Summary
LIN28 is a bipartite RNA-binding protein that post-transcriptionally inhibits the biogenesis of let-7 microRNAs to regulate development and influence disease states. However, the mechanisms of let-7 suppression remains poorly understood, because LIN28 recognition depends on coordinated targeting by both the zinc knuckle domain (ZKD)—which binds a GGAG-like element in the precursor—and the cold shock domain (CSD), whose binding sites have not been systematically characterized. By leveraging single-nucleotide-resolution mapping of LIN28 binding sites in vivo, we determined that the CSD recognizes a (U)GAU motif. This motif partitions the let-7 microRNAs into two subclasses, precursors with both CSD and ZKD binding sites (CSD+) and precursors with ZKD but no CSD binding sites (CSD−). LIN28 in vivo recognition—and subsequent 3ʹ uridylation and degradation—of CSD+ precursors is more efficient, leading to their stronger suppression in LIN28-activated cells and cancers. Thus, CSD binding sites amplify the regulatory effects of LIN28.
Keywords: LIN28, let-7 microRNA biogenesis, cold shock domain, bipartite binding, selective suppression
Introduction
MicroRNAs (miRNAs) are a class of small regulatory RNAs of ~22 nucleotide (nt) that are involved in essentially all cellular processes. To produce mature miRNAs, the primary transcripts of the miRNA genes (pri-miRNAs) are first cleaved in the nucleus into hairpin precursors (pre-miRNAs) by the Microprocessor complex containing DROSHA and the RNA binding protein (RBP) DGCR8 (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004; Lee et al., 2003), and then exported to the cytoplasm (Lund et al., 2004; Yi et al., 2003) for further processing by DICER to remove its loop region (Bernstein et al., 2001; Grishok et al., 2001; Hutvagner et al., 2001; Knight and Bass, 2001). One strand of the resulting duplex is incorporated into the RNA-induced silencing complex (RISC) to serve as a template for suppressing target mRNA through complementary base-pairing (Meister et al., 2004).
Let-7 is an ancient family of miRNAs initially discovered as a heterochronic gene in C. elegans (Reinhart et al., 2000; Slack et al., 2000) but later found in all bilateral animals (Pasquinelli et al., 2000). In mammals, the let-7 family consists of 12 members that are expressed from 8 different loci generated by genomic duplication events during evolution (Hertel et al., 2012). All members of the let-7 family contain the identical seed sequence, the major determinant of target selection, and their targets include oncogenes RAS (Johnson et al., 2005), HMGA2 (Lee and Dutta, 2007; Mayr et al., 2007), c-MYC (Sampson et al., 2007), and multiple genes involved in pluripotency maintenance (Worringer et al., 2014). Interestingly, while the levels of pri- and pre-let-7 are comparable between undifferentiated and differentiated cells, it was reported that mature let-7 are detected only after differentiation of ESCs (Thomson et al., 2006), suggesting a post-transcriptional mechanism that suppresses their biogenesis. This suppression was later found to be mediated by an RBP named LIN28 (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008).
The LIN28 protein, consisting of an N-terminal cold shock domain (CSD) and a C-terminal CCHC-type zinc knuckle domain (ZKD), is encoded by two paralogous genes LIN28A and LIN28B (Figure 1A and Figure S1A). Expression of LIN28 is mainly restricted to ESCs and certain transformed cell lines, but is reactivated in ~15% of tumors (Shyh-Chang and Daley, 2013; Viswanathan et al., 2009). The profound impact of the LIN28/let-7 pathway is highlighted by the fact that LIN28 is one of four factors sufficient to reprogram human somatic cells into induced pluripotent stem cells (Yu et al., 2007). Consequently, extensive efforts have been made to understand the underlying mechanism of LIN28-mediated let-7 suppression and multiple mechanisms have been proposed. These include blocking of DROSHA processing of pri-let-7 in the nucleus (Newman et al., 2008; Viswanathan et al., 2008); DICER processing of pre-let-7 (Rybak et al., 2008); and 3ʹ end uridylation (Hagan et al., 2009; Heo et al., 2008; Heo et al., 2009) which stimulates further degradation of pre-let-7 by the DIS3L2 exonuclease (Chang et al., 2013; Ustianenko et al., 2013).
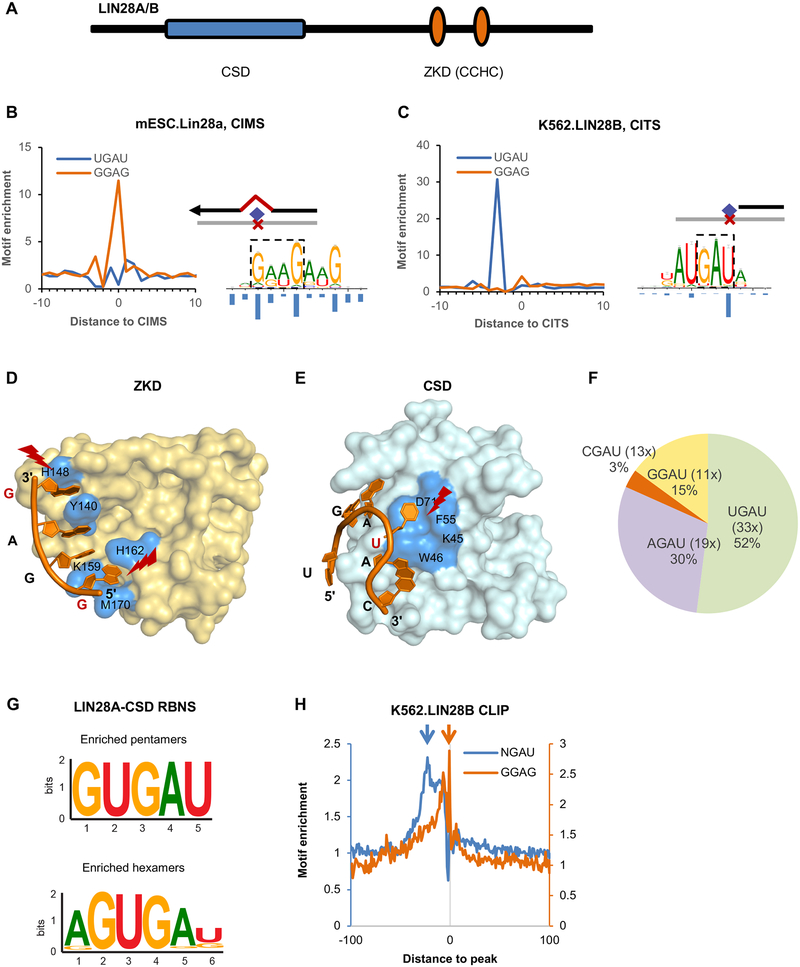
Figure 1: LIN28 cold shock domain (CSD) and zinc knuckle domain (ZKD) recognize distinct sequence motifs as defined by single-nucleotide-resolution analysis of CLIP data.
Related to Figures S1.
(A) Schematic representation of LIN28 protein domains.
(B, C) The ZKD and CSD binding motifs determined from single-nucleotide-resolution analysis of CLIP data. A GGAG-like motif was identified by modeling sequences around LIN28A CIMS derived from mouse ESCs (B, right panel), and a UGAU motif was determined by modeling sequences around LIN28B CITS derived from K562 cells (C, right panel). The frequency of crosslinking at each motif position is shown under the motif logos. The enrichment of GGAG and UGAU tetramers around CIMS or CITS is shown on the left of each panel.
(D, E) The crystal structure of LIN28A ZKD (D) and CSD (E) in complex with let-7g hairpin (PDB accession: 3TS2). Residues that are in direct contact with RNA are highlighted in blue. The crosslinked nucleotides are indicated in red and highlighted.
(F) Frequency of tetramers conforming to the NGAU consensus in LIN28B eCLIP data from K562 cells. The fold enrichment of each tetramer at the crosslink site in comparison to matched control sequences is shown in the parentheses.
(G) CSD binding motifs identified from RBNS analysis. The most enriched pentamers and hexamers after two rounds of LIN28A CSD selection are shown.
(H) Enrichment of NGAU and GGAG around LIN28 eCLIP tag cluster peaks from K562 cells.
Both the CSD and ZKD are involved in recognition of the pre-let-7 through the terminal loop structure, as demonstrated by extensive mutational analysis, in vitro miRNA processing assays, and LIN28/pre-let-7 co-crystal structure (Heo et al., 2009; Mayr et al., 2012; Nam et al., 2011; Piskounova et al., 2008). It has been well established that the ZKD recognizes a GGAG-like motif located in the stem loop structure. In mammals, this motif is present in all members but one of the let-7 family (Triboulet et al., 2015), and it is crucial for stabilizing the LIN28 and pre-let-7 complex and recruiting the terminal uridine transferase (TUTase) that uridylates pre-let-7 (Wang et al., 2017).
Multiple studies have reported that the CSD has a higher affinity to several tested pre-let-7 members than the ZKD (Mayr et al., 2012; Nam et al., 2011; Wang et al., 2017), but its sequence specificity is under debate. Analysis of the LIN28/pre-let-7 co-crystal structure revealed that the CSD interacts with the single stranded loop area of pre-let-7 hairpin and is predicted to have a preference for the GNGAY sequence (Y=pyrimidine; N=any base). However, due to variations in the loop region among the 12 let-7 family members, this motif is only present in a subset of pre-let-7. Assuming all pre-let-7 family members are uniformly suppressed by LIN28, Nam et al. proposed that the CSD has weaker sequence specificity, so that it can adopt to different substrate sequences (Nam et al., 2011). The CSD was also reported to have a preference for pyrimidine rich sequences and it was suggested that its interaction with the loop region of pre-let-7 might induce a conformational change that exposes the GGAG motif in the hairpin (Mayr et al., 2012).
In addition to let-7 miRNAs, several recent studies using crosslinking and immunoprecipitation followed by high-throughput sequencing (HITS-CLIP or CLIP-seq) demonstrated that LIN28 recognizes thousands of mRNA transcripts, and might play a role in regulating RNA splicing and translation through less characterized mechanisms (Cho et al., 2012; Graf et al., 2013; Hafner et al., 2013; Wilbert et al., 2012). Analysis of LIN28 binding sites in mRNA revealed an enrichment of GGAG-like sequences corresponding to the ZKD binding motif. However, these studies have so far provided limited insights into the sequence specificity of the CSD and its contribution to in vivo LIN28-RNA interactions, possibility due to insufficient resolution for deconvolution of the bipartite LIN28 binding motifs.
In this study, we characterized the in vivo binding specificity of LIN28 using single-nucleotide-resolution maps of thousands of LIN28 binding sites in mRNA derived from CLIP data. Our analysis confirmed that the GGAG motif is recognized by the ZKD. Importantly, we identified a distinct, high-confidence CSD binding motif—(U)GAU—which is reminiscent of the CSD-binding consensus sequence proposed based on the LIN28/pre-let-7 co-crystal structure (Nam et al., 2011). The specificity of CSD recognition of this motif was validated using in vitro binding assays and mutagenesis analysis. We further observed that LIN28 binds much more robustly to the subclass of pre-let-7 harboring (U)GAU (CSD+), than to the other subclass without the motif (CSD−), both in mouse ESCs and human cancer cell lines. Consequently, CSD+ let-7 family members are efficiently uridylated and suppressed in vivo, while the impact of LIN28 on CSD− let-7 family members is much more moderate. Differential inhibition of the two subclasses of let-7 was also observed in multiple tumor types where LIN28 expression is reactivated, implying a potential role of this selective suppression model in tumorigenesis.
Results
Identification of a specific CSD binding motif from single-nucleotide-resolution analysis
Given the bipartite nature of LIN28 RNA-binding domains, we postulated that a single-nucleotide-resolution map of LIN28-RNA interaction sites would help to better characterize its binding specificity. To this end, we took advantage of the computational approaches we previously developed to infer the precise protein-RNA crosslink sites from CLIP data by identifying crosslink-induced mutation sites (CIMS) and truncation sites (CITS) (Shah et al., 2017; Weyn-Vanhentenryck et al., 2014; Zhang and Darnell, 2011). We applied these methods to two in-depth LIN28 CLIP datasets: LIN28A HITS-CLIP performed in mouse ESCs (Cho et al., 2012), and LIN28B CLIP derived from two human cell lines K562 and HepG2 using a modified CLIP protocol named eCLIP (Van Nostrand et al., 2016) (for this study, we mainly describe results from K562 cells, as the results obtained from HepG2 cells are very similar). Due to the differences in protocols used to generate these CLIP libraries, we expected HITS-CLIP to capture only CIMS and eCLIP to be enriched in CITS (see Discussion). Following our established pipeline (Shah et al., 2017), we identified 50,292 substitution CIMS from LIN28A HITS-CLIP data and 22,673 CITS from LIN28B eCLIP data in K562 cells (Table S1). Consistent with the previous analysis (Cho et al., 2012), we observed a striking enrichment of the GGAG motif at CIMS inferred from HITS-CLIP data (11.5 fold at position 0), indicating the predominant crosslinking of the first G of the GGAG motif (Figure 1B left panel and Figure 1D). Surprisingly, we found only very moderate enrichment of the GGAG motif around CITS inferred from eCLIP data (4.2 fold at position 0 as compared to ~2 fold in neighboring positions; Figure 1C left panel), suggesting the possibility that these binding sites reflect a second mode of LIN28-RNA interaction.
To better understand the binding specificity of LIN28, we performed de novo motif analysis using an algorithm we developed to simultaneously model the binding specificity of an RBP and its crosslinking position in the sequence motif. This algorithm recovered the GGAG-like motif from sequences around CIMS, with more degeneracy allowed between the first and last guanines (Figure 1B and Table S2), which is consistent with previous structural and mutational analyses (Loughlin et al., 2011; Nam et al., 2011). Intriguingly, applying this method to sequences around CITS revealed a distinct AUGAU or GUGAU motif, with predominant crosslinking in the last uridine (Figure 1C and Table S2). The core tetramer motif UGAU is strikingly enriched in sequences around CITS (31 fold at position −3, corresponding to crosslinking to the last uridine), but not CIMS (Figure 1B and C), suggesting its potential importance for LIN28 binding to thousands of mRNA transcripts.
After careful examination, we noticed that the UGAU motif largely resembles the CSD-binding consensus GNGAY proposed from X-ray crystal structural analysis of LIN28 in complex with pre-let-7d, pre-let-7f-1 and pre-let-7g (Figure 1E). The position of the crosslink sites in the motif we identified is highly consistent with the RNA contact of the LIN28 CSD (Figure 1E). The presence of purines in the middle of the motif (UGAU) is critical for reaching the protein surface while the last pyrimidine is essential due to the steric hindrance that is imposed by the surrounding amino acids (Nam et al., 2011). Given that a uridine before GAU does not seem to be crucial for LIN28 binding to pre-let-7 (Nam et al., 2011), we examined the other variants of the GAU motif in LIN28 binding sites in mRNAs. Indeed, we observed that UGAU, AGAU, GGAU and CGAU are all enriched around CITS to a varying degree (Figure 1F), suggesting that a GAU core motif is the primary determinant of CSD binding.
In order to validate our motif identification and provide additional support for the CSD specific recognition, we performed RNA Bind-and-Seq (RBNS), a high-throughput in vitro assay to identify RNA ligands recognized by a protein of interest with high affinity (Lambert et al., 2014). For this experiment, the FLAG-tagged CSD domain of LIN28 was purified from HEK293 cells and exposed to a large library of 8-nt RNA fragments. CSD-bound high-affinity sequences were isolated, amplified and subject to a second round of selection followed by deep sequencing as well as motif enrichment analysis (see Methods for details). The most enriched pentamers and hexamers present in the final RNA pool contained a UGAU core motif that highly resembled the motif identified from LIN28 CITS (Figure 1G), supporting specific recognition of this motif by LIN28 CSD.
To further confirm the involvement of the CSD binding motif in the bipartite RNA binding of LIN28, we examined the enrichment of both GGAG and GAU motifs in sequences around the most robust LIN28 CLIP tag peaks in mRNAs independent of the identified crosslink sites. Both motifs were found enriched in HITS-CLIP as well as in eCLIP data sets, with the GAU motif being highly represented 5–30 nucleotide upstream of the GGAG motif (Figure 1H and Figure S1B,C). We also predicted LIN28 binding sites in mRNA transcripts by using our mCarts algorithm to identify likely functional clusters of conserved LIN28 motif sites (Weyn-Vanhentenryck and Zhang, 2016; Zhang et al., 2013). The clusters predicted with conserved GAU motifs better overlap the LIN28B eCLIP data than those predicted with GGAG. Critically, the best performance was achieved using a hybrid model that allows any combination of GAU and GGAG motifs (Figure S1D–F). These results indicate that the presence of both CSD and ZKD binding elements contributes to high-affinity interaction of LIN28 and mRNA targets, which is consistent with the bipartite mode of LIN28-pre-let-7 interaction. Together, our analysis suggests the sequence-specificity of both LIN28 CSD and ZKD and the importance of the bipartite binding motif for in vivo protein-RNA interaction.
Selective recognition of pre-let-7 is modulated by the CSD binding site
Since let-7 pre-miRNAs are the best known targets of LIN28, we investigated whether the CSD binding motif we identified fits the in vivo recognition pattern of LIN28 to let-7 precursors. Examination of pre-let-7 sequences suggests that the whole family can be divided into two subclasses based on the presence of the CSD binding motif (Figure 2A). Half of let-7 family members contain both GAU and GGAG-like motifs, which we denote the CSD+ subclass. These include pre-let-7b, pre-let-7d, pre-let-f-1, pre-let-7g, and mir-98. We also include pre-let-7i in the CSD+ subclass, which has GAC, a variant of GAU predicted to be compatible with the CSD structure (Nam et al., 2011); in this case the uridine before the GAC triplet also matches the CSD binding consensus we determined. The other six let-7 family members, denoted CSD− subclass, lack the GAU motif in the terminal loop region of pre-miRNA. This subclass includes pre-let-7a-1/2/3, pre-let-7c, pre-let-7e, and pre-let-7f-2. All CSD− subclass members with exception of one (pre-let-7a-3 in human) have the GGAG-like motif. Interestingly, it was previously reported that pre-let-7a-3 completely escapes LIN28-mediated suppression (Triboulet et al., 2015). However, the distinction of CSD+ and CSD− let-7 family members with respect to LIN28 binding and LIN28-mediated suppression is unclear.
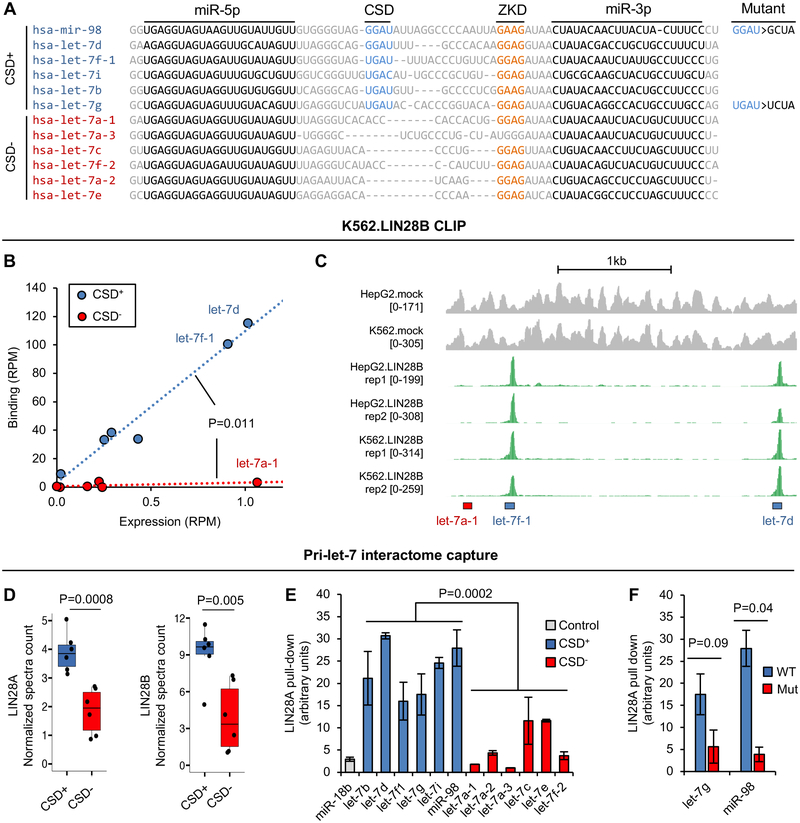
Figure 2: The cold shock domain modulates LIN28 binding to CSD+ let-7 precursors.
Related to Figure S2.
(A) Multiple sequence alignments of pre-let-7 hairpins. Sequences corresponding to mature miRNAs, and binding sites of LIN28 CSD and ZKD are indicated. Let-7 family members are divided into two subclasses, denoted CSD+ and CSD−, depending on the presence of the GAU (GAC in the case of let-7i) motif. Mutant CSD+ let-7 precursors tested in this study are also shown.
(B) Quantification of LIN28B binding to let-7 pre-miRNAs in K562 cells. The y-axis shows the total number of unique CLIP tags expressed in reads per million (RPM) that overlap with each pre-let-7. The x-axis shows the number of mock CLIP tags (input) expressed in RPM reflecting the abundance of the pre-let-7. ANOVA was used to test the difference in LIN28 binding to the CSD+ versus CSD− pre-let-7s after controlling for pre-miRNA abundance.
(C) LIN28 binding to different let-7 family members in human HepG2 and K562 cells using the let-7a-1/7f-1/7d poly-cistronic miRNA locus as an example. The number of mock (gray) and IP (green) tags in each genomic position is shown, and the locations of the pre-miRNA hairpins are indicated at the bottom.
(D) RNA-mediated LIN28A/B pull-down using different pri-let-7 family hairpins as a bait quantified by mass spectrometry. The normalized spectrum counts of mass spectrometry-identified peptides from LIN28A (left) or LIN28B (right) are shown for each bait and compared between the CSD+ and CSD− subclasses. The boxplots indicate the interquartile range of each subclass. The difference between the two subclasses was evaluated by a t-test.
(E) RNA-mediated LIN28A pull-down using different pri-let-7 family hairpins as a bait quantified by immunoblots. LIN28 intensity detected using a specific antibody was normalized using northern blot signal for each individual bait. CSD+ hairpins are shown in blue and CSD− hairpins are shown in red, respectively. pri-miR-18b is used as a negative control. Error bars represent standard error of the mean (SEM) of two replicates. Comparison of CSD+ and CSD− hairpins was performed using ANOVA of a linear mixed effect model.
(F) RNA-mediated LIN28A pull-down using wild type (WT) and mutant (Mut) pri-let-7g and pri-mi-R98 hairpins. The amount of bound LIN28 is quantified as in (E). Reduction of the LIN28 in the mutant is compared to the wild type of the corresponding miRNA precursor using a single-sided t-test. Error bars represent SEM of two replicates.
We hypothesized that if the (U)GAU motif uncovered from analysis of tens of thousands of LIN28 mRNA binding sites reflects the in vivo binding specificity of LIN28 CSD, its presence should also be important for LIN28 recognition of let-7 precursors. Since multiple studies consistently reported higher binding affinity of the CSD to pre-let-7 compared to the ZKD (Mayr et al., 2012; Nam et al., 2011; Wang et al., 2017), we predicted that LIN28 binds CSD+ pre-let-7 more robustly than CSD− pre-let-7 in vivo. To validate this prediction, we examined LIN28 binding to all pre-let-7 family members as determined by CLIP data. Intriguingly, while strong LIN28B CLIP tag clusters were found for all of the CSD+ pre-let-7’s, very few CLIP tags were detected from CSD− let-7s in both K562 and HepG2 cells. The difference of the two subclasses is statistically significant after controlling the abundance of pre-miRNA expression (P=0.011, ANOVA; Figure 2B). The distinction can be most clearly observed in let-7 family members expressed from a poly-cistronic locus as a single primary transcript (e.g., pre-let-7d and pre-let-7f-1 in CSD+ versus pre-let-7a-1 in CSD−, Figure 2C. See additional examples in Figure S2A) (Wang et al., 2011). Moreover, the same patterns were observed from LIN28A HITS-CLIP in mouse ESCs (Figure S2B), suggesting that the selective binding of LIN28 to the two subclasses of let-7 precursors modulated by the CSD is not specific for LIN28A or LIN28B, or the cellular contexts we examined.
To further validate our hypothesis and exclude the possibility that the selective binding observed from CLIP data is due to a technical bias (e.g., differences in crosslinking efficiency), we examined a recently published dataset of pri-miRNA-binding interactomes in 11 cell lines, in which pri-miRNA-hairpin-interacting proteins were captured using an RNA-mediated protein pull-down assay followed by mass spectrometry analysis (Treiber et al., 2017). A number of miRNA precursors including all members of the let-7 family were used as a bait to identify specific protein interactors. We compared the number of LIN28 peptide spectras identified in the mass spectrometry data between CSD+ and CSD− pre-let-7 family members. Both LIN28A and LIN28B showed a greater preference for binding of CSD+ pri-let-7 hairpins (P=0.0008 and 0.005, respectively, t-test; Figure 2D). The spectrum counts from the mass spectrometry data is not quantitative by nature, although a normalization procedure was performed to allow unbiased comparison of protein pull-down using different pri-miRNA-hairpin baits (Treiber et al., 2017). To obtain a more quantitative measure of the interaction between LIN28 and the two subclasses of pre-let-7 miRNAs, we performed the similar RNA-mediated protein pull-down assay using all 12 let-7 precursors and endogenous LIN28 from NTera2 teratocarcinoma cell line under harsh washing conditions; the pri-miR-18b hairpin without LIN28 binding motifs was used as a negative control. Instead of using mass-spectrometry, interaction of LIN28A was quantified by immunoblots with a specific antibody and normalized using signal from northern blots that measured the amount of coupled bait RNA. As we expected, all CSD+ pri-let-7 hairpins showed stronger interaction with LIN28 compared to CSD− pri-let-7 hairpins (P=2.1e-4, ANOVA; Figure 2E and Figure S2C). To directly evaluate the importance of the CSD binding site for LIN28 interaction, we also tested loss-of-function mutants of two pri-let-7 hairpins from the CSD+ subclass (let-7g and miR-98), where the (U)GAU motif was mutated (Figure 2A). Importantly, mutation in the CSD-binding motif greatly reduced the amount of the associated protein to the level similar to that observed from the CSD− pri-let-7 hairpins, validating the importance of the (U)GAU motif in recognition by LIN28 (Figure 2F).
To provide additional biochemical evidence for the differential recognition of let-7 precursors, we expressed and purified recombinant LIN28A containing both the CSD and the ZKD or the CSD alone, and performed electromobility shift assays (EMSA) using all let-7 precursors. Similar to our observation from CLIP data and in vitro RNA-mediated protein pull-down, LIN28 showed higher binding affinity towards all CSD+ let-7 precursors, no matter whether the longer protein or the isolated CSD was used in EMSA (Figure S2D, E and G) (apparent Kd= 7–16nM for CSD+ pre-let-7 and 13–95 for CSD− pre-let-7 when the recombinant protein with both domains was used in the assays). Mutation in the (U)GAU motif in pre-let-7g and pre-miR-98 also resulted in reduced binding compared to the wild type (Figure S2F and H), consistent with previous results from similar experiments (Mayr et al., 2012; Nam et al., 2011), although the magnitude of change is relatively moderate in our assay. Taken together, our data suggest that the (U)GAU motif modulates selective recognition of the CSD+ subclass of pre-let-7 by LIN28 both in vivo and in vitro.
CSD+ let-7 miRNAs are selectively uridylated and suppressed by LIN28 in human cells and in cancer
As a major functional outcome, the LIN28 and pre-let-7 interaction results in suppression of mature miRNA levels. One important mechanism of this suppression is LIN28-mediated recruitment of the terminal uridyltransferase TUT4 or TUT7, which modifies the 3ʹ end of the pre-miRNA with a stretch of uridines, and stimulates degradation of the polyuridylated pre-miRNA by DIS3L2 exonuclease (Chang et al., 2013; Ustianenko et al., 2013). To evaluate whether selective binding of LIN28 to CSD+ versus CSD− let-7 precursors has any impact on their suppression through the TUT4/DIS3L2 pathway, we referred to a previously published DIS3L2 CLIP analysis (Ustianenko et al., 2016). In this study, a catalytically inactive mutant of DIS3L2 exonuclease with intact RNA binding abilities was used to identify a variety of uridylated RNA transcripts in HEK293 cells. We compared the number of uridylated pre-let-7 DIS3L2 CLIP tags between the two let-7 subclasses and found that CSD+ pre-let-7s exhibit up to 20-fold greater uridylation levels compared to CSD− pre-let-7’s (P=0.005, Wilcoxon rank sum test; Figure 3A). Motivated by this finding, we compared uridylation levels of let-7 precursors detected in the LIN28B eCLIP data, and found that CSD+ precursors also exhibit significantly higher uridylation levels compared to CSD− precursors, regardless of their expression levels (P=6.3e-5, ANOVA; Figure 3B and Table S3). These observations confirmed that the high-affinity LIN28 binding in CSD+ pre-let-7 mediated by both CSD and ZKD results in their efficient uridylation in vivo.
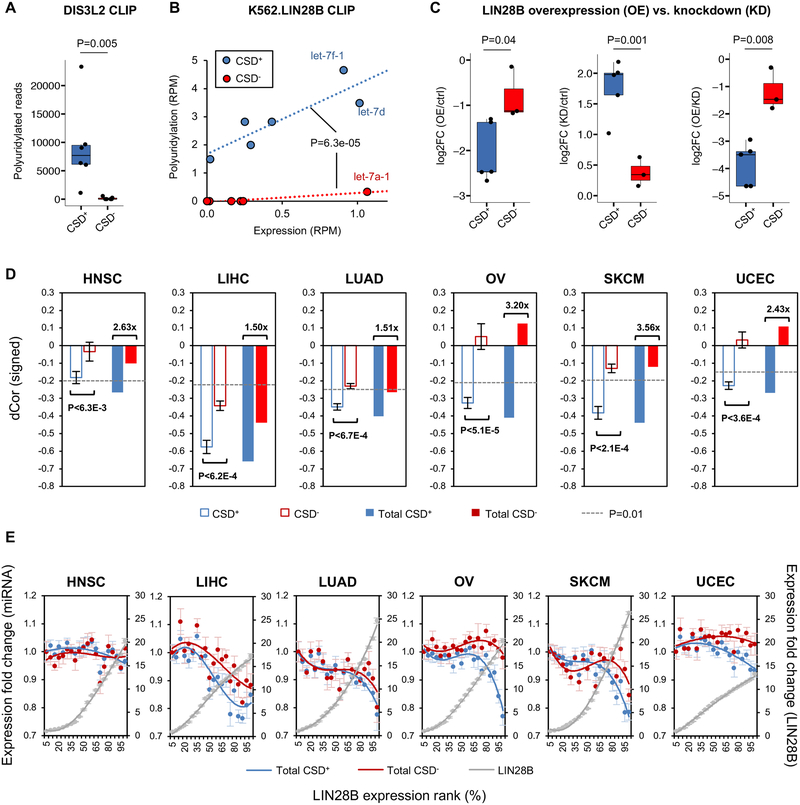
Figure 3: Selective 3ʹ polyuridylation and suppression of CSD+ let-7 in human cells and tumor samples with LIN28B reactivation.
Related to Figures S3.
(A) Boxplot showing the level of 3ʹ polyuridylation for the two subclasses of let-7 precursors from DIS3L2 CLIP in HEK293 cells. Wilcox rank sum test was used to evaluate the difference between the two subclasses.
(B) Quantification of 3ʹ polyuridylation of let-7 pre-miRNAs from LIN28B eCLIP in K562 cells. The y-axis shows the total number of unique uridylated CLIP tags expressed in reads per million (RPM) that overlap with each pre-let-7. The x-axis shows the number of mock CLIP tags (input) expressed in RPM reflecting the abundance of the pre-let-7. The difference between LIN28-mediated uridylation after controlling for pre-miRNA abundance is tested using ANOVA.
(C) Changes in the expression of mature let-7 miRNA upon perturbation of LIN28B levels (overexpression or knockdown) in HEK293 cells. The boxplots indicate the interquartile range of each subclass. The difference between the two subclasses was evaluated by a t-test.
(D) CSD+ let-7 miRNAs showed stronger downregulation by LIN28B than CSD− let-7 miRNAs in multiple types of tumor samples. For each tumor type, average distance correlation (dCor) estimated between LIN28B and miRNAs from each subclass are given on the left (hollow bars); the sign is designated by Spearman’s correlation, and p-values estimated by Mann-Whitney U test. Error bars represent SEM. Pooled reads across miRNA classes produced total expression per class and their dCor with LIN28B expression is given on the right (solid bars) for each tumor type; the p<0.01 cutoff, estimated by permutation testing, is given in broken gray lines.
(E) The response of CSD+ and CSD− let-7 miRNAs to changes in LIN28B expression in tumor samples. Samples are binned into 20 same-size bins according to LIN28B expression. Each bin is represented by the average fold change of total expression in each subclass relative to the first bin across samples in the bin, and curves were fit to a polynomial distribution with order 3. Similarly, LIN28B average expression fold changes are given on the right axis. Error bars represent SEM.
Since 3ʹ uridylated pre-let-7 is expected to be degraded by DIS3L2, we directly examined whether LIN28 selectively suppresses CSD+ versus CSD− let-7 family members. To this end, we examined the abundance of the individual let-7 family members upon manipulation of LIN28 protein levels either by overexpression or siRNA-mediated knockdown in HEK293 cells (Hafner et al., 2013). We found that overexpression of LIN28B resulted in stronger repression of CSD+ let-7 compared to CSD− let-7; conversely, knockdown of LIN28B resulted in more de-repressed CSD+ let-7 compared to CSD− let-7 (P<0.05 in all comparison, t-test; Figure 3C). The same pattern was observed in additional independent datasets derived from similar experiments (Powers et al., 2016; Wilbert et al., 2012), although the distinction between CSD+ and CSD− let-7 family members was not discussed in the original studies (see Discussion). These results confirmed that the CSD binding site in let-7 family members plays an important role in determining the efficiency of LIN28-dependent suppression of miRNA biogenesis in vivo.
Finally, LIN28 activation, followed by loss of let-7, is a hallmark of cancer etiology (Balzeau et al., 2017). To investigate the suppression of let-7 miRNAs by LIN28 in the context of tumorigenesis, we performed a pan-cancer analysis of fourteen tumor types for which both mRNA and miRNA expression was profiled by The Cancer Genome Atlas (TCGA) using deep sequencing (Table S4). In total, we found that LIN28B and LIN28A are variably expressed (mean absolute deviation greater than zero) in six and two tumor types, respectively. In each of these tumor types, the expression of let-7 miRNAs was significantly anti-correlated with that of LIN28 (Figure S3A,B), suggesting suppression of let-7 following LIN28 reactivation, which is consistent with previous studies (Viswanathan et al., 2009). Importantly, in these contexts, CSD+ and CSD− let-7’s demonstrate variable response to LIN28 activation with CSD+ miRNAs showing significantly stronger anti-correlation with both LIN28B (Figure 3D) and LIN28A (Figure S3C) expression. Furthermore, the difference was most evident in samples with high LIN28 abundance (Figure 3E and Figure S3D). Taken together, our results suggest that the CSD+ subclass of let-7 miRNAs are selectively suppressed following LIN28 reactivation in human cells and in cancer, and that the (U)GAU motif serves as an amplifier of the LIN28 regulatory effects.
Discussion
Due to the important role of the LIN28/let-7 axis in developmental biology and cancer, the mechanisms underlying post-transcriptional suppression of let-7 miRNAs by LIN28 have been a subject of intensive investigation. These studies have revealed a fascinating degree of complexity, which is derived, at least in part, from the plasticity of protein-RNA interactions. In the more general contexts, each RNA-binding domain of an RBP recognizes a short and degenerate sequence or structural motif. Therefore, specificity has to be achieved through combinations of multiple domains, which allow an expansion of the RNA pool that can be regulated in both sequence and structure-dependent manners (Lunde et al., 2007). In the case of LIN28, bipartite binding motif sites recognized by the CSD and ZKD domains are required for high-affinity interactions of LIN28 with substrate RNAs, including let-7 pre-miRNAs.
It has been a prevailing view that all mammalian let-7 miRNAs, with the exception of hsa-let-7a-3 or its homolog, are suppressed by LIN28 through a similar mechanism (Triboulet et al., 2015). This model postulates that the major determinant of specificity is the GGAG-like RNA element recognized by the ZKD (Heo et al., 2009; Mayr et al., 2012) while the CSD contacts the terminal loop of pre-let-7 with limited specificity but higher affinity (Nam et al., 2011; Wang et al., 2017). This ZKD-mediated interaction either blocks different steps of miRNA processing or leads to the degradation of pre-let-7 (Hagan et al., 2009; Heo et al., 2008; Heo et al., 2009; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). However, previous reports that investigated the general mechanisms of LIN28/let-7 interaction and LIN28-dependent let-7 biogenesis frequently tested only one or a few selected miRNAs without distinguishing between different let-7 family members. Examination and comparison of the results from multiple studies (see below) suggest that the impact of LIN28 varies across the let-7 family members. While some of these seemingly conflicting results could be due to variability of cellular contexts and experiments, we conjectured that they might also reflect unknown mechanisms that cannot be accounted for by the uniform suppression model. One possible source of variation is selective binding of pre-let-7 mediated by the LIN28 CSD, as this domain contacts the terminal loop sequence of the pre-let-7 hairpin which is substantially diverged among let-7 family members but highly conserved across different mammalian species for each member. However, this question cannot be answered without a precise understanding of the LIN28 CSD binding specificity.
Our single-nucleotide-resolution analysis of tens of thousands of LIN28 binding sites in mRNA using recent eCLIP data unexpectedly uncovered a distinct motif (U)GAU. A similar motif GNGAY was proposed to be the consensus binding site of the CSD based on the LIN28/pre-let-7 crystal structures (Nam et al., 2011). However, the previous prediction was based on a very limited number of sequences (i.e., pre-let-7d, pre-let-7f-1 and pre-let-7g, all of which contain (U)GAU, for which structures were determined), making it unclear whether this consensus precisely reflects the specificity of the CSD. Partial representation of the motif among let-7 family members is also inconsistent with the uniform suppression model, as well as other studies reporting conflicting results of the CSD binding specificity (Mayr et al., 2012). Therefore, without additional support for the significance of the GNGAY consensus, it was postulated that the binding specificity of the CSD is limited, allowing it to adopt to other variable sequences found in all let-7 family members (Nam et al., 2011; Wang et al., 2017).
Our confidence in the (U)GAU motif required for high-affinity LIN28 interaction was initially based on its striking enrichment in tens of thousands of LIN28 binding mRNA targets. So why does the eCLIP data capture a distinct LIN28 motif compared to previous CLIP analyses which only identified a GGAG-like motif (Cho et al., 2012; Graf et al., 2013; Wilbert et al., 2012)? While speculative, this discrepancy is probably due to differences in the protocols used to prepare CLIP libraries. All previous studies using LIN28 CLIP cloned immunoprecipitated RNA crosslinked to LIN28 through ligation of 3ʹ and 5ʹ RNA linkers, followed by reverse transcription and PCR amplification using primers that matches the linker sequences. Due to the irreversibility of the crosslinking, it was demonstrated that the residual amino acid-RNA adducts can interfere with reverse transcriptase (RT), sometimes resulting in premature truncation of the cDNA. Only read-through CLIP tags including a subset carrying crosslink-inducted mutations (Zhang and Darnell, 2011) were captured by these protocols, while truncated tags were lost during PCR amplification. On the other hand, eCLIP, among several other similar protocols (Lee and Ule, 2018), are able to capture both truncated and read-through tags using different cloning strategies. Whether RT enzyme predominantly stops at or reads through crosslink sites depends on the identity of the amino acid-RNA adducts as well as properties of the RT enzyme and other experimental conditions (Van Nostrand et al., 2017). For example, we previously demonstrated that another RBP, RBFOX, can be crosslinked to its binding element UGCAUG at either G2 or G6, which results in predominant read-through and premature stop, respectively. If CSD and ZKD can be crosslinked to different positions in the bipartite binding site at which they directly contact, the two crosslink sites could also affect RT differently. For example, crosslinking of the GGAG motif with the ZKD could result in frequent read-through detected in earlier CLIP assays, while crosslinking of the (U)GAU with the CSD could predominantly result in truncations that can only be detected by eCLIP and other improved CLIP protocols. A recent study closely investigated the crosslinking between LIN28 and pre-let-7f in an in vitro binding assay (Ransey et al., 2017). Interestingly, it confirmed the crosslinking at the guanines in the GGAG motif by CIMS analysis, but also found a predominant crosslink site at the last uridine of the GAU element to Phe55, a core residue of the CSD, by tandem mass-spectrometry, which is consistent with our results from genome-wide analysis of in vivo LIN28 binding sites.
As the CSD-binding motif is important for LIN28 high-affinity interaction with pre-let-7 and the refined motif is not found in all pre-let-7 family members, its presence would divide the let-7 miRNAs into two subclasses. Only the subclass that possesses both GAU and GGAG-like elements (CSD+) is predicted to be efficiently targeted by LIN28. We thus propose to modify the current uniform suppression model with a selective suppression model where the presence of the CSD binding element in CSD+ let-7 family members modulates the efficiency of LIN28-dependent suppression (Figure 4). This model has found strong support from multiple lines of evidence using datasets independently generated by different laboratories. It fits well with our observation that CSD+ let-7 family members show much stronger in vivo interaction with LIN28 than CSD− family members. The difference is reproducible across multiple CLIP datasets independent of CLIP protocol modification (HITS-CLIP versus eCLIP), cellular contexts (HepG2, K562 cells and mESCs) and the targeted protein (LIN28A versus LIN28B). The difference of the two subclasses in LIN28 binding affinity and the contribution of CSD to specific binding were also validated using in vitro binding assays including RNA-mediated interactome capture and EMSA together with mutagenesis analyses. This proposed model explains why isolated CSD does not bind, or binds only weakly, to human let-7a-1 (Nowak et al., 2017) and Xtr-pre-let-7f (Mayr et al., 2012), which do not have the GAU motif, but binds efficiently to let-7d, let-7f-1, and let-7g, which contain the GAU (Wang et al., 2017). Coincidentally, the crystal structure of LIN28 and let-7 was obtained for three CSD+ let-7 family members, but not any CSD− family members (Nam et al., 2011). Similarly, in previous studies using LIN28 CLIP assays (Cho et al., 2012; Graf et al., 2013; Wilbert et al., 2012), the pre-let-7 members shown as examples for robust LIN28 binding are all from the CSD+ subclass.
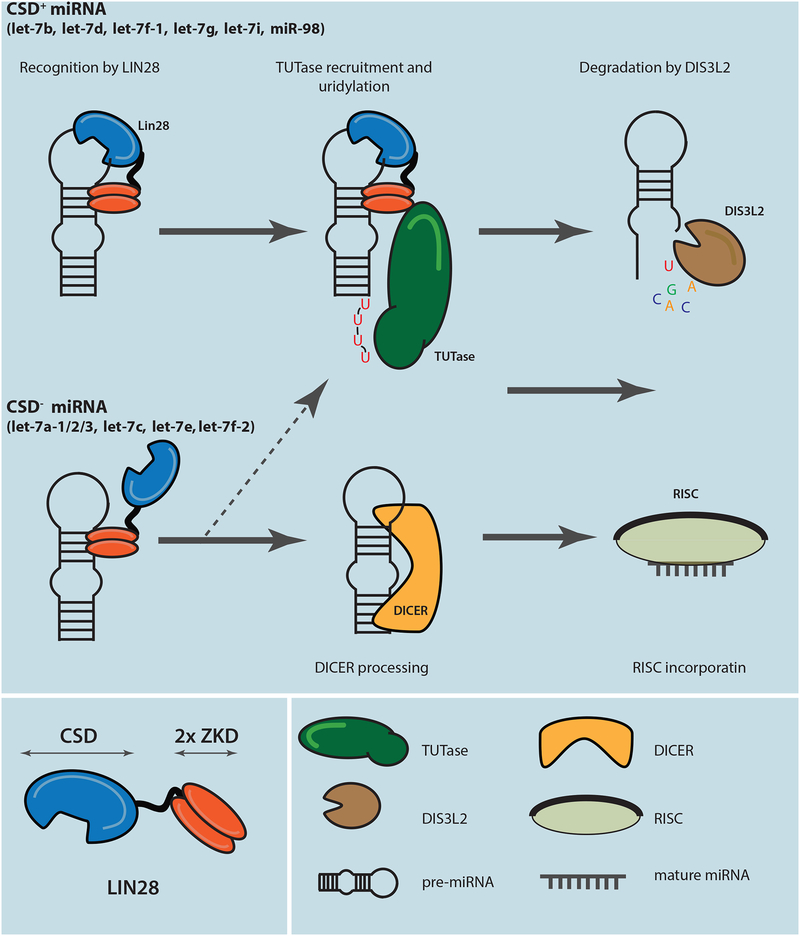
Figure 4: The proposed model of selective let-7 microRNA suppression modulated by the bipartite LIN28 binding.
Related to Figure S4.
CSD+ let-7 miRNA precursors have both CSD and ZKD binding elements, which efficiently recruit LIN28, leading to their 3ʹ uridylation by TUTase and degradation by DIS3L2 (arrows with solid line). CSD− let-7 miRNA precursors lack (U)GAU binding element and are recognized by LIN28 with lower binding affinity, leading to less efficient or partial suppression of these miRNAs by the LIN28/TUT/DIS3L2 pathway (arrows with dotted line), allowing them to enter the DICER processing and RISC incorporation.
The functional consequence of selective LIN28 binding to CSD+ versus CSD− let-7 is clearly reflected in the much more efficient 3ʹ uridylation (observed from CLIP data of LIN28 and DIS3L2) and degradation (observed in HEK293 cells upon LIN28 overexpression or knockdown (Hafner et al., 2013; Wang et al., 2017; Wilbert et al., 2012)). Similarly, depletion of LIN28B in neuroblastoma cells using Cas9 targeting showed a much greater level of de-repression of CSD+ let-7 family members compared to CSD− members (Powers et al., 2016). In these cancer cell lines, expression of let-7a members appears to be predominant among the let-7 family, despite high levels of LIN28 expression, suggesting that let-7a precursors lacking the GAU element are at least partially escaping LIN28-mediated repression (Hafner et al., 2013; Powers et al., 2016; Wilbert et al., 2012).
So what is the implication of this selective suppression model for developmental biology? On one hand, this model is consistent with the large number of let-7 family members in all bilateral animals, suggesting a strong evolutionary selection pressure to maintain diversity within the family. On the other hand, the benefit of having selective suppression remains a major, unanswered question. We propose several potential scenarios in which pluripotent stem cells might need to have a divergent subset of let-7 family members to escape LIN28 suppression. First, the selective suppression model could provide a mechanism for fine-tuning the abundance of let-7 expression. The timely and precise adjustment of the mature let-7 miRNA pool might be required to tightly control their downstream mRNA targets, including those essential regulators of stem cell pluripotency. Second, both LIN28A and LIN28B mRNAs possess let-7 binding sites on their 3ʹ UTRs and can themselves be subject to suppression by let-7 (Rybak et al., 2008). In addition, let-7 also regulates the levels of MYC, a transcriptional regulator of LIN28 (Balzeau et al., 2017). Such a complex multi-layer regulatory feedback loop might be essential for the robust maintenance of the pluripotent state in ESCs, but will require a break during transition to the differentiated state. The subset of let-7 family members (CSD−) that are capable of partially escaping from LIN28 suppression could provide such a trigger. Interestingly, during reprogramming of mouse embryonic fibroblasts towards induced pluripotent cells, greater efficiency was achieved by using let-7 antisense oligonucleotides compared to expression of LIN28 proteins, possibly due to the lack of uniform suppression of all let-7 family members by LIN28 (Worringer et al., 2014). Finally, our analysis suggests that despite their identical seed regions, each let-7 miRNA targets a unique, but not mutually exclusive, gene set (H.-S.C. and P.S., unpublished observation). Consequently, each target is potentially regulated by a set of CSD+ and CSD− let-7 miRNAs, which act as selector switches to amplify the effects of fluctuations in LIN28 abundance. The magnified response of CSD+ miRNAs to LIN28 upregulation might lead to variable effects on the post-transcriptional regulation of let-7 targets, and let-7 target expression profiles following LIN28 upregulation might vary depending on the identities and classes of the regulating miRNAs. These possibilities do not have to be mutually exclusive.
While our proposed selective suppression model effectively accounted for variability between CSD+ and CSD− miRNA responses to LIN28 dysregulation, it is nevertheless likely to be a simplification as some of the discrepancies among previously reported results remain unaddressed. For instance, several studies suggested that let-7a-1 (CSD−) levels are low in ESCs and P19 cells, and that their abundance increased upon LIN28A knockdown, indicating relatively efficient suppression by LIN28 (Heo et al., 2009; Viswanathan et al., 2008). We found that let-7a generated from pre-let-7a-1/2/3, all CSD−, is among the top 20 most abundant miRNAs in human ESCs (Wilbert et al., 2012); similarly, let-7f, generated from pre-let-7f-1 from CSD+ and pre-let-7f-2 from CSD−, is also relatively abundant, compared to other let-7 miRNAs that are expressed from the same poly-cistronic loci (Figure S4), indicating that these CSD− let-7 miRNAs at least partially escape LIN28 suppression.
Our analysis did not completely rule out the possibility that LIN28A and LIN28B might have subtle difference in selectivity for binding CSD+ and CSD− pre-miRNAs. In addition, these homologs were reported to have largely mutually exclusive expression in cell lines (with ESCs mainly expressing LIN28A) and they differ in their subcellular localizations, leading to potentially different mechanisms of action (Piskounova et al., 2011). It is also possible that additional sequence or structural features (e.g., secondary RNA structures) in pre-let-7, co-factors (including KSRP, hnRNP A1, and TRIM25 that were identified in previous studies (Choudhury et al., 2014; Michlewski and Caceres; Trabucchi et al., 2009)), and their stoichiometry could have a significant impact on LIN28-mediated suppression. This complexity is particularly worth noting because in vitro assays likely have caveats due to the difficulty to faithfully recapitulate the in vivo cellular contexts, which might have contributed to experimental variations observed across studies. For example, previous EMSA assays did not show clear difference in binding affinity of LIN28 to CSD+ versus CSD− let-7 precursors (Triboulet et al., 2015). We found that the inclusion of Mg2+ ions in the binding reaction, which may stabilize the native hairpin fold of the pre-miRNAs, resulted in increased LIN28 binding affinities and differential binding of the two subclasses (see STAR Methods). Accurate reproduction of cellular contexts may be important for modeling LIN28 binding and function.
In conclusion, we propose a selective suppression model that provides mechanistic insights into the remarkable complexity of the LIN28/let-7 axis, which may have implications in developmental biology and cancer.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-LIN28A | Abcam | AB63740 |
| Recombinant proteins | ||
| LIN28A (aa. 25–181) | This study | NA |
| LIN28A CSD (aa. 25–120) | This study | NA |
| Experimental Models: Cell Lines | ||
| Human NTera2 cells | ATCC | NA |
| Human HEK 293 cells | ATCC | NA |
| Oligonucleotides | ||
| RNA-mediated interactome capture:
3´-biotinylated 2´-O-methyl-RNA adaptor:
5´-AGGCUAGGUCUCCC-3´ |
Metabion GmbH, Planegg, Germany | NA |
| RBNS: 3´ DNA
adaptor: 5´-AAACTGGAATTCTCGGGTGCCAAGG-3´-Amino-C7 |
Metabion GmbH, Planegg, Germany | NA |
| RBNS: 5´RNA
adaptor: 5´-GUUCAGUAAUACGACUCACUAUAGGG-3´ |
Metabion GmbH, Planegg, Germany | NA |
| RBNS: RT
primer: 5´-GCCTTGGCACCCGAGAATTCCAGTTT-3´ |
Metabion GmbH, Planegg, Germany | NA |
| RBNS: forward PCR
primer: 5´-AATGATACGGCGACCACCGAGATCTACACGTTCAGTAATACGACTCACTATAGG-3´ |
Metabion GmbH, Planegg, Germany | NA |
| RBNS: reverse PCR
primer: 5´-GCCTTGGCACCCGAGAATTCCAGTTT-3 |
Metabion GmbH, Planegg, Germany | NA |
| RBNS: read 1 sequencing
primer: 5´-GATCTACACGTTCAGTAATACGACTCACTATAGGG-3´ |
Metabion GmbH, Planegg, Germany | NA |
| Datasets | ||
| LIN28B eCLIP (K562 cells) | ENCODE (Van Nostrand et al., 2016) | ENCSR970NKP |
| LIN28B eCLIP (HepG2 cells) | ENCODE (Van Nostrand et al., 2016) | ENCSR861GYE |
| LIN28 CLIP (mES cells) | (Cho et al., 2012) | GSE37114 |
| LIN28 CSD RBNS | This study | SRP149796 |
| Pre-let-7 mediated protein pull-down | (Treiber et al., 2017) | |
| DIS3L2 CLIP (HEK293 cells) | (Ustianenko et al., 2016) | |
| LIN28B knock down and overexpression (HEK293 cells) | (Hafner et al., 2013) | |
| TCGA RNA-seq data | TCGA Data Portal https://gdc.cancer.gov | |
| TCGA miRNA-seq data | Firehorse https://confluence.broadinstitute.org/display/GDAC/Dashboard-Stddata | |
| Software and Algorithms | ||
| CLIP data analysis by CTK | (Shah et al., 2017) | http://zhanglab.c2b2.columbia.edu/index.php/CTK |
| De novo motif analysis of RBNS data by Weeder 2.0 | (Pavesi et al., 2001) | http://159.149.160.51/modtools/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
CLIP data processing
To determine the binding specificity of LIN28, we used LIN28A CLIP data derived from mouse ESCs (SRP012118) (Cho et al., 2012) and LIN28B eCLIP data derived from HepG2 and K562 human cell lines as part of the ENCODE project (https://www.encodeproject.org). For each dataset, raw reads were downloaded and processed using our established analysis pipeline CLIP Tool Kit (CTK) (Shah et al., 2017).
In analysis of the eCLIP data, slight modifications were made, as recommended by the original study. Specifically, the 3ʹ adaptors were trimmed using the cutadapt program (Martin, 2011), similar to the analysis pipeline used by the ENCODE consortium (--match-read-wildcards --times 1 -e 0.1 -O 1 --quality-cutoff 6 -m 18 -a $a1 -A ATTGCTTAGATCGGAAGAGCGTCGTGT -A ACAAGCCAGATCGGAAGAGCGTCGTGT -A AACTTGTAGATCGGAAGAGCGTCGTGT -A AGGACCAAGATCGGAAGAGCGTCGTGT -A ANNNNGGTCATAGATCGGAAGAGCGTCGTGT -A ANNNNACAGGAAGATCGGAAGAGCGTCGTGT -A ANNNNAAGCTGAGATCGGAAGAGCGTCGTGT -A ANNNNGTATCCAGATCGGAAGAGCGTCGTGT; here $a1=NNNNNAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC or NNNNNNNNNNAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC, depending on the length of the degenerate barcode used for a specific library). After collapsing exact duplicates, the reads were subject to barcode removal and mapped to the reference genome (hg19) using bwa (Li and Durbin, 2009). Reads mapped to repetitive RNAs such as rRNAs and tRNAs as annotated in the RepeatMasker track were excluded. Potential PCR duplicates were further collapsed by modeling the random barcode to get unique tags. Only read2 (the read starting from 5ʹ end of the RNA tag) was used for analysis described in this paper. The unique tags were used for all downstream analysis, including visualization of read coverage in each genomic position.
To define LIN28 binding sites, replicates were combined to call CLIP tag clusters using a valley seeking algorithm (P≤0.05 after Bonferroni multiple testing correction; valley depth≥0.5). The sequences around the peak center (+/−100nt) were then extracted to evaluate the enrichment of the LIN28 consensus motif (GGAG and NGAU), using flanking sequences of the same size but 500 nt away from the peak center.
To define LIN28 binding sites at the single-nucleotide resolution, we performed crosslink-induced mutation site (CIMS) analysis on the LIN28A CLIP data, as the protocol used to generate this dataset does not capture reads truncated at the crosslink sites. CIMS based on reproducible substitutions (FDR<0.05) were reported in this study (Table S1). For the LIN28 eCLIP dataset, we performed crosslinking induced truncation site (CITS) analysis, as we observed minimal evidence of CIMS in this dataset. CITS with FDR<0.001 were reported in this study (Table S1). Sequences around CIMS and CITS (−10,+10nt) were extracted for de novo motif analysis as described below.
LIN28 de novo motif discovery
Currently, most of the software tools for de novo motif discovery use a standard model with a position-specific weight matrix (PWM) to characterize the specificity of DNA- or RNA-binding proteins (Stormo, 2000). Such a model is applied to a set of training sequences (e.g., sequences around CLIP tag peaks) to find the most over-represented sequence patterns allowing degeneracy. Since many RBPs recognize short and degenerate motifs, the reliability of this approach varies. To improve the precision of de novo motif discovery, we developed an algorithm which takes advantage of the single-nucleotide-resolution map of protein-RNA interactions from CIMS and CITS analysis. This algorithm uses a model that augments the standard PWM model by jointly modeling RBP sequence specificity and the precise protein-RNA crosslink sites at specific motif positions at single-nucleotide resolution. As a result, this method reports both the sequence specificity of an RBP and the probability of crosslinking in each position of the motif. Details of the method will be described elsewhere. We used this algorithm to determine LIN28 binding motifs using CIMS from LIN28A CLIP in mESCs and CITS from LIN28B from human cell lines. The motifs were visualized using WebLogo (Crooks et al., 2004). The complete list of motifs was summarized in Table S2.
Prediction of LIN28 binding sites in mRNA
To predict clusters of LIN28 motif sites genome-wide, we used our mCarts algorithm (Weyn-Vanhentenryck and Zhang, 2016; Zhang et al., 2013). We generated the positive training set from the significant peaks in the LIN28B eCLIP data from K562 cells, masking repeats, requiring a location within 1000 nt of an exon, and extending the peak center by 50 nt, resulting in 38,957 regions. The negative training set consisted of exonic regions extended by 1000 nt which did not overlap with any tags. mCarts was run to identify clusters containing at least 3 motif sites, with neighboring motif sites ≤30 nt apart. We generated three models: one searching for clusters of GGAGs, one searching for clusters of GAUs, and one searching for clusters with any combination of GGAGs and GAUs. These resulted in 214,152, 1,086,840, and 3,590,347 clusters, respectively. To evaluate the sensitivity of the results, we removed clusters overlapping with repetitive regions, ranked the clusters according to their score, and determined whether the cluster center overlapped with CLIP peaks (peak height region extended by 50 nt). We plotted the fraction of clusters overlapping CLIP peaks at each rank to compare the models.
LIN28 structural visualization
All structural visualization of LIN28 and its targeted RNA let-7g was performed using PyMol software (https://pymol.org). All the data was retrieved from PDB (accession: 3TS2).
Uridylation analysis using ENCODE mock and LIN28B eCLIP data
The ENCODE project assayed over 100 RBPs using eCLIP in two human cell lines HepG2 and K562 (Van Nostrand et al., 2016), and data for each RBP consists of a mock and IP experiment. The mock experiment measures all captured RNA fragments crosslinked with any RBPs, so we estimated the expression levels of each miRNA by combining all generated mock experiments (94 in HepG2 and 92 in K562 at the time of this study) and counting the number of unique tags mapping to each pre-miRNA normalized by the total number of unique tags (read per million or RPM) in each sample. We estimated polyuridylation by identifying uridylated tags in LIN28B CLIP experiments (which should contain a stretch of Ts at the end of the read). To identify uridylated tags, we began with the unmapped LIN28B reads remaining after standard CLIP data processing. Using cutadapt (Martin, 2011), we first obtained the set of unmapped reads containing ≥4 consecutive Ts on the 3ʹ end and removed the Ts. These trimmed reads were then re-mapped to the genome and collapsed to identify unique uridylated tags using the CTK pipeline as described above. We then counted the number of uridylated reads on each miRNA precursor normalized by the total number of unique tags (expressed as RPM; Table S3). Pre-let-7g contains 3 T’s around the uridylation site (Ustianenko et al., 2016), so reads mapping to let-7g were filtered to require ≥7 Ts. The coordinates of microRNA hairpins were based on miRBase R21 (June 2014) (Kozomara and Griffiths-Jones, 2014).
Let-7 expression change upon LIN28 overexpression and knockdown
To evaluate the impact of LIN28 on let-7 expression, we used a miRNA-seq dataset from a published study (Hafner et al., 2013). This dataset was derived from HEK293 cells after expressing LIN28B for 72 hrs, after mock transfection (ctrl), and after LIN28B knockdown 72 hrs post-LIN28B siRNA transfection. The fold changes of let-7 expression from pairwise comparison were obtained from the original study.
Selective suppression of let-7 by LIN28 in cancer
To investigate the correlation between LIN28 expression and let-7 expression we analyzed a panel of TCGA tumor samples of fourteen types in which LIN28A/B are sometimes reactivated (Table S4). For each of these tumor types, primary tumors were profiled using both RNA-seq (Illumina Genome Analyzer or HiSeq RNA Sequencing Version 2) and miRNA-seq (Illumina HiSeq 2000 miRNA Sequencing) by TCGA. RNA-seq data that quantify mRNA expression level of 17,792 protein-coding genes, including LIN28A/B, were downloaded from the TCGA Data Portal (level 3 normalization; retrieved on 05/12/2015). We used log2 (normalized count+1) in our analysis. miRNA expression estimates (level 3 normalization) were processed by Firehorse and downloaded from https://confluence.broadinstitute.org/display/GDAC/Dashboard-Stddata (Release: 2015_04_02 stddata Run). All the “NA” values were replaced by “0”. In our analysis, we used log2-transformed RPM (Reads Per Million miRNA mapped), and miRNA identities were taken from miRBase R21 (Kozomara and Griffiths-Jones, 2014).
To test whether let-7 miRNAs were enriched for correlation with LIN28, we performed gene set enrichment analysis of these miRNAs, in the context of all expressed miRNAs, as a function of miRNA correlation with LIN28A and LIN28B expression in tumors that showed LIN28A and LIN28B variability (median absolute deviation score > 0). GSEA (Subramanian et al., 2007) used weighted enrichment statistics and ratio of the two subclasses, with p-values computed using 1k gene-set permutations.
To evaluate the suppression of let-7 miRNAs by LIN28, we calculated the distance correlation (dCor) between LIN28A/B expression profiles and the profiles of each mature miRNA. We used dCor because of its ability to capture non-linear correlations (Szekely et al., 2007). Spearman’s correlation was used to determine the sign of dCor, which implies the direction of regulation. Only samples showing LIN28 presence, i.e., with nonzero read counts, were included for analysis. To estimate the significance of dCor, we shuffled the expression of LIN28A/B 1000 times and then calculated dCor between randomized LIN28A/B profiles and the profiles of all other protein-coding genes and mature miRNAs to produce nonparametric p-value estimates. We used a Mann-Whitney U test to compare distributions of distance correlations. We performed two types of comparisons: 1) We calculated dCor between the profiles of LIN28 and each miRNA species, including CSD+ and CSD− let-7 miRNAs; then, we obtained the distribution and the average of dCor values within each subclass. 2) We summed up normalized expression across CSD+ let-7 miRNAs and CSD− let-7 miRNAs (total expression) and calculated the dCor between total miRNA and LIN28 expression profiles.
Pri-Let-7 hairpin RNA-mediated protein pull-down
The initial analysis of LIN28 pull-down using let-7 pri-miRNA haipin baits was performed using published interactome capture data in 11 different cell lines (Treiber et al., 2017). Statistical analysis of LIN28A and LIN28B pull down using CSD+ and CSD− let-7 precursors was performed using normalized mass spectrometry-based spectrum counts derived by the original study, which were the percentage of total counts, averaged over all cell lines in which the protein was identified.
We also performed similar interactome captures with a more quantitative measure of LIN28A pull down. For each pull-down sample 50 μl of magnetic streptavidin beads (M-270, Invitrogen) were washed with lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5% (v/v) glycerol, 1 mM DTT, 1 mM AEBSF) and coupled to 4 μg of a 3´-biotinylated 2´-O-methyl-RNA adaptor (5´-AGGCUAGGUCUCCC-3´) for 1 h at 4°C in 300 μl lysis buffer. After washing twice with lysis buffer, half of the adaptor-coupled beads were removed for pre-clearing. The second half was incubated with 10 μg in vitro transcribed let-7 hairpin RNA containing a 5´ leader sequence complementary to the adaptor oligonucleotide in 300 μl lysis buffer overnight at 4°C and washed twice with lysis buffer directly before adding the cell lysate.
For the preparation of cell lysate, two 15 cm-plates of confluent NTera2 teratocarcinoma cells were harvested, resuspended in 1 ml lysis buffer and lysed by sonication. Insoluble matter was removed by centrifugation (30 min, 20000 g, 4°C) and the supernatant was subject to a pre-clearing step by adding the adaptor-coupled beads and rotating for 3–4 hours at 4°C. After removal of the beads, the supernatant was used for the pull-down experiment.
The RNA-coupled beads were incubated with the pre-cleared lysate at 4°C overnight while rotating. The beads were then washed three times with wash buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 5% (v/v) glycerol, 0.1% TritonX-100). Beads were resuspended in 60 μl SDS gel loading dye. 20 μl of eluate was separated on a 10% SDS-PAGE gel, blotted on nitrocellulose membrane (Protran 0.45μM, GE Healthcare) and detected with a LIN28A specific antibody (Abcam 63740).
To normalize for the amount of intact bait-RNA, 3μl of the pull-down eluate was diluted with 30μl 50% formamide and loaded on a 12% urea acrylamide gel (Roth) and run at 350 V with TBE as running buffer. The nucleic acid was then transferred onto a nylon membrane (Hybond-N, GE Healthcare) by semi-dry blotting (20 V, 1 h) and crosslinked with UV light. The membranes were hybridized with 32P-labelled 2´O-methyl adaptor oligo overnight at 42°C in a hybridization solution containing 5x SSC, 7% (w/v) SDS, 20 mM sodium phosphate buffer, pH 7, and 2% Denhardt´s solution. The blots were washed twice with a solution containing 5x SSC and 1% (w/v) SDS and once with a solution of 1x SSC and 1% (w/v) SDS. The radioactive signals were analyzed using storage screens and a PMI system (Biorad).
Recombinant protein expression and purification
For expression of recombinant LIN28A containing the CSD and ZKD, the sequence coding for LIN28A amino acid (aa) 25–181 was cloned into the vector pET32a and expressed as Thioredoxin-His-fusion protein containing a TEV cleavage site in front of the LIN28A sequence. For protein production, the vector was introduced into E. coli Rosetta (DE3), bacteria was grown to an OD600 of 0.6 and induced with 1mM IPTG. After induction the culture was grown overnight at 25°C.
Bacteria were harvested and resuspended in Buffer A (50mM Na-Phosphate pH 8, 1M NaCl, 10mM Imidazol) supplemented with 1mg/ml Lysozyme, and lysed by sonication. The lysate was cleared by centrifugation (50000g, 30min, 4°C) and filtration. The Trx-His-Lin28 fusion protein was bound to a Ni-IMAC Sepharose column (GE Healthcare) and eluted with Buffer B (50mM Na-Phosphate pH 8, 300mM NaCl, 500mM Imidazol). The peak fractions were pooled, supplemented with 2mM MgCl2 and incubated with 200U Benzonase overnight to digest co-purified nucleic acids. The solution was concentrated by ultrafiltration and subjected to a gel filtration on a Superdex S200 column in GPC buffer (50mM HEPES pH 7.5 200mM NaCl, 2mM DTT). The peak fractions were again pooled, supplemented with 0.1mg/ml TEV protease and dialyzed overnight against Buffer C (50mM Tris pH 7.5, 100mM NaCl, 2mM DTT). The cleaved protein was separated on a Source S ion exchange column (GE Healthcare) run with a gradient of Buffer C and Buffer D (50mM Tris pH7.5, 1M NaCl, 2mM DTT). Pure LIN28A (aa 25–181) eluted in the gradient, was concentrated to >1mg/ml and mixed with 1 volume of glycerol before freezing.
For expression of LIN28A CSD (aa 25–120) a stop codon was introduced in the abovementioned construct by targeted mutagenesis. After expression, lysis and IMAC chromatography as described above the peak fractions were concentrated and subjected to gel filtration on a Superdex S75 column (GE Healthcare) in GPC buffer. Fusion protein containing fractions were pooled and supplemented with 0.1mg/ml TEV protease. After overnight incubation at 4°C the cleaved protein was again run over the S75 gel filtration column and the peak corresponding to the isolated CSD was collected, concentrated and frozen with 50% (v/v) glycerol as cryoprotectant.
Electrophoretic Mobility Shift Assay (EMSA)
20 pmol of pre-let-7 RNA was 5´-end labeled using Polynucleotide Kinase (Thermo) and γ32P-ATP (Hartmann Analytic). After 1 h the labeling reaction was stopped by addition of 18 mM EDTA and the labeled RNA was purified with Illustra MicroSpin G25 columns (GE Healthcare).
0.4 pmol labeled RNA was combined with 5–160 nM purified LIN28A or 0.5–4μM LIN28A CSD in a 20 μl reaction containing 20mM Tris pH 7.6, 5mM MgCl2, 100mM NaCl, 10% Glycerol, 2mM DTT, and 1 μg yeast t-RNA. Reactions with the full-length LIN28 additionally contained 15 μg/ml Heparin as non-specific competitor. The binding reactions were incubated for 10 min at 4°C and separated on a 6 % PA-Gel cast in a buffer of 45 mM Tris 45 mM Borate and 5 % glycerol. The gel was run at 200 V for 2h (for full-length LIN28A) or 45 min (for CSD), then dried and exposed to a phosphoimager screen.
RNA Bind-and-Seq (RBNS)
The LIN28A CSD (aa 1–120) with an N-terminal FLAG-HA-tag was transiently expressed in HEK293 cells for 48h. Cells from two 15cm culture dishes were harvested and lysed in 1 ml IP lysis buffer (50 mM Tris-HCl, pH 7.5, 300 mM KCl, 1 mM AEBFS, 1 mM DTT, 0.5% (v/v) NP-40). Insoluble material was pelleted by centrifugation (20,000g, 4°C, 15min) and the supernatant transferred to a fresh reaction tube containing 20 μl FLAG-M2 Agarose Beads (Sigma). The binding reaction was incubated at 4°C for 2–3 hours while agitating. The beads were then washed three times with 1 ml IP wash buffer (50 mM Tris-HCl, pH 7.5, 300 mM KCl, 0.05 % (v/v) NP40). The immunoprecipitated proteins were used directly in an RNA-selection reaction by resuspending the beads in 400 μl binding buffer (25 mM Tris-Cl pH 7.5, 150 mM KCl, 3 mM MgCl2, 0.01% (v/v) NP-40, 1 mg/ml BSA, 1 mM DTT, 5% (v/v) glycerol, 0.1U/μl Ribolock) and adding 10μg of an RNA-pool of the sequence 5´-NNNNNNNNGUUU-3´. The binding reaction was incubated for 30 minutes at room temperature with agitation. Beads were then collected by centrifugation (1000 g, 2 min, 4°C) and washed three times with ice-cold binding buffer. Bound RNA was eluted in 200 μl elution buffer (10 mM Tris-Cl pH 7.0, 400 mM NaCl, 1 mM EDTA, 1% (w/v) SDS) and purified by phenol-chloroform extraction and ethanol precipitation.
The selected RNA molecules were sequentially ligated to a 3´ DNA adaptor and a 5´RNA adaptor containing a T7 promoter sequence. The ligated product was reverse transcribed using the First Strand cDNA Synthesis Kit (Thermo) and the primer 5´-GCCTTGGCACCCGAGAATTCCAGTTT-3´. A PCR reaction was used to amplify the cDNA sequence and introduce barcodes for next generation sequencing (NGS).
To separate insert containing amplification products from empty adaptor sequences, the PCR reaction was run on a 6% Urea PAGE gel and the band at 150bp corresponding to the desired product was excised. The DNA was eluted overnight in 0.4 M NaCl and precipitated with ethanol. The re-dissolved PCR product was stored for NGS analysis.
To generate RNA for a second round of selection, 50 ng of the PCR-pool was amplified using the primers 5´-AATGATACGGCGACCACCGAGATCTACACGTTCAGTAATACGACTCACTATAGG-3´ and 5´-GCCTTGGCACCCGAGAATTCCAGTTT-3´. The resulting PCR-product was purified using a PCR Clean-up Kit (Macherey Nagel) and cleaved by addition of 1.5 μl Fast digest MssI (Thermo), which recognizes the restriction site GTTTAAAC that is generated by the ligation of the RNA insert with the 3´adaptor. The cleaved DNA was transcribed with T7 polymerase, which yields a new pool of RNAs with the sequence GGGNNNNNNNNGUUU. The RNA was purified by 18% Urea PAGE, dephosphorylated with FastAP (Thermo) and monophosphorylated with polynucleotide kinase. 10 μg of the prepared RNA were used in a second selection cycle with freshly immunoprecipitated LIN28A CSD, ligated and amplified as described above.
Libraries from once and twice selected RNA were sequenced on a MiSeq instrument (Illumina) with a 150 cycle MiSeq Reagent Kit to which we added a custom Read1 sequencing primer (5´-GATCTACACGTTCAGTAATACGACTCACTATAGGG-3´).
Obtained sequence reads were barcode sorted and filtered for sequences containing the 3´ adaptor and the full Mss1-cleavage site, indicative of ligation of an intact RNA from the selection pool. After clipping of the adaptor and invariant sequence, only reads of the correct length (8 nt for first round and 11 nt for second round libraries) were used for further analysis. The first three nucleotides of the second round 11mer reads were trimmed and the resulting 8-mer sequences were analyzed for enriched sequence motifs using Weeder2 (Pavesi et al., 2001). A cloned and sequenced input library was used to generate background frequencies. Due to the short length of the input sequences, a modified version of the Weeder2 was used to search for enriched 5mer, 6mer and 7mer motifs. The enriched motifs were visualized using the WebLogo server (Crooks et al., 2004).
Supplementary Material
Supplemental Table S1: The list of CIMS and CITS identified in LIN28 CLIP data analyzed in this study. Related to Figure 1.
Supplemental Table S2: The list of motifs identified at Lin28 CIMS or CITS. Related to Figure 1.
Supplemental Table S3: Summary of miRNA abundance, LIN28 binding and 3’ uridylation in LIN28B eCLIP data derived from K562 cells. Relatd to Figures 2 and 3.
Supplemental Table S4: List of tumor types analyzed in this study. Related to Figure 3.
Acknowledgements
We thank members of the Zhang laboratory for helpful discussion and Federico Zambelli at University of Milan for a modified version of the Weeder2 software. This study was supported by grants from the National Institutes of Health (NIH) (R01NS089676, R01GM124486, R21NS098172 and R03HG009528 to CZ), the Simons Foundation Autism Research Initiative (307711 to CZ), European Union’s Horizon 2020 Research and Innovation Programme (668858 to PS) and European Research Council (ERC) (682291 to GM). High-performance computation was supported by NIH grants S10OD012351 and S10OD021764.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Balzeau J, Menezes MR, Cao S, and Hagan JP (2017). The LIN28/let-7 pathway in cancer. Front Genet 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, and Hannon GJ (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, and Gregory RI (2013). A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, and Kim VN (2012). LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765–777. [DOI] [PubMed] [Google Scholar]
- Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, and Michlewski G (2014). Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation. Cell Rep 9, 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, and Hannon GJ (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235. [DOI] [PubMed] [Google Scholar]
- Graf R, Munschauer M, Mastrobuoni G, Mayr F, Heinemann U, Kempa S, Rajewsky N, and Landthaler M (2013). Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol 10, 1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, and Shiekhattar R (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, and Mello CC (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, and Tuschl T (2013). Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 19, 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, and Gregory RI (2009). Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16, 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, and Kim VN (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18, 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, and Kim VN (2008). Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32, 276–284. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, and Kim VN (2009). TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708. [DOI] [PubMed] [Google Scholar]
- Hertel J, Bartschat S, Wintsche A, Otto C, Students of the Bioinformatics Computer, L., and Stadler PF (2012). Evolution of the let-7 microRNA family. RNA Biol 9, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, and Zamore PD (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, and Slack FJ (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647. [DOI] [PubMed] [Google Scholar]
- Knight SW, and Bass BL (2001). A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, and Griffiths-Jones S (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42, D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, and Burge CB (2014). RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell 54, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, and Tuschl T (2004). The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14, 2162–2167. [DOI] [PubMed] [Google Scholar]
- Lee FCY, and Ule J (2018). Advances in CLIP technologies for studies of protein-RNA Interactions. Mol Cell 69, 354–369. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. [DOI] [PubMed] [Google Scholar]
- Lee YS, and Dutta A (2007). The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21, 1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, and Allain FH (2011). Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol 19, 84–89. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, and Kutay U (2004). Nuclear export of microRNA precursors. Science 303, 95–98. [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, and Varani G (2007). RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17. [Google Scholar]
- Mayr C, Hemann MT, and Bartel DP (2007). Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr F, Schutz A, Doge N, and Heinemann U (2012). The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res 40, 7492–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, and Tuschl T (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15, 185–197. [DOI] [PubMed] [Google Scholar]
- Michlewski G, and Caceres JF Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol 17, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, and Sliz P (2011). Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, and Hammond SM (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JS, Hobor F, Downie Ruiz Velasco A, Choudhury NR, Heikel G, Kerr A, Ramos A, and Michlewski G (2017). Lin28a uses distinct mechanisms of binding to RNA and affects miRNA levels positively and negatively. RNA 23, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, and Pesole G (2001). An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics 17 Suppl 1, S207–214. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, and Gregory RI (2011). Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, and Gregory RI (2008). Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283, 21310–21314. [DOI] [PubMed] [Google Scholar]
- Powers JT, Tsanov KM, Pearson DS, Roels F, Spina CS, Ebright R, Seligson M, de Soysa Y, Cahan P, Theissen J, et al. (2016). Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 535, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransey E, Bjoörkbom A, Lelyveld VS, Biecek P, Pantano L, Szostak JW, and Sliz P (2017). Comparative analysis of LIN28-RNA binding sites identified at single nucleotide resolution. RNA Biol 14, 1756–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, and Ruvkun G (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, and Wulczyn FG (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10, 987–993. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, and Krueger LJ (2007). MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67, 9762–9770. [DOI] [PubMed] [Google Scholar]
- Shah A, Qian Y, Weyn-Vanhentenryck SM, and Zhang C (2017). CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 33, 566–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, and Daley GQ (2013). Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, and Ruvkun G (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5, 659–669. [DOI] [PubMed] [Google Scholar]
- Stormo GD (2000). DNA binding sites: representation and discovery. Bioinformatics 16, 16–23. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, and Mesirov JP (2007). GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251–3253. [DOI] [PubMed] [Google Scholar]
- Szekely GJ, Rizzo ML, and Bakirov NK (2007). Measuring and testing independence by correlation of distances Ann Stat 35, 2769–2794. [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, and Hammond SM (2006). Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20, 2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, and Rosenfeld MG (2009). The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Plessmann U, Harlander S, Daiss JL, Eichner N, Lehmann G, Schall K, Urlaub H, and Meister G (2017). A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol Cell 66, 270–284 e213. [DOI] [PubMed] [Google Scholar]
- Triboulet R, Pirouz M, and Gregory RI (2015). A single let-7 microRNA bypasses LIN28-mediated repression. Cell Rep 13, 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, and Vanacova S (2013). Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 19, 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, Zavolan M, and Vanacova S (2016). TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J 35, 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Meth 13, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Shishkin AA, Pratt GA, Nguyen TB, and Yeo GW (2017). Variation in single-nucleotide sensitivity of eCLIP derived from reverse transcription conditions. Methods 126, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, and Gregory RI (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 41, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Nam Y, Lee AK, Yu C, Roth K, Chen C, Ransey EM, and Sliz P (2017). LIN28 zinc knuckle domain is required and sufficient to induce let-7 oligouridylation. Cell Rep 18, 2664–2675. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X, Xie D, Shen Z, Sze J, Li K, et al. (2011). MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem 286, 39703–39714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck S, Mele A, Sun S, Yan Q, Farny N, Zhang Z, Xue C, Silver PA, Zhang MQ, Krainer AR, et al. (2014). HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, and Zhang C (2016). mCarts: genome-wide prediction of clustered sequence motifs as binding sites for RNA-binding proteins. Methods Mol Biol 1421, 215–226. [DOI] [PubMed] [Google Scholar]
- Wilbert Melissa L., Huelga Stephanie C., Kapeli K, Stark Thomas J., Liang Tiffany Y., Chen Stella X., Yan Bernice Y., Nathanson Jason L., Hutt Kasey R., Lovci Michael T., et al. (2012). LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell 48, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, and Yamanaka S (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14, 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, and Cullen BR (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17, 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhang C, and Darnell RB (2011). Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotech 29, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lee K-Y, Swanson MS, and Darnell RB (2013). Prediction of clustered RNA-binding protein motif sites in the mammalian genome. Nucleic Acids Res 41, 6793–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1: The list of CIMS and CITS identified in LIN28 CLIP data analyzed in this study. Related to Figure 1.
Supplemental Table S2: The list of motifs identified at Lin28 CIMS or CITS. Related to Figure 1.
Supplemental Table S3: Summary of miRNA abundance, LIN28 binding and 3’ uridylation in LIN28B eCLIP data derived from K562 cells. Relatd to Figures 2 and 3.
Supplemental Table S4: List of tumor types analyzed in this study. Related to Figure 3.