Abstract
Procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD3), a membrane-bound homodimeric enzyme, hydroxylates lysyl residues in collagen-like peptides; however, its role in lung cancer is unknown. This study aimed to investigate the role of PLOD3 as a pro-metastatic factor and to elucidate the underlying mechanism. First, we experimentally confirmed the release of PLOD3 in circulation in animal models, rendering it a potential serum biomarker for lung cancer in humans. Thereafter, we investigated the effects of PLOD3 overexpression and downregulation on cancer cell invasion and migration in vitro and in vivo, using human lung cancer cell lines and a mouse tumor xenograft model, respectively. Further, PLOD3 levels were determined in lung tissue samples from lung cancer patients. Functional analyses revealed that PLOD3 interacts with STAT3, thereby expressing matrix metalloproteinases (MMP-2 and MMP-9) and with urokinase plasminogen activator (uPA) to enhance tumor metastasis. PLOD3 and the STAT3 pathway were significantly correlated in the metastatic foci of lung cancer patients; PLOD3–STAT3 levels were highly correlated with a poor prognosis. These results indicate that PLOD3 promotes lung cancer metastasis in a RAS-MAP kinase pathway-independent manner. Therefore, secreted PLOD3 serves as a potent inducer of lung cancer metastasis and a potential therapeutic target to enhance survival in lung cancer.
Introduction
The incidence of lung cancer, one of the most common malignancies, has increased worldwide1,2. Despite remarkable advancements in targeted therapy for lung cancer patients, survival rates remain unchanged. More than 79% of lung cancer patients develop metastases, and the 5-year survival rate of patients with distant metastases is <5%3. Therefore, metastases account for increased mortality rates in lung cancer4,5. Despite advancements in early detection and improvements in treatment, the long-term survival of lung cancer patients remains poor. Thus, it is important to further the current understanding of the molecular underpinnings of metastatic progression in lung cancer and to apply this understanding in developing improved therapeutic alternatives. Identification of biomarkers predicting the risk of recurrence and metastasis is therefore clinically important. Recently, via proteomics analyses, we identified four proteins, namely, plasminogen activator inhibitor type-2 (PAI-2), NODAL modulator 2 (NOMO2), kinesin light chain 4 (KLC4), and PLOD3 to identify radioresistance-related genes. As these proteins have not been studied in detail6, it is important to investigate their role in lung cancer.
Collagens constitute a highly specialized family of extracellular matrix proteins, which maintain tissue architecture and regulate cellular responses7–9. Collagen production and deposition are regulated by various enzymes, including prolyl 4-hydroxylases (P4Hs), procollagen-lysine, 2-oxoglutarate 5-dioxygenases (PLODs), and lysyl oxidase (LOXs). PLOD proteins, involved in fibrosis and tissue remodeling, are also known as lysyl hydroxylases (LHs)10,11. Three PLOD isoforms have been characterized thus far: PLOD1 (LH1), PLOD2 (LH2), and PLOD3 (LH3)12. PLOD3 is a multifunctional enzyme with lysyl hydroxylase, collagen galactosyltransferase, and glucosyltransferase activity13, generating-specific glucosylgalactosylhydroxylysine (Glc-Gal-Hyl) residues in the Y position of X-Y-Gly triplets of collagens14. Lysine residues are modified in the endoplasmic reticulum (ER) lumen15. In addition to its ER localization, LH3 is present in the extracellular space of tissues and in serum16. PLOD2 specifically hydroxylates lysine residues in the telopeptide of procollagens, whereas PLOD1 hydroxylates lysine residues in the α-helical or central domain17. However, substrate specificity of PLOD3 is unknown18–21. Moreover, the extracellular function of PLOD3 is unclear, although glycosyltransferase activity of PLOD3 in the extracellular space is reportedly important for cell growth and viability. Although, the role of PLODs in cancer is unclear, their significance in other diseases has been reported. Recent studies have reported the role of PLODs in fibrotic diseases; lysine hydroxylation is impaired in Bruck syndrome20 and Ehlers–Danlos syndrome type VIA22; however, it is enhanced in fibrotic diseases23. Furthermore, recent studies have reported that PLOD2 expression is associated with an increased mortality risk in breast cancer patients24 and its inhibition suppresses metastases in sarcomas25. These results suggest that collagen-modifying enzymes, such as PLODs, may play critical roles in cancer cell metastasis. However, the physiological role of PLOD3 in lung cancer remains unknown. Hence, it is important to understand whether PLOD3 plays a role in lung cancer metastasis and its underlying mechanism.
Signal transducer and activator of transcription 3 (STAT3) belong to the family of intracellular transcription factors that mediate numerous physiological and pathological processes, such as cell proliferation, differentiation, apoptosis, and metastasis26. Enhanced STAT3 activity, reported in various human tumors, promotes cancer cell survival via therapeutic resistance27. Cytokines and growth factors promote STAT3 phosphorylation via receptor-associated Janus kinases, resulting in homo- or hetero-dimer formation and STAT3 translocation to the nucleus, where it functions as a transcriptional activator28. In response to ligand-receptor binding (interferons, epidermal growth factor, and interleukins), STAT3 is activated via phosphorylation at residues Tyr705 and Ser727 and it transduces signals from various signaling pathways by transactivating target genes involved in metastasis and proliferation29,30. STAT3 activation reportedly promotes cancer metastasis via matrix metalloproteinase (MMP)-2 and MMP-9, which are involved in cell migration and invasion31,32 owing to degradation of the basement membrane and the extracellular matrix (ECM)33. The serine protease urokinase plasminogen activator (uPA) also regulates cell migration via degradation of ECM components34. Moreover, uPA plays an important role in the maturation of other proteins, including MMPs and growth factors, via conversion of plasminogen to plasmin35.
Hence, the present study aimed to test the hypothesis that targeting of STAT3 enables cancer suppression at multiple levels via attenuation of the oncogenic potential of PLOD3 owing to upstream and downstream aberrations in cancer metastasis pathways. Our findings may underscore the importance of further elucidation of the role of PLOD3 in lung metastases via regulation of STAT3 and the associated molecular mechanisms.
Results
PLOD3 upregulation in lung cancer patients is associated with metastasis and poor prognosis
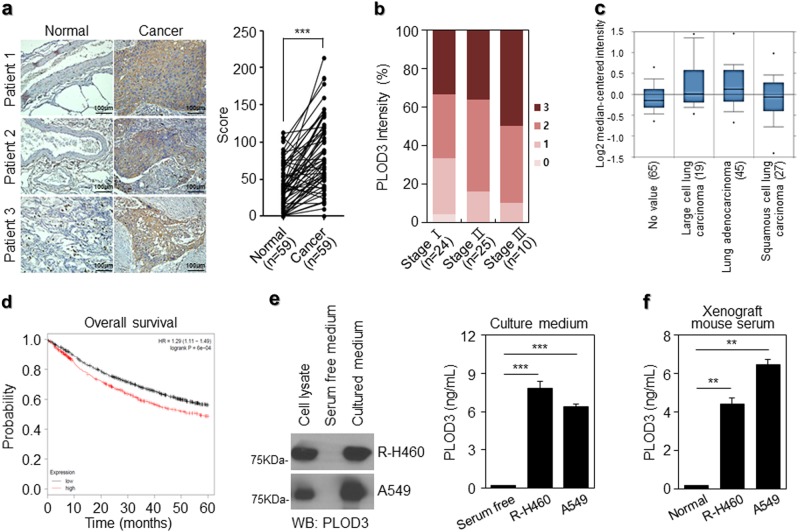
We first analyzed PLOD3 expression in tumor samples from lung cancer patients to determine the clinical relevance of PLOD3 expression in lung cancer. Notably, in 50 of 59 lung cancer patients, PLOD3 was upregulated in lung cancer tissue than in the corresponding normal tissue (Fig. 1a). Furthermore, we analyzed the association between PLOD3 expression and the pathological grade of lung cancer, using patient samples. PLOD3 was upregulated in pathological grade III (n = 10) rather than in pathological grades I (n = 24) and II (n = 25) lung cancer. Moreover, PLOD3 upregulation was significantly correlated with lung cancer stage (comparing I–II–III) (Fig. 1b). Furthermore, PLOD3 was upregulated in 37 squamous cell carcinoma patients, 15 adenocarcinoma patients, and 7 others compared to that in the corresponding tumors from the same patients (Supplementary figure 1a). To further determine the association between PLOD3 and lung cancer in clinical-pathological features, we analyzed PLOD3 expression in 156 primary lung cancer tumors relative to that in the corresponding tumors from the same patients (large cell lung carcinoma [n = 19], adenocarcinoma [n = 45], and squamous cell carcinoma [n = 27]) (Fig. 1c). In particular, ONCOMINE analysis data indicate that PLOD3 mRNA was upregulated in lung cancer (Fig. 1c) and gastric cancer patients (Supplementary figure 1b). Subsequently, PLOD3 expression levels were inversely correlated with reduced survival in lung cancer patients and in multiple human cancer types, including gastric cancers, using Kaplan–Meier plotter analysis36,37 (Fig. 1d, Supplementary figure 1c).
Fig. 1. Endogenous PLOD3 plays an important role in lung cancer.
a Representative microscopic images of lung cancers and their normal tissue counterparts stained with an anti-PLOD3 antibody (left panel, scale bar, 100 μm). Staining intensity was scored as follows in all patient samples (n = 59). The scores are calculated by multiplying the staining intensity with the percentage of stained cells in primary lung cancer relative to the paired normal tissue. Statistical significance was determined by Student’s t-test. ***P < 0.001. b PLOD3 upregulation is positively correlated with the stage of lung cancer in all patient samples (n = 59). c Box plot analysis of the PLOD3 mRNA levels obtained from ONCOMINE from lung cancer patients. Gene: PLOD3; Analysis type: cancer vs normal analysis; Data type: mRNA; Sample type: clinical specimen; Lung. d The effects of PLOD3 on the overall survival of lung cancer patients, using Kaplan–Meier plotter analysis. e R-H460 and A549 cells were cultured up to 80% confluence, followed by changing of the medium to fresh serum-free RPMI1640. After 6 h, collected supernatants were analyzed via immunoblotting with anti-PLOD3 antibody (left panel) and enzyme-linked immunosorbent assay (ELISA) at 450 nm (right panel). Statistical significance was determined by Student’s t-test. ***P < 0.001. f R-H460 and A549 cells (1 × 106 cells) were inoculated subcutaneously into the backs of nude mice. After tumorigenesis, mice were killed. Serum collected from each group was analyzed via ELISA. Statistical significance was determined by Student’s t-test. **P < 0.01
Furthermore, upon in vivo and in vitro analyses, PLOD3 was detected in the culture medium of R-H460 and A549 cells in the immunoblotting assay and sandwich ELISA (Fig. 1e). PLOD3 levels in serum-free-medium, R-H460 cell culture medium, and A549 cell culture medium were ~0.2 ng/ml, 7.8 ng/ml, and 6.4 ng/ml, respectively. We quantified PLOD3 secretion in an animal model; non-injection group (normal) and A549 and R-H460 cell groups. PLOD3 levels were quantified in serum collected after 4 weeks (Fig. 1f). PLOD3 levels in the normal group and R-H460 and A549 cell groups were ~0.19 ng/ml, 4.4 ng/ml, and 6.4 ng/ml, respectively. These results suggested that PLOD3 is secreted from lung cancer cells in vivo and in vitro, and PLOD3 is markedly correlated with a poor prognosis in lung cancer patients, playing important roles in the metastasis and lung cancer progression.
PLOD3 promotes lung cancer cell metastasis in vitro
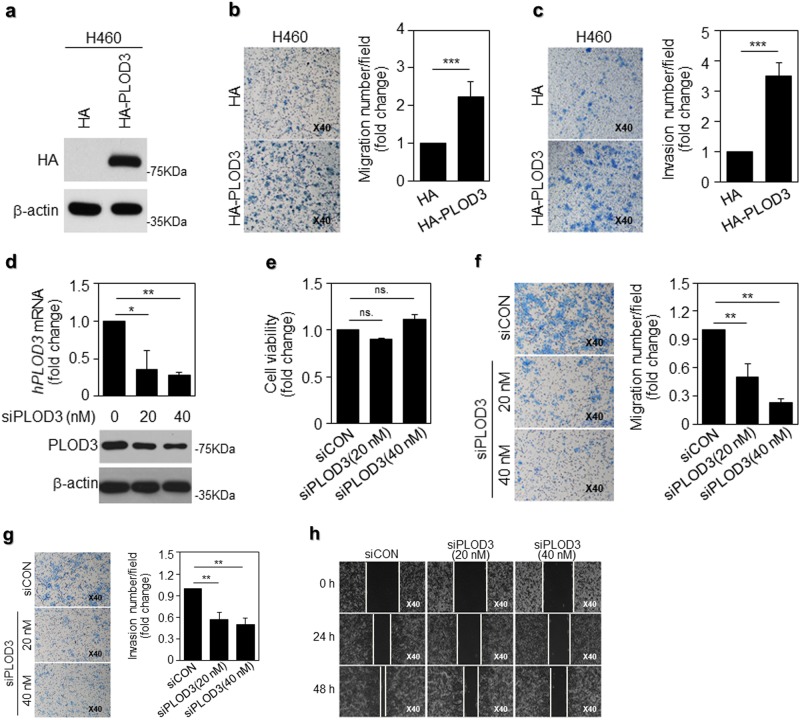
To further investigate the effect of PLOD3 on lung cancer cell metastasis, we evaluated its effects on metastasis. First, to overexpress PLOD3, we transfected HA-PLOD3 into the H460 and A549 cells, which express PLOD3 lesser than R-H460 cells6 (Fig. 2a, Supplementary figure 2a). Exogenous PLOD3 introduction into H460 and A549 cells conferred significantly enhanced metastatic potential, as indicated by the enhanced invasive and migratory ability of these cells in a chamber assay (Figs. 2b, c, Supplementary figures 2b and 2c). To further identify the genes essential for lung cancer metastasis, we generated-specific siRNA constructs to stably knockdown human PLOD3 (hPLOD3) mRNA and transfected siPLOD3 into the R-H460 cells, which express PLOD3 more than H460 cells. hPLOD3 mRNA and PLOD3 protein were downregulated in an siRNA-dose-dependent manner (Fig. 2d). We next assessed whether PLOD3 knockdown altered proliferation in R-H460 cells. Depletion of PLOD3 in R-H460 cells did influence cell proliferation under the present experimental conditions. (Fig. 2e). However, depletion of PLOD3 greatly reduced migration and invasiveness of R-H460 cells in a siRNA-dose-dependent manner (Fig. 2f, g). Results of the wound-healing assay further supported the potent pro-metastatic effects of PLOD3 (Fig. 2h). Together, these data suggest that PLOD3 promotes metastasis in lung cancer in vitro.
Fig. 2. PLOD3 possesses greater potential to promote lung cancer malignancy.
a PLOD3 protein expression was assessed via western blot analysis, following transfection of HA-PLOD3 in H460 cells for 48 h. b A transwell assay was performed to evaluate the motility of PLOD3-overexpressing H460 cells. PLOD3 displayed superior potential to promote cell migration. The relative numbers of migratory cells were determined and presented as the mean ± SD values from three independent experiments. Statistical significance was determined by Student’s t-test. ***P < 0.001. c The invasion assay revealed different cell motilities in PLOD3-overexpressing lung cancer cells. PLOD3 expression promoted the invasion of H460 cells. The relative numbers of invasive cells were determined and presented as the mean ± SD values from three independent experiments. Statistical significance was determined by Student’s t-test. ***P < 0.001. d PLOD3 protein and mRNA levels were determined via western blotting (lower) and quantitative reverse transcription polymerase chain reaction (qRT-PCR; upper), respectively, after the indicated concentration of siPLOD3 was transfected in R-H460 cells for 24 h. The qRT-PCR data are expressed as the mean ± SD values *P < 0.05, **P < 0.01. e The cell viability assay for siPLOD3-treated cells for 24 h; x-axis: siCON, siPLOD3 (concentration). f A transwell assay was conducted to evaluate the motility of PLOD3 knockdown R-H460 cells at 12 h. Statistical significance was determined by Student’s t-test. **P < 0.01. g Cell invasion assay revealed that siPLOD3 had a limited effect on pro-metastasis at 24 h in R-H460 cells. Statistical significance was determined by Student’s t-test. **P < 0.01. h R-H460 cells were treated with siPLOD3 and then incubated for 24 and 48 h. The cells were then scraped with a sterile 200-μl pipette tip for the scratch assay
PLOD3 promotes lung cancer metastasis in vivo
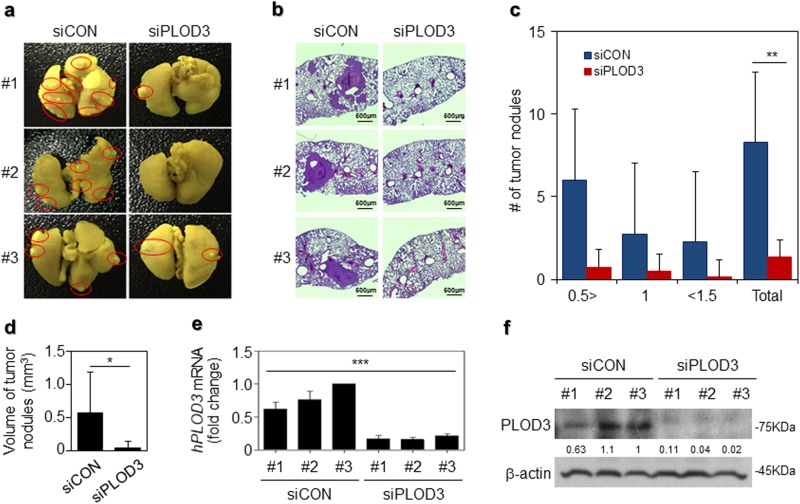
To directly evaluate the role of PLOD3 in lung metastasis in vivo, we conducted a separate analysis in nude mice via injection of control siRNA or PLOD3 siRNA in the tail vein. PLOD3 silencing reduced the incidence of lung metastasis, as determined via the nodule assay. Excised lungs after fixation in Bouin’s solution and H&E staining reduced the number of lung metastatic foci (Fig. 3a, b). PLOD3 silencing reduced the incidence of lung metastasis by decreasing the number of nodules (Fig. 3c) and tumor nodule volume (siControl (siCON)—5.8 mm3 vs siPLOD3—0.53 mm3) (Fig. 3d). Furthermore, to verify the knockdown efficiency of PLOD3, we also confirmed hPLOD3 mRNA and PLOD3 protein downregulation in the lung nodules (Fig. 3e, f). These data indicate that PLOD3 induces metastasis in an in vivo model of lung cancer.
Fig. 3. Loss of PLOD3 suppressed R-H460 tumor metastasis in vivo.
a Representative micrographs with metastatic nodules are shown via Bouin’s staining. b Hematoxylin and eosin staining were performed using xenografts. c, d The number and volume of metastatic nodules were determined microscopically in PLOD3 xenograft tumors. Statistical significance was determined by Student’s t-test. *P < 0.05, **P < 0.01. e, f Estimation of hPLOD3 mRNA and PLOD3 protein levels in the lung nodules. Statistical significance was determined by ANOVA analysis. ***P < 0.001
Expression of MMP-2/9 and uPA during PLOD3-induced metastasis
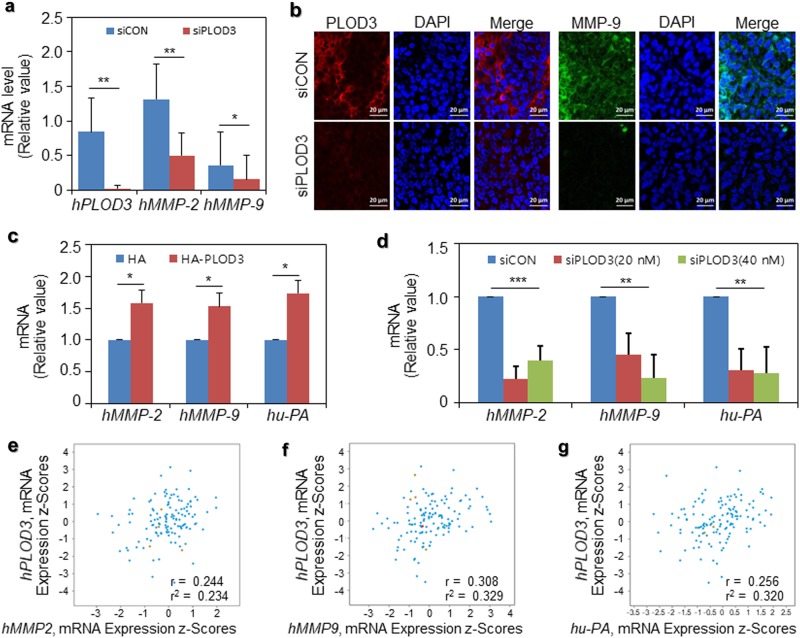
Further, we focused on signaling downstream of PLOD3 in relation to metastasis. Invasion during metastasis requires the upregulation of proteolytic enzymes, such as MMP-2/9 or uPA, to degrade the ECM, thereby facilitating cancer cell invasion into the surrounding tissues25–27. We evaluated the mRNA expression levels of metastatic genes in the R-H460 xenograft model, siCON-treated group, and siPLOD3-treated group. Human MMP-2 (hMMP-2) and human MMP-9 (hMMP-9) mRNA were markedly downregulated in siPLOD3-treated groups compared with those in the corresponding control groups in vivo (Fig. 4a). Furthermore, MMP-9 was downregulated in siPLOD3-treated tumors compared to the controls, upon immunofluorescence (IF) analysis (Fig. 4b). We then evaluated hMMP-2, hMMP-9, and human u-PA (hu-PA) mRNA expression levels in these two systems with HA-PLOD3-overexpressing or siPLOD3-knockdown cells, using qRT-PCR in vitro. As shown in Fig. 4c, hMMP-2/9 and hu-PA mRNA were significantly upregulated in HA-PLOD3-transfected cells compared with that in control cells (Fig. 4c). Consistent with the in vivo data, the opposite effect was observed in siPLOD3-treated cells (Fig. 4d). Thus, upon analyzing the publicly available microarray datasets, we further observed a strong positive correlation among hPLOD3 mRNA and hMMP-2/9 mRNA, and hu-PA mRNA levels in lung cancer patients (Fig. 4e–g). Together, the findings from in vitro and in vivo analyses and from lung cancer patient samples demonstrate the sufficient and necessary roles of PLOD3, MMP-2/9, and uPA in promoting lung cancer metastasis.
Fig. 4. MMP-2/9 and uPA expression in siPLOD3- or HA-PLOD3-treated lung cancer cells.
a hMMP-2/9 mRNA levels were determined after siPLOD3 treatment in vivo. Statistical significance was determined by Student’s t-test. *P < 0.05; **P < 0.01. b In vivo immunofluorescence of siCON- and siPLOD3-treated groups. c hMMP-2/9 and hu-PA mRNA levels after the indicated concentration of HA-PLOD3 was transfected were determined via quantitative reverse transcription polymerase chain reaction (RT-qPCR). Statistical significance was determined by Student’s t-test. *P < 0.05. d H460 cells were transfected with siCON or the indicated concentration of siPLOD3 for 24 h. hMMP-2/9 and hu-PA mRNA levels were determined via RT-qPCR. Statistical significance was determined by ANOVA analysis. **P < 0.01, ***P < 0.001. e–g Correlation of hPLOD3 with hMMP-2 or hMMP-9 or hu-PA transcripts in the cbioportal datasets of lung cancer patients
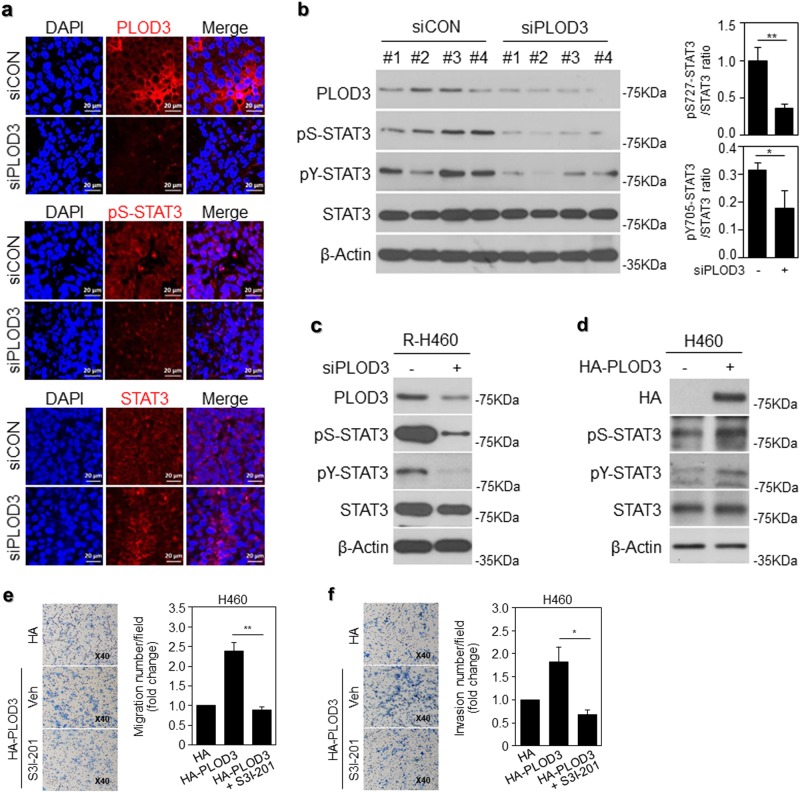
PLOD3 regulates STAT3 phosphorylation to promote lung cancer metastasis
Considering the previously established role of STAT3 in cancer metastasis, we hypothesized that PLOD3 promotes metastasis in a STAT3-dependent manner in the model system used herein. Constitutive activation of the STAT3 signaling pathway plays a critical role in tumorigenesis and cancer progression in humans via promotion of cancer cell growth, survival, migration, and invasion38. Moreover, STAT3 is an important signaling mediator in malignant diseases and is persistently activated in 22–65% of non-small-cell lung cancers39. Therefore, we investigated whether PLOD3 knockdown inhibited the constitutive phosphorylation/activation of STAT3. As shown in Fig. 5a, in siPLOD3-treated in vivo models, constitutively activated STAT3 phosphorylation at Ser727 (pS-STAT3) was dependently inhibited, as evident from immunohistochemical analysis (Fig. 5a). Further, we quantified the in vivo data from western blotting analysis and the constitutively activated STAT3 phosphorylation of STAT3 at Tyr705 (pY-STAT3) and Ser727 was markedly decreased (Fig. 5b). In siPLOD3-treated R-H460 cells, phospho-STAT3 was significantly downregulated (Fig. 5c). In contrast, when H460 cells were treated with HA-PLOD3, phospho-STAT3 was upregulated (Fig. 5d). We confirmed that JAK activity upstream of STAT3 resulted in an overall downregulation of JAK1, whereas no effects were observed in the case of JAK2 (data not shown)40. In this context, these findings strongly suggest that PLOD3 knockdown effectively suppresses the constitutive activation of STAT3 in lung cancer cells. To examine the direct effect of STAT3, we used the STAT3 inhibitor, S31–201. Cells treated with this inhibitor showed decreased migration and invasion owing to PLOD3 overexpression in chamber assays (Fig. 5e, f). Our data indicate that PLOD3 regulates lung cancer metastasis directly via STAT3 signaling.
Fig. 5. PLOD3 regulated the malignant phenotype via STAT3 signaling.
a Immunostaining of PLOD3, pS-STAT3, and STAT3 in PLOD3 knockdown xenograft tumors. b Protein levels of PLOD3, pS-STAT3, pY-STAT3, and STAT3 were determined via western blotting in PLOD3 xenograft tumors. Statistical significance was determined by Student’s t-test. *P < 0.05, **P < 0.01. c Levels of the indicated proteins were determined via western blotting after siPLOD3 treatment in R-H460 cells. d Levels of the indicated proteins were determined via western blotting in HA-PLOD3-overexpressing H460 cells. e, f The HA-PLOD3 and STAT3 inhibitor S3I-201-treated H460 cells were analyzed via a transwell assay to evaluate cell motility and invasiveness. Statistical significance was determined by Student’s t-test. *P < 0.05; **P < 0.01
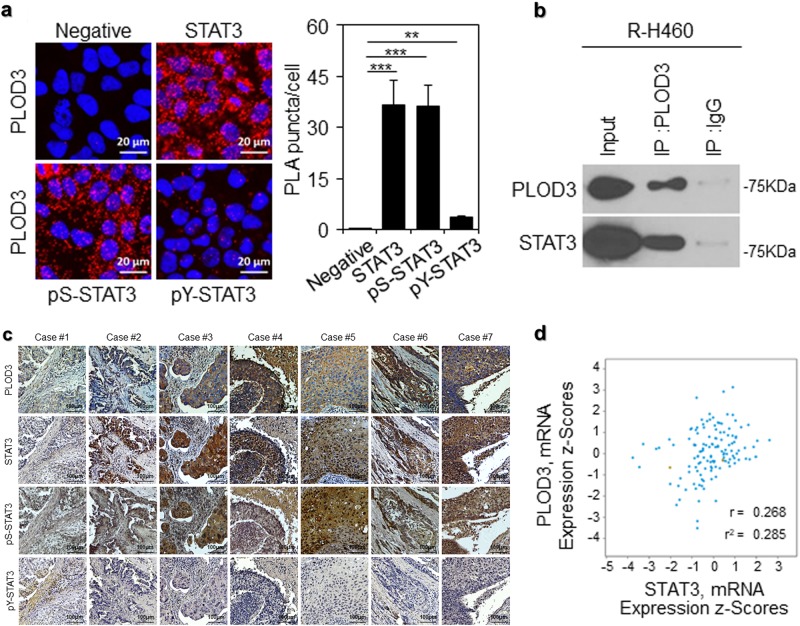
PLOD3–STAT3 interaction is associated with progression and poor prognosis in lung cancer patients
Importantly, the present results help elucidate the mechanism underlying PLOD3-mediated regulation of STAT3 function via direct interactions; PLOD3 may induce robust binding of STAT3 to its consensus-binding elements and upregulate target genes, thereby promoting lung cancer metastasis. To confirm this supposition, we analyzed STAT3 and PLOD3 interactions in R-H460 cells via an in situ proximity ligation assay (Fig. 6a). In particular, pS-STAT3 robustly interacted with PLOD3 compared with pY-STAT3. This was confirmed via immunoprecipitation analyses. As shown in Fig. 6b, PLOD3 directly interacted with STAT3. To determine whether PLOD3 and STAT3 coordinate to promote lung cancer metastasis in patients, we analyzed the potential correlation between PLOD3 and STAT3 expression levels in lung cancer patients. We performed immunohistochemical analysis of PLOD3, pS-STAT3, pY-STAT3, and STAT3 levels, using tumor tissue samples harvested from lung cancer patients (the same samples used in Fig. 1). The results in Fig. 6c indicate that pS-STAT3 levels were positively correlated with PLOD3 upregulation in lung cancer tissues. Furthermore, analysis of the publicly available microarray datasets revealed a correlation between PLOD3 and STAT3 mRNA levels in lung cancer patients (Fig. 6d). These data suggest that an activated PLOD3-STAT3 signaling pathway is correlated with a poor prognosis in lung cancer patients, and pS-STAT3 plays a major role in comparison with pY-STAT3 in regulating PLOD3 during metastasis.
Fig. 6. Correlation between the PLOD3-STAT3 signaling pathway and the survival and prognosis of lung cancer patients.
a The in situ proximity ligation assay was performed for R-H460 cells stained with STAT3, pS-STAT3, and pY-STAT3. Statistical significance was determined by Student’s t-test. **P < 0.01; ***P < 0.001. b Immunoblotting analysis of lysates after immunoprecipitation from R-H460 cells with expressing PLOD3 and STAT3. c Representative microscopic images of specimens of lung cancer patients stained with an anti-PLOD3, anti-STAT3, anti-pS-STAT3, and anti-pY-STAT3 antibodies. d Correlation between PLOD3 and STAT3 transcripts in the cbioportal datasets of lung cancer patients
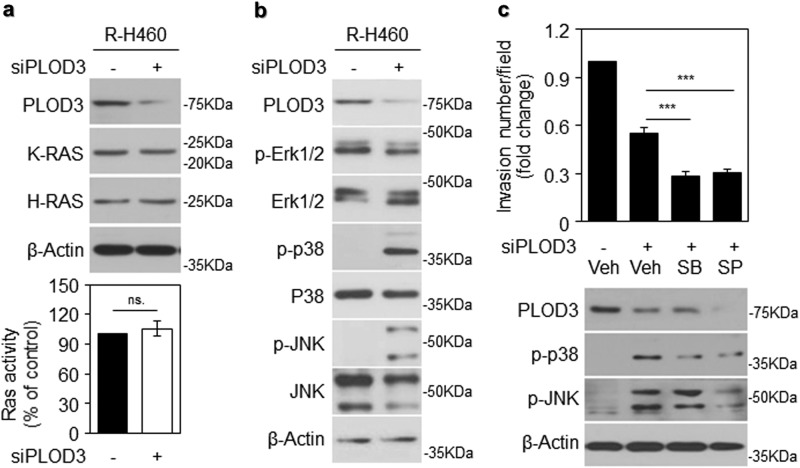
PLOD3 promotes lung cancer metastasis in a RAS-MAP kinase pathway-independent manner
Further, to determine whether PLOD3 is required for Ras-induced metastasis, we examined K-Ras and N-Ras levels via western blotting after siPLOD3 transfection in R-H460 cells. Protein expression levels in siPLOD3-treated cells were almost similar to those in control cells (Fig. 7a, upper panel). Subsequently, upon analysis of Ras activity in siPLOD3-treated cells, Ras activity was almost similar between the two groups (Fig. 7a, lower). MAP kinase activation and STAT3 signaling upregulate MMP-2/9 during the induction of metastasis41. Activation levels of MAP kinase proteins (p38 and JNK) increased after siPLOD3 transfection in R-H460 cells (Fig. 7b). Accordingly, inhibitors of MAP kinase proteins were used to distinctly identify the signal transduction pathways regulated by PLOD3. The levels of invading cells decreased significantly upon SB and SP treatment in siPLOD3-transfected cells (Fig. 7c, upper). Western blotting revealed that siPLOD3-induced upregulation of p-p38 and p-JNK was attenuated upon SB and SP treatment (Fig. 7c, lower panel), indicating that PLOD3-induced metastasis is mediated independently via MAPK signal transduction pathways.
Fig. 7. PLOD3-induced metastasis is independent of the RAS-MAPK pathway.
a Ras expression assessed via western blot analysis following transfection with siPLOD3 in R-H460 cells (upper). Ras activity was determined via an enzyme-linked immunosorbent assay-based activity assay after siPLOD3 treatment in R-H460 cells (lower, x-axis: siCON, siPLOD3). b Protein levels of MAP kinases were determined via western blotting after siPLOD3 treatment in R-H460 cells. c A cell invasion assay showing that SB and SP have a limited effect on pro-metastatic PLOD3 in R-H460 cells. (upper). Protein levels of indicated proteins determined via western blotting after treatment with siPLOD3 and MAP kinase inhibitor (p38 inhibitor, SB203580 (SB); JNK inhibitor, SP600125 (SP)) in R-H460 cells (lower). Statistical significance was determined by Student’s t-test. ***P < 0.001
Discussion
The morbidity and mortality of lung cancer patients predominantly results from primary tumor cell invasion and metastasis to secondary sites. In this context, we first identified that PLOD3 is one of the most widely upregulated proteins in human lung cancers and observed that PLOD3 upregulation is significantly associated with tumor progression and advanced stages of lung cancer. Furthermore, secreted PLOD3 in the blood of the animal model was identified for the first time in this study, suggesting an application of PLOD3 as a prognostic biomarker.
PLODs are implicated in metastasis because of their role in regulating collagen biosynthesis. Collagens provide the scaffold for ECM assembly and are considered “highways” for cancer cell migration42. Increasing evidence indicates that collagens support a barrier for migration and enhance metastasis in a collagen organization-dependent manner. Studies involving various types of human cancers have reported that components of stabilized collagen are promoted via different intra- and inter-molecular covalent cross-linkages24,25,42, and the different types of collagen organization are determined after cross-linking via hydroxylation of collagen telopeptidyl and helical Lys residues43. These changes are primarily mediated by lysyl hydroxylases, which are encoded by distinct PLOD genes25. Thus, we predicted that PLOD may be involved in cancer metastasis. Recently, Wang et al. reported that PLOD3 silencing decreases proliferation and viability in fibrosarcoma HT-1080 cells44. Moreover, PLOD3 overexpression has been reported in gastric, colorectal, and pancreatic cancers and glioma45–47. In addition, Tsai et al. and Shen et al. reported that PLOD3 overexpression promotes tumor progression and is associated with a poor prognosis in gliomas and hepatocellular carcinoma (HCC)48,49. These results indicate that PLOD3 may be an oncogene associated with tumor malignancy in glioma and HCC patients. These reports indicate that PLOD3 is associated with tumor progression, and PLOD3 mRNA upregulation is associated with an unfavorable prognosis50,51. Furthermore, PLOD2 expression has recently been reported as a useful biomarker for the effects of antiangiogenic treatment for malignant cancers42,43. Furthermore, PLOD2, induced under hypoxia, is a potential novel prognostic factor for HCC following surgery52. Although PLOD3 has been assessed in multiple cancers, its regulation in lung cancers is unclear.
Herein, PLOD3 was first confirmed to be frequently upregulated in lung cancer patients upon immunohistochemical analysis. Clinical data also revealed that PLOD3 expression was significantly correlated with the pathological grade of lung cancer. Moreover, PLOD3 upregulation is associated with reduced overall survival in lung cancer and gastric cancer patients, and PLOD3 serves as a prognostic marker in these patients. Our finding that genetic depletion of PLOD3 in mice was sufficient to selectively impair lung metastasis reveals an essential role in the physiological modification of PLOD3 during metastasis. The PLOD3 siRNA model system used herein allowed for the elucidation, for the first time, of PLOD3 as a selective marker of metastasis. Unexpectedly, the PLOD3 siRNA model system initially exhibited metastasis 12 h after siRNA transfection and apoptosis in the late period (We are currently expanding this mechanistic investigation). This is an interesting finding regarding the importance of PLOD3 upregulation, as it indicates a temporal dual function of PLOD3. Thus, interference with metastasis using siPLOD3 may represent an alternative to conventional chemotherapy for lung cancer, as most patients diagnosed with distant metastasis have a decreased survival rate.
In particular, the role of PLOD3 in promoting lung cancer metastasis has remained unclear. Initially, the correlation of PLOD3 with MMP and uPA expression was related to metastasis, upon analysis of data from a publicly available patient database. Recently, Dayer et al. reported that recruitment of MMP-9 to the fibroblast cell surface by LH3 triggers transforming growth factor-β (TGF-β) activation and fibroblast differentiation, suggesting an association between PLOD3 and MMP-953. To our knowledge, the present study is the first to report that PLOD3 may inhibit lung cancer invasion by decreasing MMP-2, MMP-9, and uPA activity. However, the mechanism underlying PLOD3-mediated upregulation of MMPs and the signaling pathway by which PLOD3 regulates lung cancer cell metastasis remain unknown. Furthermore, the results from mechanistic studies, based on the observed selective regulation of lung cancer metastasis upon PLOD3 depletion, identified STAT3 as the binding partner of PLOD3. STAT3 is a constitutively upregulated and activated signaling molecule in cancer, which plays an essential role in cancer cell invasion upstream of MMPs in humans25. STAT3 plays an important role in metastasis in different cancer models, including lung cancer, by participating in multiple steps of metastasis26–28. STAT3 is extensively regulated via phosphorylation and transduces signals from various signaling pathways, including the transactivation of the target genes mediating metastasis and proliferation28,31. Importantly, the present results show that the mRNAs of STAT3 target genes involved in metastasis, including MMP-2 and MMP-9, were markedly downregulated upon PLOD3 depletion. Moreover, STAT3 inhibition greatly attenuated invasion by exogenous PLOD3-expressing H460 cells. These findings elucidate the mechanism underlying PLOD3-induced metastasis and indicate an indispensable role of PLOD3 in the regulation of the STAT3 pathway in this process.
These results render PLOD3 as the first upstream coactivator of STAT3 and increase the possibility that the commonly observed STAT3 upregulation in human lung cancer may be mechanistically associated with the upregulation of PLOD3 oncogene. The mechanistic study clearly revealed that PLOD3 knockdown inhibits STAT3 activation, suggesting that inactivation of STAT3 signaling by PLOD3 may serve as an effective therapeutic approach for lung cancer. The mechanism underlying the MMP upregulation and promotion of invasiveness in PLOD3-overexpressing lung cancer cells warrants further investigation. To completely elucidate the mechanisms underlying PLOD3-induced lung cancer cell migration and invasion, we further investigated the MAPK-associated signaling pathways in cellular invasion. Many studies have assessed the association between the MAPK signaling pathway and cancer metastasis and invasion54. However, the present study reports that activation of the Ras/MAPK signaling pathway is not essential for PLOD3 knockdown-induced invasion of lung cancer cells. Furthermore, hydroxylation of the monovalent protein can modulate protein stability, thus regulating STAT3-Y705 phosphorylation by reducing JAK1 stability upon PLOD3 inhibition. In addition, hydroxylation can modulate the protein–protein interactions, thereby regulating STAT3 phosphorylation by regulating the binding of STAT3 to its corresponding kinase40. Hence, it is anticipated that hydroxylation regulates STAT3 phosphorylation via STAT3 and PLOD3 in the complex by binding in the cytoplasm. In addition, PLOD3 knockdown is expected to downregulate JAK1; therefore, we consider that the JAK-PLOD3-STAT3 axis plays an important signaling role in PLOD3-induced lung cancer metastasis.
Thus far, the mechanisms underlying tumor cell migration and invasion involving PLOD3 have been unknown. EMT, characterized by downregulation of epithelial markers, particularly E-cadherin, and upregulation of mesenchymal markers, particularly vimentin, N-cadherin, and several key EMT transcription factors, e.g., snail and slug, is crucial for cancer cell migration and invasion in various cancer types55. Therefore, to elucidate the precise mechanisms underlying PLOD3-induced cell migration and invasion, the effects of PLOD3 on EMT-associated proteins require further investigation. Further studies are required to investigate the clinical relevance of quantifying the expression of secreted PLOD3 to develop a new target biomarker. Furthermore, PLOD3 and rapid analysis of biopsy specimens via pathological examination (picrosirius red and Masson’s trichrome staining) will provide a credible prognostic signature for the clinical evaluation of genes, such as EGFR, KRAS, and ALK56,57.
In conclusion, the present study identified PLOD3 as a novel pro-metastatic factor in lung cancer and as a novel tumor oncogene with an essential role in inducing metastasis, potentially serving as a promising prognostic biomarker for lung cancer. Additionally, verification of the serum levels of secreted PLOD3 confirmed the value of the model and the strategy for the identification of novel candidate prognostic biomarkers with clinical potential. We suggest that monitoring of serum PLOD3 levels may, in most cases, provide a more sensitive method for early detection of potentially curable cases of lung cancer metastasis.
Materials and methods
Cell culture and treatment
Human lung cancer cell lines (H460 and A549) were purchased from the ATCC (Manassas, VA, USA), and we generated a radioresistant H460 (R-H460) cell line derived from parental radiosensitive H460 lung cancer cells treated cumulatively with 2-Gy radiation twice a week for 20 weeks20. H460, A549, and R-H460 cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum. Cells were irradiated using a 137Cs-ray source (Atomic Energy of Canada, Ltd.) at a dose rate of 3.81 Gy/min. Cells were treated with SB203580 or SP600125 to block MAP kinase activation or S3I-201 as a STAT3 inhibitor.
Immunohistochemistry
Lung cancer tissue arrays (Cat No. CC5, CCN5) were obtained from Superbiochips Laboratories (Seoul, Korea). Tissue arrays contained 59 sections prepared from tumor tissue obtained from 59 lung cancer patients. Immunohistochemical staining was performed using anti-PLOD3 rabbit polyclonal antibody (Proteintech Group; 1:50), followed by the avidin–biotin–peroxidase method in accordance with the manufacturer’s instructions (Invitrogen). The results were categorized on the basis of the staining intensity: “negative” when the total score was 0 and “positive” when the total score was ≥1. The percentage of staining was analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Data mining using ONCOMINE database, Kaplan–Meier plotter database, and cBioportal
PLOD3 gene expression was analyzed using microarray gene expression datasets from the ONCOMINE database (https://www.oncomine.org). The cancer type was defined as lung cancer; data type, mRNA; analysis type, cancer vs normal analysis. Kaplan–Meier survival curves relative to PLOD3 expression were generated for lung cancer. Data were analyzed using the KM-plotter (http://kmplot.com/). Co-expression data for PLOD3 and MMP-2, MMP-9, and uPA were extracted from the cancer TCG RNA-seq data in the cBioportal database (https://www.cbioportal.org, Lung Squamous Cell Carcinoma, TCGA, Provisional).
PLOD3 secretion assay
R-H460 or A549 cells were cultured at a density of 80%, and then the medium was changed to serum-free RPMI1640. Cells were incubated for 6 h and centrifuged at <centrifuge speed> for <duration> at <temperature>; thereafter, the supernatants were collected for the PLOD3 secretion assay via western blotting (WB) and an enzyme-linked immunosorbent assay (ELISA) (MyBioSource, Cat No. MBS2516046). Mouse blood samples were collected in a tube containing K2EDTA (BD Microtainer) and centrifuged at 3000 rpm for 30 min at 4 °C. The supernatants were collected for the PLOD3 secretion assay via ELISA.
Western blot analysis
Western blot analyses were performed as described previously6, using the following primary antibodies: anti-PLOD3 (Proteintech Group, Inc., Chicago, IL, USA); anti-HA, anti-ERK1/2, anti-p38, anti-K-RAS, anti-H-RAS (Santa Cruz Biotechnology Inc.); anti-p-anti-ERK1/2, anti-p-p38, anti-p-JNK, anti-JNK, anti-STAT3, anti-p-STAT3 (Y705), and anti-p-STAT3 (S727) (Cell Signaling Technology Inc.) antibodies. β-actin (Sigma) was used as a loading control.
Plasmid construction and transfections
Plasmids were constructed via standard recombinant cloning techniques and all changes were verified via DNA sequencing. Human PLOD3 cDNA (wild type) purchased from OriGene (Cat No. SC324563) was amplified via polymerase chain reaction (PCR) and cloned into pcDNA3.1-HA vectors. Wild-type PLOD3 was generated using primers 5ʹ-GATGGATCCATGACCTCCTCGGGGCCTGGA-3ʹ (BamHI) and 5ʹ-TGAATTCTCAGGGGTCGACAAAGGAC-3ʹ (EcoRI). Thereafter, PLOD3 was inserted between BamHI and XhoI restriction sites in pHA vectors. Transfection with these plasmids was performed using Mirus 2020 Reagent in accordance with the manufacturer's guidelines.
PLOD3 siRNA transfections
The following human-specific siRNAs, synthesized in accordance with the manufacturer’s instructions, were used: PLOD3 siRNA, 5ʹ-GGAAGUACAAGGAUGAUGAUGACGACGA-3ʹ (IDT). Transfection with these siRNA duplexes was performed using the Lipofectamine®RNAiMAX Reagent in accordance with the manufacturer's instructions.
Transwell migration and invasion assays
Transwell migration and invasion assays were conducted as described previously6. A 6.5-mm Transwell® with 8.0-μm pore polycarbonate membrane-coated inserts was purchased from Corning (NY, USA). Cells were seeded in 100-mm dishes (1 × 106 cells) and incubated for 24 h. After transfection, cells were cultured in complete medium for an additional 8 h. Cell density was adjusted to 5 × 105 cells/ml to account for non-adhered cells. For the invasion assay, 5 × 104 cells in 100 μl of serum-free RPMI1640 medium was seeded in the upper chamber of the insert, with 10% Matrigel on the upper chamber membrane. Thereafter, 600 μl of RPMI1640 supplemented with 0.1% BSA was added to the lower chamber, followed by incubation for 18 h. The medium and cells were then eliminated from the upper chamber using cotton swabs. The cells were fixed with 800 μl of methanol for 1 min, stained with hematoxylin for 3 min, washed thrice with distilled water, and enumerated using a microscope.
Cell viability assay
Cells were seeded at a density of 5000 cells per well in a 96-well plate and incubated for 24 h, in accordance with the indicated experimental conditions. To quantify cell viability, an equal volume of culture media containing Cyto XTM Reagent (LPS solution) was added to the cells and incubated for 4 h. Cell viability was then quantified using Multiskan EX (Thermo) at 450 nm.
In vitro wound-healing assay
An in vitro wound-healing assay was performed as described previously58. Cells were seeded in 100-mm dishes (1 × 106 cells) and incubated for 24 h. After transfection, cells were cultured in complete medium for an additional 8 h. Cells were cultured in a six-well plate until confluent monolayers were formed. Each cellular monolayer was then wounded or scratched with a 200-μl tip. Cells were rinsed twice with PBS and incubated with fresh media, and images were captured at 0 h, using an OLYMPUS CKX41 inverted phase-contrast microscope at ×10 magnification.
Tumor xenograft animal model
Five-week-old male BALB/c nude mice were purchased from Orient Bio Inc. (Gapyeong, Korea), permitted to acclimate under laboratory conditions for 1 week, and provided ad libitum access to a non-purified commercial mouse diet (Superfeed Co., Wonju, Korea) and water. Thereafter, 1 × 106 R-H460 cells were subcutaneously injected into nude mice aged 6 weeks. Seven days after injection of R-H460 cells, the mice were injected with control siRNA or PLOD3 siRNA (40 μg of siRNA/mouse) via the tail vein every 2 days. Ten days after injection of R-H460 cells, 6-Gy X-ray radiation was applied to the R-H460 xenograft tumors. The tumor cells were allowed to proliferate for 3 weeks. Tumor growth was evaluated by measuring the length and width with electronic calipers, and tumor volume was calculated using the following formula: Volume = (Length × Width2 × 3.14)/6. Three weeks later, the injected nude mice were killed, and the tumor tissues were excised, weighed, and fixed in 4% paraformaldehyde solution for further analysis. Tumor growth was evaluated on the basis of tumor volume (mean ± SD), which was plotted against time (KIRAMS 2015–0070).
Quantitative reverse transcription-PCR (RT-qPCR) analysis
Total RNA was isolated using an RNeasy mini kit (QIAGEN). Quantitative real-time PCR was performed in triplicate, using a PIKOREAL 96 (Thermo) and SYBR Premix Ex Taq (Takara Bio, Shiga, Japan). A two-temperature thermocycling program was used, with 42 cycles of 95 °C (denaturation) and 55 °C (annealing). Target mRNA expression levels were normalized to that of GAPDH in the same reaction.
In situ proximity ligation assay
The in situ proximity ligation assay (PLA) was performed as described previously58. Paraformaldehyde-fixed R-H460 cells were permeabilized with 0.2% Triton X-100, washed, and blocked with blocking solution (Olink Bioscience). Mouse monoclonal anti-PLOD3 antibody (Proteintech Group, Inc.) and rabbit polyclonal anti-STAT3 antibody (Cell Signaling) were used for the proximity ligation reaction. The assay was performed using the Duolink Detection Kit with a pair of nucleotide-labeled secondary antibodies (rabbit PLA probe MINUS and mouse PLA probe PLUS; Olink Bioscience) in accordance with the manufacturer’s protocol. Nuclei were stained with Hoechst obtained from the PLA reagent kit. Amplified PLA signals, represented as red fluorescent dots, were analyzed using a confocal laser-scanning microscope (Leica Microsystems, Heidelberg, Germany).
Immunoprecipitation
R-H460 cells were transfected with the vector under the experimental conditions for 48 h. Cells were washed twice with PBS, collected, and lysed for 30 min in NP-40 buffer (50 mM Tris-HCl (pH 8), 150 mM NaCl, 1% NP-40, and protease inhibitor (×100)) containing protease and phosphatase inhibitors. After centrifugation (10 min at 15,000 × g) to eliminate particulate material, the supernatant was incubated with antibodies (1:100) with constant agitation at 4 °C. The immunocomplexes were precipitated with protein A-sepharose (Sigma) and analyzed via immunoblotting.
Statistical analyses
Cell culture experiments were performed at least in triplicate. All data are expressed as mean ± standard deviation values. Statistical differences between groups were assessed using Student’s t-test (two-tailed) and ANOVA analysis. P-values were interpreted as follows; not significant (ns), *P < 0.05, **P < 0.01, and ***P < 0.001.
Electronic supplementary material
Acknowledgements
This study was supported by a National Research Foundation of Korea (NRF) grant (No. 2017 M2A2A7A01019417) and a grant from the Korea Institute of Radiological and Medical Sciences (KIRAMS), which was funded by the Ministry of Science, ICT (MSIP) Republic of Korea (50531–2018).
Author contributions
S.H. and E.K. conceived and designed the study and the experimental setup and wrote the manuscript. J.B., H.Y., G.K., J.K., C.L., J.S., H.U., C.K., J.P., and J.K. performed the experiments. S.H. and E.K analyzed the data. All authors read and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by C. Munoz-Pinedo
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Eun Ho Kim, Sang-Gu Hwang
Contributor Information
Eun Ho Kim, Email: eh140149@kirams.re.kr.
Sang-Gu Hwang, Email: sgh63@kcch.re.kr.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-1186-5).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–1137. doi: 10.1002/cncr.21324. [DOI] [PubMed] [Google Scholar]
- 3.Chen VW, et al. Analysis of stage and clinical/prognostic factors for lung cancer from SEER registries: AJCC staging and collaborative stage data collection system. Cancer. 2014;120:3781–3792. doi: 10.1002/cncr.29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, et al. AZD3759, a BBB-penetrating EGFR inhibitor for the treatment of EGFR mutant NSCLC with CNS metastases. Sci. Transl. Med. 2016;8:368ra172–368ra172. doi: 10.1126/scitranslmed.aag0976. [DOI] [PubMed] [Google Scholar]
- 5.Wong MC, Lao XQ, Ho KF, Goggins WB, Shelly L. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci. Rep. 2017;7:14300. doi: 10.1038/s41598-017-14513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yun HS, et al. Radiotherapy diagnostic biomarkers in radioresistant human H460 lung cancer stem-like cells. Cancer Biol. Ther. 2016;17:208–218. doi: 10.1080/15384047.2016.1139232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reichenberger Ernst, Olsen Bjorn R. Collagens as organizers of extracellular matrix during morphogenesis. Seminars in Cell & Developmental Biology. 1996;7(5):631–638. doi: 10.1006/scdb.1996.0077. [DOI] [Google Scholar]
- 8.Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol. Cell. 1997;1:13–23. doi: 10.1016/S1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 9.Shrivastava A, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol. Cell. 1997;1:25–34. doi: 10.1016/S1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- 10.Hausmann E. Cofactor requirements for the enzymatic hydroxylation of lysine in a polypeptide precursor of collagen. Biochim. Biophys. Acta. 1967;133:591–593. doi: 10.1016/0005-2795(67)90566-1. [DOI] [PubMed] [Google Scholar]
- 11.Rhoads RE, Udenfriend S. Decarboxylation of alpha-ketoglutarate coupled to collagen proline hydroxylase. Proc. Natl. Acad. Sci. USA. 1968;60:1473–1478. doi: 10.1073/pnas.60.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salo AM, et al. The lysyl hydroxylase isoforms are widely expressed during mouse embryogenesis, but obtain tissue-and cell-specific patterns in the adult. Matrix Biol. 2006;25:475–483. doi: 10.1016/j.matbio.2006.08.260. [DOI] [PubMed] [Google Scholar]
- 13.Heikkinen J, et al. Lysyl hydroxylase 3 is a multifunctional protein possessing collagen glucosyltransferase activity. J. Biol. Chem. 2000;275:36158–36163. doi: 10.1074/jbc.M006203200. [DOI] [PubMed] [Google Scholar]
- 14.Myllylä R, et al. Expanding the lysyl hydroxylase toolbox: new insights into the localization and activities of lysyl hydroxylase 3 (LH3) J. Cell Physiol. 2007;212:323–329. doi: 10.1002/jcp.21036. [DOI] [PubMed] [Google Scholar]
- 15.Gelse K, Pöschl E, Aigner T. Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Salo AM, et al. Lysyl hydroxylase 3 (LH3) modifies proteins in the extracellular space, a novel mechanism for matrix remodeling. J. Cell Physiol. 2006;207:644–653. doi: 10.1002/jcp.20596. [DOI] [PubMed] [Google Scholar]
- 17.Gilkes D, et al. Procollagen lysyl hydroxylase 2 is essential for breast cancer metastasis. Mol. Cancer Res., molcanres. 2013;0629:2012. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Steinmann B, Raghunath M. Delayed helix formation of mutant collagen. Science. 1995;267:258–259. doi: 10.1126/science.7809634. [DOI] [PubMed] [Google Scholar]
- 19.Yeowell HN, Walker LC, Murad S, Pinnell SR. A common duplication in the lysyl hydroxylase gene of patients with Ehlers Danlos syndrome type VI results in preferential stimulation of lysyl hydroxylase activity and mRNA by hydralazine. Arch. Biochem. Biophys. 1997;347:126–131. doi: 10.1006/abbi.1997.0319. [DOI] [PubMed] [Google Scholar]
- 20.van der Slot AJ, et al. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 2003;278:40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 21.Takaluoma K, Lantto J, Myllyharju J. Lysyl hydroxylase 2 is a specific telopeptide hydroxylase, while all three isoenzymes hydroxylate collagenous sequences. Matrix Biol. 2007;26:396–403. doi: 10.1016/j.matbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Steinmann B, Eyre DR, Shao P. Urinary pyridinoline cross-links in Ehlers-Danlos syndrome type VI. Am. J. Hum. Genet. 1995;57:1505. [PMC free article] [PubMed] [Google Scholar]
- 23.Brinckmann J, et al. Altered x-ray diffraction pattern is accompanied by a change in the mode of cross-link formation in lipodermatosclerosis. J. Invest. Dermatol. 1996;107:589–592. doi: 10.1111/1523-1747.ep12582991. [DOI] [PubMed] [Google Scholar]
- 24.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Eisinger-Mathason TK, et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3:1190–1205. doi: 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy DE, Darnell J., Jr Signalling: Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 28.Barton BE, Karras JG, Murphy TF, Barton A, Huang HF. Signal transducer and activator of transcription 3 (STAT3) activation in prostate cancer: direct STAT3 inhibition induces apoptosis in prostate cancer lines. Mol. Cancer Ther. 2004;3:11–20. doi: 10.1186/1476-4598-3-11. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, et al. Mitochondrial-localized Stat3 promotes breast cancer growth via phosphorylation of serine 727. J. Biol. Chem. 2013;M113:505057. doi: 10.1074/jbc.M113.505057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 31.Xie Tx, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 32.Jia ZH, et al. Phosphorylation of STAT3 at Tyr705 regulates MMP-9 production in epithelial ovarian cancer. PLoS One. 2017;12:e0183622. doi: 10.1371/journal.pone.0183622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itoh Y, Nagase H. Matrix metalloproteinases in cancer. Essays Biochem. 2002;38:21–36. doi: 10.1042/bse0380021. [DOI] [PubMed] [Google Scholar]
- 34.Danø, K. et al. Plasminogen Activators, Tissue Degradation, and Cancer. Adv Cancer Res.44,139–266 (1985). [DOI] [PubMed]
- 35.Bacchiocchi R, et al. Prognostic value analysis of urokinase-type plasminogen activator receptor in oral squamous cell carcinoma: an immunohistochemical study. Bmc. Cancer. 2008;8:220. doi: 10.1186/1471-2407-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajanbabu A, Ahmad SZ, Vijaykumar DK, Pavithran K, Kuiakose S. The significance of the site of origin in primary peritoneal carcinosarcoma: case report and literature review. Ecancermedicalscience. 2013;7:295. doi: 10.3332/ecancer.2013.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lung Cancer, http://kmplot.com/analysis/index.php?p=service&cancer=lung
- 38.Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastas.-. Rev. 2005;24:315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- 39.Harada D, Takigawa N, Kiura K. The role of STAT3 in non-small cell lung cancer. Cancers. 2014;6:708–722. doi: 10.3390/cancers6020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zurlo G, Guo J, Takada M, Wei W, Zhang Q. New insights into protein hydroxylation and its important role in human diseases. Biochim. Biophys. Acta. 2016;1866:208–220. doi: 10.1016/j.bbcan.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravichandran K, Tyagi A, Deep G, Agarwal C, Agarwal G. Interleukin-1-induced iNOS expression in human lung carcinoma A549 cells: involvement of STAT and MAPK pathways. Indian J. Exp. Biol. 2011;49:840–847. [PubMed] [Google Scholar]
- 42.Gilkes DM, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73:3285–3296. doi: 10.1158/0008-5472.CAN-12-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, et al. Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. J. Clin. Invest. 2015;125:1147–1162. doi: 10.1172/JCI74725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, et al. The glycosyltransferase activities of lysyl hydroxylase 3 (LH3) in the extracellular space are important for cell growth and viability. J. Cell Mol. Med. 2009;13:508–521. doi: 10.1111/j.1582-4934.2008.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng L, et al. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med. Genom. 2012;5:14. doi: 10.1186/1755-8794-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicastri A, et al. N-glycoprotein analysis discovers new up-regulated glycoproteins in colorectal cancer tissue. J. Proteome Res. 2014;13:4932–4941. doi: 10.1021/pr500647y. [DOI] [PubMed] [Google Scholar]
- 47.Schiarea S, et al. Secretome analysis of multiple pancreatic cancer cell lines reveals perturbations of key functional networks. J. Proteome Res. 2010;9:4376–4392. doi: 10.1021/pr1001109. [DOI] [PubMed] [Google Scholar]
- 48.Tsai CK, et al. Overexpression of PLOD3 promotes tumor progression and poor prognosis in gliomas. Oncotarget. 2018;9:15705. doi: 10.18632/oncotarget.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Q, et al. Barrier to autointegration factor 1, procollagen‐lysine, 2‐oxoglutarate 5‐dioxygenase 3, and splicing factor 3b subunit 4 as early‐stage cancer decision markers and drivers of hepatocellular carcinoma. Hepatology. 2018;67:1360–1377. doi: 10.1002/hep.29606. [DOI] [PubMed] [Google Scholar]
- 50.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med. Genom. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noda T, et al. PLOD2 induced under hypoxia is a novel prognostic factor for hepatocellular carcinoma after curative resection. Liver Int. 2012;32:110–118. doi: 10.1111/j.1478-3231.2011.02619.x. [DOI] [PubMed] [Google Scholar]
- 53.Dayer C, Stamenkovic I. Recruitment of matrix metalloproteinase-9 (MMP-9) to the fibroblast cell surface by lysyl hydroxylase 3 (LH3) triggers transforming growth factor-β (TGF-β) activation and fibroblast differentiation. J. Biol. Chem. 2015;290:13763–13778. doi: 10.1074/jbc.M114.622274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho HJ, et al. Ascofuranone suppresses PMA-mediated matrix metalloproteinase-9 gene activation through the Ras/Raf/MEK/ERK-and Ap1-dependent mechanisms. Carcinogenesis. 2006;28:1104–1110. doi: 10.1093/carcin/bgl217. [DOI] [PubMed] [Google Scholar]
- 55.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial–mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindeman NI, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 2013;8:823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narita Y, et al. Cost-effectiveness analysis of EGFR mutation testing and gefitinib as first-line therapy for non-small cell lung cancer. Lung Cancer. 2015;90:71–77. doi: 10.1016/j.lungcan.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Kim JS, Kim EJ, Oh JS, Park IC, Hwang SG. CIP2A modulates cell-cycle progression in human cancer cells by regulating the stability and activity of Plk1. Cancer Res. 2013;73:6667–6678. doi: 10.1158/0008-5472.CAN-13-0888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.