Abstract
Core fucosylation plays a critical role in modulating the effector functions of therapeutic antibodies such as the antibody-dependent cellular cytotoxicity (ADCC) through adversely affecting the affinity of antibodies for Fcγ receptors. Thus, a facile method for Fc defucosylation of antibodies is important both for functional studies and for an enhanced therapeutic efficacy. In this chapter, we describe a detailed protocol for chemoenzymatic defucosylation of antibodies using Herceptin (trastuzumab) as a model system. The protocol includes (a) Fc deglycosylation using endoglycosidase S2 (Endo-S2); (b) enzymatic defucosylation of the resulting Fucα1,6GlcNAc-Herceptin using two distinct bacterial α-fucosidases, AlfC and BfFuc; (c) transglycosylation of the GlcNAc-Herceptin using an Endo-S2 mutant (Endo-S2 D184M) as the enzyme and a complex N-glycan oxazoline as the donor substrate; and (d) SPR analysis of the binding of antibody glycoforms with the FcγIIIA receptor. The protocol of enzymatic defucosylation of Herceptin should be equally applicable for the Fc glycan engineering of other mAbs.
Keywords: Therapeutic antibody, Chemoenzymatic synthesis, Glycoengineering, α-Fucosidase, Defucosylation, Transglycosylation, ADCC
1. Introduction
Monoclonal antibodies (mAbs) of the immunoglobulin G (IgG) class are an important class of therapeutic agents widely used for the treatment of cancer, autoimmune diseases, and infectious diseases [1, 2]. The IgG-type antibodies consist of two identical Fab domains responsible for antigen binding and the Fc domain that interacts with Fc receptors to mediate antibody effector functions. The Fc domain of all IgGs possesses a conserved N-glycosylation site to which a biantennary complex-type glycan is typically attached, but significant structural heterogeneity arises from terminal and core modifications, including variations in terminal galactosylation and sialylation, addition of bisecting N-acetylglucosamine (GlcNAc), and core fucosylation [3]. Compelling data have demonstrated that corefucosylation, the attachment of an α1,6-linked fucose to the innermost GlcNAc moiety of the Fc N-glycans, significantly decreases the affinity of antibodies for the FcγIIIA receptor on effector cells, leading to dramatically reduced antibody-dependent cell-mediated cytotoxicity (ADCC) [4–6]. However, antibodies produced from various host expression systems are usually core-fucosylated. Thus one way to enhance the ADCC-based therapeutic efficacy is to produce antibodies with low Fc fucose content. Recently, two gly-coengineered mAbs with low fucose content were approved by the FDA that showed enhanced therapeutic efficacy in anticancer treatment. One is the mogamulizumab (POTELIGEO) produced in an engineered CHO cell line in which the α1,6-fucosyltransferase (FUT8) responsible for core fucosylation was knocked out [7]. The other is the obinutuzumab (Gazyva) produced from the CHO cell lines overexpressing the β1,4-GlcNAc transferase III (GnT III), which overexpresses a bisecting GlcNAc moiety that prevents further core fucosylation of the Fc N-glycans [8].
As an alternative approach to Fc defucosylation through the engineering of host expression systems, Wang and co-workers have recently developed a chemoenzymatic glycan remodeling method that allows an efficient in vitro Fc glycan engineering of existing intact therapeutic antibodies to produce homogeneous glycoforms [9–11]. The method consists of two key steps: the Fc deglycosyla- tion of an antibody with an endoglycosidase such as the endogly- cosidase S (Endo-S) from Streptococcus pyogenes and the subsequent enzymatic reglycosylation using a glycosynthase mutant such as Endo-S D233A to afford a homogeneous glycoform of the antibody [9–11]. More recently, novel glycosynthase mutants were generated from Endo-S2 that demonstrated much enhanced catalytic efficiency over the Endo-S enzyme. In combination with the use of an α-fucosidase, non-fucosylated glycoforms of antibodies could be generated, that showed much higher ADCC than the original antibodies [12, 13].
We have previously described the general protocols for the chemoenzymatic glycoengineering of intact antibodies to produce homogeneous antibody glycoforms [14, 15]. In this paper, we describe a detailed protocol of the chemoenzymatic defucosylation of Herceptin (i.e., trastuzumab, a mAb drug widely used for the treatment of breast cancer) as a model system with the focus on the comparison of defucosylation by two bacterial α-fucosidases. One is the α-fucosidase AlfC from Lactobacillus casei that shows both hydrolysis and transglycosylation activity [16–18]. The other is the α-fucosidase BfFuc from Bacteroides fragilis that demonstrates very broad substrate specificity [12,13]. The schematic procedures are shown in Fig. 1. The protocol includes (1) Fc deglycosylation using the endoglycosidase S2 (Endo-S2); (2) enzymatic defucosylation of the resulting Fuca1,6GlcNAc-Herceptin by two distinct bacterial α-fucosidases, the AlfC and BfFuc; (3) reglycosylation of the GlcNAc-Herceptin using an Endo-S2 glycosynthase mutant (Endo-S2 D184M) as the enzyme and a complex N-glycan oxazo-line as the donor substrate; and (4) SPR analysis of the binding of antibody glycoforms with the FcγlllA receptor. The comparative studies showed that the α-fucosidase AlfC was much more efficient for defucosylation of the Fc than the α-fucosidase BfFuc and the non-fucosylated Fc glycoform demonstrated significantly higher affinity for the FcγlllA receptor than that of the commercial Herceptin.
Fig. 1.
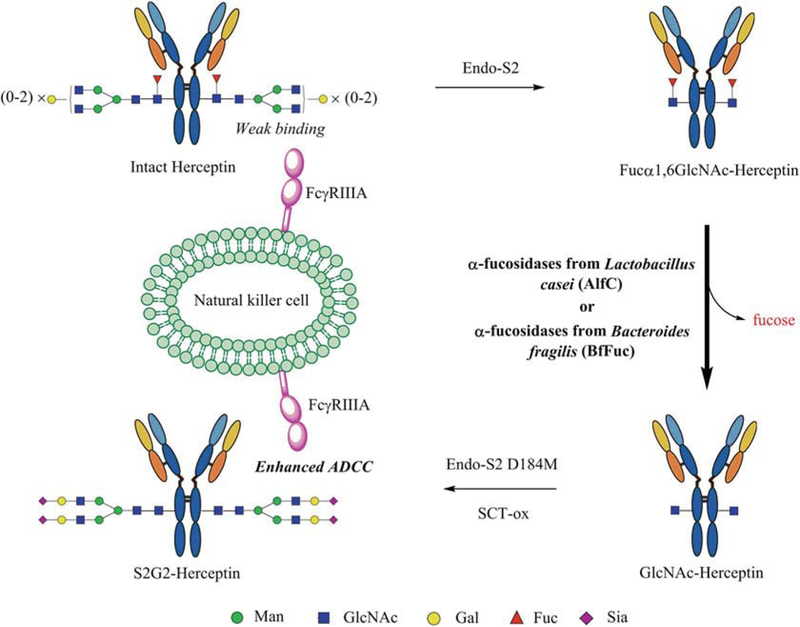
Chemoenzymatic glycan remodeling of monoclonal antibody Herceptin. The procedures include enzymatic defucosylation of the therapeutic antibody by a combined use of endoglycosidase S2 and an α-fucosidase, followed by enzymatic transfer of an N-glycan using glycosynthase Endo-S2 D184M to afford a non-fucosylated homogeneous glycoform of Herceptin
2. Materials
All chemicals, reagents, and solvents were purchased from Sig-ma-Aldrich and TCI and used as received unless specially noted. The α-fucosidases AlfC from Lactobacillus casei BL23 (GenBankaccession no. CAQ67984.1) and BfFuc from Bacteroides fragilis NCTC 9343 (GenBank accession no. CAH08937.1) were cloned and overexpressed as previously described [13,18,19]. The recombinant proteins Endo-S2 WT and Endo-S2 D184M were prepared according to the procedures described previously [10]. The sugar oxazolines used in this protocol were prepared using previously reported methods [20].
2.1. Antibody, Enzymes, and Substrates
Recombinant monoclonal antibody (mAb) Herceptin® (tras-tuzumab) (Genentech, Inc.) (see Note 1).
Recombinant α-fucosidases AlfC from Lactobacillus casei (see Note 2).
Recombinant α-fucosidases BfFuc from Bacteroides fragilis (see Note 3).
Recombinant Endo-S2 WT and D184M (see Note 4).
Sialo biantennary complex-type N-glycan oxazoline (SCT-ox) (see Note 5).
2.2. Buffers, Columns, and Miscellaneous
100 mM phosphate-buffered saline (PBS) buffer, pH 7.4.
100 mM PBS buffer, pH 7.0.
Protein A column washing buffer: 100 mM PBS buffer, pH 7.4.
Protein A column elution buffer: 100 mM citric acid, pH 3.5 (see Note 6).
Protein A column neutralization buffer: 1 M Trizma® buffer, pH 9.0.
LC-MS mobile phase (Buffer A): 0.1% formic acid (v/v) in ddH2O.
LC-MS mobile phase (Buffer B): 0.1% formic acid (v/v) in acetonitrile.
SPR running buffer: 10 mM HEPES [N-(2-hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid)], 150 mM NaCl, 0.05% (v/v) surfactant P20 (polyoxyethylene sorbitan), pH 7.4 (GE Healthcare).
SPR chip regeneration buffer: 10 mM glycine HCl buffer, pH 1.5.
Sephadex G-15 (GE Healthcare).
HiTrap Q XL, 5 mL (GE Healthcare).
HisTrap FF column, 1 mL (GE Healthcare).
HiTrap Protein A column HP, 1 mL (GE Healthcare).
XBridge™ BEH300 C4, 2.1 × 50 mm, 3.5 μm (Waters).
10 kDa MWCO centrifuge filter unit (EMD Millipore).
Isopropyl β-D-1-thiogalactopyranoside, IPTG (GoldBio).
Triethylamine, TEA (TCI).
2-Chloro-1,3-dimethylimidazolinium chloride, DMC (Sigma-Aldrich).
2.3. Equipment and Software
Model 366 incubator (Thermo Scientific, Waltham, MA).
AKTA FPLC protein purification system with UPC-900 and P-920 (GE Healthcare).
Exactive™ Plus Orbitrap Mass Spectrometer (Thermo Scientific).
Dionex UltiMate 3000 (RS Pump, RS Column Compartment, RS Autosampler) (Thermo Scientific).
NanoDrop 2000c (Thermo Scientific).
Biacore™ T200 system (GE Healthcare).
CM5 biosensor chip (GE Healthcare) (see Note 7).
Xcalibur for raw MS data analysis (Thermo Scientific).
MagTran for deconvolution of MS data.
Biacore™ T200 evaluation software.
3. Methods
3.1. Purification and LC-ESI-MS Analysis of Whole Herceptin
Buffer exchange of commercial Herceptin: Apply 20 mg of Herceptin to the centrifuge filter unit (10 kDa MWCO), and centrifuge at 4000 x g. Wash five times with 4 mL of PBS buffer, and resuspend the antibody into PBS buffer (pH 7.4, 10 μg/μL, final).
Purification of Herceptin using Protein A column: Protein A affinity chromatography was used to purify Herceptin. Wash the column with 5 mL of Protein A column elution buffer, and equilibrate the column with 10 mL of Protein A column washing buffer at a flowrate of 1 mL/min. Apply 20 mg of Herceptin onto the column, and wash with 15 mL of the washing buffer. Add 100 μL of Protein A column neutralization buffer into fraction tubes (1 mL for each) on the collector. Elute the bound Herceptin with 20 mL of the elution buffer. Pool the fractions containing Herceptin. Transfer the combined fractions to a centrifuge filter unit (10 kDa MWCO), and perform the buffer exchange as described above.
LC-MS analysis of antibody: Add 1 μL of Herceptin (1 μg/μL) into 19 μL of ddH2O containing 0.1% formic acid (v/v) in a MS sample vial (see Note 8). Inject 10 μL of the sample by RS Autosampler system. Elute antibody with 5–90% buffer B within 6 min on an XBridge™ BEH300 C4 column; then wash the column with 90% buffer B for 3 min. Equilibrate the column with 5% buffer B for 2 min. Under the above condition, the whole antibody (Herceptin) elutes at around 3.5 min. Perform mass spec analysis of the antibody with collision- induced dissociation (CID) method (Fig. 2a) (see Note 9).
Analyze the MS data of whole antibody using Xcalibur Qual Browser. Process the raw data in MagTran to get the deconvo- luted mass spectra of the whole antibody (see Note 10).
Fig. 2.
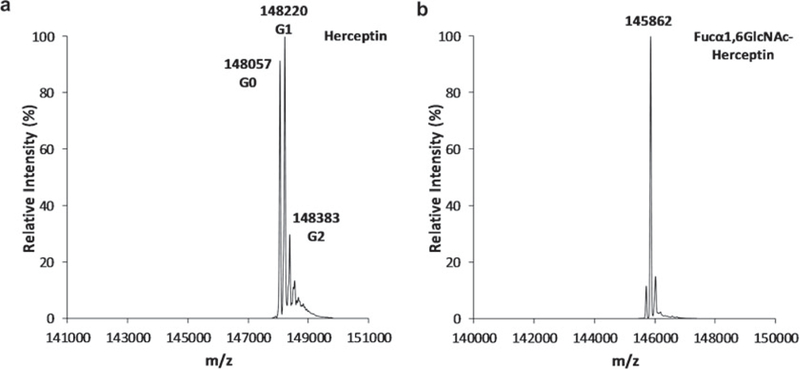
LC-ESI-MS analysis of intact Herceptin and the deglycosylated Herceptin. (a) Intact commercial Herceptin; (b) Deglycosylated Herceptin (Fucα1,6GlcNAc-Herceptin, F2-Herceptin) obtained from Endo-S2 treatment
3.2. Deglycosylation of Intact Herceptin Using Endo-S2 WT
The cloning, overexpression, and purification of Endo-S2 WT (MW = 110 kDa) follow the previously reported methods (see Note 4). Perform the buffer exchange as described above against PBS buffer (pH 7.0) (see Subheading 3.1).
Add 2 μL of Endo-S2 WT (10 μg/μL) to 20 mg of Herceptin in 2 mL of PBS buffer (pH 7.4). Incubate the reaction mixture at 30 °C for 30 min.
Monitor the deglycosylation process of antibody by LC-ESI-MS (see Subheading 3.1). Deglycosylation of intact Herceptin should be complete within 30 min (Fig. 2b).
Purify the Fucα1,6GlcNAc-Herceptin by Protein A affinity column (see Subheading 3.1) (see Note 11).
3.3. Defucosylation of Fucα1,6GlcNAc-Herceptin Using the AlfC α-Fucosidase
The cloning, overexpression, and purification of AlfC α-fucosidase (MW = 66 kDa) follow the previously reported methods (see Note 2). Perform the buffer exchange as described above against PBS buffer (pH 7.0) (see Subheading 3.1).
Defucosylation of Fucα1,6GlcNAc-Herceptin: Apply 10 μL of AlfC α-fucosidase (1.35 nmol, 6.1 μg/μL) into 10 mg of Fucα1,6GlcNAc-Herceptin in 0.5 mL of PBS buffer (pH 7.0) (see Note 12). Incubate the reaction mixture at 37 °C overnight (see Note 13).
Monitor the defucosylation process of Fucα1,6GlcNAc-Her-ceptin by LC-ESI-MS (see Subheading 3.1) (Fig. 3a). Defucosylation of Fucα1,6GlcNAc-Herceptin should complete within 15 h (Fig. 3c) (see Note 14).
Purify the GlcNAc-Herceptin by Protein A affinity column (see Subheading 3.1) (see Note 15).
Fig. 3.
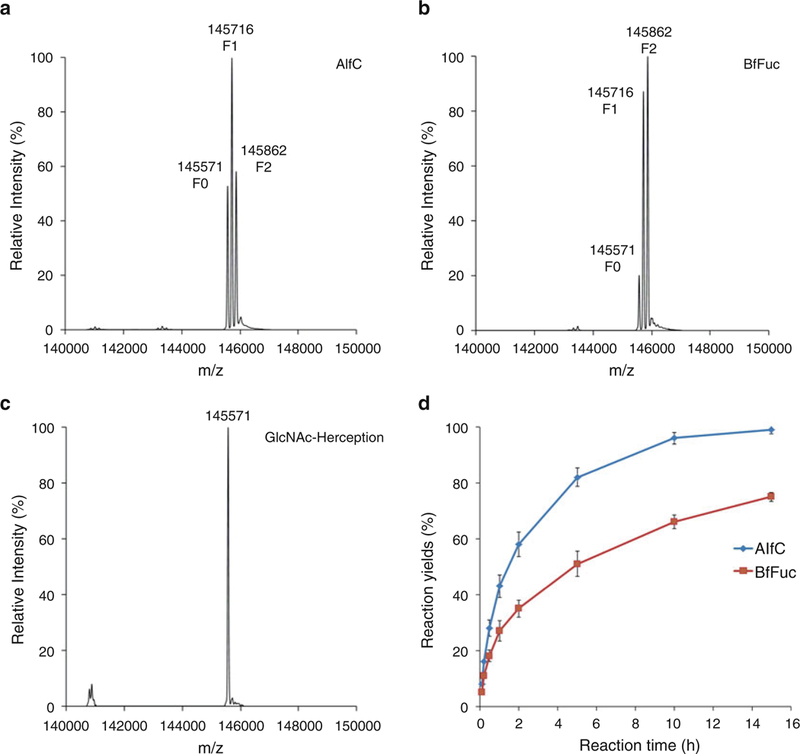
Comparison of the enzymatic defucosylation by the two α-fucosidases. (a) LC-ESI-MS analysis of the defucosylation of Fucα1,6GlcNAc-Herceptin by the α-fucosidase AlfC in 5 h; (b) LC-ESI-MS analysis of the defucosylation of Fucα1,6GlcNAc-Herceptin by the α-fucosidase BfFuc in 5 h; (c) LC-ESI-MS analysis of the final defucosylated product, GlcNAc-Herceptin; (d) Comparison of the time course of enzymatic defucosylation by the two α-fucosidases, AlfC and BfFuc
3.4. Defucosylation of Fucα1,6GlcNAc-Herceptin Using the BfFuc α-Fucosidase
The cloning, overexpression, and purification of BfFuc α-fucosidase (MW = 5G kDa) follow the previously reported methods (see Note 3). Do the buffer exchange as described above against PBS buffer (pH 7.0) (see Subheading 3.1).
Defucosylation of Fucα1,6GlcNAc-Herceptin: Apply 10 μL of BfFuc α-fucosidase (1.35 nmol, 6.75 μg/μL) into 10 mg of Fucα1,6GlcNAc-Herceptin in 0.5 mL of PBS buffer (pH 7.G) (see Note 13). Incubate the reaction mixture at 37 °C overnight.
Monitor the defucosylation process of Fucα1,6GlcNAc-Herceptin by LC-ESI-MS (see Subheading 3.1) (Fig. 3b). Defucosylation of Fucα1,6GlcNAc-Herceptin should complete within 30 h.
Purify the GlcNAc-Herceptin by Protein A affinity column (see Subheading 3.1).
3.5. Transglycosylation from Complex Type N-Glycan to GlcNAc-Herceptin
The cloning, overexpression, and purification of Endo-S2 D184M (MW = 11G kDa) follow the previously reported methods (see Note 4). Perform the buffer exchange as described above against PBS buffer (pH 7.0) (see Subheading 3.1).
Transfer sialo complex-type N-glycan to GlcNAc-Herceptin: Add 10 μL of Endo-S2 D184M mutant (20 μg/μL) and 5.4 mg of substrate SCT-ox (40 equiv.) to 10 mg of GlcNAc-Herceptin (1 equiv.) in 1 mL of PBS buffer (pH 7.4) (see Note 16).
Monitor the transglycosylation process of GlcNAc-Herceptin by LC-ESI-MS (see Subheading 3.1). Transglycosylation of GlcNAc-Herceptin should complete within 1 h (Fig. 4).
Purify the sialo biantennary complex-type N-glycosylated Her-ceptin (S2G2-Herceptin) by Protein A affinity column (see Subheading 3.1) (see Note 17).
Fig. 4.
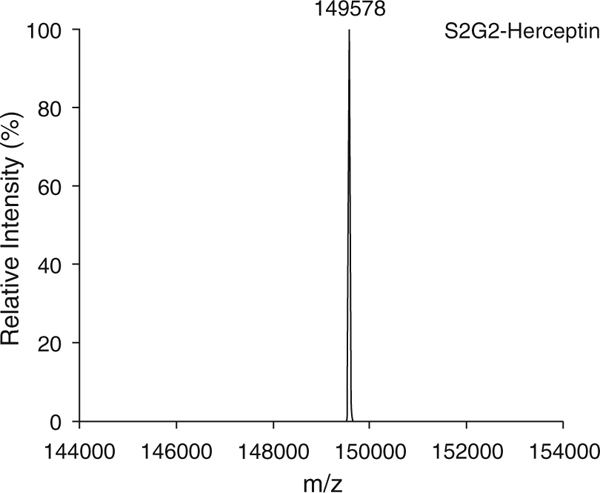
LC-ESI-MS analysis of the non-fucosylated homogeneous glycoform of Herceptin (S2G2-Herceptin)
3.6. Surface Plasmon Resonance (SPR) Binding Assay Between FcγRIIIA and Various Glycoforms of Herceptin
Couple protein A on all four flow cells of a CM5 sensor chip using the amine coupling kit, as described in previous report [21]. About 5000 RU of Protein A can be coupled to each of the four flow cells.
Dilute antibodies to 2 μg/mL in the running buffer, and inject on flow cells at 20 μL/min to reach a capture level of 200 RU (see Note 18). Dilute the soluble recombinant FcγRIIIAV158 (2000 nM) in the running buffer (assuming a molecular weight of 45 kDa), and further prepare serial dilution from 2000 to 7.8125 nM (nine concentrations). Inject each of the nine concentrations for 90 s each at a flow rate of 30 μL/min followed by a 300 s dissociation. Inject the running buffer four times to serve as blank. After each cycle, regenerate the surface by injecting a glycine HCl buffer (10 mM, pH 1.5) (see Note 19).
After subtracting blank sensorgrams (2–1, 4–3), fit the corrected data using the Biacore T200 evaluation software with the Langmuir (1:1) model (Fig. 5) (see Note 20).
Fig. 5.
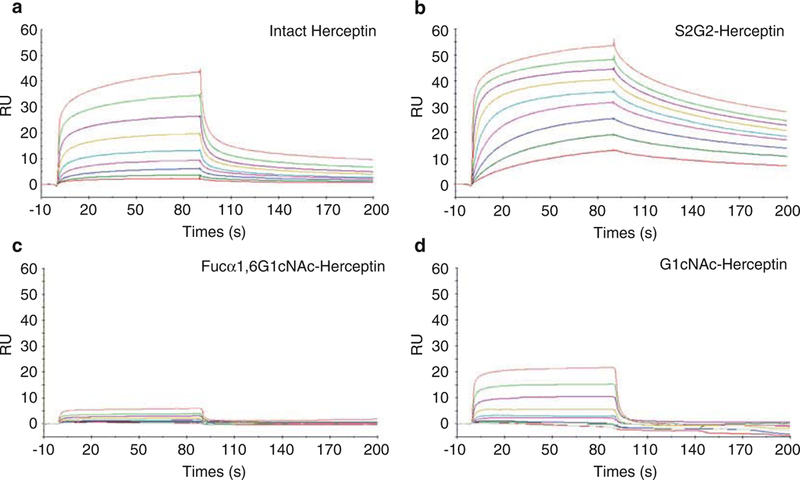
SPR analysis of the binding between FcγRIIIA-V158 and various glycoforms of Herceptin. The antibody was immobilized on a Protein A chip, and the Fc receptor was run as analytes at 2X serial dilutions starting at 2 μM. (a) Binding with intact commercial Herceptin; (b) binding with non-fucosylated sialyl complex-type Herceptin (S2G2-Herceptin); (c) binding with Fucα1,6GlcNAc-Herceptin; (d) binding with GlcNAc-Herceptin
4. Notes
In the present study, we describe the detailed procedure of the chemoenzymatic defucosylation of monoclonal antibodies for enhanced effector functions using Herceptin as a model. The protocol is also applicable for producing other non-core-fuco- sylated antibodies.
The DNA sequence encoding AlfC α-fucosidase from L. casei was inserted into pET22b-CPD vector, and enzyme was overexpressed in E. coli BL21 (DE3) as a fusion protein with a C-terminal cysteine protease domain (CPD) carrying a 10 x His tag at 20 °C for 16 h (induced by 0.2 mM IPTG). The recombinant enzyme can be purified with HisTrap FF column (1 mL, GE Healthcare) to yield pure proteins with a mass of 66 kDa (>40 mg/L). AlfC α-fucosidase should be stored at 4 °C with a concentration among 5–10 μg/μL and good for 1 month. It is found that the enzyme is not very stable at —20 °C as it might loss some activities. Also too high or too low concentration may cause some precipitation during storage. Usually the enzyme should be diluted for use just before the reaction.
The DNA sequence encoding BfFuc α-fucosidase was cloned from B. fragilis genomic DNA (ATCC® 25285D-5™) and inserted into pET28(a) vector. The enzyme carrying a 6x His tag was overexpressed in E. coli BL21 (DE3) at 20 °C for 16 h (induced by 0.2 mM IPTG) and purified with HisTrap FF column (1 mL, GE Healthcare) to yield pure proteins with a mass of 50 kDa (≈30 mg/L). BfFuc α-fucosidase can be stored at 4 or —20 °C with a concentration among 1–20 μg/μL and good for several months. Usually the enzyme should be diluted for use just before the reaction.
The DNA sequence encoding Endo-S2 WT or D184M mutant was cloned into a pET22b-CPD vector, and enzyme was overexpressed in E. coli BL21 (DE3) as fusion proteins with a C-terminal cysteine protease domain (CPD) carrying a 10x His tag at 20 °C for 16 h (induced by 0.5 mM IPTG). The recombinant enzyme can be purified with HisTrap FF column (1 mL, GE Healthcare) to yield pure proteins with a mass of 110 kDa (≈ 20 mg/L). The proteins could be stored at 4 or —20 ° C and good for 1 year.
Crude SCT N-glycanwas obtained by Endo-S catalyzed hydrolysis of egg yolk sialylglycopeptide (SGP) and purified with HiTrap Q XL column. SCT glycan can be readily converted into SCT-ox by treating with TEA (45 equiv.) and DMC (20 equiv.) in aqueous solution at 4 °C for 30 min. The resulting SCT-ox is purified by Sephadex G-15 column. Sugaroxazolines (including SCT-ox) are not stable in acidic or neutral pH conditions. They are dissolved in ddH2O containing NaOH (10 mM) immediately after purification and frozen at —80 °C for longtime storage. Usually sugar oxazolines solution should be directly diluted for use in the reaction mixture to avoid spontaneous hydrolysis.
This buffer is not very stable. It should be stored at 4 °C and good for 1 month.
The CM5 chip immobilized with Protein A should be stored at 4 °C to keep the activity of protein and good for longtime use.
Samples should be spun down before injection into LC-ESI-MS system. Adjust injection amount to the Orbitrap MS system, as too much injection (>5 μg) could contaminate the C4 column with antibody residuals and mislead the analysis of MS spectra.
The retention time of the whole Herceptin depends on the analytic conditions, including column, temperature, glycoform of antibody, flowrate, and mobile phase of LC system.
With collision-induced dissociation (CID) applied on the Orbitrap Mass Spectrometer, the whole antibody can be detected with signals of different glycoforms of antibodies on the ESI-MS spectra. The addition of respective N-glycans to the intact Herceptin account for a mass of 148,057 Da for G0 (with no galactose moiety), 148,220 Da for G1 (with 1 galactose moiety), and 148,383 Da for G2 (with 2 galactose moieties). There are still some other species following those peaks on the MS spectrum, indicating more complex glycoforms of Herceptin as minor species (Fig. 2a).
The deglycosylated antibody (e.g., Herceptin) could be less stable than intact Herceptin, so it would be better to do the next steps (defucosylation and transglycosylation) within 1 week to prevent potential degradation or aggregation due to the deglycosylation. The Protein A purification in this step is essential and must be extensive, as the transglycosylation would be diminished by even a trace amount of Endo-S2 WT.
The activity of the two α-fucosidases can be determined using commercially available p-nitrophenyl α-L-fucoside, as both α-fucosidases are very active on it.
The defucosylation can be carried out at 37 °C or 30 °C to prevent the potential inactivation of Herceptin, although the optimal temperature for AlfC α-fucosidase is 42 °C as previously reported.
The kinetics of Herceptin defucosylation depends on the reaction conditions, including α-fucosidase, enzyme activity, Herceptin concentration, temperature, and buffer pH. Usually highly concentrated Herceptin (10–20 μg/μL) solution is good for efficient defucosylation, and low concentration (<1 μg/μL) can significantly decrease the hydrolysis efficiency of the two α-fucosidases. Under the test condition, the defucosylation activity of AlfC is much higher than that of BfFuc (Fig. 3).
The difference of 146 Da among the species indicates status of fucosylation of the Fc domains: 145571 Da for GlcNAc-Herceptin (F0), 145,716 Da for GlcNAc-Herceptin with one fucose moiety (F1), and 145,862 Da for Fucα1,6GlcNAc-Herceptin (F2) carrying two fucose moieties (Fig. 3).
The kinetics of Herceptin transglycosylation depends on the reaction conditions, including enzyme dosage, substrate ratio (usually 20 equiv. sugar oxazoline for each glycosylation site of the whole antibody), Herceptin concentration, temperature, and buffer pH (Fig. 4). Usually relatively high concentration of Herceptin (10–20 μg/μL) solution is good for transglycosylation, and low concentration (<1 μg/μL) can significantly decrease the transfer efficiency by Endo-S2 D184M. Too high amount of sugar oxazoline (e.g., >100 equiv.) should be avoided to prevent potential nonenzymatic glycosylation of the antibody.
The flow through from the Protein A column can be collected and analyzed to recover the excess SCT glycan oxazoline (in the form of the free N-glycan after hydrolysis). The final purified glycoengineered Herceptin by the methods described here may be contaminated by potential bacterial endotoxin, as all the enzymes are overexpressed in E. coli expressing system. Thus for the purposes of cell-based or any in vivo studies in which contaminated endotoxin may interfere with the biological assays, endotoxin activity should be tested, and the potential endotoxin contamination could be removed by endotoxin removal kit.
Antibodies are captured on flow cells 2 or 4 (Fc-2, Fc-4). Each flow cell captures a different antibody glycoform. No antibodies are captured on flow cells 1 and 3 (Fc-1 and Fc-3) as they serve as references for flow cells 2 and 4, respectively.
The regeneration also removes the captured antibody so antibody capture is performed at the beginning of each cycle.
The defucosylated Herceptin (S2G2-Herceptin) showed much higher affinity than that of intact Herceptin (fully core-fucosylated forms). As estimated by the dissociation constant (KD), the affinity of S2G2-Herceptin (KD = 3.6 nM) for FcγRIIIA V158 was about 52-fold higher than that of the intact Herceptin (KD = 189 nM) (Fig. 5a, b). This result was consistent with our previous reported binding data. The twodeglycosylated Herceptin (Fucα1,6GlcNAc-Herceptin and GlcNAc-Herceptin) showed much lower binding affinity than that of the glycosylated Herceptin (Fig. 5c, d). Herceptin with only the innermost GlcNAc (GlcNAc-Herceptin) showed a moderate affinity for FcγRIIIA V158 with a KD of 320 nM, while the corresponding fucosylated Herceptin (Fucα1,6Glc-NAc-Herceptin) only had a marginal affinity, and the KD was not determined.
Acknowledgment
We thank other members of the Wang Lab for technical assistance and helpful discussions. This work was supported by the National Institutes of Health (NIH grants R01GM080374 and R01GM096973 to LXW).
References
- 1.Ecker DM, Jones SD, Levine HL (2015) The therapeutic monoclonal antibody market. MAbs 7(1):9–14. https://doi.org/10.4161/19420862.2015.989042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams GP, Weiner LM (2005) Monoclonal antibody therapy of cancer. Nat Biotechnol 23 (9):1147–1157 [DOI] [PubMed] [Google Scholar]
- 3.Jefferis R(2005) Glycosylation of recombinant antibody therapeutics. Biotechnol Prog 21 (1):11–16. https://doi.org/10.1021/bp040016j [DOI] [PubMed] [Google Scholar]
- 4.Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng yG , Weikert SH, Presta LG(2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody- dependent cellular toxicity. J Biol Chem 277 (30):26733–26740. https://doi.org/10.1074/jbc.M202069200 [DOI] [PubMed] [Google Scholar]
- 5. Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, Hanai N, Shitara K(2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem 278 (5):3466–3473. https://doi.org/10.1074/jbc.M210665200 [DOI] [PubMed] [Google Scholar]
- 6.Nimmerjahn F, Ravetch JV (2005) Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science 310 (5753):1510–1512. https://doi.org/10.1126/science.1118948 [DOI] [PubMed] [Google Scholar]
- 7.Beck A, Reichert JM (2012) Marketing approval of mogamulizumab. MAbs 4 (4):419–425. https://doi.org/10.4161/mabs.20996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagez AL, Cartron G (2014) Obinutuzumab: a new class of anti-CD20 monoclonal antibody. Curr Opin Oncol 26(5):484–491. https://doi.org/10.1097/CCO.0000000000000107 [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX (2012) Chemoenzymatic glycoengi-neering of intact IgG antibodies for gain of functions. J Am Chem Soc 134 (29):12308–12318. https://doi.org/10.1021/ja3051266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Tong X, Yang Q, Giddens JP, Wang LX (2016) Glycosynthase mutants ofendoglycosi-dase S2 show potent transglycosylation activity and remarkably relaxed substrate specificity for antibody glycosylation remodeling. J Biol Chem 291(32):16508–16518. https/doi.org/10.1074/jbc.M116.738765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LX, Amin MN (2014) Chemical and chemoenzymatic synthesis of glycoproteins for deciphering functions. Chem Biol 21 (1):51–66. https://doi.org/10.1016/j.chembiol.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsai TI, Li ST, Liu CP, Chen KY, Shivatare SS, Lin CW, Liao SF, Lin CW, Hsu TL, Wu YT, Tsai MH, Lai MY, Lin NH, Wu CY, Wong CH (2017) An effective bacterial fucosidase for glycoprotein remodeling. ACS Chem Biol 12 (1):63–72. https://doi.org/10.1021/acschembio.6b00821 [DOI] [PubMed] [Google Scholar]
- 13.Lin CW, Tsai MH, Li ST, Tsai TI, Chu KC, Liu YC, Lai MY, Wu CY, Tseng YC, Shivatare SS, Wang CH, Chao P, Wang SY, Shih HW, Zeng YF, You TH, Liao JY, Tu YC, Lin YS, Chuang HY, Chen CL, Tsai CS, Huang CC, Lin NH, Ma C, Wong CH (2015) A common glycan structure on immunoglobulin G for enhancement of effector functions. Proc Natl Acad Sci USA 112(34):106 11–10616. https/doi.org/10.1073/pnas.1513456112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giddens JP, Wang LX (2015) Chemoenzy-matic Glyco-engineering of monoclonal antibodies. Methods Mol Biol 1321:375–387. https://doi.org/10.1007/978-1-4939-2760-9_25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q, Wang LX (2017) Chemoenzymatic glycan remodeling of natural and recombinant glycoproteins. Methods Enzymol 597:265–281. https://doi.org/10.1016/bs.mie.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becerra JE, Coll-Marques JM, Rodriguez-Diaz J, Monedero V, Yebra MJ (2015) Preparative scale purification of fucosyl-N-acetylglu-cosamine disaccharides and their evaluation as potential prebiotics and antiadhesins. Appl Microbiol Biotechnol 99(17):7165–7176. https://doi.org/10.1007/s00253-015-6666-2 [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez-Diaz J, Carbajo RJ, Pineda-Lucena A, Monedero V, Yebra MJ (2013) Synthesis of fucosyl-N-acetylglucosamine disaccharides by transfucosylation using alpha-L-fucosidases from lactobacillus casei. Appl Environ Microbiol 79(12):3847–3850. https://doi.org/10.1128/AEM.00229-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Diaz J, Monedero V, Yebra MJ (2011) Utilization of natural fucosylated oligosaccharides by three novel alpha-L-fucosidases from a probiotic Lactobacillus casei strain. Appl Environ Microbiol 77(2):703–705. https://doi.org/101128/AEM.01906-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Zhu S, Ma C, Wang LX (2017) Designer alpha1,6-fucosidase mutants enable direct core fucosylation of intact N-glycopeptides and N-glycoproteins. J Am Chem Soc 139 (42):15074–15087. https://doi.org/10.1021/jacs.7b07906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W, Li J, Wang LX (2011) Unusual transglycosylation activity of Flavobacterium meningosepticum Endoglycosidases enables convergent chemoenzymatic synthesis of core fucosylated complex N-glycopeptides. Chem-biochem 12(6):932–941. https://doi.org/10.1002/cbic.201000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Champion T, Beck A (2013) Capture of the human IgG1 antibodies by protein A for the kinetic study of h-IgG/FcgammaR interaction using SPR-based biosensor technology. Methods Mol Biol 988:331–343. https://doi.org/10.1007/978-1-62703-327-5_21 [DOI] [PubMed] [Google Scholar]


