Abstract
This study investigated the prevalence of Listeria monocytogenes in slaughterhouses, and determined serovars and genotypes, and antibiotic resistance of the isolates obtained from slaughterhouses and humans in Korea. Two hundred ninety samples were collected from feces (n=136), carcasses [n=140 (cattle: n=61, swine: n=79)], and washing water (n=14) in nine slaughterhouses. Eleven human isolates were obtained from hospitals and the Korea Center for Disease Control and Prevention. Listeria monocytogenes was enriched and identified, using polymerase chain reaction (PCR) and 16S rRNA sequencing. Serovars and presence of virulence genes were determined, and genetic correlations among the isolates were evaluated by the restriction digest patterns of AscI. Antibiotic resistance of L. monocytogenes isolates were examined against 12 different antibiotics. Of 290 slaughterhouse samples, 15 (5.17%) carcass samples were L. monocytogenes positive. Most L. monocytogenes isolates possessed all the virulence genes, while polymorphisms in the actA gene were found between carcass and human isolates. Serovars 1/2a (33.3%) and 1/2b (46.7%) were the most frequent in carcass isolates. Genetic correlations among the isolates from carcass and clinical isolates were grouped within serotypes, but there were low geographical correlations. Most L. monocytogenes isolates were antibiotic resistant, and some strains showed resistance to more than four antibiotics. These results indicate that L. monocytogenes are isolated from carcass and human in Korea, and they showed high risk serotypes and antibiotic resistance. Therefore, intensive attentions are necessary to be aware for the risk of L. monocytogenes in Korea.
Keywords: Listeria monocytogenes, slaughterhouse, epidemiological analysis, molecular typing, antibiotic
Introduction
Listeria monocytogenes is a Gram-positive zoonotic pathogen, responsible for listeriosis (Fagerlund et al., 2016; Pérez-Trallero et al., 2014). The pathogen is ubiquitous in the environment, and has been detected in water and soil as well as animal feces. Cross-contamination of carcasses can occur in slaughterhouses or meat processing plants, resulting in transmission to consumers, and it causes fatal illness, especially in the susceptible populations such as the elderly, pregnant women, and fetuses (Sofos and Geornaras, 2010). To treat listeriosis patients, ampicillin, gentamycin, or streptomycin are primarily used, but the resistance of L. monocytogenes has been increased to the antibiotics used for therapeutic purposes in human (Gerzova et al., 2015).
Meloni et al. (2013) investigated the prevalence of L. monocytogenes in swine carcasses in Italy from 2008 to 2011, and they reported that 33% of swine carcasses were L. monocytogenes positive with only two serotypes (1/2a and 1/2c). Khen et al. (2015) investigated L. monocytogenes contamination in beef carcasses in the Republic of Ireland from July 2007 to June 2009, and reported that 27% of bovine hide and 14% of pre-chill carcasses were contaminated with L. monocytogenes, and the most commonly isolated serotype groups were 1/2a and 4b. In the US, the prevalence of L. monocytogenes in cattle carcasses was 0%–23.2% depending on slaughterhouse and season (Guerini et al., 2007).
L. monocytogenes outbreaks in the US have recently been linked to consumption of packaged salad, soft cheese and cantaloupes, and the pathogen caused 292 deaths or fetal losses from 2009 to 2011, with a mortality rate of approximately 21% (CDC, 2013; CDC, 2016). In the EU member states, L. monocytogenes outbreaks were linked to the intake of ready-to-eat meat and poultry, cheese and fishery products (EFSA, 2013a). In 2011, 0.32 cases per 10,000 individuals were reported in the EU member states (EFSA, 2007; EFSA, 2013b). In Japan, the first L. monocytogenes outbreak was reported in 2001, caused by consumption of contaminated cheese (Makino et al., 2005). The incidence rate in Japan was estimated to be 0.65 cases per million (83 cases per year), which is lower than that in the US and EU member states (Okutani et al., 2004). In Korea, L. monocytogenes is considered not threatening human because there are no reported L. monocytogenes outbreaks. However, L. monocytogenes has been isolated from various foods such as sausages, smoked duck, and smoked salmon in Korea. In addition, the statistics of HIRAS (2016) showed that the number of listeriosis cases has increased from 16 in 2012 to 33 in 2014. These results indicate that there should be serious attention on L. monocytogenes in Korea.
The antibiotics consumption through self-prescription by farmers for prevention and treatment of animal disease increased by 25% from 2003 to 2012 (Lim et al., 2014b). Since then, many studies reported the higher prevalence of antibiotic resistant Escherichia coli, Salmonella spp., Campylobacter spp. and Staphylococcus aureus in Korea (Chae et al., 2011; Lim et al., 2011; Lim et al., 2014a; Kim et al., 2011). Although L. monocytogenes rarely acquired antibiotic resistance, the first antibiotic resistant L. monocytogenes was found in 1988 (Altuntas et al., 2012), and recent studies reported the antibiotic resistances of L. monocytogenes to ampicillin, amoxicillin, gentamicin, chloramphenicol, erythromycin, tetracycline, and vancomycin (Chen et al., 2009; Ennaji et al., 2008; Pesavento et al., 2010; Yücel et al., 2005). However, there are only limited Korean studies on the prevalence of antibiotic resistant L. monocytogenes.
The objective of this study was to investigate the prevalence of L. monocytogenes in slaughterhouses and to determine serotype, genetic correlations, and antibiotic resistances of L. monocytogenes isolated from slaughterhouses and humans in Korea.
Materials and Methods
Sample collection
Samples were collected from nine slaughterhouses (four in central area, three in southeastern area, and two in southwestern area) in Korea. In each slaughterhouse, three swine farms and three cattle farms were randomly selected, and three carcasses from each farm were randomly selected for sampling. After evisceration, fecal samples (n=136) were collected aseptically by cutting the cecum with a sterile scalpel, and these samples were placed in plastic vials. To obtain carcass samples (n=140) which were identical carcasses for collected fecal samples, 10 cm×10 cm sections of the rump areas of carcasses were swabbed with sterile gauze pads rehydrated with 40 mL buffered peptone water (BPW; Becton, Dickinson and Company, Sparks, MD, USA). Fourteen samples of the water used to wash the carcass were collected in conical tubes as environment samples. All slaughterhouse samples were stored at cold temperature until analyzed. In addition, 11 human isolates of L. monocytogenes were obtained from hospitals and the Korean Centers for Disease Control and Prevention.
L. monocytogenes isolation
Carcass samples were homogenized with 50 mL Listeria enrichment broth (LEB; Becton, Dickinson and Company) in a pummeler (BagMixer® 400, Interscience, Saint Nom, France). The homogenates, fecal (25 g) and water (10 mL) samples in 50 mL LEB were enriched at 30°C for 24 h. For secondary enrichment, 1 mL aliquot of the primary enrichment were inoculated in 9 mL Fraser broth (Becton Dickinson and Company) supplemented with Fraser broth supplement (Becton, Dickinson and Company) and incubated at 37°C for 24–48 h. A loopful aliquot of the secondary enrichments was streaked on a Palcam agar (Becton, Dickinson and Company), followed by incubation at 30°C for 48 h. Presumptive L. monocytogenes colonies on the Palcam agar plates were inoculated in 10 mL tryptic soy broth plus 0.6% yeast extract (TSBYE; Becton, Dickinson and Company), followed by incubation at 30°C for 48 h.
A loopful aliquot of the cultures was then streaked on brain-heart infusion (BHI) agar (Becton, Dickinson and Company) and incubated at 30°C for 24 h. The isolated colonies on the plates were identified by polymerase chain reaction (PCR) analysis with prs (specific for Listeria spp.) primers [F (5’ GCTGAAGAGATTGCGAAAGAAG 3’) and R (5’ CAAAGAAACCTTGGATTTGCGG 3’)] (Doumith et al., 2004) and hlyA (specific for L. monocytogenes) primers [F (5’CCT AAC ATA TCC AGG TGC TCT C 3’) and R (5’ CTG ATT GCG CCG AAG TTT AC 3’)] (Burall et al., 2011). PCR amplification was conducted using Phire Hot Start II DNA Polymerase Kit (Thermo Fisher, Waltham, MA, US) and performed by Rotor-Gene Q (Qiagen, Hilden, Germany). After an initial stage of 98°C for 30 sec, 35 cycles were performed with denaturation at 98°C for 5 sec, annealing at 60°C for 5 sec, and extension at 72°C for 10 sec. A final 1-min extension was performed at 72°C.
The hlyA-positive samples were further-analyzed by 16S rRNA sequencing to identify L. monocytogenes. The primers 27F (5' AGA GTT TGA TCM TGG CTC AG 3') and 1492R (5' TAC GGY TAC CTT GTT ACG ACT T 3') were used for the 16S rRNA sequencing PCR (Lane, 1991; Weisburg et al., 1991). The PCR reaction was performed with 20 ng genomic DNA as the template in a 30-μL reaction mixture by using EF-Taq (Solgent, Daejeon, Korea). Activation of Taq polymerase was performed at 95°C for 2 min, followed by 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min and a final stage of 72°C for 10 min. The amplified products were purified with a multiscreen filter plate (Millipore Corp., Bedford, MA, USA). The DNA samples containing the extension products were added to Hi-Di formamide (Applied Biosystems, Foster City, CA, USA); the mixture was incubated at 95°C for 5 min and on ice for 5 min, and then analyzed using a ABI Prism 3730XL DNA analyzer (Applied Biosystems).
Polymerase chain reaction (PCR) analysis of virulence factors
Five virulence genes (actA, inlA, inlB, plcB, and hlyA) were detected in isolated colonies from Palcam agar plates, using PCR analysis with the primers listed in Table 1. The colonies in Palcam agar plates were suspended in 50 μL of 0.05N NaOH (Samchun, Gyeonggi, Korea) with 0.25% sodium dodecyl sulfate (SDS). One hundred microliters of sterile dH2O were added to the suspension, which was incubated at 99°C for 15 min. For PCR amplification, this mixture (2 μL) was mixed with Phire Hot Start II DNA Polymerase Kit (Thermo Fisher), mixed Taq DNA polymerase (20 mM Tris-HCl, pH 7.4 at 25°C; 0.1 mM EDTA; 1 mM DTT; 100 mM KCl; 200 μg/mL BSA; and 50% glycerol), 1× reaction buffer [1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTP; Promega Corporation, Madison, USA)], and 0.5 μM each primer. PCR was performed by Rotor-Gene Q (Qiagen) with initial denaturation at 98°C for 30 sec, followed by 35 cycles of 98°C for 5 sec, 60°C for 5 sec, and 72°C for 10 sec and a final extension at 72°C for 1 min. Twenty microliters of reaction PCR products were mixed with 4 μL loading star (Dyne Bio, Gyeonggi, Korea), followed by electrophoresis analysis with a 1.5% agarose gel.
Table 1. Primers used for PCR amplification of virulence genes from Listeria monocytogenes isolates.
| Primer | Size (bp) | Sequence (5´ to 3´) | Reference |
|---|---|---|---|
| inlA | 255 | F :
CCTAGCAGGTCTAACCGCAC R : TCGCTAATTTGGTTATGCCC |
Mathakiya et al., 2009 |
| inlB | 146 | F :
AAAGCACGATTTCATGGGAG R : ACATAGCCTTGTTTGGTCGG |
Corantin et al., 2005 |
| hlyA | 174 | F :
GCATCTGCATTCAATAAAGA R : TGTCACTGCATCTCCGTGGT |
Wesley et al., 2002 |
| actA | 268(385) | F :
GACGAAAATCCCGAAGTGAA R : CTAGCGAAGGTGCTGTTTCC |
Jaradat et al., 2002 |
| plcB | 261 | F :
GGGAAATTTGACACAGCGTT R : ATTTTCGAAGGTAGTCCGCTTT |
Corantin et al., 2005 |
PCR, Polymerase chain reaction.
L. monocytogenes serotyping
Serotypes for L. monocytogenes isolates were determined by both agglutination and multiplex-PCR analysis. Common results for serotypes from both analyses were used as the serotypes for the isolates. For agglutination analysis, Listeria antisera (Denka Seiken, Tokyo, Japan) were used to identify O antigen and H antigen according to the manufacturer’s instruction. Multiplex-PCR analysis was performed according to the previous methods with lmo0737, lmo1118, ORF2819, ORF2110, and prs gene target primers (Doumith et al., 2004). Amplification reactions for all isolates were performed using the Qiagen multiplex PCR kit (Qiagen) in Rotor-Gene Q with pre-incubation at 94°C for 15 min, followed by 35 cycles at 94°C for 30 sec, 57°C for 90 sec, and 72°C for 60 sec and terminal elongation at 60°C for 30 min. The final multiplex PCR products mixed with loading star were resolved on 1.5% agarose within 1× TAE buffer for 20 min. Subsequently, the serotypes of L. monocytogenes were determined by multiplex PCR amplification patterns.
Pulsed-field gel electrophoresis
L. monocytogenes isolates were analyzed using pulsed-field gel electrophoresis (PFGE) according to the standard protocol by Graves and Swaminathan (2001) with some modifications. L. monocytogenes cultures were embedded in 1% SeaKem gold agarose (Cambrex, NJ, USA) and lysed with cell lysis buffer (50 mM Tris, 50 mM EDTA, 1% Sarcosine) and proteinase K for 2 h at 54–55°C. Molds of the samples were washed with TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) for seven times, followed by digestion with restriction enzyme AscI (Enzynomics, Korea) at 37°C for 2 h. Size separation of restricted DNA fragments was performed in 0.5× Tris Borate EDTA (TBE) by using a CHEF MAPPER XA system (Bio-Rad, Richmond, CA, USA) with 1% SeaKem gold agarose gel at 14°C and 6.0 V/cm angle of 120° with a linear ramping factor of 2.2–54 sec for 19 h. XbaI-digested Salmonella enterica-serotype Braenderup H9812 DNA was used as a standard to compare each band of L. monocytogenes isolates. After electrophoresis, the agarose gel was stained with SYBR gold (Molecular Probes, Eugene, USA) and destained with dH2O for 1 h. Pattern images were captured using an LAS-3000 imaging system (Fujifilm, Japan), and PFGE patterns were analyzed using Gelcompar II 6.6 software (Applied Maths, Sint-Matins-Latem, Belgium). Similarity clustering analyses of all isolates were performed by UPGMA (unweighted pair group method with arithmetic mean) with Dice correlation coefficient and 1% tolerance.
Antibiotic resistance analysis
Antibiotic disc diffusion test was performed according to Clinical Laboratory Standards Institute procedure to evaluate antibiotic resistances (CLSI, 2014). A single isolated colony of L. monocytogenes was transferred into 10 mL TSBYE, incubated at 30°C for 24 h. After the incubation, the culture was diluted with 5 mL phosphate buffered saline (PBS, pH 7.4; KH2PO4, 0.2 g; Na2HPO4, 1.5 g; NaCl, 8.0 g; KCl, 0.2 g in 1 L of dH2O) to a turbidity equivalent to 0.2 at OD620 (approximately 108 CFU/mL). Sterile swab was dampened with the diluent, and spread onto the surface of a Muller-Hinton agar plate (MHA; Becton, Dickinson and Company). The MHA plates were dried at room temperature for 10 min to allow adsorption of the liquid. Antibiotic discs (Oxoid, Basingstoke, UK) were then placed on the surface of the MHA plates. The 12 antibiotics (amounts) were examined as following; gentamycin (10 μg), penicillin G (10 units), tetracycline (30 μg), spectinomycin (100 μg), kanamycin (30 μg), erythromycin (15 μg), tigecycline (15 μg), ampicillin (10 μg), streptomycin (10 μg), vancomycin (30 μg), chloramphenicol (30 μg), and rifampicin (5 μg) (Oxoid). After incubation at 30°C for 24 h, the diameters of the clear zones were measured and sensitivities (susceptible, intermediates, and resistance) were determined according to the standards of the Clinical Laboratory Standard Institute (CLSI, 2014).
Results
Prevalence of L. monocytogenes
Fifteen samples (5.17%) of 290 samples such as carcasses, feces, and water samples from nine slaughterhouses were L. monocytogenes positive. The pathogen was isolated only from carcass samples, and not from fecal or water samples. Of 140 carcass samples, 15 samples (10.71%) were contaminated with the pathogen, which could be considered high prevalence (Table 2). L. monocytogenes was isolated from six (66.7%) of nine slaughterhouses (data not shown). Of 75 swine carcass samples, 11 samples (14.7%) were contaminated with L. monocytogenes, and four (6.6%) of 61 cattle carcass samples were contaminated with the pathogen (data not shown), indicating that the swine carcasses were more frequently contaminated with the pathogen than cattle carcasses in Korea.
Table 2. Prevalence of Listeria monocytogenes in the slaughterhouse.
| Slaughterhouse | Sample | Total (%) | ||
|---|---|---|---|---|
| Feces | Carcass (%) | Environment | ||
| Central 1 | 0/18 | 5/18(27.78) | 0/1 | 5/37(13.51) |
| Central 2 | 0/18 | 1/18(5.56) | 0/2 | 1/38(2.63) |
| Central 3 | 0/18 | 2/18(11.11) | 0/2 | 2/38(5.26) |
| Central 4 | 0/3 | 0/6 | 0 | 0/9 |
| South west 1 | 0/18 | 4/18(22.22) | 0/2 | 4/38(10.53) |
| South west 2 | 0/18 | 1/19(5.26) | 0/2 | 1/39(2.56) |
| South east 1 | 0/15 | 2/15(13.33) | 0/2 | 2/32(6.25) |
| South east 2 | 0/19 | 0/19 | 0/2 | 0/40 |
| South east 3 | 0/9 | 0/9 | 0/1 | 0/19 |
| Total | 0/136 | 15/140(10.71) | 0/14 | 15/290(5.17) |
Serotypes of L. monocytogenes isolates
Fifteen isolates from carcasses and 11 human isolates obtained from hospitals and KCDC in Korea were further analyzed for serotypes by multiplex PCR and agglutination assay. Multiplex PCR analysis classified L. monocytogenes isolates into four groups. Group 1 (1/2a and 3a), Group 2 (1/2c and 3c), Group 3 (1/2b, 3b, and 7), and Group 4 (4b, 4d, and 4e) of serovars as determined by lineage-specific and serogroup-specific sequences for L. monocytogenes strains according to the method by Doumith (2004). This method identified several serotypes rather than one serotype, and thus, an agglutination assay was necessary; however, the result of agglutination assay could be affected by researcher subjectivity. Therefore, common serotypes identified in both methods were considered the serotypes of the isolate. The results showed that the human isolates were three 1/2b isolates (27.3%), three 4b isolates (27.3%), and two 1/2a isolates (18.2%) (Table 3), which are known to be the most common serotypes for foodborne disease (Althaus et al., 2014). While slaughterhouse isolates were five 1/2a isolates (33.3%), seven 1/2b isolates (46.7%), and three 1/2c isolates (20%) from cattle and swine carcasses (Table 4). These results indicate that serotypes between slaughterhouse and human isolates are similar, and most strains were foodborne disease-related serotypes.
Table 3. Serotypes and information of Listeria monocytogenes isolates from humans.
| Strains | Sources | Serotype |
|---|---|---|
| SMFM-CI-1 | Male, 18 year-old | 4b |
| SMFM-CI-2 | Human, blood, 1/2a | 1/2a |
| SMFM-CI-3 | Female, blood, 82 year-old | 1/2b |
| SMFM-CI-4 | Female, blood, 72 year-old | 1/2b |
| SMFM-CI-5 | Male, blood, 67 year-old | 3b |
| SMFM-CI-6 | Male, blood, 71 year-old | 1/2b |
| SMFM-CI-7 | Male, blood, 73 year-old | 1/2a |
| SMFM-CI-8 | Male, stomach cancer, 66 year-old | 1/2c |
| SMFM-CI-9 | Female, bacterial meningitis, 61 year-old | 4d |
| SMFM-CI-10 | Male, bacterial meningitis, 78 year-old | 4b |
| SMFM-CI-11 | Female, hepatic abscess, 81 year-old | 4b |
Table 4. Serotypes and information of Listeria monocytogenes isolates from carcasses.
| Strains | Sampling location1) | Animal | Serotype |
|---|---|---|---|
| SMFM-SI-1 | A | Swine | 1/2b |
| SMFM-SI-2 | A | Swine | 1/2b |
| SMFM-SI-3 | A | Swine | 1/2b |
| SMFM-SI-4 | A | Swine | 1/2b |
| SMFM-SI-5 | A | Swine | 1/2b |
| SMFM-SI-6 | E | Cattle | 1/2a |
| SMFM-SI-7 | E | Cattle | 1/2c |
| SMFM-SI-8 | E | Cattle | 1/2c |
| SMFM-SI-9 | E | Cattle | 1/2c |
| SMFM-SI-10 | B | Cattle | 1/2a |
| SMFM-SI-11 | C | Swine | 1/2a |
| SMFM-SI-12 | C | Swine | 1/2a |
| SMFM-SI-13 | D | Swine | 1/2b |
| SMFM-SI-14 | D | Swine | 1/2b |
| SMFM-SI-15 | F | Swine | 1/2a |
1) A, slaughterhouse-1 in central area; B, slaughterhouse-2 in central area; C, slaughterhouse-3 in central area; D, slaughterhouse-1 in south west area; E, slaughterhouse-1 in south west area; F, slaughterhouse-2 in south west area.
Virulence characterization of L. monocytogenes
Five virulence genes (actA, inlA, inlB, hlyA, and plcB) were observed for L. monocytogenes slaughterhouse and human isolates, except for inlB in L. monocytogenes SMFM-SI-15 (Fig. 1). The strain did not contain inlB, which is involved in mediating invasion into mammalian cells (Carvalho et al., 2014). Interestingly, 10 of 11 human isolates (90.9%) had a 268-bp amplicon of actA, but most of 15 slaughterhouse isolates (86.7%) had a 385-bp amplicon of actA (Fig. 2).
Fig. 1. PCR amplification of InlB from Listeria monocytogenes isolates (SI) obtained from carcasses.

PCR amplification showed that L. monocytogenes isolate SI-15 has no inlB, which plays a role in invasion.
Fig. 2. PCR amplification of actA from Listeria monocytogenes isolates obtained from humans (A; CI) and carcasses (B; SI).
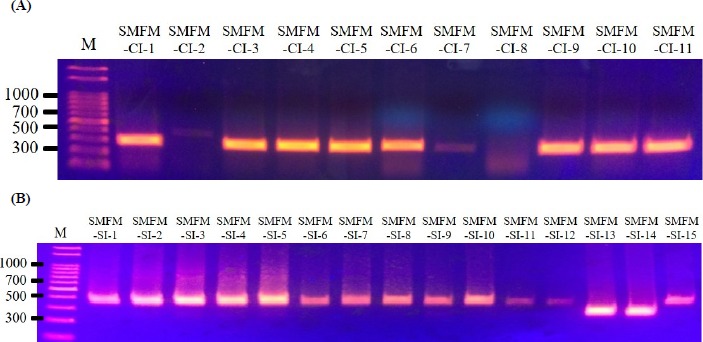
ActA gene showed generally different size of base pair between slaughterhouse and human isolates. 90.9% of human isolates had 268 bp size of actA, and 86.7% of slaughterhouse isolates had 385 bp.
PFGE genotype of L. monocytogenes isolates
Twenty-six L. monocytogenes slaughterhouse and human isolates were genotyped using PFGE (Fig. 3). The application of DNA macro restriction with AscI revealed major four groups with 70% similarity and 12 clusters with 90% similarity. All L. monocytogenes isolates could be divided into two major patterns at approximately 55.3% similarity, which were serotyped as 1/2a and 1/2c (59.3% similarity between groups) and 1/2b-4 (64.7% similarity). Group 1 (more than 80.4 % similarity) included five isolates of serotype 1/2c (n=4) and 1/2a. Group 2 included five isolates of serotype 1/2a; L. monocytogenes SMFM-CI-2 human isolate had 93.3 % similarity with L. monocytogenes strains SMFM-SI-6 (south west 1-swine) and SMFM-SI-10 (central 2-cattle). Group 3 included 11 isolates of serotype 1/2b (n=10) and 3b (n=1), which were divided into clusters based on actA size and geographical characterization. In Group 3, although isolation sources were different, the carcass isolates (L. monocytogenes strains SMFM-SI-13 and SMFM-SI-14) containing 268-bp actA showed more similarity (81.2%) with human isolates containing 268-bp actA, than carcass isolates (77.7%) with 385-bp actA. Group 4 contained serotype 4b and 4d isolates. Some L. monocytogenes isolates shared 100% similarity isolates from different locations.
Fig. 3. PFGE patterns observed for Listeria monocytogenes isolates from carcasses and humans.

Dendrogram (UPGMA clustering based on the Dice correlation coefficient) of L. monocytogenes AscI PFGE patterns for 26 clinical and slaughterhouse isolates. Region 1: central area, Region 2: south west area, Region, 3: south east area. PFGE, pulsed-field gel electrophoresis; UPGMA, unweighted pair group method with arithmetic mean.
Antibiotic resistance
All fifteen L. monocytogenes strains isolated from slaughterhouses were susceptible to the antibiotics, including gentamicin, bacitracin, spectinomycin, kanamycin, erythromycin, tigecycline, vancomycin, chloramphenicol, and rifampicin. However, the resistance rates of the isolates to penicillin G, tetracycline, ampicillin, and streptomycin resistances were 93.3%, 20%, 26.7%, and 60%, respectively (Table 5). L. monocytogenes strains SMFM-SI-13 and SMFM-SI-14 exhibited multiple resistances to four antibiotics such as tetracycline, penicillin, ampicillin, and streptomycin. L. monocytogenes SMFM-SI-12 also showed multiple resistance to penicillin G, ampicillin, and streptomycin. L. monocytogenes strains SMFM-SI-1, SMFM-SI-5, and SMFM-SI-15 had resistance only penicillin G, L. monocytogenes SMFM-SI-11 had resistance to streptomycin, and L. monocytogenes SMFM-SI-8 had no resistance for used antibiotics in this study. In clinical isolates, all 11 L. monocytogenes strains isolated from clinical patients were susceptible to the antibiotics such as gentamycin, spectinomycin, kanamycin, tigecycline, and chloramphenicol (Table 6). Six L. monocytogenes isolates (54.5%) from clinical patients had resistance more than three antibiotics, especially, two L. monocytogenes isolates (L. monocytogenes strains SMFM-CI-4 and SMFM-CI-7) showed multiple resistances to six antibiotics such as bacitracin, penicillin G, erythromycin, ampicillin, streptomycin, and rifampicin. These results indicate that clinical isolates had multiple antibiotic resistance more than the isolates from slaughterhouses.
Table 5. Antibiotic sensitivity of Listeria monocytogenes strains isolated from slaughterhouses (n=15).
| Antibiotics | Strains isolated from slaughterhouse | Sensitivity of the isolates (n, %) 1) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMFM-SI-1 | SMFM-SI-2 | SMFM-SI-3 | SMFM-SI-4 | SMFM-SI-5 | SMFM-SI-6 | SMFM-SI-7 | SMFM-SI-8 | SMFM-SI-9 | SMFM-SI-10 | SMFM-SI-11 | SMFM-SI-12 | SMFM-SI-13 | SMFM-SI-14 | SMFM-SI-15 | Susceptible | Intermediate | Resistant | |
| Gentamycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Bacitracin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Penicillin G | R | R | R | R | R | R | R | S | R | R | R | R | R | R | R | 1 (6.7) |
14 (93.3) |
|
| Tetracycline | S | S | S | S | S | S | S | S | R | S | S | S | R | R | S | 12 (80) |
3 (20) |
|
| Spectinomycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Kanamycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Erythro-mycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Tigecycline | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Ampicillin | S | I | I | S | S | I | S | S | S | R | S | R | R | R | I | 7 (46.6) |
4 (26.7) |
4 (26.7) |
| Strepto-mycin | I | R | R | R | I | R | R | I | I | I | R | R | R | R | I | 6 (40) |
9 (60) |
|
| Chloram-phenicol | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Rifampicin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | 15 (100) |
||
| Susceptible (n, %) |
10 (83.3) |
9 (75) |
9 (75) |
10 (83.3) |
10 (83.3) |
9 (75) |
10 (83.3) |
11 (91.7) |
9 (75) |
9 (75) |
11 (91.7) |
9 (75) |
8 (66.7) |
8 (66.7) |
9 (75) |
|||
| Intermediate (n, %) |
1 (8.3) |
1 (8.3) |
1 (8.3) |
0 | 1 (8.3) |
1 (8.3) |
0 | 1 (8.3) |
1 (8.3) |
1 (8.3) |
0 | 0 | 0 | 0 | 2 (16.7) |
|||
| Resistant (n, %) |
1 (8.3) |
2 (16.7) |
2 (16.7) |
2 (16.7) |
1 (8.3) |
2 (16.7) |
2 (16.7) |
0 | 2 (16.7) |
2 (16.7) |
1 (8.3) |
3 (25) |
4 (33.3) |
4 (33.3) |
1 (8.3) |
|||
According to CLSI guidelines using the breakpoints of Staphylococcus species resistance because no resistance criteria exist for Listeria susceptibility testing in the CLSI guidelines (Byrne et al., 2016).
Table 6. Antibiotic sensitivity of Listeria monocytogenes strains obtained from human (n=11).
| Antibiotics | Strains obtained from human | Sensitivity of the isolates (n, %)1) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMFM-CI-1 | SMFM-CI-2 | SMFM-CI-3 | SMFM-CI-4 | SMFM-CI-5 | SMFM-CI-6 | SMFM-CI-7 | SMFM-CI-8 | SMFM-CI-9 | SMFM-CI-10 | SMFM-CI-11 | Susceptible | Intermediate | Resistant | ||
| Gentamycin | S | S | S | S | S | S | S | S | S | S | S | 11 (100) |
|||
| Bacitracin | S | R | S | R | S | S | R | S | S | S | S | 8 (72.7) |
3 (27.3) |
||
| Penicillin G | R | R | R | R | R | R | R | R | R | R | R | 11 (100) |
|||
| Tetracycline | S | S | R | S | R | R | S | S | S | S | S | 8 (72.7) |
3 (27.3) |
||
| Spectinomycin | S | S | S | S | S | S | S | S | S | S | S | 11 (100) |
|||
| Kanamycin | S | S | S | S | S | S | S | S | S | S | S | 11 (100) |
|||
| Erythromycin | S | I | S | R | S | S | R | S | S | S | S | 8 (72.7) |
1 (9.1) |
2 (18.2) |
|
| Tigecycline | S | S | S | S | S | S | S | S | S | S | S | 11 (100) |
|||
| Ampicillin | S | R | S | R | S | S | R | S | S | S | S | 8 (72.7) |
3 (27.3) |
||
| Streptomycin | I | R | R | R | R | R | R | S | R | I | I | 1 (9.1) |
3 (27.3) |
7 (63.6) |
|
| Chloramphenicol | S | S | S | S | S | S | S | S | S | S | S | 11 (100) |
|||
| Rifampicin | S | R | S | R | S | S | R | S | S | S | S | 8 (72.7) |
3 (27.3) |
||
| Susceptible (n, %) |
10 (83.3) |
6 (50) |
9 (75) |
6 (50) |
9 (75) |
9 (75) |
6 (50) |
11 (91.7) |
10 (83.3) |
10 (83.3) |
10 (83.3) |
||||
| Intermediate (n, %) |
1 (8.3) |
1 (8.3) |
1 (8.3) |
1 (8.3) |
|||||||||||
| Resistant (n, %) |
1 (8.3) |
5 (41.7) |
3 (25) |
6 (50) |
3 (25) |
3 (25) |
6 (50) |
1 (8.3) |
2 (16.7) |
1 (8.3) |
1 (8.3) |
||||
1) According to CLSI guidelines using the breakpoints of Staphylococcus species resistance because no resistance criteria exist for Listeria susceptibility testing in the CLSI guidelines (Byrne et al., 2016).
Discussion
Although only a few cases of L. monocytogenes infection in humans are reported in Korea, it must be considered a major public health problem because of its ubiquitous distribution and presence in many animals, food products, raw materials, and environments. More information for the epidemiology, antibiotic resistant profiles and the genetic relationships of L. monocytogenes isolates from production chains would be helpful in understanding the cause of listeriosis and aspect of L. monocytogenes antibiotic resistance, and determining an appropriate approach for prevention of contamination.
In this study, 290 samples of carcasses, feces, and washing water from nine slaughterhouses in Korea were collected. Fifteen L. monocytogenes isolates were obtained, only from carcasses, not from feces and water samples. It suggests that pathogens on the carcasses may be cross-contaminated by workers or physical contact between carcasses in a slaughterhouse rather than cross-contamination from feces. This result was supported by a study of Muhterem-Uyar et al. (2015), suggesting that L. monocytogenes can be cross-contaminated onto meat carcasses from cutting board and packaging rooms due to unhygienic evisceration equipment. Thus, in our study it can be also suggested that L. monocytogenes on the carcasses might be from unhygienic environments.
Notably, at least 95% of L. monocytogenes strains isolated from listeriosis patients belonged to serotypes 1/2a, 1/2b, and 4b (Kathariou, 2002). The most frequent L. monocytogenes serotypes in the carcasses were 1/2a and 1/2b (80%), followed by 1/2c (20%), which has low pathogenic potential but sporadically causes listeriosis in animals (Hadjilouka et al., 2014). This result indicates that L. monocytogenes serotypes 1/2a, 1/2b, and 4b are the most prevalent isolates in carcasses and humans in Korea, which are the predominant serotypes causing human infections are 1/2a, 1/2b, and 4b (Swaminathan and Gerner-Smidt, 2007). In this study, most isolates harbored virulence genes (actA, inlA, inlB, hlyA, and plcB), indicating that they are pathogenic. Ten of the 11 clinical L. monocytogenes isolates showed 268 bp size of actA, rather than 385 bp size of actA which was the size of the gene in most of the slaughterhouse isolates. Jacquet el al. (2002) found that the low molecular mass of actA was due to a deletion in actA, and it is probably not in the amino-terminal region, which is essential for F-actin assembly and movement. The deletion likely occurred in the internal proline-rich regions or carboxy-terminal domain (Sokolovic et al., 1996). Wiedmann et al. (1997) suggested that the length of actA is related to pathogenic properties. Ha et al. (2018) reported that the invasion efficiency of L. monocytogenes strains with the 268 bp size of actA was 1–2.5 times lower than those of the strains with the 385 bp size of actA in healthy cells, but the invasion efficiency became similar in senescent cells. Thus, this actA size may be related to pathogenicity.
PFGE of the DNA of the 26 isolates digested with AscI showed 7–16 fragments of approximately 30 to 1,130 kb (Neves et al., 2008). Generally, AscI and ApaI are considered the most useful restriction enzymes for Listeria spp. (Howard et al., 1992; Jang et al., 2005). However, Carriere et al. (1991) reported that the DNA of many Listeria serovar 1/2c isolates was not digested by ApaI and NotI. Therefore, we chose AscI as it is more efficient for serovar 1/2c and 4c (Brosch et al., 1994; Jang et al., 2006). When the DI value quoted by Simpson’s discriminatory index was calculated (Hunter and Gaston, 1988), twenty six L. monocytogenes isolates from our study had 0.93 of DI. The isolates were divided into four groups with at least 70% similarity, and the serogroups clustered according to their pulsotypes: 1/2c (Group 1), 1/2a (Group 2), 1/2b (Group 3), and 4 types (Group 4). At the 0.90–1.00 similarity level, some clusters were diverse according to geographical origins and isolation origins. Thus, the results of PFGE indicates that L. monocytogenes serotypes are genetically correlated, and L. monocytogenes can be cross-contamination between different geographical locations, which may be caused by collecting animals from different farms but slaughtering in same slaughterhouse.
Although many foodborne pathogens were concerned because of their antibiotic resistance, the antibiotic resistance of L. monocytogenes has not been issued yet. To treat listeriosis various antibiotics such as penicillin, vancomycin, erythromycin, tetracycline, and chloramphenicol are currently used (Pesavento et al., 2010). Altuntas et al. (2012) reported that 7.1% L. monocytogenes strain isolate from animal food showed antibiotic resistance to streptomycin. In our study, the L. monocytogenes isolates showed the resistances to some of the therapeutic antibiotics such as for penicillin G (93.3%) and tetracycline (20%) isolated from slaughterhouses. Also, L. monocytogenes isolates from clinical patients have the resistances to penicillin G (100%) and streptomycin (63.6%). In addition, some isolates were multidrug resistant to more than three antibiotics. Twenty percent L. monocytogenes isolates from slaughterhouses and 54.5% isolates from clinical patients showed multidrug resistant. These results suggest that the antibiotic resistances of L. monocytogenes have been increased, especially to therapeutic antibiotics of listeriosis treatment for human, and thus, L. monocytogenes infection may cause worse pathological results than the past (Conter et al., 2009; Morvan et al., 2010; Srinivasan et al., 2005).
In conclusion, L. monocytogenes were isolated from carcasses in slaughterhouses in Korea, and the pathogens may be cross-contaminated in slaughterhouses. There were also human listeriosis patients, and the serotypes of the isolates from carcasses and humans were the one related to the foodborne illness such as 1/2a, 1/2b and 4b serotypes. In addition, the prevalence of multiantibiotic resistant L. monocytogenes was high, especially in human isolates. These results suggest that the risk of L. monocytogenes could be considered high in Korea. Therefore, intensive attention and control plans for L. monocytogenes are necessary.
Acknowledgment
This research was supported by a grant (Z-1543081-2014-15-01) from the Animal and Plant Quarantine Agency in 2014–2015.
References
- Althaus D, Lehner A, Brisse S, Maury M, Tasara T, Stephan R. Characterization of Listeria monocytogenes strains isolated during 2011–2013 from human infections in Switzerland. Foodborne Pathog Dis. 2014;11:753–758. doi: 10.1089/fpd.2014.1747. [DOI] [PubMed] [Google Scholar]
- Altuntas EG, Kocan D, Cosansu S, Ayhan K, Juneja VK, Materon L. Antibiotic and bacteriocin sensitivity of Listeria monocytogenes strains isolated from different foods. Food Nutr Sci. 2012;3:363–368. doi: 10.4236/fns.2012.33052. [DOI] [Google Scholar]
- Brosch R, Chen J, Luchansky JB. Pulsed-field fingerprinting of listeriae: Identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl Environ Microbiol. 1994;60:2584–2592. doi: 10.1128/aem.60.7.2584-2592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burall LS, Simpson AC, Datta AR. Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multiplex PCR assay for identifying the major serotypes of Listeria monocytogenes. J Food Prot. 2011;74:403–409. doi: 10.4315/0362-028X.JFP-10-355. [DOI] [PubMed] [Google Scholar]
- Byrne VDV, Hofer E, Vallim DC, Almeida RCDC. Occurrence and antimicrobial resistance patterns of Listeria monocytogenes isolated from vegetables. Braz J Microbiol. 2016;47:438–443. doi: 10.1016/j.bjm.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere C, Allardet-Servent A, Bourg G, Audurier A, Ramuz M. DNA polymorphism in strains of Listeria monocytogenes. J Clin Microbiol. 1991;29:1351–1355. doi: 10.1128/jcm.29.7.1351-1355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F, Sousa S, Cabanes D. How Listeria monocytogenes organizes its surface for virulence. Front Cell Infect Microbiol. 2014;4:48. doi: 10.3389/fcimb.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centersfor Disease Control and Prevention (CDC) Vital signs: Listeria illnesses, deaths, and outbreaks-United States, 2009-2011. 2013 Available from: http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6222a4.htm#tab1. Accessed at April 4, 2016. [PMC free article] [PubMed]
- Centersfor Disease Control and Prevention (CDC) Listeria outbreaks. 2016 Available from: http://www.cdc.gov/listeria/outbreaks/index.html. Accessed at March 6, 2016.
- Chae MH, Nam HM, Jang GC, Kim HJ, Kim SR, Jung SC, Kang DJ, Kim JK, Lim SK. Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli isolated from food animals and raw meats in slaughterhouse in Korea during 2010. Kor J Vet Publ Hlth. 2011;35:239–245. [Google Scholar]
- Chen J, Luo X, Jiang L, Jin P, Wei W, Liu D, Fang W. Molecular characteristics and virulence potential of Listeria monocytogenes isolates from Chinese food systems. Food Microbiol. 2009;26:103–111. doi: 10.1016/j.fm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement (M100-S24) 2014 [Google Scholar]
- Conter M, Paludi D, Zanardi E, Ghidini S, Vergara A, Ianieri A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int J Food Microbiol. 2009;128:497–500. doi: 10.1016/j.ijfoodmicro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Corantin H, Quessy S, Gaucher ML, Lessard L, Leblanc D, Houde A. Effectiveness of steam pasteurization in controlling microbiological hazards of cull cow carcasses in a commercial plant. Can J Vet Res. 2005;69:200–207. [PMC free article] [PubMed] [Google Scholar]
- Doumith M, Bucherieser C, Glaser P, Jaquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennaji H, Timinouni M, Ennaji MM, Hassar M, Cohen N. Characterization and antibiotic susceptibility of Listeria monocytogenes isolated from poultry and red meat in Morocco. Infect Drug Resist. 2008;1:45–50. doi: 10.2147/idr.s3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EuropeanFood Safety Authority (EFSA) The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. 2007 Available from: http://www.efsa.europa.eu/en/efsajournal/pub/130r. Accessed at March 27, 2016.
- European Food Safety Authority (EFSA) Analysis of the baseline survey on the prevalence of Listeria monocytogenes in certain ready-to-eat foods in the EU 2010–2011 Part A: Listeria monocytogenes prevalence estimates. EFSA J. 2013a;11:3241. doi: 10.2903/j.efsa.2013.3241. [DOI] [Google Scholar]
- European Food Safety Authority (EFSA) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 2013b;11:3129. doi: 10.2903/j.efsa.2013.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund A, Langsrud S, Schirmer BCT, Møretrø T, Heir E. Genome analysis of Listeria monocytogenes sequence type 8 strains persisting in salmon and poultry processing environments and comparison with related strains. Plos One. 2016;11:e0151117. doi: 10.1371/journal.pone.0151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerzova L, Babak V, Sedlar K, Faldynova M, Videnska P, Cejkova D, Jensen AN, Denis M, Kerouanton A, Ricci A, Cibin V, Österberg J, Rychlik I. Characterization of antibiotic resistance gene abundance and microbiota composition in feces of organic and conventional pigs from four EU countries. Plos One. 2015;10:e0132892. doi: 10.1371/journal.pone.0132892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LM, Swaminathan B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol. 2001;65:55–62. doi: 10.1016/s0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- Guerini MN, Brichta-Harhay DM, Shackelford SD, Arthur TM, Bosilevac JM, Kalchayanand N, Wheeler TL, Koohmaraie M. Listeria prevalence and Listeria monocytogenes serovar diversity at cull cow and bull processing plants in the United States. J Food Prot. 2007;70:2578–2582. doi: 10.4315/0362-028x-70.11.2578. [DOI] [PubMed] [Google Scholar]
- Ha J, Oh H, Kim S, Lee J, Lee S, Lee H, Choi Y, Moon SS, Choi KH, Yoon Y. Effect of gene actA on the invasion efficiency of Listeria monocytogenes, as observed in healthy and senescent intestinal epithelial cells. J Microbiol Biotechnol. 2018;28:59–64. doi: 10.4014/jmb.1710.10026. [DOI] [PubMed] [Google Scholar]
- Hadjilouka A, Andritsos ND, Paramithiotis S, Mataragas M, Drosinos EH. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J Food Prot. 2014;77:2115–2120. doi: 10.4315/0362-028X.JFP-14-072. [DOI] [PubMed] [Google Scholar]
- Health Insurance Review and Assessment Service (HIRAS) Disease statistics. 2016 Available from: http://opendata.hira.or.kr/op/opc/olap4thDsInfo.do. Accessed at April 4, 2016.
- Howard PJ, Harsono KD, Luchansky JB. Differentiation of Listeria monocytogenes, Listeria innocua, Listeria ivanovii, and Listeria seeligeri by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1992;58:709–712. doi: 10.1128/aem.58.2.709-712.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: An application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet C, Gouin E, Jeannel D, Cossart P, Rocourt J. Expression of actA, ami, inlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl Environ Microbiol. 2002;68:616–622. doi: 10.1128/AEM.68.2.616-622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SS, Choo E, Han K, Miyamoto T, Heu S, Ryu S. Antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from chicken carcasses in Korea. J Microbiol Biotechnol. 2006;16:1276–1284. [Google Scholar]
- Jang SS, Fleet GH, Cox JM. Pulsed-field gel electrophoresis for subtyping of Listeria monocytogenes. J Korean Soc Appl Biol Chem. 2005;48:58–64. [Google Scholar]
- Jaradat ZW, Schutze GE, Bhunia AK. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int J Food Microbiol. 2002;76:1–10. doi: 10.1016/S0168-1605(02)00050-8. [DOI] [PubMed] [Google Scholar]
- Kathariou S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot. 2002;65:1811–1829. doi: 10.4315/0362-028X-65.11.1811. [DOI] [PubMed] [Google Scholar]
- Khen BK, Lynch OA, Carroll J, McDowell DA, Duffy G. Occurrence, antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef chain in the republic of Ireland. Zoonoses Public Hlth. 2015;62:11–17. doi: 10.1111/zph.12106. [DOI] [PubMed] [Google Scholar]
- Kim SR, Nam HM, Jang GC, Kim AR, Kang MS, Chae MH, Jung SC, Kang DJ, Kim JK, Lim SK. Antimicrobial resistance in Salmonella isolates from food animals and raw meats in Korea during 2010. Kor J Vet Publ Hlth. 2011;35:246–254. [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. In Nucleic acid techniques in bacterial systematics. John Wiley and Sons; Chichester, UK: 1991. pp. 115–175. [Google Scholar]
- Lim SK, Byun JR, Lee HS, Moon DC, Jang GC, Jung SC. Antimicrobial resistance of Escherichia coli strains isolated from pigs and their farm environment in Korea. Prev Vet Med. 2014a;38:61–68. doi: 10.13041/jpvm.2014.38.3.61. [DOI] [Google Scholar]
- Lim SK, Lee JE, Lee HS, Nam HM, Moon DC, Jang GC, Park YJ, Jung YG, Jung SC, Wee SH. Trends in antimicrobial sales for livestock and fisheries in Korea during 2003–2012. Korean J Vet Res. 2014b;54:81–86. [Google Scholar]
- Lim SK, Nam HM, Jang GC, Kim SR, Chae MH, Jung SC, Kang DJ, Kim JK. Antimicrobial resistance in Staphylococcus aureus isolated from raw meats in slaughterhouse in Korea during 2010. Kor J Vet Publ Hlth. 2011;35:231–238. [Google Scholar]
- Makino SI, Kawamoto K, Takeshi K, Okada Y, Yamasaki M, Yamamoto S, Igimi S. An outbreak of food-borne listeriosis due to cheese in Japan, during 2001. Int J Food Microbiol. 2005;104:189–196. doi: 10.1016/j.ijfoodmicro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Mathakiya RA, Roy A, Nandasana KN, Koringa PG, Joshi CG. Evaluation of a rapid molecular method for detection of Listeria monocytogenes directly from broth culture. Vet World. 2009;2:177–178. [Google Scholar]
- Meloni D, Piras F, Mureddu A, Fois F, Consolati SG, Lamon S, Mazzette R. Listeria monocytogenes in five Sardinian swine slaughterhouses: Prevalence, serotype, and genotype characterization. J Food Prot. 2013;76:1863–1867. doi: 10.4315/0362-028X.JFP-12-505. [DOI] [PubMed] [Google Scholar]
- Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, Lecuit M, Courvalin P, Monnier AL. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob Agents Chemother. 2010;54:2728–2731. doi: 10.1128/AAC.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhterem-Uyar M, Dalmasso M, Bolocan AS, Hernandez M, Kapetanakou AE, Kuchta T, Manios SG, Melero B, Minarovicova J, Nicolau AI, Rovira J, Skandamis PN, Jordan K, Rodriguez-Lazaro D, Stessl B, Wagner M. Environmental sampling for Listeria monocytogenes control in food processing facilities reveals three contamination scenarios. Food Control. 2015;51:94–107. [Google Scholar]
- Neves E, Lourenco A, Silva AC, Coutingo R, Brito L. Pulsed-field gel electrophoresis (PFGE) analysis of Listeria monocytogenes isolates from different sources and geographical origins and representative of the twelve serovars. Syst Appl Microbiol. 2008;31:387–392. doi: 10.1016/j.syapm.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Okutani A, Okada Y, Yamamoto S, Igimi S. Nationwide survey of human Listeria monocytogenes infection in Japan. Epidemiol Infect. 2004;132:769–772. doi: 10.1017/s0950268804001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Trallero E, Zigorraga C, Artieda J, Alkorta M, Marimón JM. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg Infect Dis. 2014;20:2155–2157. doi: 10.3201/eid2012.140993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento G, Ducci B, Nieri D, Comodo N, Nostro AL. Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control. 2010;21:708–713. [Google Scholar]
- Sofos JN, Geornaras I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Esherichia coli O157:H7 in nonintact, and Listeria monocytogenes in ready-to-eat, meat products. Meat Sci. 2010;86:2–14. doi: 10.1016/j.meatsci.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sokolovic Z, Schüller S, Bohne J, Baur A, Rdest U, Dickneite C, Nichterlein T, Goebel W. Differences in virulence and in expression of PrfA and PrfA-regulated virulence genes of Listeria monocytogenes strains belonging to serogroup 4. Infect Immun. 1996;64:4008–4019. doi: 10.1128/iai.64.10.4008-4019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V, Nam HM, Nguyen LT, Tamilselvam B, Murinda SE, Oliver SP. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog Dis. 2005;2:201–211. doi: 10.1089/fpd.2005.2.201. [DOI] [PubMed] [Google Scholar]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley IV, Harmon KM, Dickson JS, Schwartz AR. Application of a multiplex polymerase chain reaction assay for the simultaneous confirmation of Listeria monocytogenes and other Listeria species in turkey sample surveillance. J Food Prot. 2002;65:780–785. doi: 10.4315/0362-028x-65.5.780. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yücel N, Çitak S, Önder M. Prevalence and antibiotic resistance of Listeria species in meat products in Ankara, Turkey. Food Microbiol. 2005;22:241–245. [Google Scholar]


