Abstract
STUDY QUESTION
Can genome-wide haplotyping increase success following preimplantation genetic testing for a monogenic disorder (PGT-M) by including zygotes with absence of pronuclei (0PN) or the presence of only one pronucleus (1PN)?
SUMMARY ANSWER
Genome-wide haplotyping 0PNs and 1PNs increases the number of PGT-M cycles reaching embryo transfer (ET) by 81% and the live-birth rate by 75%.
WHAT IS KNOWN ALREADY
Although a significant subset of 0PN and 1PN zygotes can develop into balanced, diploid and developmentally competent embryos, they are usually discarded because parental diploidy detection is not part of the routine work-up of PGT-M.
STUDY DESIGN, SIZE, DURATION
This prospective cohort study evaluated the pronuclear number in 2229 zygotes from 2337 injected metaphase II (MII) oocytes in 268 cycles. PGT-M for 0PN and 1PN embryos developing into Day 5/6 blastocysts with adequate quality for vitrification was performed in 42 of the 268 cycles (15.7%). In these 42 cycles, we genome-wide haplotyped 216 good quality embryos corresponding to 49 0PNs, 15 1PNs and 152 2PNs. The reported outcomes include parental contribution to embryonic ploidy, embryonic aneuploidy, genetic diagnosis for the monogenic disorder, cycles reaching ETs, pregnancy and live birth rates (LBR) for unaffected offspring.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Blastomere DNA was whole-genome amplified and hybridized on the Illumina Human CytoSNP12V2.1.1 BeadChip arrays. Subsequently, genome-wide haplotyping and copy-number profiling was applied to investigate the embryonic genome architecture. Bi-parental, unaffected embryos were transferred regardless of their initial zygotic PN score.
MAIN RESULTS AND THE ROLE OF CHANCE
A staggering 75.51% of 0PN and 42.86% of 1PN blastocysts are diploid bi-parental allowing accurate genetic diagnosis for the monogenic disorder. In total, 31% (13/42) of the PGT-M cycles reached ET or could repeat ET with an unaffected 0PN or 1PN embryo. The LBR per initiated cycle increased from 9.52 to 16.67%.
LIMITATIONS, REASONS FOR CAUTION
The clinical efficacy of the routine inclusion of 0PN and 1PN zygotes in PGT-M cycles should be confirmed in larger cohorts from multicenter studies.
WIDER IMPLICATIONS OF THE FINDINGS
Genome-wide haplotyping allows the inclusion of 0PN and 1PN embryos and subsequently increases the cycles reaching ET following PGT-M and potentially PGT for aneuploidy (PGT-A) and chromosomal structural rearrangements (PGT-SR). Establishing measures of clinical efficacy could lead to an update of the ESHRE guidelines which advise against the use of these zygotes.
STUDY FUNDING/COMPETING INTEREST(S)
SymBioSys (PFV/10/016 and C1/018 to J.R.V. and T.V.), the Horizon 2020 WIDENLIFE: 692065 to J.R.V., T.V., E.D., A.D. and M.Z.E. M.Z.E., T.V. and J.R.V. co-invented haplarithmisis (‘Haplotyping and copy-number typing using polymorphic variant allelic frequencies’), which has been licensed to Agilent Technologies. H.M. is fully supported by the (FWO) (ZKD1543-ASP/16). The authors have no competing interests to declare.
Keywords: 1PN and 0PN zygotes, whole-genome embryo haplotyping, preimplantation genetic testing, karyomapping, haplarithmisis
Introduction
Zygotes with phenotypes deviating from the normal morphometric standards, including a-, mono-, tri- and multi-pronucleation (0PN, 1PN, 3PN, ≥3PN) combined with the presence/absence of the second polar body (PB2) or the fragmentation of both polar bodies, are treated as evidence of failed or abnormal fertilization and as proxies of suboptimal gamete quality. As a consequence, in current clinical practice, it is not recommended to transfer zygotes that do not have two pronuclei at the time of visual assessment (Eshre Guideline Group on Good Practice in IVF Labs et al., 2016).
The observation of three pronuclei in zygotes (3PN) is considered proof of triploidy and IVF practitioners dispose of the 3PN zygotes at the start of the IVF cycle. This globally adopted practice aims to avoid the risk of adverse developmental outcomes including abnormal foetal development, molar pregnancy, miscarriage, neonatal death and congenital anomalies which are associated with triploid and 2n/3n mixoploid embryonic development (Montgomery et al., 1993; Redline et al., 1998; Zaragoza et al., 2000; van de Laar et al., 2002; Golubovsky, 2003; Philipp et al., 2004; Devriendt, 2005). Interpretation of the absence of pronuclei (0PN) or the presence of only one pronucleus (1PN) into zygote/embryo ploidy is not so straightforward and the use of these zygotes in IVF has been a topic of debate for the past 20 years. Numerous cytogenetic analyses have revealed that a significant subset of them are diploid and developmentally competent. The inefficiency of PN number scoring as a criterion for euploid embryo selection was demonstrated first by Lim et al. (2000), who showed that a staggering 66% diploid 0PN and 1PN-derived embryos are discarded at the zygote stage due to erroneous assignment of ploidy based on pronuclear numbers FISH analyses have shown that 57–62% of the embryos developing from 0PNs are diploid (Manor et al., 1996; Lim et al., 2000) and 4.4–30.3% of 0PN zygotes cleave into good quality blastocysts (Li et al., 2015; Yao et al., 2016; Yin et al., 2016). More importantly in cycles where only 0PN-derived embryos were available their transfer yielded pregnancies, which resulted in the birth of healthy babies (Manor et al., 1996; Yin et al., 2016). Similarly to 0PNs, successful development to term has been reported following transfers of 1PN-derived embryos (Staessen et al., 1993; Dasig et al., 2004; Itoi et al., 2015; Bradley et al., 2017; Mateo et al., 2017b) and previous cytogenetic analyses report the detection of 1PN diploid embryos albeit with high variabilty (2.2–80.5%, reviewed in Rosenbusch (2014)). Several groups have thus advocated that a considerable loss of zygotes can be overcome by implementing a genetic analysis technology to confirm bi-parental diploidy in the embryos developing from 0PN and 1PN zygotes (Levron et al., 1995; Li et al., 2015; Bradley et al., 2017; Capalbo et al., 2017; Mateo et al., 2017a, b).
Establishing genome-wide diploidy by means of FISH is challenging mainly because the technique analyses a subset of the genome and cannot provide information regarding the parental origin of diploidy. Genome-wide technologies such as array comparative genomic hybridization (aCGH) or low pass next generation sequencing (NGS) which are routinely used for embryonic aneuploidy testing (PGT-A) cannot map ploidy anomalies because normalization eliminates ploidy information. Therefore, recent exploratory studies opted for complementary strategies (first line and second line) such as the combined use of aCGH/NGS and short tandem repeat (STR) analysis or targeted NGS coupled with quantitative PCR (qPCR) to obtain proof of bi-parental diploidy (Bradley et al., 2017; Capalbo et al., 2017). Both studies provide further evidence that PN number scoring is an inefficient tool for embryonic ploidy prediction. However, these approaches require the optimization, development and implementation of more than one method to establish a diagnostic result, this being prohibitive for generalized adoption. As a consequence, current standard practice discards 0PN and 1PN zygotes.
Recently, the development of single-cell haplotyping methods has enabled genome-wide linkage analysis in embryos from couples who are carriers of pathogenic variants. The performance and dynamic range of these technologies, such as Haplarithmisis and Karyomapping, have been demonstrated in both clinical and research settings (Natesan et al., 2014; Zamani Esteki et al., 2015; Destouni et al., 2016; Ottolini et al., 2016; Dimitriadou et al., 2017; McCoy et al., 2018). These methods are ‘generic’ and standalone because they can detect an inherited haplotype block harbouring a pathogenic variant at any genomic locus by interogating 300 K highly polymorphic single nucleotide polymorphisms (SNPs) spanning evenly the entire human genome. In parallel to genome-wide haplotyping, Haplarithmisis can also uncover ploidy as a function of the parental haplotype copies inherited by the embryo by making use of haplotyped SNP B-allele frequencies, which are segmented into parental haplotype blocks. This analytical component disclosed events such as maternal metaphase II (MII) triploidy in human embryos and uni-parental ploidy and triploidy caused by dispermic fertilization or maternal genome segregation errors during meiosis II, as demonstrated in dissociated whole bovine embryos (Destouni et al., 2016; Dimitriadou et al., 2017; Tsuiko et al., 2017).
We hypothesized that genome-wide haplotyping can close the existing gap between routine clinical practice whereby 0 and 1PNs are discarded and the knowledge that a substantial proportion of 0PN and 1PN zygotes can develop into normal diploid embryos. We present the performance of this strategy in a good prognosis cohort, who opt for IVF in the context of preimplantation genetic testing for a monogenic disorder (PGT-M). We demonstrate that Haplarithmisis allows the inclusion of 0PN and 1PN embryos by simultaneously excluding the presence of mutant haplotypes and by confirming bi-parental diploidy across the embryonic genome in PGT-M cycles. With this approach we increase success in 31% of the cycles, which would otherwise not have available embryos for transfer (no-ET cycles) and would not have a second ET following the implantation failure of blastocysts developing from 2PN zygotes. Furthermore, we report the birth of unaffected offspring for the interrogated single gene disorder (SGD) following the transfer of 0PN and 1PN-derived embryos. This outcome is an added measure of clinical efficiency as accurate mapping of bi-parental diploidy is deterministic for the exclusion of an affected carrier status in the embryo. Based on our observations we propose that genome-wide haplotyping is a standalone, generic technology which allows routine inclusion of 0PN and 1PN-derived blastocysts in clinical practice.
Materials and Methods
Characteristics of the 0PN and 1PN PGT-M cohort
Between May 2016 and March 2018 we recruited 204 couples at risk of transmitting variants causing a monogenic disorder to their offspring and who were referred to the University Hospitals of Leuven (UZLeuven) for PGT-M. Participants received counselling and signed an informed consent in compliance with the hospital, local and federal regulatory requirements, prior to the initiation of the cycle. PGT-M in 0PN and 1PN-derived embryos was performed in 42 cycles from 38 couples (of the total 204) who had embryos of adequate quality for both a Day-3 biopsy and subsequent blastocyst vitrification. All analyses were performed by comparing outcomes between the 0PN, 1PN and the 2PNs generated in the 42 cycles of this PGT-M cohort. The cycle characteristics are presented in Supplementary Table SI.
IVF, pronuclear scoring and embryo handling
Female partners underwent ovarian stimulation, oocyte retrieval and ICSI as previously described (Debrock et al., 2010). On Day 1 (16–20 h after injection) the number of pronuclei was assessed. Zygotes with 2PN as well as 0PN and 1PN were further cultured. Cleavage stage day-3 embryos fulfilling the in-house morphology standards were biopsied as previously described (Debrock et al., 2010; Paternot et al., 2013). A single blastomere was aspirated from each embryo, and whole-genome amplified (WGAed) with the Repli-G single cell kit (Qiagen, Hilden, Germany). The embryos were immediately transferred to fresh medium and further cultured to Day 5/6 post injection. Those that formed blastocysts of sufficient quality were vitrified (Gardner and Schoolcraft, 1999). Blastocysts were vitrified in all cases except PGD148, where Day-3 embryos were frozen due to clinic logistics. Single embryo transfers were performed following warming of the cryopreserved blastocysts that were unaffected on genetic analysis. Pregnancy was determined as positive serum hCG levels (≥ 25 IU/L). The clinical implantation rate per embryo transferred was defined as the presence of a interuterine gestational sac on ultrasound at 6–8 weeks of pregnancy. Secondary outcomes included ongoing pregnancy rate (at 12 weeks of pregnancy) and the live birth rate (LBR) per embryo transferred. Live birth was defined as the live birth of an unaffected child beyond 24 weeks of gestation (Zegers-Hochschild et al., 2009).
Single-cell genome-wide haplotyping and concurrent copy number/ploidy profiling for embryo selection
siCHILD/Haplarithmisis (Zamani Esteki et al., 2015) was applied to analyse the embryonic genome-wide haplotype and copy-number landscapes by testing a single Day-3 biopsied blastomere. The clinical protocol was implemented as previously published (Dimitriadou et al., 2017). Case work-up involves the analysis of the genomic DNA obtained from the couple and the phasing reference(s) (either an affected/unaffected offspring or grandparents) to establish the family specific informative SNP metrics and hence the efficiency of haplotyping per case. WGAed DNA from embryo biopsies were hybridized on the Human CytoSNP-12v2.1 SNP arrays (Illumina, Inc., San Diego, CA, USA) and the raw genotype data exported by the Genome Studio software (Illumina, Inc., San Diego, CA, USA) were further analysed by siCHILD/Haplarithmisis as previously described (Zamani Esteki et al., 2015). In principle, siCHILD/Haplarithmisis reconstructs the inherited parental embryonic genome-wide haplotypes and by assigning embryo (single-cell) B-allele frequencies to haplotype blocks the pipeline can ‘count’ the copies of each inherited parental haplotype block across the embryonic genome (Fig. 1A). This analytical feature detects the mechanistic origin of whole chromosome and segmental aneuploidies by mapping which parental haplotype block is missing or is in excess. When an entire parental genome is contributing to ploidy anomalies as in dispermic/gynogenetic triploidy and uni-parental ploidy, the additional copy(ies) of the parental haplotypes are mapped along the genome (Fig 1B).
Figure 1.
Genome-wide haplotyping and haplotype block copy number analysis in PGT-M. The figure has been adapted from Vermeesch et al. (2016). In this schematic we describe the principles for the simultaneous haplotyping and copy number analysis of a single-cell applied in the context of preimplantation genetic testing for a monogenic disorder (PGT-M). These principles are implemented in a series of algorithmic modules comprising siCHILD/Haplarithmisis. (A) (i) Shown here is a pedigree where the father is affected by an autosomal dominant disorder caused by a variant, which is passed on to the affected offspring. Haplotype phase (the string of linked informative single nucleotide polymorphisms (SNPs) belonging to one homologue) can be established with the use of the affected offspring genotypes or other relatives such as grandparents, parental siblings etc. To simplify the description we present only phasing of the paternal haplotypes but the same principles apply to the maternal ones. Informative paternal SNPs are those that are heterozygous (AB) in the father and homozygous in the mother. The offspring genotypes are used to phase the paternal SNPs and obtain the paternal haplotypes. Following this step, the single-cell genotypes corresponding to a biopsied blastomere, can be allocated to the inherited paternal haplotypes and the haplotype block harbouring the disease variant can be traced in the embryo (ii). This analytical procedure makes use of discrete genotypes (letters A and B) and it is implemented in Karyomapping. (iii) siCHILD/Haplarithmisis entails additional steps whereby the single-cell B-allele frequencies (BAF) (embryo) which correspond to the paternal informative SNP loci are assigned to paternal haplotype sub-categories. These ‘haplotype-assigned’ BAFs are segmented into blocks. The output of the analysis represents the copy number state (frequency) of each paternal haplotype block inherited by the embryo. This analysis is performed at the genome-wide level and provides the genome-wide copy-number state of haplotype blocks inherited by the embryo. (B) Schematic examples of haplarithm profiles corresponding to different ploidy scenarios. Shown here are single-chromosomes but in case of genome-wide ploidy anomalies the signature is detected along the entire genome. With siCHILD/Haplarithmisis, reciprocal haplotype block signatures are obtained for each parental genome. This feature increases the accuracy of copy number aberration detection. It also provides insights into the mechanism of aneuploidy or genome-wide ploidy detection. For example in the triploid digynic signature, a mitotic error contributed to the extra maternal chromosome because both maternal haplotypes are similar (i.e. the same breakpoint is detected in the maternal haplarithm = haplotype block copies). SGD = single gene disorder; Pat = paternal; Mat = maternal.
The embryo selection criteria implemented at UZLeuven have been previously published (Dimitriadou et al., 2017). Embryos are selected based on two major genomic criteria: pathogenic variant carrier status and incidental aneuploidy status. Embryos bearing signatures corresponding to common viable aneuploidies (T13, T18, T21) are not selected for transfer regardless of their single-gene disorder carrier status, which might be determined by a locus residing on a different chromosome. Embryos are also selected against when a structural or numerical anomaly affects the disease-causing locus and when a meiotic trisomy is detected. Genome-wide aberrations, such as chaotic copy number signatures spanning the entire genome (gross aneuploidy), and anomalies of embryonic ploidy (triploidy, haploidy) are also indications for designating an embryo as unsuitable for transfer (Dimitriadou et al., 2017).
Statistics and data visualization
Categorical data are presented as percentages. Frequencies of observed outcomes were compared with Fisher’s exact and Chi-square test with significance set at P < 0.05 (GraphPAD Prism v.6, GraphPad Software, La Jolla, CA, USA).
Results
Genome-wide ploidy and haplotyping outcomes in 0PN, 1PN and 2PN derived embryos
There were 301 0PN, 132 1PN and 1796 2PN embryos in culture representing 12.9, 5.6 and 76.9% of the total 2337 successfully injected MIIs in the study (Table I). Significantly less 0PNs and 1PNs develop into good quality blastocysts compared to 2PNs (16.3%, 11.4 versus 53.3%, Chi-square P < 0.0001) (Table I). PGT-M was performed in 49 0PN and 15 1PN embryos in 42 of the 268 initiated cycles (15.7%) (Fig. 2).
Table I.
Proportion of cultured, biopsied and vitrified embryos developing from 0PN, 1PN and 2PN zygotes.
| Total cycles, n = 268 | |||
|---|---|---|---|
| Total successfully injected MIIs | |||
| n = 2337 | 0PN | 1PN | 2PN |
| N embryos in culture | 301 | 132 | 1796 |
| Embryos in culture/successfully injected MII (%) | 12.9 | 5.6 | 76.9 |
| N embryos biopsied (Day-3 p.i.) | 86 | 70 | 1557 |
| Embryos biopsied/ embryos in culture (%) | 28.6 | 53.0 | 86.7 |
| N embryos biopsied and vitrified (Day-5/6 p.i.) | 49 | 15 | 957 |
| Embryos biopsied and vitrified/ embryos in culture (%) | 16.3 | 11.4 | 53.3 |
PN: pronucleus/pronuclei, MII: metaphase II arrested oocytes, p.i.: post injection
Figure 2.
Flow diagram of the study. PN = pronucleus/ei; MII = metaphase II; DBP = diploid bi-parental; ET = embryo transfer. *The embryos in one case were not cultured to the blastocyst stage due to clinic logistics.
Confirmation of embryonic bi-parental diploidy is a prerequisite for accurate PGT-M. Since the absence of a pronucleus (0PN) or the presence of one pronucleus (1PN) are treated as indications of abnormal parental ploidy (i.e. maternal or paternal haploidy), we first investigated the presence of both parental haplotypes along the embryonic genome. We score these embryos as diploid bi-parental (DBP). Balanced DBPs are further categorized as ‘euploid’. DBPs bearing segmental and/or a single whole chromosome anomalies were categorized as ‘aneuploid’ DBPs. Embryos with genome-wide, gross chromosome anomalies were scored as ‘abnormal’ and were further categorized into the following sub-classes: chaotic, if more than five chromosomes were affected; grossly aneuploid if more than two but less than five chromosomes were affected; and genome-wide ploidy anomalies (uni-parental signatures and triploidy).
The majority of the analysed 0PN embryos are DBPs (37/49, 75.51%) allowing genetic diagnosis. Genetic diagnosis was not performed in the 0PN embryos that were scored as abnormal (n = 12). The abnormal group involved embryos with signatures of bi-maternal triploidy, gynogenesis, and chaotic and gross aneuploidy (Fig. 2). Of the 14 analysed 1PN embryos, six were DBPs (42.86%) whereas the remaining eight were all found to be gynogenetic (Fig. 2). Of the 151 analysed 2PN embryos 121 (80.13%) were scored as DPBs with the remaining 30 having chaotic and grossly aneuploid profiles. In this cohort none of the abnormal 2PNs were gynogenetic but one embryo was triploid. The frequencies of DPBs are significantly different between 1PNs and 2PNs (42.86 versus 80.13% Fisher’s exact, P = 0.0042) but not between 0PNs and 2PNs (75.51 versus 80.13%, Fisher’s exact, P = 0.5457). As abnormal PN number is considered an indication of abnormal parental ploidy (e.g. 1PN = absence of one parental genome) we analysed the frequency of parental ploidy anomalies by comparing gynogenetic and triploid embryos in each zygote category. This analysis shows that the rates of gynogenesis and triploidy differ significantly between 0PN (8.61%), 1PN (57.14%) and 2PNs (0.66%) (2PN versus 1PN Fisher’s exact P < 0.0001; 0PN versus 1PN Fisher’s exact P = 0.0003; 0PN versus 2PN Fisher’s exact P = 0.0134).
We further investigated if 0PNs and 1PNs have significantly different frequencies of euploidy than the presumably normal 2PNs. For this comparison we grouped embryos with simple aneuploidies and abnormal profiles in a single category. We found that euploid embryo rates are not significantly different between the 0PN, 1PN and 2PNs (55.10% 0PN, 35.71% 1PN and 47.02% 2PN, Chi-square P > 0.05).
The gynogenetic and triploid profiles obtained for the 0PNs, 1PNs and 2PNs are presented in Supplementary Figs S1 and S2.
Cycle outcomes following the inclusion of 0PN and 1PN
Diagnosis was achieved in DBP embryos because both parental genome-wide haplotypes could be faithfully reconstructed. The majority of the genetic diseases under investigation are inherited in an autosomal dominant mode (Supplementary Table SI). Unaffected, DBP embryos were prioritized for replacement regardless of their initial PN score.
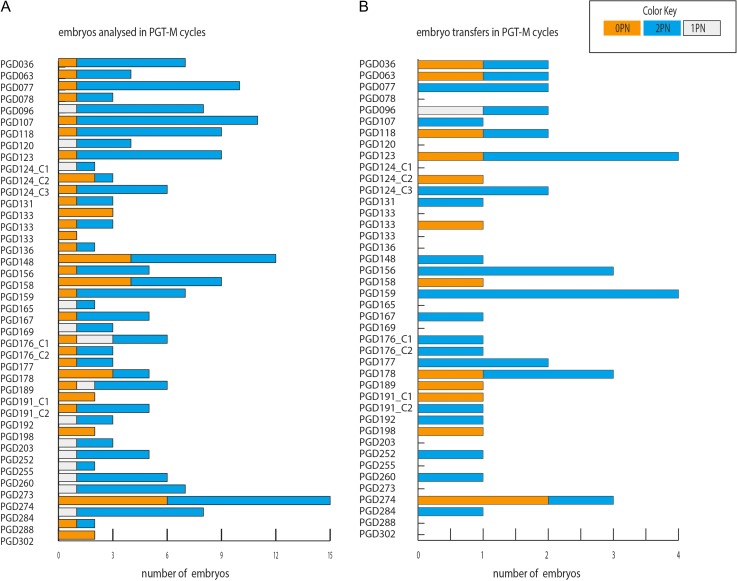
At least one-third of the analyzed embryos developed from 0PN and 1PN zygotes in 24 of the 42 cycles (57.14%) (Fig. 3A). In five PGT-M cycles only 0PN embryos were available for genetic analysis (Fig. 3A). Unaffected embryos were transferred in 29 of the 42 cycles (69.04%) (Fig. 3B). Only because 0PN and 1PN embryos were diagnosed as unaffected, 13 of the 29 PGT-M cycles (44.87%) could reach ET or repeat ET. The increase of cycles reaching ET following PGT-M in 0PN and 1PN embryos is statistically significant (ET with 2PNs only (n = 16) versus ET with 0PN+1PNs (n = 29), Fisher’s exact P = 0.0083). Six cycles (20.7%) proceeded to ET with 0PNs only and seven cycles would not have the chance to repeat ET had 0PN and 1PN blastocysts not been available for analysis (Fig. 3B).
Figure 3.
Distribution of 0PN, 1PN and 2PN derived embryos in the reported PGT-M cycles up to ET.
Of the 34 2PN transferred embryos, eight yielded pregnancies in PGD077, PGD107, PGD123, PGD156, PGD176_C2, PGD177, PGD178 and PGD284. Of these, four resulted in the delivery of healthy babies and one is still ongoing (24 weeks of gestation, no adverse events reported at time of writing). The remaining three were biochemical pregnancies. Four of the 13 0PN transferred embryos implanted and established a pregnancy. Of the four pregnancies, one spontaneously miscarried at 9 weeks of gestation, two resulted in live births and one is ongoing (uncomplicated at 29 weeks of gestation). The pregnancy in cycle PGD096 following the replacement of a 1PN blastocyst resulted in the birth of a healthy baby. The LBR per initiated cycle increased from 9.52 to 16.67% (Fisher’s exact, P > 0.05) corresponding to a 75% increase in cycle success.
Discussion
In this study, we demonstrate that genome-wide haplotyping of developmentally competent 0PN and 1PN embryos allows their routine inclusion in PGT-M cycles. This procedure increases the cycles reaching and repeating ET by a staggering 81%. The diagnostic accuracy of the method is confirmed by the live-birth of unaffected babies for the tested monogenic disorder.
The rate of 0PN, 1PN and 2PN zygotes per injected MII oocytes was 23.2, 7.1 and 60.1%, respectively (Fig. 2). This translates to a one-third decrease in the number of embryos available for genotyping and to a considerable PGT-M cycle failure had 0PN and 1PN zygotes been discarded after PN evaluation. Here, four cycles would have no embryos available for genotyping and six cycles would fail to reach ET (Fig. 3). In total, 13 of the 42 cycles (31%) could be rescued (reach ET or repeat ET) because 0PNs and 1PNs were accurately diagnosed as diploid, bi-parental and unaffected for the genetic disorder. This increase led to higher LBRs per initiated cycle (from 9.52 to 16.67%). Therefore, the inclusion of 0PN and 1PN embryos into the diagnostic pipeline is beneficial for PGT-M couples whose transferrable pool of embryos is significantly reduced by the embryonic monogenic disorder associated genotype.
The observed increase of transferrable embryos in the 42 PGT-M cycles is a function of the high diagnostic efficiency of the method (diagnosis failure rate = 0.93%) and the high blastocyst rate in these 42 cycles (41.6% for 0PNs and 45.5% for 1PNs) (Fig. 2). This indicates that in cycles with 0PN and 1PN good quality blastocysts, genome-wide haplotyping can benefit families where the number of transferrable embryos is very limited by the underlying genetic status, for example in HLA-SGD cycles where only 11.6% of the embryos is suitable for transfer (Kakourou et al., 2018).
The present study contributes the first genome-wide dataset regarding the parental ploidy of 0PN, 1PN and 2PN embryos in ICSI cycles. Gynogenesis occurred in more than half (57.14%) of the 1PN embryos, 6.1% of the 0PNs and none of the 2PNs. The 1PN gynogenesis rates are in line with the 45.5% reported by van der Heijden in ICSIed 1PN zygotes (van der Heijden et al., 2009). Although triploidy is not predicted based on the 0PN and 2PN classification, it was detected at very low frequencies in 0PN and 2PN embryos (2 and 0.6%, respectively). Our observations conform with a recent study showing a 2.5–3% digynic triploidy rate in a cohort of 1061 2PN embryos from PGT-A cycles (McCoy et al., 2018). These studies strengthen the notion that PN number is not an accurate proxy of parental ploidy. In our cohort, we did not detect any triploid 1PN embryos. This is in contrast to Capalbo et al., who reported two triploid 1PN blastocysts. We also found that 0PNs are more likely to be DBP than 1PNs. Accordingly, Capablo et al. (2017) reported the analysis of eight fast cleaving ICSI zygotes scored as 0PNs in PGT-A cycles and found that all were diploid bi-parental in contrast to the 69.2% 1PNs. These findings suggest that ICSIed 0PNs should not be neglected from further investigations in favour of 1PNs as they are probably misclassified zygotes due to faster cell cycle progression likely leading to early pronuclear breakdown (Manor et al., 1996; Feenan and Herbert, 2006; Liu et al., 2016).
Although the limitations of static fertilization check can be overcome by the use of time-lapse microscope coupled incubators (TLM), the data of Capalbo et al. (2017) show that even when PN state is assigned by TLM, four out of five 1PNs are diploid (Capalbo et al., 2017). This suggests that pronuclear evolution might not always reflect bi-parental diploidy and that even when PN scoring (coupled with PB scoring) is done with TLM, a subset of embryos will still be misclassified. Recent data from a study of 1PNs from conventional IVF cycles showed that 78% are diploid and could be non-invasively detected by measuring the pronuclear diameter and monitoring PN disappearance (Kai et al., 2018). Although this approach is promising, live-imaging showed that the bi-parental 1PNs were prone to abnormal mitosis leading to the unequal segregation of parental genomes (Kai et al., 2018). Considering the above, we therefore propose that the combined implementation of TLM with genomic technologies will provide further insights into the true genomic status of 0PNs and 1PNs and into how pronuclear evolution dynamics correlate with spindle function during mitosis and the embryonic ploidy.
Overall, we found that euploid 0PNs, 1PNs and 2PNs blastocyst frequencies are not significantly different. This observation is in line with the reported results from Lee et al. (2013) showing no significant differences in euploidy rates between Day-3 embryos from 0PNs, 1PNs and 2PNs. More recent findings show that aneuploidy rates did not differ significantly between 1PN and 2PN blastocysts (39.3 versus 36.5%, respectively) (Bradley et al., 2017) and no significant difference between euploid 0PN and 2PN PGT-M blastocysts (Yao et al., 2016). Collectively, these observations further strengthen the argument in favour of their use in IVF.
Importantly, the entire reported diagnostic procedure relies on a single generic technology with high diagnostic efficacy rates, which does not require the optimization of complementary strategies. To date, recently published approaches analysed 1PNs in the context of PGT-A but not PGT-M. These involve combinations of aCGH and NGS with STR or qPCR with a targeted NGS panel of 40 to ~2 K SNPs to generate low resolution ploidy estimations and were applied in PGT-A cases (Bradley et al., 2017; Capalbo et al., 2017). Although, these studies contributed significant data in favour of analysing abnormally PN zygotes, they rely on additional laboratory steps each requiring separate subsequent analysis. In Bradley et al., the group uses a copy number variation detection technology (aCGH or NGS) and selects only euploid female embryos for STR analysis, which is performed on aliquoted SurePlex material. Taking into account the estimated allelic drop out of 10.7 ± 6.1% for multi-cell samples (Vander Plaetsen et al., 2017) and that four of the 10 markers in the panel were not fully informative, only six STRs can predict embryonic parental ploidy in targeted loci. Capalbo et al., implemented a first line qPCR for comprehensive aneuploidy testing and enriched this with a panel of 40 SNPs for ploidy analysis. In only 63% (17/27) of embryos could both aneuploidy and parental ploidy be analysed. Ploidy measurement required a blastocyst re-biopsy and a subsequent test using a panel of ~2 K SNPs in 18/27 of the reported embryos. Further to the methodological complications, since only allelic ratios are calculated but not assigned to parental haplotypes, ploidy testing does not provide information regarding the parental origin of an underlying ploidy anomaly (e.g. gynogenetic triploidy, gynogenesis, androgenesis). A recent study validated the use of a SNP array platform to obtain a heterozygosity threshold for the selection of bi-parental 1PNs for PGT-A (Xie et al., 2018). However, its validation was based on prior predictions of ploidy anomalies and as such aimed only at the exclusion of parthenogenesis (either full or hybrid) without taking into account digynic triploids, which might produce a heterozygosity score above the selection cut-off. The unbiased operation of siCHILD/Haplarithmisis is based on the analysis of genome-wide haplotype copy states, which discriminate true abnormal fertilization events (e.g. as shown here gynogenesis and digynic triploidy) from erroneous assignment of ploidy based on PN scoring during a ‘snapshot’ fertilization check.
The use of 0PN and 1PN zygotes in IVF practice is still not recommended by ESHRE (Eshre Guideline Group on Good Practice in IVF Labs et al., 2016) despite accumulating evidence demonstrating that a significant subset can be euploid and developmentally competent. The reason for preserving these recommendations is the current lack of procedures which would accurately and comprehensively detect genome-wide parental ploidy in clinical routine such as PGT-M, PGTA, PGT for chromosomal structural rearrangements (PGT-SR) and IVF cycles. With this study we provide evidence that haplotyping can determine the genome-wide parental ploidy in 0PN and 1PN embryos and as a consequence eliminate their ongoing loss due to static PN misclassification. In principle, Karyomapping is also poised to detect diploid, bi-parental embryos developing from 0PN and 1PN zygotes. To date, haplotyping requires DNA from relatives for phasing the parental genotypes. While this is a standard procedure for PGT-M cycles, it currently limits a more generalized adoption for PGT-A or for replacing static fertilization check in IVF cycles. However, we anticipate that couples with cycles failing to yield 2PN blastocysts would consent to providing samples and that novel sequencing technologies allowing direct phasing will soon render genome-wide haplotyping a comprehensive PGT technology.
We propose that genome-wide haplotyping holds promise as a personalized medicine feature in PGT with a potential revision of the ESHRE guidelines to achieve optimal outcomes in the management of reproductive problems. This would be achieved following larger multicenter studies which will provide accurate measures of clinical efficacy.
Supplementary Material
Acknowledgements
The authors would like to thank the couples who participated in the study.
Authors’ roles
A.D., Ef.D., H.M. and J.R.V. conceived and designed the study. M.Z.E. and T.V. developed haplarithmisis. Ch.M., K.P. T.d.R., El.D. and E.L. counselled the patients. S.D. and K.P. performed the IVF–PGT associated procedures. Ef.D., K.V.D.B and Ci.M. performed PGT-M sample analysis, embryo scoring, annotation of findings and genetic diagnosis. J.D. implemented Haplarithmisis on the clinical samples. A.D. and S.D. performed data and statistical analysis. A.D. wrote the original version of the article. A.D., Ef.D., H.M., S.D, K.P. and J.R.V. edited the article. J.R.V. supervised the study. All co-authors read and approved the final version of the article.
Funding
SymBioSys (PFV/10/016 and C1/018 to J.R.V. and T.V.), the Horizon 2020 WIDENLIFE: 692065 to J.R.V., T.V., E.D., A.D. and M.Z.E. H.M. is fully supported by FWO (ZKD1543-ASP/16).
Conflict of interest
The authors have no competing interests to declare. M.Z.E, T.V. and J.R.V. co-invented haplarithmisis (‘Haplotyping and copy-number typing using polymorphic variant allelic frequencies’) licensed to Agilent Technologies.
References
- Bradley CK, Traversa MV, Hobson N, Gee AJ, McArthur SJ. Clinical use of monopronucleated zygotes following blastocyst culture and preimplantation genetic screening, including verification of biparental chromosome inheritance. Reprod Biomed Online 2017;34:567–574. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Treff N, Cimadomo D, Tao X, Ferrero S, Vaiarelli A, Colamaria S, Maggiulli R, Orlando G, Scarica C et al. Abnormally fertilized oocytes can result in healthy live births: improved genetic technologies for preimplantation genetic testing can be used to rescue viable embryos in in vitro fertilization cycles. Fertil Steril 2017;108:1007–1015.e3. [DOI] [PubMed] [Google Scholar]
- Dasig D, Lyon J, Behr B, Milki AA. Monozygotic twin birth after the transfer of a cleavage stage embryo resulting from a single pronucleated oocyte. J Assist Reprod Genet 2004;21:427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrock S, Melotte C, Spiessens C, Peeraer K, Vanneste E, Meeuwis L, Meuleman C, Frijns JP, Vermeesch JR, D’Hooghe TM. Preimplantation genetic screening for aneuploidy of embryos after in vitro fertilization in women aged at least 35 years: a prospective randomized trial. Fertil Steril 2010;93:364–373. [DOI] [PubMed] [Google Scholar]
- Destouni A, Zamani Esteki M, Catteeuw M, Tsuiko O, Dimitriadou E, Smits K, Kurg A, Salumets A, Van Soom A, Voet T et al. Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res 2016;26:567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K. Hydatidiform mole and triploidy: the role of genomic imprinting in placental development. Hum Reprod Update 2005;11:137–142. [DOI] [PubMed] [Google Scholar]
- Dimitriadou E, Melotte C, Debrock S, Esteki MZ, Dierickx K, Voet T, Devriendt K, de Ravel T, Legius E, Peeraer K et al. Principles guiding embryo selection following genome-wide haplotyping of preimplantation embryos. Hum Reprod 2017;32:687–697. [DOI] [PubMed] [Google Scholar]
- Eshre Guideline Group on Good Practice in IVF Labs, De los Santos MJ, Apter S, Coticchio G, Debrock S, Lundin K, Plancha CE, Prados F, Rienzi L, Verheyen G, Woodward B et al. Revised guidelines for good practice in IVF laboratories (2015). Hum Reprod 2016;31:685–686. [DOI] [PubMed] [Google Scholar]
- Feenan K, Herbert M. Can ‘abnormally’ fertilized zygotes give rise to viable embryos? Hum Fertil 2006;9:157–169. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Schoolcraft WB. Culture and transfer of human blastocysts. Curr Opin Obstet Gynecol 1999;11:307–311. [DOI] [PubMed] [Google Scholar]
- Golubovsky MD. Postzygotic diploidization of triploids as a source of unusual cases of mosaicism, chimerism and twinning. Hum Reprod 2003;18:236–242. [DOI] [PubMed] [Google Scholar]
- Itoi F, Asano Y, Shimizu M, Honnma H, Murata Y. Birth of nine normal healthy babies following transfer of blastocysts derived from human single-pronucleate zygotes. J Assist Reprod Genet 2015;32:1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai Y, Moriwaki H, Yumoto K, Iwata K, Mio Y. Assessment of developmental potential of human single pronucleated zygotes derived from conventional in vitro fertilization. J Assist Reprod Genet 2018;35:1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakourou G, Kahraman S, Ekmekci GC, Tac HA, Kourlaba G, Kourkouni E, Sanz AC, Martin J, Malmgren H, Gimenez C et al. The clinical utility of PGD with HLA matching: a collaborative multi-centre ESHRE study. Hum Reprod 2018;33:520–530. [DOI] [PubMed] [Google Scholar]
- Lee C, Yap W, Low S, Lim Y. P-23 euploidy rates for day 3 apronuclear (0PN) and unipronuclear (1PN) embryos. Reprod Biomed Online 2013;26:S36. [Google Scholar]
- Levron J, Munne S, Willadsen S, Rosenwaks Z, Cohen J. Male and female genomes associated in a single pronucleus in human zygotes. Biol Reprod 1995;52:653–657. [DOI] [PubMed] [Google Scholar]
- Li M, Lin S, Chen Y, Zhu J, Liu P, Qiao J. Value of transferring embryos that show no evidence of fertilization at the time of fertilization assessment. Fertil Steril 2015;104:607–611.e602. [DOI] [PubMed] [Google Scholar]
- Lim AS, Goh VH, Su CL, Yu SL. Microscopic assessment of pronuclear embryos is not definitive. Hum Genet 2000;107:62–68. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang XL, Zhang X, Shen CY, Zhang Z. Live births resulting from 0PN-derived embryos in conventional IVF cycles. J Assist Reprod Genet 2016;33:373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor D, Kol S, Lewit N, Lightman A, Stein D, Pillar M, Itskovitz-Eldor J. Undocumented embryos: do not trash them, FISH them. Hum Reprod 1996;11:2502–2506. [DOI] [PubMed] [Google Scholar]
- Mateo S, Vidal F, Coll L, Veiga A, Boada M. Chromosomal analysis of blastocyst derived from monopronucleated ICSI zygotes: approach by double trophectoderm biopsy. JBRA Assist Reprod 2017. a;21:203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo S, Vidal F, Parriego M, Rodriguez I, Montalvo V, Veiga A, Boada M. Could monopronucleated ICSI zygotes be considered for transfer? Analysis through time-lapse monitoring and PGS. J Assist Reprod Genet 2017. b;34:905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Newnham LJ, Ottolini CS, Hoffmann ER, Chatzimeletiou K, Cornejo OE, Zhan Q, Zaninovic N, Rosenwaks Z, Petrov DA et al. Tripolar chromosome segregation drives the association between maternal genotype at variants spanning PLK4 and aneuploidy in human preimplantation embryos. Hum Mol Genet 2018;27:2573–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery EA, Roberts EF, Conran RM, Hitchcock CL. Triploid abortus presenting as an ectopic pregnancy. Arch Pathol Lab Med 1993;117:652–653. [PubMed] [Google Scholar]
- Natesan SA, Bladon AJ, Coskun S, Qubbaj W, Prates R, Munne S, Coonen E, Dreesen JC, Stevens SJ, Paulussen AD et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med 2014;16:838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini CS, Capalbo A, Newnham L, Cimadomo D, Natesan SA, Hoffmann ER, Ubaldi FM, Rienzi L, Handyside AH. Generation of meiomaps of genome-wide recombination and chromosome segregation in human oocytes. Nat Protoc 2016;11:1229–1243. [DOI] [PubMed] [Google Scholar]
- Paternot G, Debrock S, De Neubourg D, D’Hooghe TM, Spiessens C. Semi-automated morphometric analysis of human embryos can reveal correlations between total embryo volume and clinical pregnancy. Hum Reprod 2013;28:627–633. [DOI] [PubMed] [Google Scholar]
- Philipp T, Grillenberger K, Separovic ER, Philipp K, Kalousek DK. Effects of triploidy on early human development. Prenat Diagn 2004;24:276–281. [DOI] [PubMed] [Google Scholar]
- Redline RW, Hassold T, Zaragoza MV. Prevalence of the partial molar phenotype in triploidy of maternal and paternal origin. Hum Pathol 1998;29:505–511. [DOI] [PubMed] [Google Scholar]
- Rosenbusch B. The chromosomal constitution of embryos arising from monopronuclear oocytes in programmes of assisted reproduction. Int J Reprod Med 2014;2014:418198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staessen C, Janssenswillen C, Devroey P, Van Steirteghem AC. Cytogenetic and morphological observations of single pronucleated human oocytes after in-vitro fertilization. Hum Reprod 1993;8:221–223. [DOI] [PubMed] [Google Scholar]
- Tsuiko O, Catteeuw M, Zamani Esteki M, Destouni A, Bogado Pascottini O, Besenfelder U, Havlicek V, Smits K, Kurg A, Salumets A et al. Genome stability of bovine in vivo-conceived cleavage-stage embryos is higher compared to in vitro-produced embryos. Hum Reprod 2017;32:2348–2357. [DOI] [PubMed] [Google Scholar]
- van de Laar I, Rabelink G, Hochstenbach R, Tuerlings J, Hoogeboom J, Giltay J. Diploid/triploid mosaicism in dysmorphic patients. Clin Genet 2002;62:376–382. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, van den Berg IM, Baart EB, Derijck AA, Martini E, de Boer P. Parental origin of chromatin in human monopronuclear zygotes revealed by asymmetric histone methylation patterns, differs between IVF and ICSI. Mol Reprod Dev 2009;76:101–108. [DOI] [PubMed] [Google Scholar]
- Vander Plaetsen AS, Deleye L, Cornelis S, Tilleman L, Van Nieuwerburgh F, Deforce D. STR profiling and Copy Number Variation analysis on single, preserved cells using current Whole Genome Amplification methods. Sci Rep 2017;7:17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeesch JR, Voet T, Devriendt K. Prenatal and pre-implantation genetic diagnosis. Nature Reviews Genetics 2016;17:643–656. [DOI] [PubMed] [Google Scholar]
- Xie PY, Tang Y, Hu L, Ouyang Q, Gu YF, Gong F, Leng LZ, Zhang SP, Xiong B, Lu GX et al. Identification of biparental and diploid blastocysts from monopronuclear zygotes with the use of a single-nucleotide polymorphism array. Fertil Steril 2018;110:545–554.e545. [DOI] [PubMed] [Google Scholar]
- Yao G, Xu J, Xin Z, Niu W, Shi S, Jin H, Song W, Wang E, Yang Q, Chen L et al. Developmental potential of clinically discarded human embryos and associated chromosomal analysis. Sci Rep 2016;6:23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin BL, Hao HY, Zhang YN, Wei D, Zhang CL. Good quality blastocyst from non-/mono-pronuclear zygote may be used for transfer during IVF. Syst Biol Reprod Med 2016;62:139–145. [DOI] [PubMed] [Google Scholar]
- Zamani Esteki M, Dimitriadou E, Mateiu L, Melotte C, Van der Aa N, Kumar P, Das R, Theunis K, Cheng J, Legius E et al. Concurrent whole-genome haplotyping and copy-number profiling of single cells. Am J Hum Genet 2015;96:894–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza MV, Surti U, Redline RW, Millie E, Chakravarti A, Hassold TJ. Parental origin and phenotype of triploidy in spontaneous abortions: predominance of diandry and association with the partial hydatidiform mole. Am J Hum Genet 2000;66:1807–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K, Sullivan E, Vanderpoel S. International Committee for Monitoring Assisted Reproductive T, World Health O. International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil Steril 2009;92:1520–1524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.