Key Points
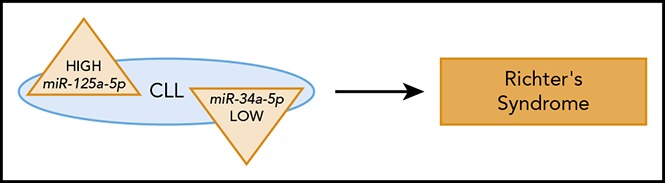
High expression of miR-125a-5p or low expression of miR-34a-5p can predict ∼50% of RS with a false positive rate of ∼9%.
miR-125a-5p and miR-34a-5p can be valuable markers to predict RS development in CLL patients.
Abstract
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia. It is characterized by the accumulation of CD19+/CD5+ lymphocytes and can have variable outcomes. Richter syndrome (RS) is a lethal complication in CLL patients that results in aggressive B-cell lymphomas, and there are no tests to predict its occurrence. Because alterations in microRNA expression can predict the development and progression of several cancers, we investigated whether dysregulation of specific microRNAs can predict RS in CLL patients. Thus, we compared microRNA expression levels in samples from 49 CLL patients who later developed RS with samples from 59 CLL patients who did not. We found that high expression of miR-125a-5p or low expression of miR-34a-5p can predict ∼50% of RS with a false positive rate of ∼9%. We found that CLL patients predicted to develop RS show either an increase of miR-125a-5p expression (∼20-fold) or a decrease of miR-34a-5p expression (∼21-fold) compared with CLL patients that are not predicted to develop RS. Thus, miR-125a-5p and miR-34a-5p can be valuable predictor markers of RS and have the potential to provide physicians with information that can indicate the best therapeutic strategy for CLL patients.
Visual Abstract
Introduction
Richter syndrome (RS) is a lethal complication of chronic lymphocytic leukemia (CLL) that occurs in 1% to 15% of patients. Frequently, RS develops as a diffuse large B-cell lymphoma,1-3 and most RS diffuse large B-cell lymphomas show clonal similarity with underlying CLL.2,3 No effective treatment for RS has been developed.2,3 Several studies determined clinical features and molecular mechanisms of RS, including inactivation of TP53 and CDKN2a gene, advanced Rai stage, NOTCH1 mutation, and IGHV4-39 usage.4-7 Multiple rounds of therapy also have been associated with a higher risk for RS.2,3 To our knowledge, there is no molecular marker able to predict which CLL patients will develop RS. Because microRNAs were reported as markers for cancer development and progression,8-10 we investigated whether alteration of microRNA expression can be associated with RS development in CLL patients.
Methods
Samples
This study was carried out in accordance with the protocol approved by the Institutional Review Board of The Ohio State University. A total of 116 samples was obtained from 108 CLL patients enrolled in the CLL Research Consortium upon written informed consent. Patients were grouped as follows: 49 patients developed RS within 5 years (47 patients) or 12 years (2 patients) from sample collection, and 59 patients did not develop RS in ≥5 years (53 patients) or 3 years (6 patients) from sample collection (supplemental Tables 1-5, available on the Blood Web site). Six RS patients were assayed at different time points for a total of 57 RS samples. Because RS is an aggressive disease, for the control cohorts we accrued only samples from CLL patients in a clinically active and aggressive stage selected for immediate treatment.
Results and discussion
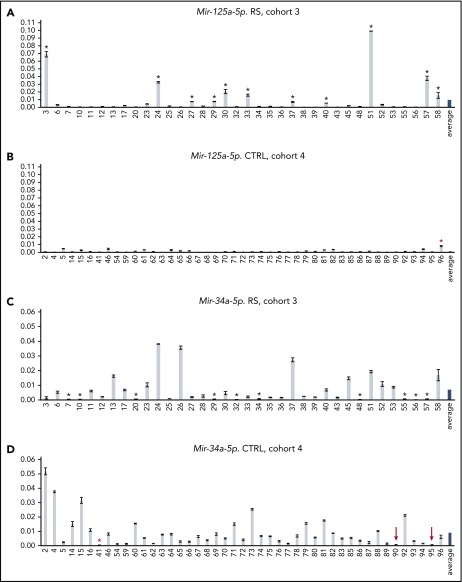
To study whether microRNAs can be markers for RS development, we compared microRNA expression of CLL patients who did not develop RS with that of CLL patients who did develop RS. MicroRNA expression was assessed using NanoString technology in a training set of samples that were divided into 2 groups: cohort 1 contained 21 CLL samples from patients who developed RS 0.5 to 5 years after sample collection, with the exception of #1600 and #1632 (RS samples) (supplemental Table 1), and cohort 2 consisted of 14 CLL samples from patients with no RS development in ≥3 years of follow-up after sample collection, with the exception of #1526 and #1544 (control samples) (supplemental Table 2). We identified a signature of 23 microRNAs differentially expressed between these 2 cohorts that could predict RS development in CLL patients (Table 1). We then analyzed 12 microRNAs by real-time reverse-transcription polymerase chain reaction using all 21 samples from cohort 1 and 8 samples from cohort 2 and determined that miR-125a-5p and miR-34a-5p can be predictors of RS (Table 1). We found that high expression of miR-125a-5p or low expression of miR-34a-5p could predict RS (supplemental Figure 1). Indeed, 8 of 21 and 10 of 21 RS samples had high expression of miR-125a-5p and low expression of miR-34a-5p, respectively, whereas 3 of 8 control samples were false positives (supplemental Figure 1). To confirm our results, we analyzed a validation set: 36 CLL samples from patients taken 0.5 to 5 years before RS (cohort 3, chosen on availability) (supplemental Table 3) and 45 CLL samples from patients with no RS development during ≥5 years of follow-up after sample collection (cohort 4) (supplemental Table 4). Results are shown in Figure 1. By applying a classification algorithm (decision tree) (supplemental Figure 2), we extracted the expression thresholds for miR-125a-5p and miR-34a-5p, maximizing the number of samples correctly classified. Specifically, with miR-125a-5p expression ≥ 5.1 × 10−3 (expressed as 2−ΔCt), 11 RS samples were correctly classified, whereas 1 control sample was not. Similarly, with miR-34a-5p expression < 6.9 × 10−4 (expressed as 2−ΔCt), 10 RS samples were correctly classified, whereas 1 control sample was not. Therefore, we selected the correctly classified samples in association with such thresholds to assess the significance of their differential expression compared with all control samples. We calculated a linear fold change of 20.11 (P < .013) for the set of 11 RS samples having miR-125a-5p expression above the threshold and a linear fold change of 21.34 (P < 1.66 × 10−6) for the set of 10 RS samples having miR-34a-5p expression below the threshold. Two RS samples (#29 and #57) are predicted by both microRNAs. Thus, 19 of 36 (53%) RS samples showed high expression of miR-125a-5p or low expression of miR-34a-5p, and 2 of 45 (4.4%) control samples were false positives (Figure 1). Hence, using these criteria, we can predict ∼50% of RS 0.5 to 5 years before it occurs with a false positive rate of ∼5%. Because the expression of miR-34a-5p in control samples #90 and #95 is very close to the threshold, we adjusted the false positive rate to ∼9%.
Table 1.
Signature of 23 microRNAs differentially expressed in RS vs control samples
| Gene name | Control | RS | Linear FC: control vs RS | P (control vs RS) | Predictors |
|---|---|---|---|---|---|
| hsa-miR-145-5p | 4.69 | 23.56 | −5.02 | .0064 | |
| hsa-miR-125a-5p | 6.67 | 31.88 | −4.78 | .0028 | Selected |
| hsa-miR-548al | 6.78 | 22.81 | −3.36 | .0135 | |
| hsa-miR-365a-3p+hsa-miR-365b-3p | 16.98 | 51.25 | −3.02 | .0014 | |
| hsa-miR-181a-5p | 156.83 | 466.26 | −2.97 | .0000 | Not selected* |
| hsa-miR-199a-3p+hsa-miR-199b-3p | 28.57 | 78.04 | −2.73 | .0164 | |
| hsa-miR-223-3p | 6293.79 | 16 777.22 | −2.67 | .0011 | Not selected |
| hsa-miR-582-5p | 18.97 | 50.19 | −2.65 | .0417 | |
| hsa-miR-221-3p | 21.87 | 53.66 | −2.45 | .0492 | |
| hsa-miR-126-3p | 138.26 | 322.04 | −2.33 | .0302 | Not selected |
| hsa-miR-451a | 525.34 | 1 161.25 | −2.21 | .0445 | Not selected |
| hsa-miR-337-3p | 101.18 | 49.08 | 2.06 | .0021 | |
| hsa-miR-32-5p | 401.21 | 176.95 | 2.27 | .0061 | |
| hsa-miR-1260b | 33.1 | 14.47 | 2.29 | .0032 | |
| hsa-miR-486-3p | 76.1 | 33.17 | 2.29 | .0159 | |
| hsa-miR-630 | 81.35 | 35.09 | 2.32 | .0387 | |
| hsa-miR-335-5p | 90.02 | 38.14 | 2.36 | .0074 | Not selected |
| hsa-miR-601 | 64.24 | 24.89 | 2.58 | .0138 | Not selected |
| hsa-miR-502-5p | 108.82 | 37.92 | 2.87 | .0003 | Not selected |
| hsa-miR-195-5p | 36.24 | 11.35 | 3.19 | .0010 | Not selected |
| hsa-miR-142-5p | 119.03 | 36.68 | 3.25 | .0036 | Not selected |
| hsa-miR-34a-5p | 86.95 | 17.08 | 5.09 | .0031 | Selected |
| hsa-miR-570-3p | 22.64 | 3.78 | 5.99 | .0001 | Not selected |
Samples from Cohort 1 (21 RS) and Cohort 2 (14 controls) were assayed using NanoString technology. Mir-125a-5p and miR-34a-5p were selected for further studies in an independent validation set. The Student t test was applied for differential expression analysis.
FC, fold change.
Mir-181a-5p was initially selected but not confirmed in the validation experiment.
Figure 1.
Expression of miR-125a-5p and miR-34a-5p in cohorts 3 and 4. RS was predicted in samples indicated with black asterisks. (A) RS is predicted by high expression of miR-125a-5p in 11 out of 36 samples from cohort 3. (B) Low expression of miR-125a-5p in control cohort 4. (C) RS is predicted by low expression of miR-34a-5p in 10 out of 36 samples from cohort 3. (D) High expression of miR-34a-5p in control cohort 4. False positives are indicated by red asterisks. Samples showing expression of miR-34a-5p very close to the established threshold are indicated with a red arrow.
Based on the data presented in Figure 1, we hypothesized that miR-125a-5p and miR-34a-5p may also play a role in RS. TP53 is a well-known target of miR-125a-5p,11 and it is also involved in RS development.5 Thus, we investigated the expression of p53 in 6 RS samples with high expression of miR-125a-5p and in 6 controls samples with low expression of miR-125a-5p (from cohorts 3 and 4, respectively). Supplemental Figure 3 shows a dramatic decrease in p53 expression in RS samples with high miR-125a-5p, suggesting the importance of the miR-125a-5p/TP53 pathway in RS development. To determine whether other genes can predict RS, we performed a gene-expression analysis using NanoString technology on 24 samples from cohorts 3 and 4 grouped as follows: 6 RS samples with high expression of miR-125a-5p, 6 RS samples with low expression of miR-34a-5p, 6 control samples with low expression of miR-125a-5p, and 6 control samples with high expression of miR-34a-5p (supplemental Table 6). We found that PAX5 (a predicted target of miR-125a-5p) and SETBP1 can be used as additional markers to predict RS. Furthermore, mRNA expression of PAX5 and SETBP1 in control samples is consistently higher than that of RS samples (supplemental Table 6). Pax5 is a transcription factor that is essential for commitment of lymphoid progenitors to the B-cell lineage and is dysregulated in a subset of leukemias and non-Hodgkin lymphomas.12 Thus, we investigated the level of Pax5 by performing a western blot analysis of the samples previously tested for p53. We found that, similarly to p53, high expression of miR-125a-5p strongly correlates with low expression of Pax5 (supplemental Figure 3). Indeed, low expression of PAX5 could predict RS in 6 samples (#7, #12, #24, #27, #29, #51), with sample 12 predicted only by PAX5. Low expression of SETBP1 could predict RS in 6 samples (#27, #29, #7, #25, #12, #17), with sample #17 predicted only by SETPB1 (supplemental Tables 3 and 4).
In conclusion, our results show that high expression of miR-125a-5p or low expression of miR-34a-5p can predict ∼50% of RS in CLL patients, starting as early as 5 years before transformation, with a false positive rate of ∼9%. Furthermore, low expression of PAX5 and/or SETBP1 can serve as additional predictive markers of RS.
With the development of new therapies, such as idelalisib, ibrutinib, and venetoclax, it is not clear whether treatment with these drugs vs traditional chemotherapy is associated with a higher incidence of RS.2,3 We anticipate that these data will emerge in the near future; when they become available, prediction of RS by expression of miR-125a-5p and miR-34a-5p can indicate the best therapeutic strategy for CLL patients.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Tuan Tran and Monica Spydell for excellent clinical and technical assistance and Andrew W. Greaves and Elvin Chu for database management.
This work was supported in part by National Institutes of Health, National Cancer Institute grant P01 CA81534 of the CLL Research Consortium (T.J.K., C.M.C., L.Z.R.) and in part by National Institutes of Health, National Cancer Institute grant R35 CA197706.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: V.B., L.T., and Y.P. designed the study, performed the research, analyzed data, and wrote the manuscript; D.V. and G.N. analyzed data and contributed to scientific discussions, data interpretation, and manuscript revision; L.Z.R., H.-Y.W., and T.J.K. provided patient samples and contributed to scientific discussions, data interpretation, and manuscript revision; J.A.T. performed experiments; Y.P. supervised the study; and C.M.C. supervised the study, designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo M. Croce, The Ohio State University, Comprehensive Cancer Center, Biomedical Research Tower, Room 1082, 460 West 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu; and Yuri Pekarsky, The Ohio State University, Comprehensive Cancer Center, Biomedical Research Tower, Room 1090, 460 West 12th Ave, Columbus, OH 43210; e-mail: pekarsky.yuri@osumc.edu
REFERENCES
- 1.Sgambati M, Linet M, Devesa S Chronic lymphocytic leukemia, epidemiological, familial, and genetic aspects. In: Bruce C, ed. Chronic Lymphocytic Leukemias, 2nd ed., revised and expanded. New York, NY: Marcel Dekker, Inc.; 2001:33-62. [Google Scholar]
- 2.Khan M, Siddiqi R, Thompson PA. Approach to Richter transformation of chronic lymphocytic leukemia in the era of novel therapies. Ann Hematol. 2018;97(1):1-15. [DOI] [PubMed] [Google Scholar]
- 3.Eyre TA, Schuh A. An update for Richter syndrome - new directions and developments. Br J Haematol. 2017;178(4):508-520. [DOI] [PubMed] [Google Scholar]
- 4.Fabbri G, Khiabanian H, Holmes AB, et al. . Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med. 2013;210(11):2273-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chigrinova E, Rinaldi A, Kwee I, et al. . Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122(15):2673-2682. [DOI] [PubMed] [Google Scholar]
- 6.Rossi D, Spina V, Deambrogi C, et al. . The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117(12):3391-3401. [DOI] [PubMed] [Google Scholar]
- 7.Rossi D, Spina V, Cerri M, et al. . Stereotyped B-cell receptor is an independent risk factor of chronic lymphocytic leukemia transformation to Richter syndrome. Clin Cancer Res. 2009;15(13):4415-4422. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, et al. . A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volinia S, Galasso M, Costinean S, et al. . Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20(5):589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Ferracin M, Cimmino A, et al. . A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793-1801. [DOI] [PubMed] [Google Scholar]
- 11.Le MT, Teh C, Shyh-Chang N, et al. . MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463-470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.