Abstract
Aflatoxins (AFs) are a group of carcinogenic and immunosuppressive mycotoxins that threaten global food safety. Globally, over 4.5 billion people are exposed to unmonitored levels of AFs. Aspergillus flavus is the major source of AF contamination in agricultural crops. One approach to reduce levels of AFs in agricultural commodities is to apply a non-aflatoxigenic competitor, e.g., Afla-Guard, to crop fields. In this study, we demonstrate that the food fermenting Aspergillus oryzae M2040 strain, isolated from Korean Meju (a brick of dry-fermented soybeans), can inhibit aflatoxin B1 (AFB1) production and proliferation of toxigenic A. flavus in lab culture conditions and peanuts. In peanuts, 1% inoculation level of A. oryzae M2040 could effectively displace the toxigenic A. flavus and inhibit AFB1 production. Moreover, cell-free culture filtrate of A. oryzae M2040 effectively inhibited AFB1 production and A. flavus growth, suggesting A. oryzae M2040 secretes inhibitory compounds. Whole genome-based comparative analyses indicate that the A. oryzae M2040 and Afla-Guard genomes are 37.9 and 36.4 Mbp, respectively, with each genome containing ~100 lineage specific genes. Our study establishes the idea of using A. oryzae and/or its cell-free culture fermentate as a potent biocontrol agent to control A. flavus propagation and AF contamination.
Introduction
Aflatoxins (AFs) are a group of small molecular weight fungal toxins that threaten world food safety by contaminating ~25% of the world’s crops1. AFs are considered to be an unavoidable contaminant in human food and animal feed by the US Food and Drug Administration (FDA)2. Among AFs, aflatoxin B1 (AFB1) is the most potent carcinogen present in nature and is produced mainly by the ubiquitous soil filamentous fungus Aspergillus flavus3. AFs are acutely toxic, carcinogenic, mutagenic, teratogenic, and immunosuppressive, and are classified as group 1 carcinogens in human (Fig. 1)4. Of the 550,000–600,000 new liver cancer cases worldwide each year, it is estimated that 25,200–155,000 may be attributed to AF exposure5,6. Due to their high toxicity and carcinogenicity, over 120 countries have set maximum limits of AFs in foods (4~30 ppb) and feeds (20~300 ppb)1. In the U.S. corn industry, AF contamination could cause losses ranging from $52.1 million to $1.68 billion annually as was reported in year 20127.
Figure 1.
Schematic presentation summarizing the major AFB1 and AFM1 contamination/exposure routes and adverse health effects to human.
AFs can frequently contaminate cereals, oilseeds, spices, tree nuts, corn, groundnuts (peanuts), pistachios, chilies, black pepper, dried fruit and fig, raising global health and economy concerns1. Furthermore, animal by-products such as milk, meat, and egg can be indirect sources of AF exposure8,9. Human milk can have aflatoxin M1 (AFM1), a hydroxylated metabolite of AFB1, and pose a serious threat for infants10,11 (Fig. 1). In certain areas in Africa and Asia, AFs are considered as the leading cause of serious acute illnesses and deaths each year12. Improper storage of the crops, nuts, and grains further contributes to increased levels of AFs. Unfortunately, over half of the global population is exposed to high, unmonitored levels of AFs13.
Previous research has demonstrated that both water availability and temperature affect A. flavus growth and the expression of genes in the AF biosynthesis gene cluster14. Predictions associated with global warming suggest that A. flavus is likely to infect more crop plants, and will show increase expression of the AF biosynthetic genes (e.g., aflD and aflR)15, enhancing the risk of crop contamination by AF. With a 2 °C temperature increase, AFB1 is predicted to become a food safety issue in the European maize production16. Several strategies have been developed to reduce AF contamination, including the use of a non-toxigenic strain of A. flavus to outcompete and displace toxigenic strains17,18. This effective biological control method results in greatly reduced AF levels in a diversity of harvested agricultural products and has been applied worldwide19,20. Commercially available non-toxigenic A. flavus isolates include K49 (NRRL 30797, isolated from Maize), Afla-Guard (NRRL 21882, isolated from peanuts), and AF36 (NRRL 18543, isolated from Cottonseed)21.
Here, we investigated the potential of using Aspergillus oryzae, the food grade non-toxigenic domesticated ecotype of A. flavus, as a biocontrol agent for inhibiting AFB1 production and growth of the toxigenic A. flavus strain NRRL 3357. A. oryzae is used for food fermentation (e.g. sake, miso, soy sauce, meju) and is classified as a Generally Recognized As Safe (GRAS) organism by the FDA and the WHO22,23. Fermented soy pastes produced with A. oryzae are rarely contaminated with AFs. Thus, we hypothesize that there is a strong anti-mycotic potential of A. oryzae to outcompete A. flavus in soy-based food. In this study, we have found that A. oryzae M2040 (designated as M2040 hereafter) isolated from Korean Meju (a soy brick used to make soybean paste called Doen-Jang in Korea) inhibits growth and AFB1 production by A. flavus significantly better than the widely used commercial biocontrol isolate Afla-Guard. To quantify the competitive effects of M2040, we generated a GFP-labeled A. flavus NRRL 3357 strain and used to quantify the competitive displacement of A. flavus by M2040 in peanuts. Importantly, inoculum level of M2040 as low as 1% was effective for controlling of AFB1 production and A. flavus proliferation. Additionally, cell-free culture filtrate of M2040 grown in potato dextrose broth (PDB) inhibited A. flavus germination, propagation, and AFB1 production, suggesting the presence of anti-mycotic compound(s) in the M2040 fermentate. Whole genome sequencing and comparative analyses revealed the presence of an additional 1.5 Mbp in the M2040 genome (37.9 Mbp) compared to Afla-Guard (36.4 Mbp). We identified 111 M2040 lineage specific genes arranged in several clusters that may play a role in the observed phenotypes. This report provides a systematic investigation and strong basis for the use of the GRAS fungus A. oryzae as a potential biocontrol agent for AFB1 contamination in food, and corroborates the expired patent for using certain strains of A. oryzae and A. sojae as biocontrol agents (US6027724A).
Results
Inhibition of AFB1 production by M2040
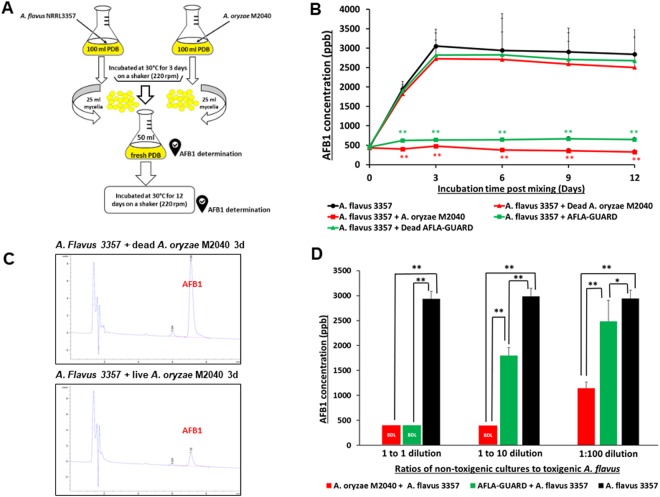
To test the central hypothesis that M2040 inhibits AFB1 production, co-culture experiments of M2040 and A. flavus NRRL 3357, and Afla-Guard and A. flavus NRRL 3357 in PDB were performed as shown in Fig. 2A. We tested various media and found that PDB resulted in equal growth rates for M2040 and A. flavus, and high level production of AFB1. As controls, a set of the 3-day old cultures of M2040 and Afla-Guard were autoclaved (dead) and mixed with the 3-day old culture of A. flavus NRRL 3357. The mixed cultures were further incubated for up to 12 additional days and the amount of AFB1 was measured every 3 days. As shown in Fig. 2B, mixing live cells of both M2040 and Afla-Guard with A. flavus 3357 effectively blocked accumulation of AFB1 throughout the incubation. HPLC chromatograms of AFB1 in 3-day post mixing cultures clearly demonstrate the differences of AFB1 levels between co-culture of A. flavus 3357 with live and dead M2040 (Fig. 2C). AFB1 inhibition rates of M2040 were 98.8% and 100% at 3 and 12 days of incubation, respectively. Afla-Guard showed AFB1 inhibition rates of 93.0% and 94% at 3 and 12 days of incubation, respectively. Autoclaved (dead) cells of M2040 and Afla-Guard did not reduce AFB1 accumulation, resulting in accumulation up to 3000 ppb. These data indicate that M2040 can inhibit AFB1 production in PDB when co-cultured.
Figure 2.
Inhibitory effects of A. oryzae M2040 on AFB1 production by A. flavus. (A) Experimental design. (B) Levels of AFB1 accumulation in a liquid co-culture media. *P < 0.05; **P < 0.01. (C) HPLC chromatograms of AFB1 at 3-day incubation of A. flavus vs dead and live M2040. Note the differences in the AFB1 peak size. (D) AFB1 accumulation and in peanut co-inoculated with M2040 and Afla-Guard and A. flavus NRRL3357 at different ratios.
To corroborate the control of AFB1 contamination by M2040 on food matrix, we inoculated 1:1, 1:10, and 1:100 ratios of M2040 vs A. flavus NRRL 3357, and Afla-Guard vs A. flavus NRRL 3357 on peanuts and examined AFB1 levels at day 5 (Fig. 2D). At a 1:1 inoculation ratio, both M2040 and Afla-Guard blocked accumulation of AFB1 (Fig. 2D) compared to A. flavus NRRL 3357 alone (black bar). Surprisingly however, at 1:10 ratio (10% of a biocontrol strain), M2040 completely inhibited AFB1 accumulation while Afla-Guard allowed AFB1 accumulation to reach ~1,750 ppb. Furthermore, even at 1:100 ratio (1% of biocontrol strain), M2040 led to over 61% inhibition of AFB1 accumulation (Fig. 2D), whereas Afla-Guard allowed only about 15% inhibition compared to A. flavus NRRL 3357 alone. Collectively, these data indicate that M2040 has a very strong biocontrol potential that is comparable or superior to Afla-Guard.
Quantification of A. flavus displacement by M2040
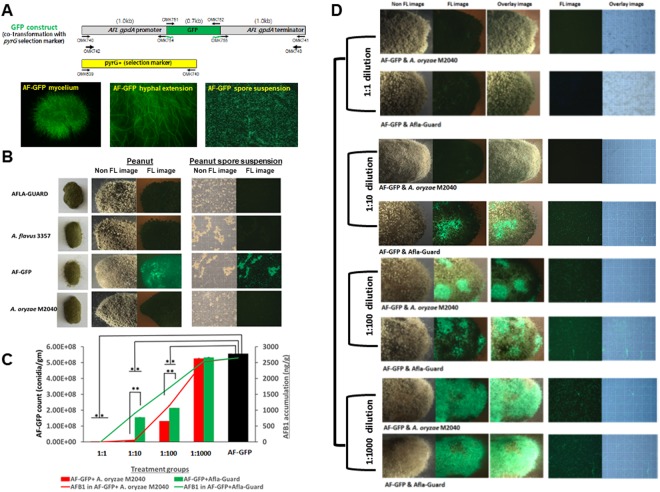
In the aforementioned experiments AFB1 inhibition experiments, it was impossible to distinguish the target and control strains when they were mixed due to their morphological similarities. Thus, in order to quantify the growth rates of isolates in co-culture, we generated several GFP labeled A. flavus NRRL 3357 strains by co-transformation, and confirmed that these transformants produced AFB1 similar to wildtype. GFP levels of one strain named AF-GFP is depicted in Fig. 3A,B.
Figure 3.
Quantitation of A. flavus displacement by A. oryzae M2040 and Afla-Guard on peanuts. (A) The GFP construct and 5 day old culture of AF-GFP showing highly fluorescent mycelia, hyphae, and conidial suspension. (B) Fluorescence (FL) and non-fluorescence images representing inoculation of control groups observed at 5 days of incubation. (C) AF-GFP conidial count and AFB1 accumulation in peanut samples co-inoculated with varying ratios of M2040 or Afla-Guard. *P < 0.05; **P < 0.01. (D) Fluorescence (FL) and non-fluorescence images of peanuts representing the treatment groups observed at 5 days of incubation. Photographs were taken in a dark room with a 1-2s exposure time.
With the successful generation of AF-GFP, we performed co-inoculation experiments with AF-GFP and M2040, and AF-GFP with Afla-Guard on peanuts with 1:1, 1:10, 1:100, and 1:1,000 ratios. As shown in Fig. 3B–D, when co-inoculated with AF-GFP at a ratio of 1:1, both M2040 and Afla-Guard completely blocked AFB1 accumulation (100% inhibition rate), and essentially no AF-GFP spores were detected at 5 days of incubation (Fig. 3D, top panel). Importantly, M2040 exhibited 99% and 65% inhibition rates at 1:10 and 1:100 ratio, respectively, whereas Afla-Guard showed inhibition rates of 60% and 51% at ratio of 1:10 and 1:100, respectively. At 1:1000 ratio, both M2040 and Afla-Guard failed to inhibit AFB1 accumulation. Importantly, the recovery of AF-GFP spores and AFB1 levels were proportional throughout these experiments. At 1:1000 ratio, the peanuts were fully covered by AF-GFP (Fig. 3D, bottom panel). This experiment resulted in the first quantitative measure for A. flavus displacement by biocontrol strains, and suggests that M2040 can outcompete a toxigenic A. flavus strain even when present at only 1%.
Cell-free culture broth of M2040 inhibits AFB1 accumulation
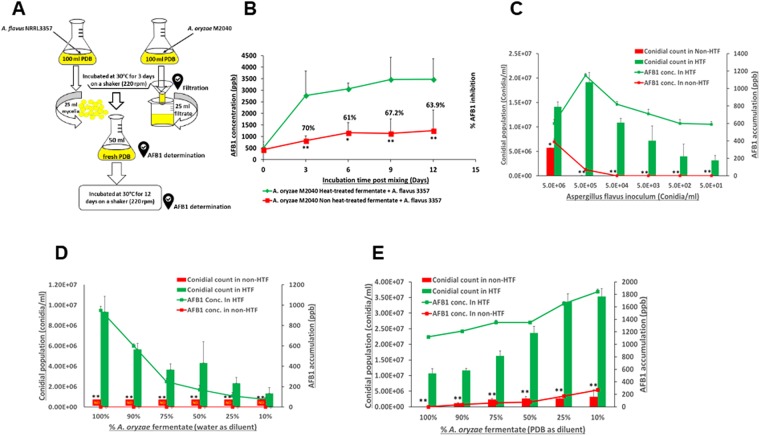
To test whether M2040 secretes unknown compound(s) into medium that confer AFB1 inhibition, we filter-sterilized (0.45 μm filter) the 8-day old culture of M2040 grown in PDB, and combined the cell-free culture with 3 day old culture of A. flavus 3357 and 50 ml of fresh PDB (Fig. 4A). As a control, a set of M2040 PBD culture filtrates were autoclaved (designated as heat-treated) and mixed with the 3 day old A. flavus culture. Autoclaving abolished the inhibitory effects of the cell-free culture (Fig. 4). Levels of AFB1 in the mixed medium were determined at 3, 6, 9 and 12 days post mixing. M2040 non-heat-treated culture filtrate was able to inhibit AFB1 accumulation at the inhibition rate of 60–70% compared to the control (heat-treated) group even at 25% levels (25 ml + 75 ml culture and PDB) (Fig. 4B).
Figure 4.
Effects of cell-free culture filtrate of A. oryzae M2040. (A) Experimental design. (B) Time course of the AFB1 accumulation in a mixed liquid of M2040 cell-free culture fermentate and A. flavus mycelial cells. *P < 0.05. **P < 0.01. (C) Conidial numbers and AFB1 production in HT and non HT M2040 fermentate inoculated with different conidial numbers of A. flavus. **P < 0.05. **P < 0.01. (D) Conidial count and AFB1 production in different concentrations of HT and non HT A. oryzae fermentate inoculated with 5 × 105 A. flavus conidia/ml. *P < 0.05. **P < 0.01. ND: Conidia were not detected under a microscope. Fermentate was diluted in sterile distilled water. (E) Conidial count and AFB1 production in different concentrations of HT and non HT M2040 fermentate inoculated with 5 × 105 A. flavus conidia/ml. **P < 0.05. **P < 0.01. Fermentate was diluted in fresh PDB.
To test the extent of the cell free culture filtrates ability to inhibit propagation and AFB1 production in A. flavus, we inoculated varying numbers (5 million to 50) of A. flavus NRRL 3357 spores into 2 mL of 100% culture filtrates, non-heat-treated, and heat-treated groups, and quantified the recovery of A. flavus spores and AFB1 levels (Fig. 4C). Non-heat-treated cell-free culture of M2040 effectively inhibited A. flavus growth and propagation as indicated by low conidial counts compared to the control group. It is important to note that active M2040 culture filtrate could effectively block the growth and AFB1 production even with 500,000 inoculated A. flavus spores (Fig. 4C). The maximum dilution levels of M2040 filtrate in water and PDB were further examined for inhibition of 500,000 A. flavus spores. As shown in Fig. 4D,E, the non-heat-treated culture fermentate in concentrations of 10%, 25%, 50%, 75%, and 100% (V:V in water or PDB) significantly inhibited A. flavus NRRL 3357 spore recovery and AFB1 production. These results indicate that the food grade (PDB) culture broth of the GRAS fungus M2040 can be effectively used as a safe agent to control A. flavus contamination and AF production.
Whole genome sequencing and comparative analyses of M2040 and Afla-Guard
In order to verify that M2040 is indeed an A. oryzae strain lacking the ability to produce AFs, genomic DNA of M2040 was isolated, and genome sequencing was performed as previously described24. Though a draft genome sequence of Afla-Guard is available25 we sequenced its genome to obtain sufficient and comparable coverage for our comparative analysis. These genomes were also used to investigate potential genetic differences between A. oryzae M2040 and A. flavus Afla-Guard that may underlie their varying ability to inhibit AF production and proliferation of AF producing isolates. The M2040 and Afla-Guard genomes were both sequenced to >50X coverage and assembled into 1,479 and 1,766 scaffolds, with cumulative assembly sizes of 37.9 and 36.4 Mb, and N50 values of 135.8 Kb and 47.4 Kb, respectively. In silico gene prediction yielded 11,782 and 11,489 gene models for the M2040 and Afla-Guard genomes, respectively. Using antiSMASH 3.0, 46 and 42 secondary metabolic gene clusters were predicted in the M2040 and Afla-Guard genomes, respectively.
Phylogenetic analysis
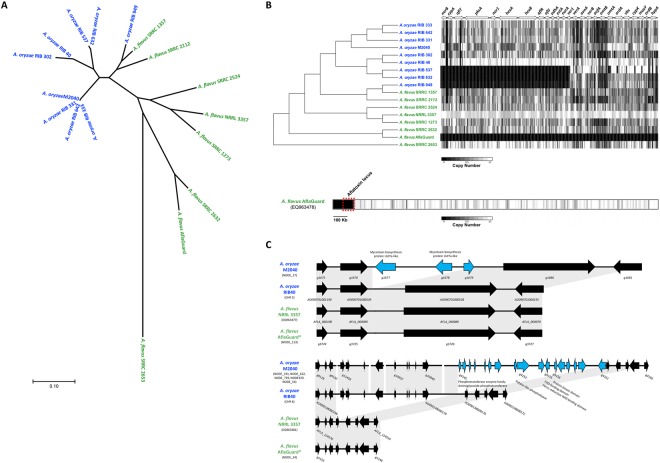
The phylogeny of the 17 A. oryzae and A. flavus genomes was inferred using 305,543 whole-genome single nucleotide polymorphisms (SNPs: Fig. 5A). Consistent with earlier work26, our analysis suggests that the A. oryzae isolates, including A. oryzae M2040, are monophyletic and that A. flavus SRRC 1357 and SRRC 2112 show a closer relationship to A. oryzae than to other A. flavus isolates (Fig. 5A). Our results suggest that A. oryzae M2040 is extremely closely related to three Japanese sake derived isolates of A. oryzae, while A. flavus Afla-Guard is most closely related to A. flavus SRRC 2632 (Fig. 5A); a strain capable of producing cyclopiazonic acid, AFB1, and AFB2.
Figure 5.
Comparative genome analyses of A. oryzae M2040 and Afla-Guard. (A) Phylogenetic relationship of A. oryzae and A. flavus isolates. An unrooted phylogeny was generated using the Maximum Likelihood method from 305,543 SNPs across the entire genome. Branch lengths represent the number of substitutions per site. All bootstrap values were ≥94%. Blue and green taxa labels represent A. oryzae and A. flavus, respectively. (B) Deletion profiles in the AF gene cluster. The chromosomal architecture of the AFB1 gene cluster relative to the A. flavus NRRL 3357 genome is shown above the heatmap, where arrows represent genes, and their orientations represents the direction of transcription. The heatmap represents copy number estimates for each non overlapping 100 bp bin across the AF gene cluster. Black and white represent copy numbers of 0 and ≥1, respectively. Bottom bar shows the Afla-Guard heatmap depicting deletions relative to the AF gene cluster containing A. flavus NRRL 3357 EQ963478 scaffold. Windows represent copy number estimates for each non-overlapping 10 kb bin across the scaffold. The chromosomal region containing the AF cluster is outlined with a red box. (C) Genome architecture of examples M2040 lineage specific genes clusters. Microsynteny of regions covering a three gene (top) and 17 gene (bottom) cluster unique to the M2040 genome in comparison to A. oryzae RIB 40, A. flavus NRRL 3357 and Afla-Guard. For each cluster arrows represent genes, and their orientations represents the direction of transcription. Genes colored black are conserved in at least 2 isolates, while genes colored light blue are unique to the M2040 genome. Gray blocks represent genomic regions exhibiting sequence similarity between isolates. Chromosome, or scaffold identifiers containing these loci are listed under each isolate. Gene identifiers are listed for each gene in panel A, and for the range of genes in panel.
AF gene cluster variation
Using a read depth approach, we estimated copy number for each non-overlapping 100 bp bin across the AF biosynthetic gene cluster. A. oryzae M2040 possesses a number of deletions, including the intergenic region between the divergently transcribed norB and cypA genes27. norB is an aryl alcohol dehydrogenase, and cypA is a cytochrome P450 monooxygenase28. Both genes are involved in aflatoxin G formation. Isolates with this deletion do not express norB or cypA26. The entire AF biosynthetic gene cluster is deleted in A. flavus Afla-Guard (Fig. 5B). Closer examination of the A. flavus Afla-Guard genome reveals a ~155 Kb deletion beginning at the AF gene cluster and extending to the end of the chromosome (Fig. 5C).
Identification of A. oryzae M2040 lineage specific genes
Using a conservative BLAST based approach, lineage specific genes were identified in A. oryzae RIB 40, A. oryzae M2040, A. flavus NRRL 3357, and A. flavus Afla-Guard, with special focus given to A. oryzae M2040. We identified 111, 58, 140, and 111 lineage specific genes in the A. oryzae M2040, A. oryzae RIB 40, A. flavus NRRL 3357, and A. flavus Afla-Guard genomes, respectively. PFAM domains were predicted for 55 of the 111 A. oryzae M2040 genes. Interestingly, we identified two genes encoding proteins with heterokaryon incompatibility domains, one gene encoding a protein with an aminoglycoside phosphotransferase domain which confers resistance to various aminoglycosides, as well as several other genes encoding transporters, transcription factors, and protein kinases (Table 1).
Table 1.
PFAM and InterPro annotation for A. oryzae M2040 lineage specific genes. The A. oryzae M2040 lineage specific genes and predicted domains.
| Scaffold | Gene ID | PFAM Domain | InterPro Classification |
|---|---|---|---|
| NODE_14 | g1437 | Carbon-nitrogen hydrolase | Carbon-nitrogen hydrolase |
| NODE_17 | g1677 | Domain of unknown function (DUF3328) | Mycotoxin biosynthesis protein UstYa-like |
| NODE_17 | g1678 | Domain of unknown function (DUF3328) (x2) | Mycotoxin biosynthesis protein UstYa-like (x2) |
| NODE_35 | g3027 | Ankyrin repeat (x2) | Ankyrin repeat (x2) |
| Ankyrin repeats (many copies) | AAA + ATPase domain | ||
| AAA domain | Ankyrin repeat-containing domain | ||
| Ankyrin repeats (3 copies) | |||
| NODE_54 | g4145 | Phosphotransferase enzyme family | Aminoglycoside phosphotransferase |
| NODE_54 | g4153 | Patatin-like phospholipase | Patatin-like phospholipase domain |
| NODE_54 | g4155 | FAD-binding domain | FAD-binding 8 |
| Ferric reductase NAD binding domain | Ferric reductase, NAD binding domain | ||
| NODE_54 | g4156 | Protein kinase domain | Protein kinase domain |
| NODE_54 | g4157 | Protein of unknown function (DUF3723) | Protein of unknown function DUF3723 |
| NODE_54 | g4161 | Protein of unknown function (DUF3435) | Protein of unknown function DUF3435 |
| NODE_73 | g5191 | WD domain, G-beta repeat (x8) | WD40 repeat (x8) |
| NODE_193 | g9143 | SET domain | SET domain |
| NODE_194 | g9174 | AAA domain (x2) | |
| NODE_194 | g9176 | Argonaute linker 1 domain | Piwi domain |
| Piwi domain | Argonaute, linker 1 domain | ||
| PAZ domain | PAZ domain | ||
| Helicase conserved C-terminal domain | Helicase, C-terminal | ||
| N-terminal domain of argonaute | Protein argonaute, N-terminal | ||
| Dicer dimerisation domain | Dicer dimerisation domain | ||
| Ribonuclease III domain (x2) | Ribonuclease III domain (x2) | ||
| NODE_289 | g10541 | Protein kinase domain | Protein kinase domain |
| Ulp1 protease family, C-terminal catalytic domain | Ulp1 protease family, C-terminal catalytic domain | ||
| NODE_290 | g10546 | Man1-Src1p-C-terminal domain | Man1-Src1p-C-terminal domain |
| NODE_326 | g10914 | Ferric reductase like transmembrane component | Ferric reductase transmembrane component-like domain |
| FAD-binding domain | FAD-binding 8 | ||
| NODE_326 | g10915 | Phosphorylase superfamily | Nucleoside phosphorylase domain |
| NODE_326 | g10916 | Protein kinase domain | Protein kinase domain |
| NODE_341 | g11019 | Protein of unknown function (DUF3645) | Protein of unknown function DUF3645 |
| Protein of unknown function (DUF3638) | Protein of unknown function DUF3638 | ||
| NODE_369 | g11222 | Heterokaryon incompatibility protein (HET) | Heterokaryon incompatibility |
| NODE_371 | g11231 | Shwachman-Bodian-Diamond syndrome (SBDS) protein | Ribosome maturation protein SBDS, N-terminal |
| ATP synthase alpha/beta chain, C terminal domain | ATP synthase, alpha subunit, C-terminal | ||
| ATP synthase alpha/beta family, beta-barrel domain | ATPase, F1/V1/A1 complex, alpha/beta subunit, N-terminal domain | ||
| ATP synthase alpha/beta family, nucleotide-binding domain | ATPase, F1/V1/A1 complex, alpha/beta subunit, nucleotide-binding domain | ||
| NODE_371 | g11232 | V-type ATPase 116 kDa subunit family | V-type ATPase, V0 complex, 116 kDa subunit family |
| NODE_378 | g11269 | Man1-Src1p-C-terminal domain | Man1-Src1p-C-terminal domain |
| NODE_378 | g11273 | Methyltransferase domain | Methyltransferase type 11 |
| NODE_384 | g11303 | Heterokaryon incompatibility protein (HET) | Heterokaryon incompatibility |
| NODE_404 | g11391 | ATPase family associated with various cellular activities (AAA) | ATPase, AAA-type, core |
| NODE_416 | g11434 | Glycosyl hydrolase family 10 (x2) | Glycoside hydrolase family 10 domain (x2) |
| NODE_417 | g11436 | Glycosyl hydrolases family 32 N-terminal domain | Glycosyl hydrolase family 32, N-terminal |
| NODE_417 | g11437 | Sugar (and other) transporter | Major facilitator, sugar transporter-like |
| NODE_437 | g11494 | Alcohol dehydrogenase GroES-like domain | Alcohol dehydrogenase, N-terminal |
| Zinc-binding dehydrogenase | Alcohol dehydrogenase, C-terminal | ||
| NODE_437 | g11495 | Eukaryotic elongation factor 5 A hypusine, DNA-binding OB fold | Translation elongation factor, IF5A C-terminal |
| Elongation factor P (EF-P) KOW-like domain | Translation elongation factor, KOW-like | ||
| NODE_450 | g11526 | Cation transporter/ATPase, N-terminus | Cation-transporting P-type ATPase, N-terminal |
| NODE_450 | g11527 | MAC/Perforin domain | Membrane attack complex component/perforin (MACPF) domain |
| NODE_454 | g11536 | Ankyrin repeats (3 copies) (x3) | Ankyrin repeat-containing domain (x3) |
| Ankyrin repeats (many copies) | |||
| Fungal specific transcription factor domain | Transcription factor domain, fungi | ||
| AAA domain | AAA + ATPase domain | ||
| NODE_468 | g11564 | Protein of unknown function (DUF3435) | Protein of unknown function DUF3435 |
| NODE_468 | g11565 | Protein kinase domain | Protein kinase domain |
| NODE_472 | g11572 | Protein of unknown function (DUF3435) | Protein of unknown function DUF3435 |
| NODE_476 | g11576 | NmrA-like family | NmrA-like domain |
| NODE_479 | g11582 | Glycosyltransferase family 25 (LPS biosynthesis protein) | Glycosyl transferase, family 25 |
| NODE_479 | g11583 | Glycosyltransferase sugar-binding region containing DXD motif | Glycosyltransferase, DXD sugar-binding motif |
| NODE_486 | g11594 | Ankyrin repeats (3 copies) (x2) | Ankyrin repeat-containing domain (x2) |
| NODE_486 | g11595 | Glycosyl transferase family 8 | Glycosyl transferase, family 8 |
| NODE_494 | g11605 | Phosphotransferase enzyme family | Aminoglycoside phosphotransferase |
| NODE_498 | g11612 | NACHT domain | |
| NODE_501 | g11617 | Clostridium epsilon toxin ETX/Bacillus mosquitocidal toxin MTX2 | Clostridium epsilon toxin ETX/Bacillus mosquitocidal toxin MTX2 |
| NODE_513 | g11632 | Haloacid dehalogenase-like hydrolase | |
| Cation transporting ATPase, C-terminus | Cation-transporting P-type ATPase, C-terminal | ||
| NODE_541 | g11662 | T5orf172 domain | Bacteriophage T5, Orf172 DNA-binding |
| NODE_573 | g11683 | LysM domain (x3) | LysM domain (x3) |
| NODE_578 | g11688 | Sodium/hydrogen exchanger family | Cation/H+ exchanger |
| NODE_612 | g11709 | Sugar (and other) transporter | Major facilitator, sugar transporter-like |
| NODE_654 | g11732 | Phosphorylase superfamily | Nucleoside phosphorylase domain |
| NODE_691 | g11743 | Peptidase family M49 | |
| NODE_713 | g11751 | BTB/POZ domain | BTB/POZ domain |
| NODE_860 | g11779 | E1-E2 ATPase |
Genes containing 2, 3, and 8 identical PFAM or InterPro domain are denoted as x2, x3, and x8, respectively. Genes g1679, g4146, g4147, g4148, g4149, g4150, g4151, g4152, g4154, g4158, g4159, g4160, g9173, g9175, g9177, g10449, g10540, g10542, g10543, g10544, g10545, g10547, g10913, g10943, g10944, g10945, g10946, g10947, g11018, g11020, g11270, g11271, g11272, g11307, g11438, g11487, g11503, g11504, g11566, g11588, g11660, g11673, g11674, g11681, g11682, g11699, g11707, g11714, g11716, g11729, g11733, g11736, g11760, g11765, g11777, and g11780 contained no predicted PFAM or InterPro classification.
In A. oryzae M2040, 63% of lineage specific genes were found in clusters of two or more genes, with an average cluster size of ~4 genes and the largest cluster containing 17 genes. Of note, we identified a cluster of three lineage specific genes in A. oryzae M2040 in which two of the genes contain mycotoxin biosynthesis protein UstYa-like domains (Fig. 5C). This protein domain is involved in the production of toxic cyclic peptides. Additionally, we identified a 17 gene cluster with varying genome architecture between A. oryzae M2040, A. oryzae RIB 40, and the two A. flavus isolates (Fig. 5C). Most of these genes encode proteins annotated as hypothetical proteins in other organisms, though BLAST searches against the RefSeq non-redundant protein database, PFAM domain prediction, and InterPro classification revealed proteins annotated as phosphotransferase family protein, patatin/phospholipase A2-related, tyrosine-protein kinase, and casein kinase family protein.
Discussion
The use of atoxigenic A. flavus to control AFs was demonstrated in 1990s (Peter Cotty USDA Agricultural Research Service) in Arizonian cotton fields18. However, studies dated back to 1965 found that AF levels can be reduced by co-cultivating toxigenic A. flavus strains with certain fungi or bacteria29,30. This biocontrol strategy is currently the most widely used method for reducing contamination levels of AFs in some crops. Nevertheless, there are many challenges facing this strategy at both short and long-term31,32. One of the major drawbacks of the biocontrol strategy is a potential risk of introducing a heavy dose of A. flavus strains that could alter the soil microbiome populations especially with global warming. The inherent diversity of A. flavus populations makes a biocontrol strategy more difficult because A. flavus populations differ in their abilities to produce AFs and other toxic secondary metabolites31. To address these potential issues, we tested the potential use of A. oryzae M2040 as a biocontrol agent.
Prior to the beginning of the studies, we tested the inhibitory activity of 10 A. oryzae strains and found that M2040 most significantly inhibited AF production and A. flavus growth. Our data indicates that AFB1 is not detected in PDB when equal number of spores of A. oryzae and A. flavus are co-inoculated. Therefore, in order to test the precise inhibitory effects, we cultured each strain separately for 3 days and mixed equal volume (25 mL each) of the fungal cell aggregates (mycelia) in new PDB medium (50 mL) and quantified submerged growth and AFB1 production. Additionally, Afla-Gurad® was selected as a positive control based on its effective use in vitro and in the field to control AFs contamination. Both live cells of A. oryzae and Afla-Gurad® significantly inhibited AFB1 accumulation over a range of laboratory conditions indicating that fungal cell-cell interactions may act as an additional control factor mediating the inhibitory process.
To track the competitive effect of nontoxigenic strains, we co-inoculated M2040 and Afla-Guard at varying inoculum levels with a transgenic A. flavus NRRL 3357 (AF-GFP) strain expressing GFP on peanut samples. AFB1 was measured as well. Previous experiments indicated that Afla-Guard can result in more than 85% reduction in AFs33,34. At 1:1 ratio, the AF-GFP fluorescence in the co-inoculated peanut kernels was significantly reduced, and AFB1 was not detected by when co-cultured with M2040 or Afla-Guard. Since the inoculum size is the same for both non-toxigenic and toxigenic strains, the faster growing isolate (biocontrol strains) should outcompete the other isolate (toxin-producing strain). However, when both biocontrol agents were further diluted to 10% levels, M2040 clearly showed superior reduction (87%) in total AFB1 accumulation compared to Afla-Guard (60.6%). Abbas et al.21 reported 58% and 83% in AF reduction when they applied the non-aflatoxigenic A. flavus K49 strain in the two-corn fields at a ratio of 10-fold dilution. However, when K49 was applied at equal inoculum size with toxigenic one, AFs were reduced by 86% and 96%. It has been hypothesized that displacement of the toxigenic strains occurs simply by the biocontrol strains superior ability to better sequester nutrients. However, using different biocontrol strains has resulted in variable reduction rates of AFs, suggesting that other unknown factors may be involved for the biocontrol mechanism such as the production of extracellular compounds that might inhibit AFB1 production. M2040 showed significant activity for AFB1 reduction (50%) even at 100-fold dilution compared to Afla-Guard (26%), however, at 1000-fold dilution, both biocontrol agents resulted in AFB1 reduction of less than 10%.
When cell free culture fermentate of M24040 was tested for inhibiting the mycelial growth and AFB1 production of A. flavus, it was found to be highly effective against AFB1 production by reducing levels by more than 70%. Furthermore, our broth microdilution experiments revealed that the fermentate was found to be highly effective in inhibiting conidial germination and AFB1 production. The inhibitory activity of the culture fermentate was observable even at 10% concentration and these effects were abolished by autoclaving, but not by treating with proteinase K. These results suggest the non-protein nature of the active substances in the fermentate.
To better understand the phenotypic differences between M2040 and Afla-Guard through a broader evolutionary context, we sequenced the genomes of both isolates. This analysis yielded several key findings. First, we note that Afla-Guard is nested within an aflatoxigenic clade of A. flavus, and most closely related to the clinical isolate A. flavus SRRC 2632 which is capable of producing AF and cyclopiazonic acid (CPA) (Fig. 5A). Our read depth analysis confirms that the Afla-Guard genome lacks the ability to produce AF and CPA because of a ~155 Kb deletion spanning the gene clusters encoding both of these secondary metabolites35 (Fig. 5B). Additionally, our phylogenetic analysis revealed that M2040 is nearly identical to three Japanese strains isolated from miso (RIB 331 and RIB 333) and sake (642) (Fig. 5A).
Genome sequencing of M2040 and Afla-Guard also allowed us to conduct comparative genomic analysis to identify potential gain-of-function genes that might be associated with the inhibitory ability of M2040. We hypothesized that certain genes involved in unknown anti-fungal products may curtail growth and AF production of A. flavus. We identified 111 lineage-specific genes in the M2040 genome, including several genes that may be involved in biosynthesis of toxin-like products. For instance, gene g11617 was annotated by a “Clostridium epsilon toxin ETX/Bacillus mosquitocidal toxin MTX2” domain (Table 1). BLAST searches against the NCBI non-redundant protein database reveal the presence of this gene in the A. oryzae BCC7051 genome36, along with only two other significant hits to the Talaromyces cellulolyticus Y-94 genome37 (E-value = 7e−17) and the Botrytis cinerea BcDW1 (E-value = 8e−12)38. We also identified two neighboring genes (g1677 and g1678) that have the Interpro classification “Mycotoxin biosynthesis protein UstYa-like” (Fig. 5C). ustYa is an oxidase involved in the production of ustiloxins and homologs have been identified in A. flavus NRRL 335739 and A. oryzae RIB 40. However, homologs to the A. oryzae M2040 genes g1677 and g1678 are not present in A. oryzae RIB 40, A. flavus NRRL 3357, or A. flavus Afla-Guard, and BLAST searches against the NCBI non-redundant protein database show a patchy distribution across Ascomycota including A. oryzae BCC705136, A. bombycis40, A. udagawae41, Xylona heveae42, Penicillium oxalicum43, and Penicillium nordicum44. Moving forward, defining the transcriptional landscape of M2040 in co-culture will allow us to further narrow in on the genes involved in inhibiting AF production and growth.
In conclusion, our studies raise the idea of potentially using a food grade A. oryzae strain as a potent biocontrol agent to reduce A. flavus growth and AF contamination. Additionally, A. oryzae cell free PDB fermentate could be employed as a valuable biocontrol agent. To the best of our knowledge, our report is the first study systematically showing the advantages of using GRAS fungus, and its fermentate to control A. flavus growth and AFs contamination. These results, along with further studies, will eventually provide a GRAS product(s) that can be used as a natural anti-fungal and anti-AF agent.
Materials and Methods
Fungal strains and culture conditions
The aflatoxigenic strain A. flavus NRRL3357 was used as a high AFB1 producer45. A. flavus NRRL3357 labeled with green fluorescent protein (AF-GFP) was developed and used in this study. Bright fluorescence was observed for mycelia and hyphae of the AF-GFP (Fig. 3A). A. flavus NRRL 21882 (Afla-Guard) and A. oryzae M2040 (isolated from Meju, Korea) were used as non-aflatoxigenic strains. All strains were maintained on potato dextrose agar (PDA) medium (containing 4 g potato starch, 20 g glucose, and 15 g agar in 1 L of distilled water) at 4 °C. To prepare inoculum, Aspergillus cultures were grown on PDA for 7 days at 30 ± 2 °C. Spores were harvested from individual cultures on PDA using 0.1% Tween-80 solution. Asexual spores (conidia) were counted with a hemocytometer and numbers were adjusted to 5 × 107 conidia/mL with water. Fungal spore suspensions were stored at 4 °C and used within 1 week of preparation.
Generation of GFP labeled A. flavus NRRL 3357 strains
The oligonucleotides used in this study are listed in Table 2. Double joint PCR (DJ-PCR) was used to generate AF-GFP strains46. To generate the PCR amplicon of the GFP Open Reading Frame (ORF), the primer pair OMK751; OMK752 was used from pFNO3 (vectors provided by Dr. P. N Keller) plasmid DNA. Both 5′ and 3′ flanking regions of the glyceraldehyde phosphate dehydrogenase (gpdA) gene was amplified from genomic DNA of A. flavus NRRL 3357 using OMK740; OMK754 and OMK755; OMK741. The final DJ-PCR of AF-GFP construct was amplified with OMK742; OMK743. The A. flavus pyrG+ marker was amplified with the primer pair OMK639; OMK640. The final DJ-PCR construct and the A. flavus pyrG+ marker amplicons were co-introduced into A. flavus NRRL3357.5 (pyrG-). Protoplasts were generated using the Vinoflow FCE lysing enzyme as described (Novozymes)47. Fungal transformants were isolated and confirmed by PCR followed by restriction enzyme digestion46. At least three independent AF-GFP strains were isolated and confirmed for AFB1 production.
Table 2.
Oligonucleotides used in this study.
| Name | Sequence (5′ → 3′) | Purpose |
|---|---|---|
| OMK740 | TCACTGAAAAAGAGCTAAGACTA | 5′ flanking of AFL gpdA |
| OMK741 | TCCCATGACAGTGTCTTCGT | 3′ flanking of AFL gpdA |
| OMK742 | ACCCCAGTACAGTTTCATGCAA | 5′ nest of AFL gpdA |
| OMK743 | TTGCGCAGAAGCCTAGACAAGTC | 3′ nest of AFL gpdA |
| OMK751 | ATGAGTAAAGGAGAAGAACTT | 5′ GFP ORF |
| OMK752 | TTTGTATAGTTCATCCATGC | 3′ GFP ORF |
| OMK754 | AAGTTCTTCTCCTTTACTCATTGTTTAGATGTGTCTGTTG | 3′ AFL gpdA with GFP taila |
| OMK755 | TGGCATGGATGA ACTATACAAAAAGTCATACCTAACAAGTGCT | 5′ AFL gpdA with GFP taila |
| OMK639 | TCGAGAGATGAGGGCTGCCAGCA | 5′ AFL pyrG marker |
| OMK640 | CAGAAGAAAAGGATGATCAATAC | 3′ AFL pyrG marker |
aTail sequence is in italic.
Reagents and chemicals
AFB1 standard was purchased from Sigma-Aldrich (U.S.). Water, methanol, acetonitrile chloroform were purchased from Fisher Chemical (U.S.). All solvents were of HPLC-grade. Membrane filters (47 mm × 0.45 μm) and syringe filters (13 mm × 0.2 μm) were obtained from Millipore (U.S.).
HPLC analysis of AFB1
Extraction of AFB1 from liquid culture media
AFB1 was extracted from submerged media by liquid-liquid extraction. Briefly, 2 ml of the fungal culture broth was mixed with equal volume of chloroform in 15-ml centrifuge tube and vortexed for 60 sec, left at room temperature for 5 min, then vortexed again for 60 sec. The mixture was then centrifuged for 5 min at 5000 g. Two ml of the lower layer was transferred to a new glass vial. The chloroform extracts were evaporated to complete dryness under a gentle stream of air. The dried extracts were reconstituted with 1 ml methanol. All samples were filtered into HPLC vials through 0.2 μm syringe filter prior to HPLC analysis.
Measurement of conidia and AFB1 from peanut
Extraction of AFB1 from peanuts was performed as described48 with slight modifications. Two peanut cotyledons were placed in a 50-ml Falcon tube containing 5 ml of 0.1% Tween 80 and each tube was vortexed thoroughly. To check the conidial number, 1 mL was transferred from each sample into an Eppendorf tube and spores were counted. Next, 5 mL of acetone was added to the remaining samples, followed by shaking for 15 min in a rotary shaker at 150 rpm. Samples were kept standing for 5 min at room temperature, 5 ml of chloroform was then added, and samples were agitated for 15 min at 150 rpm. Samples were left to stand for an additional 5 min at room temperature. The organic lower layer was collected by centrifugation of samples for 10 min at 5000 g, transferred to a new tube and dried under gentle stream of air. The dried extracts were reconstituted with 1 ml methanol. All samples were filtered into HPLC vials through 0.22 μm disposable syringe filter prior to HPLC analysis.
Chromatographic conditions
Samples were analyzed for AFB1 using a model 1100 HPLC system consisting of a degasser, an autosampler, a quaternary pump, and a diode array (DAD) detector (Agilent). Samples were eluted at a wave length of 365 nm with a mobile phase of H2O:CH3OH:CH3CN (50:40:10) at a flow rate of 0.8 ml/min. The mobile was degassed and filtered through membrane filter (47 mm × 0.45 μm) prior to use. The separation was performed via a Zorbax Eclipse XDB-C18 4.6 mm × 150 mm, 3.5 μm column. The injection volume was 10 μl. AFB1 peaks area were recorded and integrated using ChemStation software (Agilent). The limit of AFB1 detection was 1 ppb.
Determining the effect of A. oryzae M2040 on AFB1 production in submerged culture
Conidia (5 × 107) of A. oryzae M2040 and A. flavus NRRL 3357 were separately inoculated into PDB (100 ml) in 250 ml Erlenmeyer flask and incubated at 30 ± 2 °C with shaking at 220 rpm. After 3 days, 25 ml of A. oryzae and 25 ml of A. flavus mycelia were transferred to a new 250 ml Erlenmeyer flask containing 50 ml of fresh PDB. This co-cultured mixture (100 ml) was incubated for 12 days under the same culture conditions mentioned above. AFB1 concentration in the culture medium was assayed every three days. The following three controls were used in this study; 1) Co-cultured mixture of A. flavus and dead cells of A. oryzae M2040 heat-treated by autoclaving at 121 °C and 15 psi for 20 min, 2) A. flavus NRRL 3357 only, 3) Co-cultured mixture of A. flavus NRRL 3357 and A. flavus Afla-Guard. All treatments were tested in triplicate flasks and the experiment was performed three time.
Determining the effect of A. oryzae M2040 on A. flavus growth and AFB1 production in peanuts
We tested the effect of varying inoculation ratios of A. oryzae M2040, A. flavus Afla-Guard, and our toxigenic GFP-labeled strain of A. flavus NRRL 3357 (AF-GFP) on AFB1 accumulation and sporulation 5 days after inoculation. This experiment was performed in triplicate for each treatment group and repeated twice.
Preparation of peanut samples
Peanut infection procedure was performed as described previously49 with some modifications. Mature peanut seeds were obtained by local market and prepared by removing the exterior layer. Two cotyledons were separated, and the embryo was removed gently. Then, cotyledons (0.5 g) were surface sterilized by placing them in beaker containing 0.05% NaClO in sterile water for 2 min. Then the cotyledons were washed by placing them in a new beaker containing sterile distilled water for 1 min, followed by a 10-second wash with 70% ethanol in a new beaker. A final washing step with sterile distilled water for 2 min was performed to ensure complete removal of detergents. The cotyledons were dried completely for at least 2 hours under aseptic condition until the time of infection.
Peanut infection
Peanut cotyledons were allocated into four treatment groups. Group 1 was inoculated with 100,000 spores of AF-GFP alone, which served as a control. Group 2 was co-inoculated with 1:1 (105:105), 1:10 (104:105), 1:100 (103:105), and 1:1000 (102:105) of A. oryzae M2040 relative to 105 spores of AF-GFP. Group 3 was inoculated with 1:1 (105:105), 1:10 (104:105), 1:100 (103:105), and 1:1000 (102:105) of A. flavus Afla-Guard relative to 105 spores of AF-GFP. Group 4 was treated with water (mock inoculation). For all treatments, 10 peanut cotyledons were used for each plate in triplicate. Cotyledons were placed in petri dishes lined with 3 pieces of moist filter paper and a water reservoir (lid of a 50 ml centrifuge tube containing 2 ml of sterile water) to maintain high humidity. Cotyledons were incubated for 5 days at 30 °C.
Determination of conidia number and AFB1 accumulation
AFB1 was extracted from peanut cotyledons as described above and calculated for all treatment groups. In order to determine the extent of AF-GFP growth in peanut and the percentage of toxigenic strain reduction, conidia of AF-GFP were counted in all groups by hemocytometer and the microscopy images were taken using a Zeiss Axio Observer D4 Fluorescence Microscope with Achromat S 1, 5x FWD 28 mm lenses (Carl Zeiss, Germany).
Determining the effects of cell free culture fermentate of A. oryzae M2040 on A. flavus growth and AFB1 production
The effects of cell free culture fermentate on AFB1 production by A. flavus was studied in 250 ml Erlenmeyer flasks under submerged culture condition. Conidia (5 × 107) of A. oryzae and A. flavus were inoculated separately in 100 ml PDB and incubated at 30 ± 2 °C with shaking at 220 rpm. After 3 days, 25 ml of A. oryzae M2040 cell free culture fermentate was transferred to a new flask containing 50 ml of fresh PDB and, at this time, 25 ml of A. flavus mycelia was also added to this flask. This mixture was incubated for 12 days under the same culture conditions mentioned above and the AFB1 concentrations in the culture medium were assayed every 3 days of incubation. Autoclaved (heat-treated) cell free culture fermentate was used as a control. All treatments were tested in triplicate and the experiment was repeated twice.
Production of the cell free culture fermentate
Conidia (5 × 108) of A. oryzae M2040 were inoculated into 1000 mL of PDB in a 2 L Erlenmeyer flasks and incubated at 30 ± 2 °C for 8 days with shaking at 220 rpm. The mycelia were separated from the culture broth by filtration with four layers of Miracloth (MilliporeSigma) and the cell free culture fermentate was obtained by filtering through a 0.2 μm PES filter unit (Thermo Scientific, USA). The fermentate was kept at 4 °C and used within one month of production. Portions of the fermentate were treated with proteinase-K (BDH Biochemicals).
Testing the effect of cell-free culture fermentate on the growth and germination of A. flavus at different conidia counts
Using a 24-well microdilution plate, 2 ml of the fermentate was loaded into 6 wells and A. flavus conidia were kept in the first well at 5 × 106 conidia/ml final concentration. Ten-fold serial dilutions were made to 5 × 101 conidia/ml. Heat treated fermentate served as a control. The plate was incubated at 30 ± 2 °C for 5 days. Samples were harvested, conidia counted with a hemocytometer, and AFB1 levels were estimated. All treatments were tested in triplicate wells and repeated at least three times.
Testing different concentrations of cell-free culture fermentate on the growth and germination of A. flavus
Cell-free culture fermentate was diluted in sterile distilled water or PDB to give final concentration (v/v) of 100%, 90%, 75%, 50%, 25%, and 10%. Heat-treated cell-free culture fermentate was prepared as a control. Two ml of treatment groups were loaded into the well and A. flavus was inoculated in all wells at a final concertation of 5 × 105 conidia/ml. The plate was incubated at 30 ± 2 °C for 5 days. Samples were harvested, conidia were counted with a hemocytometer, and AFB1 levels were quantified. All treatments were tested in triplicate wells and repeated at least three times.
Genome Sequencing and Assembly of A. oryzae M2040 and Afla-Guard
Genomic DNA was extracted as previously described24. A paired-end 152-bp Illumina library was constructed from genomic DNA and sequenced at ProteinCT (Madison, Wisconsin). Illumina sequence data were first deduplicated using Tally50. Next, trim_galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to trim reads at bases with quality scores < 30. Trimmed read pairs, with at least 1 read < 50 bp were discarded. Trim_galore was also used to removed residual adapter sequences from reads, using the conservative parameter “stringency = 1”. Lastly, this set of deduplicated, quality trimmed, and adapter trimmed reads were error corrected using SPAdes version 3.10.051, resulting in a high-quality dataset of 15,376,723 paired-end reads, representing ~115X coverage for M2040, and 8,042,928 paired-end reads, representing ~53X coverage for Afla-Guard. Both genomes were assembled using SPAdes version 3.10.051 using the “careful” mismatch mode and k-mers sizes of 25, 35, 45, 55, 65, 78, 85, and 95. antiSMASH 3.0 was used to predict secondary metabolic encoding gene clusters in the A. oryzae M2040 and A. flavus Afla-Guard genomes52.
Phylogenetic analysis
We examined the evolutionary relationships of A. oryzae M2040, Afla-Guard, and previously sequenced isolates26 using an alignment of 305,543 SNPs collected across the entire genome. SNPs were collected using the Phylogenetic and Molecular Evolution (PhaME) analysis tool on assembled genomes. All genome assemblies were performed using SPAdes version 3.10.051 as described above. The A. oryzae RIB 40 and A. flavus NRRL 3357 genomes were obtained from FungiDB53. The A. oryzae RIB 40 genome was used as the reference during SNP prediction with PhaME. A phylogenetic tree was constructed using the Maximum Likelihood method based on the Hasegawa-Kishino-Yano model54 with 100 bootstrap replicates in MEGA755.
AF gene cluster variation analysis
A number of deletions responsible for limiting AFB1 production have been previously characterized27,56. We used a read depth approach to better characterize large scale deletions in the AF biosynthetic gene cluster in the A. oryzae M2040 and A. flavus Afla-Guard genomes. For all isolates, reads were mapped against the reference A. flavus NRRL 3357 genome using the “sensitive” pre-set parameters in bowtie257. SAM alignment files were converted into sorted BAM format using the view and sort functions in SAMtools58. The SAMtools depth function was then used to estimate average coverage across the entire genome. Average coverage values for each non-overlapping 100 bp portion of the AF gene cluster were then divided by the average coverage across the entire genome to estimate copy number.
To further test the M2040’s inability to produce AFs, the strain was cultured in submerged and solid-state media. PDB and PDA are considered the best media for optimal AFs production. One milliliter (5 × 107) of A. oryzae M2040 spore suspension was inoculated into 100 ml of medium and incubated for 10 days at 30 °C with shaking at 220 rpm. For testing the AFB1 production in solid media, 0.1 ml (5 × 106) of A. oryzae M2040 spore suspension was streaked on PDA plates and incubated for 7 days at 30 °C. The amount of AFB1 in the liquid and solid culture medium was analyzed by HPLC and TLC.
Identification of lineage specific genes
We conservatively identified lineage specific genes in A. oryzae M2040, A. flavus Afla-Guard, A. oryzae RIB 4022, and A. flavus NRRL 335759. Gene models were predicted in the A. oryzae M2040 and A. flavus Afla-Guard genome assembly using Augustus v2.5.560 with the following parameters: “strand = both”, “genemodel = complete”, and “species = aspergillus_oryzae”. Lineage specific genes were conservatively identified in each genome by pairwise BLAST searches of each isolate’s gene models against each genome61. Gene models with BLAST scores > 1e-6 were considered unique to each respective isolate. phmmer and hmmscan were used to annotate lineage specific genes62.
Statistical analysis of the data
Statistical significance was determined using student’s t-test with a 2-tailed distribution. Difference was considered significant as P < 0.05. Error bars correspond to the standard deviation.
Acknowledgements
Ahmad Alshannaq was supported by the Islamic Developmental Bank (IDB) Merit Scholarship Programme for High Technology (MSP) and by the Jordan Food & Drug Administration (JFDA) and by Food Research Institute (FRI) E Michael and Winona Foster Wisconsin Distinguished Fellowship Award. This work was primarily supported by the Rural Development of Administration grant of Korea (PJ012304), and the Intelligent Synthetic Biology Center of Global Frontier Project (2011-0031955) funded by the Ministry of Education, Science and Technology grants, and Food Research Institute research fund of UW-Madison to JHY.
Author Contributions
A.A., J.G.G., K.H.H., S.B.H., and J.H.Y. designed the study. A.A., J.G.G., and M.K.L. performed the experiments. A.A., J.G.G., K.H.H., S.B.H., and J.H.Y. analyzed the data. A.A., J.G.G., M.K.L., and J.H.Y. wrote the manuscript. All authors reviewed the manuscript.
Data Availability
The genome sequence data are available under the BioProject ID (PRJNA483302) and the SRA Run IDs (SRR7615261 for A. oryzae M2040 and SRR7615262 for A. flavus Afla-Guard). https://www.ncbi.nlm.nih.gov/Traces/study/?acc = SRP155606.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alshannaq, A. & Yu, J. H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Env. Res. Public Health. 14 (2017). [DOI] [PMC free article] [PubMed]
- 2.Wood GE. Mycotoxins in foods and feeds in the United States. J. Anim. Sci. 1992;70:3941–3949. doi: 10.2527/1992.70123941x. [DOI] [PubMed] [Google Scholar]
- 3.Klich MA. Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 2007;8:713–722. doi: 10.1111/j.1364-3703.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- 4.Ostry V, Malir F, Toman J, Grosse Y. Mycotoxins as human carcinogens-the IARC Monographs classification. Mycotoxin Res. 2017;33:65–73. doi: 10.1007/s12550-016-0265-7. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Wu F. Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ. Health Perspect. 2010;118:818–824. doi: 10.1289/ehp.0901388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchese Silvia, Polo Andrea, Ariano Andrea, Velotto Salvatore, Costantini Susan, Severino Lorella. Aflatoxin B1 and M1: Biological Properties and Their Involvement in Cancer Development. Toxins. 2018;10(6):214. doi: 10.3390/toxins10060214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell NJ, Bowers E, Hurburgh C, Wu F. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2016;33:540–550. doi: 10.1080/19440049.2016.1138545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal SZ, Nisar S, Asi MR, Jinap S. Natural incidence of aflatoxins, ochratoxin A and zearalenone in chicken meat and eggs. Food Control. 2014;43:98–103. doi: 10.1016/j.foodcont.2014.02.046. [DOI] [Google Scholar]
- 9.Armorini S, Altafini A, Zaghini A, Roncada P. Occurrence of aflatoxin M1 in conventional and organic milk offered for sale in Italy. Mycotoxin Res. 2016;32:237–246. doi: 10.1007/s12550-016-0256-8. [DOI] [PubMed] [Google Scholar]
- 10.Iha MH, Barbosa CB, Heck AR, Trucksess MW. Aflatoxin M1 and ochratoxin A in human milk in Ribeirão Preto-SP, Brazil. Food Control. 2014;40:310–313. doi: 10.1016/j.foodcont.2013.12.014. [DOI] [Google Scholar]
- 11.Cantú-Cornelio F, et al. Occurrence and factors associated with the presence of aflatoxin M1 in breast milk samples of nursing mothers in central Mexico. Food Control. 2016;62:16–22. doi: 10.1016/j.foodcont.2015.10.004. [DOI] [Google Scholar]
- 12.Ssepuuya, G. et al. Mycotoxin contamination of sorghum and its contribution to human dietary exposure in four sub-Saharan countries. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess., 1–10 (2018). [DOI] [PubMed]
- 13.CDC, Centeres for Disease control and prevention. Aflatoxin (2012).
- 14.Medina A, Rodriguez A, Magan N. Effect of climate change on Aspergillus flavus and aflatoxin B1 production. Front. Microbiol. 2014;5:348. doi: 10.3389/fmicb.2014.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battilani P, et al. Aflatoxin B1 contamination in maize in Europe increases due to climate change. Sci. Rep. 2016;6:24328. doi: 10.1038/srep24328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Assuncao, R. et al. Climate change and the health impact of aflatoxins exposure in Portugal - an overview. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess., 1–12 (2018). [DOI] [PubMed]
- 17.Dorner JW, Cole RJ, Blankenship PD. Effect of Inoculum Rate of Biological Control Agents on Preharvest Aflatoxin Contamination of Peanuts. Biological Control. 1998;12:171–176. doi: 10.1006/bcon.1998.0634. [DOI] [Google Scholar]
- 18.Damann K., Jr. Atoxigenic Aspergillus flavus biological control of aflatoxin contamination: what is the mechanism? World Mycotoxin J. 2014;8:235–244. doi: 10.3920/WMJ2014.1719. [DOI] [Google Scholar]
- 19.Mauro Antonio, Garcia-Cela Esther, Pietri Amedeo, Cotty Peter, Battilani Paola. Biological Control Products for Aflatoxin Prevention in Italy: Commercial Field Evaluation of Atoxigenic Aspergillus flavus Active Ingredients. Toxins. 2018;10(1):30. doi: 10.3390/toxins10010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alaniz Zanon MS, Chiotta ML, Giaj-Merlera G, Barros G, Chulze S. Evaluation of potential biocontrol agent for aflatoxin in Argentinean peanuts. Int. J. Food Microbiol. 2013;162:220–225. doi: 10.1016/j.ijfoodmicro.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Abbas HK, et al. Comparison of major biocontrol strains of non-aflatoxigenic Aspergillus flavus for the reduction of aflatoxins and cyclopiazonic acid in maize. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2011;28:198–208. doi: 10.1080/19440049.2010.544680. [DOI] [PubMed] [Google Scholar]
- 22.Machida M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 23.Abe K, Gomi K, Hasegawa F, Machida M. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153. doi: 10.1007/s11046-006-0049-2. [DOI] [PubMed] [Google Scholar]
- 24.Lee MK, Park HS, Han KH, Hong SB, Yu JH. High molecular weight genomic DNA mini-prep for filamentous fungi. Fungal Genet. Biol. 2017;104:1–5. doi: 10.1016/j.fgb.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Weaver, M. A. et al. Genome Sequences of Three Strains of Aspergillus flavus for the Biological Control of Aflatoxin. Genome Announc. 5 (2017). [DOI] [PMC free article] [PubMed]
- 26.Gibbons JG, et al. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Current microbiology. 2012;22:1403–1409. doi: 10.1016/j.cub.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang PK, Horn BW, Dorner JW. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005;42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Ehrlich KC, Chang PK, Yu J, Cotty PJ. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol. 2004;70:6518–6524. doi: 10.1128/AEM.70.11.6518-6524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashworth LJ, Jr, Schroeder HW, Langley BC. Aflatoxins: Environmental Factors Governing Occurrence in Spanish Peanuts. Science. 1965;148:1228–1229. doi: 10.1126/science.148.3674.1228. [DOI] [PubMed] [Google Scholar]
- 30.Weckbach LS, Marth EH. Aflatoxin production by Aspergillus parasiticus in a competitive environment. Mycopathologia. 1977;62:39–45. doi: 10.1007/BF00491994. [DOI] [PubMed] [Google Scholar]
- 31.Ehrlich KC. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations. Front. Microbiol. 2014;5:50. doi: 10.3389/fmicb.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrlich KC, Moore GG, Mellon JE, Bhatnagar D. Challenges facing the biological control strategy for eliminating aflatoxin contamination. World Mycotoxin J. 2014;8:225–233. doi: 10.3920/WMJ2014.1696. [DOI] [Google Scholar]
- 33.Dorner JW, Lamb MC. Development and commercial use of afla-Guard((R)), an aflatoxin biocontrol agent. Mycotoxin Res. 2006;22:33–38. doi: 10.1007/BF02954555. [DOI] [PubMed] [Google Scholar]
- 34.Dorner JW. Efficacy of a biopesticide for control of aflatoxins in corn. J. Food Prot. 2010;73:495–499. doi: 10.4315/0362-028X-73.3.495. [DOI] [PubMed] [Google Scholar]
- 35.Abbas HK, et al. Selection of Aspergillus flavus isolates for biological control of aflatoxins in corn. Toxin Reviews. 2001;30:59–70. doi: 10.3109/15569543.2011.591539. [DOI] [Google Scholar]
- 36.Thammarongtham C, et al. Genome Characterization of Oleaginous Aspergillus oryzae BCC7051: A Potential Fungal-Based Platform for Lipid Production. Current microbiology. 2018;75:57–70. doi: 10.1007/s00284-017-1350-7. [DOI] [PubMed] [Google Scholar]
- 37.Fujii, T., Koike, H., Sawayama, S., Yano, S. & Inoue, H. Draft Genome Sequence of Talaromyces cellulolyticus Strain Y-94, a Source of Lignocellulosic Biomass-Degrading Enzymes. Genome Announc. 3 (2015). [DOI] [PMC free article] [PubMed]
- 38.Blanco-Ulate, B., Allen, G., Powell, A. L. & Cantu, D. Draft Genome Sequence of Botrytis cinerea BcDW1, Inoculum for Noble Rot of Grape Berries. Genome Announc. 1 (2013). [DOI] [PMC free article] [PubMed]
- 39.Umemura M, et al. Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol. 2014;68:23–30. doi: 10.1016/j.fgb.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Moore GG, Mack BM, Beltz SB, Gilbert MK. Draft Genome Sequence of an Aflatoxigenic Aspergillus Species, A. bombycis. Genome Biol Evol. 2016;8:3297–3300. doi: 10.1093/gbe/evw238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kusuya, Y., Takahashi-Nakaguchi, A., Takahashi, H. & Yaguchi, T. Draft Genome Sequence of the Pathogenic Filamentous Fungus Aspergillus udagawae Strain IFM 46973T. Genome Announc. 3 (2015). [DOI] [PMC free article] [PubMed]
- 42.Gazis R, et al. The genome of Xylona heveae provides a window into fungal endophytism. Fungal biology. 2016;120:26–42. doi: 10.1016/j.funbio.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, et al. Efficient enzymatic hydrolysis and simultaneous saccharification and fermentation of sugarcane bagasse pulp for ethanol production by cellulase from Penicillium oxalicum EU2106 and thermotolerant Saccharomyces cerevisiae ZM1-5. Biomass and Bioenergy. 2015;77:53–63. doi: 10.1016/j.biombioe.2015.03.020. [DOI] [Google Scholar]
- 44.Wingfield BD, et al. IMA Genome-F 5: Draft genome sequences of Ceratocystis eucalypticola, Chrysoporthe cubensis, C. deuterocubensis, Davidsoniella virescens, Fusarium temperatum,Graphilbum fragrans, Penicillium nordicum, and Thielaviopsis musarum. IMA fungus. 2015;6:493–506. doi: 10.5598/imafungus.2015.06.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nierman, W. C. et al. Genome Sequence of Aspergillus flavus NRRL 3357, a Strain That Causes Aflatoxin Contamination of Food and Feed. Genome Announc. 3 (2015). [DOI] [PMC free article] [PubMed]
- 46.Yu JH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Szewczyk E, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 48.Amaike S, Keller NP. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell. 2009;8:1051–1060. doi: 10.1128/EC.00088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kale SP, et al. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 2008;45:1422–1429. doi: 10.1016/j.fgb.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber T, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stajich JE, et al. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2012;40:D675–681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tominaga M, et al. Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Appl. Environ. Microbiol. 2006;72:484–490. doi: 10.1128/AEM.72.1.484-490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England). 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Payne GA, et al. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006;44:9–11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 60.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 62.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data are available under the BioProject ID (PRJNA483302) and the SRA Run IDs (SRR7615261 for A. oryzae M2040 and SRR7615262 for A. flavus Afla-Guard). https://www.ncbi.nlm.nih.gov/Traces/study/?acc = SRP155606.