a roadmap for its use in other organisms.
MATERIALS AND METHODS
Identification of candidate TFs
This workflow combined the previously published machine-learning algorithm, TFpredict, with biological knowledge to identify candidate TFs for experimental validation. TFpredict was originally trained to predict whether a eukaryotic protein was a TF (30). In brief, this algorithm takes a protein sequence as input and outputs a quantified score in the range [0,1] that represents the likelihood of the protein being a TF, based on sequence homology; zero is unlikely, one is very likely. In this study, TFpredict was applied to predict candidate TFs in E. coli K-12 MG1655. To assess whether the algorithm translates well from the eukaryotic realm to bacteria, the data from the proteobacteria were compiled for training. The details about the training data are described below.
The proteobacteria protein sequences in UniProt were filtered to meet the following criteria (31): (i) the protein sequences have functional annotation for DNA-binding; (ii) the proteins were reviewed as non-hypothetical; (iii) the proteins were annotated by Gene Ontology (GO) term as being related to the regulation of transcription or nucleic acid binding transcription factor activity (32) and (iv) the proteins were filtered out to exclude any protein sequences that were annotated with non-TF keywords: kinase, ubiquitin, actin, antigen, biotin, histone, chaperone, tubulin, transmembrane protein, endonuclease, exonuclease, translation initiation factor (Supplementary Figure S1). TFpredict was trained to rank order the candidate TFs. To evaluate whether the algorithm generalizes from eukaryotes to prokaryotes, model accuracy was rigorously assessed by cross-validation (Supplementary Figures S2 and S3). The results from the proteobacteria training set were comparable to those obtained from the eukaryotic training set, with a slight decrease in the area under the curve (AUC) due to the much smaller size of training data used. The large eukaryotic training sets result in similar prediction performance for all validated machine learning approaches. Hence, it is sufficient to choose the output of either approach. For the proteobacteria training set, performance varied. Instead of choosing one of approaches, we used a consensus of the output from all of the available approaches to make the final prediction.
Additionally, the memory and run-time efficiency for TFpredict were improved, and all code and data have been made freely available on GitHub (https://github.com/draeger-lab/TFpredict/tree/prokaryote). The example section in the README summarizes the settings we used to execute TFpredict. Documentation and installation instructions are provided on the GitHub page.
The uncharacterized protein sequences of E. coli K-12 MG1655 were the input data for TFpredict. The output from TFpredict was a rank-ordered list of proteins with confidence scores; some sequences could not be assigned a confidence score due to a lack of homologs. The primary selection was made based on the confidence scores from TFpredict. Next, further classification of the primary selection was made based on the predicted interactions between candidates and DNA sequences. This process not only removed some false positives but also provided group-specific strategies to predict the experimental conditions for ChIP-exo. Finally, an initial subset of 16 candidates was selected for experimental validation.
Bacterial strains, media, and growth conditions
All strains used in this study are E. coli K-12 MG1655 and its derivatives, deletion strains and myc-tagged strains (Dataset S2). For ChIP-exo experiments, the E. coli strains harboring 8-myc were generated by a λ red-mediated site-specific recombination system targeting the C-terminal region as described previously (33). For expression profiling by RNA-seq, deletion strains ΔydcI, ΔyeiE, ΔyafC, ΔyiaJ, ΔyheO, ΔybaO, ΔybaQ, ΔybiH, ΔyddM and ΔyieP were also constructed by a λ red-mediated site-specific recombination system (34). For ChIP-exo experiments, glycerol stocks of E. coli strains were inoculated into M9 minimal media (47.8 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4 and 0.1 mM CaCl2) with 0.2% (w/v) glucose. M9 minimal media was also supplemented with 1 mL trace element solution (100×) containing 1 g EDTA, 29 mg ZnSO4.7H2O, 198 mg MnCl2.4H2O, 254 mg CoCl2.6H2O, 13.4 mg CuCl2 and 147 mg CaCl2. The culture was incubated at 37°C overnight with agitation and then was used to inoculate the fresh media (1/200 dilution). The volume of the fresh media was 150 ml for each biological replicate. The fresh culture was incubated at 37°C with agitation to the mid-log phase (OD600 ≈ 0.5). For RNA-seq expression profiling, glycerol stocks of E. coli strains were inoculated into M9 minimal media with the same carbon sources as used in the ChIP-exo experiment for each candidate TF. The concentration of carbon sources was 0.2% (w/v). M9 minimal media was also supplemented with 1 ml trace element solution (100×). The culture was incubated at 37°C overnight with agitation and then was used to inoculate the fresh media. The fresh culture was incubated at 37°C with agitation to the mid-log phase (OD600 ≈ 0.5).
Measurement of bacterial growth
The effects of iron-limited conditions on cell growth were examined by growing E. coli K-12 MG1655 and yeiE deletion strain under four media treatments: (i) M9 minimal glucose medium; (ii) M9 minimal glucose medium containing 0.2 mM of the iron chelating agent 2,2′-dipyridyl (DPD) (Fluka); (iii) M9 minimal glucose medium containing 0.3 mM of DPD; (iv) M9 minimal glucose medium containing 0.4 mM DPD. Cells grown overnight on M9 minimal glucose medium at 37°C with agitation were inoculated into these four kinds of fresh media. Aliquots of overnight cell culture were diluted 1:200 into four types of fresh media, then were incubated at 37°C with agitation.
Similarly, to measure growth rate on low pH or acetate medium, the culture was incubated at low pH or acetate medium at 37°C overnight with agitation and then was used to inoculate the fresh media (1/200 dilution). The volume of the fresh media was 150 ml. The fresh culture was incubated at 37°C with agitation.
To measure growth on l-ascorbate, cells were grown anaerobically in medium containing l-ascorbate as described by the literature (35). Briefly, E. coli strains were grown overnight on M9 minimal glucose medium, and the cells were suspended in M9 salts medium. The aliquots were adjusted to 1.0, and 100 μL aliquots were inoculated into 10 ml culture tubes (Fisher Scientific) that were filled to the top with M9 minimal medium with a concentration of 20 mM l-ascorbate. Then the culture tubes were capped, sealed with parafilm, and then incubated at 37°C. All of growth curves were measured by six independent experiments at least and recorded by OD600 using Thermo BIOMATE 3S UV-visible spectrophotometer. The growth rate was calculated with GrowthRates 2.0 (36). The significant difference between wild type and deletion strain was determined by the Student's t test, P < 0.01.
ChIP-exo experiment
ChIP-exo experimentation was performed following the procedures previously described (37). In brief, to identify each candidate TF binding maps in vivo, the DNA bound to each candidate TF from formaldehyde cross-linked E. coli cells were isolated by chromatin immunoprecipitation (ChIP) with the specific antibodies that specifically recognize myc tag (9E10, Santa Cruz Biotechnology), and Dynabeads Pan Mouse IgG magnetic beads (Invitrogen) followed by stringent washings as described previously (38). ChIP materials (chromatin-beads) were used to perform on-bead enzymatic reactions of the ChIP-exo method (17). Briefly, the sheared DNA of chromatin-beads was repaired by the NEBNext End Repair Module (New England Biolabs) followed by the addition of a single dA overhang and ligation of the first adaptor (5′-phosphorylated) using dA-Tailing Module (New England Biolabs) and NEBNext Quick Ligation Module (New England Biolabs), respectively. Nick repair was performed by using PreCR Repair Mix (New England Biolabs). Lambda exonuclease- and RecJf exonuclease-treated chromatin was eluted from the beads and overnight incubation at 65°C reversed the protein-DNA cross-link. RNAs- and Proteins-removed DNA samples were used to perform primer extension and second adaptor ligation with following modifications. The DNA samples incubated for primer extension as described previously were treated with dA-Tailing Module (New England Biolabs) and NEBNext Quick Ligation Module (New England Biolabs) for second adaptor ligation. The DNA sample purified by GeneRead Size Selection Kit (Qiagen) was enriched by polymerase chain reaction (PCR) using Phusion High-Fidelity DNA Polymerase (New England Biolabs). The amplified DNA samples were purified again by GeneRead Size Selection Kit (Qiagen) and quantified using Qubit dsDNA HS Assay Kit (Life Technologies). Quality of the DNA sample was checked by running Agilent High Sensitivity DNA Kit using Agilent 2100 Bioanalyzer (Agilent) before sequenced using HiSeq 2500 (Illumina) following the manufacturer's instructions. The antibody (NT63, Biolegend) that specifically recognize RNA polymerase β was used to conduct the ChIP-exo experiment to detect the binding sites of RNA polymerase in E. coli K-12 MG1655. Each modified step was also performed following the manufacturer's instructions. ChIP-exo experiments were performed in biological duplicate.
RNA-seq expression profiling
Three milliliters of cells from mid-log phase culture were mixed with 6 ml RNAprotect Bacteria Reagent (Qiagen). Samples were mixed immediately by vortexing for 5 s, incubated for 5 min at room temperature, and then centrifuged at 5000g for 10 min. The supernatant was decanted and any residual supernatant was removed by inverting the tube once onto a paper towel. Total RNA samples were then isolated using RNeasy Plus Mini kit (Qiagen) following the manufacturer's instruction. Samples were then quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and quality of the isolated RNA was checked by running RNA 6000 Pico Kit using Agilent 2100 Bioanalyzer (Agilent). Paired-end, strand-specific RNA-seq library was prepared using KAPA RNA Hyper Prep kit (KAPA Biosystems), following the instruction (39,40). Resulting libraries were analyzed on an Agilent Bioanalyzer DNA 1000 chip (Agilent). Sequencing was performed on a Hiseq 2500 sequencer at the Genomics Core facility of University of California, San Diego.
Peak calling for ChIP-exo dataset
Peak calling was performed as previously described (37). Sequence reads generated from ChIP-exo were mapped onto the reference genome (NC_000913.2) using bowtie with default options to generate SAM output files (Dataset S3) (41). MACE program was used to define peak candidates from biological duplicates for each experimental condition with sequence depth normalization (42). To reduce false-positive peaks, peaks with signal-to-noise (S/N) ratio <1.5 were removed. The noise level was set to the top 5% of signals at genomic positions because top 5% makes a background level in a plateau and top 5% intensities from each ChIP-exo replicates across conditions correlate well with the total number of reads (37,43,44). The calculation of S/N ratio resembles the way to calculate ChIP-chip peak intensity where IP signal was divided by Mock signal. Then, each peak was assigned to the nearest gene. Genome-scale data were visualized using MetaScope (http://systemsbiology.ucsd.edu/Downloads/MetaScope).
Motif search from ChIP-exo peaks
The sequence motif analysis for TFs and σ-factors was performed using the MEME software suite (45). For YdcI, YbiH, YbaQ, YeiE, YddM and YieP, sequences in binding regions were extracted from the reference sequence (NC_000913.2).
Calculation of differentially expressed gene
Sequence reads generated from RNA-seq were mapped onto the reference genome (NC_000913.2) using bowtie with the maximum insert size of 1000 bp, and two maximum mismatches after trimming 3 bp at 3′ ends (Dataset S4) (41). SAM files generated from bowtie were then used for Cufflinks (http://cufflinks.cbcb.umd.edu) to calculate fragments per kilobase of exon per million fragments (FPKM) (46). Cufflinks was run with default options with the library type of dUTP RNA-seq and the default normalization method (classic-fpkm). Expression with log2 fold change ≥ log2(1.5) and q-value ≤ 0.05 or log2fold change ≤ –log2(1.5) and q-value ≤ 0.05 was considered as differentially expressed. Genome-scale data were visualized using MetaScope.
COG functional enrichment
The regulons were categorized according to their annotated clusters of orthologous groups (COG) category (47). Functional enrichment of COG categories in the target genes was determined by performing a hypergeometric test, and P-value < 0.05 was considered significant.
Structural analysis of candidate TFs
Homology models of the candidate transcription factors YdcI, YeiE and YiaJ were constructed using the SWISS-MODEL pipeline, which also carries out a prediction of the oligomeric state of the enzyme (48). Multiple templates were analyzed, and inference of the oligomeric state was based on the reported interface conservation scores to existing complexes of similar sequence identity. The structures were annotated using information in UniProt (31) and visualized with VMD (49).
Phylogenetic tree analysis
The homolog sequences of candidate TF YdcI across common Gram-negative strains were searched in NCBI databases, to show the shared origin of them. The phylogenetic tree (neighbor-joining without distance corrections) was generated by MUSCLE (50).
RESULTS
Establishing a workflow to discover uncharacterized transcription factors
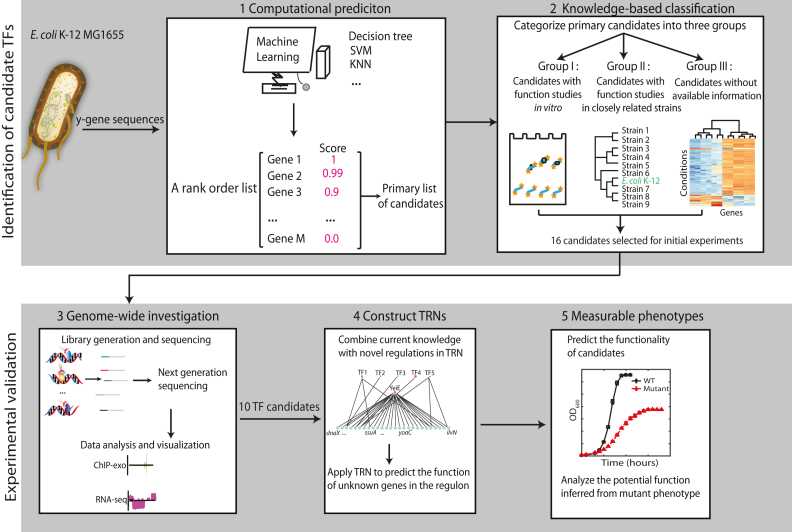
This workflow consisted of computational prediction, knowledge-based classification, and experimental validation of candidate TFs at the genome-scale (Figure 1). TFpredict is a machine learning algorithm that uses sequence homology to predict whether a given protein is a TF (30). The uncharacterized protein sequences in E. coli K-12 MG1655 were evaluated using the model trained by TFpredict with the data from the proteobacteria (see Materials and Methods). The output from TFpredict was a rank-ordered list of uncharacterized candidates based on the likelihood of their being a TF (Dataset S1). The initial output from TFpredict was reduced down to 474 primary candidates by excluding the lowest confidence scores (arbitrary cutoff value of 0.05) and proteins with no homologs (and therefore no prediction from TFpredict).
Figure 1.
The scheme of the systematic workflow for discovering uncharacterized transcription factors in E. coli K-12 MG1655. This workflow consists of computational prediction, knowledge-based classification, and experimental validation. The uncharacterized gene sequences of E. coli K-12 MG1655 are the input data for TFpredict. The output is a rank order list of genes with confidence scores. The primary selection is made based on the confidence scores from TFpredict. Subsequently, the primary list of genes is categorized into three groups based on the confidence level of biochemical/biological roles. An initial subset of 16 candidates was selected for experimental validation.Next, genome-wide binding sites were identified by ChIP-exo, and differential expression of their target genes was analyzed by RNA-seq. Finally, hypothesized functions of selected candidate TFs were inferred by comparing phenotypes between wild type and TF knockout mutants.
To further exclude false positives, the primary candidates were categorized into three groups based on the predicted interactions between candidates and DNA sequences (Supplementary Table S1). The first group contains candidates whose interactions with DNA were studied in vitro with gel shift assays (51) or SELEX (6,52), yet their in vivo biological functions remain largely unknown. The second group consists of candidates whose interactions with DNA could be predicted according to a well-studied homologous TF in a closely related strain. The third group includes candidates without available information about the protein-DNA interactions.
Considering that uncharacterized TFs are likely to be expressed at low levels, especially at non-active conditions (53), it is necessary to predict the conditions under which uncharacterized TFs are active. This classification also suggests the group-specific strategies to predict experimental conditions for the downstream ChIP-exo experiment (Supplementary Table S1). The activating conditions for candidate TFs could be inferred based on the characterization of the interaction between candidates and DNA. For the first group, the conditions were inferred based on biochemical features of binding targets, e.g., a previous study showed that yiaJ might be involved in the catabolism of rare carbon sources (54). The conditions for the second group were inferred from functional studies of a homologous TF in a closely related strain. For example, ydcI is a highly conserved gene and is responsible for pH stress response in Salmonella enterica serovar Typhimurium (55), thus it is likely to function at similar conditions in E. coli K-12, though it may play multiple biological roles. For the third group, the conditions were inferred based on expression profiling data from the NCBI GEO repository (Supplementary Figure S4) (56,57). If the expression level of the candidate TF is relatively high under a particular condition, it might be inferred as a test condition. The data showed that yeiE is highly expressed in glucose medium compared to other conditions. In this study, the characteristics of candidates were analyzed, and their active conditions were predicted accordingly.
To prioritize the candidate TFs for experimental validation, an initial subset of 16 candidates was chosen from three groups (Table 1). Next, to examine whether selected candidates have DNA-binding peaks at the genome-scale, ChIP-exo experiments were conducted at predicted conditions. For those candidates having DNA-binding sites, the expression profiles upon deletion of each TF were further investigated. Combining DNA bindings from ChIP-exo with gene expression, the hypotheses for the regulatory functions of candidate TFs were formed. To further test the hypotheses, mutant phenotypes were measured and then analyzed under active conditions (Figure 1).
Table 1.
Category of uncharacterized transcription factors in this study
| Candidate TFs | Locus | Family typea | Protein size (# amino acids) | TFpredict score | Target genes identified previouslyb | Target genes identified in this study |
|---|---|---|---|---|---|---|
| Group I | ||||||
| YiaJ | b3574 | IclR | 282 | 1.0 | 157 binding sites | Supplementary Figure S7 |
| YagI | b0272 | IclR | 252 | 1.0 | yagA, yagE | N/A |
| YbiH | b0796 | TetR | 223 | 0.98 | ybiH, rhlE | Supplementary Figure S7 |
| Group II | ||||||
| YdcI | b1422 | LysR | 307 | 1.0 | N/A | Supplementary Figure S8 |
| YbaO | b0447 | AsnC | 152 | 0.06 | tdcG, yfdV, yfjI, yhaO | Supplementary Figure S8 |
| Group III | ||||||
| YeiE | b2157 | LysR | 293 | 1.0 | N/A | Supplementary Figure S9 |
| YjhU | b4295 | N/A | 328 | 1.0 | N/A | N/A |
| YafC | b0208 | LysR | 304 | 0.96 | N/A | Supplementary Figure S9 |
| YihY | b3886 | N/A | 290 | 0.91 | N/A | N/A |
| YieP | b3755 | GntR | 230 | 0.38 | N/A | Supplementary Figure S9 |
| YddM | b1477 | Xre | 94 | 0.34 | N/A | Supplementary Figure S9 |
| YiaG | b3555 | Xre | 96 | 0.31 | N/A | N/A |
| YbaQ | b0483 | Xre | 113 | 0.06 | N/A | Supplementary Figure S9 |
| YjdC | b4135 | TetR | 191 | 0.05 | N/A | N/A |
| YchA | b1214 | N/A | 269 | 0.05 | N/A | N/A |
| YheO | b3346 | Xre | 240 | 0.06 | N/A | Supplementary Figure S9 |
acandidate TFs are classified into 54 families based on the DNA-binding motifs (6);
bTarget genes identified previously are from Regulon DB and TEC database (https://shigen.nig.ac.jp/ecoli/tec/top/);
Abbreviations: N/A: no available information.
Capturing a genome-wide distribution of uncharacterized transcription factors (TFs)
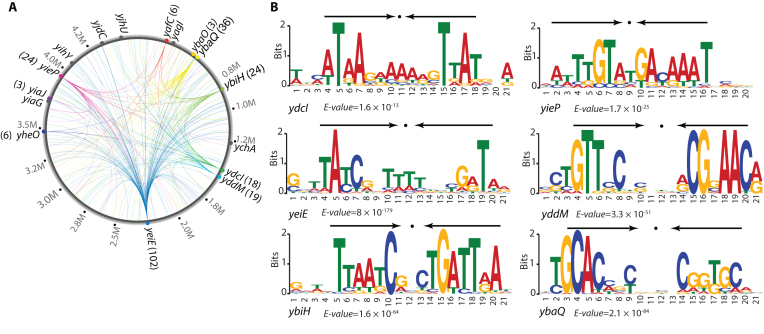
To validate the in silico predictions of candidate TFs, the ChIP-exo experiment was employed to determine the in vivo genome-wide DNA-binding events of each candidate during growth under active conditions. The global binding profiles for all candidates were examined using the peak calling algorithm MACE (58) and confirmed that 10 out of 16 were DNA-binding proteins (Figure 2A). A total of 255 reproducible binding peaks were identified for 241 unique binding sites (Dataset S5). Of the six unconfirmed candidates, YagI and YjhU had high confidence scores (score > 0.8). Therefore, it is possible that these proteins are TFs, but are not active under the basal condition used here.
Figure 2.
A global landscape of DNA binding events for uncharacterized TFs during growth at active conditions. (A) Binding sites identified by ChIP-exo. Verified uncharacterized TFs were labeled with colored circles. The numbers in the parentheses represent the number of identified binding sites for individual uncharacterized TFs. Gray circles represent uncharacterized TFs without binding peaks under the growth conditions used, which include YjhU, YjdC, YihY, YiaG, YagI, and YchA. (B) The sequence motifs for six uncharacterized TFs. The height of the letters (in bits on the y-axis) represents the degree of conservation at a given position within the aligned sequence set, with perfect conservation being 2 bits. Arrows above motif indicate the presence of palindromic sequences.
Compared to known global TFs, these ten uncharacterized TFs exhibit some interesting regulatory features. First, they have more intragenic binding peaks and fewer peaks located within putative regulatory regions. The binding sites from these confirmed TFs showed that only 41% (98 of 241) were located in putative regulatory regions (promoters and 5′-proximal to coding regions). Second, individual uncharacterized TFs had fewer binding peaks than those of global TFs such as CRP, Lrp, Fnr, and ArcA (12,59,60). Most of the uncharacterized TFs have 3–25 binding sites under active conditions, while global TFs in E. coli usually have more than 40 binding sites. Third, the uncharacterized TFs bind to more genes with putative functions (Supplementary Figure S5). Finally, the average expression level of these uncharacterized TFs is relatively lower than the majority of global TFs. These observations are consistent with the previous study showing that TF position in the TRN hierarchy network is correlated with its expression levels, its number of target genes, and its scope of regulatory function (61). TFs in the top hierarchy usually have high protein concentration in the cell, and regulate a significant number of genes of diverse functions. On the contrary, these candidate TFs are likely located in the lower levels of the E. coli hierarchical TRN, and may regulate local specific physiological functions instead of broad biological roles.
Next, for six of the ten confirmed TFs, the conserved binding motifs were further analyzed using the MEME algorithm (E-value < 10−10) (Figure 2B) (62). Interestingly, the consensus binding motifs were palindromic, suggesting a dimeric protein conformation. Specifically, the transcriptional factor binding sites (TFBS) of YdcI and YbiH enclose AT-rich inverted repeats separated by 7-nt. This finding is consistent with the structural predictions (Supplementary Table S2 and Supplementary Figure S6) that these candidate TFs likely form homodimers or tetramers, which facilitate tight binding to DNA molecules in the cell.
Interactions between uncharacterized TFs and RNA polymerase (RNAP)
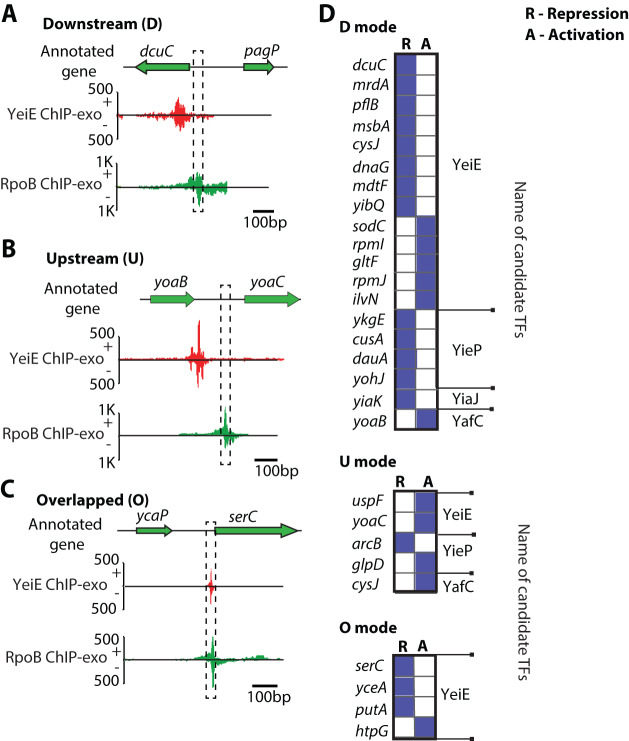
A transcriptional repressor down-regulates transcription by steric exclusion of RNAP from the promoter regions. To determine the interaction between the uncharacterized TFs and RNAP, the binding sites of the uncharacterized TFs were compared with the –10 and –35 promoter elements occupied by RNAP. Three interaction modes were observed based on their relative location: (i) downstream (D): TF binds downstream of the –10 and –35 promoter region (Figure 3A); (ii) upstream (U): TF binds upstream of the –10 and –35 promoter region (Figure 3B) and (iii) overlap (O): TF binding site coincides with the –10 and –35 promoter region (Figure 3C). To further illustrate how different TF-RNAP interaction modes may affect TF function, the regulatory effects on the target genes were characterized by their differential expression in ΔTF strain with respect to WT.
Figure 3.
Transcriptional regulation by the position of uncharacterized TFs relative to the binding of RNA polymerase (RNAP), using binding sites from YeiE, YieP, YiaJ, and YafC as representatives. (A) In the case of dcuC, YeiE-binding is located downstream of the promoter. (B) YeiE binds to the upstream site of the yoaC. (C) YeiE-binding region upstream of serC overlaps with the promoter occupied by RNAP. (D) The binding positions of YeiE, YieP, YiaJ, and YafC from the promoter are categorized according to the gene regulation. The abbreviations, D, U, and O indicate the downstream, upstream, and overlapped position, respectively. R and A indicate the regulation modes: repression and activation, respectively.
To demonstrate how binding sites of uncharacterized TFs interact with RNAP in vivo, four candidate TFs (YeiE, YieP, YiaJ, YafC) were used as representatives, since they showed a large number of binding sites. The most common binding mode for these transcription factors is downstream of the RNAP binding region. This binding mode commonly results in the repression of the target gene (13/19 or 68%). For example, YeiE represses and binds downstream of the RNAP binding region of the gene dcuC (Figure 3A). However, the upstream binding mode is more commonly activated, as shown by yoaC (Figure 3B). Three of the four binding sites that overlap with the RNAP binding location lead to the repression of the target genes serC, yceA, and putA (Figure 3C). Genes having upstream, downstream, or overlap modes from these four representatives mentioned earlier were determined (Figure 3D). This data suggested that transcriptional regulation by uncharacterized TFs are likely mediated by using steric exclusion mechanisms, though this pattern is not always true, as in gltF, rpmI, ilvN, and htpG. It is possible that other TFs are involved in the regulation of these target genes (Supplementary Figure S7) (63–66). Together, these data demonstrated that different sets of uncharacterized TFs have similar regulatory mechanisms, though they may have different biological functions.
To confirm the regulatory roles of candidate TFs, three of ten candidates identified by ChIP-exo (YiaJ, YdcI, YeiE) from three different groups were selected for further analysis, respectively. These three case studies illustrate how experimental observations from ChIP-exo and RNA-seq can be used to infer regulatory functions of a candidate TF. The binding sites of YiaJ and YdcI directly indicated their potential functions, so mutant phenotypes were used to validate biological roles. The genome-wide binding sites for YeiE showed that it is involved in diverse biological processes. Therefore, integration of expression profiling data with ChIP-exo was used to infer its potential roles in addition to mutant phenotype validation.
Case I: YiaJ regulates genes that are responsible for the utilization of l-ascorbate
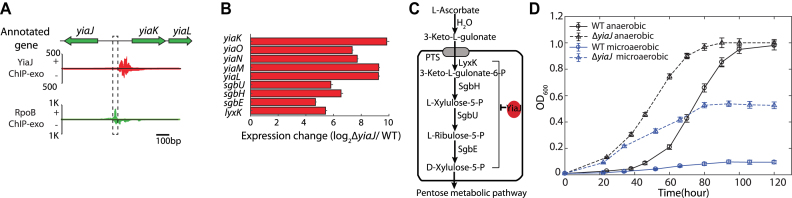
Group I contains candidates whose biochemical activities were studied in vitro, yet their in vivo biological functions still remain unclear. One of the candidates is YiaJ, which has been studied in vitro by gel mobility shift assays (35,67). However, in vivo analysis of direct interactions between YiaJ and DNA in E. coli has not been reported. In this study, we found that there were two binding peaks between the yiaJ and yiaKLMNOPQRS (yiaK-yiaL-yiaM-yiaN-yiaO-lyxK-sgbH-sgbU-sgbE) operon (Figure 4A and Supplementary Figure S8). One binding peak suggested autogenous regulation and the other showed that YiaJ binds to a promoter region of the yiaK-S operon, which occupied the position of RNAP. We compared the expression data of the yiaK-S operon in the wild type and yiaJ deletion strain (Figure 4B) and found that the expression of the operon yiaK-S was highly up-regulated in the deletion strain. This result suggests the repression function of YiaJ on the yiaK-S operon. A previous study showed that YiaJ might be involved in the utilization of an uncommon carbon sugar (54). To further identify the substrate catabolized by the yiaK-S operon, we compared the products of the yiaK-S operon with the known operon ulaABCDEF encoding for catabolic enzymes in the utilization of l-ascorbate, and found that the yiaK-S operon encodes similar catabolic enzymes in the l-ascorbate degradation pathway. Thus, we proposed the regulatory role of YiaJ in E. coli, based on the products of the yiaK-S operon (Figure 4C). When l-ascorbate is imported and converted to l-ascorbate-6-phosphate by the phosphotransferase system (PTS) in E. coli K-12 MG1655, expression of YiaJ would be repressed. Subsequently, lyxK, sgbH, sgbU, and sgbE encode four metabolic enzymes, l-xylulose kinase, gulonate-6-phosphate, l-xyluose-5-phosphate-3-epime, and l-ribulose-5-phosphate-4-epime, respectively. They can eventually metabolize l-ascorbate-6-phosphate to d-xylulose-5-phosphate. Thus, E. coli could ferment l-ascorbate using a branch of the pentose metabolic pathway (35).
Figure 4.
The regulatory role of the uncharacterized TF YiaJ is involved in the utilization of l-ascorbate in E. coli K-12 MG1655. (A) YiaJ binding sites at the promoter region between yiaJ and the yiaKLMNO-lyxK-sgbH-sgbU-sgbE operon. (B) Expression changes for genes in the yiaJ deletion strain in the yiaKLMNO-lyxK-sgbH-sgbU-sgbE operon compared to the wild type strain. (C) The proposed function of YiaJ is to repress the ascorbate utilization pathway, therefore regulating the level of d-xylulose-5-P that feeds into the pentose phosphate pathway. (D) Growth curve of wild type and yiaJ deletion strains at ascorbate as the carbon source under anaerobic and microaerobic conditions, respectively.
To verify the function of the repressor YiaJ, the growth profiles of the wild type and the yiaJ deletion strain were measured in l-ascorbate medium. The data showed that the deletion of gene yiaJ allowed more rapid utilization of L-ascorbate and reduced the lag phase compared to wild type (Figure 4D). Furthermore, growth profiles suggested that the yiaJ deletion strain allowed cells to grow on l-ascorbate medium under microaerobic conditions, while the wild type could not. This confirmed that YiaJ is a repressor of operon yiaK-S and that it influenced growth under microaerobic conditions.
Case II: YdcI is a transcription factor involved in pH homeostasis and acetate metabolism
Group II consists of highly conserved candidate TFs, which were studied in a closely related species. The regulatory function of the LysR-type regulator YdcI in E. coli K-12 MG1655 has not been studied with experimental approaches (6). Thus, a ydcI myc-tagged strain was constructed to detect 18 binding sites using ChIP-exo (Supplementary Figure S9).
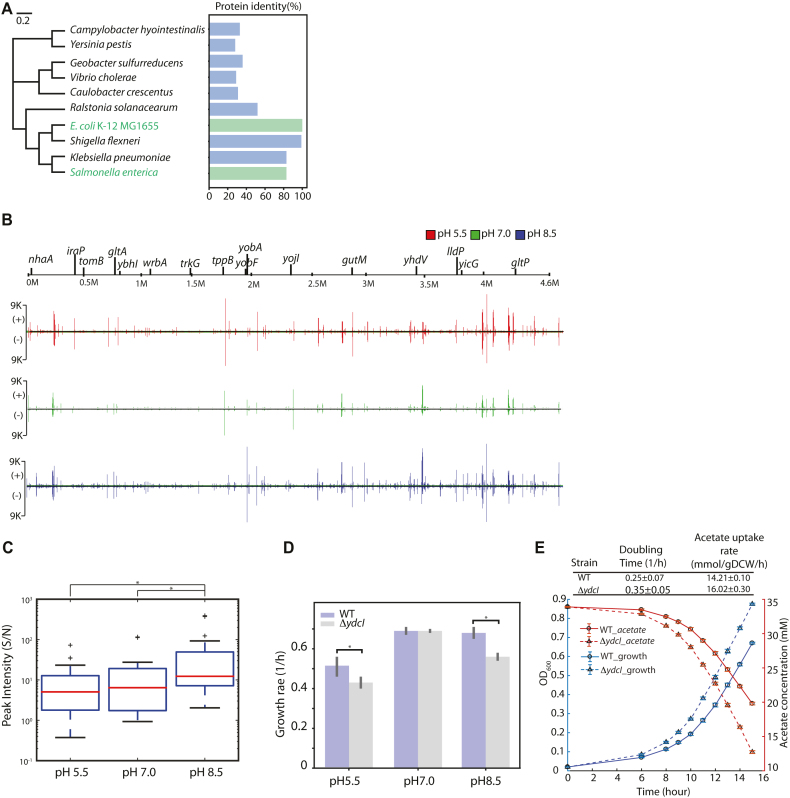
Previous studies showed that YdcI is responsible for acid stress resistance in Salmonella enterica (55). The protein identity of YdcI was analyzed among multiple strains across Gram-negative bacteria, which showed that YdcI encodes a highly conserved protein with related homologs present in a range of Gram-negative bacterial genera (E. coli K-12 MG1655, S. enterica, K. pneumoniae, and S. flexneri) (Figure 5A, Supplementary Figure S10). Notably, YdcI from E. coli K-12 MG1655 shares 80% of its identity with that from Salmonella enterica. Given that the function of a protein is tightly associated with its sequence, we can hypothesize that YdcI has similar biological roles in E. coli K-12 MG1655.
Figure 5.
The regulatory role of the uncharacterized TF YdcI is involved in proton and acetate metabolism in E. coli K-12 MG1655. (A) Phylogenetic trees displaying the relatedness of YdcI from E. coli K-12 MG1655 and from Salmonella enterica. (B) Genome-wide YdcI DNA binding. YdcI binding across the genome was compared under different pH conditions in E. coli K-12 MG1655 by ChIP-exo. (C) Peak intensity (Signal/Noise) of YdcI ChIP-exo binding sites at pH 5.5, pH 7.0, and pH 8.5. Among the three different pH conditions, peak intensity was most active at pH 8.5 (* indicates rank sum test P-value < 0.05). (D) The growth rate of wild type and ydcI deletion strain at low pH, neutral pH, and high pH media. (E) Growth and acetate uptake rates of wild type and ydcI deletion strains in acetate growth medium.
To test our hypothesis, ChIP-exo experiments for YdcI were conducted at different pH conditions (Figure 5B). Under low pH conditions, YdcI bound to 16 locations, and two-thirds of these binding peaks were found in intergenic regions. Under neutral or high pH conditions, YdcI bound to all sites identified at low pH conditions but had differential binding intensity. Thus, the ratio of signal to noise (S/N) was analyzed. The data showed that YdcI had the highest average binding intensity at high pH medium (Figure 5C). More important, we found that four of the intergenic target genes (nhaA, dtpA, lldP, and gltP) encode proton transporters, which play important roles in the acidic/alkaline conditions. Especially, as a major cation/proton antiporter, NhaA reveals a prominent role in alkaline pH homeostasis (68). Therefore, the growth phenotypes of the ydcI deletion strain were examined at low pH, neutral pH, and high pH media (Figure 5D). At pH 5.5 or pH 8.5, the ydcI deletion strain showed significant growth defects compared to the wild type. However, there was no defect observed at neutral pH conditions. These data confirmed that YdcI is required to maintain physiological activity at acidic/alkaline conditions in E. coli.
Additionally, there were two binding sites in the proximity of known ncRNAs (Supplementary Figure S11A and B). In panel A, YdcI binding regulates the transcription of nhaA encoding Na:H+ antiporter. There is a small RNA sokC annotated as antisense RNA sokC blocking mokC and hokC (69). In panel B, YdcI binds to the promoter region of yobA. At the downstream of the binding event, there is a small RNA sdsR, which is the base-pair with some part of mutS coding region. Deletion of sdsR decreases ampicillin-induced mutagenesis (70). Overexpression of SdsR decreases biofilm development and swarming motility (71). This data showed that no YdcI binding was found upstream of ncRNAs, indicating that YdcI does not directly regulate ncRNAs.
YdcI has another important binding site at the gene gltA, which encodes a citrate synthase in E. coli K-12 MG1655. It is induced and becomes the rate-limiting step for the TCA cycle when acetate is the sole carbon source (72,73). A previous study hypothesized that YdcI may regulate the carbon flux in the TCA cycle through gltA expression (74). To test this hypothesis, the growth of E. coli WT and the ydcI deletion strain were compared in acetate medium (Figure 5E). The ydcI deletion strain grew significantly faster than the wild type, showing that YdcI represses the gene gltA. The acetate uptake rate increased upon ydcI deletion compared to WT using high-performance liquid chromatography (HPLC), which confirmed that YdcI is also involved in regulating the carbon flux in the TCA cycle.
Case III: YeiE is a transcription factor that is involved in iron homeostasis
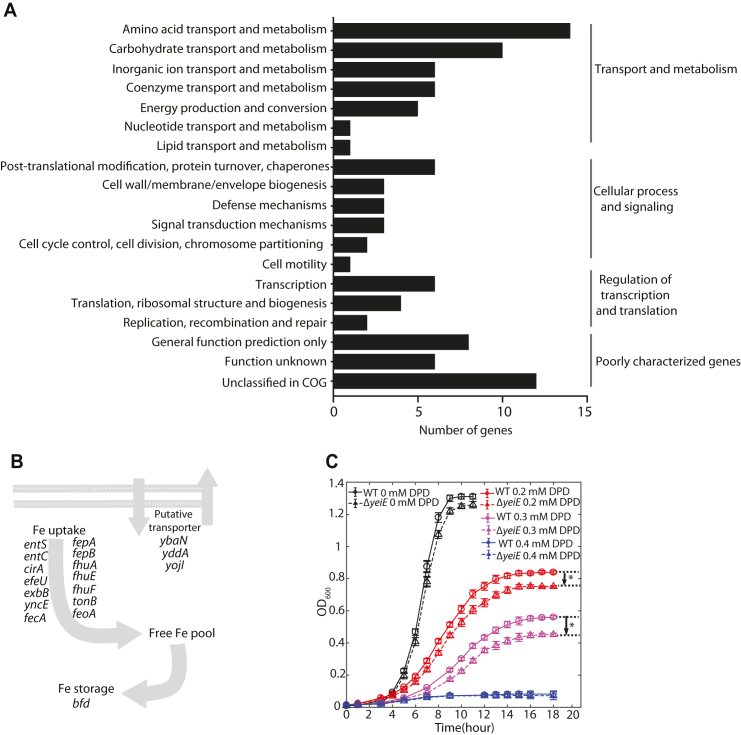
Group III includes candidates with neither biochemical characterization nor biological function prediction. Among them, the global binding profile of LysR-type YeiE showed over 100 binding sites across the E. coli K-12 MG1655 genome (Supplementary Figure S12) (6). Target genes of YeiE are involved in diverse biological processes, including transport and metabolism, cell wall/membrane biogenesis, signal transduction, and transcriptional regulation (Figure 6A). Furthermore, functional classification showed that approximately 42% (43 /102) of YeiE bindings are involved in main transport processes, including amino acids, carbohydrate, and inorganic ions, though it is not significantly enriched in any functional group. This data suggests that YeiE may play multiple biological roles in E. coli K-12 MG1655. To further investigate the potential functions of YeiE, the expression profiles of the ΔyeiE strain were examined. Three Clusters of Orthologous Groups (COGs) functional groups were significantly (P-value < 0.01) associated with the functions of YeiE: energy production and conversion, amino acid transport and metabolism, and inorganic ion transport and metabolism (Supplementary Figure S13). Notably, many metal ion homeostasis-related genes, such as entS, entC, cirA, fhuA, fhuF, fepB, and feoA, were down-regulated in the yeiE deletion strain (Figure 6B). These results suggest that YeiE may be involved in the iron-uptake regulation pathway.
Figure 6.
The regulatory role of the uncharacterized TF YeiE is involved in maintaining iron homeostasis under iron-limited conditions in E. coli K-12 MG1655. (A) Functional classification of target genes from YeiE genome-wide bindings. The enriched functions are in three groups: amino acid transport/metabolism, carbohydrate transport/metabolism, and inorganic ion transport/metabolism. (B) The proposed regulatory roles of genes down-regulated in the yeiE deletion strain. (C) Growth profile of wild-type and yeiE deletion strain in iron-free M9 minimal medium supplemented with 0, 0.2 mM, 0.3 mM, 0.4 mM of 2,2′-dipyridyl (DPD) (an iron chelator), respectively. The growth curves were determined from at least six independent cultures and significant differences in the stationary phase between wild type and yeiE deletion strain were determined by the Student's t test, P < 0.01.
To examine the role of YeiE in inorganic ion transport and metabolism, the growth profiles of the wild type and yeiE deletion strain were measured in M9 medium with or without iron chelator (Figure 6C). There was no appreciable difference between the growth profiles of the two strains in the iron-rich condition without iron chelator. With 0.2 mM of the iron chelator (2,2′-dipyridyl, DPD), the yeiE deletion strain grew slower than the wild type in the early-mid log phase. As cells entered into late log phase, different growth curves were observed. The differences in the stationary phase between the wild-type and yeiE deletion strain increased with the concentration of iron chelator in the media. When the concentration of iron chelator reached 0.4 mM, neither strain could enter the log phase. The fact that this growth defect was only observed under iron-limited conditions suggested that YeiE is involved in iron-uptake pathways under these conditions.
DISCUSSION
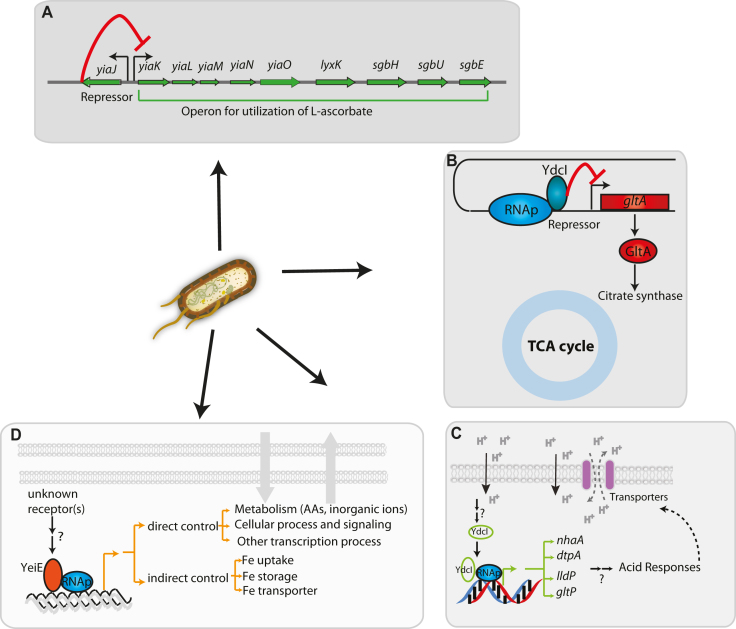
The characterization of a transcriptional regulatory network (TRN) is an essential step in our understanding of organism function and evolution. A critical limitation of this step is that a complete set of characterized TFs for an individual organism does not exist. Here, this was addressed by the development of an integrated bioinformatic and experimental workflow. This workflow was applied to E. coli K-12 MG1655, one of the most well-studied organisms. Ten previously uncharacterized TFs were discovered in vivo. The regulon for each novel TF was reconstructed, and the physiological roles for three of them were determined; YiaJ is involved in the utilization of l-ascorbate (Figure 7A), YdcI is involved in proton and acetate metabolism (Figure 7B and C), and YeiE is involved in iron uptake under iron-limited conditions (Figure 7D). Also, in vivo binding patterns of YbiH and YbaO were consistent with the genomic SELEX results, though the genome-wide binding profile of YbiH showed some extra target genes (Supplementary Figure S8) (75,76). This suggests that the binding patterns of some regulators are very consistent between in vivo and in vitro methods. The results of this study have several notable implications.
Figure 7.
The model for the regulatory network integrating three candidate TFs (YiaJ, YdcI, and YeiE) and their biological functions in E. coli K-12 MG1655. (A) YiaJ is a regulator that controls the operon yiaK-yiaL-yiaM-yiaN-yiaO-lyxK-sgbH-sgbU-sgbE in the catabolism pathway. (B) YdcI inhibits the transcription of target gene gltA, resulting in the down-regulation of citrate synthase that is required in the TCA cycle. (C) YdcI binds to genomic DNA and activates target genes nhaA, dtpA, lldP, and gltP that are responsible for proton transfer. (D) YeiE affects multiple transporters (amino acids, inorganic ions, lipids) and metabolic processes, maintaining iron homeostasis at iron-limited conditions.
First, the ten newly identified TFs represent a 6% increase to the 185 already known TFs. Furthermore, new knowledge about the co-binding of candidate TFs and known TFs was provided at the genome-scale (Supplementary Figure S7). This TF discovery workflow enables the systematic examination of the remaining putative TFs identified by the initial computational step of the workflow. In this study, six of the examined candidate TFs were not found to have any binding sites at test conditions. This failure to identify binding sites could have happened for two reasons: (i) our conditions did not activate these TFs (e.g., YagI was recently identified as a regulator of xylonate catabolism using the SELEX method in vitro (77)); and (ii) current prediction algorithm methods may generate false-positive candidates. Recently, the annotations of YihY and YchA have been updated to putative inner membrane protein and transglutaminase-like/TPR repeat-containing protein, respectively, though their physiological functions remain unclear (3,78). However, it is still necessary to develop a systematic workflow to predict and validate TFs and improve our knowledge of the TRN.
Second, differential expression data between wild type and uncharacterized TF deletion strains allowed us to reconstruct new regulons. A regulatory network containing 47 new regulatory interactions was reconstructed between candidate TFs and their target genes (Supplementary Figure S7). Specifically, more regulatory information was added for 25 target genes that previously had no known regulator. The reconstructed regulons suggest functional associations between both characterized and uncharacterized genes (Supplementary Figure S14). For instance, as a periplasmic protein, the physiological role of GltF in E. coli is still unknown (3). Functional enrichment suggests that it may transport inorganic ions or other metabolites. Future experimental studies are needed to discover the functions of these uncharacterized genes.
Third, detailing the functions of three of the ten regulons adds to our understanding of the TRN in E. coli. The iron response is a crucial characteristic in most enterobacteria, as well as bacteria in general. Although Fur is a well-known TF for the iron response, the discovery of YeiE as an active TF under low iron conditions adds to our understanding of the overall iron response (Figure 7D). Low iron levels are especially important in understanding the interactions between pathogens and hosts (79,80). Transcriptional regulation of ascorbate metabolism has been largely unknown, and the discovery of the role that YiaJ plays helps fill this knowledge gap (Figure 7A). The transcriptional repressor YiaJ belongs to the IclR family and controls the hypothetical ascorbate transport system (named yiaMNO) and four genes (lyxK-sgbH-sgbU-sgbE) encoding ascorbate catalytic enzymes (6,81).
Although the strengths of the presented workflow were demonstrated in the study, there is room for improvement to broaden the applicability of the workflow. More uncharacterized TFs will be discovered after further experimental validation. The characterization of more TFs in databases would allow for a larger training set, improving the predictive power of machine learning methods like TFpredict. On the other hand, while ChIP-exo is commonly used for the mapping of TF-DNA interactions, its application to the elucidation of regulon function is limited by the knowledge of suitable conditions that activate a target TF. For non-model bacteria, the lack of biochemical/biological function studies may limit the possibility of directly inferring the active conditions from the functional studies. To address potential issues with predicting experimental conditions under which a TF is expressed, previous studies have used conservation analysis, expression profiling data, fitness scores, and investigated basal conditions to predict the conditions for candidate TFs (Supplementary Figure S15) (56,82–84). Furthermore, next-generation sequencing (NGS) technology has led to an explosion of genomic data, annotations, and expression studies (85), which would expand the availability of the data resources.
In this study, we have presented a workflow for the systematic discovery of uncharacterized TFs, which enables the reconstruction of their regulons. A study of an initial set of 16 candidate TFs demonstrated that the workflow could systematically elucidate TF functions in E. coli. This workflow also provides a path for the discovery of uncharacterized gene functions that were found in the newly discovered regulons. As more data is made available, the workflow presented here may pave the way towards a more robust discovery of uncharacterized TFs.
DATA AVAILABILITY
The whole dataset of ChIP-exo and RNA-seq has been deposited to GEO with the accession number of GSE111095. All code for the TFpredict algorithm trained on data from proteobacteria is freely available on GitHub (https://github.com/draeger-lab/TFpredict/tree/prokaryote).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Szubin for help with ChIP-exo and RNA-seq library sequencing. We thank Zachary A. King, Justin Tan, and Amitesh Anand for helpful discussions. We thank Marc Abrams for reviewing and editing the manuscript.
Author Contributions: Y.G., D.K. and B.O.P. designed the study. Y.G. and D.K. performed experiments. J.T.Y., A.D. and J.E. performed computational analysis. Y.G., D.K., S.W.S., I.K., A.V.S. and X.F. did data analysis. K.C. and N.M. contributed to the protein structure analysis. Y.G., J.T.Y., B.K.C., D.K. and B.O.P. wrote the manuscript, with contributions from all other authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation [NNF10CC1016517]; Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education [NRF-2017R1C1B2002441]; Ministry of Food and Drug Safety [17162MFDS601]. A.D. acknowledges support from the National Institutes of General Medical Sciences (NIH/NIGMS) grant [R01 GM070923]. Funding for open access charge: Novo Nordisk Foundation [NNF10CC1016517].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cannon W., Claverie-Martin F., Austin S., Buck M.. Core RNA polymerase assists binding of the transcription factor σ; 54 to promoter DNA. Mol. Microbiol. 1993; 8:287–298. [DOI] [PubMed] [Google Scholar]
- 2. Alkema W.B.L., Lenhard B., Wasserman W.W.. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 2004; 14:1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keseler I.M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martínez C., Caspi R., Fulcher C., Gama-Castro S., Kothari A., Krummenacker M. et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017; 45:D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keseler I.M., Collado-Vides J., Santos-Zavaleta A., Peralta-Gil M., Gama-Castro S., Muñiz-Rascado L., Bonavides-Martinez C., Paley S., Krummenacker M., Altman T. et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011; 39:D583–D590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A. et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2015; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ishihama A., Shimada T., Yamazaki Y.. Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res. 2016; 44:2058–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galperin M.Y., Koonin E.V.. From complete genome sequence to ‘complete’ understanding. Trends Biotechnol. 2010; 28:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liolios K., Chen I.-M.A., Mavromatis K., Tavernarakis N., Hugenholtz P., Markowitz V.M., Kyrpides N.C.. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010; 38:D346–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang Y.-C., Hu Z., Rachlin J., Anton B.P., Kasif S., Roberts R.J., Steffen M.. COMBREX-DB: an experiment centered database of protein function: knowledge, predictions and knowledge gaps. Nucleic Acids Res. 2016; 44:D330–D335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minchin S.D., Busby S.J.W.. Analysis of mechanisms of activation and repression at bacterial promoters. Methods. 2009; 47:6–12. [DOI] [PubMed] [Google Scholar]
- 11. Ogawa N., Biggin M.D.. High-throughput SELEX determination of DNA sequences bound by transcription factors in vitro. Methods Mol. Biol. 2012; 786:51–63. [DOI] [PubMed] [Google Scholar]
- 12. Cho B.-K., Barrett C.L., Knight E.M., Park Y.S., Palsson B.Ø.. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19462–19467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H., Yin Y., Olman V., Xu Y.. Genomic arrangement of regulons in bacterial genomes. PLoS One. 2012; 7:e29496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elmas A., Wang X., Samoilov M.S.. Reconstruction of novel transcription factor regulons through inference of their binding sites. BMC Bioinformatics. 2015; 16:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho B.-K., Federowicz S.A., Embree M., Park Y.-S., Kim D., Palsson B.Ø.. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011; 39:6456–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shimada T., Ogasawara H., Ishihama A.. Single-target regulators form a minor group of transcription factors in Escherichia coli K-12. Nucleic Acids Res. 2018; 46:3921–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rhee H.S., Pugh B.F.. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr. Protoc. Mol. Biol. 2012; doi:10.1002/0471142727.mb2124s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beauchene N.A., Myers K.S., Chung D., Park D.M., Weisnicht A.M., Keleş S., Kiley P.J.. Impact of anaerobiosis on expression of the Iron-Responsive Fur and RyhB regulons. MBio. 2015; 6:e01947-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beauchene N.A., Mettert E.L., Moore L.J., Keleş S., Willey E.R., Kiley P.J.. O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12261–12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D., Seo S.W., Gao Y., Nam H., Guzman G.I., Cho B.-K., Palsson B.O.. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. Nucleic Acids Res. 2018; 46:2901–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu Y., Jarboe L.R., Dickerson J.A.. Reconstructing genome-wide regulatory network of E. coli using transcriptome data and predicted transcription factor activities. BMC Bioinformatics. 2011; 12:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang X., Sastry A., Mih N., Kim D., Tan J., Yurkovich J.T., Lloyd C.J., Gao Y., Yang L., Palsson B.O.. Global transcriptional regulatory network for Escherichia coli robustly connects gene expression to transcription factor activities. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:10286–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zare H., Sangurdekar D., Srivastava P., Kaveh M., Khodursky A.. Reconstruction of Escherichia coli transcriptional regulatory networks via regulon-based associations. BMC Syst. Biol. 2009; 3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faria J.P., Overbeek R., Xia F., Rocha M., Rocha I., Henry C.S.. Genome-scale bacterial transcriptional regulatory networks: reconstruction and integrated analysis with metabolic models. Brief. Bioinform. 2014; 15:592–611. [DOI] [PubMed] [Google Scholar]
- 25. Seo S.W., Kim D., Latif H., O’Brien E.J., Szubin R., Palsson B.O.. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014; 5:4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seo S.W., Kim D., Szubin R., Palsson B.O.. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 2015; 12:1289–1299. [DOI] [PubMed] [Google Scholar]
- 27. Seo S.W., Gao Y., Kim D., Szubin R., Yang J., Cho B.-K., Palsson B.O.. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci. Rep. 2017; 7:2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seo S.W., Kim D., O’Brien E.J., Szubin R., Palsson B.O.. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 2015; 6:7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho S., Cho Y.-B., Kang T.J., Kim S.C., Palsson B., Cho B.-K.. The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Res. 2015; 43:3079–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eichner J., Topf F., Dräger A., Wrzodek C., Wanke D., Zell A.. TFpredict and SABINE: sequence-based prediction of structural and functional characteristics of transcription factors. PLoS One. 2013; 8:e82238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Consortium The UniProt. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho B.-K., Knight E.M., Palsson B.O.. PCR-based tandem epitope tagging system for Escherichia coli genome engineering. BioTechniques. 2006; 40:67–72. [DOI] [PubMed] [Google Scholar]
- 34. Datta S., Costantino N., Court D.L.. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006; 379:109–115. [DOI] [PubMed] [Google Scholar]
- 35. Yew W.S., Gerlt J.A.. Utilization of L-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 2002; 184:302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall B.G., Acar H., Nandipati A., Barlow M.. Growth rates made easy. Mol. Biol. Evol. 2014; 31:232–238. [DOI] [PubMed] [Google Scholar]
- 37. Seo S.W., Kim D., Latif H., O’Brien E.J., Szubin R., Palsson B.O.. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014; 5:4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cho B.-K., Kim D., Knight E.M., Zengler K., Palsson B.O.. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014; 12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ross M.G., Russ C., Costello M., Hollinger A., Lennon N.J., Hegarty R., Nusbaum C., Jaffe D.B.. Characterizing and measuring bias in sequence data. Genome Biol. 2013; 14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quail M.A., Otto T.D., Gu Y., Harris S.R., Skelly T.F., McQuillan J.A., Swerdlow H.P., Oyola S.O.. Optimal enzymes for amplifying sequencing libraries. Nat. Methods. 2011; 9:10. [DOI] [PubMed] [Google Scholar]
- 41. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang L., Chen J., Wang C., Uusküla-Reimand L., Chen K., Medina-Rivera A., Young E.J., Zimmermann M.T., Yan H., Sun Z. et al. MACE: model based analysis of ChIP-exo. Nucleic Acids Res. 2014; 42:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seo S.W., Kim D., Szubin R., Palsson B.O.. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 2015; 12:1289–1299. [DOI] [PubMed] [Google Scholar]
- 44. Ogasawara H., Ohe S., Ishihama A.. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol. Lett. 2015; 362:1–8. [DOI] [PubMed] [Google Scholar]
- 45. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V.. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000; 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 50. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hellman L.M., Fried M.G.. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2007; 2:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Riley T.R., Slattery M., Abe N., Rastogi C., Liu D., Mann R.S., Bussemaker H.J.. SELEX-seq: a method for characterizing the complete repertoire of binding site preferences for transcription factor complexes. Methods Mol. Biol. 2014; 1196:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Janga S.C., Contreras-Moreira B.. Dissecting the expression patterns of transcription factors across conditions using an integrated network-based approach. Nucleic Acids Res. 2010; 38:6841–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ibañez E., Campos E., Baldoma L., Aguilar J., Badia J.. Regulation of expression of the yiaKLMNOPQRS operon for carbohydrate utilization in Escherichia coli: involvement of the main transcriptional factors. J. Bacteriol. 2000; 182:4617–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jennings M.E., Quick L.N., Soni A., Davis R.R., Crosby K., Ott C.M., Nickerson C.A., Wilson J.W.. Characterization of the Salmonella enterica serovar Typhimurium ydcI gene, which encodes a conserved DNA binding protein required for full acid stress resistance. J. Bacteriol. 2011; 193:2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang L., Chen J., Wang C., Uusküla-Reimand L., Chen K., Medina-Rivera A., Young E.J., Zimmermann M.T., Yan H., Sun Z. et al. MACE: model based analysis of ChIP-exo. Nucleic Acids Res. 2014; 42:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Federowicz S., Kim D., Ebrahim A., Lerman J., Nagarajan H., Cho B.-K., Zengler K., Palsson B.. Determining the control circuitry of redox metabolism at the genome-scale. PLoS Genet. 2014; 10:e1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grainger D.C., Hurd D., Harrison M., Holdstock J., Busby S.J.W.. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17693–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Janga S.C., Salgado H., Martínez-Antonio A.. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009; 37:3680–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lestienne P., Plumbridge J.A., Grunberg-Manago M., Blanquet S.. Autogenous repression of Escherichia coli threonyl-tRNA synthetase expression in vitro. J. Biol. Chem. 1984; 259:5232–5237. [PubMed] [Google Scholar]
- 64. Hommais F., Krin E., Coppée J.-Y., Lacroix C., Yeramian E., Danchin A., Bertin P.. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology. 2004; 150:61–72. [DOI] [PubMed] [Google Scholar]
- 65. Lemke J.J., Sanchez-Vazquez P., Burgos H.L., Hedberg G., Ross W., Gourse R.L.. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5712–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Friden P., Tsui P., Okamoto K., Freundlich M.. Interaction of cyclic AMP receptor protein with the ilvB biosynthetic operon in E. coli. Nucleic Acids Res. 1984; 12:8145–8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ibañez E., Campos E., Baldoma L., Aguilar J., Badia J.. Regulation of expression of theyiaKLMNOPQRS operon for carbohydrate utilization inEscherichia coli: Involvement of the main transcriptional factors. J. Bacteriol. 2000; 182:4617–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krulwich T.A., Sachs G., Padan E.. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011; 9:330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Franch T., Thisted T., Gerdes K.. Ribonuclease III processing of coaxially stacked RNA helices. J. Biol. Chem. 1999; 274:26572–26578. [DOI] [PubMed] [Google Scholar]
- 70. Gutierrez A., Laureti L., Crussard S., Abida H., Rodríguez-Rojas A., Blázquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J. et al. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013; 4:1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bak G., Lee J., Suk S., Kim D., Young Lee J., Kim K.-S., Choi B.-S., Lee Y.. Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci. Rep. 2015; 5:15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Walsh K., Schena M., Flint A.J., Koshland D.E. Jr. Compensatory regulation in metabolic pathways–responses to increases and decreases in citrate synthase levels. Biochem. Soc. Symp. 1987; 54:183–195. [PubMed] [Google Scholar]
- 73. Walsh K., Koshland D.E. Jr. Characterization of rate-controlling steps in vivo by use of an adjustable expression vector. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:3577–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nishio Y., Suzuki T., Matsui K., Usuda Y.. Metabolic control of the TCA cycle by the YdcI transcriptional regulator in Escherichia coli. J. Microb. Biochem. Technol. 2013; 5:59–67. [Google Scholar]
- 75. Shimada T., Tanaka K., Ishihama A.. Transcription factor DecR (YbaO) controls detoxification of L-cysteine in Escherichia coli. Microbiology. 2016; 162:1698–1707. [DOI] [PubMed] [Google Scholar]
- 76. Yamanaka Y., Shimada T., Yamamoto K., Ishihama A.. Transcription factor CecR (YbiH) regulates a set of genes affecting the sensitivity of Escherichia coli against cefoperazone and chloramphenicol. Microbiology. 2016; 162:1253–1264. [DOI] [PubMed] [Google Scholar]
- 77. Shimada T., Momiyama E., Yamanaka Y., Watanabe H., Yamamoto K., Ishihama A.. Regulatory role of XynR (YagI) in catabolism of xylonate in Escherichia coli K-12. FEMS Microbiol. Lett. 2017; 364. [DOI] [PubMed] [Google Scholar]
- 78. Brinza L., Calevro F., Charles H.. Genomic analysis of the regulatory elements and links with intrinsic DNA structural properties in the shrunken genome of Buchnera. BMC Genomics. 2013; 14:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kortman G.A.M., Boleij A., Swinkels D.W., Tjalsma H.. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One. 2012; 7:e29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Skaar E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010; 6:e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang Z., Aboulwafa M., Smith M.H., Saier M.H. Jr. The ascorbate transporter of Escherichia coli. J. Bacteriol. 2003; 185:2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Moretto M., Sonego P., Dierckxsens N., Brilli M., Bianco L., Ledezma-Tejeida D., Gama-Castro S., Galardini M., Romualdi C., Laukens K. et al. COLOMBOS v3.0: leveraging gene expression compendia for cross-species analyses. Nucleic Acids Res. 2016; 44:D620–D623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T. et al. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015; 43:D1113–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Price M.N., Wetmore K.M., Waters R.J., Callaghan M., Ray J., Liu H., Kuehl J.V., Melnyk R.A., Lamson J.S., Suh Y. et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature. 2018; 557:503–509. [DOI] [PubMed] [Google Scholar]
- 85. Tripathi R., Sharma P., Chakraborty P., Varadwaj P.K.. Next-generation sequencing revolution through big data analytics. Front. Life Sci. 2016; 9:119–149. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole dataset of ChIP-exo and RNA-seq has been deposited to GEO with the accession number of GSE111095. All code for the TFpredict algorithm trained on data from proteobacteria is freely available on GitHub (https://github.com/draeger-lab/TFpredict/tree/prokaryote).